Abstract
TAL1 is a helix-loop-helix transcription factor that is essential for hematopoiesis. In vitro DNA binding site selection experiments have previously identified the preferred binding site for TAL1 heterodimers as AACAGATGGT. TAL1 homodimers do not bind DNA with significant affinity. A subset of other E-box sequences is also bound by TAL1 heterodimers. Here, we present an analysis of TAL1 heterodimer DNA binding specificity, using E-boxes derived from genomic clones, which were isolated by immunoadsorption of K562 erythroleukemia cell chromatin with a TAL1 antibody. We show that TAL1 heterodimer binding to a CAGATG E-box is strongly modulated by nucleotides flanking the E-box. A 10 base pair element consisting of the CAGATG E-box and two flanking nucleotides in both the 5′ and 3′ direction is sufficient for high-affinity binding. Certain mutations of nucleotides in either the 5′ (−1 and −2) or 3′ (+1 and +2) direction strongly inhibit binding. The importance of flanking nucleotides also exists in the context of nonpreferred E-boxes recognized by TAL1 heterodimers. Although there are no known target genes for TAL1, the regulatory regions of several genes involved in hematopoiesis contain the preferred E-box CAGATG. However, based on our results, the E-boxes in these potential target genes contain flanking sequences that would be expected to significantly reduce TAL1 heterodimer binding in vitro. Thus, additional stabilizing forces, such as protein-protein interactions between TAL1 heterodimers and accessory factors, may be required to confer high-affinity TAL1 heterodimer binding to such sequences.
Keywords: TAL1, SCL, Helix-loop-helix, Transcription factor, Hematopoiesis, E-box, Erythroid, Leukemogenesis, T-ALL
ONE quarter of the cases of T-cell acute lymphoblastic leukemia (T-ALL) involve a chromosomal translocation or deletion that activates expression of the transcription factor TAL1 (4,10). TAL1 is not normally expressed in T-cells. By contrast, malignant T-cells from the majority of patients with T-ALL, even those without tumor-specific rearrangement involving TAL1, express TAL1 (4). Based on these observations, it has been hypothesized that ectopic expression of TAL1 within immature T-cells is a causative factor in this type of leukemia.
Mouse embryonic stem cells homozygous for a targeted disruption of Tal1 are unable to differentiate along any hematopoietic lineage in both in vitro and in vivo assays (38). Mice homozygous for such a targeted disruption of Tal1 fail in embryonic development around day 9.5 of gestation. Yolk sac blood islands, the site of embryonic hematopoiesis, are completely absent in the Tal1 mutant embryos (40,42). These observations indicate that the TAL1 gene product is essential for hematopoiesis and functions in the specification or maintenance of early hematopoietic progenitor cells. TAL1 also seems to be involved later in hematopoietic development, during differentiation along myeloid and erythroid lineages (1,21,27,36,45).
TALI is a member of the basic helix-loop-helix (bHLH) family of transcription factors (4). Functional bHLH dimers regulate transcription through binding to a consensus DNA sequence known as the E-box (CANNTG) (26). TAL1 forms functional heterodimers with other bHLH proteins, including E12 and E47, the ubiquitously expressed products of the E2A gene (22,23). These TAL1 heterodimers bind DNA with high affinity and sequence specificity (22–24,31,47), whereas TAL1 homodimers appear to lack DNA binding activity (22).
Using in vitro binding site selection methodology, Hsu et al. (24) reported that the optimal binding site for TAL1 heterodimers is AACAGATGGT. This consensus was derived from experiments using heterodimers containing recombinant TAL1 and binding partners synthesized either in vitro or from leukemic cell extract. A strong preference was observed for the E-box core; 77–93% of the E-boxes sequenced were CAGATG (24). By contrast, a significant but more modest preference was observed for the nucleotides flanking the E-box: an A two nucleotides 5′ of the E-box (A−2), an A 5′ of the E-box (A−1), a G two nucleotides 3′ of the E-box (G+2), a T 3′ of the E-box (T+1) (24). The preferred nucleotide at each of these flanking positions was present at a frequency of 56–80%. However, only 17% of the E-boxes had the preferred nucleotide at all four flanking positions. The binding site specificity of TALI complexes containing different heterodimeric partners appeared to be identical (24).
Sequences resembling the preferred TAL1 binding site are present in the promoters/enhancers of several genes that are candidates for regulation by TAL1. These include the erythroid bridging factor LM02/RBTN2 (AGCAGATGAT) (41), the T-cell-specific tyrosine kinase Lck (CCCAGATGCA) (43), the hematopoietic stem cell antigen CD34 (TCCAGATGCC) (4), the erythropoietin receptor (EpoR) (TACAGATGAG) (33), the erythroid transcription factor GATA-1 (GTCAGATGGC) (51), and the β-globin locus (CCCAGATGTT). At present, none of these genes have been shown to be regulated directly by TAL1.
We have initiated experiments using chromatin immunoadsorption to identify TAL1 target genes. This assay demands a physiological interaction between TAL1 and regulatory sequences of the target genes. Previously, chromatin immunoadsorption has been used to identify genes regulated by the thyroid hormone receptor (7,8), c-Myc (20), and NF-E2 (15). Analysis of the binding of TAL1 heterodimers to cloned DNA fragments obtained by chromatin immunoadsorption led us to analyze further the DNA binding specificity of TAL1 heterodimers. The experiments described herein confirm and extend the previous work (24) showing that the DNA binding of TAL1 heterodimers in vitro is critically dependent on the nucleotides flanking an optimal E-box core.
MATERIALS AND METHODS
Production and Characterization of Anti-Human TAL1 Polyclonal Antisera
Polyclonal antibodies specific for TAL1 were generated by immunizing rabbits with purified GST-TAL1, a glutathione-S-transferase fusion protein that contains amino acids 151–331 of TAL1 (2), encoded by a truncated form of the human TAL1 cDNA. To determine the specificity of the antibodies, Western blot analysis was performed with crude serum and antibodies that had been affinity purified on a resin containing immobilized GST-TAL1.
Nuclear extracts prepared from K562 human erythroleukemia cells and MEL mouse erythroleukemia cells (5 μl) were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12%). Gels were transfered to Immobilon P PVDF membrane (Millipore). Transfers were performed at 15 V for 35 min using a semidry transfer apparatus (BioRad) in 48 mM Tris, 39 mM glycine, 0.037% SDS, and 20% methanol. Blots were incubated overnight in 25 mM Tris-HCl (pH 7.4), 137 mM NaCl, 2.7 mM KCl, 0.1% Tween-20 (Sigma) (TBS-Tween), and 5% Block (Amersham). Primary antibodies were diluted in TBS-Tween plus 1% BSA to a final concentration of 1:1500 for crude serum or 1:100 for affinity-purified material. Blots were incubated with primary antibody for 1 h. Unbound primary antibody was removed by washing the blots three times for 5 min each with TBS-Tween. Blots were then incubated for 1 h with a donkey-anti-rabbit secondary antibody conjugated with horseradish peroxidase (Amersham), diluted 1:1000 in TBS-Tween with 1% BSA. Blots were then washed three times for 7 min each in TBS-Tween. Incubations and washes were carried out at room temperature with constant rotation. Blots were developed using the ECL system (Amersham) and exposed to X-ray film.
Chromatin Immunoadsorption Assay
Immunoadsorptions were performed using a modification of the method of Bigler and Eisenman (7,8). Nuclei from K562 cells (3 × 107) were isolated and resuspended in 200 μl of a solution containing 10 mM Tris-HCl (pH 7.9), 10 mM MgCl2, 6 mM DTT, 5 μg/ml leupeptin, and 0.1 mM phenylmethylsulfonyl fluoride (PMSF). Samples were then incubated with 500 units of HindIII restriction endonuclease at 37°C for 45 min. After digestion, nuclei were pelleted at 2000 × g for 2 min. Protein–DNA complexes were eluted by incubation in 200 μl 12 mM Tris (pH 7.6), 3 mM EDTA, 5 mM DTT, 5 μg/ml leupeptin, and 0.1 mM PMSF at 4°C for 1 h with constant rotation. Nuclear debris was pelleted by centrifugation at 2000 × g for 2 min. Aliquots of the supernatant (200 μl) were transfered to a tube containing 20 μl of 50% protein A-Sepharose and 2 μl preimmune serum. NaCl and BSA were added to final concentrations of 100 mM and 1 mg/ml, respectively. The samples were then incubated at 4°C for 30 min with constant rotation. Following removal of the Sepharose pellet by centrifugation, 100 μl of the supernatant was incubated with 15 μg of affinity-purified TAL1 antisera or preimmune serum for 2 h at 4°C with constant rotation. The samples were then incubated with 10 μl of 50% protein A-Sepharose for 1 h at 4°C with constant rotation. The Sepharose pellets were washed twice with 12 mM Tris-HCl (pH 7.6), 100 mM NaCl, 1 mg/ml BSA, 5 μg/ml leupeptin, and 0.1 mM PMSF and resuspended in 50 μl of 10 mM Tris-HCl (pH 8.0), 1 mm EDTA (TE). After the addition of 10 μg of yeast tRNA, samples were extracted once with phenol/chloroform. The organic phase was extracted a second time with TE. Pooled aqueous phases were precipitated with ethanol, and the nucleic acid pellet was resuspended in 20 μl TE (pH 7.6). The resuspended genomic DNA was ligated into HindIII linearized pB2SK+ vector (Stratagene) and used to transform high-efficiency Escherichia coli JM109 competent cells (Promega).
DNA Sequence Analysis
Sequencing reactions were performed using an AmpliTaq dye terminator cycle sequencing kit (Perkin Elmer). The reactions were run on an ABI DNA sequencer and the results analyzed using EditView software (Perkin Elmer). Genbank searches were performed at NCBI using the BLAST program.
Synthesis of TAL1 and E12 by In Vitro Transcription/Translation
For synthesis of E12, the plasmid E12*pBS was used. This plasmid is derived from the pBluescript vector and encodes the full-length E12 polypeptide driven from the T3 promoter (R. Baer, unpublished data). For the in vitro transcription, the plasmid was linearized just 3′ of the coding sequence with KpnI. For the synthesis of TAL1, the tal1M1/pTM3326 plasmid was used (24). This plasmid encodes the full-length TAL1 polypeptide under control of the T7 promoter. As the plasmid contains a T7 transcription terminator, it was not necessary to linearize it for the in vitro transcription reactions.
The in vitro transcription and translation reactions were performed using the TNT Coupled Reticulocyte Lysate System (Promega). RNA is synthesized and then translated in a single reaction containing rabbit reticulocyte lysate as instructed by the manufacturer. The 20–25 μl reactions contained 0.1 μg/μl of the E12 plasmid and/or the TAL1 plasmid. Labeled reactions were performed in the presence of [35S]methionine (Dupont, >1000 Ci/mmol). The labeled reactions were analyzed by SDS-PAGE (10%). For electrophoretic mobility shift assays (EMSAs), the reactions were performed with unlabeled methionine.
Oligonucleotide Probes
Single-stranded oligonucleotides were purified from 12% denaturing polyacrylamide gels. DNA duplexes were generated by annealing two complementary single-stranded oligonucleotides in 10 mM Tris (pH 8.0) with 50 mM KCl. Double-stranded oligonucleotides were end-labeled with [γ-32P]ATP (Dupont, 6000 Ci/mmol) by T4 polynucleotide kinase. The labeled probes were purified on Sephadex G-25 spin columns (Boehringer Mannhiem).
Electrophoretic Mobility Shift Assay
Aliquots of the transcription/translation reactions (3 μl) or fractionated MEL cell nuclear extract (4 μl) were incubated with 10% glycerol, 10 mM Tris-HCl (pH 7.9), 20 mM HEPES (pH 7.9), 16 mM NaCl, 1.2 mM MgCl2, 1.25 μg poly(dI-dC), 7.5 pg BSA, and 1 mM DTT in a final reaction volume of 25 μl for 20 min at room temperature. For supershift experiments, 1 μl of preimmune or immune serum was added to the binding reactions and incubated for another 15 min at room temperature. The protein-DNA complexes were resolved on 6.5% nondenaturing polyacrylamide gels in 0.75 × TAE running buffer [30 mM Tris acetate (pH 8.0), 0.75 mM EDTA] at 200 V for 2 h at 4°C.
The intensity of bands representing protein-DNA complexes and free DNA were quantitated on a phosphorImager (Molecular Dynamics) using ImageQuant software. The concentration of bound DNA was determined relative to known amounts of free DNA. In the quantitative DNA binding experiments, nonlinear regression analysis was performed using the Kaleidagraph program (Synergy Software) to estimate the equilibrium binding constant (K d) and the concentration of molecules competent for DNA binding (B m).
Nuclear Extract Preparation
Isolation of nuclei and preparation of K562 and MEL cell nuclear extracts was performed as described previously (29). MEL cell nuclear extract was fractionated on a Resource-S cation exchange column with a Pharmacia FPLC system as described previously (30). Because the binding activity was low, fractions containing TAL1 were concentrated 13-fold using a Centricon 30 microconcentrator (Amicon). TAL1 DNA binding activity was assayed by EMSA with the concentrated material (4 μl).
RESULTS
Charaterization of TAL1 Antisera
Antibodies directed against a transcription factor that react with the DNA-bound factor without disrupting the protein-DNA complex can be used in chromatin immunoprecipitation assays to isolate genomic DNA fragments bound by the factor in intact cells. The assay, which has been used to identify target genes for several factors (7,8,15,20), involves immunoprecipitating restriction enzyme solubilized chromatin fragments with an antibody directed against a transcription factor. To identify DNA sequences bound by TAL1, we generated rabbit polyclonal antibodies against a GST-TAL1 fusion protein. The specificity of the antiserum was determined by Western blot analysis using K562 and MEL cell nuclear extracts as well as purified GST-TAL1. As shown in Fig. 1A, the antiserum reacted strongly with GST-TAL1 (lane 4) and detected a series of ∼43-kDa polypeptides in the nuclear extracts (lanes 2 and 3). The detection of multiple TAL1 species is consistent with previous reports of TAL1 isoforms resulting from phosphorylation (13,17,39).
FIG. 1.
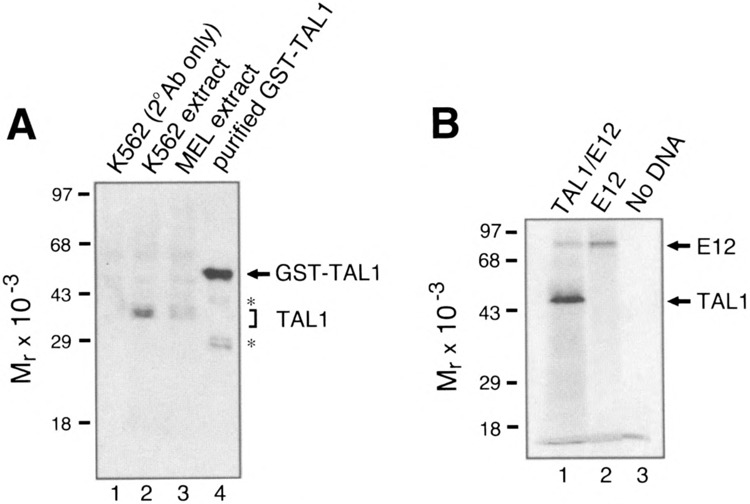
(A) Immunodetection of TAL1 in K562 and MEL cell nuclear extracts. Nuclear extracts were resolved by SDS-PAGE and analyzed by Western blotting with rabbit anti-human TAL1 antibody. The position of endogenous TAL1 polypeptides in K562 and MEL cell nuclear extracts is indicated by the bracket. The purified GST-TAL1 fusion protein is indicated by an arrow. The proteolytic fragments of the fusion protein are indicated by asterisks. (B) SDS-PAGE of TAL1 and E12 generated by in vitro transcription and translation. Lane 1, synthesis of TAL1 and E12; lane 2, synthesis of E12 alone; lane 3, control reaction in which no DNA was added. The positions of TAL1 and E12 are indicated by arrows.
The ability of the antiserum to react with the TAL1-DNA complex was measured by EMS A (Fig. 2). DNA binding assays were performed with recombinant TAL1/E12 heterodimers synthesized by in vitro transcription/translation in a rabbit reticulocyte lysate and a previously described oligonucleotide probe (TAL1con) containing the preferred site for TAL1 heterodimer binding (AACAGATGGT) (24). Figure 1B shows an SDS-PAGE analysis of TAL1 and E12 proteins synthesized in the presence of [35S]methionine. For EMSAs, TAL1, E12, or TAL1/ E12 heterodimers were synthesized with nonradioactive methionine.
FIG. 2.
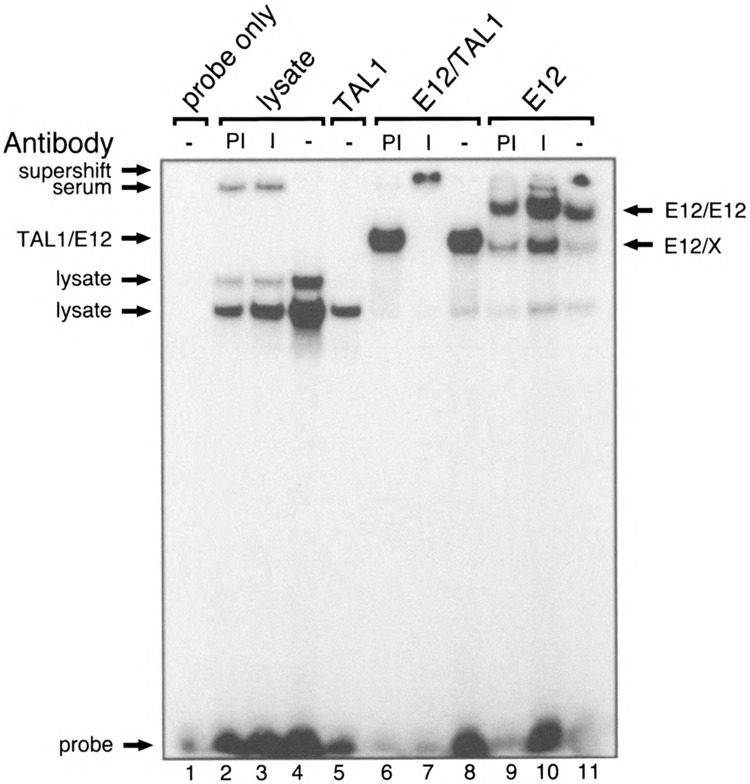
Binding of recombinant TAL1/E12 heterodimers to the preferred TAL1 recognition site. TAL1 and E12 were transcribed and translated in vitro and incubated with the TAL1con probe. EMSAs were performed as described in Materials and Methods, except that the gel was run for 3 h. The components added to each reaction and the presence of preimmune serum (PI), immune serum (I), or no serum (−) are indicated at the top. Complexes resulting from the binding of proteins endogenous to the lysate to the probe are indicated as lysate. E12/X may represent a complex formed by the binding of a heterodimer between E12 and a protein in the lysate to the probe. A protein-DNA complex formed by the binding of a protein in preimmune and immune serum to the probe is indicated as serum. The positions of E12/E12, TAL1/E12 complexes, and the probe are indicated.
As shown in Fig. 2, the TAL1 antiserum can “supershift” TAL1-DNA complexes. Transcription/translation reactions in which no DNA template was added were used as a negative control. These control assays revealed proteins endogenous to the reticulocyte lysate that bound the probe (lanes 2–4). No additional bands were detected in the reactions containing TAL1 alone (lane 5). This observation is consistent with previous reports, suggesting that TAL1 homodimers lack DNA binding activity (24). By contrast, reactions containing E12 alone generated two additional complexes (lanes 9–11). The relative mobility of the lower mobility complex is consistent with that expected for E12 homodimers, which bind E-box-containing sequences (3). The faster migrating species (E12/X) may represent a heterodimer between E12 and an unknown protein in the reticulocyte lysate. The TAL1 antibody does not affect the mobility of this complex (lane 10). However, we cannot exclude the possibility that this complex contains rabbit TAL1, as the ability of the TAL1 antiserum to cross-react with rabbit TAL1 is unknown. A complex that migrated slightly slower than the E12/X complex was unique to the reactions containing both TAL1 and E12 (lanes 6–8). The relative mobility of this complex, and the fact that it was supershifted by the TAL1 antibodies (lane 7), suggests that it represents a TAL1/E12 heterodimer bound to the DNA.
TAL1 Binding to E-Boxes From Clones Isolated by Chromatin Immunoadsorption
To identify genes regulated by TAL1, we used a chromatin immunoadsorption assay, similar to that described previously (7,8). Anti-TAL1 antibodies were used to immunoadsorb native protein-DNA complexes released from nuclei following restriction endonuclease digestion. After deproteinization, the DNA fragments were cloned and sequenced. Although none of the sequenced clones were identical to any known sequence, most of the clones had numerous potential binding sites for both erythroid and ubiquitous transcription factors. Several cloned DNA fragments contained E-boxes, some of which might be recognized by TAL1. A detailed characterization of the immunoadsorption results will be presented elsewhere.
To determine whether TAL1 heterodimers can bind to E-boxes in the clones, we generated oligonucleotide probes based on the E-box sequences of the clones (Fig. 3A). EMSAs were performed using in vitro synthesized TAL1/E12. We observed binding of TAL1/E12 to several of these oligonucleotides (Fig. 3B, lanes 2, 5, 8, 11). The identity of the protein-DNA complexes was confirmed by addition of the TAL1 antiserum, which retarded complex migration (Fig. 3B, lanes 3, 6, 9, 12). Oligonucleotide probes bound by TAL1 /E12 competed effectively for heterodimer binding to the TAL1con probe (data not shown). The TAL1/E12 interaction with the probes was E-box dependent, as mutation of the E-box abolished the ability to both bind and compete (data not shown). Each of the E-boxes tested was distinct from the preferred site of TAL1con, indicating that TAL1 heterodimers can interact with a variety of E-boxes. Probes derived from clones with the E-box sequences, AACACATGAG and GGCATATGAT, were also examined and showed no evidence for TAL1 /E12 binding (data not shown).
FIG. 3.
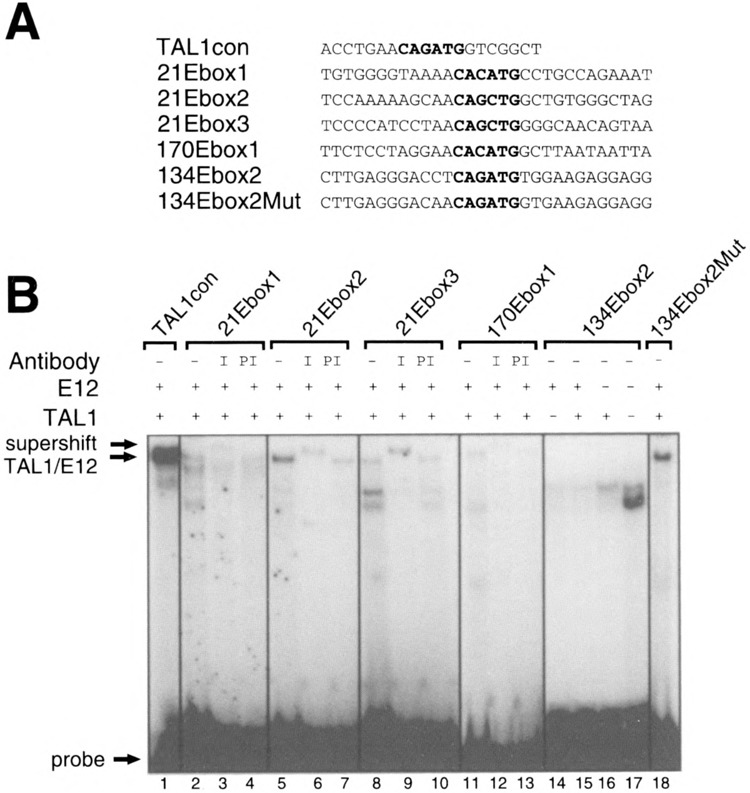
EMSA analysis of TAL1/E12 binding to sequences from clones isolated by immunoadsorption. (A) One strand of the double-stranded oligonucleotide probes is shown in the 5′ to 3′ orientation. (B) The presence (+) or absence (−) of TAL1 and E12 in the in vitro transcription/translation reactions and presence of preimmune serum (PI), immune serum (I), or no serum (−) in the binding reactions is shown at the top of the gel. The positions of the TAL1/E12 complexes and the probes are indicated.
Surprisingly, we observed no binding of TAL1/ E12 to the 134Ebox2 probe (Fig. 3B, lane 15), which contained an E-box identical to the TAL1con probe. Moreover, in competition experiments, a 400-fold excess of the unlabeled 134Ebox2 probe did not compete efficiently for TAL1/E12 heterodimer binding to the labeled TAL1con probe (data not shown). These observations suggest that nucleotides flanking the E-box core can strongly modulate TAL1/E12 binding and that the core itself is insufficient for high-affinity binding.
Importance of Nucleotides Flanking the E-Box for TAL1 Binding
In the in vitro binding site selection experiments that defined the preferred binding site of TALI heterodimers, some preference was observed for the nucleotides 5′ (−1, −2) and 3′ (+1.+2) of the E-box core (24). The diversity of flanking sequences obtained in the selection experiments suggested that these positions were less important determinants of binding than the core sequences (24). However, our observation that TAL1 heterodimers fail to bind probes containing the preferred E-box core, which are divergent at the nucleotides flanking the E-box, suggested that these flanking nucleotides are critical determinants of binding.
Comparison of the sequence of the 134Ebox2 probe to that of the TAL1con probe revealed that these oligonucleotides differ in numerous positions beyond the two nucleotides flanking the E-box. To determine whether nucleotides extending beyond the +2 and −2 positions relative to the E-box are important factors in TAL1/E12 binding, we mutated the two bases flanking each side of the E-box of probe 134Ebox2, so that they were identical to those in the TAL1con probe (Fig. 3A). Indeed, TAL1/E12 heterodimers bound this probe, 134Ebox2Mut (Fig. 3B, lane 18).
Quantitative EMSAs were used to estimate the affinity of TAL1/E12 heterodimers for the TAL1con (Fig. 4B) and 134Ebox2Mut probes (Fig. 5B). Titrations were performed with a constant amount of in vitro translated TAL1/E12 and increasing concentrations of the labeled probes. After quantitation on a PhosphorImager, nonlinear regression analysis was used to estimate the equilibrium binding constant (K d), a measure of affinity. TAL1 heterodimers bound the TAL1con probe with an apparent K d of 4.2 nM (Fig. 4C). A K d of 3.7 nM was estimated for binding to the 134Ebox2Mut probe (Fig. 5C). The fact that these K d values are similar indicates that TAL1 heterodimers have a roughly equivalent affinity for both probes. These K d values indicate high-affinity protein-DNA interactions and resemble K d values for other bHLH-DNA interactions, including those involving E12 heterodimers (9,14). Together, these data suggest that a 10 base pair element consisting of the E-box core and two flanking nucleotides in both the 5′and 3′ directions is sufficient for high-affinity TAL1/E12 binding. In addition, nucleotides outside of the 10 base pair element do not measurably effect binding.
FIG. 4.
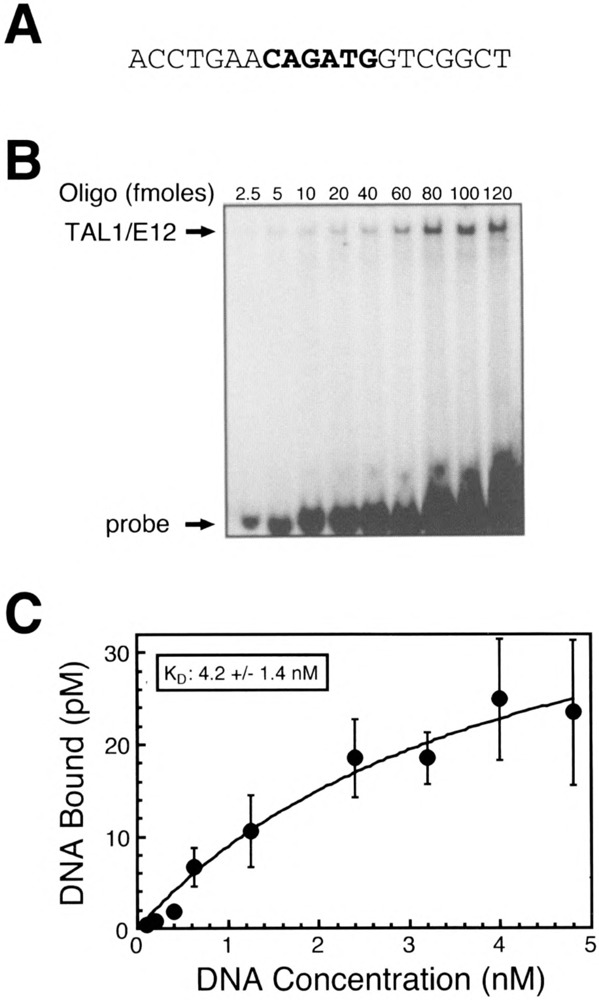
Quantitative EMSA analysis of recombinant TAL1/E12 heterodimer binding to the TAL1con probe. (A) Sequence of one strand of the double-stranded TAL1con oligonucleotide probe is shown in the 5′ to 3′ orientation. (B) EMSA analysis of binding reactions in which increasing amounts of the labeled probe were incubated with recombinant TAL1/E12 heterodimers as described in Materials and Methods. The positions of the TAL1/E12 complexes and the probe are indicated. (C) The concentration of the TAL1/E12-DNA complexes (pM) was determined by quantitation with a Phosphorlmager and plotted as a function of the total DNA concentration (nM) in the binding assay. Nonlinear regression analysis was used to estimate the equilibrium binding constant (K d = 4.2 ± 1.4 nM) and the concentration of molecules competent for DNA binding (B m = 47.3 ± 9.2) (mean ± S E, n = 4).
FIG. 5.
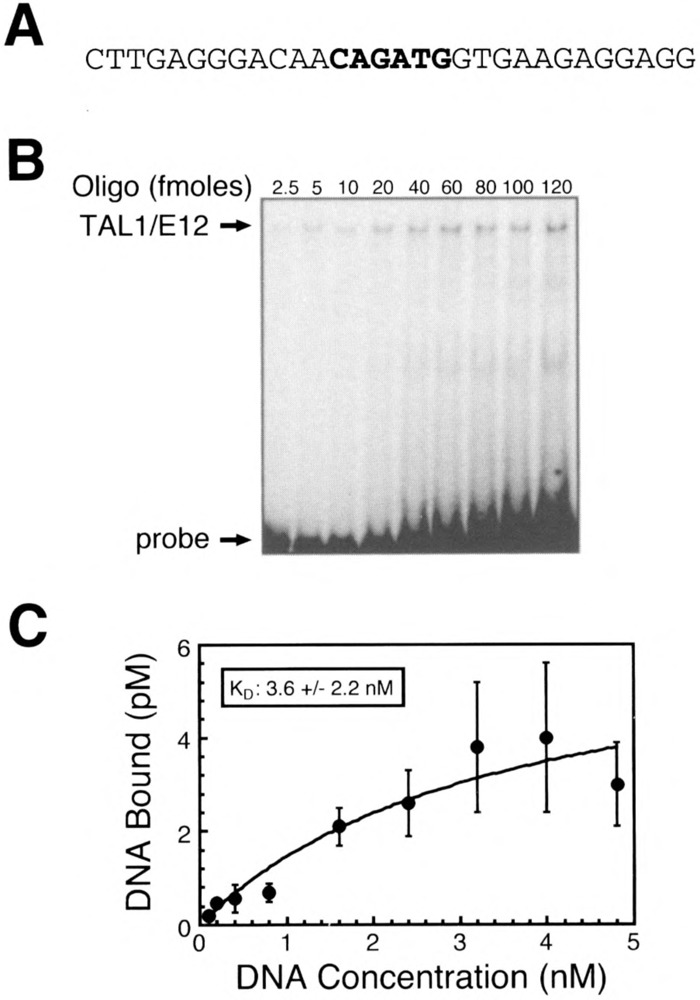
Quantitative EMSA analysis of recombinant TAL1/E12 heterodimer binding to the 134Ebox2Mut probe. (A) Sequence of one strand of the double-stranded TAL1con oligonucleotide probe is shown in the 5′ to 3′ orientation. (B) EMSA analysis of binding reactions in which increasing amounts of the labeled probe were incubated with recombinant TAL1/E12 heterodimers as described in Materials and Methods. The positions of the TAL1/E12 complexes and the probe are indicated. (C) The concentration of the TAL1/E12-DNA complexes (pM) was determined by quantitation with a phosphorlmager and plotted as a function of the total DNA concentration (nM) in the binding reactions. Nonlinear regression analysis was used to estimate the equilibrium binding constant (K d = 3.6 ± 2.2 nM) and the concentration of molecules competent for DNA binding (B m = 6.5 ± 2.2) (mean ± SE, n = 4).
To examine in more detail the preference for nucleotides flanking the E-box, we generated a series of oligonucleotide probes based on the 134Ebox2Mut sequence. Each oligonucleotide contained a base pair substitution mutation in a single flanking nucleotide (Figs. 6A, 7A). EMSAs were performed with increasing concentrations of DNA probe to assess the ability of TAL1 heterodimers to bind these probes relative to the 134Ebox2Mut probe (Figs. 6B, 7B). Substitution of the A−1, nucleotide 5′ of the E-box with a T resulted in almost complete abolition of DNA binding (Fig. 6C). By contrast, substitution at this position with a C only had a minimal effect on binding (Fig. 6C). The oligonucleotide probe containing a G substitution displayed intermediate binding (Fig. 6C). Substitution of the A nucleotide at the −2 position relative to the E-box with either a C or T resulted in a strong reduction in TAL1/E12 binding, although binding was still detectable (Fig. 6C). The presence of a T at this position resulted in a significant loss of binding relative to the 134Ebox2Mut probe, but binding was consistently higher than with other substitutions at this position (Fig. 6C).
FIG. 6.
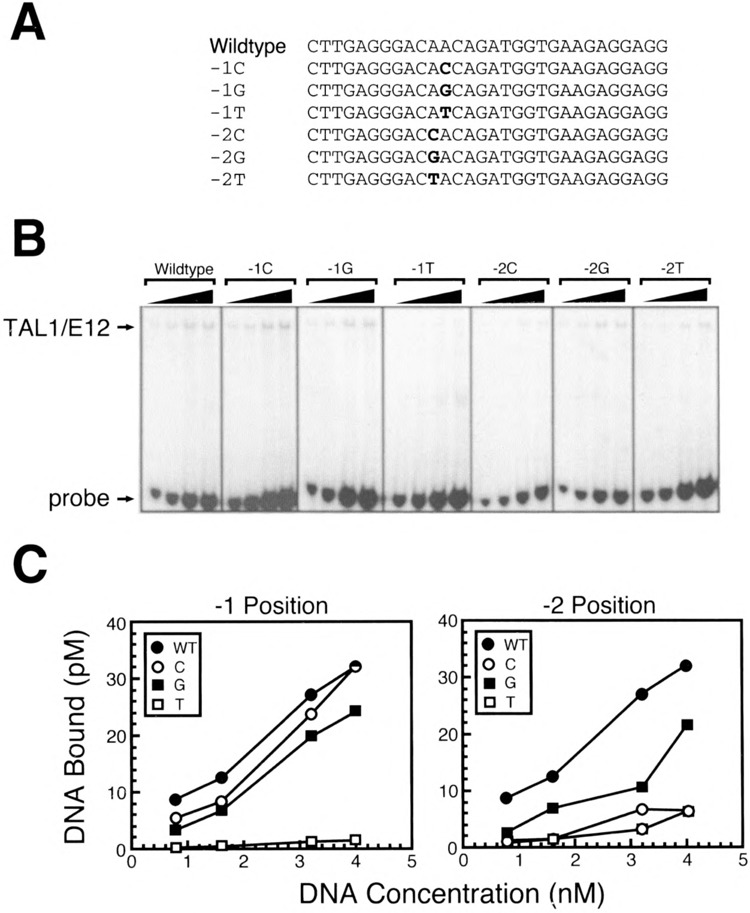
Importance of nucleotides 5′ of the preferred CAGATG E-box for TAL1/E12 binding. (A) Sequences of one strand of each of the double-stranded oligonucleotide probes used is shown in the 5′ to 3′ orientation. (B) EMSA analysis of binding reactions in which increasing amounts of labeled probe were incubated with recombinant TAL1/E12 heterodimers as described in Materials and Methods. The positions of the TAL1/E12 complexes and the probes are indicated. (C) The concentration of the TAL1/E12-DNA complexes (pM) was determined by quantitation with a PhosphorImager and plotted as a function of the total DNA concentration (nM) in the binding reactions so that the relative binding to different probes could be directly compared.
On the 3′ side of the E-box, substitution of the G nucleotide adjacent to the E-box with an A or a C virtually eliminated TAL1/E12 binding (Fig. 7C). Introduction of a T at this position only slightly affected binding (Fig. 7C). The substitution of the T at the +2 position with a C had no effect on binding, whereas an A or a G at this position strongly inhibited binding (Fig. 7C). Thus, certain substitutions of each nucleotide flanking the E-box core can result in a profound loss of binding. Most nucleotide substitutions significantly reduced the amount of the TAL1/ E12-DNA complex formed. These results support a major role for the nucleotides in each of the four positions flanking the E-box in determining high-affinity TAL1 binding.
FIG. 7.
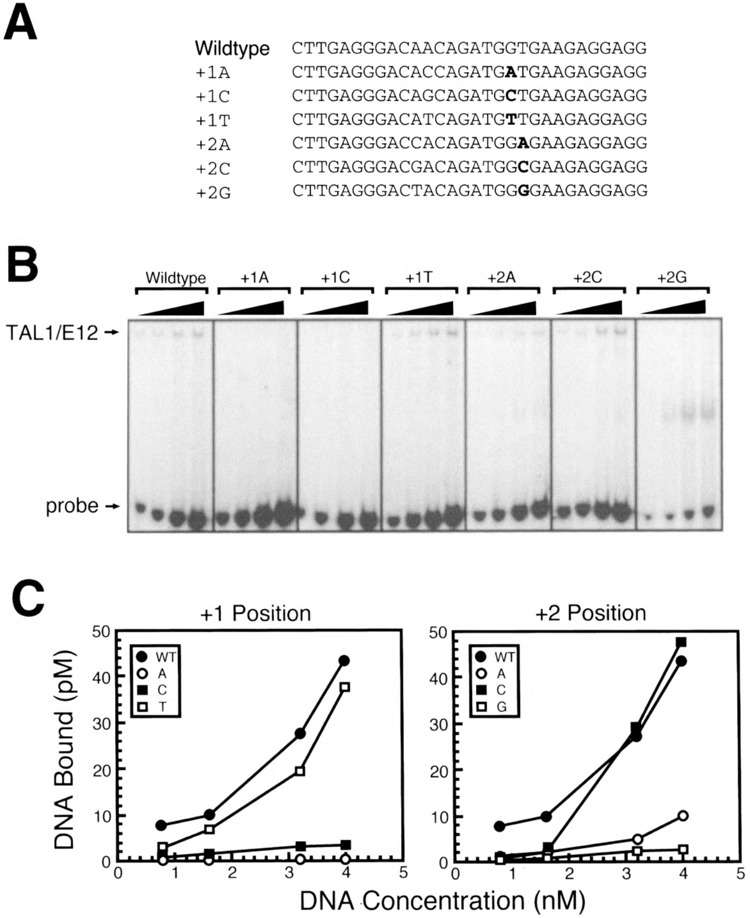
Importance of nucleotides 3′ of the preferred CAGATG E-box for TAL1/E12 binding. (A) Sequences of one strand of each of the double-stranded oligonucleotide probes used is shown in the 5′ to 3′ orientation. (B) EMSA analysis of binding reactions in which increasing amounts of labeled probe were incubated with recombinant TAL1/E12 heterodimers as described in Materials and Methods. The positions of the TAL1/E12 complexes and the probes are indicated. (C) The concentration of the TAL1/E12-DNA complexes (pM) was determined by quantitation with a PhosphorImager and plotted as a function of the total DNA concentration (nM) in the binding reactions so that the relative binding to different probes could be directly compared.
Importance of Nucleotides Flanking the E-Box for TAL1 Binding to Nonpreferred E-Boxes
Previously, we observed binding of TAL1/E12 heterodimers to oligonucleotide probes 170Ebox1 and 21Ebox3, which contained E-boxes differing from the preferred core (Fig. 3). To estimate the affinity of these interactions, we performed quantitative DNA binding analysis as described previously (Figs. 8, 9). However, the interaction between these oligonucleotides and TAL1/E12 heterodimers appeared to be of low affinity, as the binding was not saturated over a wide range of oligonucleotide concentrations. Consequently, we were unable to estimate K d values. The low-affinity nature of these interactions is consistent with the observation that TAL1 heterodimers exhibit a strong preference for the CAGATG E-box (24). Three of the four nucleotides flanking the E-box in probe 170Ebox1 were identical to those in the optimal binding site. However, this probe contained a C in the +2 position rather than a T (Fig. 8A). On the 134Ebox2Mut probe, a T to C substitution at this position only minimally affected binding (Fig. 6C). Therefore, we postulated that the low affinity of TAL1/E12 binding to this probe may simply reflect the E-box core preference.
FIG. 8.
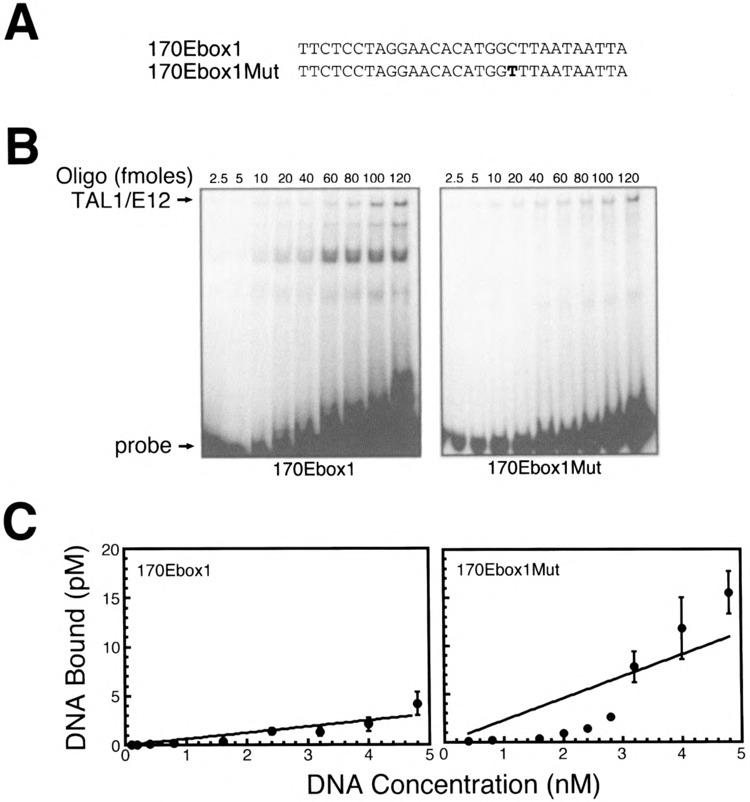
Importance of nucleotides flanking the nonpreferred CACATG E-box for TAL1/E12 binding. (A) Sequences of one strand of the double-stranded 170Ebox1 and 170Ebox1Mut oligonucleotide probes. (B) EMSA analysis of binding reactions in which increasing amounts of the labeled probe were incubated with recombinant TAL1/E12 heterodimers as described in Materials and Methods. The positions of the TAL1/E12 complexes and the probes are indicated. (C) The concentration of the TAL1/E12-DNA complexes (pM) was determined by quantitation with a PhosphorImager and plotted as a function of the total DNA concentration (nM) in the binding reactions (mean ± SE, n = 4).
FIG. 9.
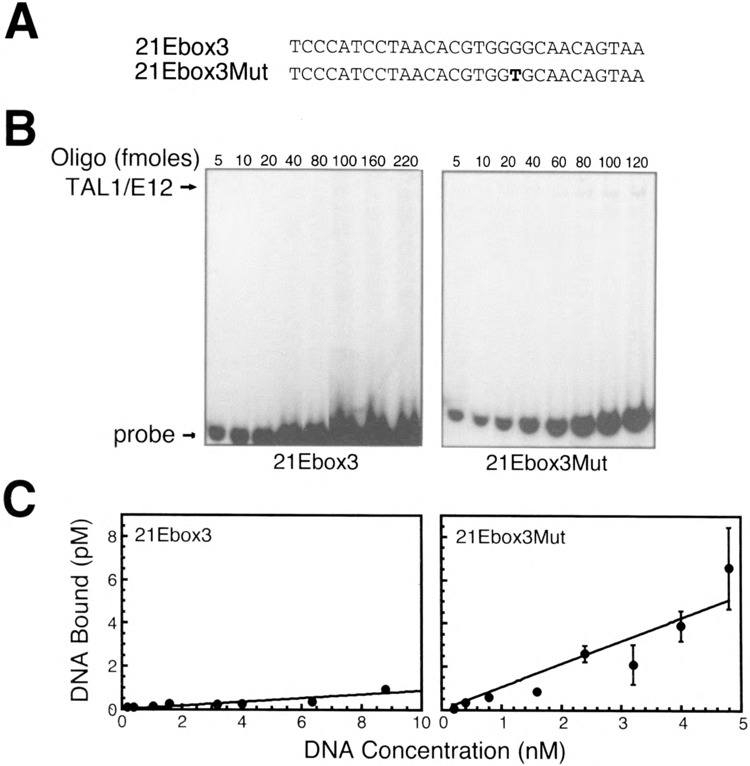
Importance of nucleotides flanking the nonpreferred CACGTG E-box for TAL1/E12 binding. (A) Sequences of one strand of the double-stranded 21Ebox3 and 21Ebox3Mut oligonucleotide probes. (B) EMSA analysis of binding reactions in which increasing amounts of the labeled probe were incubated with recombinant TAL1/E12 heterodimers as described in Materials and Methods. The positions of the TAL1/El2 complexes and the probes are indicated. (C) The concentration of the TAL1/E12-DNA complexes (pM) was determined by quantitation with a PhosphorImager and plotted as a function of the total DNA concentration (nM) in the binding reactions (mean ± SE, n = 4).
To test this hypothesis, we generated an oligonucleotide, 170Ebox lMut, which was identical to 170Ebox1, except that it contained a T at the +2 position (Fig. 8A). TAL1 heterodimers bound to the 170Ebox1Mut probe (Fig. 8B). As expected, this interaction was of low affinity and did not differ significantly from that of 170Ebox1 (Fig. 8C). In the 21Ebox3 oligonucleotide, three of the four flanking nucleotides differed from the optimal sequence (Fig. 9A). Very low levels of binding were observed with the 21Ebox3 probe (Fig. 9B). To examine the effects of flanking sequences on TAL1 heterodimer binding to this E-box, we generated the oligonucleotide probe 21Ebox3Mut. This probe was identical to 21Ebox3, except that it contained optimal flanking sequences (Fig. 9A). These substitutions resulted in a significant increase in the amount of TAL1 heterodimer-DNA complex formed (Fig. 9B, C). However, the presence of optimal flanking sequences surrounding a suboptimal E-box was insufficient to generate a high-affinity interaction. Nonetheless, it is clear that the preference for specific flanking nucleotides exists even in the context of different E-box core sequences.
The analyses described above were performed with in vitro-synthesized TAL1 heterodimers. To determine whether binding of endogenous TAL1 complexes exhibited a specificity similar to recombinant TAL1/E12, EMS As were performed with TAL1 heterodimers from fractionated MEL cell nuclear extract (data not shown). Resource S cation exchange column fractions containing TAL1 heterodimers were identified by EMSA using the TAL1con probe. A TAL1 complex, which was disrupted by addition of TAL1 antisera, eluted from the column at approximately 200 mM NaCl. The mobility of the complex as identical to that observed with the in vitro-generated TAL1/E12 heterodimers, suggesting that the TAL1 heterodimeric partner in this MEL cell complex may be E12. The amount of complex formed with endogenous TAL1 heterodimers was very low, which likely reflects the low abundance of TAL1 in the fractions. However, the sequence specificity of the endogenous TAL1 complex and the recombinant TAL1/E12 heterodimers appeared to be similar.
TAL1 Binding to E-Boxes in Regulatory Regions of Putative TAL1 Target Genes
Certain single base deviations from the preferred flanking sequences result in poor binding of TAL1/ E12. For example, the 134EBox2 probe, which exhibited no TAL1 heterodimer binding, contained nucleotides at three of the four flanking positions (C−2, T−1, G+2), which would be expected to significantly reduce binding. Thus, by extension, we predict that TAL1 heterodimers would interact weakly in vitro with E-boxes in regulatory regions of the putative TAL1 target genes, LMO2, Lck CD34, EpoR, GATA-1, and the β-globin locus. In each case, at least one of the nucleotides flanking the CAGATG E-box would be predicted to significantly inhibit binding. For the β-globin locus control region (LCR), the CCCAGATGTT sequence within hypersensitive site 2 differs from the preferred sequence at three of the four flanking nucleotides. Two of these differences, C−1 and T+1, would not be predicted to strongly reduce binding. However, the presence of a C at the −2 position would be expected to have a major impact on binding. As predicted, TAL1/E12 heterodimers bound weakly to this sequence (Fig. 10, lane 2). The binding of endogenous TAL1 heterodimers in MEL cell nuclear extract to the LCR sequence has been reported previously (16). The CCCAGATGAT sequence from the Lck promoter differed from the β-globin LCR sequence at the +1 (C vs. T) and +2 (A vs. T) flanking positions. This sequence was recognized specifically by TAL1 heterodimers, but with low affinity (Fig. 10, lane 3). Thus, if TAL1 recognizes these sequences in vivo, additional factors may be required to stabilize the interaction.
FIG. 10.
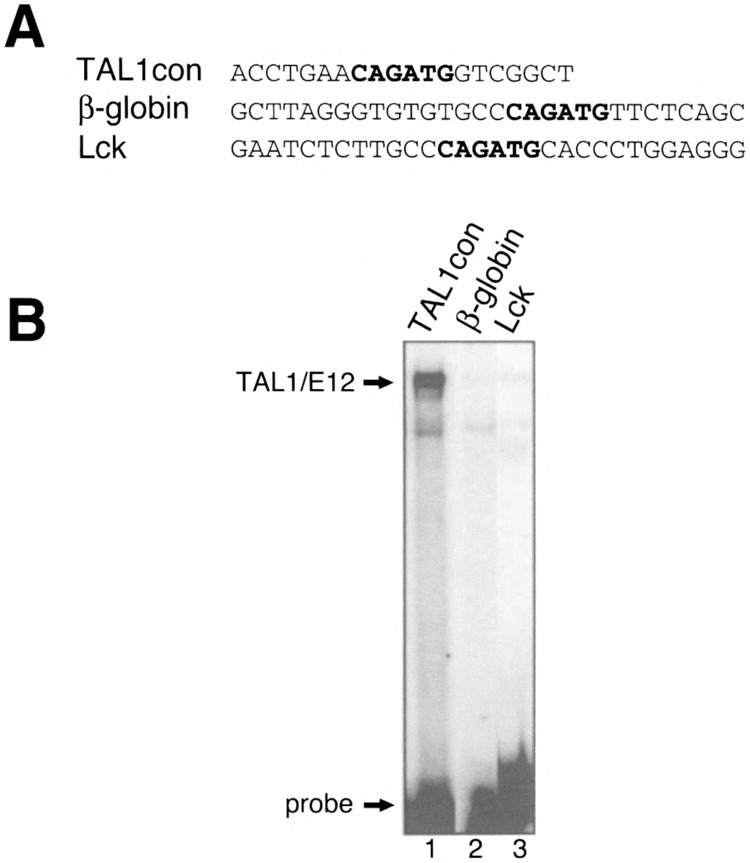
Binding of recombinant TAL1/E12 to E-boxes from potential TAL1 target genes. (A) One strand of each double-stranded oligonucleotide probe is shown in the 5′ to 3′ orientation. (B) TAL1 and E12.were transcribed and translated in vitro and incubated with each of the labeled probes. EMSA analysis was performed as described in Materials and Methods. The positions of the TAL1/E12 complexes and the probes are indicated.
DISCUSSION
Here, we show that the binding of TAL1 heterodimers to a CAGATG E-box is strongly modulated by nucleotides flanking the E-box. Previous studies have assessed the importance of nucleotides flanking an E-box in sequence-specific DNA binding by other bHLH proteins (12,18,32). This phenomenon has been studied for Myc, Max, and USF, three bHLH proteins that bind preferentially to CACGTG elements. Sequence variation flanking this common E-box has been shown to differentially affect the binding of hetero- and homodimers (6,19). For example, the presence of a T at the 5′ side of the E-box significantly inhibited the binding of Myc/Max heterodimers, but not Max or USF homodimers (6,19). These observations suggested that flanking nucleotides play a major role in the ability of these transcription factors to discriminate between CACGTG binding sites. Likewise, nucleotides flanking CAGATG E-boxes may contribute to differential binding of the related proteins TAL1, TAL2 (50), LYL1 (35), and other factors that bind preferentially to this E-box.
Mechanisms of binding site discrimination by structurally related transcription factors, based on variable flanking sequences surrounding a common core recognition motif, appear to be a common theme. For example, the GATA transcription factors, which regulate hematopoiesis and other aspects of development [reviewed in (49)], comprise a group of homologous zinc finger proteins that recognize the core sequence GATA. Subtle variations in both the core motif and flanking sequences confer preferential binding by the individual GATA proteins (28,34). By combining a cell- and tissue-specific expression pattern, distinct sequence preferences for DNA recognition, and potentially distinct coactivators (44), a considerable number of target genes can be regulated by a limited set of related proteins.
The results described herein may have a significant impact on the ability to evaluate potential TAL1 heterodimer binding sites and target genes. We have not found the 10 base pair high-affinity TAL1 binding site, AACAGATGGT, in regulatory regions of any hematopoietic genes. The fact that the E-boxes on regulatory regions of hematopoietic genes indicated above are low-affinity sites may reflect a requirement for additional factors to confer high-affinity TAL1 heterodimer binding. In this regard, if a high-affinity TAL1 site was present, TAL1 heterodimers might bind stably and constitutively. This scenario would be avoided if physiological targets of TAL1 have low-affinity sites, and additional factors are required to generate a stable nucleoprotein complex. Such a layer of regulation may be necessary, because TAL1 heterodimers are relatively resistant to inhibition by Id proteins, which negatively regulate many bHLH transcription factors (25,37).
A requirement for protein-protein interactions to stabilize TAL1 association with a nonoptimal E-box was shown recently (48). In these studies, a large multimeric complex containing TAL1, E47, GATA-1, LMO2, and LDB1/NL1 bound a CAGGTG E-box, which was part of the preferred binding site for the complex. Interestingly, however, LDB1 was shown recently to oppose the stimulatory effect of TAL1 on hematopoiesis (46). It is unclear whether this antihematopoietic activity is related to the modulation of TAL1 DNA binding specificity through formation of a multimeric complex. The identification of additional TAL1-interacting proteins that modulate the affinity and/or specificity of TAL1 DNA binding, accompanied by functional studies to measure the influence of these proteins on transactivation by TAL1, should facilitate our understanding of how TAL1 controls hematopoiesis.
ACKNOWLEDGMENTS
We gratefully acknowledge Dr. R. Baer for providing the E12 and TAL1 expression vectors and Dr. I. Kirsch for the GST-TAL1 expression construct. We acknowledge Fang Dong for technical assistance in purification of the GST-TAL1 fusion protein. We also thank the members of the Bresnick lab for helpful discussions. These studies were supported by a Leukemia Society of America Scholar Award (E.H.B.), a Leukemia Society of America Postdoctoral Fellow Award (K.A.G.), a Shaw Scholar Award from the Milwaukee Foundation (E.H.B.), and a Leukemia Research Foundation Grant (E.H.B.).
REFERENCES
- 1. Aplan P. D.; Begley C. G.; Bertness V.; Nussmeier M.; Ezquerra A.; Coligan J.; Kirsch I. R. The SCL gene is formed from a transcriptionally complex locus. Mol. Cell. Biol. 10:6426–6435; 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aplan P. D.; Nakahara K.; Orkin S. H.; Kirsch I. R. The SCL gene product: A positive regulator of erythroid differentiation. EMBO J. 11:4073–4081; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bain G.; Gruenwald S.; Murre C. E2A and E2–2 are subunits of B-cell-specific E2-box DNA-binding proteins. Mol. Cell. Biol. 13:3522–3529; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bash R. O.; Hall S.; Timmons C. F.; Crist W. M.; Amylon M.; Smith R. G.; Baer R. Does activation of the TAL1 gene occur in the majority of patients with T-cell acute lymphoblastic leukemia? Blood 86:666–676; 1995. [PubMed] [Google Scholar]
- 5. Begley C. G.; Apian P. D.; Denning S. M.; Haynes B. F.; Waldmann T. A.; Kirsch I. R. The gene SCL is regulated during early hematopoiesis and encodes a differentiation-related DNA binding motif. Proc. Natl. Acad. Sci. USA 86:10128–10132; 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bendall A. J.; Molloy P. L. Base preferences for DNA binding by the bHLH-Zip protein USF: Effects of MgCl2 on specificity and comparison with binding of Myc family. Nucleic Acids Res. 22:2801–2810; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bigler J.; Eisenman R. N. Isolation of a thyroid hormone responsive gene by immunoprecipitation of thyroid hormone receptor–DNA complexes. Mol. Cell. Biol. 14:7621–7632; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bigler J.; Eisenman R. N. Novel location and function of a thyroid hormone response element. EMBO J. 14:5710–5723; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bresnick E. H.; Felsenfeld G. Evidence that the transcription factor USF is a component of the human beta-globin locus control region heteromeric protein complex. J. Biol. Chem. 268:18824–18834; 1993. [PubMed] [Google Scholar]
- 10. Brown L.; Cheng J. T.; Chen Q.; Siciliano M. J.; Crist W.; Buchanan G.; Baer R. Site-specific recombination of the tal-1 gene is a common occurrence in T cell leukemia. EMBO J. 9:3343–3351; 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Burn T. C.; Satterthwaite A. B.; Tenen D. G. The human CD34 hematopoietic stem cell promoter and a 3′ enhancer direct hematopoietic expression in tissue culture. Blood 80:3051–3059; 1992. [PubMed] [Google Scholar]
- 12. Catron K. M.; Iler N.; Abate C. Nucleotides flanking a conserved TAAT core dictate the DNA binding specificity of three murine homeodomain proteins. Mol. Cell. Biol. 13:2354–2365; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cheng J. T.; Hsu H. L.; Hwang L. Y.; Baer R. Products of the TAL1 oncogene: Basic helix–loop–helix proteins phosphorylated at serine residues. Oncogene 8:677–683; 1993. [PubMed] [Google Scholar]
- 14. Czernik P. J.; Peterson C. A.; Hurlburt B. K. Preferential binding of MyoD-E12 versus myogenin-E12 to the murine sarcoma virus enhancer in vitro . J. Biol. Chem. 271:9141–9149; 1996. [DOI] [PubMed] [Google Scholar]
- 15. Deveaux S.; Cohen-Kaminsky S.; Shivdasani R. A.; Andrews N. C.; Filipe A.; Kuzniak I.; Orkin S. H.; Romeo P. H.; Mignotte V. p45 NF-E2 regulates expression of thromboxane synthase in megakaryocytes. EMBOJ. 16:5654–5661; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Elnitski L.; Miller W.; Hardison R. Conserved E-boxes function as part of the enhancer in hypersensitive site 2 of the beta-globin locus control region: Role of basic helix–loop–helix proteins. J. Biol. Chem. 272:369–378; 1997. [DOI] [PubMed] [Google Scholar]
- 17. Elwood N. J.; Green A. R.; Melder A.; Begley C. G.; Nicola N. The SCL protein displays cell-specific heterogeneity in size. Leukemia 8:106–114; 1994. [PubMed] [Google Scholar]
- 18. Fisher F.; Goding C. R. Single amino acid substitutions alter helix–loop–helix protein specificity for bases flanking the core CANNTG motif. EMBO J. 11:4103–109; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fisher F.; Crouch D. H.; Jayaraman P. S.; Clark W.; Gillespie D. A. F.; Grading C. R. Transcription activation by Myc and Max: Flanking sequences target activation to a subset of CACGTG motifs in vivo. EMBO J. 12:5075–5082; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Grandori C. J. M.; Siebelt F.; Ayer D. E.; Eisenman R. N. Myc-Max heterodimers activate a DEAD box gene and interact with multiple E-box related sites in vivo. EMBO J. 15:4344–4357; 1996. [PMC free article] [PubMed] [Google Scholar]
- 21. Green A. R.; Salvaris E.; Begley C. G. Erythroid expression of the helix–loop–helix gene scl. Oncogene 6:475–179; 1991. [PubMed] [Google Scholar]
- 22. Hsu H. L.; Cheng J. T.; Chen Q.; Baer R. Enhancer binding activity of the tal-1 oncoprotein in association with the E47/E12 helix–loop–helix proteins. Mol. Cell. Biol. 11:3037–3042; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hsu H. L.; Wadman I.; Baer R. Formation of in vivo complexes between the TAL1 and E2A polypeptides of leukemic T-cells. Proc. Natl. Acad. Sci. USA 91:3181–3185; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hsu H. L.; Huang L.; Tsan J. T.; Funk W.; Wright W. E.; Hu J. S.; Kingston R. E.; Baer R. Preferred sequences for DNA recognition by the TAL1 helix–loop–helix proteins. Mol. Cell. Biol. 14:1256–1265; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hsu H. L.; Wadman I.; Tsan J. T.; Baer R. Positive and negative transcriptional control by the TALI helix–loop–helix protein. Proc. Natl. Acad. Sci. USA 91:5947–5951; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kadesch T. Consequences of heteromeric interactions among helix–loop–helix proteins. Cell Growth Differ. 4:49–55:1993. [PubMed] [Google Scholar]
- 27. Kallianpur A. R.; Jordan J. E.; Brandt S. J. The SCL/TAL-1 gene is expressed in progenitors of both the hematopoietic and vascular systems during embryogenesis. Blood 83:1200–1208; 1994. [PubMed] [Google Scholar]
- 28. Ko L. J.; Engel J. D. DNA-binding specificities of the GATA transcription factor family. Mol. Cell. Biol. 13:4011–1022; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lam L.; Bresnick E. H. Evidence for distinct DNA binding forms of the erythroid-specific transcription factor NF-E2. Biochemistry 34:16347–16357; 1995. [DOI] [PubMed] [Google Scholar]
- 30. Lam L. T.; Bresnick E. H. A novel DNA binding protein HS2NF5 interacts with a functionally important sequence of the human beta-globin locus control region. J. Biol. Chem. 271:32421–32429; 1996. [DOI] [PubMed] [Google Scholar]
- 31. Larson R. C.; Lavenir I.; Larson T. A.; Baer R.; Warren A. J.; Wadman I.; Nottage K.; Rabbitts T. H. Protein dimerization between Lmo2 (Rbtn2) and Tall alters thymocyte development and potentiates T cell tumorigenesis in transgenic mice. EMBO J. 15:1021–1027; 1996. [PMC free article] [PubMed] [Google Scholar]
- 32. Lundin M.; Nehlin J. O.; Ronne H. Importance of a flanking AT-rich region in target site recognition by the GC box-binding zinc finger protein MIG1. Mol. Cell. Biol. 14:1979–1985; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maouche L.; Tournamille C.; Hattab C.; Boffa G.; Cartron J. P.; Chretien S. Cloning of the gene encoding the human erythropoietin receptor. Blood 78:2557–2563; 1991. [PubMed] [Google Scholar]
- 34. Merika M.; Orkin S. H. DNA-binding specificity of GATA family transcription factors. Mol. Cell. Biol. 13:3999–4010; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Miyamoto A.; Cui X.; Naumovski L.; Cleary M. L. Helix–loop–helix proteins LYL1 and E2a form heterodimeric complexes with distinctive DNA-binding properties in hematolymphoid cells. Mol. Cell. Biol. 16:2394–2401; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Murrell A. M.; Bockamp E. O.; Gottgens B.; Chan Y. S.; Cross M. A.; Heyworth C. M.; Green A. R. Discordant regulation of SCL/TAL-1 mRNA and protein during erythroid differentiation. Oncogene 11:131–139; 1995. [PubMed] [Google Scholar]
- 37. Nielsen A. L.; Norby P. L.; Pedersen F. S.; Jorgensen P. E-box sequence and context-dependent TALI/SCL modulation of basic helix–loop–helix protein-mediated transcriptional activation. J. Biol. Chem. 271:31463–31469; 1996. [DOI] [PubMed] [Google Scholar]
- 38. Porcher C.; Swat W.; Rockwell K.; Fujiwara Y.; Alt F. W.; Orkin S. H. The T-cell leukemia oncoprotein SCL/ta1-1 is essential for development of all hematopoietic lineages. Cell 86:47–57; 1996. [DOI] [PubMed] [Google Scholar]
- 39. Pulford K.; Lecointe N.; Leroy-Viard K.; Jones M.; Mathieu-Mahul D.; Mason D. Y. Expression of TAL-1 proteins in human tissues. Blood 85:675–684; 1995. [PubMed] [Google Scholar]
- 40. Robb L.; Lyons I.; Li R.; Hartley L.; Kontgen F.; Harvey R. P.; Metcalf D.; Begley C. G. Absence of yolk sac hematopoiesis from mice with a targeted disruption of the scl gene. Proc. Natl. Acad. Sci. USA 92:7075–7079; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Royer-Pokora B.; Loos U.; Ludwig W. D. TTG-2 a new gene encoding a cysteine-rich protein with a LIM motif, is overexpressed in acute T-cell leukemia with the t(11;14)(p13;q11) translocation. Oncogene 6:1887–1893; 1991. [PubMed] [Google Scholar]
- 42. Shivdasani R. A.; Mayer E. L.; Orkin S. H. Absence of blood formation in mice lacking the T-cell leukemia oncoprotein SCL/tal-1. Nature 373:432–434; 1995. [DOI] [PubMed] [Google Scholar]
- 43. Takadera T.; Leung S.; Gernone A.; Koga Y.; Takihara Y.; Miyamoto N. G.; Mak T. W. Structure of the two promoters of the human lck gene: Differential accumulation of two classes of lck transcripts in T-cells. Mol. Cell. Biol. 9:2173–2180; 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tsang A. P.; Visvader J. E.; Turner C. A.; Fujiwara Y.; Yu C.; Weiss M. J.; Crossley M.; Orkin S. H. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell 90:109–119; 1997. [DOI] [PubMed] [Google Scholar]
- 45. Visvader J.; Begley C. G.; Adams J. M. Differential expression of the LYL, SCL, and E2A helix–loop–helix genes within the hematopoietic system. Oncogene 6:187–194; 1991. [PubMed] [Google Scholar]
- 46. Visvader J. E.; Mao X.; Fujiwara Y.; Hahm K.; Orkin S. H. The LIM-domain binding protein Ldb1 and its partner LMO2 act as negative regulators of erythroid differentiation. Proc. Natl. Acad. Sci. USA 94:13707–13712; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wadman I.; Li J.; Bash R. O.; Forster A.; Osada H.; Rabbitts T. H.; Baer R. Specific in vivo association between the bHLH and LIM proteins implicated in T-cell leukemia. EMBO J. 13:4831–4839; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wadman I. A.; Osada H.; Grutz G. G.; Agulnick A. D.; Westphal H.; Forster A.; Rabbitts T. H. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldbl/NLI proteins. EMBO J. 16:3145–3157; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Weiss M. J.; Orkin S. H. GATA transcription factors: Key regulators of hematopoiesis. Exp. Hematol. 23:99–107; 1995. [PubMed] [Google Scholar]
- 50. Xia Y.; Hwang L.; Cobb M. H.; Baer R. Products of the TAL2 oncogene in leukemic T cells: bHLH phosphoproteins with DNA-binding activity. Oncogene 9:1437–1446; 1994. [PubMed] [Google Scholar]
- 51. Zon L. I.; Orkin S. H. Sequence of the human GATA-1 promoter. Nucleic Acids Res. 20:1812; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]


