Abstract
Cysteine dioxygenase 1 (CDO1) converts cysteine to cysteine sulfinic acid, which can be further converted by cysteine sulfinic acid decarboxylase (CSAD) to hypotaurine for taurine production. This cysteine catabolic pathway plays a major role in regulating hepatic cysteine homeostasis. Furthermore, taurine is used for bile acid conjugation, which enhances bile acid solubility and physiological function in the gut. Recent studies show that this cysteine catabolic pathway is repressed by bile acid signaling, but the molecular mechanisms have not been fully elucidated. The mechanisms of bile acid and farnesoid X receptor (FXR) regulation of hepatic CSAD expression were studied in mice and hepatocytes. We showed that hepatocyte nuclear factor 4α (HNF4α) bound the mouse CSAD proximal promoter and induced CSAD transcription. FXR-induced small heterodimer partner (SHP) repressed mouse CSAD gene transcription via interacting with HNF4α as a repressor. Consistent with this model, cholic acid feeding, obeticholic acid administration, and liver HNF4α knockdown reduced hepatic CSAD expression, while liver SHP knockout and apical sodium-dependent bile acid transporter (ASBT) inhibitor treatment induced hepatic CSAD expression in mice. Furthermore, TNF-α also inhibited CSAD expression, which may be partially mediated by reduced HNF4α in mouse hepatocytes. In contrast, bile acids and GW4064 did not inhibit CSAD expression in human hepatocytes. This study identified mouse CSAD as a novel transcriptional target of HNF4α. Bile acids and cytokines repress hepatic CSAD, which closely couples taurine production to bile acid synthesis in mice. The species-specific regulation of CSAD reflects the differential preference of bile acid conjugation to glycine and taurine in humans and mice, respectively.
Key words: Farnesoid X receptor (FXR), Small heterodimer partner (SHP), Nuclear receptor, Cholestasis, Cytokine
INTRODUCTION
Bile acids are synthesized from cholesterol in hepatocytes and circulate between the liver and the small intestine via a process called enterohepatic circulation1. In addition to facilitating nutrient absorption in the gut, bile acids play important roles in regulating metabolic homeostasis, immune response, and cell proliferation in physiological and pathological conditions1. Bile acids exert strong feedback inhibition on hepatic bile acid synthesis via transcriptional repression of the CYP7A1 gene encoding the rate-limiting enzyme cholesterol 7α-hydroxylase (CYP7A1)1. Bile acids activate the nuclear receptor farnesoid X receptor (FXR) to induce the atypical nuclear receptor small heterodimer partner (SHP) in hepatocytes, which acts as a repressor of hepatocyte nuclear factor 4α (HNF4α) and liver receptor homolog-1 (LRH-1) on the CYP7A1 gene promoter2,3. Bile acids in the intestine activate FXR to induce an endocrine hormone fibroblast growth factor 15 to transcriptionally repress the CYP7A1 gene in hepatocytes4. After synthesis, the majority of the bile acids are efficiently conjugated to glycine or taurine to form N-acyl amidates in hepatocytes5. Bile acid amidation is a two-step reaction mediated by bile acid–CoA synthase (BACS) and bile acid–CoA:amino acid N-acetyltransferase (BAAT). Humans and some nonhuman primates can use both glycine and taurine for bile acid conjugation, and the human bile acid pool contains about two times more glycine-conjugated bile acids than taurine-conjugated bile acids6,7. In contrast, mice and rats primarily use taurine for bile acid conjugation and have little glycine-conjugated bile acids6,7. Bile acid conjugation to amino acids increases bile acid solubility in the physiological environment and enhances their digestive function in the small intestine. Humans with defective bile acid-conjugating enzymes show vitamin deficiency, growth delay, and cholangiopathy8,9.
In the liver, cysteine dioxygenase 1 (CDO1) catalyzes the irreversible conversion of cysteine to cysteine sulfinic acid, which is further converted by cysteine sulfinic acid decarboxylase (CSAD) to hypotaurine, which leads to the synthesis of taurine10. This pathway is highly active in the liver and serves two important functions: cysteine elimination and taurine synthesis10–12. Elevated cellular cysteine concentration increases CDO1 mRNA expression and decreases CDO1 protein degradation, which serves as a feedforward mechanism to prevent intracellular cysteine accumulation10. We recently reported that this cysteine elimination pathway was under the negative regulation by bile acids and FXR, and increased cysteine flux through this pathway due to loss of bile acid repression of CDO1 decreased intracellular cysteine availability, which impaired GSH synthesis and sensitized liver to acetaminophen toxicity13. It has also been reported previously that hepatic CSAD was strongly repressed by bile acids in mice14, which suggested that hepatic taurine production and bile acid synthesis may be coordinately regulated by bile acids. Currently, the molecular mechanism of bile acid regulation of CSAD expression remains incompletely understood. To better characterize the bile acid-regulated hepatic amino acid metabolic pathways, here we further elucidated the differential regulation of hepatic CSAD transcription by bile acids and nuclear receptors in mouse livers and human hepatocytes.
MATERIALS AND METHODS
Reagents
Anti-CSAD (ab91016) antibody was purchased from Abcam (Cambridge, MA, USA). Anti-HNF4α antibody (PP-H1415) and recombinant human TNF-α protein were purchased from R&D Systems (Minneapolis, MN, USA). Anti-actin antibody and sodium cholate were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Mice
Male C57BL/6J mice (The Jackson Lab) 10–12 weeks old were used for the study. Sodium cholate (0.5%, w/w) was mixed with food. Obeticholic acid (OCA) and GSK2330672 (MedChem Express, South Brunswick, NJ, US) were prepared in 1% methylcellulose and administered by oral gavage. OCA was given at 20 mg/kg/day for 2 weeks. GSK2330672 was given at 2 mg/kg twice a day for 1 week. SHP-floxed mice on a C57BL/6J background were a gift from Drs. Johan Auwerx and Kristina Schoonjans (the Ecole Polytechnique de Lausanne). Liver-specific SHP KO mice (L-SHP KO) were generated by breeding SHP-floxed mice with Albumin-Cre mice (Jackson Laboratory). Littermates that do not express the Cre recombinase were used as wild-type (WT) controls. Bile duct ligation (BDL) in male C57BL/6J mice was performed as previously described15. All animal protocols were approved by the Institutional Animal Care and Use Committee.
Recombinant Adenovirus
Ad-Null was purchased from Vector Biolabs Inc. (Philadelphia, PA, USA). Ad-shHNF4α was a gift from Dr. Yanqiao Zhang (Northeast Ohio Medical University, Rootstown, OH, USA). Adenovirus was purified from HEK293A cells by CsCl centrifugation. Adenovirus titer was determined with an Adeno-X rapid titer kit from Clontech (Mountain View, CA, USA). Mice were injected 1 × 109 pfu/mouse adenovirus via the tail vein. Hepatic gene expression was analyzed 7 days postinjection in overnight-fasted mice.
Cell Culture
Primary mouse and human hepatocytes were obtained from the Cell Isolation Core at KUMC. Male 10- to 12-week-old WT C57BL/6J mice were used for hepatocyte isolation. Primary hepatocytes were plated in collagen-coated plates, and culture medium was replaced 3 h later after the cells were attached. Treatments were initiated after 16 h. During the treatments, cells were cultured in serum-free DMEM supplemented with 1% penicillin–streptomycin. For HNF4α knockdown experiments, Ad-scramble, and Ad-shHNF4α (MOI = 10) were added to culture medium 3 h after the hepatocytes were plated. TNF-α treatment was initiated after 16 h. Similarly, Ad-Null and Ad-cre were added 3 h after hepatocytes from the SHP-floxed mice were plated, and TNF-α treatment was initiated after 16 h. AML12 cells were provided by Dr. Yanqiao Zhang (Northeast Ohio Medical University).
Electrophoretic Mobility Shift Assay (EMSA)
An EMSA Kit with SYBR Green detection (Thermo Fisher Scientific, Grand Island, NY, USA) was used to perform this assay following the manufacturer’s instruction. Recombinant human HNF4α protein (TP316588) was purchased from OriGene (Rockville, MD, USA). DNA probes were chemically synthesized. Images were acquired with a LI-COR Odyssey Imaging System.
Western Blotting
Cells or livers were homogenized in 1× RIPA buffer containing 1% SDS and protease inhibitors, incubated for 1 h on ice. Supernatant after centrifugation was used for SDS-PAGE and immunoblotting. Densitometry was performed using ImageJ software and normalized to loading controls.
Real-Time PCR
Total RNA was isolated with TRI Reagent (Sigma-Aldrich). Reverse transcription was performed with SuperScript III Reverse Transcriptase (Thermo Fisher Scientific). SYBR Master Mix was used in real-time PCR. Amplification of 18S was used for normalization. Relative mRNA expression was calculated using the comparative CT (Ct) method and expressed as 2−ΔΔCt. Real-time PCR was performed with a Bio-Rad CFX384 real-time PCR detection system.
Luciferase Reporter Assay
Mouse CSAD promoter fragments were generated by PCR and cloned into PGL3-basic vector (Promega, Madison, WI, USA). Mutations were introduced using a QuickChange Site-Directed Mutagenesis Kit (Agilent Technologies Inc., Santa Clara, CA, USA). Expression plasmids were described previously16,17. Luciferase reporter constructs and expression plasmids were transfected into AML12 cells with Lipofectamine 3000 reagent following the manufacturer’s instruction (Thermo Fisher Scientific). Luciferase activity was measured with the Bright-Glo Luciferase Assay System (Promega), and β-galactosidase activity was measured with the β-Galactosidase Enzyme Assay System (Promega) at 48 h after transfection. Luciferase activity was normalized to β-galactosidase activity and expressed as relative luciferase activity. Results of triplicate assays were expressed as mean ± SD. A representative assay of three independent experiments was shown.
Chromatin Immunoprecipitation (ChIP) Assay
Freshly isolated nuclei from mouse livers were cross-linked in formaldehyde. ChIP assay was performed with a ChIP assay kit (Millipore, Bellerica, MA, USA) as described previously18. Anti-HNF4α antibody (PP-H1415; R&D Systems) was used in immunoprecipitation. The ChIP assay real-time PCR primer pairs amplify the −57/+60 Csad chromatin region (relative to the transcriptional start site as +1).
Statistical Analysis
Results were expressed as mean ± SE or mean ± SD as noted. Statistical analysis was performed by either two-way ANOVA followed by Tukey post hoc test or Student’s t-test. A value of p < 0.05 was considered statistically significant.
RESULTS
Bile Acids Repress CSAD Gene Transcription in Mice
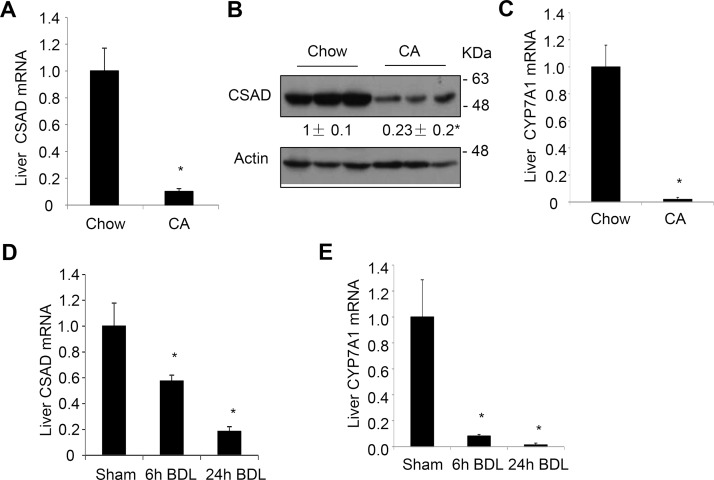
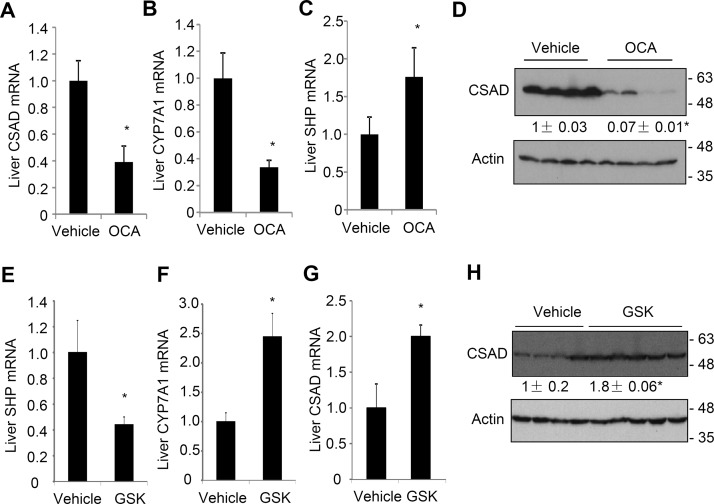
Feeding mice a chow diet containing 0.5% cholic acid for 7 days significantly decreased hepatic CSAD mRNA and protein (Fig. 1A and B). As positive controls, cholic acid-fed mice also showed reduced CYP7A1 mRNA (Fig. 1C) and increased SHP mRNA15. We next turned to BDL mice, which we have recently shown to have intrahepatic bile acid accumulation and induction of liver SHP and TNF-α expression15. BDL time-dependently reduced hepatic CSAD and CYP7A1 mRNA expression at the 6- and 24-h time points in mice (Fig. 1D and E). To further determine if the bile acid-repressive effect on CSAD is mediated by FXR, we treated mice with the FXR agonist OCA. Results showed that OCA significantly reduced hepatic CSAD and CYP7A1 mRNA and induced hepatic SHP mRNA (Fig. 2A–C). Consistently, CSAD protein was significantly reduced by OCA treatment in mouse livers (Fig. 2D). Finally, we treated mice with an intestine-restricted apical sodium-dependent bile acid transporter (ASBT) inhibitor GSK2330672 (GSK672) to block intestine bile acid reuptake in mice. As expected, GSK672 decreased hepatic SHP and induced CYP7A1 mRNA (Fig. 2E and F), suggesting a reduced hepatic FXR activation by bile acids. Hepatic CSAD mRNA and protein were significantly increased in mice treated with GSK672 (Fig. 2G and H). Taken together, these results suggest that bile acids activate FXR to inhibit hepatic CSAD expression.
Figure 1.
Bile acids repress hepatic cysteine sulfinic acid decarboxylase (CSAD) expression in mice. (A–C) Male C57BL/6J mice were fed a chow diet or a chow diet containing 0.5% cholic acid for 7 days. Mice were fasted overnight, and tissues were collected. Hepatic CSAD mRNA and protein were determined. The mRNA results were expressed as mean ± SE. n = 5. *Statistical significance versus chow-fed mice. (D, E) Male C57BL/6J mice were subjected to sham operation or bile duct ligation (BDL). Liver CSAD and CYP7A1 mRNA was measured. The mRNA results were expressed as mean ± SE. n = 4–7. *Statistical significance versus sham. Densitometry was performed using ImageJ software and normalized to loading controls.
Figure 2.
Obeticholic acid (OCA) represses hepatic CSAD, while an apical sodium-dependent bile acid transporter (ASBT) inhibitor induces hepatic CSAD in mice. (A–D) Male C57BL/6J mice fed a chow diet were gavaged with OCA (20 mg/kg/day) or vehicle for 2 weeks (n = 4). Mice were fasted overnight, and tissues were collected. Liver mRNA and protein were measured. (E–H) Male C57BL/6J mice were treated with GSK672 (2 mg/kg twice a day for 1 week) via oral gavage (n = 5). Mice were fasted overnight, and tissues were collected. Liver mRNA and protein were measured. All mRNA results were expressed as mean ± SE. *Statistical significance versus vehicle. Densitometry was performed using ImageJ software and normalized to loading controls.
Mouse CSAD Is a Novel Target Gene of HNF4α
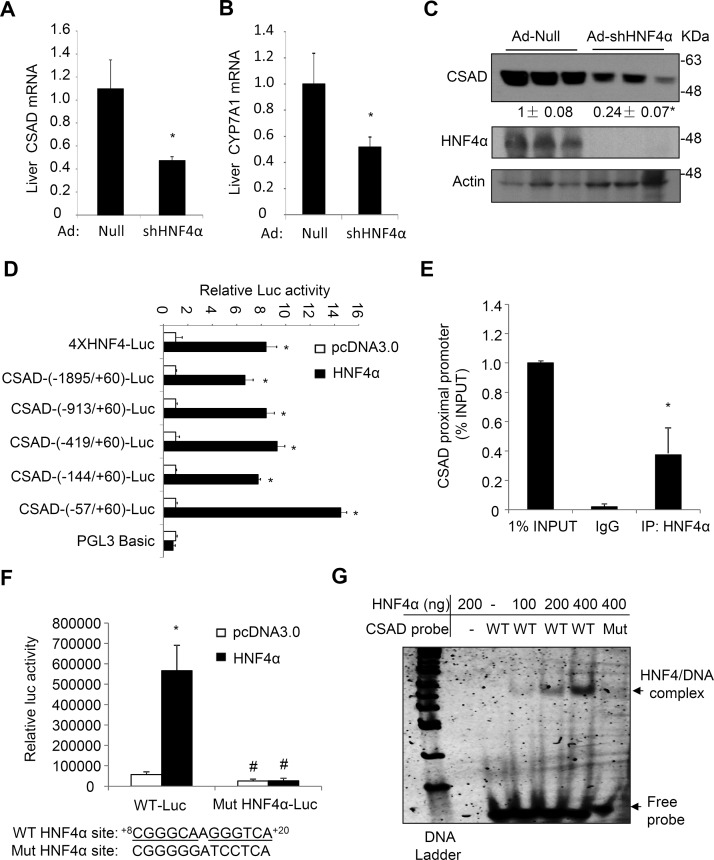
The molecular mechanism by which bile acids and FXR activation repressed CSAD expression in mouse livers was still not clear. HNF4α is known to induce a large number of liver-enriched genes19,20. Furthermore, FXR-induced SHP is known to act as a repressor of HNF4α to inhibit many HNF4α target genes21–23. Analysis of a published mouse liver HNF4α ChIP-seq data set revealed HNF4α binding peaks in the CSAD promoter chromatin region24, suggesting that CSAD may be regulated by HNF4α. Indeed, adenovirus-mediated knockdown of liver HNF4α in mice markedly decreased hepatic CSAD mRNA and protein expression (Fig. 3A and C). As a positive control, hepatic CYP7A1 mRNA was also reduced upon HNF4α knockdown in mice (Fig. 3B)25,26. To determine if HNF4α directly induces CSAD transcription, we constructed a series of mouse CSAD promoter luciferase reporter constructs and performed reporter assays in mouse liver AML12 cells. Results showed that cotransfection of HNF4α strongly induced the luciferase reporter activity driven by various CSAD promoter fragments in mouse liver AML12 cells, and the HNF4α-responsive region was mapped to the CSAD proximal promoter region (Fig. 3D). As a positive control, HNF4α cotransfection induced luciferase reporter activity driven by an artificial promoter containing four copies of the consensus HNF4α binding sites (Fig. 3D). The previously published ChIP-seq data set showed one HNF4α binding peak in the CSAD proximal promoter chromatin region24. ChIP assay was then used to confirm HNF4α occupancy of the proximal CSAD promoter chromatin region in mouse livers (Fig. 3E). Sequence analysis identified a putative HNF4α binding site containing the CAAAG-like core sequence in this region (Fig. 3F)20. Mutations introduced into this sequence abolished HNF4α induction of the CSAD promoter reporter activity (Fig. 3F). EMSA further confirmed direct HNF4α binding to the WT CSAD DNA probe containing the putative HNF4α binding site, which was abolished when the HNF4α binding site mutant DNA probe was used in EMSA (Fig. 3G).
Figure 3.
Hepatocyte nuclear factor 4α (HNF4α) binds mouse CSAD promoter to induce its gene transcription. (A–C) C57BL/6J male mice (n = 5) were injected with Ad-Null or Ad-shHNF4α at a dose of 1 × 109 pfu/mouse. After 7 days, mice were fasted overnight, and tissues were collected. Liver mRNA and protein were measured. The mRNA results are expressed as mean ± SEM. *Statistical significance versus Ad-Null. Densitometry was performed using ImageJ software and normalized to loading controls. (D) Luciferase reporter constructs (0.2 μg), β-gal expression construct (0.05 μg), and 0.1 μg of pcDNA3.0 or HNF4α expression plasmid were cotransfected into AML12 cells. Luciferase and β-gal activities were measured 48 h later. Controls (pcDNA3.0) for different reporter constructs were normalized to “1” for comparison. *Statistical significance versus pcDNA3.0. The indicated promoter length is relative to the transcriptional start site of the Csad gene as “+1”. The “ATG” start codon of the Csad gene starts at +169 bp. (E) Chromatin immunoprecipitation (ChIP) assay detection of HNF4α occupancy to the proximal promoter region of the Csad chromatin in mouse livers. The ChIP assay real-time PCR primer pairs amplify the −57/+60 Csad chromatin region. (F) Wild-type (WT) and HNF4α mutant CSAD-(−913/+60)-luc constructs (0.2 μg), β-gal expression construct (0.05 μg), and 0.1 μg of pcDNA3.0 or HNF4α expression plasmid were cotransfected into AML12 cells. Luciferase and β-gal activities were measured 48 h later. The putative HNF4α binding site and mutant sequences are shown below the bar graph. *Statistical significance versus pcDNA3.0. #Statistical significance versus WT-Luc. (G) EMSA detection of HNF4α binding to the WT but not the mutant HNF4α binding site in the CSAD promoter DNA probe. Mut, CSAD probe with mutations introduced into the HNF4α binding site as shown in (C). All reporter assay results are shown as mean ± SD of triplicate assays.
SHP and Cytokines Target HNF4α to Repress Mouse CSAD Expression
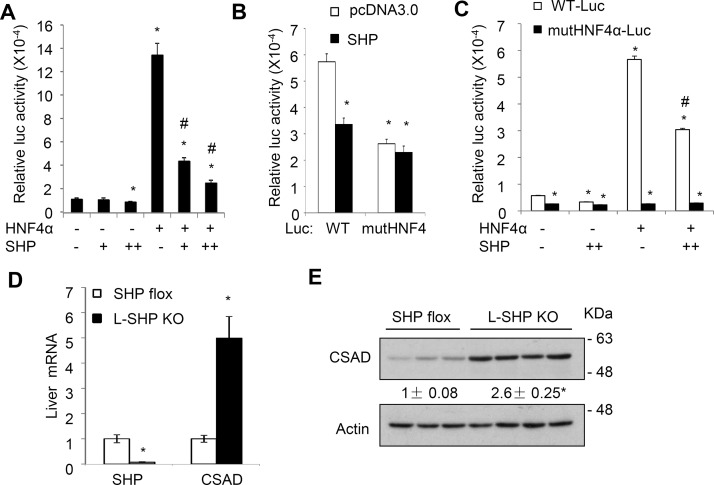
Coexpression of SHP dose-dependently decreased basal and HNF4α-stimulated activity of the mouse CSAD reporter containing the functional HNF4α binding sites (Fig. 4A). Furthermore, mutations of the HNF4α binding site significantly decreased the basal CSAD promoter activity and further abolished the repressive effect of SHP on the mouse CSAD promoter reporter activity (Fig. 4B and C), suggesting that SHP repressed CSAD reporter activity mainly via inhibition of the HNF4α trans-activating activity. Consistent with these in vitro results, we also found that hepatic CSAD mRNA and protein expression were significantly increased in L-SHP KO mice, which suggested that hepatic SHP negatively regulated hepatic CSAD expression in mice in vivo (Fig. 3D and E).
Figure 4.
Small heterodimer partner (SHP) represses HNF4α transactivation of the mouse CSAD reporter activity. (A–C) WT or mutant CSAD-(−1,895/+60)-luc construct (0.2 μg), β-gal expression construct (0.05 μg), and 0.1 μg (+) or 0.2 μg (++) of HNF4α and/or SHP expression plasmids were cotransfected into AML12 cells. The total amount of transfected plasmid DNA in each condition was adjusted by pcDNA3.0 plasmid. Luciferase and β-gal activities were measured 48 h later. *Statistical significance versus WT-Luc control + pcDNA3.0. #Statistical significance versus WT-Luc + HNF4α. All results are shown as mean ± SD of triplicate assays. (D, E) CSAD mRNA and protein expression in liver-specific SHP KO mice and controls. Results of mRNA expression were expressed as mean ± SE (n = 3–4). *Statistical significance versus WT. Densitometry was performed using ImageJ software and normalized to loading controls.
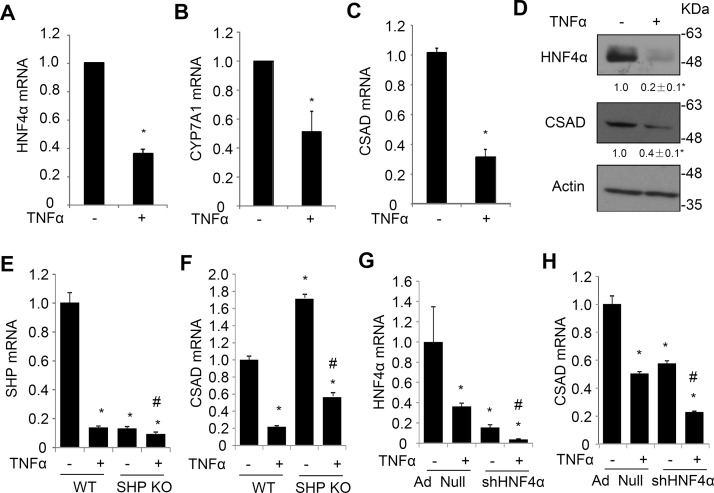
In addition to bile acids and FXR-induced SHP, proinflammatory cytokines such as TNF-α have been shown to inhibit HNF4α, leading to strong repression of CYP7A1 in vivo and in vitro26–28. To further substantiate the role of HNF4α in regulating hepatic CSAD expression, the effect of TNF-α on CSAD expression was further studied in primary mouse hepatocytes. Results showed that TNF-α treatment strongly repressed HNF4α mRNA and protein expression in primary mouse hepatocytes (Fig. 5A and D). Reduced HNF4α was associated with significantly reduced CSAD and CYP7A1 in TNF-α-treated mouse hepatocytes (Fig. 4B–D). We found that SHP KO hepatocytes showed elevated CSAD expression compared to WT hepatocytes (Fig. 5E and F), while TNF-α reduced CSAD mRNA to similar extent in WT and SHP KO hepatocytes (Fig. 5F). TNF-α also strongly reduced SHP mRNA in WT hepatocytes. These results suggest that TNF-α repression of CSAD is likely independent of SHP. Next, we knocked down HNF4α in mouse hepatocytes and treated the cells with TNF-α. Knockdown of HNF4α by ∼85% reduced CSAD mRNA expression by ∼45% (Fig. 5G and H). TNF-α treatment reduced CSAD by ∼50% in control hepatocytes and further reduced HNF4α by ∼75% and CSAD by ∼60% in HNF4α knockdown hepatocytes (Fig. 5H). Taken together, these results suggest that HNF4α is a key transcriptional stimulator that maintains basal hepatic CSAD expression and mediates bile acids and cytokine repression of CSAD in mouse livers.
Figure 5.
Inflammatory cytokine TNF-α inhibits HNF4α and CSAD expression in mouse hepatocytes. (A–D) Primary mouse hepatocytes were treated with TNF-α (50 ng/ml) for 24 h. The mRNA results are representative of two independent preparations of mouse hepatocytes. *Statistical significance versus control. Western blot is a representative of three independent experiments. Densitometry was performed using ImageJ software and normalized to loading controls. (E, F) WT and SHP KO hepatocytes were prepared as described in Materials and Methods. Cells were treated with TNF-α (50 ng/ml) for 24 h. SHP and CSAD mRNA were measured in triplicates and expressed as mean ± SD. (G, H) Mouse hepatocytes infected with Ad-Null or Ad-shHNF4α (MOI = 10) were treated with TNF-α (50 ng/ml) for 24 h as described in Materials and Methods. HNF4α and CSAD mRNA were measured in triplicates and expressed as mean ± SD. *Statistical significance versus WT or Ad-Null. #Statistical significance versus SHP KO or Ad-shHNF4α without TNF-α treatment.
HNF4α Overexpression and FXR Activation Do Not Affect CSAD Expression in Human Hepatocytes
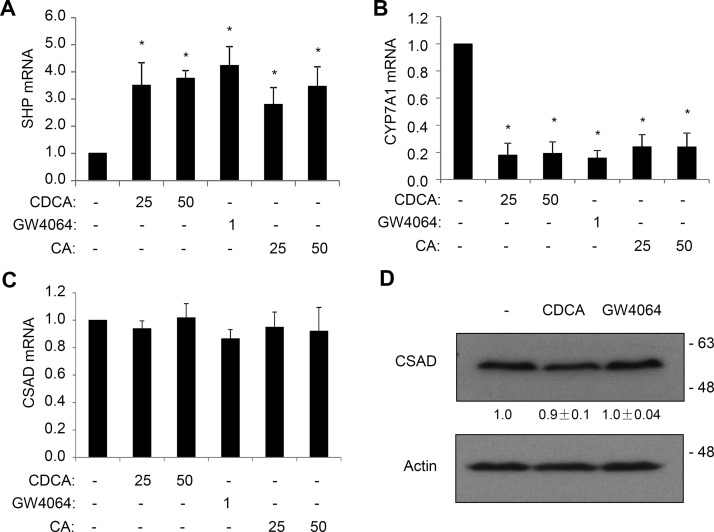
Currently, it is still not clear if bile acids also repress hepatic CSAD expression in humans. To test this, we next treated human hepatocytes with FXR agonist GW4064, CA, or chenodeoxycholic acid (CDCA). Interestingly, GW4064 and bile acids strongly induced SHP mRNA expression and repressed CYP7A1 mRNA expression but did not affect CSAD mRNA expression in human hepatocytes (Fig. 6A–C). In addition, CSAD protein abundance was not affected by GW4064 or bile acids (Fig. 6D). These results suggest that human CSAD may not be regulated by bile acids or FXR agonist.
Figure 6.
Bile acids and farnesoid X receptor (FXR) activation do not repress CSAD in human hepatocytes. (A–C) Primary human hepatocytes were treated with bile acids (25 or 50 μM) or GW4064 (1 μM) for 16 h. CSAD, CYP7A1, and SHP mRNA expression was measured. Results are expressed as mean ± SE of seven hepatocyte preparations. *Statistical significance versus controls. (D) Primary human hepatocytes were treated with 50 μM CDCA or 1 μM GW4064 for 16 h. A representative blot is shown. The values below the CSAD bands represent the mean ± SE of four hepatocyte preparations. Densitometry was performed with ImageJ software, and CSAD band intensity was normalized to that of actin bands.
DISCUSSION
Liver is a major organ that synthesizes taurine, which is used for bile acid conjugation10–12. Studies showed that hepatic CSAD expression was highly sensitive to bile acid repression, suggesting that hepatic taurine production is closely coupled to hepatic bile acid synthesis rate in mice14. However, the molecular mechanisms of bile acid inhibition of hepatic CSAD expression is still not clear. HNF4α is known to play an important role in maintaining the relatively high basal expression of many hepatic-enriched genes, and hepatic HNF4α deficiency resulted in profound changes in metabolic homeostasis and cellular proliferation in mice20,29,30. In this study, we present new findings to show that HNF4α plays a key role in stimulating hepatic CSAD expression in mice, which can be evidenced by the results showing that hepatic HNF4α knockdown caused significant reduction in CSAD expression in mouse livers and hepatocytes, and cotransfection of HNF4α strongly induced mouse CSAD promoter luciferase reporter activity. Furthermore, we showed that HNF4α served as a downstream target to mediate bile acid repression of CSAD expression. Our results collectively support a model whereby bile acid-activated FXR induces SHP, which acts as a repressor to inhibit HNF4α transactivation of CSAD transcription in mice. The role of SHP in this regulatory cascade was further supported by the significantly increased hepatic CSAD expression in L-SHP KO mice. Induction of CSAD upon ASBT inhibition also suggests that hepatic CSAD expression is under bile acid inhibition under physiological condition. It should be further noted that HNF4α is also a key inducer of hepatic CYP7A1 and CYP8B120,22,23,31. In obstructive cholestasis, several bile acid-initiated signaling pathways including the FXR/SHP axis and elevated proinflammatory cytokine signaling have been shown to target HNF4α to repress CYP7A1 and CYP8B126,27,32. These signaling pathways likely contributed to the strong hepatic CSAD repression in BDL mice. However, we found that despite reduced basal CSAD expression, TNF-α still repressed CSAD in HNF4α knockdown hepatocytes. On the one hand, this could be partially due to TNF-α repression of the remaining HNF4α in HNF4α knockdown cells, and on the other hand, TNF-α may inhibit CSAD via redundant HNF4α-independent mechanisms.
The parallel regulation of hepatic CYP7A1 and CSAD by nuclear receptors and bile acid signaling implies that a major purpose of hepatic taurine synthesis is to meet the need for bile acid conjugation. This may be because decreased bile acid conjugation can be associated with impaired physiological function of bile acids in the gut and potentially toxic effect elicited by unconjugated bile acids in the biliary tract. This can be evidenced by fat-soluble vitamin deficiency, growth retardation, and, in some cases, cholangiopathy reported in pediatric patients lacking functional BACS or BAAT8,9,33. In hepatocytes, newly synthesized bile acids are efficiently conjugated before they are secreted into the bile. Under normal physiology, hepatic de novo bile acid synthesis is maintained at relatively low levels because the fecal loss of bile acids is minimal. Furthermore, the amount of unconjugated bile acids circulating back to the liver from the small and large intestines is also relatively small. The strong inhibition of CSAD expression by bile acids and FXR provides a mechanism that preserves cysteine for use by other cellular pathways when bile acid synthesis is repressed and ensures sufficient taurine is available when de novo bile acid synthesis is induced. This is consistent with previous reports that the basal cysteine flux to taurine in hepatocytes occurs at a very low rate but can be induced34,35. Another mechanism by which FXR regulates bile acid conjugation is through upregulation of hepatic expression of human BACS and BAAT via direct binding to the promoter or intron regions of the two genes36. Under conditions with a significantly expanded bile acid pool, the unconjugated bile acids returning from the intestine to the liver may increase in hepatocytes despite inhibited hepatic de novo bile acid synthesis. In this scenario, induction of bile acid conjugation enzymes provides a mechanism to stimulate bile acid conjugation in response to elevated hepatocellular bile acid load.
Our study conducted in human hepatocytes suggested that human CSAD may not be repressed by bile acids and FXR. Humans and other nonhuman primates can use both glycine and taurine for bile acid conjugation6,7. In humans, about two thirds of the bile acids are glycine conjugates, which is likely a result of the substrate specificity of the BAAT enzyme toward glycine over taurine and also the relative subcellular taurine concentration in the peroxisomes37–39. Therefore, humans rely less on hepatic taurine production to maintain the conjugated bile acid content in the pool. However, it should also be noted that the overall rate of cysteine catabolism is considered to be predominantly controlled at the upstream CDO1 level10. Increased CDO1 was sufficient to drive cysteine flux to taurine without the need for CSAD induction35,40. In contrast, CSAD downstream of CDO1 may further increase taurine synthesis by controlling the partition of cysteine sulfinic acid between taurine synthesis and the production of pyruvate and sulfate. Recently, we reported that bile acids repressed CDO1 expression in both mouse livers and human hepatocytes, suggesting conserved bile acid repression of hepatic cysteine catabolism in humans and mice13. We also showed that loss of bile acid repression of this pathway in cholestyramine-fed mice increased hepatic taurine production and depleted hepatic cysteine and impaired GSH synthesis13, which may underlie the hepatotoxicity associated with the use of bile acid sequestrants in hyperlipidemia patients41–44. In addition to being used for bile acid conjugation, taurine has many other biological functions in different organ systems and cell types45. Csad knockout mice had defective taurine synthesis and showed neonatal mortality, which was rescued by dietary taurine supplement46. In contrast, growth defects, postnatal lethality, and many other pathological abnormalities in mice lacking CDO1 could not be fully rescued by dietary taurine supplement47,48, further suggesting that altered expression of CDO1 or CSAD may result in distinct metabolic consequences in the liver.
In summary, this study revealed that the liver-enriched transcriptional factor HNF4α is a strong transcriptional activator of the mouse CSAD gene, which is consistent with previous findings that the liver is the major taurine-producing organ and has markedly higher CSAD expression than nonhepatic tissues in mice10–12. Furthermore, bile acids, via the FXR/SHP axis, target HNF4α to repress hepatic CSAD, which thus couples taurine production to CYP7A1 expression and bile acid synthesis in mice. In addition, we report differential regulation of hepatic CSAD in humans and mice by bile acids and FXR. The loss of FXR repression of CSAD in humans may possibly reflect the different preference for taurine or glycine in the bile acid amidation reaction.
ACKNOWLEDGMENTS
This work was supported in part by an American Diabetes Association Junior Faculty Award (T.L.), the National Institutes of Health (NIH) grant 1R01DK102487-01 (T.L), the National Center for Research Resources (5P20RR021940-07), and the National Institute of General Medical Sciences (8 P20 GM103549-07) of the NIH.
Footnotes
The authors declare no conflicts of interests.
REFERENCES
- 1. Li T, Chiang JY. Bile acid signaling in metabolic disease and drug therapy. Pharmacol Rev. 2014;66(4):948–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Goodwin B, Jones SA, Price RR, Watson MA, McKee DD, Moore LB, Galardi C, Wilson JG, Lewis MC, Roth ME, and others. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell 2000;6(3):517–26. [DOI] [PubMed] [Google Scholar]
- 3. Lu TT, Makishima M, Repa JJ, Schoonjans K, Kerr TA, Auwerx J, Mangelsdorf DJ. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell 2000;6(3):507–15. [DOI] [PubMed] [Google Scholar]
- 4. Inagaki T, Choi M, Moschetta A, Peng L, Cummins CL, McDonald JG, Luo G, Jones SA, Goodwin B, Richardson JA, and others. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2(4):217–25. [DOI] [PubMed] [Google Scholar]
- 5. Shonsey EM, Wheeler J, Johnson M, He D, Falany CN, Falany J, Barnes S. Synthesis of bile acid coenzyme a thioesters in the amino acid conjugation of bile acids. Methods Enzymol. 2005;400:360–73. [DOI] [PubMed] [Google Scholar]
- 6. Hardison WG, Grundy SM. Effect of bile acid conjugation pattern on bile acid metabolism in normal humans. Gastroenterology 1983;84(3):617–20. [PubMed] [Google Scholar]
- 7. Hafkenscheid JC, Hectors MP. An enzymic method for the determination of the glycine/taurine ratio of conjugated bile acids in bile. Clin Chim Acta 1975;65(1):67–74. [DOI] [PubMed] [Google Scholar]
- 8. Carlton VE, Harris BZ, Puffenberger EG, Batta AK, Knisely AS, Robinson DL, Strauss KA, Shneider BL, Lim WA, Salen G, and others. Complex inheritance of familial hypercholanemia with associated mutations in TJP2 and BAAT. Nat Genet. 2003;34(1):91–6. [DOI] [PubMed] [Google Scholar]
- 9. Setchell KD, Heubi JE, Shah S, Lavine JE, Suskind D, Al-Edreesi M, Potter C, Russell DW, O’Connell NC, Wolfe B, and others. Genetic defects in bile acid conjugation cause fat-soluble vitamin deficiency. Gastroenterology 2013;144(5):945–55 e6; quiz e14-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stipanuk MH, Dominy JE Jr., Lee JI, Coloso RM. Mammalian cysteine metabolism: New insights into regulation of cysteine metabolism. J Nutr. 2006;136(6 Suppl):1652S–9S. [DOI] [PubMed] [Google Scholar]
- 11. Pasantes-Morales H, Chatagner F, Mandel P. Synthesis of taurine in rat liver and brain in vivo. Neurochem Res. 1980;5(4):441–51. [DOI] [PubMed] [Google Scholar]
- 12. Stipanuk MH, Londono M, Lee JI, Hu M, Yu AF. Enzymes and metabolites of cysteine metabolism in nonhepatic tissues of rats show little response to changes in dietary protein or sulfur amino acid levels. J Nutr. 2002;132(11):3369–78. [DOI] [PubMed] [Google Scholar]
- 13. Wang Y, Li J, Matye D, Zhang Y, Dennis K, Ding WX, Li T. Bile acids regulate cysteine catabolism and glutathione regeneration to modulate hepatic sensitivity to oxidative injury. JCI Insight 2018;3(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kerr TA, Matsumoto Y, Matsumoto H, Xie Y, Hirschberger LL, Stipanuk MH, Anakk S, Moore DD, Watanabe M, Kennedy S, and others. Cysteine sulfinic acid decarboxylase regulation: A role for farnesoid X receptor and small heterodimer partner in murine hepatic taurine metabolism. Hepatol Res. 2014;44(10):E218–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li J, Woolbright BL, Zhao W, Wang Y, Matye D, Hagenbuch B, Jaeschke H, Li T. Sortilin 1 loss-of-function protects against cholestatic liver injury by attenuating hepatic bile acid accumulation in bile duct ligated mice. Toxicol Sci. 2018;161(1):34–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li T, Chiang JY. Rifampicin induction of CYP3A4 requires pregnane X receptor cross talk with hepatocyte nuclear factor 4alpha and coactivators, and suppression of small heterodimer partner gene expression. Drug Metab Dispos. 2006;34(5):756–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Song KH, Li T, Chiang JY. A Prospero-related homeodomain protein is a novel co-regulator of hepatocyte nuclear factor 4alpha that regulates the cholesterol 7alpha-hydroxylase gene. J Biol Chem. 2006;281(15):10081–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li T, Matozel M, Boehme S, Kong B, Nilsson LM, Guo G, Ellis E, Chiang JY. Overexpression of cholesterol 7alpha-hydroxylase promotes hepatic bile acid synthesis and secretion and maintains cholesterol homeostasis. Hepatology 2011;53(3):996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol. 2001;21(4):1393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bolotin E, Liao H, Ta TC, Yang C, Hwang-Verslues W, Evans JR, Jiang T, Sladek FM. Integrated approach for the identification of human hepatocyte nuclear factor 4alpha target genes using protein binding microarrays. Hepatology 2010;51(2):642–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stroup D, Chiang JY. HNF4 and COUP-TFII interact to modulate transcription of the cholesterol 7alpha-hydroxylase gene (CYP7A1). J Lipid Res. 2000;41(1):1–11. [PubMed] [Google Scholar]
- 22. Yamagata K, Daitoku H, Shimamoto Y, Matsuzaki H, Hirota K, Ishida J, Fukamizu A. Bile acids regulate gluconeogenic gene expression via small heterodimer partner-mediated repression of hepatocyte nuclear factor 4 and Foxo1. J Biol Chem. 2004;279(22):23158–65. [DOI] [PubMed] [Google Scholar]
- 23. Zhang M, Chiang JY. Transcriptional regulation of the human sterol 12a-hydroxylase gene (CYP8B1): Roles of hepatocyte nuclear factor 4a (HNF4a) in mediating bile acid repression. J Biol Chem. 2001;276:41690–9. [DOI] [PubMed] [Google Scholar]
- 24. Alder O, Cullum R, Lee S, Kan AC, Wei W, Yi Y, Garside VC, Bilenky M, Griffith M, Morrissy AS, and others. Hippo signaling influences HNF4A and FOXA2 enhancer switching during hepatocyte differentiation. Cell Rep. 2014;9(1):261–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chiang JY, Stroup D. Identification and characterization of a putative bile acid-responsive element in cholesterol 7 alpha-hydroxylase gene promoter. J Biol Chem. 1994;269(26):17502–7. [PubMed] [Google Scholar]
- 26. Li T, Jahan A, Chiang JY. Bile acids and cytokines inhibit the human cholesterol 7 alpha-hydroxylase gene via the JNK/c-jun pathway in human liver cells. Hepatology 2006;43(6):1202–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jahan A, Chiang JY. Cytokine regulation of human sterol 12alpha-hydroxylase (CYP8B1) gene. Am J Physiol Gastrointest Liver Physiol. 2005;288(4):G685–95. [DOI] [PubMed] [Google Scholar]
- 28. Miyake JH, Wang SL, Davis RA. Bile acid induction of cytokine expression by macrophages correlates with repression of hepatic cholesterol 7alpha-hydroxylase. J Biol Chem. 2000;275(29):21805–8. [DOI] [PubMed] [Google Scholar]
- 29. Walesky C, Edwards G, Borude P, Gunewardena S, O’Neil M, Yoo B, Apte U. Hepatocyte nuclear factor 4 alpha deletion promotes diethylnitrosamine-induced hepatocellular carcinoma in rodents. Hepatology 2013;57(6):2480–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yin L, Ma H, Ge X, Edwards PA, Zhang Y. Hepatic hepatocyte nuclear factor 4alpha is essential for maintaining triglyceride and cholesterol homeostasis. Arterioscler Thromb Vasc Biol. 2011;31(2):328–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Crestani M, Sadeghpour A, Stroup D, Galli G, Chiang JY. Transcriptional activation of the cholesterol 7alpha-hydroxylase gene (CYP7A) by nuclear hormone receptors. J Lipid Res. 1998;39(11):2192–200. [PubMed] [Google Scholar]
- 32. Li T, Chiang JY. A novel role of transforming growth factor beta1 in transcriptional repression of human cholesterol 7alpha-hydroxylase gene. Gastroenterology 2007;133(5):1660–9. [DOI] [PubMed] [Google Scholar]
- 33. Chong CP, Mills PB, McClean P, Gissen P, Bruce C, Stahlschmidt J, Knisely AS, Clayton PT. Bile acid-CoA ligase deficiency—A new inborn error of bile acid metabolism. J Inherit Metab Dis. 2012;35(3):521–30. [DOI] [PubMed] [Google Scholar]
- 34. Stipanuk MH, Coloso RM, Garcia RA, Banks MF. Cysteine concentration regulates cysteine metabolism to glutathione, sulfate and taurine in rat hepatocytes. J Nutr. 1992;122(3):420–7. [DOI] [PubMed] [Google Scholar]
- 35. Bagley PJ, Stipanuk MH. The activities of rat hepatic cysteine dioxygenase and cysteinesulfinate decarboxylase are regulated in a reciprocal manner in response to dietary casein level. J Nutr. 1994;124(12):2410–21. [DOI] [PubMed] [Google Scholar]
- 36. Pircher PC, Kitto JL, Petrowski ML, Tangirala RK, Bischoff ED, Schulman IG, Westin SK. Farnesoid X receptor regulates bile acid-amino acid conjugation. J Biol Chem. 2003;278(30):27703–11. [DOI] [PubMed] [Google Scholar]
- 37. Falany CN, Johnson MR, Barnes S, Diasio RB. Glycine and taurine conjugation of bile acids by a single enzyme. Molecular cloning and expression of human liver bile acid CoA:amino acid N-acyltransferase. J Biol Chem. 1994;269(30):19375–9. [PubMed] [Google Scholar]
- 38. Falany CN, Fortinberry H, Leiter EH, Barnes S. Cloning, expression, and chromosomal localization of mouse liver bile acid CoA:amino acid N-acyltransferase. J Lipid Res. 1997;38(6):1139–48. [PubMed] [Google Scholar]
- 39. Hardison WG. Hepatic taurine concentration and dietary taurine as regulators of bile acid conjugation with taurine. Gastroenterology 1978;75(1):71–5. [PubMed] [Google Scholar]
- 40. Bagley PJ, Stipanuk MH. Rats fed a low protein diet supplemented with sulfur amino acids have increased cysteine dioxygenase activity and increased taurine production in hepatocytes. J Nutr. 1995;125(4):933–40. [DOI] [PubMed] [Google Scholar]
- 41. A multicenter comparison of lovastatin and cholestyramine therapy for severe primary hypercholesterolemia. The Lovastatin Study Group III. JAMA 1988;260(3):359–66. [PubMed] [Google Scholar]
- 42. Singhal R, Harrill AH, Menguy-Vacheron F, Jayyosi Z, Benzerdjeb H, Watkins PB. Benign elevations in serum aminotransferases and biomarkers of hepatotoxicity in healthy volunteers treated with cholestyramine. BMC Pharmacol Toxicol. 2014;15:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Goldfine AB, Fonseca VA, Jones MR, Wang AC, Ford DM, Truitt KE. Long-term safety and tolerability of colesevelam HCl in subjects with type 2 diabetes. Horm Metab Res. 2010;42(1):23–30. [DOI] [PubMed] [Google Scholar]
- 44. Sirmans SM, Beck JK, Banh HL, Freeman DA. Colestipol-induced hepatotoxicity. Pharmacotherapy 2001;21(4):513–6. [DOI] [PubMed] [Google Scholar]
- 45. Huxtable RJ. Physiological actions of taurine. Physiol Rev. 1992;72(1):101–63. [DOI] [PubMed] [Google Scholar]
- 46. Park E, Park SY, Dobkin C, Schuller-Levis G. Development of a novel cysteine sulfinic acid decarboxylase knockout mouse: Dietary taurine reduces neonatal mortality. J Amino Acids 2014;2014:346809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ueki I, Roman HB, Valli A, Fieselmann K, Lam J, Peters R, Hirschberger LL, Stipanuk MH. Knockout of the murine cysteine dioxygenase gene results in severe impairment in ability to synthesize taurine and an increased catabolism of cysteine to hydrogen sulfide. Am J Physiol Endocrinol Metab. 2011;301(4):E668–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Roman HB, Hirschberger LL, Krijt J, Valli A, Kozich V, Stipanuk MH. The cysteine dioxgenase knockout mouse: Altered cysteine metabolism in nonhepatic tissues leads to excess H2S/HS(-) production and evidence of pancreatic and lung toxicity. Antioxid Redox Signal. 2013;19(12):1321–36. [DOI] [PMC free article] [PubMed] [Google Scholar]