Abstract
Arrhythmogenic Cardiomyopathy (ACM) is an inherited cardiac disease characterized by arrhythmias and fibro-fatty replacement in the ventricular myocardium. Causative mutations are mainly reported in desmosomal genes, especially in plakophilin2 (PKP2). Here, using a virus-free reprogramming approach, we generated induced pluripotent stem cells (iPSCs) from skin fibroblasts of one ACM patient carrying the frameshift heterozygous PKP2 mutation c.2569_3018del50. The iPSC line (EURACi004-A) showed the typical morphology of pluripotent cells, possessed normal karyotype and exhibited pluripotency markers and trilineage differentiation potential, including cardiomyogenic capability. Thus, this line can represent a human in vitro model to study the molecular basis of ACM.
Resource table
| Unique stem cell line identifier | EURACi004-A |
| Alternative name(s) of stem cell line | N/A |
| Institution | Institute for Biomedicine, Eurac Research, Bolzano, Italy |
| Contact information of distributor | Alessandra Rossini (alessandra.rossini@eurac.edu) |
| Type of cell line | iPSCs |
| Origin | Human |
| Additional origin info | Age: 34-year-old Sex: Male Ethnicity: Caucasian |
| Cell source | Skin fibroblasts |
| Clonality | Clonal |
| Method of reprogramming | Electroporation of episomal vectors (pCXLE hOCT3/4-shp53-F, pCXLE-hSK, and pCXLE-hUL) |
| Genetic modification | YES |
| Type of modification | Spontaneous mutation |
| Associated disease | Arrhythmogenic Cardiomyopathy |
| Gene/locus | Heterozygous PKP2 c.2569_3018del50 |
| Method of modification | N/A |
| Name of transgene or resistance | N/A |
| Inducible/constitutive system | N/A |
| Date archived/stock date | January 2015 |
| Cell line repository/bank | N/A |
| Ethical approval | Skin fibroblasts were collected after patient informed consent and after approval from Centro Cardiologico Monzino – IRCCS Ethical Committee (12/06/2012). The research project was also reviewed and approved by Ethical Committee of the Province of South Tyrol (Nr.1/2014, 12/03/2014) |
Resource utility
The established iPSC line (EURACi004-A) provides an unlimited source of in vitro human cardiomyocytes that can be a useful model to investigate the molecular basis of arrhythmia generation in ACM. This might help to identify pharmacological approaches to ameliorate the arrhythmogenic and/or metabolic phenotypes associated with ACM.
Resource details
Arrhythmogenic Cardiomyopathy (ACM) is a cardiac genetic disorder characterized by potentially lethal ventricular arrhythmias, progressive loss of cardiomyocytes and fibro-fatty replacement in the myocardium, mainly in the right ventricle. The disease usually has an autosomal dominant transmission with variable penetrance and expressivity. Most mutations have been found in genes encoding desmosomal proteins and PKP2 is the most common causal gene (Awad et al., 2008). ACM is a complex disease and the associated pathogenic mechanisms are still unclear. Given the difficulty to obtain isolated human cardiomyocytes for in vitro studies, patient-specific iPSCs constitute an unlimited source of human cardiomyocytes for disease modelling, thus allowing the elucidation of ACM pathophysiology.
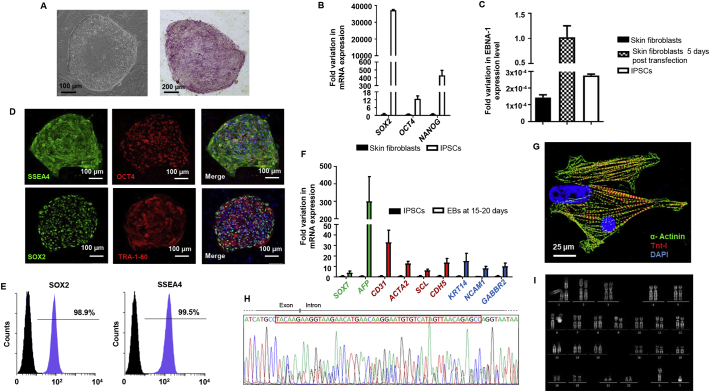
Here, we generated iPSCs from skin fibroblasts of a 34 years old male carrying a heterozygous frameshift mutation in PKP2 gene (c.2569_3018del50), leading to a premature termination on the PKP2 protein, already described as associated with ACM (Antoniades et al., 2006). Three different iPSC clones were generated from this patient and the EURACi004-A iPSC clone reported in this work was randomly selected for the characterization reported in Table 1. In detail, skin fibroblasts were reprogrammed into iPSCs using electroporation of episomal plasmids carrying OCT3/4, SOX2, KLF4, and L-MYC (Meraviglia et al., 2016). The selected EURACi004-A iPSC was amplified on mouse embryonic fibroblasts (MEFs). iPSC colonies showed a typical human embryonic-stem cell morphology and displayed high positivity for alkaline phosphatase (Fig. 1A). Undifferentiated iPSCs expressed endogenous pluripotency genes (i.e. SOX2, OCT4, NANOG) at higher levels compared to parental skin fibroblasts (Fig. 1B).
Table 1.
Characterization and validation.
| Classification | Test | Result | Data |
|---|---|---|---|
| Morphology | Light microscopy | Normal morphology and alkaline phosphatase positivity | Fig. 1 panel A |
| Phenotype | Immunocytochemistry | Expression of pluripotency markers: SSEA4, OCT4, TRA-1-60, SOX2 | Fig. 1 panel D |
| Flow cytometry and gene expression analysis by RT-qPCR | Fold change for pluripotency genes (RT-qPCR): SOX2 = 37,189 ± 430 OCT4 = 13 ± 2 NANOG = 456 ± 64% of positive cells (flow cytometry analysis): SOX2 98.1% ± 1.1% SSEA4 99% ± 0.9% |
Fig. 1 panel B (gene expression analysis) and Fig. 1 panel E (flow cytometry) | |
| Genotype | Karyotype (Q-banding) Resolution: 300–400 bands | Normal karyotype: 46, XY | Fig. 1 panel I |
| Identity | Microsatellite PCR (mPCR) | Not performed | |
| STR analysis | 21 markers tested: Amelogenin (for gender identification), D3S1358, D1S1656, D6S1043, D13S317, Penta E, D16S539, D18S51, D2S1338, CSF1PO, Penta D, TH01, vWA, D21S11, D7S820, D5S818, TPOX, D8S1179, D12S391, D19S433 and FGA with 100% match |
Available with the authors | |
| Mutation analysis (IF APPLICABLE) | Sequencing | Heterozygous PKP2 c.2569_3018del50 | Fig. 1 panel H |
| Southern Blot OR WGS | N/A | N/A | |
| Microbiology and virology | Mycoplasma | Mycoplasma testing by luminescence: Negative | Supplementary Fig. S1 |
| Differentiation potential | Embryoid body formation | Expression of genes of the three germ layers in embryoid bodies (SOX7 and AFP for endoderm; CD31, ACTA2, SCL and CDH5 for mesoderm; KRT14, NCAM1 and GABBR2 for ectoderm) and immunocytochemistry for spontaneous differentiation into cardiomyocytes |
Fig. 1 panel F (gene expression analysis) and Fig. 1 panel G (immunocytochemistry for cardiac markers) |
| Donor screening (OPTIONAL) | HIV 1 + 2 Hepatitis B, Hepatitis C | N/A | N/A |
| Genotype additional info (OPTIONAL) | Blood group genotyping | N/A | N/A |
| HLA tissue typing | N/A | N/A |
Fig. 1.
Generation and characterization of iPSC line EURACi004-A obtained from skin fibroblasts of one ACM patient.
(A) Representative images showing the iPSC colony morphology (scale bar 100 μm) and positive staining for alkaline phosphatase (scale bar 200 μm). (B) Gene expression analysis indicating the re-expression of endogenous pluripotency genes. (C) qPCR analysis shows no genome-integration of episomal vectors using episomal-specific primers (EBNA-1) (relative to control FBOX15 primers). (D) Representative immunofluorescence staining showing significant expression of pluripotency proteins SSEA-4, OCT4, SOX-2 and TRA-1-60. Nuclei are counterstained with DAPI; scale bar 100 μm. (E) Expression of pluripotency markers SSEA4 ad SOX2 evaluated by flow citometry analysis. (F) qRT-PCR analysis of three germ layer genes after 15-20 days of iPSC differentiation via embryoid body formation. (G) Immunofluorescence images for cardiac sarcomeric proteins α-Actinin and Troponin I (Tn-I) in single-cell dissociated beating areas; scale bar 25 μm. (H) Sanger sequencing results highlighting PKP2 gene region containing the heterozygous deletion. (I) Representative picture of normal karyogramm by Q-banding karyotype analysis.
Of note, EURACi004-A iPSC clone did not show the expression of episomal transgenes, evaluated by performing qPCR using primers for the episomal specific EBNA-1 gene, thus confirming successful activation of endogenous pluripotency genes without genome integration of exogenous plasmids (Fig. 1C).
The expression of transcription factors OCT4, SOX2 and surface markers SSEA4, TRA-1-60 associated with pluripotency was evaluated by immunofluorescence analysis (Fig. 1D), while the percentage of positive cells for SOX2 (98%) and SSEA4 (99%) was assessed by flow cytometry (Fig. 1E). The potential to differentiate into three germ lineages was evaluated after spontaneous differentiation through embryoid body (EB) formation by qRT-PCR on endodermal (SOX7 and AFP, green), mesodermal (CD31, ACTA2, SCL and CDH5, red) and ectodermal (KTR14, NCAM1 and GABRR2, blue) genes (Fig. 1F). Cardiomyogenic differentiation occurred spontaneously through embryoid body (EB) formation, as evident by the clear striated pattern of α-Actinin and Troponin I (Fig. 1G). As indicated in Fig. 1H, Sanger sequencing confirmed the presence of the PKP2 mutation and EURACi004-A iPSC line showed a normal karyotype at passage 33(Fig. 1I). STR analysis indicated that the newly created EURACi004-A iPSC clone yielded a 100% match with the skin fibroblast counterpart, confirming that they are correctly derived from the donor (Table 1, available with the authors). Finally, iPSCs resulted negative for mycoplasma contamination (Supplementary Fig. S1).
Supplementary Fig. S1.
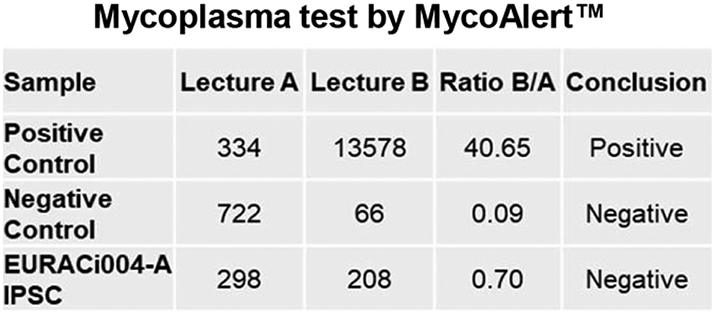
Mycoplasma detection.
Materials and methods
Cell culture and reprogramming
Skin fibroblasts were isolated from skin biopsy and amplified as previously described (Meraviglia et al., 2016). IPSCs from skin fibroblasts were generated using the episomal vectors pCXLE-hOCT3/4-shp53-F, pCXLE-hSK, and pCXLE-hUL (Addgene). Specifically, 7.5 × 105 skin fibroblasts were electroporated with 1 μg of each episomal vector using the Neon System (Thermo Fisher Scientific) at the program: 1650 V, 10 msec, 3 pulses and plated onto a plastic tissue culture dish for seven days in standard culture medium composed of Iscove's Modified Dulbecco's Medium (IMDM) (Thermo Fisher Scientific), 20% FBS Defined (Hyclone), 1% Penicillin/Streptomycin, 1% l-Glutamine (all from Thermo Fisher Scientific). Then, transfected fibroblasts were trypsinized, plated on 0.1% gelatin-coated plate with irradiated MEF feeder layer and cultured in stem cell medium containing knockout DMEM, 20% KO-Serum Replacement (KOSR), 1 mM NEAAs, 1% Penicillin/Streptomycin, 1% l-Glutamine, 0.1 mM β-mercaptoethanol (all from Thermo Fisher Scientific) and 10 ng/ml bFGF (Merck-Millipore), until the first iPSC colonies appeared. Then, iPSCs on MEF were cultured in the same stem cell medium and expanded once a week by enzymatic dissociation using 1 mg/ml Collagenase IV (Thermo Fisher Scientific) (ratio 1:4). IPSCs adapted to feeder-free condition were cultured in StemMACS™ iPS-Brew XF (Miltenyi Biotec) on Matrigel matrix (Corning) and amplified using TrypLE™ (Thermo Fisher Scientific) twice a week with a split ratio of 1:6. All the cells were incubated at 37 °C, 5% CO2 in a humidified incubator.
qRT-PCR
Total RNA from skin fibroblasts, iPSCs and EBs was extracted using TRIzol® Reagent. Reverse transcription of RNA (1 μg) was performed using SuperScript VILO cDNA Synthesis Kit following manufacturer's instruction (all from Thermo Fisher Scientific). All-in-One SYBR® Green qPCR Mix (GeneCopoeia) was used for cDNA amplification on CFX96 Real-Time PCR Detection System (BioRad). Table 2 reports primer sequences.
Table 2.
Reagents details.
| Antibodies used for immunocytochemistry/flow-citometry | |||
|---|---|---|---|
| Antibody | Dilution | Company Cat # and RRID | |
| Pluripotency Markers (Immunocytochemistry) | Rabbit anti-OCT4 | 1:100 | Thermo Fisher Scientific Cat# A24867, RRID:AB_2650999 |
| Pluripotency Markers (Immunocytochemistry) | Mouse anti-SSEA4 (IgG3) | 1:100 | Thermo Fisher Scientific Cat# A24866, RRID:AB_2651001 |
| Pluripotency Markers (Immunocytochemistry) | Rat anti-SOX2 | 1:100 | Thermo Fisher Scientific Cat# A24759, RRID:AB_2651000 |
| Pluripotency Markers (Immunocytochemistry) | Mouse anti-TRA-1-60 (IgM) | 1:100 | Thermo Fisher Scientific Cat# A24868, RRID:AB_2651002 |
| Secondary antibodies (Immunocytochemistry) | Alexa Fluor® 555 Donkey Anti-Rabbit | 1:250 | Thermo Fisher Scientific Cat# A24869, RRID:AB_2651006 |
| Secondary antibodies (Immunocytochemistry) | Alexa Fluor® 488 Goat Anti-Mouse IgG3 | 1:250 | Thermo Fisher Scientific Cat# A24877, RRID:AB_2651008 |
| Secondary antibodies (Immunocytochemistry) | Alexa Fluor® 488 Donkey Anti-Rat | 1:250 | Thermo Fisher Scientific Cat# A24876, RRID:AB_2651007 |
| Secondary antibodies (Immunocytochemistry) | Alexa Fluor® 555 Goat Anti-Mouse IgM | 1:250 | Thermo Fisher Scientific Cat# A24871, RRID:AB_2651009 |
| Pluripotency Markers (Flow cytometry) | PE-SOX2 Mouse IgG2A | 1:20 | R&D System Cat# IC2018P, RRID:AB_357273 |
| Pluripotency Markers (Flow cytometry) | CFS-SSEA-4 Mouse IgG3 | 1:20 | R&D System Cat# FAB1435F, RRID:AB_952015 |
| Pluripotency Markers (Flow cytometry) | PE Isotype control- mouse IgG2A | 1:20 | R&D System Cat# IC003P, RRID:AB_357245 |
| Pluripotency Markers (Flow cytometry) | CFS Isotype control- Mouse IgG3 | 1:20 | R&D System Cat# IC007F, RRID:AB_952037 |
| Cardiomyocyte markers (Immunocytochemistry) |
Mouse anti-Sarcomeric actinin | 1:250 | Sigma Aldrich Cat# A7732, RRID:AB_2221571 |
| Cardiomyocyte markers (Immunocytochemistry) |
Rabbit anti-Troponin I (H-170) | 1:500 | Santa Cruz Biotechnology Cat# sc-15,368, RRID:AB_793465 |
| Secondary antibodies (Immunocytochemistry) |
Alexa Fluor® 488 Goat Anti-Mouse IgG | 1:1000 | Thermo Fisher Scientific Cat# A11029, RRID:AB_2534088 |
| Secondary antibodies (Immunocytochemistry) |
Alexa Fluor® 555 Goat Anti-Rabbit IgG | 1:1000 | Thermo Fisher Scientific Cat# A21429, RRID:AB_2535850 |
| Primers | ||
|---|---|---|
| Target | Forward/Reverse primer (5′-3′) | |
| Pluripotency Markers (qRT-PCR) | SOX2 | GGGAAATGGGAGGGGTGCAAAAGAGG/TTGCGTGAGTGTGGATGGGATTGGTG |
| Pluripotency Markers (qRT-PCR) | OCT4 | GACAGGGGGAGGGGAGGAGCTAGG/CTTCCCTCCAACCAGTTGCCCCAAAC |
| Pluripotency Markers (qRT-PCR) | NANOG | TGCAAGAACTCTCCAACATCCT/ATTGCTATTCTTCGGCCAGTT |
| Three germ layer markers (endoderm) (qRT-PCR) | SOX7 | TGAACGCCTTCATGGTTTG/AGCGCCTTCCACGACTTT |
| Three germ layer markers (endoderm) (qRT-PCR) | AFP | GTGCCAAGCTCAGGGTGTAG/CAGCCTCAAGTTGTTCCTCTG |
| Three germ layer markers (mesoderm) (qRT-PCR) | CD31 | ATGCCGTGGAAAGCAGATAC/CTGTTCTTCTCGGAACATGGA |
| Three germ layer markers (mesoderm) (qRT-PCR) | ACTA2 | GTGATCACCATCGGAAATGAA/TCATGATGCTGTTGTAGGTGGT |
| Three germ layer markers (mesoderm) (qRT-PCR) | SCL | CCAACAATCGAGTGAAGAGGA/CCGGCTGTTGGTGAAGATAC |
| Three germ layer markers (mesoderm) (qRT-PCR) | CDH5 | GAGCATCCAGGCAGTGGTAG/CAGGAAGATGAGCAGGGTGA |
| Three germ layer markers (ectoderm) (qRT-PCR) | KRT14 | CACCTCTCCTCCTCCCAGTT/ATGACCTTGGTGCGGATTT |
| Three germ layer markers (neuro-ectoderm) (qRT-PCR) | NCAM1 | CAGATGGGAGAGGATGGAAA/CAGACGGGAGCCTGATCTCT |
| Three germ layer markers (neuro-ectoderm) (qRT-PCR) | GABRR2 | CTGTGCCTGCCAGAGTTTCA/ACGGCCTTGACGTAGGAGA |
| House-Keeping Gene (qRT-PCR) | GAPDH | CCACCCATGGCAAATTCC/TCGCTCCTGGAAGATGGTG |
| Episomal plasmid Gene (episome silencing) | EBNA-1 | ATCAGGGCCAAGACATAGAGATG/GCCAATGCAACTTGGACGTT |
| House-Keeping Gene (episome silencing) | FBXO15 | GCCAGGAGGTCTTCGCTGTA/AATGCACGGCTAGGGTCAAA |
| Mutation analysis (PCR) | PKP2 | GAACACCCACAGGCCGC/TTTCTTGGGCTGGGTAGTAGAAA |
| Mutation analysis (sequencing) | PKP2 | GCCTCACTCATTCTCCCTGATCTCAG/TAGGCTTTGGCAGTCCGGCTGTTG |
Immunofluorescence staining
Pluripotent Stem Cell 4-Marker Immunocytochemistry Kit (Thermo Fisher Scientific) was used for the detection of pluripotency markers on undifferentiated iPSCs, according to manufacturer's instructions (Table 2). Cardiomyocytes at 30–35 days of differentiation were fixed in PFA 4% for 10 min and permeabilized with 0.2% of Triton-100× at room temperature for 7 min. Blocking buffer (5% goat serum) was added for 1 h at room temperature. Primary antibodies were incubated overnight at 4 °C, while secondary antibodies were incubated for 1 h at 37 °C (Table 2). Nuclei were counterstained with DAPI. All the images have been acquired using Leica SP8-X confocal microscope.
Flow cytometry
SOX2 and SSEA4 expression in iPSCs adapted to feeder-free condition was analyzed by flow cytometry using Multi-Color Flow Cytometry Kit (R&D System) following manufacturer's instruction. Samples were analyzed using the S3e Cell Sorter (BioRad). Flow cytometry data analysis was performed using Flowing Software 2.5.1.
Embryoid body formation
IPSCs were differentiated as embryoid bodies (EBs) by detaching colonies and growing them in ultra-low attachment plates for 7 days in EB 20% medium composed of KO-DMEM (Thermo Fisher Scientific) with 20% FBS Defined (Hyclone), 1 mM NEAAs, 1% Penicillin/Streptomycin, 1% l-Glutamine, 0.1 mM β-mercaptoethanol (all from Thermo Fisher Scientific). EBs were then plated onto 0.1% gelatin-coated dishes and cultured in EB 20% medium for further 15–20 days. Approximately at day 25 of differentiation, spontaneous beating clusters, indicating iPSC spontaneous cardiomyogenic ability, were manually microdissected, dissociated at single cell level using TrypLE and plated onto gelatin-coated plates in EB 2% medium (same EB medium except for supplementation with only 2% FBS) for additional 5–10 days, in order to avoid fibroblast over-growth.
Karyotyping
Karyotype evaluation was assessed on iPSC adapted to feeder-free condition after 33 in vitro passages by cytogenetic Q-banding analysis as previously described (Meraviglia et al., 2015). Specifically, 20 metaphases were analyzed and all metaphases showed a normal karyogramm.
Sequencing and STR analysis
Genomic DNA used for PKP2 sequencing and STR analysis was isolated using DNeasy Blood and Tissue kit (Qiagen), following manufacturer's instructions. PKP2 mutation analysis was performed on a PCR product obtained by genomic DNA amplification using Kapa High Fidelity DNA Polymerase (Kapa Biosystems), following manufacturer's instructions (see primers listed in Table 2, mutation analysis PCR). The PCR reaction has been performed on Mastercycler® pro S instrument (Eppendorf) using the following conditions: 95 °C 5 min/98 °C 20 s; 60 °C 20 s; 72 °C 30 s for 35 cycles/72 °C 1 min 30 s;4 °C 10 min; 16 °C ∞. The amplification product was purified using Agencourt AMPure PCR purification system (Beckman Coulter) and PCR product size was inspected using agarose-gel electrophoresis (agarose gel 1%). Then, PKP2 mutation was confirmed on iPSCs and their skin fibroblast counterpart on the amplification product by Sanger sequencing performed by Eurofins Genomics (Germany), according to ANSI/ATCC standard ASN-0002 (Table 1), using a combined set of forward and reverse primers (Table 2, see mutation analysis sequencing). Specifically, cell identity was analyzed on genomic DNA, evaluating 21 independent loci by PCR-single-locus-technology (Promega, PowerPlex 21 PCR Kit).
Mycoplasma test
MycoAlert™ Mycoplasma Detection Kit (Lonza) was used to test mycoplasma contamination, following manufacturer's instructions (Table 2).
The following are the supplementary data related to this article.
Acknowledgements
This study was supported by the Telethon Grant GGP16001 to Giulio Pompilio and Alessandra Rossini and by the Department of Innovation, Research and Universities of the Autonomous Province of Bolzano-South Tyrol (Italy).
Contributor Information
Viviana Meraviglia, Email: viviana.meraviglia@gmail.com.
Alessandra Rossini, Email: alessandra.rossini@eurac.edu.
References
- Antoniades L., Tsatsopoulou A., Anastasakis A., Syrris P., Asimaki A., Panagiotakos D., Zambartas C., Stefanadis C., McKenna W.J., Protonotarios N. Arrhythmogenic right ventricular cardiomyopathy caused by deletions in plakophilin-2 and plakoglobin (Naxos disease) in families from Greece and Cyprus: genotype-phenotype relations, diagnostic features and prognosis. Eur. Heart J. 2006;27:2208–2216. doi: 10.1093/eurheartj/ehl184. [DOI] [PubMed] [Google Scholar]
- Awad M.M., Calkins H., Judge D.P. Mechanisms of disease: molecular genetics of arrhythmogenic right ventricular dysplasia/cardiomyopathy. Nat. Clin. Pract. Cardiovasc. Med. 2008;5:258–267. doi: 10.1038/ncpcardio1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraviglia V., Zanon A., Lavdas A.A., Schwienbacher C., Silipigni R., Di Segni M., Chen H.S., Pramstaller P.P., Hicks A.A., Rossini A. Generation of induced pluripotent stem cells from frozen buffy coats using non-integrating episomal plasmids. J. Vis. Exp. 2015:e52885. doi: 10.3791/52885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraviglia V., Wen J., Piacentini L., Campostrini G., Wang C., Florio M.C., Azzimato V., Fassina L., Langes M., Wong J., Miragoli M., Gaetano C., Pompilio G., Barbuti A., DiFrancesco D., Mascalzoni D., Pramstaller P.P., Colombo G.I., Chen H.S., Rossini A. Higher cardiogenic potential of iPSCs derived from cardiac versus skin stromal cells. Front. Biosci. 2016;21:719–743. doi: 10.2741/4417. Landmark Ed. [DOI] [PubMed] [Google Scholar]