Abstract
The incidence of serious fungal infections is increasing rapidly, and yet the rate of new drugs becoming available to treat them is slow. The limited therapeutic armamentarium is a challenge for clinicians, because the available drugs are often toxic, expensive, difficult to administer, ineffective or a combination of all four. Given this setting, the emergence of resistance is especially concerning, and a review of the topic is timely. Here we discuss antifungal drug resistance in Candida spp. and Aspergillus spp. with reference to the most commonly used first-line antifungal agents – azoles and echinocandins. We review the resistance mechanisms of the leading pathogens, how resistance can be identified in the diagnostic lab and the clinical implications of resistance once detected.
The global incidence of fungal disease has increased dramatically in recent years. Current (likely under-) estimates suggest that there are approximately 300 million life-threating fungal infections annually, resulting in 1.6 million deaths [1]. Health impacts worldwide include high morbidity, an overall mortality of 30–80% and a multibillion dollar annual economic burden. The limitations of current therapies – including one or more of toxicity, poor bioavailability and relative inefficacy – are now further amplified by the emergence and escalation of drug resistance.
In considering the impact of drug resistance in the clinical setting, it is important to note that resistance does not always equate with clinical failure. Many factors contribute to clinical outcome. In particular, host factors such as an impaired immune system, other comorbidities, site of invasive fungal disease; therapeutic factors such as dosage regimen, compliance and drug toxicity; and ancillary factors such as source control all impact clinical outcomes. Nevertheless, there is evidence that rapid initiation of appropriate antifungal therapy reduces mortality in invasive candidiasis in the critically ill and in invasive aspergillosis in at-risk hematology patients [2]. Detection and characterization of drug resistance in vitro can assist clinicians to select the best antifungal regimen. A review of how this can be achieved is timely.
This review focuses on Candida and Aspergillus. The yeast Candida is a normal part of human skin flora, but can cause serious invasive infections. Aspergillus, a mold, is associated with serious invasive infections, and allergic conditions. Azole resistance in Candida and Aspergillus and echinocandin resistance in Candida are among the most challenging problems in clinical mycology. We will discuss the drivers of antifungal resistance and the clinical value of characterizing resistance. We will discuss different laboratory methods for detection of resistance and whether rapid detection can improve outcomes in outbreaks or for individual patients. In addition, we will discuss strategies for management of resistant fungal infections.
Epidemiology of azole resistance
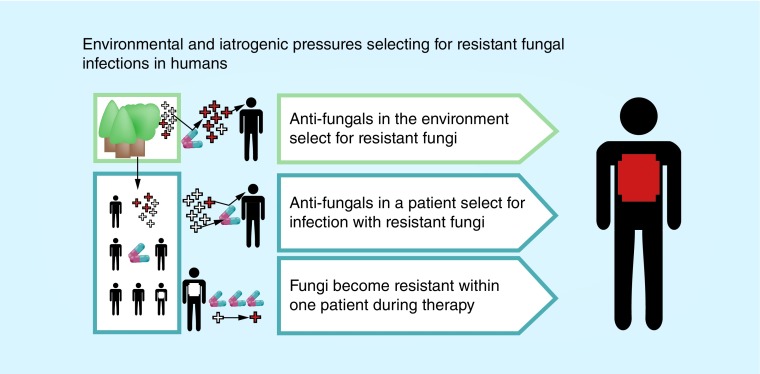
Antifungal resistance, whether intrinsic (primary) or acquired (secondary), can result in worse clinical outcomes or clinical failure. Pressure exerted by the use of antifungals may select for infection with intrinsically resistant strains, or strains may acquire resistance during prolonged therapy. Furthermore, environmental organisms with intrinsic or acquired resistance may infect antifungal naive patients. See Figure 1.
Figure 1. . Illustration of the three broad pathways via which patients can acquire resistant fungal infections.
White crosses indicate susceptible organisms; red crosses indicate resistant organisms. Pills indicate antifungals, used either in human health or agriculture.
Epidemiology of azole resistance in Candida
In Candida, susceptibility to azoles varies by species. A global surveillance study showed 92% of all cases of candidemia were caused by: Candida albicans (65.3%), Candida glabrata (11.3%), Candida tropicalis (7.2%), Candida parapsilosis (6%) and Pichia kudriavzevii (formerly and more commonly known as Candida krusei) (2.4%) [3]. These species display important differences in susceptibility. In the surveillance period 2005–2007, 98 and 98.5% of C. albicans were susceptible to fluconazole and voriconazole, respectively. For C. glabrata, these figures were 68 and 83%, C. tropicalis 91 and 90% and C. parapsilosis 93 and 97%. Only 9% of C. krusei were susceptible to fluconazole, while 83% were susceptible to voriconazole. The sixth commonest species, Meyerozyma guillermondii (previously Candida guillermondii), was only responsible for 0.7% of candidemia cases, but 74% of those showed reduced susceptibility to fluconazole and 91% displayed reduced voriconazole susceptibility [3].
Although it remains an uncommon pathogen, particular concern has been raised about Candida auris as a cause of nosocomial outbreaks since its emergence in multiple countries in 2009 [4]. Candida auris is resistant to multiple antifungal agents; only 7 and 46% of isolates are susceptible to fluconazole and voriconazole, respectively.
The frequency of different Candida species in clinical isolates (and therefore the anticipated susceptibility patterns) varies with geographic location and patient characteristics. For example, C. glabrata is over-represented in North America, occurring in 21% of patients [3]. Similarly, C. krusei, is over-represented in Europe (3.4%) and North America (3.1%). Meyerozyma guillermondii ranks in the top five causes in Latin America, being responsible for 2.2% of cases. Finally, C. tropicalis is notably more common in Asia Pacific (11.7%) and Latin America (13.2%) than the global average.
Variation in species distribution with patient age was noted in the SENTRY surveillance study [5]. Candidemia due to C. albicans and C. parapsilosis decreased with advancing age, whereas that due to C. glabrata increased. The influence of underlying diseases was demonstrated in an Australian study of hematology and oncology patients. While C. albicans was the leading cause of candidemia in both patient groups (causing 51 and 33% of episodes in oncology and hematology patients, respectively), there were differences in the proportion of non-albicans species [6]. For example, C. glabrata was the second commonest in oncology patients (causing 19.3% of episodes) and fourth in hematology patients (causing 12.3% of episodes). Candida krusei caused <1% of episodes in oncology patients, but 16.6% in hematology patients. This study also highlighted that prior antifungal therapy reduced the proportion of infections caused by C. albicans and increased the proportions due to C. glabrata, C. parapsilosis or C. krusei. Forty-four percent of patients receiving more than 6 days of fluconazole therapy prior to the appearance of candidemia were infected with C. krusei, compared with 11% of those who had received less than 6 days of therapy. Furthermore, the odds of having a fluconazole-resistant isolate were significantly increased by prior azole therapy.
Epidemiology of azole resistance in Aspergillus
Azoles are the first-line treatment for most infections caused by Aspergillus species [7]. Globally, Aspergillus fumigatus is the most frequent cause of invasive aspergillosis, causing over 90% of such infections [8], followed by Aspergillus flavus, Aspergillus niger and Aspergillus terreus. Distribution varies by geographic location, for example, A. flavus is isolated at least as frequently as A. fumigatus in subtropical and tropical regions [9,10].
Intrinsic resistance to azoles varies by Aspergillus species. They are all intrinsically resistant to fluconazole and ketoconazole, because of a naturally occurring amino acid substitution in 14-α sterol demethylase A, encoded by CYP51A [11]. However, they are generally considered intrinsically susceptible to other azoles. Aspergillus fumigatus is actually a species complex containing up to 63 members, many of which can only be distinguished by molecular methods [12]. Aspergillus fumigatus sensu stricto is the commonest species in the complex, accounting for approximately 95% of all isolates [13]. Although it is intrinsically susceptible to all Aspergillus-active azoles, some of its sibling species, including Aspergillus lentulus, Aspergillus thermomutatus and Aspergillus udagawae, have caused disease refractory to azole therapy [13]. Furthermore, within the A. niger species complex, itraconazole resistance has been observed in 25–50% of clinical isolates, with some evidence of cross-resistance with voriconazole and posaconazole [14–16]. Both cases highlight the importance of molecular epidemiology.
Mechanisms of azole resistance
Azoles inhibit the enzyme lanosterol 14α-demethylase, preventing the synthesis of ergosterol, a fungal cell-membrane component. In Candida and Aspergillus, the major causes of azole resistance are changes in this target enzyme (reduced drug–target affinity, upregulated production or both) or upregulation of efflux pumps [17].
Mechanisms of azole resistance in Candida
Two mechanisms are responsible for most azole resistance in Candida species. These are reduced affinity for the target enzyme, lanosterol 14α-demethylase, and overexpression of efflux pumps.
Mutations in lanosterol 14α-demethylase, encoded by the gene ERG11, were first described in 1997 [18]. Many point mutations have been identified subsequently and have been reviewed in detail [19–21]. ERG11 mutations are responsible for intrinsic fluconazole resistance in C. krusei [22] and have been reported in C. albicans, C. parapsilosis and C. tropicalis [21]. In addition, upregulation of lanosterol 14α-demethylase production can overcome drug concentrations achievable within fungal cells and has caused resistance in C. albicans and C. tropicalis [21].
Two families of efflux pump, the ATP-binding cassette pumps (encoded by CDR genes) and major facilitator superfamily pumps (encoded by MDR genes) [23] are both involved in antifungal resistance. Increased expression of CDR genes occurs in C. albicans, C. glabrata and C. krusei and is generally associated within a broader spectrum of antifungal resistance [19]. Increased expression of MDR genes has only been described in C. albicans and C. parapsilosis [21].
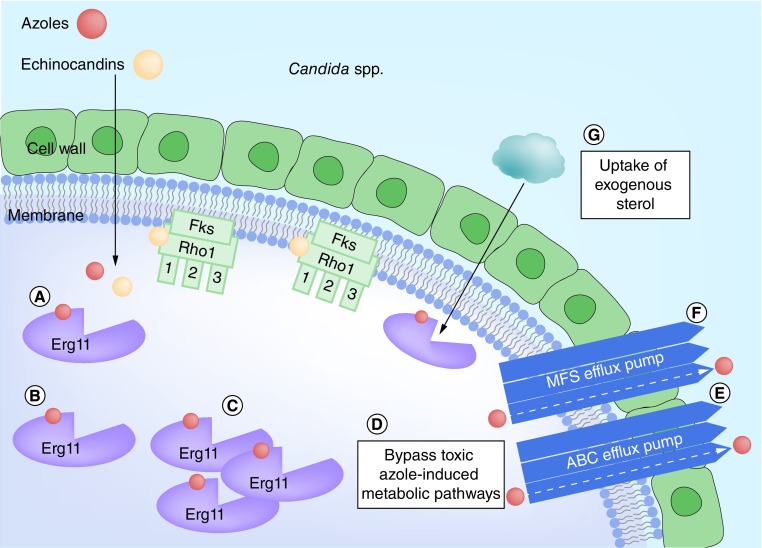
Azoles both prevent the formation of ergosterol and cause intracellular accumulation of toxic 14α-methyl- 3, 6-diol. Resistance in Candida can result from bypassing these metabolic pathways. Isolates with an inactive ERG3 gene do not form 14α-methyl-3,6-diol from 14α-methylfecosterol [24]. The accumulated 14α-methylfecosterol provides a functional substitute for ergosterol in the cell membrane, thus negating the impact of the azole. This mechanism occurs in C. albicans and C. tropicalis [21]. A related mechanism, whereby exogenous sterols are taken up by the cell, has been described in C. glabrata [21]. See Figure 2.
Figure 2. . Overview of the major resistance mechanisms in Candida sp.
(A) Shows normal binding of azoles (red) to lanosterol 14α-demethylase (Erg11); and normal binding of echinocandins (peach) to β-(1,3)-D-glucan synthase (Fks, including the three functional subunits Fks1, Fks2 and Fks3, and the regulatory subunit Rho1). (B) Shows reduced azole binding due to conformational changes in lanosterol 14α-demethylase. (C) Shows reduced azole efficacy due to increased production of lanosterol 14α-demethylase (+/- conformational changes). (D) Reduced azole efficacy secondary to bypassing metabolic toxicities. (E & F) Show reduced azole efficacy due to increased efflux pump activity. (G) Illustrates reduced azole efficacy due to uptake of replacement sterols. (H) Shows conformational changes to either Fks1 or Fks2 subunit of β-(1,3)-D-glucan synthase leading to reduced echinocandin binding.
Mechanisms of azole resistance in Aspergillus
The problem of acquired azole resistance in A. fumigatus has been well-described. Resistance acquired following prolonged therapy has been recognized since the late 1980s. Contributory mechanisms include increased efflux pump activity and decreased target enzyme affinity [25].
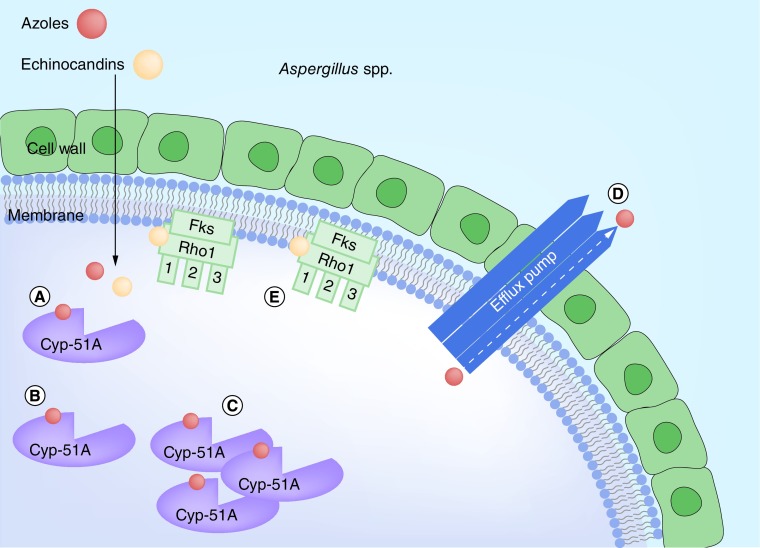
However, there has been growing interest in acquired resistance in environmental isolates of A. fumigatus over the past decade, which may have emerged through exposure to agricultural azoles. Such isolates have been associated with infection in azole naive patients [26,27]. In a global surveillance study, over 50% of azole-resistant clinical isolates shared common resistance mechanisms, with point mutations in CYP51A and variable tandem repeat insertions in the promoter region (TR34/L98H or TR46/Y121F/T289A). These mechanisms confer multi-azole resistance, and have been frequently identified in environmental samples [28]. See Figure 3. The mechanism of intrinsic azole resistance in the cryptic species described above is yet to be clarified.
Figure 3. . Overview of the major resistance mechanisms in Aspergillus sp.
(A) Shows normal binding of azoles (red) to 14-α-sterol demethylase (Cyp-51A); and normal binding of echinocandins (peach) to β-(1,3)-D-glucan synthase (Fks, including the three functional subunits Fks1, Fks2, and Fks3, and the regulatory subunit Rho1). (B) Shows reduced azole binding due to conformational changes in 14-α-sterol demethylase. (C) Shows reduced azole efficacy due to increased production of 14-α-sterol demethylase (+/- conformational changes). (D) Shows reduced azole efficacy due to increased efflux pump activity. (E) Shows conformational changes in the Fks1 subunit of β-(1,3)-D-glucan synthase leading to reduced echinocandin binding.
Echinocandin resistance
Echinocandins inhibit the membrane-bound enzyme β-(1,3)-D-glucan synthase and prevent formation of β-(1,3)-D-glucan, a vital cell-wall component. The enzyme complex is comprised of three functional (Fks1, Fks2 and Fks3) subunits and one regulatory (Rho1) subunit [29]. The specific targets for echinocandins are the Fks1 and Fks2 subunits [17], and resistance arises from conformational changes in these target sites [30]. Upregulation of efflux pumps has not been implicated in echinocandin resistance [31].
Echinocandin resistance in Candida
Candida develops resistance to echinocandins through point mutations in the FKS1 or FKS2 genes, although the exact biochemical mechanism is yet to be described [20]. Candida parapsilosis and C. guillermondii naturally contain mutations in FKS1, which are responsible for their reduced susceptibility to echinocandins [32,33], although the clinical impact of this mutation has not been established. Of concern, one study reported echinocandin resistance in 7% of C. auris isolates [4]. Acquired mutations in FKS1 arising after prolonged drug exposure, and associated with breakthrough infections, have been observed in C. albicans, C. tropicalis, C. krusei, and C. glabrata [34]. Acquired resistance mutations in FKS2 have, thus far, only been observed in C. glabrata [35].
Echinocandin resistance in Aspergillus
None of the pathogenic Aspergillus species are intrinsically resistant to echinocandins. Indeed, the only noted variation between the species complexes is that A. niger is hypersusceptible to caspofungin [36]. Development of echinocandin resistance in Aspergillus, by similar mechanisms to those described for Candida, is possible. There is laboratory evidence that mutations in FKS can reduce susceptibility in Aspergillus, but so far only a handful of clinical cases have been reported [37].
Laboratory methods to detect drug resistance in Candida & Aspergillus spp.
In vitro antifungal susceptibility (AFST) testing aims to detect drug resistance or the probability that antifungal treatment will fail [38]. Clinical microbiology laboratories rely on AFST to guide therapeutic choice and to develop an understanding of local resistance patterns [39]. AFST measures the ability of a specific organism to grow in the presence of a particular drug in vitro, generating a minimum inhibitory concentration (MIC) [40,41]. As the echinocandin activity against molds is fungistatic, end points are not clear-cut and the minimum effective concentration (MEC) is used instead. The MEC is the lowest concentration of a drug resulting in growth of small, rounded, compact hyphal balls compared with the filamentous hyphal growth seen in control wells [42].
Reference antifungal susceptibility testing methods
The Clinical and Laboratory Standards Institute (CLSI) [43,44] and the European Committee on Antibiotic Susceptibility Testing (EUCAST) [45,46] have developed standardized reference methods for the AFST of yeasts and filamentous molds. Both employ variations on broth microdilution methods, where the growth of isolates is observed in concentrations of an antifungal agent, prepared by serial dilution with growth medium. There are some technical differences between CLSI and EUCAST methods, but both reliably discriminate between susceptible ‘wild-type’ strains and resistant strains [38,47–48].
Test standardization has enabled the generation of clinical break points (CBPs) and/or epidemiologic cut-off values (ECVs) for the triazoles, echinocandins (and amphotericin B) against the main Candida spp. [42,44–45] and Aspergillus spp. [43,46]. The CBP is the concentration of drug which categorizes an organism as susceptible, susceptible dose-dependent/intermediate or resistant. CBPs are based on a combination of MIC values, pharmacokinetic/pharmacodynamic (PK/PD) parameters and clinical outcome data, in other words, an MIC value predictive of whether a microorganism will respond in vivo [41]. In contrast, the ECV is the upper limit of the wild-type population and usually includes 90–95% of a given population for a specific agent [42], in other words, an MIC separating a population into those with and without resistance based on their phenotype. ECVs can be generated against any species provided there are MIC data from enough isolates (typically ≥100) [41], but they do not reliably predict clinical response. The CLSI and EUCAST methods produce MICs which can differ by as much as three dilutions, so results cannot be used interchangeably [41,47]. However, the two methods generally agree on susceptibility categorization when testing azoles and echinocandins against Candida spp. [49,50] and azoles against Aspergillus species [51].
Commercial antifungal susceptibility assays
Because the ‘gold standard’ reference techniques for AFST are labor intensive and complex [52], many clinical laboratories use commercial tests. These are convenient, but are not fully standardized against the reference tests for all drug–fungus combinations and can result in the misclassification of susceptibility results [53].
The Sensititre YeastOne® (Trek Diagnostics Systems, OH, USA) is a commercial broth microdilution assay, based on CLSI methodology, which determines the MIC (or MEC) of nine antifungal drugs. It has been approved by the US FDA for use in clinical care with Candida spp. [38]. For Candida spp., several multicenter evaluations of YeastOne have demonstrated excellent reproducibility and results comparable with both reference methods [48–49,54]. Species-specific YeastOne ECVs of echinocandins have been defined for eight common Candida species. These correctly classified 88.9–93.8% of 81 well-characterized FKS mutant strains of five Candida spp., demonstrating the potential of the YeastOne test to help monitor the emergence of echinocandin resistance in routine diagnostic laboratories [55]. There are fewer studies of YeastOne among mold isolates, however high levels of agreement with reference methodologies have been observed for amphotericin B and the azoles with Aspergillus spp. [56–58], including detection of azole resistance in all A. fumigatus isolates (n = 10) with a CYP51A mutant genotype [59]. Data are limited for echinocandins, although a recent study demonstrated detection of A. fumigatus echinocandin-resistant isolates after 24 h incubation using YeastOne [60].
The Vitek 2® system (bioMérieux, Marcy d'Etoile, France) is a fully automated identification and AFST system, which provides results in 14–18 h [54]. Seven antifungal agents are included, although only fluconazole, caspofungin and voriconazole are approved by the FDA [48]. Results are accurate and reproducible when compared with CLSI/EUCAST reference methods [49,54,61–62]. Of note, the range of reported MIC values using the Vitek 2 system is limited, making interpretation of intermediate results difficult [26]. For caspofungin, the lowest concentration tested (0.25 μg/ml) is above the CLSI susceptible break point for C. glabrata (≤0.12 μg/ml) [63] and therefore misclassifies C. glabrata isolates with known mechanisms of resistance (i.e., FKS hotspot mutations) as susceptible [64]. Additionally, this method resulted in higher MICs than the CLSI reference method, which could lead to inappropriate therapy recommendations [65]. The Vitek 2 system is not suitable for antifungal susceptibility testing of molds.
Another frequently used commercial method for AFST is the agar-based Etest (bioMérieux, Marcy d'Etoile, France). Agreement between the Etest and both reference methods has been reported to be >90% [54]. However, in one study, 13% of reference method-susceptible Candida isolates (n = 497) were misclassified as intermediate or resistant by Etest [66]. The majority of those misclassified were C. krusei (73.1%) and C. glabrata (33.1%). Etest-specific ECVs were recently published for Candida spp. to amphotericin B and the echinocandins. Using these ECVs, 92% of the Candida FKS mutants were classified as non-wild-type (non-WT) for anidulafungin. However, the performance was inferior for caspofungin (75%) and micafungin (84%). These results suggest anidulafungin Etest ECVs may serve as a useful screen for echinocandin resistance in Candida spp. [67].
Etest-specific ECVs for the four most common Aspergillus species complexes (i.e., A. fumigatus, A. flavus, A. niger and A. terreus) have only been determined for amphotericin B and no CBPs are available. Preliminary data evaluating an isavuconazole Etest strip revealed that it showed promise in separating wild-type (n = 40) from CYP51A mutant (n = 39) A. fumigatus strains [68]. To date, the FDA has not approved any Etest strip for clinical use in mold infections.
Four-well azole supplemented screening plates (Balis Laboratorium VOF, Boven Leeuwen, The Netherlands) containing itraconazole (4 mg/l), posaconazole (0.5 mg/l) and voriconazole (1 mg/l) are a convenient and inexpensive, rapid screening method for azole-resistance in A. fumigatus [28]. They cannot provide an exact MIC value, but identify potentially resistant isolates for further investigation.
Molecular methods to detect azole resistance in Candida spp.
AFST of non-glabrata Candida species against the azoles remains a core function of the clinical laboratory, as interpretative CBPs predict outcomes [52,69]. However, molecular assays have the advantage that they can determine the underlying genetic basis of azole resistance [70]. These molecular methods are not standardized and individual mutations do not necessarily correlate with MICs – since multiple mechanisms can lead to reduced azole susceptibility [52]. Molecular methods to detect azole resistance must be multiplexed and capable of determining the presence and upregulation of housekeeping genes.
Point mutations in the three ‘regional hotspots’ of the ERG11 gene (amino acid positions 105–165, 266–287 and 405–488) and the ERG3 gene (T330A and A351V) can reliably and easily be detected with high-throughput DNA sequence analysis, allele-specific real-time molecular probes, high resolution melt curve analysis and DNA microarray technology, as reported by Perlin [70,71]. These techniques are robust and can be used with amplified DNA from primary specimens, but they are technically demanding. Whether such mutations in the ERG11 gene confer phenotypic resistance to the azoles requires additional experiments to demonstrate causal effect – not all amino acid substitutions have been validated [72,73].
Most recently, next generation sequencing (NGS) has been used to detect alterations in genes associated with azole resistance in Candida spp. (ERG11, ERG3, TAC1 and CgPDR1) [4,74–75]. NGS has the ability to detect novel mutations implicated in phenotypic resistance of clinical isolates that may otherwise be missed by targeted DNA sequence analysis [75]. Although the practicability and cost (∼60 USD sample) of NGS mean that it is currently beyond the scope of most routine clinical laboratories [76], rapid technological advances will reduce these barriers and make routine application of NGS more likely.
Molecular methods to detect echinocandin resistance in Candida spp.
AFST is unable to reliably distinguish WT isolates from echinocandin-resistant (FKS mutant) isolates, particularly for caspofungin, due to substantial interlaboratory variability in MIC results [77]. FKS genotype analysis has been deemed a better predictor of resistance than AFST alone because: an FKS mutation is an independent risk factor for therapeutic failure; the number of Candida strains exhibiting resistance in vitro is low; and there is a limited spectrum of mutations conferring resistance [52,70,78,79]. Echinocandin resistance can be assessed by real-time PCR, DNA sequencing or NGS, [70,74,75,80–82], with the latter being more suited (and potentially more cost effective) to accommodate the larger number of mutations in multiple FKS genes seen with C. glabrata.
In one recent study, ECVs established by CLSI were used to select isolates for screening for FKS mutations by PCR and DNA sequencing. Among the C. glabrata isolates selected, 73% (11/15) harbored mutations in FKS1 (S663P, S629P or F625S) and/or FKS2 (F659S/Y) genes. No known FKS mutations were detected in the remaining four, highlighting that some isolates may be categorized as non-WT due to elevated MICs in the absence of known resistance mechanisms [74].
FKS mutations have been detected using NGS in clinical isolates of C. glabrata with high MICs to the echinocandins [75,82]. Biswas et al. [82] recently used NGS to retrospectively study three strain pairs of C. glabrata from three patients where antifungal resistance developed during treatment. Two of three isolate pairs developed a >60-fold increase in the MICs to all echinocandins and NGS detected mutations in either the FKS1 (S629P) or FKS2 (S663P) genes of the resistant isolates [82].
Molecular methods to detect azole resistance in Aspergillus spp.
The application of molecular testing for azole resistance in Aspergillus spp. has been more straightforward than in Candida spp. as there are fewer resistance mechanisms. Resistance-associated mutations have been detected in Aspergillus spp. by real-time PCR, with molecular beacon probes, sequencing, high resolution melt curve analysis and most recently NGS [70,83,84]. Although a method for detection of CYP51A gene mutations has not been standardized, the most commonly used is a simple PCR amplification of the entire coding and promoter regions, followed by Sanger sequencing [85]. Ahmad et al. [86] developed a simple PCR-restriction fragment length polymorphism assay for simultaneous detection of TR34 and TR40 repeats (and likely others), as well as the L98H substitution. NGS, however, has the ability to determine the genome-wide basis of azole resistance. Hagiwara et al. [83] used NGS to identify nonsynonymous mutations in A. fumigatus strains sequentially isolated from each of two patients, which would not have been detected by traditional methods such as PCR-restriction fragment length polymorphism or microsatellite genotyping.
Although routine AFST is recommended for all clinically significant isolates of Aspergillus spp., to monitor for resistance [87], this is dependent on fungal culture and invasive aspergillosis is frequently diagnosed by culture-independent methods. Thus it is possible that azole resistance is underdiagnosed [85]. To overcome this problem, non-culture-based molecular assays that identify Aspergillus spp. and detect azole resistance directly from clinical samples have been developed. These methods combine high sensitivity with high specificity to ensure amplification of the small amounts of Aspergillus DNA present in biological samples. Most formats are PCR-based with subsequent DNA sequence analysis to detect the mutations [85]. Two commercially available multiplex real-time PCR assays are available; AsperGenius® (PathoNostics BV, Maastricht, The Netherlands) and MycoGENIE (Ademtech, Pessac, France). AsperGenius detects DNA from A. fumigatus and A. terreus. It uses melt curve analysis, rather than DNA sequencing, to identify TR34/L98H and Y121F/T289A CYP51A substitutions. It has performed well on bronchoalveolar lavage fluid (including culture-negative fluid) and blood samples [88,89]. The MycoGENIE detects only A. fumigatus DNA and specific TR34 and L98H mutations in CYP51A. Its sensitivity and specificity were 92.9 and 90.1%, respectively, using respiratory samples (n = 88) and 100 and 84.6%, respectively with serum samples [90].
Management of drug-resistant Candida infections
The intrinsic susceptibility patterns of various Candida species have been discussed above. Perspectives on the treatment of invasive fungal diseases caused by C. glabrata, and selected species which are less susceptible to antifungal agents (including the more recently-described C. auris), are outlined below.
Candida glabrata
The rising incidence of invasive C. glabrata infections should prompt a revisit of the antifungal treatment algorithms used in hospitals. This is especially pertinent given reported resistance rates to azoles of 11–13% [91] and to echinocandins of >13% [30]. Knowledge of local susceptibility patterns is essential for clinical decision-making.
Echinocandins are first line therapy for C. glabrata infections and are recommended for empiric therapy of invasive candidiasis in many clinical settings [92]. This is appropriate unless there is a strong suspicion of echinocandin resistance in high risk individuals, for example, in patients with previous echinocandin exposure, a previous episode of invasive candidiasis, or known azole resistance [93]. Concerningly, co-resistance to the azoles and echinocandins has been reported [94]. In such cases, treatment should be initiated with a different class of antifungal such as a polyene [92].
Candida auris
Candida auris is increasingly recognized worldwide as a cause of nosocomial candidemia and other forms of invasive candidiasis, with a high mortality [4,95–97]. It is often misidentified on conventional tests and is typically resistant to fluconazole and variably susceptible to the other azoles, amphotericin B and the echinocandins. Lockhart et al. in a US CDC-based survey reported that of 54 isolates, 93% were fluconazole-resistant, 35% were amphotericin B-resistant and 7% were echinocandin-resistant [4]. Of note, 41 and 4% of isolates were resistant to 2 and 3 drug classes, respectively. These data were derived using CBPs established for related Candida species as species-specific CBPs for C. auris have not yet been endorsed.
At present, first-line antifungal therapy for C. auris candidiasis remains an echinocandin [93]. Following AFST, treatment should be tailored to a drug against which this isolate is susceptible. Some clinicians favor combining antifungal drugs despite limited evidence favoring combination therapy. Fahkim et al. demonstrated in vitro synergy between micafungin and voriconazole against multidrug-resistant C. auris although combinations of caspofungin and fluconazole or voriconazole were indifferent [98]. This is consistent with findings in mice with invasive candidiasis, where clinically relevant doses of micafungin were more effective than fluconazole and amphotericin B [99].
The recommended duration of antifungal therapy is similar to that for other Candida spp. Where feasible, indwelling devices such as central venous catheters should be removed [93]. Critically, as C. auris can persist in the environment and colonize patients and clinical staff, aggressive infection control measures are recommended. These include daily and terminal cleaning of patient rooms, elimination of the organism from colonized body sites using chlorhexidine body washes, from the environment using disinfectants and from hands of personnel by compliance with hand hygiene guidelines [100].
Management of drug-resistant A. fumigatus infections
The Infectious Diseases Society of America (IDSA) in 2016 updated guidelines for management of invasive, chronic and allergic aspergillosis [7]. Azoles remain the preferred first-line therapy, for both treatment and prevention [7]. However, experimental evidence to guide treatment of azole-resistant A. fumigatus infections is scarce and recommendations have not been included in the management guidelines.
The choice of empiric antifungal therapy will be guided by the prevalence of azole resistance in the country, region or preferably, in the treating institution. Where an elevated MIC to an azole is demonstrated by a validated test method (discussed above), treatment with one or more alternative antifungal agents is recommended. As the number of effective drugs remains limited, regimens to extend azole effectiveness and overcome decreased susceptibility by optimizing drug exposure have also been explored [101].
Alternatives to azole monotherapy
Minimal prevalence of azole resistance
In countries, or at the institutional level, where there is reliable evidence of minimal (<3–5%) or no azole resistance, antifungal guidelines recommend voriconazole as initial therapy for almost all patients with confirmed or suspected invasive aspergillosis [7,102–104].
In infections caused by A. fumigatus confirmed to be voriconazole-resistant, expert consensus recommends discontinuation of voriconazole as monotherapy and either substitution of a different class of antifungal agent in its place or combining the azole with an echinocandin [102]. Despite the absence of large systematic studies correlating in vitro azole resistance with clinical outcome, observations of higher mortality in patients infected with azole-resistant strains compared with those affected by azole-susceptible A. fumigatus (88 vs 30–50%) are concerning [105]. Alternative regimens such as voriconazole plus an echinocandin or in combination with liposomal amphotericin B (L-AMB) have been used without clinical efficacy studies to support either [102]. Echinocandin monotherapy is another option but success rates in invasive aspergillosis have been only in the vicinity of 50% or less [106]. As a single agent, in vitro studies indicate comparable efficacy of L-AMB against invasive aspergillosis due to azole-susceptible and azole-resistant Aspergillus, consistent with the absence of cross-resistance between these drug classes [107]. Hence, L-AMB monotherapy remains an important therapeutic option in the setting of azole resistance.
Where the laboratory can perform AFST, this is recommended. Although differential azole susceptibility may occur, frequently isolates are resistant to more than one azole including isavuconazole. Hence substitution with a different azole should be undertaken with caution.
High (>10%) prevalence of azole resistance
Many experts consider a prevalence of azole resistance >10% to be ‘high’ and would recommend against primary treatment with voriconazole. Either a combination of voriconazole plus an echinocandin or L-AMB have been recommended as empiric therapy [102] prior to obtaining susceptibility results; when these are known, therapy can be tailored accordingly.
Where infection due to azole-resistant Aspergillus is confirmed, antifungal therapy should be continued using either of the above options or with an agent to which the isolate is susceptible. If the isolate is shown to be susceptible to voriconazole, voriconazole monotherapy can be instituted [102]. Where voriconazole cannot be used or is not tolerated by the patient, then posaconazole or isavuconazole may be considered. In all instances, therapeutic drug monitoring is desirable to ensure that azole serum levels are adequate.
In the absence of a cultured isolate, the choice of antifungal relies on clinical judgement and local epidemiology. One approach may be to employ initial therapy with 2–4 weeks of L-AMB followed by step down oral therapy with an azole.
Intermediate (5–10%) prevalence of azole resistance
Opinions differ regarding preferred empiric treatment in settings with intermediate rates of resistance. Unnecessary fear of voriconazole (or other azole) resistance may lead to use of another agent with lower efficacy. Voriconazole, a combination of voriconazole and echinocandin, or L-AMB monotherapy could be considered for initial use. No head-to-head comparative clinical studies between L-AMB and voriconazole have been performed. However observational reports have shown that the efficacy of voriconazole in invasive aspergillosis is about 15–20% higher than that of all amphotericin B formulations and echinocandins (reviewed in [102]).
CNS aspergillosis
International guidelines recommend the use of voriconazole as first-line therapy for CNS aspergillosis [7] despite limited clinical data comparing it with other antifungal treatments. Schwarz et al. [108] showed that the use of voriconazole led to more favorable clinical outcomes compared with the polyenes. Unlike the other mold active azoles itraconazole and posaconazole, which cannot be detected in appreciable amounts in the cerebrospinal fluid (CSF), voriconazole has favorable CNS PK properties, including penetration into brain tissue [109].
If the use of voriconazole is precluded due to drug resistance, alternatives are limited, as neither itraconazole, posaconazole, nor lipid formulations of amphotericin B penetrate well into the CSF (although concentrations in the brain are often adequate to achieve a clinical response). Based on PK data in animals, experts prefer L-AMB over other amphotericin B formulations [102,110].
Clinical outcomes in patients with CNS aspergillosis due to azole-resistant strains have been poor, despite combination antifungal therapy [105]. Many experts recommend combining amphotericin B with a second agent, specifically, 5-flucytosine [7,102]. The agent 5-flucytosine achieves good CSF drug levels. Echinocandins achieve only very low CSF levels and are not recommended in CNS aspergillosis.
Increasing exposure to azoles
Depending on specific gene mutations corresponding to a particular resistant phenotype, loss of activity of an azole may be complete or partial. For isolates in which MICs have increased only by a few twofold dilutions, escalating the drug dose may translate to a clinical response. Such an approach must take into account PK/PD parameters and binding of the drug to the target site. Seyedmousavi et al. reviewed the use of azole monotherapy for azole-resistant aspergillosis [101]; based on in vitro susceptibility, PK/PD studies, and experimental models of infection, they concluded that there may be a case for voriconazole dose escalation. Where the MIC is 2 mg/l (intermediate susceptibility), plasma drug levels should exceed 1.03 mg/l; where the MIC is 4 mg/l (resistant), a plasma level of >/= 2.65 mg/l is recommended. In the latter case, the likelihood of drug toxicity may be increased, and intravenous administration is required. For isolates with an MIC of >4 mg/l, very high levels of drug are required (plasma level > 5.3 mg/l). At present, there appears to be no role for posaconazole when the MIC of the isolate is outside the WT, although data using the intravenous formulation have not yet been published.
Newer antifungal agents for Candida & Aspergillus
Several newer antifungals are in development for use against Candida. T-2307 (Toyoma Chemical Company) is active in vitro against azole- and echinocandin-resistant Candida including C. glabrata, and has good in vivo efficacy in animal models (reviewed in [111]). CD101 (biafungin, Cidara Therapeutics, CA, USA), an echinocandin under development, is a long-acting compound with good anti-Candida activity [111]; against 16 C. auris isolates, the MIC90 was 0.25 mg/l [112]. CD101 is also effective against Candida species in biofilms [113].
The new oral glucan synthase inhibitor SCY-078 (Scynexis, Inc., NJ, USA) has potent activity against Candida species, which includes those in biofilms and isolates with known FKS1 and FKS2 mutations. MIC values against C. auris range from 0.5 to 2 mg/l, with an MIC90 of 1 mg/l [114].
The drug F901318, developed by F2G Ltd (Manchester, UK) (http://www.f2g.com/), is a promising antifungal agent, now entering Phase III clinical studies. It selectively inhibits the fungal dihydro-orotate dehydrogenase enzyme resulting in perturbed pyrimidine synthesis. In vitro, F901318 is highly active against azole-resistant Aspergillus isolates (MICs against A. fumigatus 0.06–0.12 mg/l) including against isolates with CYP51A mutations known to confer azole resistance (summarized in [115]).
Other promising anti-Aspergillus agents include the arylamidine T-2307 (Toyoma Chemical Company, Tokyo, Japan) which selectively targets fungal mitochondria, E121010/APX001 (Amplyx Pharmaceuticals, CA, USA) which inhibits glycosylphosphatidylinositol-dependent anchoring of the fungal cell wall, and ASP2397 (Astellas Pharma, Tokyo, Japan) which interferes with fungal siderophore transport [115].
Conclusion & future perspective
Here we have discussed current knowledge of, and controversies regarding, antifungal resistance, with a focus on Candida and Aspergillus. Coordinated surveillance of fungal pathogens including antifungal resistance remains inadequate. Since many routine diagnostic laboratories, especially in lower and middle-income settings, lack the capacity to perform even routine species identification, it is important that clinicians everywhere have access to local epidemiology and susceptibility patterns from reference laboratories to guide treatment choices.
At the same time, development of diagnostic capacity for fungal infections is a priority. A recent review outlined a ‘roadmap’, calling for immediate access to point of care testing for endemic fungal infections, followed by staged development of microscopy, culture and ultimately molecular techniques for low and middle-income countries [116] and we endorse that roadmap. NGS is a new technology that can be used to identify emerging threats; its value has been illustrated in identifying and tracking azole resistance in A. fumigatus. As molecular technologies continue to evolve and their widespread use becomes more financially viable, molecular data can be integrated into clinical management guidelines.
Finally, we note the importance of new drug development. Increasing azole and echinocandin resistance heightens the urgency of the need for a larger antifungal armamentarium to improve the availability, efficacy and tolerability of therapeutic options for these serious but neglected diseases.
Executive summary.
The incidence of fungal infections is increasing, but the availability of new agents to treat them is not.
Although results of antifungal susceptibility testing do not correlate precisely with treatment response in vivo, they can be used to guide treatment choices.
This review focuses on azole and echinocandin resistance in Candida spp. and Aspergillus spp.
Azole resistance in Candida spp. & Aspergillus spp.
Intrinsic resistance patterns vary by species. The likelihood of a particular species causing disease in a given patient is affected by geographic location and patient characteristics.
The principle mechanisms by which Candida spp. acquire resistance to azoles are structural alterations to lanosterol 14α-demethylase resulting from mutations in its encoding gene ERG11, upregulated production of lanosterol 14α-demethylase and upregulation of ATP-binding cassette or major facilitator's superfamily efflux pumps.
Cryptic Aspergillus species within complexes such as Aspergillus fumigatus and Aspergillus niger, which can usually only be identified by molecular testing, are frequently azole-resistant. This complicates treatment recommendations, and reinforces the importance of performing antifungal susceptibility testing.
Most azole resistance in Aspergillus arises from mutations in the CYP51A gene. The alarming emergence of TR34/L98H and TR46/Y121F/T289A as the dominant global mutation has raised the possibility of environmental/agricultural selection pressure.
Echinocandin resistance in Candida spp. & Aspergillus spp.
Intrinsic reduced susceptibility to echinocandins is uncommon and of uncertain significance in Candida spp. other than Candida auris. Most species can acquire resistance through mutations in the FKS1 and FKS2 genes, leading to alterations in subunits of the target enzyme β-(1,3)-D-glucan synthase, and refractory infections. Clinically relevant echinocandin resistance in Aspergillus is rare.
Laboratory methods to detect drug resistance in Candida & Aspergillus spp.
Clinical and Laboratory Standards Institute and European Committee on Antibiotic Susceptibility Testing reference broth dilution methods for both Candida and Aspergillus spp. are discussed, including some of the commercial kits that have been developed from these methods.
Molecular methods, including next generation sequencing, are also discussed. They are useful for describing resistance mechanisms at the epidemiological level, and as diagnostic tests in the routine laboratory.
In particular, the value of identifying FKS mutations in Candida is highlighted as an area where molecular methods may better predict echinocandin treatment failure than standard antifungal susceptibility testing.
Management of drug resistant Candida & Aspergillus spp. infections
Treatment strategies are discussed with particular reference to Candida glabrata and C. auris. Strategies covered include combination, substitution, dose-escalation and new antifungal agents, and which indicators to consider when selecting empiric therapy.
Azoles are first line therapy for Aspergillus infections. Empiric management of Aspergillus spp. infections are discussed in light of known low, high or intermediate rates of azole resistance.
CNS infections with Aspergillus are especially challenging to manage, even in the absence of azole resistance. Echinocandins are not recommended, and treatment relies on combination therapy with amphotericin and flucytosine.
New mold active agents are also discussed.
Conclusion & future perspective
Enhanced global surveillance efforts are required to better understand the prevalence and drivers of resistance in fungal pathogens, and to ensure that they are identified early.
Lab capacity, especially in low and middle income settings where disease burden is highest, needs development. Point-of-care testing for mycoses should be made available rapidly, while molecular testing is likely to be key for both diagnostics and epidemiological surveillance in coming years.
New drugs are required to improve therapeutic options, especially in light of emerging antifungal resistance.
Footnotes
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
Open access
This work is licensed under the Attribution-NonCommercial-NoDerivatives 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Editorial. Stop neglecting fungi. Nat. Microbiol. 2017;2(8):17120. doi: 10.1038/nmicrobiol.2017.120. [DOI] [PubMed] [Google Scholar]
- 2.Colombo AL, de Almeida Júnior JN, Slavin MA, Chen SC-A, Sorrell TC. Candida and invasive mould diseases in non-neutropenic critically ill patients and patients with haematological cancer. Lancet Infect. Dis. 2017;17(11):e344–e356. doi: 10.1016/S1473-3099(17)30304-3. [DOI] [PubMed] [Google Scholar]
- 3.Pfaller MA, Diekema DJ, Gibbs DL, et al. Results from the artemis disk global antifungal surveillance study, 1997 to 2007: a 10.5-year analysis of susceptibilities of Candida species to fluconazole and voriconazole as determined by CLSI standardized disk diffusion. J. Clin. Microbiol. 2010;48(4):1366–1377. doi: 10.1128/JCM.02117-09. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Large global study of species distribution and azole susceptibility in Candida spp.
- 4.Lockhart SR, Etienne KA, Vallabhaneni S, et al. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin. Infect. Dis. 2017;64(2):134–140. doi: 10.1093/cid/ciw691. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Fascinating paper providing evidence for the recent simultaneous emergence of independent clades of Candida auris in multiple global locations, rather than global spread of one strain.
- 5.Pfaller M a, Diekema DJ, Jones RN, Messer S a, Hollis RJ SENTRY Participants Group. Trends in antifungal susceptibility of Candida spp. isolated from pediatric and adult patients with bloodstream infections: SENTRY Antimicrobial Surveillance Program, 1997 to 2000. J. Clin. Microbiol. 2002;40(3):852–856. doi: 10.1128/JCM.40.3.852-856.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slavin MA, Sorrell TC, Marriott D, et al. Candidaemia in adult cancer patients: risks for fluconazole-resistant isolates and death. J. Antimicrob. Chemother. 2010;65(5):1042–1051. doi: 10.1093/jac/dkq053. [DOI] [PubMed] [Google Scholar]
- 7.Patterson TF, Thompson GR, Denning DW, et al. Practice guidelines for the diagnosis and management of aspergillosis: 2016 update by the infectious diseases society of America. Clin. Infect. Dis. 2016;63(4):e1–e60. doi: 10.1093/cid/ciw326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taccone F, Van den Abeele A-M, Bulpa P, et al. Epidemiology of invasive aspergillosis in critically ill patients: clinical presentation, underlying conditions, and outcomes. Crit. Care. 2015;19(1):7. doi: 10.1186/s13054-014-0722-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pasqualotto AC. Differences in pathogenicity and clinical syndromes due to Aspergillus fumigatus and Aspergillus flavus . Med. Mycol. 2009;47(Suppl. 1):S261–S270. doi: 10.1080/13693780802247702. [DOI] [PubMed] [Google Scholar]
- 10.Chawla K, Kosaraju K, Rayasam S, Mukhopadhyay C. Clinico-microbiological profile of chronic pulmonary aspergillosis from a tertiary care centre in southern India. J. Clin. Diagn. Res. 2013;7(12):2712–2715. doi: 10.7860/JCDR/2013/6314.3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leonardelli F, Macedo D, Dudiuk C, Cabeza MS, Gamarra S, Garcia-Effron G. Aspergillus fumigatus intrinsic fluconazole resistance is due to the naturally occurring T301I substitution in Cyp51Ap. Antimicrob. Agents Chemother. 2016;60(9):5420–5426. doi: 10.1128/AAC.00905-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frisvad JC, Larsen TO. Extrolites of Aspergillus fumigatus and other pathogenic species in aspergillus section fumigati. Front. Microbiol. 2016;6:1–14. doi: 10.3389/fmicb.2015.01485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamoth F. Aspergillus fumigatus-related species in clinical practice. Front. Microbiol. 2016;7:1–8. doi: 10.3389/fmicb.2016.00683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howard SJ, Harrison E, Bowyer P, Varga J, Denning DW. Cryptic species and azole resistance in the Aspergillus niger complex. Antimicrob. Agents Chemother. 2011;55(10):4802–4809. doi: 10.1128/AAC.00304-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Espinel-Ingroff A, Cuenca-Estrella M, Fothergill A, et al. Wild-type MIC distributions and epidemiological cutoff values for amphotericin B and aspergillus spp. for the CLSI broth microdilution method (M38-A2 document) Antimicrob. Agents Chemother. 2011;55(11):5150–5154. doi: 10.1128/AAC.00686-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagiwara D, Watanabe A, Kamei K, Goldman GH. Epidemiological and genomic landscape of azole resistance mechanisms in Aspergillus fungi. 2016;7:1–14. doi: 10.3389/fmicb.2016.01382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perea S, Patterson TF. Antifungal resistance in pathogenic fungi. Clin. Infect. Dis. 2002;35(9):1073–1080. doi: 10.1086/344058. [DOI] [PubMed] [Google Scholar]
- 18.Löffler J, Kelly SL, Hebart H, Schumacher U, Lass-Flörl C, Einsele H. Molecular analysis of cyp51 from fluconazole-resistant Candida albicans strains. FEMS Microbiol. Lett. 1997;151(2):263–268. doi: 10.1111/j.1574-6968.1997.tb12580.x. [DOI] [PubMed] [Google Scholar]
- 19.Kanafani ZA, Perfect JR. Resistance to antifungal agents: mechanisms and clinical impact. Clin. Infect. Dis. 2008;46(1):120–128. doi: 10.1086/524071. [DOI] [PubMed] [Google Scholar]
- 20.Kołaczkowska A, Kołaczkowski M. Drug resistance mechanisms and their regulation in non-albicans Candida species. J. Antimicrob. Chemother. 2016;71(6):1438–1450. doi: 10.1093/jac/dkv445. [DOI] [PubMed] [Google Scholar]
- 21.Whaley SG, Berkow EL, Rybak JM, Nishimoto AT, Barker KS, Rogers PD. Azole antifungal resistance in Candida albicans and emerging non-albicans Candida species. Front. Microbiol. 2016;7:2173. doi: 10.3389/fmicb.2016.02173. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Clear, concise and up to date review of molecular mechanisms for azole resistance in Candida sp. (not including C. auris).
- 22.Orozco AS, Higginbotham LM, Hitchcock CA, et al. Mechanism of fluconazole resistance in Candida krusei . Antimicrob. Agents Chemother. 1998;42(10):2645–2649. doi: 10.1128/aac.42.10.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Albertson GD, Niimi M, Cannon RD, Jenkinson HF. Multiple efflux mechanisms are involved in Candida albicans fluconazole resistance. Antimicrob. Agents Chemother. 1996;40(12):2835–2841. doi: 10.1128/aac.40.12.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelly SL, Lamb DC, Kelly DE, et al. Resistance to fluconazole and cross-resistance to amphotericin B in Candida albicans from AIDS patients caused by defective sterol delta5,6-desaturation. FEBS Lett. 1997;400(1):80–82. doi: 10.1016/s0014-5793(96)01360-9. [DOI] [PubMed] [Google Scholar]
- 25.Denning DW, Venkateswarlu K, Oakley KL, et al. Itraconazole resistance in Aspergillus fumigatus . Antimicrob. Agents Chemother. 1997;41(6):1364–1368. doi: 10.1128/aac.41.6.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snelders E, Van Der Lee HAL, Kuijpers J, et al. Emergence of azole resistance in Aspergillus fumigatus and spread of a single resistance mechanism. PLoS Med. 2008;5(11):1629–1637. doi: 10.1371/journal.pmed.0050219. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• First paper to identify the dominance of one resistance mechanism in Dutch Aspergillus isolates, and to begin raising questions about an environmental source.
- 27.Snelders E, Huis In't Veld RAG, Rijs AJMM, Kema GHJ, Melchers WJG, Verweij PE. Possible environmental origin of resistance of Aspergillus fumigatus to medical triazoles. Appl. Environ. Microbiol. 2009;75(12):4053–4057. doi: 10.1128/AEM.00231-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Linden JWM, Arendrup MC, Warris A, et al. Prospective multicenter international surveillance of azole resistance in Aspergillus fumigatus . Emerging Infect. Dis. 2015;21(6):1041–1044. doi: 10.3201/eid2106.140717. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Detailed survey highlighting, among other things, the issue of azole resistance in Aspergillus fumigatus cryptic sibling species.
- 29.Mazur P, Baginsky W. In vitro activity of 1,3-beta-D-glucan synthase requires the GTP-binding protein Rho1. J. Biol. Chem. 1996;271(24):14604–14609. doi: 10.1074/jbc.271.24.14604. [DOI] [PubMed] [Google Scholar]
- 30.Alexander BD, Johnson MD, Pfeiffer CD, et al. Increasing echinocandin resistance in Candida glabrata: Clinical failure correlates with presence of FKS mutations and elevated minimum inhibitory concentrations. Clin. Infect. Dis. 2013;56(12):1724–1732. doi: 10.1093/cid/cit136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niimi K, Maki K, Ikeda F, et al. Overexpression of Candida albicans CDR1, CDR2, or MDR1 does not produce significant changes in echinocandin susceptibility. Antimicrob. Agents Chemother. 2006;50(4):1148–1155. doi: 10.1128/AAC.50.4.1148-1155.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dudiuk C, Macedo D, Leonardelli F, et al. Molecular confirmation of the relationship between Candida guilliermondii Fks1p naturally occurring amino acid substitutions and its intrinsic reduced echinocandin susceptibility. Antimicrob. Agents Chemother. 2017;61(5):1–5. doi: 10.1128/AAC.02644-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia-Effron G, Katiyar SK, Park S, Edlind TD, Perlin DS. A naturally occurring proline-to-alanine amino acid change in Fks1p in Candida parapsilosis, Candida orthopsilosis, and Candida metapsilosis accounts for reduced echinocandin susceptibility. Antimicrob. Agents Chemother. 2008;52(7):2305–2312. doi: 10.1128/AAC.00262-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perlin DS. Resistance to echinocandin-class antifungal drugs. Drug Resist. Updat. 2007;10(3):121–30. doi: 10.1016/j.drup.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia-Effron G, Lee S, Park S, Cleary JD, Perlin DS. Effect of Candida glabrata FKS1 and FKS2 mutations on echinocandin sensitivity and kinetics of 1,3-β-D-glucan synthase: implication for the existing susceptibility breakpoint. Antimicrob. Agents Chemother. 2009;53(9):3690–3699. doi: 10.1128/AAC.00443-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Imhof A, Balajee SA, Marr KA. New methods to assess susceptibilities of Aspergillus isolates to caspofungin. J. Clin. Microbiol. 2003;41(12):5683–5688. doi: 10.1128/JCM.41.12.5683-5688.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiménez-Ortigosa C, Moore C, Denning DW, Perlin DS. Emergence of echinocandin resistance due to a point mutation in the FKS1 gene of Aspergillus fumigatus in a patient with chronic pulmonary aspergillosis. Antimicrob. Agents Chemother. 2017;61(12):e01277-17. doi: 10.1128/AAC.01277-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pfaller MA. Antifungal drug resistance: mechanisms, epidemiology, and consequences for treatment. Am. J. Med. 2012;125(Suppl. 1):S3–S13. doi: 10.1016/j.amjmed.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 39.Alastruey-Izquierdo A, Melhem MSC, Bonfietti LX, Rodriguez-Tudela JL. Susceptibility test for fungi: clinical and laboratorial correlations in medical mycology. Rev. Inst. Med. Trop. Sao Paulo. 2015;57(Suppl. 19):57–64. doi: 10.1590/S0036-46652015000700011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alcazar-Fuoli L, Mellado E. Current status of antifungal resistance and its impact on clinical practice. Br. J. Haematol. 2014;166(4):471–484. doi: 10.1111/bjh.12896. [DOI] [PubMed] [Google Scholar]
- 41.Lockhart SR, Ghannoum MA, Alexander BD. Establishment and use of epidemiological cutoff values for molds and yeasts by use of the clinical and laboratory standards institute M57 standard. J. Clin. Microbiol. 2017;55(5):1262–1268. doi: 10.1128/JCM.02416-16. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Excellent overview of difference between epidemiological cut-off values and clinical break points and how they are developed by the Clinical and Laboratory Standards Institute for both yeasts and moulds.
- 42.Clinical and Laboratory Standards Institute. Principles and procedures for the development of epidemiological cutoff values for antifungal susceptbility testing. 2016. https://clsi.org/standards/products/microbiology/documents/m57/
- 43.Clinical and Laboratory Standards Institute. Reference method for broth dilution antifungal susceptibity testing of filamentous fungi (3rd Edition) 2017. https://clsi.org/standards/products/microbiology/documents/m38/ M38-A3.
- 44.Clinical and Laboratory Standards Institute. Reference method for broth dilution antifungal susceptibility testing of yeasts (4th Edition) 2012. https://clsi.org/standards/products/microbiology/documents/m27/ M27-A4.
- 45.EUCAST. EUCAST Definitive Document E.DEF 7.3.1. Method for the determination of broth dilution minimum inhibitory concentrations of antifungal agents for yeasts. 2017. www.eucast.org/ast_of_fungi/publications_in_journals/
- 46.EUCAST. EUCAST DEFINITIVE DOCUMENT E.DEF 9.3.1. Method for the determination of broth dilution minimum inhibitory concentration of antifungal agents for conidia forming moulds. 2017. www.eucast.org/ast_of_fungi/publications_in_journals/ [DOI] [PubMed]
- 47.Espinel-Ingroff A, Cuenca-Estrella M, Cantón E. EUCAST and CLSI: working together towards a harmonized method for antifungal susceptibility testing. Curr. Fungal Infect. Rep. 2013;7(1):59–67. [Google Scholar]
- 48.Albataineh MT, Sutton DA, Fothergill AW, Wiederhold NP. Update from the laboratory: clinical identification and susceptibility testing of fungi and trends in antifungal resistance. Infect. Dis. Clin. North Am. 2016;30(1):13–35. doi: 10.1016/j.idc.2015.10.014. [DOI] [PubMed] [Google Scholar]
- 49.Pfaller MA, Castanheira M, Messer SA, Rhomberg PR, Jones RN. Comparison of EUCAST and CLSI broth microdilution methods for the susceptibility testing of 10 systemically active antifungal agents when tested against Candida spp. Diagn. Microbiol. Infect. Dis. 2014;79(2):198–204. doi: 10.1016/j.diagmicrobio.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 50.Arendrup MC, Prakash A, Meletiadis J, Sharma C, Chowdhary A. Comparison of EUCAST and CLSI reference microdilution MICs of eight antifungal compounds for Candida auris and associated tentative epidemiological cutoff values. Antimicrob. Agents Chemother. 2017;61(6):e00485-17. doi: 10.1128/AAC.00485-17. [DOI] [PMC free article] [PubMed] [Google Scholar]; • The first paper to propose epidemiological cut-off values using European Committee on Antibiotic Susceptibility Testing or Clinical and Laboratory Standards Institute reference methods for the emerging multidrug resistant yeast C. auris.
- 51.Pfaller M, Boyken L, Hollis R, et al. Comparison of the broth microdilution methods of the European committee on antimicrobial susceptibility testing and the clinical and laboratory standards institute for testing itraconazole, posaconazole, and voriconazole against Aspergillus isolates. J. Clin. Microbiol. 2011;49(3):1110–1112. doi: 10.1128/JCM.02432-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cuenca-Estrella M. Antifungal drug resistance mechanisms in pathogenic fungi: from bench to bedside. Clin. Microbiol. Infect. 2014;20(Suppl. 6):54–59. doi: 10.1111/1469-0691.12495. [DOI] [PubMed] [Google Scholar]
- 53.Arendrup MC. Update on antifungal resistance in Aspergillus and Candida . Clin. Microbiol. Infect. 2014;20(Suppl. 6):42–48. doi: 10.1111/1469-0691.12513. [DOI] [PubMed] [Google Scholar]
- 54.Cuenca-Estrella M, Gomez-Lopez A, Alastruey-Izquierdo A, et al. Comparison of the Vitek 2 antifungal susceptibility system with the clinical and laboratory standards institute (CLSI) and European Committee on Antimicrobial Susceptibility Testing (EUCAST) Broth Microdilution Reference Methods and with the Sensititre Ye. J. Clin. Microbiol. 2010;48(5):1782–1786. doi: 10.1128/JCM.02316-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Espinel-Ingroff A, Alvarez-Fernandez M, Canton E, et al. Multicenter study of epidemiological cutoff values and detection of resistance in Candida spp. to anidulafungin, caspofungin, and micafungin using the Sensititre YeastOne colorimetric method. Antimicrob. Agents Chemother. 2015;59(11):6725–6732. doi: 10.1128/AAC.01250-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Espinel-Ingroff a. Comparison of three commercial assays and a modified disk diffusion assay with two broth microdilution reference assays for testing zygomycetes, Aspergillus spp., Candida spp., and Cryptococcus neoformans with posaconazole and amphotericin B. J. Clin. Microbiol. 2006;44(10):3616–3622. doi: 10.1128/JCM.01187-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Patel R, Mendrick C, Knapp CC, Grist R, McNicholas PM. Clinical evaluation of the Sensititre YeastOne plate for testing susceptibility of filamentous fungi to posaconazole. J. Clin. Microbiol. 2007;45(6):2000–2001. doi: 10.1128/JCM.00287-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Talbot JJ, Kidd SE, Martin P, Beatty JA, Barrs VR. Azole resistance in canine and feline isolates of Aspergillus fumigatus . Comp. Immunol. Microbiol. Infect. Dis. 2015;42:37–41. doi: 10.1016/j.cimid.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 59.Mello E, Posteraro B, Vella A, et al. Susceptibility testing of common and uncommon Aspergillus species against posaconazole and other mold-active antifungal azoles using the sensititre method. Antimicrob. Agents Chemother. 2017;61(6):e00168-17. doi: 10.1128/AAC.00168-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Siopi M, Pournaras S, Meletiadis J. Comparative evaluation of sensititre YeastOne and CLSI M38-A2 reference method for antifungal susceptibility testing of Aspergillus spp. against Echinocandins. J. Clin. Microbiol. 2017;55(6):1714–1719. doi: 10.1128/JCM.00044-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peterson JF, Pfaller MA, Diekema DJ, Rinaldi MG, Riebe KM, Ledeboer NA. Multicenter comparison of the VITEK 2 antifungal susceptibility test with the CLSI broth microdilution reference method for testing caspofungin, micafungin, and posaconazole against Candida spp. J. Clin. Microbiol. 2011;49(5):1765–1771. doi: 10.1128/JCM.02517-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cretella D, Barber KE, King ST, Stover KR. Comparison of susceptibility patterns using commercially available susceptibility testing methods performed on prevalent Candida spp. J. Med. Microbiol. 2016;65(12):1445–1451. doi: 10.1099/jmm.0.000383. [DOI] [PubMed] [Google Scholar]
- 63.Clinical and Laboratory Standards Institute. Performance Standards for Antifungal Susceptibility Testing of Yeasts (1st Edition) 2017. https://clsi.org/standards/products/microbiology/documents/m60/ CLSI supplement M60. [Google Scholar]
- 64.Astvad KM, Perlin DS, Johansen HK, Jensen RH, Arendrup MC. Evaluation of caspofungin susceptibility testing by the new Vitek 2 AST-YS06 yeast card using a unique collection of FKS wild-type and hot spot mutant isolates, including the five most common Candida species. Antimicrob. Agents Chemother. 2013;57(1):177–182. doi: 10.1128/AAC.01382-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kathuria S, Singh PK, Sharma C, et al. Multidrug-resistant Candida auris misidentified as Candida haemulonii: characterization by matrix-assisted laser desorption ionization–time of flight mass spectrometry and DNA sequencing and its antifungal susceptibility profile variability by Vitek 2, CL. J. Clin. Microbiol. 2015;53(6):1823–1830. doi: 10.1128/JCM.00367-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arendrup MC, Pfaller MA Group the DFS. Caspofungin etest susceptibility testing of Candida species: risk of misclassification of susceptible isolates of C. glabrata and C. krusei when adopting the revised CLSI caspofungin breakpoints. Antimicrob. Agents Chemother. 2012;56(7):3965–3968. doi: 10.1128/AAC.00355-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Espinel-Ingroff A, Arendrup M, Cantón E, et al. Multicenter study of method-dependent epidemiological cutoff values for detection of resistance in Candida spp. and Aspergillus spp. to Amphotericin B and Echinocandins for the Etest agar diffusion method. Antimicrob. Agents Chemother. 2017;61(1):e01792-16. doi: 10.1128/AAC.01792-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arendrup MC, Verweij P, Nielsen HV. Evaluation of MIC strip isavuconazole test for susceptibility testing of wild-type and non-wild-type Aspergillus fumigatus isolates. Antimicrob. Agents Chemother. 2017;61(1):e01659-16. doi: 10.1128/AAC.01659-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van Hal SJ, Chen SC, Sorrell TC, Ellis DH, Slavin M, Marriott DM. Support for the EUCAST and revised CLSI fluconazole clinical breakpoints by Sensititre YeastOne for Candida albicans: a prospective observational cohort study. J. Antimicrob. Chemother. 2014;69(8):2210–2214. doi: 10.1093/jac/dku124. [DOI] [PubMed] [Google Scholar]
- 70.Perlin DS. Antifungal drug resistance: do molecular methods provide a way forward? Curr. Opin. Infect. Dis. 2009;22(6):568–573. doi: 10.1097/QCO.0b013e3283321ce5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Martel CM, Parker JE, Bader O, et al. Identification and characterization of four azole-resistant erg3 mutants of Candida albicans . Antimicrob. Agents Chemother. 2010;54(11):4527–4533. doi: 10.1128/AAC.00348-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jensen RH. Resistance in human pathogenic yeasts and filamentous fungi: prevalence, underlying molecular mechanisms and link to the use of antifungals in humans and the environment. Dan. Med. J. 2016;63(10):B5288. [PubMed] [Google Scholar]; • Comprehensive review of the prevalence and molecular mechanisms of resistance for Candida and Aspergillus spp.
- 73.Jiang C, Dong D, Yu B, et al. Mechanisms of azole resistance in 52 clinical isolates of Candida tropicalis in China. J. Antimicrob. Chemother. 2013;68(4):778–785. doi: 10.1093/jac/dks481. [DOI] [PubMed] [Google Scholar]
- 74.Castanheira M, Deshpande LM, Davis AP, Rhomberg PR, Pfaller MA. Monitoring antifungal resistance in a global collection of invasive yeasts and molds: application of CLSI epidemiological cutoff values and whole-genome sequencing analysis for detection of azole resistance in Candida albicans . Antimicrob. Agents Chemother. 2017;61(10):e00906-17. doi: 10.1128/AAC.00906-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Garnaud C, Botterel F, Sertour N, et al. Next-generation sequencing offers new insights into the resistance of Candida spp. to echinocandins and azoles. J. Antimicrob. Chemother. 2015;70(9):2556–2565. doi: 10.1093/jac/dkv139. [DOI] [PubMed] [Google Scholar]; •• One of the first publications to demonstrate the value of next generation sequencing to detect genetic mutations involved in antifungal resistance in a variety of Candida spp.
- 76.Halliday CL, Slavin MA, Chen SC-A. Antifungal susceptibility testing of Candida and Cryptococcus species and mechanisms of resistance: implications for clinical laboratories. Curr. Fungal Infect. Rep. 2017;11(3):124–133. [Google Scholar]
- 77.Arendrup MC, Cuenca-Estrella M, Lass-Florl C, Hope WW. Breakpoints for antifungal agents: an update from EUCAST focussing on echinocandins against Candida spp. and triazoles against Aspergillus spp. Drug Resist. Updat. 2013;16(6):81–95. doi: 10.1016/j.drup.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 78.Shields RK, Nguyen MH, Press EG, et al. The presence of an FKS mutation rather than MIC is an independent risk factor for failure of echinocandin therapy among patients with invasive candidiasis due to Candida glabrata . Antimicrob. Agents Chemother. 2012;56(9):4862–4869. doi: 10.1128/AAC.00027-12. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Landmark study documenting the importance and clinical relevance of FKS gene mutations in Candida glabrata where their presence was independently linked to clinical failure and echinocandin.
- 79.Perlin DS. Echinocandin resistance in Candida . Clin. Infect. Dis. 2015;61(Suppl. 6):S612–S617. doi: 10.1093/cid/civ791. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Detailed review of epidemiology, mechanisms and molecular detection of echinocandin resistance in Candida spp.
- 80.Grosset M, Desnos-Ollivier M, Godet C, Kauffmann-Lacroix C, Cazenave-Roblot F. Recurrent episodes of Candidemia due to Candida glabrata, Candida tropicalis and Candida albicans with acquired echinocandin resistance. Med. Mycol. Case Rep. 2016;14:20–23. doi: 10.1016/j.mmcr.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dudiuk C, Gamarra S, Leonardeli F, et al. Set of classical PCRs for detection of mutations in Candida glabrata FKS genes linked with echinocandin resistance. J. Clin. Microbiol. 2014;52(7):2609–2614. doi: 10.1128/JCM.01038-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Biswas C, Chen SC, Halliday C, et al. Identification of genetic markers of resistance to echinocandins, azoles and 5-fluorocytosine in Candida glabrata by next-generation sequencing: a feasibility study. Clin. Microbiol. Infect. 2017;23(9):676, e7–676, e10. doi: 10.1016/j.cmi.2017.03.014. [DOI] [PubMed] [Google Scholar]
- 83.Hagiwara D, Takahashi H, Watanabe A, et al. Whole-genome comparison of Aspergillus fumigatus strains serially isolated from patients with Aspergillosis. J. Clin. Microbiol. 2014;52(12):4202–4209. doi: 10.1128/JCM.01105-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Meletiadis J, Mavridou E, Melchers WJG, Mouton JW, Verweij PE. Epidemiological cutoff values for Azoles and Aspergillus fumigatus based on a novel mathematical approach incorporating cyp51A sequence analysis. Antimicrob. Agents Chemother. 2012;56(5):2524–2529. doi: 10.1128/AAC.05959-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dudakova A, Spiess B, Tangwattanachuleeporn M, et al. Molecular tools for the detection and deduction of azole antifungal drug resistance phenotypes in Aspergillus species. Clin. Microbiol. Rev. 2017;30(4):1065–1091. doi: 10.1128/CMR.00095-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ahmad S, Khan Z, Hagen F, Meis JF. Simple low-cost molecular assays for TR34/L98H mutations in the cyp51A gene for rapid detection of triazole-resistant Aspergillus fumigatus isolates. J. Clin. Microbiol. 2014;52(6):2223–2227. doi: 10.1128/JCM.00408-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.European Centre for Disease Prevention and Control. European Centre for Disease Prevention and Control; Stockholm, Sweden: 2013. Risk assessment on the impact of environmental usage of triazoles on the development and spread of resistance to medical triazoles in Aspergillus species.www.ecdc.europa.eu/sites/portal/files/media/en/publications/Publications/risk-assessment-impact-environmental-usage-of-triazoles-on-Aspergillus-spp-resistance-to-medical-triazoles.pdf [Google Scholar]
- 88.Chong GL, van de Sande WW, Dingemans GJ, et al. Validation of a new Aspergillus real-time PCR assay for direct detection of Aspergillus and azole resistance of Aspergillus fumigatus on bronchoalveolar lavage fluid. J. Clin. Microbiol. 2015;53(3):868–874. doi: 10.1128/JCM.03216-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.White PL, Posso RB, Barnes RA. An analytical and clinical evaluation of the PathoNostics AsperGenius® Assay for the detection of invasive aspergillosis and resistance to azole antifungal drugs direct from plasma samples. J. Clin. Microbiol. 2017;55(8):2356–2366. doi: 10.1128/JCM.00411-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dannaoui E, Gabriel F, Gaboyard M, et al. Molecular diagnosis of invasive aspergillosis and detection of Azole resistance by a newly commercialized PCR kit. J. Clin. Microbiol. 2017;55(11):3210–3218. doi: 10.1128/JCM.01032-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cleveland AA, Farley MM, Harrison LH, et al. Changes in incidence and antifungal drug resistance in candidemia: results from population-based laboratory surveillance in Atlanta and Baltimore, 2008–2011. Clin. Infect. Dis. 2012;55(10):1352–1361. doi: 10.1093/cid/cis697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pappas PG, Kauffman CA, Andes DR, et al. Executive summary: clinical practice guideline for the management of candidiasis: 2016 update by the Infectious Diseases Society of America. Clin. Infect. Dis. 2016;62(4):409–417. doi: 10.1093/cid/civ1194. [DOI] [PubMed] [Google Scholar]
- 93.Vallabhaneni S, Cleveland AA, Farley MM, et al. Epidemiology and risk factors for echinocandin nonsusceptible Candida glabrata bloodstream infections: data from a large multisite population-based Candidemia Surveillance Program, 2008-2014. Open forum Infect. Dis. 2015;2(4):ofv163. doi: 10.1093/ofid/ofv163. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Detailed review of epidemiology, mechanisms and molecular detection of echinocandin resistance in Candida spp.
- 94.Pfaller MA, Castanheira M, Lockhart SR, Ahlquist AM, Messer SA, Jones RN. Frequency of decreased susceptibility and resistance to echinocandins among fluconazole-resistant bloodstream isolates of Candida glabrata . J. Clin. Microbiol. 2012;50(4):1199–1203. doi: 10.1128/JCM.06112-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rudramurthy SM, Chakrabarti A, Paul RA, et al. Candida auris candidaemia in Indian ICUs: analysis of risk factors. J. Antimicrob. Chemother. 2017;72(6):1794–1801. doi: 10.1093/jac/dkx034. [DOI] [PubMed] [Google Scholar]
- 96.Calvo B, Melo ASA, Perozo-Mena A, et al. First report of Candida auris in America: clinical and microbiological aspects of 18 episodes of candidemia. J. Infect. 2016;73(4):369–74. doi: 10.1016/j.jinf.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 97.Chowdhary A, Sharma C, Meis JF. Candida auris: a rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLoS Pathog. 2017;13(5):e1006290. doi: 10.1371/journal.ppat.1006290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fakhim H, Chowdhary A, Prakash A, et al. In vitro interactions of echinocandins with triazoles against multidrug-resistant Candida auris . Antimicrob. Agents Chemother. 2017;61(11):e01056-17. doi: 10.1128/AAC.01056-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lepak AJ, Zhao M, Berkow EL, Lockhart SR, Andes DR. Pharmacodynamic optimization for treatment of invasive Candida auris infection. Antimicrob. Agents Chemother. 2017;61(8):e00791–17. doi: 10.1128/AAC.00791-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Biswal M, Rudramurthy SM, Jain N, et al. Controlling a possible outbreak of Candida auris infection: lessons learnt from multiple interventions. J. Hosp. Infect. 2017;97(4):363–370. doi: 10.1016/j.jhin.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 101.Seyedmousavi S, Mouton JW, Melchers WJG, Brüggemann RJM, Verweij PE. The role of azoles in the management of azole-resistant aspergillosis: from the bench to the bedside. Drug Resist. Updat. 2014;17(3):37–50. doi: 10.1016/j.drup.2014.06.001. [DOI] [PubMed] [Google Scholar]; • In depth, excellent discussion of the rationale for and against use of azoles in the setting of azole-resistant aspergillosis.
- 102.Verweij PE, Ananda-Rajah M, Andes D, et al. International expert opinion on the management of infection caused by azole-resistant Aspergillus fumigatus . Drug Resist. Updat. 2015;21–22:30–40. doi: 10.1016/j.drup.2015.08.001. [DOI] [PubMed] [Google Scholar]; •• International consensus recommendations for managing azole-resistant Aspergillus infections.
- 103.Blyth CC, Gilroy NM, Guy SD, et al. Consensus guidelines for the treatment of invasive mould infections in haematological malignancy and haemopoietic stem cell transplantation, 2014. Intern. Med. J. 2014;44(12b):1333–1349. doi: 10.1111/imj.12598. [DOI] [PubMed] [Google Scholar]
- 104.Maertens J, Marchetti O, Herbrecht R, et al. European guidelines for antifungal management in leukemia and hematopoietic stem cell transplant recipients: summary of the ECIL 3–2009 update. Bone Marrow Transplant. 2011;46(5):709–718. doi: 10.1038/bmt.2010.175. [DOI] [PubMed] [Google Scholar]
- 105.van der Linden JWM, Snelders E, Kampinga GA, et al. Clinical implications of azole resistance in Aspergillus fumigatus, The Netherlands, 2007-2009. Emerg. Infect. Dis. 2011;17(10):1846–1854. doi: 10.3201/eid1710.110226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.van Ingen J, van der Lee HA, Rijs TAJ, et al. Azole, polyene and echinocandin MIC distributions for wild-type, TR34/L98H and TR46/Y121F/T289A Aspergillus fumigatus isolates in The Netherlands. J. Antimicrob. Chemother. 2015;70(1):178–181. doi: 10.1093/jac/dku364. [DOI] [PubMed] [Google Scholar]
- 107.Seyedmousavi S, Melchers WJG, Mouton JW, Verweij PE. Pharmacodynamics and dose-response relationships of liposomal amphotericin B against different azole-resistant Aspergillus fumigatus isolates in a murine model of disseminated aspergillosis. Antimicrob. Agents Chemother. 2013;57(4):1866–1871. doi: 10.1128/AAC.02226-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schwartz S, Ruhnke M, Ribaud P, et al. Improved outcome in central nervous system aspergillosis, using voriconazole treatment. Blood. 2005;106(8):2641–2645. doi: 10.1182/blood-2005-02-0733. [DOI] [PubMed] [Google Scholar]
- 109.Kethireddy S, Andes D. CNS pharmacokinetics of antifungal agents. Expert Opin. Drug Metab. Toxicol. 2007;3(4):573–581. doi: 10.1517/17425225.3.4.573. [DOI] [PubMed] [Google Scholar]
- 110.Groll a H, Giri N, Petraitis V, et al. Comparative efficacy and distribution of lipid formulations of amphotericin B in experimental Candida albicans infection of the central nervous system. J. Infect. Dis. 2000;182(1):274–282. doi: 10.1086/315643. [DOI] [PubMed] [Google Scholar]
- 111.Wiederhold NP. Antifungal resistance: current trends and future strategies to combat. Infect. Drug Resist. 2017;10:249–259. doi: 10.2147/IDR.S124918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ghannoum M, Larkin E, Long L. 27th European Congress on Clinical Microbiology and Infectious Diseases. Vienna, Austria: 22 April 2017. Susceptibility of recent Candida auris isolates to the novel echinocandin CD101 and comparator antifungal agents. Presented at. [Google Scholar]
- 113.Chandra J, Ghannoum MA. CD101, a novel echinocandin, possesses potent antibiofilm activity against early and mature Candida albicans biofilms. Antimicrob. Agents Chemother. 2018;62(2):e01750-17. doi: 10.1128/AAC.01750-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Larkin E, Hager C, Chandra J, et al. The emerging pathogen Candida auris: growth phenotype, virulence factors, activity of antifungals, and effect of SCY-078, a novel glucan synthesis inhibitor, on growth morphology and biofilm formation. Antimicrob. Agents Chemother. 2017;61(5):e02396-16. doi: 10.1128/AAC.02396-16. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Up to date review of the pertinent biology of emergent C. auris and drug therapeutic approached in managing infections caused by this pathogen.
- 115.Osherov N, Kontoyiannis DP. The anti-Aspergillus drug pipeline: is the glass half full or empty? Med. Mycol. 2017;55(1):118–124. doi: 10.1093/mmy/myw060. [DOI] [PubMed] [Google Scholar]; • Up to date review of new anti-Aspergillus drugs on the horizon and their feasibility of use.
- 116.Cole DC, Govender NP, Chakrabarti A, Sacarlal J, Denning DW. Improvement of fungal disease identification and management: combined health systems and public health approaches. Lancet Infect. Dis. 2017;17(12):e412–e419. doi: 10.1016/S1473-3099(17)30308-0. [DOI] [PubMed] [Google Scholar]