Abstract
The liver and pancreas are closely associated organs that share a common embryological origin. They display amphicrine properties and have similar exocrine organization with parenchymal cells, namely, hepatocytes and acinar cells, secreting bile and pancreatic juice into the duodenum via a converging network of bile ducts and pancreatic ducts. Here we compare and highlight the similarities of molecular mechanisms leading to liver and pancreatic cancer development. We suggest that unraveling tumor development in an organ may provide insight into our understanding of carcinogenesis in the other organ.
Key words: Hepatocellular carcinoma, Intrahepatic cholangiocarcinoma, Extrahepatic cholangiocarcinoma, Hepatic fibrosis and cirrhosis animal models, Embryonic and postnatal liver development lineage tracing
INTRODUCTION
The liver and pancreas originate from adjacent regions of the definitive endoderm, with liver development being initiated by formation of a tissue bud on the ventral side of the distal foregut, and the pancreas arising from ventral and dorsal endodermal buds located caudally to the liver. Formation of the organ buds and their outgrowth from the distal foregut endoderm are controlled by mesodermal signals, which specify endoderm cells to hepatic and pancreatic fates and promote proliferation of the budding cells1,2. Cell proliferation rapidly generates a population of hepatic and pancreatic multipotent progenitors that differentiate to several mature epithelial cell types, namely, hepatocytes and cholangiocytes in the liver, and endocrine, acinar, and ductal cells in the pancreas3–6. Importantly, the extrahepatic biliary tract, namely, the gallbladder, hepatic, cystic, and common bile ducts, develop from the same endodermal region as the ventral pancreas7. However, as soon as the ventral pancreas starts rotating around the gut to merge with the dorsal pancreas, the extrahepatic biliary tract develops separately from the pancreas while maintaining a connection with the common pancreatic duct.
At the end of organogenesis, the liver and pancreas have developed a tissue organization that allows them to function as amphicrine glands. The endocrine function is ensured by the hepatocytes in the liver and the islets of Langerhans in the pancreas. In both organs, a dense network of fenestrated capillaries enables secretion of hormones and growth factors into the bloodstream. Exocrine functions are exerted by hepatocytes and acinar cells, which contribute roughly to 80% of the mass of their respective organs. They produce bile and digestive enzymes (e.g., amylase, lipases) that are discharged into the duodenum via a ductal network. Ducts are lined by cholangiocytes in the liver and by ductal cells in the pancreas. Notably, hepatocytes secrete most of their protein products directly into the plasma, secreting only bile components to the biliary ducts. In contrast, acinar cells secrete all their protein products through the pancreatic duct system.
In view of the common developmental origin and histological similarities of the two organs, it is not surprising that common transcriptional regulators control the ontogenesis and homeostasis of the liver and pancreas. Thus, members of the Forkhead box A (FoxA), Onecut, SRY-related HMG box (Sox), and hepatocyte nuclear factor (HNF) families are essential in developing adult liver and pancreas8–12. The similarities between the pancreatic and hepatic developmental programs are also reflected in the fact that acinar cells can transdifferentiate into hepatocytes13–15.
The liver and pancreas are affected by cancers characterized by poor survival rates. Considering the histological and molecular similarities of fetal and adult liver and pancreas, we reasoned that some mechanisms driving carcinogenesis might be analogous in the two organs. Here we summarize basic concepts about tumorigenesis in the liver and pancreas and discuss how tumor development in an organ may provide insight into our understanding of carcinogenesis in the other organ.
HEPATIC AND PANCREATIC CARCINOGENESIS
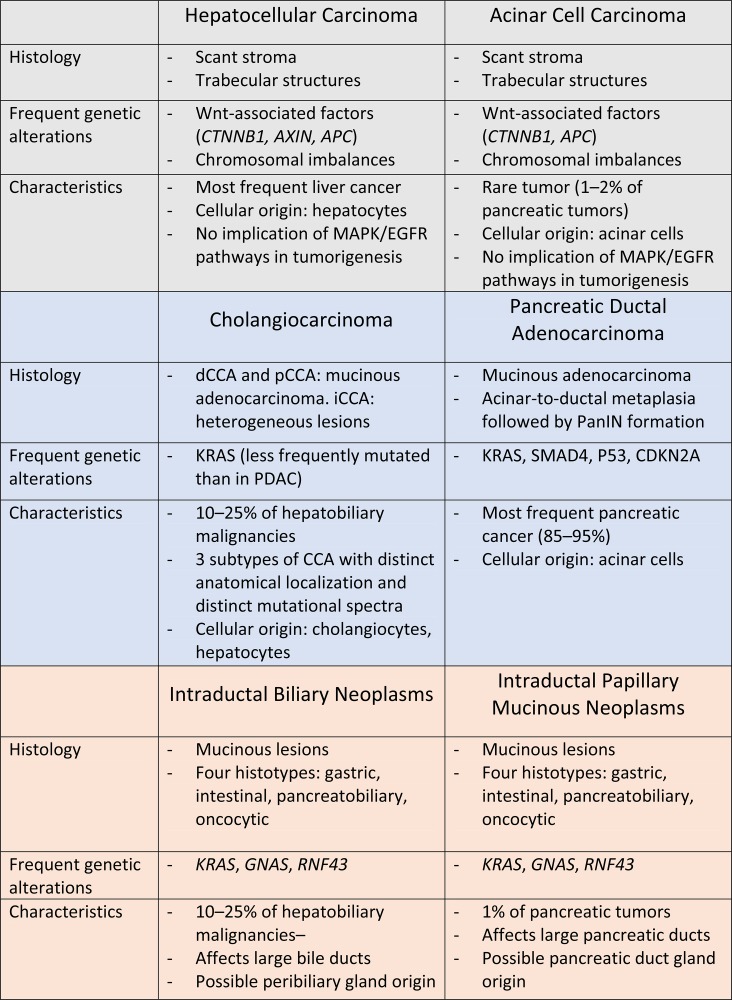
In the liver, the most frequent cancers are hepatocellular carcinoma (HCC). Cholangiocarcinoma (CCA) is the second most frequent type of liver cancer, and it accounts for about 10%–25% of all hepatobiliary malignancies. In the pancreas, pancreatic ductal adenocarcinoma (PDAC) is the most frequent pancreatic tumor (85%–95% of cases)16. Acinar cell carcinomas (ACCs) are less frequent: they are detected in 1%–2% of adult patients and about 15% of pediatric patients17. Pancreatic cystic tumors represent only 1% of cases, but nevertheless account for about 15% of pancreatic cancer resections18. These three pancreatic tumor types derive from the exocrine compartment; endocrine tumors are not discussed here. Owing to the similarities discussed above, we here make pairwise comparisons between HCC and ACC, CCA and PDAC, as well as CCA and pancreatic cystic tumors, and highlight the similarities between these cancers at the histological level and with regard to their mutational landscape.
HEPATOCELLULAR CARCINOMA AND ACINAR CELL CARCINOMA
HCC is frequently associated with cirrhosis caused by hepatitis B and C, alcohol abuse, or non-alcoholic fatty liver disease19. Several reports indicate that HCC develops from hepatocytes20, although it has also been suggested that HCC originates from liver stem cells21. Macroscopically, HCCs appear as nodular or infiltrative tumors. Generally, nodular tumors are well circumscribed, with trabecular formations composed of cells resembling hepatocytes, whereas infiltrative tumors are poorly differentiated22. Pancreatic ACC may originate from acinar cells, as suggested by the expression of acinar-specific enzymes, such as trypsin, lipase, amylase, and carboxyl ester lipase in tumor tissue23,24. Histological features are similar to those of HCC: trabecular structures and cells resembling the acinar cell type of origin are observed; poorly differentiated cells are found in some cases. In both HCC and ACC, scant stroma is detected.
Interestingly, in addition to those shared histological features, HCC and ACC are often driven by dysfunctional Wnt/β-catenin pathway. Genomic alterations have been extensively studied in HCC, revealing that 30%–50% of HCCs display perturbed Wnt/β-catenin (CTNNB1) signaling, mostly resulting from mutations in CTNNB1, APC, and AXIN1 25–28. Genomic studies of ACC are less numerous because of the rarity of cases. However, they reveal that CTNNB1 and APC alterations (mutations, gene loss, and/or promoter hypermethylation) are detected in up to 56% in patients29,30. Chromosomal imbalances are also recurrent in HCC and ACC, possibly as a consequence of APC loss25,31,32. The most frequent amplifications of chromosomal regions are found on 1q, 8q, and 20q in both lesions. This includes amplification of c-MYC at the 8q24 locus.
Another striking similarity between HCC and ACC relates to the KRAS oncogene. Indeed, despite that KRAS is the most frequently mutated oncogene in human cancers, being mutated in 22% of tumors, the frequency of KRAS mutations in HCC and ACC is surprisingly low (0% and 1%–2%, respectively)25,32,33. This observation extends to the downstream effector of KRAS, BRAF, as well as the epidermal growth factor receptor (EGFR), a receptor whose signaling critically depends on KRAS activity32,34. Altogether, this indicates that in both cancer types, the EGFR/KRAS/ERK pathway is not predominantly involved in tumorigenesis, in contrast to many other cancer types.
CHOLANGIOCARCINOMA AND PANCREATIC DUCTAL ADENOCARCINOMA
CCA has a 5-year survival rate of about 10%, and this remains unchanged in the last 20 years35. As for HCC, chronic liver inflammation is a common risk factor. In Southeast Asia, CCA development is often associated with infection by Opisthorchis viverini or Clonorchis sinensis. Based on the anatomical localization, CCAs are classified according to their anatomical localization as intrahepatic CCA (iCCA), perihilar CCA (pCCA), and distal (or extrahepatic) CCA (dCCA)36. While pCCA and dCCA are mainly mucinous adenocarcinomas, iCCAs are histologically heterogeneous: bile ductular type (mixed) arises from small intrahepatic bile ducts, and bile duct type (mucinous) develops from large intrahepatic bile ducts37. Not surprisingly, the diverse anatomical locations of CCA are associated with heterogeneity in the mutational landscape of these tumors38,39.
PDAC is one of the deadliest cancers, and the mean survival period after diagnosis has not significantly improved over the last decades. Lifestyle factors, such as cigarette smoking, excessive alcohol consumption, and obesity, increase the risk of PDAC, likely resulting from chronic pancreatitis40. A striking characteristic of PDAC is the prevalence of oncogenic KRAS mutations, which varies between 88% and 100%41,42. This very high mutation rate suggests that KRAS plays a key role in pancreatic tumorigenesis and prompted the development of mouse models with induction of mutated Kras in the pancreas43. These models revealed that PDAC could originate from acinar cells and not necessarily from ductal cells as suggested by the histological features of the tumors44–46. However, the presence of a Kras mutation in the acinar genome is not sufficient to initiate tumor development and evolution toward malignancy. Tumor development requires association of a KRAS mutation and inflammation45,47,48 and occurs in a stepwise manner starting with inflammation-induced acinar-to-ductal metaplasia (ADM), during which acinar cells acquire a ductal-like phenotype49. When a Kras mutation occurs in this inflammatory context, the metaplastic acinar cell transforms into a neoplastic lesion called pancreatic intraepithelial neoplasia (PanIN), which evolves to cancer upon accumulation of other gene mutations, mainly p53, p16, and Smad4 50–53. Still, this scheme has yet to be confirmed in humans.
In CCA, even though KRAS still appears as the most frequently mutated oncogene, its mutation rate is much less important than the prevalence of KRAS mutations in PDAC16 and varies considerably according to the CCA localization38,54. Like in mouse models of PDAC, the mere presence of a Kras mutation in hepatocytes or cholangiocytes is not sufficient to transform these cells55,56. By analogy with the pancreas, it would be interesting to couple the presence of a Kras mutation in the liver with the presence of inflammation. Supporting the need for such experiments, bile ductular-type CCA is frequently associated with chronic liver diseases (viral hepatitis or cirrhosis) in which inflammation plays an important part in disease progression and cancer initiation. It should be noted that independent of the presence of inflammation, coupling a Kras mutation with mutation in the tumor-suppressor Pten results in iCCA development55.
A central question about CCA development refers to the identity of the cell type of origin. Murine models have shown that iCCA can derive from cholangiocytes or hepatocytes57–59. In the latter case, hepatocyte-to-cholangiocyte transdifferentiation requires activation of the Notch pathway, which is comparable to Notch-controlled ADM in the pancreas60. Progression to iCCA depends on the combination of Notch activation with hepatotoxin-induced liver fibrosis or activation of the AKT pathway59,58, whereas activated Notch and activating Kras mutations synergistically induce PanIN formation from ADM44. Future work will determine the extent of similarities between hepatocyte-derived iCCA and acinar-derived PDAC and whether similar mechanisms operate in humans.
CHOLANGIOCARCINOMA AND PANCREATIC CYSTIC TUMORS
A small proportion of CCA and pancreatic cancers arise from neoplastic lesions called intraductal papillary neoplasm of the bile duct (IPNB) and intraductal papillary mucinous neoplasm of the pancreas (IPMN). Both lesions appear as papillary tumors within dilated duct lumens and are usually associated with the production of mucinous secretions and the presence of cysts61,62. IPNB can be detected both in extra- and large intrahepatic bile ducts, and IPMNs are localized in the main pancreatic duct, the branch ducts, or both locations63–65. IPNB and IPMN are classified into four identical histotypes: gastric, intestinal, pancreatobiliary, and oncocytic types. IPMN evolves into invasive carcinoma in 20% to 40% of cases and IPNB in 4% to 38% of cases61,66,67. Gastric and pancreatobiliary types give rise to tubular adenocarcinoma, whereas intestinal types form mucinous/colloid carcinoma66,67.
Beyond these histological similarities, IPMN and IPNB share common genetic alterations. In humans, KRAS is the most frequently mutated oncogene in both lesions. On average, KRAS is mutated in 29%–46% of IPNB and in 61% of IPMN68,69. GNAS, which encodes the G-protein α stimulatory subunit (Gαs) of heterotrimeric G proteins, is also frequently mutated in IPNB (50%) and IPMN (56%)68,70. Interestingly, KRAS mutations are predominant in gastric (65%–73%) and pancreatobiliary subtypes (64%–100%) compared to intestinal subtype (44%–46%), whereas GNAS mutations are more typically detected in the intestinal subtype (59%–100%) compared to gastric and pancreatobiliary subtypes (46%–65% and 0%–43%, respectively)68,71–73, suggesting that mutated GNAS promotes intestinal differentiation of IPNB and IPMN. Similarities in the mutational profiles of IPNB and IPMN extend beyond KRAS and GNAS mutations. Indeed, the third most frequently mutated gene in IPMN, namely, the gene coding for ubiquitin ligase RNF43, with 23% of the mutated cases, also appears mutated at a high frequency in IPNB (12%)68,74. Full understanding of the role of KRAS, GNAS, and RNF43 in IPNB and IPMN development requires the development of new murine transgenic models. A murine model coupling Kras and Gnas mutations in the pancreas was generated75. These mice develop IPMN, supporting a role of Kras and Gnas mutations in the development of these lesions. However, the mutations were induced during embryogenesis. Considering the sensitivity of embryonic cells to oncogenic stimuli and the resistance of mature pancreatic cells to such injury, Gnas and Kras mutations should ideally be induced in adult mouse pancreas to strengthen the conclusions.
Similar mutational profiles in IPNB and IPMN also suggest that the two lesions originate from the same cell type. Histopathological observations suggest that IPNB and IPMN arise from the biliary and pancreatic ductal cells. However, peribiliary glands (PBGs) and pancreatic duct glands (PDGs) are associated with the large biliary or pancreatic ducts76,77. PBG and PDG are proposed to contain stem/progenitor cells78–80 and may give rise to IPNB and IPMN61. The analysis of mouse models supports the hypothesis that IPMN originates from the PDG81 and that dCCA can originate from the PBG, yet in the latter model, dCCA appeared as an undifferentiated adenocarcinoma, not as a cystic tumor82.
In conclusion, current data support the notion that the mechanisms of tumorigenesis in the liver and pancreas might significantly overlap (summarized in Fig. 1). Development of new transgenic mouse models, especially to explore the possibility that PBGs and PDGs are a source of IPNB and IPMN and subsequently of cancer, as well as studies of patient-derived xenografts and liver and pancreas organoids83,84, will enable us to determine to what exact extent pancreas and liver tumorigenesis can be considered similar.
Figure 1.
Summary of similarities and differences between liver and pancreatic cancers reported in this article.
ACKNOWLEDGMENTS
This work was supported by grants from the Fondation contre le Cancer (Belgium, #2014-125 to F.P.L. and #2016-089 to P.J.), the Université catholique de Louvain (FSR # ID.21000.040 to F.P.L.), Télévie (#7.4502.16 to F.P.L. and P.J.), and the Centre du Cancer (Cliniques universitaires St-Luc). E.G. holds a Télévie fellowship (#7.4600.115). P.J. is a senior research associate at the FRS-FNRS (Belgium).
Footnotes
The authors declare no conflicts of interests.
REFERENCES
- 1. Si-Tayeb K, Lemaigre FP, Duncan SA. Organogenesis and development of the liver. Dev Cell 2010;18(2):175–89. [DOI] [PubMed] [Google Scholar]
- 2. Shih HP, Wang A, Sander M. Pancreas organogenesis: From lineage determination to morphogenesis. Annu Rev Cell Dev Biol. 2013;29:81–105. [DOI] [PubMed] [Google Scholar]
- 3. Lemaigre FP. Mechanisms of liver development: Concepts for understanding liver disorders and design of novel therapies. Gastroenterology 2009;137(1):62–79. [DOI] [PubMed] [Google Scholar]
- 4. Stanger BZ, Hebrok M. Control of cell identity in pancreas development and regeneration. Gastroenterology 2013;144(6):1170–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Goessling W, Stainier DY. Endoderm specification and liver development. Methods Cell Biol. 2016;134:463–83. [DOI] [PubMed] [Google Scholar]
- 6. Bastidas-Ponce A, Scheibner K, Lickert H, Bakhti M. Cellular and molecular mechanisms coordinating pancreas development. Development 2017;144(16):2873–88. [DOI] [PubMed] [Google Scholar]
- 7. Spence JR, Lange AW, Lin SC, Kaestner KH, Lowy AM, Kim I, Whitsett JA, Wells JM. Sox17 regulates organ lineage segregation of ventral foregut progenitor cells. Dev Cell 2009;17(1):62–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vanhorenbeeck V, Jenny M, Cornut JF, Gradwohl G, Lemaigre FP, Rousseau GG, Jacquemin P. Role of the Onecut transcription factors in pancreas morphogenesis and in pancreatic and enteric endocrine differentiation. Dev Biol. 2007;305(2):685–94. [DOI] [PubMed] [Google Scholar]
- 9. Kropp PA, Gannon M. Onecut transcription factors in development and disease. Trends Dev Biol. 2016;9:43–57. [PMC free article] [PubMed] [Google Scholar]
- 10. Yin C. Molecular mechanisms of Sox transcription factors during the development of liver, bile duct, and pancreas. Semin Cell Dev Biol. 2017;63:68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee CS, Friedman JR, Fulmer JT, Kaestner KH. The initiation of liver development is dependent on Foxa transcription factors. Nature 2005;435(7044):944–7. [DOI] [PubMed] [Google Scholar]
- 12. Gao N, LeLay J, Vatamaniuk MZ, Rieck S, Friedman JR, Kaestner KH. Dynamic regulation of Pdx1 enhancers by Foxa1 and Foxa2 is essential for pancreas development. Genes Dev. 2008;22(24):3435–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shen CN, Tosh D. Transdifferentiation of pancreatic cells to hepatocytes. Methods Mol Biol. 2010;640:273–80. [DOI] [PubMed] [Google Scholar]
- 14. Lardon J, De Breuck S, Rooman I, Van Lommel L, Kruhoffer M, Orntoft T, Schuit F, Bouwens L. Plasticity in the adult rat pancreas: Transdifferentiation of exocrine to hepatocyte-like cells in primary culture. Hepatology 2004;39(6):1499–507. [DOI] [PubMed] [Google Scholar]
- 15. Rao MS, Reddy JK. Hepatic transdifferentiation in the pancreas. Semin Cell Biol. 1995;6(3):151–6. [DOI] [PubMed] [Google Scholar]
- 16. Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med. 2014;371(11):1039–49. [DOI] [PubMed] [Google Scholar]
- 17. Klimstra DS, Adsay V. Acinar neoplasms of the pancreas—A summary of 25 years of research. Semin Diagn Pathol. 2016;33(5):307–18. [DOI] [PubMed] [Google Scholar]
- 18. La Rosa S, Sessa F, Capella C. Acinar cell carcinoma of the pancreas: Overview of clinicopathologic features and insights into the molecular pathology. Front Med. (Lausanne) 2015;2:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet 2012;379(9822):1245–55. [DOI] [PubMed] [Google Scholar]
- 20. Mu X, Espanol-Suner R, Mederacke I, Affo S, Manco R, Sempoux C, Lemaigre FP, Adili A, Yuan D, Weber A, and others. Hepatocellular carcinoma originates from hepatocytes and not from the progenitor/biliary compartment. J Clin Invest. 2015;125(10):3891–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Roskams T. Liver stem cells and their implication in hepatocellular and cholangiocarcinoma. Oncogene 2006;25(27):3818–22. [DOI] [PubMed] [Google Scholar]
- 22. Paradis V. Histopathology of hepatocellular carcinoma. Recent Results Cancer Res. 2013;190:21–32. [DOI] [PubMed] [Google Scholar]
- 23. Klimstra DS, Heffess CS, Oertel JE, Rosai J. Acinar cell carcinoma of the pancreas. A clinicopathologic study of 28 cases. Am J Surg Pathol. 1992;16(9):815–37. [DOI] [PubMed] [Google Scholar]
- 24. La Rosa S, Adsay V, Albarello L, Asioli S, Casnedi S, Franzi F, Marando A, Notohara K, Sessa F, Vanoli A, and others. Clinicopathologic study of 62 acinar cell carcinomas of the pancreas: Insights into the morphology and immunophenotype and search for prognostic markers. Am J Surg Pathol. 2012;36(12):1782–95. [DOI] [PubMed] [Google Scholar]
- 25. Kan Z, Zheng H, Liu X, Li S, Barber TD, Gong Z, Gao H, Hao K, Willard MD, Xu J, and others. Whole-genome sequencing identifies recurrent mutations in hepatocellular carcinoma. Genome Res. 2013;23(9):1422–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, Calderaro J, Bioulac-Sage P, Letexier M, Degos F, and others. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44(6):694–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schulze K, Imbeaud S, Letouze E, Alexandrov LB, Calderaro J, Rebouissou S, Couchy G, Meiller C, Shinde J, Soysouvanh F, and others. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47(5):505–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Woo HG, Kim SS, Cho H, Kwon SM, Cho HJ, Ahn SJ, Park ES, Lee JS, Cho SW, Cheong JY. Profiling of exome mutations associated with progression of HBV-related hepatocellular carcinoma. PLoS One 2014;9(12):e115152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Furlan D, Sahnane N, Bernasconi B, Frattini M, Tibiletti MG, Molinari F, Marando A, Zhang L, Vanoli A, Casnedi S, and others. APC alterations are frequently involved in the pathogenesis of acinar cell carcinoma of the pancreas, mainly through gene loss and promoter hypermethylation. Virchows Arch. 2014;464(5):553–64. [DOI] [PubMed] [Google Scholar]
- 30. Abraham SC, Wu TT, Hruban RH, Lee JH, Yeo CJ, Conlon K, Brennan M, Cameron JL, Klimstra DS. Genetic and immunohistochemical analysis of pancreatic acinar cell carcinoma: Frequent allelic loss on chromosome 11p and alterations in the APC/beta-catenin pathway. Am J Pathol. 2002;160(3):953–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Guan XY, Fang Y, Sham JS, Kwong DL, Zhang Y, Liang Q, Li H, Zhou H, Trent JM. Recurrent chromosome alterations in hepatocellular carcinoma detected by comparative genomic hybridization. Genes Chromosomes Cancer 2000;29(2):110–6. [PubMed] [Google Scholar]
- 32. Bergmann F, Aulmann S, Sipos B, Kloor M, von Heydebreck A, Schweipert J, Harjung A, Mayer P, Hartwig W, Moldenhauer G, and others. Acinar cell carcinomas of the pancreas: A molecular analysis in a series of 57 cases. Virchows Arch. 2014;465(6):661–72. [DOI] [PubMed] [Google Scholar]
- 33. Hoorens A, Lemoine NR, McLellan E, Morohoshi T, Kamisawa T, Heitz PU, Stamm B, Ruschoff J, Wiedenmann B, Kloppel G. Pancreatic acinar cell carcinoma. An analysis of cell lineage markers, p53 expression, and Ki-ras mutation. Am J Pathol. 1993;143(3):685–98. [PMC free article] [PubMed] [Google Scholar]
- 34. Tannapfel A, Sommerer F, Benicke M, Katalinic A, Uhlmann D, Witzigmann H, Hauss J, Wittekind C. Mutations of the BRAF gene in cholangiocarcinoma but not in hepatocellular carcinoma. Gut 2003;52(5):706–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tyson GL, El-Serag HB. Risk factors for cholangiocarcinoma. Hepatology 2011;54(1):173–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rizvi S, Khan SA, Hallemeier CL, Kelley RK, Gores GJ. Cholangiocarcinoma—Evolving concepts and therapeutic strategies. Nat Rev Clin Oncol. 2018;15(2):95–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Banales JM, Cardinale V, Carpino G, Marzioni M, Andersen JB, Invernizzi P, Lind GE, Folseraas T, Forbes SJ, Fouassier L, and others. Expert consensus document: Cholangiocarcinoma: Current knowledge and future perspectives consensus statement from the European Network for the Study of Cholangiocarcinoma (ENS-CCA). Nat Rev Gastroenterol Hepatol. 2016;13(5):261–80. [DOI] [PubMed] [Google Scholar]
- 38. Nakamura H, Arai Y, Totoki Y, Shirota T, Elzawahry A, Kato M, Hama N, Hosoda F, Urushidate T, Ohashi S, and others. Genomic spectra of biliary tract cancer. Nat Genet. 2015;47(9):1003–10. [DOI] [PubMed] [Google Scholar]
- 39. Rizvi S, Gores GJ. Emerging molecular therapeutic targets for cholangiocarcinoma. J Hepatol. 2017;67(3):632–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vincent A, Herman J, Schulick R, Hruban RH, Goggins M. Pancreatic cancer. Lancet 2011;378(9791):607–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, Johns AL, Miller D, Nones K, Quek K, and others. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015;518(7540):495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Witkiewicz AK, McMillan EA, Balaji U, Baek G, Lin WC, Mansour J, Mollaee M, Wagner KU, Koduru P, Yopp A, and others. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun. 2015;6:6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gopinathan A, Morton JP, Jodrell DI, Sansom OJ. GEMMs as preclinical models for testing pancreatic cancer therapies. Dis Model Mech. 2015;8(10):1185–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. De La O J, Emerson LL, Goodman JL, Froebe SC, Illum BE, Curtis AB, Murtaugh LC. Notch and Kras reprogram pancreatic acinar cells to ductal intraepithelial neoplasia. Proc Natl Acad Sci USA 2008;105(48):18907–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Guerra C, Schuhmacher AJ, Canamero M, Grippo PJ, Verdaguer L, Perez-Gallego L, Dubus P, Sandgren EP, Barbacid M. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell 2007;11(3):291–302. [DOI] [PubMed] [Google Scholar]
- 46. Kopp JL, von Figura G, Mayes E, Liu FF, Dubois CL, Morris JPt, Pan FC, Akiyama H, Wright CV, Jensen K, and others. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell 2012;22(6):737–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. De La O J, Murtaugh LC. Notch and Kras in pancreatic cancer: At the crossroads of mutation, differentiation and signaling. Cell Cycle 2009;8(12):1860–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Morris JPt, Cano DA, Sekine S, Wang SC, Hebrok M. Beta-catenin blocks Kras-dependent reprogramming of acini into pancreatic cancer precursor lesions in mice. J Clin Invest. 2010;120(2):508–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Prévot PP, Augereau C, Simion A, Van den Steen G, Dauguet N, Lemaigre FP, Jacquemin P. Let-7b and miR-495 stimulate differentiation and prevent metaplasia of pancreatic acinar cells by repressing HNF6. Gastroenterology 2013;145(3):668–78 e3. [DOI] [PubMed] [Google Scholar]
- 50. Aguirre AJ, Bardeesy N, Sinha M, Lopez L, Tuveson DA, Horner J, Redston MS, DePinho RA. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17(24):3112–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bardeesy N, Cheng KH, Berger JH, Chu GC, Pahler J, Olson P, Hezel AF, Horner J, Lauwers GY, Hanahan D, and others. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev. 2006;20(22):3130–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hill R, Calvopina JH, Kim C, Wang Y, Dawson DW, Donahue TR, Dry S, Wu H. PTEN loss accelerates KrasG12D-induced pancreatic cancer development. Cancer Res. 2010;70(18):7114–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, Rustgi AK, Chang S, Tuveson DA. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 2005;7(5):469–83. [DOI] [PubMed] [Google Scholar]
- 54. Sohal DP, Shrotriya S, Abazeed M, Cruise M, Khorana A. Molecular characteristics of biliary tract cancer. Crit Rev Oncol Hematol. 2016;107:111–8. [DOI] [PubMed] [Google Scholar]
- 55. Ikenoue T, Terakado Y, Nakagawa H, Hikiba Y, Fujii T, Matsubara D, Noguchi R, Zhu C, Yamamoto K, Kudo Y, and others. A novel mouse model of intrahepatic cholangiocarcinoma induced by liver-specific Kras activation and Pten deletion. Sci Rep. 2016;6:23899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ray KC, Bell KM, Yan J, Gu G, Chung CH, Washington MK, Means AL. Epithelial tissues have varying degrees of susceptibility to Kras(G12D)-initiated tumorigenesis in a mouse model. PLoS One 2011;6(2):e16786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Guest RV, Boulter L, Kendall TJ, Minnis-Lyons SE, Walker R, Wigmore SJ, Sansom OJ, Forbes SJ. Cell lineage tracing reveals a biliary origin of intrahepatic cholangiocarcinoma. Cancer Res. 2014;74(4):1005–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fan B, Malato Y, Calvisi DF, Naqvi S, Razumilava N, Ribback S, Gores GJ, Dombrowski F, Evert M, Chen X, and others. Cholangiocarcinomas can originate from hepatocytes in mice. J Clin Invest. 2012;122(8):2911–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sekiya S, Suzuki A. Intrahepatic cholangiocarcinoma can arise from Notch-mediated conversion of hepatocytes. J Clin Invest. 2012;122(11):3914–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Miyamoto Y, Maitra A, Ghosh B, Zechner U, Argani P, Iacobuzio-Donahue CA, Sriuranpong V, Iso T, Meszoely IM, Wolfe MS, and others. Notch mediates TGF alpha-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer Cell 2003;3(6):565–76. [DOI] [PubMed] [Google Scholar]
- 61. Ohtsuka M, Shimizu H, Kato A, Yoshitomi H, Furukawa K, Tsuyuguchi T, Sakai Y, Yokosuka O, Miyazaki M. Intraductal papillary neoplasms of the bile duct. Int J Hepatol. 2014;2014:459091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nakanuma Y, Miyata T, Uchida T. Latest advances in the pathological understanding of cholangiocarcinomas. Expert Rev Gastroenterol Hepatol. 2016;10(1):113–27. [DOI] [PubMed] [Google Scholar]
- 63. Rocha FG, Lee H, Katabi N, DeMatteo RP, Fong Y, D’Angelica MI, Allen PJ, Klimstra DS, Jarnagin WR. Intraductal papillary neoplasm of the bile duct: A biliary equivalent to intraductal papillary mucinous neoplasm of the pancreas? Hepatology 2012;56(4):1352–60. [DOI] [PubMed] [Google Scholar]
- 64. Furukawa T, Kloppel G, Volkan Adsay N, Albores-Saavedra J, Fukushima N, Horii A, Hruban RH, Kato Y, Klimstra DS, Longnecker DS, and others. Classification of types of intraductal papillary-mucinous neoplasm of the pancreas: A consensus study. Virchows Arch. 2005;447(5):794–9. [DOI] [PubMed] [Google Scholar]
- 65. Nakanuma Y, Kakuda Y, Uesaka K, Miyata T, Yamamoto Y, Fukumura Y, Sato Y, Sasaki M, Harada K, Takase M. Characterization of intraductal papillary neoplasm of bile duct with respect to histopathologic similarities to pancreatic intraductal papillary mucinous neoplasm. Hum Pathol. 2016;51:103–13. [DOI] [PubMed] [Google Scholar]
- 66. Yamada S, Fujii T, Shimoyama Y, Kanda M, Nakayama G, Sugimoto H, Koike M, Nomoto S, Fujiwara M, Nakao A, and others. Clinical implication of morphological subtypes in management of intraductal papillary mucinous neoplasm. Ann Surg Oncol. 2014;21(7):2444–52. [DOI] [PubMed] [Google Scholar]
- 67. Mino-Kenudson M, Fernandez-del Castillo C, Baba Y, Valsangkar NP, Liss AS, Hsu M, Correa-Gallego C, Ingkakul T, Perez Johnston R, Turner BG, and others. Prognosis of invasive intraductal papillary mucinous neoplasm depends on histological and precursor epithelial subtypes. Gut 2011;60(12):1712–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lee JH, Kim Y, Choi JW, Kim YS. KRAS, GNAS, and RNF43 mutations in intraductal papillary mucinous neoplasm of the pancreas: A meta-analysis. Springerplus 2016;5(1):1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Gordon-Weeks AN, Jones K, Harriss E, Smith A, Silva M. Systematic review and meta-analysis of current experience in treating IPNB: Clinical and pathological correlates. Ann Surg. 2016;263(4):656–63. [DOI] [PubMed] [Google Scholar]
- 70. Sasaki M, Matsubara T, Nitta T, Sato Y, Nakanuma Y. GNAS and KRAS mutations are common in intraductal papillary neoplasms of the bile duct. PLoS One 2013;8(12):e81706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wu J, Matthaei H, Maitra A, Dal Molin M, Wood LD, Eshleman JR, Goggins M, Canto MI, Schulick RD, Edil BH, and others. Recurrent GNAS mutations define an unexpected pathway for pancreatic cyst development. Sci Transl Med. 2011;3(92):92ra66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kuboki Y, Shimizu K, Hatori T, Yamamoto M, Shibata N, Shiratori K, Furukawa T. Molecular biomarkers for progression of intraductal papillary mucinous neoplasm of the pancreas. Pancreas 2015;44(2):227–35. [DOI] [PubMed] [Google Scholar]
- 73. Hosoda W, Sasaki E, Murakami Y, Yamao K, Shimizu Y, Yatabe Y. GNAS mutation is a frequent event in pancreatic intraductal papillary mucinous neoplasms and associated adenocarcinomas. Virchows Arch. 2015;466(6):665–74. [DOI] [PubMed] [Google Scholar]
- 74. Tsai JH, Liau JY, Yuan CT, Cheng ML, Yuan RH, Jeng YM. RNF43 mutation frequently occurs with GNAS mutation and mucin hypersecretion in intraductal papillary neoplasms of the bile duct. Histopathology 2017;70(5):756–65. [DOI] [PubMed] [Google Scholar]
- 75. Taki K, Ohmuraya M, Tanji E, Komatsu H, Hashimoto D, Semba K, Araki K, Kawaguchi Y, Baba H, Furukawa T. GNAS(R201H) and Kras(G12D) cooperate to promote murine pancreatic tumorigenesis recapitulating human intraductal papillary mucinous neoplasm. Oncogene 2016;35(18):2407–12. [DOI] [PubMed] [Google Scholar]
- 76. Strobel O, Rosow DE, Rakhlin EY, Lauwers GY, Trainor AG, Alsina J, Fernandez-Del Castillo C, Warshaw AL, Thayer SP. Pancreatic duct glands are distinct ductal compartments that react to chronic injury and mediate Shh-induced metaplasia. Gastroenterology 2010;138(3):1166–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Nakanuma Y, Katayanagi K, Terada T, Saito K. Intrahepatic peribiliary glands of humans. I. Anatomy, development and presumed functions. J Gastroenterol Hepatol. 1994;9(1):75–9. [DOI] [PubMed] [Google Scholar]
- 78. Carpino G, Renzi A, Cardinale V, Franchitto A, Onori P, Overi D, Rossi M, Berloco PB, Alvaro D, Reid LM, and others. Progenitor cell niches in the human pancreatic duct system and associated pancreatic duct glands: An anatomical and immunophenotyping study. J Anat. 2016;228(3):474–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lanzoni G, Cardinale V, Carpino G. The hepatic, biliary, and pancreatic network of stem/progenitor cell niches in humans: A new reference frame for disease and regeneration. Hepatology 2016;64(1):277–86. [DOI] [PubMed] [Google Scholar]
- 80. DiPaola F, Shivakumar P, Pfister J, Walters S, Sabla G, Bezerra JA. Identification of intramural epithelial networks linked to peribiliary glands that express progenitor cell markers and proliferate after injury in mice. Hepatology 2013;58(4):1486–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Yamaguchi J, Mino-Kenudson M, Liss AS, Chowdhury S, Wang TC, Fernandez-Del Castillo C, Lillemoe KD, Warshaw AL, Thayer SP. Loss of Trefoil Factor 2 from pancreatic duct glands promotes formation of intraductal papillary mucinous neoplasms in mice. Gastroenterology 2016;151(6):1232–44 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Nakagawa H, Suzuki N, Hirata Y, Hikiba Y, Hayakawa Y, Kinoshita H, Ihara S, Uchino K, Nishikawa Y, Ijichi H, and others. Biliary epithelial injury-induced regenerative response by IL-33 promotes cholangiocarcinogenesis from peribiliary glands. Proc Natl Acad Sci USA 2017;114(19):E3806–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Broutier L, Mastrogiovanni G, Verstegen MM, Francies HE, Gavarro LM, Bradshaw CR, Allen GE, Arnes-Benito R, Sidorova O, Gaspersz MP, and others. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat Med. 2017;23(12):1424–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Baker LA, Tiriac H, Clevers H, Tuveson DA. Modeling pancreatic cancer with organoids. Trends Cancer 2016;2(4):176–90. [DOI] [PMC free article] [PubMed] [Google Scholar]