Abstract
Glucose-regulated transcription of the L-type pyruvate kinase (L-PK) gene is mediated through its glucose response element (G1RE/L4 box) composed of two degenerated E-boxes. Upstream stimulatory factor (USF) is a component of the transcriptional glucose response complex built up on the G1RE. Cooperation of the G1RE with the contiguous binding site (L3 box) for the orphan nuclear receptor hepatocyte nuclear factor 4 (HNF4) has also been suggested. We compared by transient transfection assays the effects of USF2a and other basic helix-loop-helix leucine zipper (bHLH-LZ) factors (TFE3, c-Myc, SREBP/ADDl) on the activity and glucose responsiveness of a minimal L-PK promoter directed by oligomerized glucose response units (L4L3 boxes). We found that: (i) although USF2a is intrinsically a moderate transcriptional activator, it has a strong stimulatory effect on the activity of the L4L3-based reporter construct in hepatocyte-derived cells and interferes with the glucose responsiveness; (ii) despite its potent ability as a transactivator, TFE3 alone is barely active on the G1RE in hepatocyte-derived cells; (iii) TFE3 as USF2a acts synergistically with HNF4 and abolishes glucose responsiveness of the promoter when overexpressed; (iv) in contrast, overexpression of HNF4 alone stimulates activity of the promoter without interfering with glucose responsiveness; (v) SREBP/ADDl has a very weak activity on the L4L3 elements, only detectable in the presence of HNF4, and c-Myc does not interact with the G1RE of the L-PK promoter. Our studies indicate that different bHLH-LZ transcription factors known to recognize CACGTG-type E-boxes are not equivalent in acting through the L-PK glucose response element, with USF proteins being especially efficient in hepatocyte-derived cells.
Keywords: Upstream stimulatory factor, Basic helix-loop-helix leucine zipper, L-Type pyruvate kinase, Glucose
IT is only recently that the regulatory role of carbohydrates upon hepatic gene transcription has been clearly documented and much about the molecular mechanisms responsible for this transcriptional response to glucose still remains to be investigated (31, 38,40).
Among the genes encoding glycolytic and lipo-genic enzymes whose transcription is controlled by glucose (31,38,40), two have been extensively studied: the L-type pyruvate kinase (L-PK) gene and the spot 14 (S14) gene. We have identified in the promoter region of the L-PK gene a glucose response element termed G1RE (3,12) and Towle’s group has also described the carbohydrate response element (ChoRE) of the spot 14 gene (33,35). The functional role of the G1RE (also termed L4 box) was first established by transient transfections in hepatocytes in primary culture and in hepatocyte-like mhAT3F cells and was further demonstrated by using transgenic mice (11,12,23). This element, closely related to the ChoRE, presents a unique structure of two palin-dromic and degenerated E-boxes separated by 5 base-pairs (3,32), suggesting that transcription factors of the basic helix-loop-helix leucine zipper (bHLH-LZ) family, whose binding sites are E-boxes, can be involved in the transcriptional response to glucose. Indeed, the widely expressed upstream stimulatory factor (USF) protein belonging to the bHLH-LZ factors family is able to interact with the GIRE/ChoRE (12,16,41). Purification of the USF binding activity from HeLa cells has revealed the presence of a heterogeneous complex of two polypeptides (43 and 44 kDa) referred to as USF1 and USF2, respectively (34). USF1 cDNA was first cloned from human and then from Xenopus and sea urchin (15,17,22). Later, murine and human USF2a cDNAs were cloned (26,43). Functional domains mediating transcriptional activation were first mapped in USF1 cDNA (21) and recently we and others have described the original composite activation domains of USF2 (29) and the presence of an auxiliary hexapeptide, required to transmit the action of the different subdo-mains to the transcriptional machinery, and conserved between USF1 and USF2 (14).
We have established by transient transfection experiments in hepatocyte-derived cells that the USF protein is a part of the glucose response complex, indispensable to the transcriptional regulation of the L-PK gene by glucose (23). However, USF proteins are able to activate various promoters that are not regulated by glucose (7,8), and another group has suggested that the presence of USF protein on the G1RE would not be essential to confer a glucose responsiveness in the context of the natural glucose-responsive promoters (18). More recently, by disruption of the USF2 locus by homologous recombination and microinjection of anti-USF2 antibodies in insu-linoma cells, we have presented further evidence that USF2 protein is indeed required for a normal glucose responsiveness of the L-PK and spot 14 genes in vivo as well as ex vivo (19,39). Nevertheless, the formal possibility that different bHLH-LZ factors, also able to interact with the G1RE, may play a role in the transcriptional response of these promoters to glucose deserved further consideration. Several bHLH-LZ transcription factors can be listed: TFE3/TFEB, the immunoglobulin enhancer binding proteins (1,5); the proto-oncogene c-Myc and its binding partner Max (4); and SREBP/ADD1, which regulates multiple genes involved in cholesterol metabolism and which is involved in the adipocyte differentiation (37,44, 46).
In this study, we have compared the transactivating properties of these bHLH-LZ factors on the G1RE and their effects on the glucose responsiveness, investigating a possibility that an interaction of other bHLH-LZ factors with the G1RE may contribute to its activity. We show that Myc and SREBP/ADD1 factors are essentially inactive on the L-PK G1RE and that USF2a, although intrinsically a lower transacti-vator than TFE3, is much more efficient than this factor in the context of the L4L3 elements of the L-PK gene and in hepatocyte-derived cells.
MATERIALS AND METHODS
Plasmid Constructions
All plasmids were constructed by standard cloning procedures. The different L-PK/CAT constructs (-96PK/CAT, -54PK/CAT) and four tandemly repeated USE/MLP (upstream stimulatory element/ade-novirus major late promoter) or L4 fused to −54PK/ CAT have been previously described (3,23). To prepare the (L4L3)4-54PK/CAT or (L4L3)3-96PK/CAT constructs, L4L3 DNA fragments were PCR amplified, using the -283PK/CAT construct as a template and the following oligonucleotides as primers:
5′-gggaggatccAGCATGGGCGCA-3′
5′-ccaacggatccGTACACTGGGGG-3′.
The DNA fragments were digested with BamHI and ligated each other, and then ligated into the BamHl site of the-54PK/CAT or-96PK/CAT constructs. The resulting constructs were checked by DNA sequencing. The expression plasmids for human TFE3 [pSV2A-X3 (1)], mouse c-Myc, and mouse Max [pLTR-c-Myc and pLTR-Max (36)] were kind gifts from T. Kadesch and C. Kahana, respectively. pLTR-c-Myc was digested with Hindlll and EcoRl, and the resulting fragment containing c-Myc cDNA was reinserted into the Hindlll and EcoRl fragment of pCMV (expression vector driven by the cytomegalovirus immediate early promoter/enhancer) vector (Invitro-gene). The Xhol restriction fragment of pLTR-Max was inserted into the Xhol site of pCMV. Human USF2a cDNA and HNF4 cDNA were cloned in our laboratory as previously described (23,42,43). Rat SREBP (SREBP1/ADD1) cDNA fragment was obtained by PCR amplification of the reverse transcribed rat liver mRNA using the following oligonucleotides as primers:
5′-CGGACGACGGAGCCATGGAT-3′
5′-TGAGCCGGCGTCTGAGGGTG-3′.
The resulting cDNA fragment was directly introduced into the TA cloning site of the pCMV vector (Invitrogene). Protein product from this vector corresponds to the activated, proteolytically cleaved SREBP/ADD1, which directly enters the nucleus (44). The construct was checked by DNA sequencing and the protein product was verified by Western blotting using antibodies purchased from Santa Cruz.
Cell Cultures, Transfections, and CAT Assays
The hepatocyte-derived mhAT3F cells (25) were cultured in Ham F12-DMEM (GIBCO) medium supplemented with penicillin, streptomycin, 0.1 μM insulin, 1 μM dexamethasone, 1 μM triiodothyronin, and 5% fetal calf serum (FCS). COS cells were cultured in DMEM medium containing 5% FCS, penicillin, and streptomycin. The CAT reporter construct (5 μg) and various amounts of the expression plas-mids were cotransfected either by calcium phosphate coprecipitation procedure for COS cells or by lipo-fection for mhAT3F cells as described previously (23). In each experiment, total amounts of DNA were adjusted to 10 μg with the empty cytomegalovirus vector or SV40 vector.
Twenty-four hours before the transfection mhAT3F cells were plated and cultured for 16 h. Then the medium was removed and replaced by a serum-free, glucose-free medium containing 0.1 μM insulin, 1 (μM dexamethasone, 1 μM triiodothyronin, 10 μg/ml transferrin, 100 μg/ml albumin, and 10 mM lactate. Eight hours later, transfection was performed. Sixteen hours of culture after the transfection, the medium was replaced either by 10 mM lactate medium or by the medium containing 17 mM glucose and cells were harvested after a culture of 24 h. COS cells were plated 24 h before transfection and the medium was replaced fresh 2 h before transfection. Cells were harvested 36 h after transfection. CAT assays were performed as described (3).
Gel Retardation Assay
The DNA binding reactions were performed at 4°C in binding buffer as described previously (14) in the presence of whole-cell extracts (2 μg) used in CAT assays, 5 μg of poly(dl-dC) and 0.1 ng of end-radiolabeled appropriate double stranded oligonucleotide MLP (5′-AGGTGTAGGCCACGT-GACCGGGTGTTCC) or L4 (5′-ATGGGCGCCAC-GGGGCACTCCCGTGGTTCCTGGT). In competition experiments, proteins were added last, mixed with radiolabeled probe MLP or L4 and increasing amounts of unlabeled double-stranded oligonucleo-tides (2.5, 5,1 10, and 20 ng). DNA binding complexes separated on nondenaturating polyacrylamide gels were quantified by 1using a Phosphorimager (Molecular Dynamics).
RESULTS
Transactivation Efficiency of Different bHLH-LZ Factors on Canonical CACGTG E-Boxes
First, we examined and compared the intrinsic activities of four bHLH-LZ factors (TFE3, proto-oncogene c-Myc and its binding partner Max, SREBP/ ADD1, USF2a) on a reporter plasmid driven by oligomerized canonical CACGTG E-boxes. COS cells were transiently transfected with an expression vector for one of the bHLH-LZ factors tested and the (MLP)4-54PK/CAT reporter, which is composed of the multimerized high-affinity USE/MLP motif in front of the minimal L-PK promoter. Because the expression vectors used are different, directed by promoters/enhancers of different strengths, we established for each of them a dose-response curve (Fig. 1). As expected, the CMV-directed vectors gave maximal efficacies for amount of transfected plasmid 20- to 100-fold lower than the SV40-TFE3 vector. We observed by gel shift assays using whole-cell extracts of transfected COS cells that the binding activities observed with 0.10 ng of the USF2a vector and 10 pg of the TFE3 vector were approximately equal (Fig. 2A; data not shown). As shown in Fig. 1, USF2a is a weak activator of the canonical E-boxes as the maximal level of activation is less than threefold. In contrast, TFE3 activated the reporter construct more than 70-fold, consistent with the previous report of TFF3 as a potent activator of the E-box-based reporters (1,6). With the TFE3 expression vector, the squelching phenomenon was only observed for plasmid amounts higher than 10 ng (not shown). The differences between transactivation efficacies of USF2a and TFE3 did not reflect significant differences in the affinities for MLP motif, as shown by competition experiments using whole-cell extracts (Fig. 2B). Rather, either the transactivation domain of TFE3 is more active than that of USF2a, or TFE3 interferes in COS cells with more active coactivators than USF2a, or both. C-Myc showed only a three- to fourfold maximal activation of the reporter and, even when cotransfected with the Max expression vector, no pronounced activation was observed (data not shown). SREBP/ADD1 is known to bind to and activate several consensus elements such as SRE1 (44), SRE3 (13), and E-boxes (20). In this study, overex-pression of SREBP/ADD1 resulted in a maximal sevenfold activation of the reporter that contains the canonical E-box. We verified that none of the expression vectors modified activation of the minimal -54PK promoter (not shown). These results demonstrate that, in COS cells, TFE3 is intrinsically a much more potent transactivator than USF2a, c-Myc, and SREBP/ADD1.
FIG. 1.
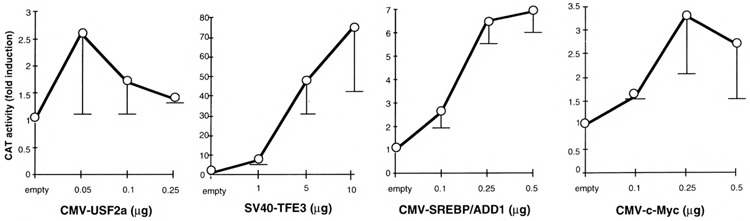
Dose-response action of various bHLH-LZ factors through oligomerized canonical MLP E-boxes. Transfections were performed in COS cells with the (MLP)4-54PK/CAT reporter and the indicated amounts of the expression plasmids for the bHLH-LZ factors. Activities are expressed as fold induction over the activity of the reporter cotransfected with each insertless vector. This activity is the same whatever the dose of each empty vector used (data not shown). In these experiments, 0.1 μg of the CMV promoter-driven expression vectors without cDNA and 1.0 μg of the SV40 promoter-driven vector without cDNA were used as control. Each bar represents the mean ± SD of at least three independent transfections.
FIG. 2.
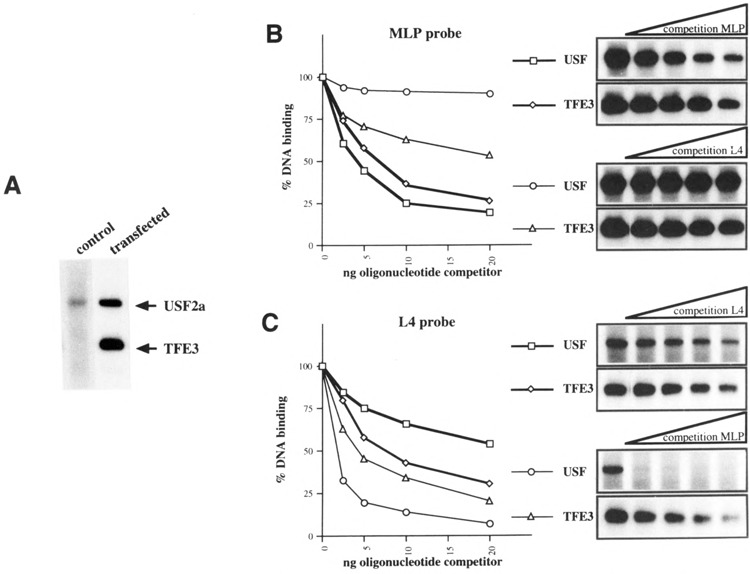
Comparison of the relative affinities of USF2a and TFE3 for MLP and L4 binding sites. (A) Representative EMSA resolving the migrating complexes generated in presence of the radiolabeled MLP oligonucleotide and whole COS cell extracts after transfection with an empty vector (control) or a mix of whole-cell extracts after transfection with the expression vector for USF2a (0.1 μg) or TFE3 (10 μg) (transfected). (B, C) The relative DNA binding affinities of USF2a and TFE3 for MLP and L4 were evaluated by competition in gel shift assays with increasing amounts of unlabeled MPL and L4 oligonucleotides, respectively, as shown on the right. The percentage of residual DNA binding is plotted on the left.
Transactivation Efficacy of Different bHLH-LZ Factors on the L-PK Glucose Response Element L4
The effects of the four bHLH-LZ factors tested were then studied on the G1RE (L4 box). COS cells were cotransfected with one of the expression vectors for USF2a, TFE3, SREBP/ADD1, or c-Myc, and the (L4)4-54PK/CAT reporter (Fig. 3). The amounts of expression vectors transfected were determined from dose-response curves as shown for the (MLP)4-54PK/CAT reporter and correspond to the maximal nonsquelching doses. In contrast with the poor activating effect of USF2a on the (MLP)4-54PK/CAT reporter, overexpression of USF2a resulted in a strong activation, higher than 100-fold, of the (L4)4-54PK/CAT reporter. This result is consistent with a previous report of a strong activation of the same reporter by ectopic USF in mhAT3F hepatoma cells (23). TFE3 also exhibited a strong effect on the same reporter. Competition experiments performed with whole-cell extracts from transfected cells demonstrated that both USF2a and TFE3 display a much lower affinity for L4 than for MLP, the affinity of TFE3 for L4 being nevertheless higher than that of USF2a (Fig. 2B, C). Therefore, the elective transactivation efficiency of USF2a on L4 cannot be explained by an elective binding affinity. On the other hand, stimulation of the reporter activity by SREBP/ADD1 was relatively modest (approximately 10-fold) and, surprisingly, c-Myc was inactive. Therefore, USF2a, a weak transactivator through the canonical MLP E-box is, in the same COS cells, a very strong activator through the L-PK G1RE, becoming even slightly more efficient than the intrinsically strong transactivator TFE3. In comparison, SREBP/ADD1 is weakly active and Myc/Max essentially inactive.
FIG. 3.
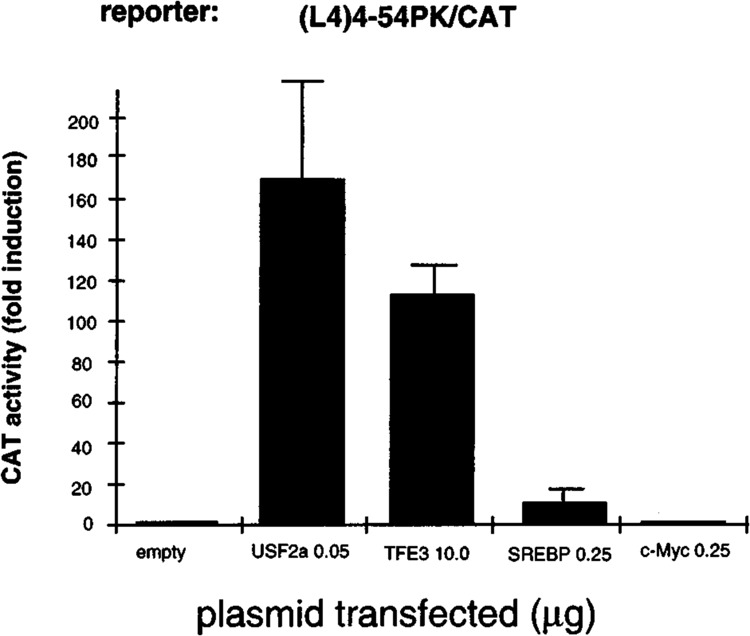
Effects of overexpression of the bHLH-LZ proteins on the G1RE. Transfections were performed in COS cells with the (L4)4-54PK/CAT reporter and the indicated amounts of the expression plasmids for the bHLH-LZ factors. The amount of the transfected expression plasmids was determined from the dose-effect experiments represented in Fig. 1. The amounts of TFE3 and SREBP/ ADD1 expression vectors (10 and 0.25 μg) were optimal. Activities are expressed as fold induction over the activity of the reporter cotransfected with the respective amount of insertless vector. Each bar represents the mean ± SD of at least three independent transfections.
Cooperation Between HNF4 and bHLH-LZ Factors
We have demonstrated that the L4 element of the L-PK promoter is by itself a G1RE when multimer-ized, but acts physiologically in close cooperation with the contiguous element (L3 box) in conferring an effective glucose responsiveness (3,27,28). Thus, we have produced a new reporter construct containing the multimerized L4L3 elements in front of the minimal -54PK/CAT promoter to characterize the effects of the bHLH-LZ factors in cooperation with the orphan nuclear receptor HNF4 (hepatocyte nuclear factor 4) binding to the L3 element. COS cells devoid of liver-enriched transcription factors such as HNF4 or HNF1 (hepatocyte nuclear factor 1) were transiently transfected with expression vectors for USF2a, TFE3, or SREBP/ADD1 and the (L4L3)4-54PK/CAT construct. Figure 4A shows that the maximal level of activation was 25-fold with USF2a and 10-fold with TFE3 expression vectors, whereas the SREBP/ADD1 vector was inactive. The weaker activation observed with the (L4L3)4-54PK/CAT reporter compared with the (L4)4-54PK/CAT plasmid could indicate that cooperative transactivation due to binding of the factors to L4 depends on the spacing of these elements, different in the two constructs. In addition, the L3 element could be occupied by ubiquitous factors of the COUP-TFI/Ear3 and COUP-TFII/Arpl type, which have been shown to behave as transcriptional inhibitors (12). Then, COS cells were transfected with the optimal amount of bHLH-LZ expression vectors, the (L4L3)4-54PK/CAT reporter and the HNF4 expression vector (Fig. 4B). Coexpres-sion of TFE3 and HNF4 resulted in a strong syner-gistic activation of the reporter, as much as 270-fold. Because the synergy was not observed on the minimal -54PK/CAT reporter and coexpression of TFE3 with Arpl (apoAI regulatory protein 1) did not stimulate the activity of the (L4L3)4-54PK/CAT reporter (data not shown), the cooperation appears to be specific to the L4L3 elements and HNF4 dependent. A synergy was also observed between USF2a and HNF4, although less spectacular than for TFE3. SREBP/ADD1, inactive by itself, also seemed able to synergize with HNF4. These results confirm that USF2a is, by itself, an especially effective transactivator through the L4 G1RE. The weaker efficiency of TFE3 can be boosted by binding of HNF4 to the element L3. Figure 2C shows that affinity of both USF2a and TFE3 for the element L4 is weak; however, a binding cooperation could exist with HNF4. Alternatively, the synergy could act at the level of transcriptional activation.
FIG. 4.
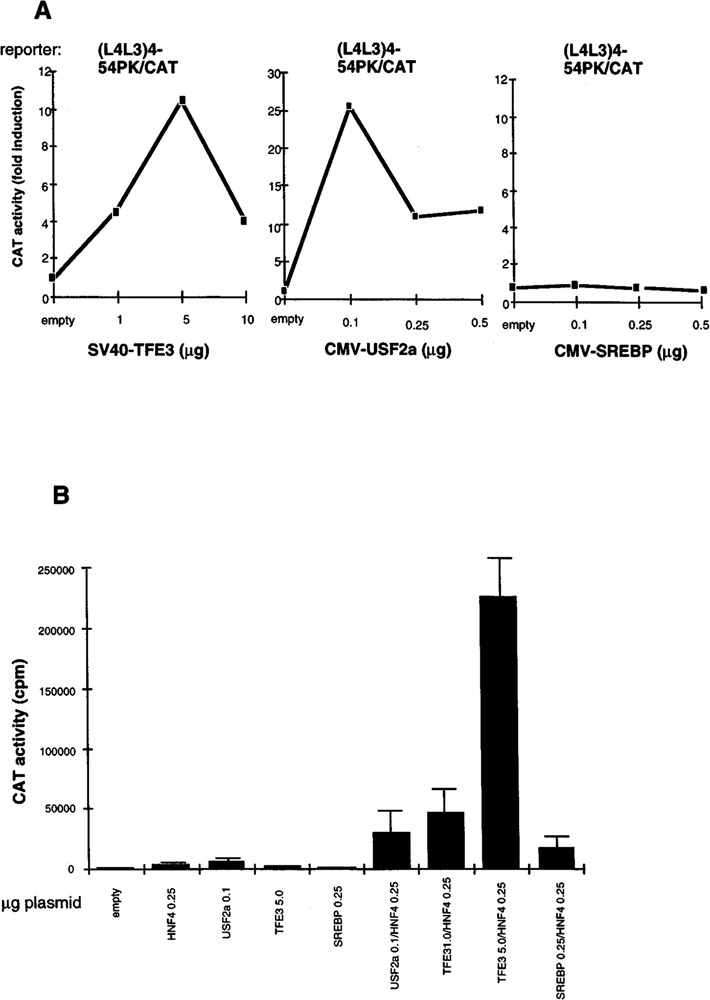
Cooperation of bHLH-LZ proteins and HNF4 on the L4L3 elements. (A) Dose-response effects of TFE3, USF2a, and SREBP ectopic expression on the L4L3-based reporter activity. Five micrograms of the (L4L3)4-54PK/CAT plasmid was cotransfected with increasing amounts of each expression vector in COS cells. As control 0.1 μg of the CMV promoter-driven empty vector and 1.0 μg of the SV40 promoter-driven empty vector were used. The results are the mean of two independent transfections. (B) Synergistic effects of HNF4 and bHLH-LZ proteins on the L4L3-based reporter. Transfections were performed in COS cells with 5 μg of the (L4L3)4-54PK/CAT plasmid and the various amounts of the expression vectors corresponding to the optimal stimulating dose indicated in Fig. 3A. The results show the mean ± SD of at least three independent transfections.
Elective Action of USF2a on the L-PK Glucose Response Unit in Hepatocyte-Derived Cells
We analyzed the transcriptional properties of USF2a, TFE3, and SREBP/ADD1 proteins through the G1RE by transient transfection of mhAT3F cells, a hepatocyte-derived cell line allowing for the glucose responsiveness of the L-PK gene. In the absence of glucose (lactate condition), overexpression of TFE3 had no detectable effect on the activity of the (L4L3)4-54PK/CAT reporter (Fig. 5A). However, co-transfection of the cells with both HNF4 and TFE3 expression vectors, whose optimal transactivating doses have been determined (see Fig. 5B), resulted in a substantial activation of the reporter (ninefold stimulation). This result confirms that TFE3 needs high HNF4 expression to become able to transacti-vate a reporter gene driven by the L-PKL4L3 elements, in COS cells (Fig. 4) as well as in hepatocyte-derived cells (Fig. 5A, B). However, TFE3 was much more active in COS cells than in mhAT3F cells. In contrast, overexpression of USF2a alone in mhAT3F cells cultured under lactate conditions led to a strong stimulation of the reporter, as much as 34-fold, and the coexpression of HNF4 and USF2a resulted in a slight additive activation. As in COS cells, SREBP/ ADD1 was practically inactive, even in the presence of HNF4. HNF4 binding to L3 seemed to slightly increase the glucose responsiveness of the (L4L3)4-54PK/CAT plasmid (induction of threefold without HNF4 and fivefold with HNF4), consistent with a cooperation between the L3 element and the contiguous L4 G1RE (3,27,28). In fact, HNF4 is practically inactive in stimulating the (L4L3)4-54PK/CAT construct in the absence of glucose and active in the presence of glucose (Fig. 5B), consistent with our previous results indicating that the G1RE is a silencer in the absence of glucose, capable of extinguishing the action of contiguous positive cw-acting elements (3,30). However, binding of excess USF2a to the G1RE interferes with the glucose responsiveness, as previously reported (24). Similarly, interaction of TFE3 with L4 in the presence of HNF4 binding to L3 reduced the response to glucose.
FIG. 5.
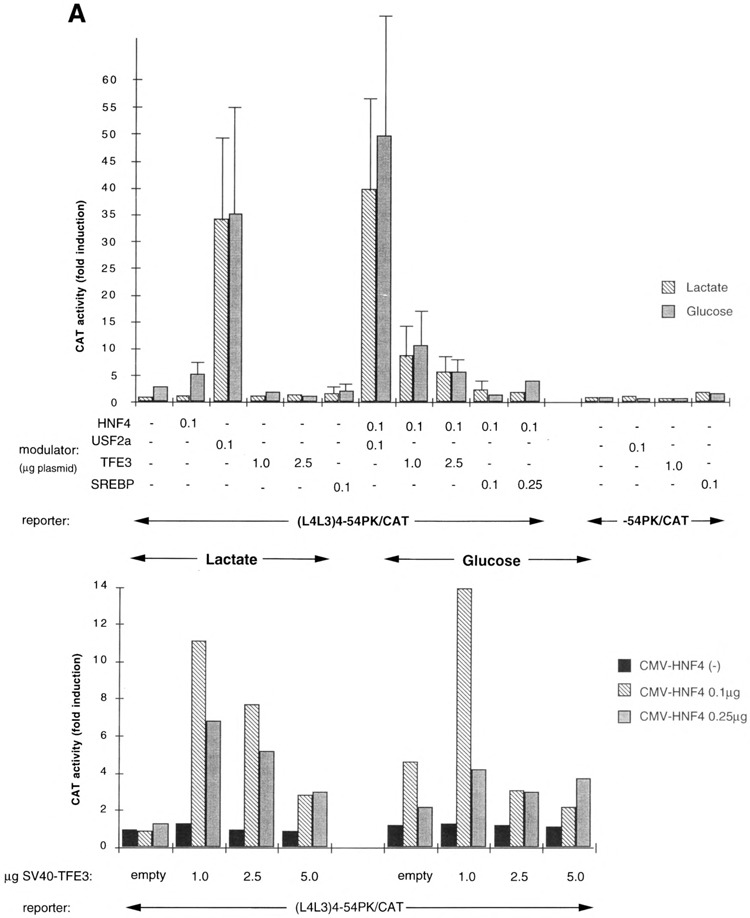
Influence of the bHLH-LZ proteins on the transcriptional response to glucose. (A) Effects of the bHLH-LZ proteins on the transcriptional response to glucose of the (L4L3)4-54PK/CAT reporter construct or the -54PK/CAT plasmid (minimal promoter). Hepatocyte-derived mhAT3F cells were cotransfected with 5 μg of the (L4L3)4-54PK/CAT or -54PK/CAT reporter plasmids and the indicated amounts of the expression vectors encoding the bHLH-LZ proteins. The amounts of the transfected plasmids were determined according to the results of the dose-response experiments. Total amounts of the expression vectors were equalized with appropriate amounts of the empty (containing no cDNA) CMV vector or SV40 vector. Cells were grown as described in Materials and Methods. Each bar represents the mean ± SD of at least four independent transfections. (B) Dose-response effects of the combinatorial ectopic expression of TFE3 and HNF4 on the transcriptional response to glucose of the (L4L3)4-54PK/CAT plasmid. Cotransfection of 5 p.g of the reporter plasmid and the indicated amounts of the expression vectors encoding TFE3 and HNF4 was performed and cells were grown in the conditions already described (21).
DISCUSSION
Our previous studies have characterized and established the role of the glucose response element of the L-PK promoter in the transcriptional regulation by glucose, and also demonstrated that USF protein is a real component of the glucose response complex assembled on the G1RE of the L-PK promoter (19,23,39). However, association with different factors regulating the glucose responsiveness is most likely required for the function of the glucose response machinery, because USF proteins have been described to interact with various promoters that are not regulated by glucose and are not themselves regulated by glucose (7,8).
In fact, we hypothesize that the glucose response complex consists of a transcriptional activator, which seems to be the USF1/USF2 heterodimer, whose transactivating efficiency is regulated by a glucose sensor protein that we are currently looking for. However, several bHLH-LZ factors are known to bind to E-boxes of the CACGTG-type [i.e., similar to E-boxes described in the GIRE/ChoRE of the L-PK and spot 14 genes (16,41)], for instance, Myc and its partners (4), TFE3, TFEB (1,5) and, most recently described, SREBP/ADD1 (46). Therefore, we asked whether USF plays a specific role in the response complex, or whether it could be replaced by any bHLH-LZ factor able to interact with CACGTG-type E-boxes. We chosed to compare in detail the activity on the G1RE of four bHLH-LZ factors: USF2a, TFE3, Myc/Max, and SREBP/ADD1. TFE3 was investigated because it has been described as a very strong transactivator with similar binding specificity to that of USF (6). Myc had a special interest in the context of the glucose responsiveness because the c-myc gene has been reported to be regulated by diet and hormones in the liver (10) and transgenic mice expressing the c-myc oncogene in the liver have been described, characterized by increased glycolysis and, in particular, L-PK gene expression (9). Finally, the SREBP/ADD1 factor has been shown to interact not only with sterol response elements but also with E-boxes, in particular the spot 14 carbohydrate response element (20). In addition, sterol has been shown to be able to regulate expression of the L-PK and spot 14 genes in hepatocytes in primary culture (38).
In fact, a potential role of Myc/Max and SREBP/ ADD1 on the L-PK G1RE function can be ruled out. Indeed, although these factors are relatively weak transactivators through oligomerized canonical MLP E-boxes, they are pratically devoid of any effect on promoters directed by oligomerized L4 or L4L3 elements. Studies on Myc DNA binding specificity have allowed to define the role of the flanking sequences. The presence of a T at -4 position (Y1) of the consensus RY1CACGTGRY2 leads to an inhibitory effect on the binding of c-Myc (2). Indeed, the L4 binding sequence (G1RE = C TCCCGTGGT) containing at the crucial position a T (also present in the ChoRE = CTCACGTGGT) may contribute to the discrimination between USF and c-Myc.
Concerning TFE3 and USF, we confirm here that, acting through a high-affinity binding site (the MLP E-box), TFE3 is a considerably stronger transactiva-tor than USF, as this has been studied by others with the pE3 binding site of the p-cell-specific immuno-globin enhancer (1). However, when MLP was replaced by the L4 G1RE, USF revealed a transactivat-ing efficacy as high as TFE3. These results could not be explained by different binding activities of USF2a and TFE3: both bind with the same affinity to the MLP motif whereas the affinity of TFE3 for L4, although low, is higher than that of USF2a. Instead, our findings point to the essential role of the exact E-box sequence and arrangement in discrimination between different factors acting through similar elements. In addition, TFE3 became still less active by itself in the context of the construct directed by oligo-merized L4L3 elements. We can hypothesize that the low efficiency of TFE3 on L4 is compensated by cooperative interaction on oligomerized (L4)4, whereas this cooperation could be inhibited by the presence of intervened L3 elements in the (L4L3)4 context. In hepatocyte-derived cells, USF2a became a very strong activator and TFE3 alone was totally inactive. Liver-specific coactivators of USF, absent in COS cells, may explain this exquisite specificity of the G1RE for USF2a in hepatocyte-derived cells.
The low efficiency of TFE3 on oligomerized L4L3 elements is compensated in COS cells by coex-pression of HNF4, indicating that some sort of interaction between TFE3 and HNF4 exist, allowing TFE3 to bind the G1RE and strongly activate the reporter gene. However, because hepatocyte-derived mhAT3F cells could lack a coactivator of TFE3, synergy between TFE3 and HNF4 is much weaker in these cells than in COS cells. Because TFE3 is mainly a B-cell factor and HNF4 a liver-specific factor, it is obvious that the synergy observed between these factors is circumstantial and nonphysiologic. In any case, our results point to a specific efficiency of USF proteins on the G1RE in liver cells compared to other bHLH-LZ factors. Indeed, (i) USFs represent the most abundant CACGTG-type E-box binding proteins in liver nuclear extracts (43); (ii) although intrinsically a weak transactivator compared to TFE3, USF exhibits a special specificity to the L-PK G1RE compared to the canonical MLP E-box; (iii) this specificity is still enhanced in liver cells, suggesting that liver-specific USF coactivators could exist.
These results are fully consistent with our proposal that USF proteins are the functional transactivator whose activity on the glucose response element is controlled by an as yet unknown glucose sensor protein. Efficiency of the glucose response complex is enhanced by cooperation with HNF4 binding to the contiguous L3 element. Recently, it has been reported that the element L3 can also bind the ubiquitous NF1 factor, HNF4 and NF1 binding sites being overlapping and their occupancy mutually exclusive. Over-expression of NF1 would have an inhibitory effect (45), whereas we confirm here that HNF4, but not NF1, can be considered as a putative auxiliary factor of the glucose response complex assembled on the G1RE/L4 element. In contrast, binding of excess bHLH-LZ proteins (USF, but also TFE3 in the presence of HNF4) to L4 disturbs the glucose response complex, perhaps by displacing the glucose sensor protein, giving a glucose-independent transcriptional activation.
In conclusion, we have found that USF proteins are not only the most abundant bHLH-LZ factors in the liver, but also are especially effective on the L-PK G1RE and in the context of liver-derived cells. Consequently, the transactivator present in the glucose response complex indeed consists most likely of USF dimers, probably the USFl/USF2a heterodimers (43).
ACKNOWLEDGMENTS
We are grateful to T. Kadesh and C. Kahana for providing TFE3 and c-Myc, Max expression vectors. We thank M. Cognet-Vasseur and B. Viollet for critical reading of the manuscript. This work was supported by the Institut National de la Santé et de la Recherche Medicalé. L. Gourdon is fellow of the Ministè;re de l’Enseignement Supérieur et de la Recherche.
REFERENCES
- 1. Beckmann H.; Su L. K.; Kadesch T. TFE3: A helix-loop-helix protein that activates transcription through the immunoglobulin enhancer muE3 motif. Genes Dev. 4:167–179; 1990. [DOI] [PubMed] [Google Scholar]
- 2. Bendall A. J.; Molloy P. L. Base preferences for DNA binding by the bHLH-Zip protein USF: Effects of MgCl2 on specificity and comparison with binding of Myc family members. Nucleic Acids Res. 22:2801–2810; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bergot M. O.; Diaz-Guerra M. J.; Puzenat N.; Raymondjean M.; Kahn A. Cis-regulation of the L-type pyruvate kinase gene promoter by glucose, insulin and cyclic AMP. Nucleic Acids Res. 20:1871–1877; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blackwood E. M.; Eisenman R. N. Max: A helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science 251:1211–1217; 1991. [DOI] [PubMed] [Google Scholar]
- 5. Carr C. S.; Sharp P. A. A helix-loop-helix protein related to the immunoglobulin E box-binding proteins. Mol. Cell. Biol. 10:4384–1388; 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carter R. S.; Ordentlich P.; Kadesch T. Selective utilization of basic helix-loop-helix-leucine zipper proteins at the immunoglobulin heavy-chain enhancer. Mol. Cell. Biol. 17:18–23; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carthew R. W.; Chodosh L. A.; Sharp P. A. An RNA polymerase II transcription factor binds to an upstream element in the adenovirus major late promoter. Cell 43:439–448; 1985. [DOI] [PubMed] [Google Scholar]
- 8. Carthew R. W.; Chodosh L. A.; Sharp P. A. The major late transcription factor binds to and activates the mouse metallothionein I promoter. Genes Dev. 1:973–980; 1987. [DOI] [PubMed] [Google Scholar]
- 9. Cartier N.; Miquerol L.; Tulliez M.; Lepetit N.; Levrat F.; Grimber G.; Briand P.; Kahn A. Diet-dependent carcinogenesis of pancreatic islets and liver in transgenic mice expressing oncogenes under the control of the L-type pyruvate kinase gene promoter. Oncogene 7:1413–1422; 1992. [PubMed] [Google Scholar]
- 10. Corcos D.; Vaulont S.; Denis N.; Lyonnet S.; Simon M. P.; Kitzis A.; Kahn A.; Kruh J. Expression of c-myc is under dietary control in rat liver. Oncogene Res. 1:193–199; 1987. [PubMed] [Google Scholar]
- 11. Cuif M. H.; Porteu A.; Kahn A.; Vaulont S. Exploration of a liver-specific, glucose/insulin-responsive promoter in transgenic mice. J. Biol. Chem. 268:13769–13772; 1993. [PubMed] [Google Scholar]
- 12. Diaz Guerra M. J.; Bergot M. O.; Martinez A.; Cuif M. H.; Kahn A.; Raymondjean M. Functional characterization of the L-type pyruvate kinase gene glucose response complex. Mol. Cell. Biol. 13:7725–7733; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ericsson J.; Jackson S. M.; Lee B. C.; Edwards P. A. Sterol regulatory element binding protein binds to a cis element in the promoter of the farnesyl diphosphate synthase gene. Proc. Natl. Acad. Sci. USA 93:945–950; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gourdon L.; Lefrançois-Martinez A. M.; Viollet B.; Martinez A.; Kahn A.; Raymondjean M. An auxiliary peptide required for the function of two activation domains in upstream stimulatory factor 2 (USF2) transcription factor. Genes Function 1:87–97; 1997. [DOI] [PubMed] [Google Scholar]
- 15. Gregor P. D.; Sawadogo M.; Roeder M. G. The adenovirus major late transcription factor USF is a member of the helix-loop-helix group of regulatory proteins and binds to DNA as a dimer. Genes Dev. 4:1730–1740; 1990. [DOI] [PubMed] [Google Scholar]
- 16. Harmon J. S.; Mariash C. N. Identification of a carbohydrate response element in rat S14 gene. Mol. Cell. Endocrinol. 123:37–44; 1996. [DOI] [PubMed] [Google Scholar]
- 17. Kaulen H.; Pognonec P.; Gregor P. D.; Roeder R. G. The Xenopus B1 factor is closely related to the mammalian activator USF and is implicated in the developmental regulation of TFIIIA gene expression. Mol. Cell. Biol. 11:412–24; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kaytor E. N.; Shih H.; Towle H. C. Carbohydrate regulation of hepatic gene expression. Evidence against a role for the upstream stimulatory factor. J. Biol. Chem. 272:7525–7531; 1997. [DOI] [PubMed] [Google Scholar]
- 19. Kennedy H. J.; Viollet B.; Rafiq I.; Kahn A.; Rutter G. A. Upstream stimulatory factor-2 (USF2) activity is required for glucose stimulation of L-pyruvate kinase promoter activity in single living islet beta-cells. J. Biol. Chem. 272:20636–20640; 1997. [DOI] [PubMed] [Google Scholar]
- 20. Kim J. B.; Spotts G. D.; Halvorsen Y. D.; Shih H. M.; Ellenberger H.; Towle H. C.; Spiegelman B. M. Dual DNA binding specificity of ADD1/SREBP1 controlled by a single amino acid in the basic helix-loop-helix domain. Mol. Cell. Biol. 15:2582–2588; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kirschbaum B. J.; Pognonec P.; Roeder R. G. Definition of the transcriptional activation domain of recombinant 43-kilodalton USF. Mol. Cell. Biol. 12:5094–5101; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kozlowski M. T.; Gan L.; Venuti J. M.; Sawadogo M.; Klein. W. H. Sea urchin USF: A helix-loop-helix protein active in embryonic ectoderm cells. Dev. Biol. 148:625–4630; 1991. [DOI] [PubMed] [Google Scholar]
- 23. Lefrancois-Martinez A. M.; Diaz-Guerra M. J.; Vallet M. J.; Kahn A.; Antoine B. Glucose-dependent regulation of the L-pyruvate kinase gene in a hepatoma cell line is independent of insulin and cyclic AMP. FASEB J. 8:89–96; 1994. [DOI] [PubMed] [Google Scholar]
- 24. Lefrancois-Martinez A. M.; Martinez A.; Antoine B.; Raymondjean M.; Kahn A. Upstream stimulatory factor proteins are major components of the glucose response complex of the L-type pyruvate kinase gene promoter. J. Biol. Chem. 270:2640–2643; 1995. [DOI] [PubMed] [Google Scholar]
- 25. Levrat F.; Vallet V.; Berbar T.; Miquerol L.; Kahn A.; Antoine B. Influence of the content in transcription factors on the phenotype of mouse hepatocyte-like cell lines (mhAT). Exp. Cell Res. 209:307–316; 1993. [DOI] [PubMed] [Google Scholar]
- 26. Lin Q.; Luo X.; Sawadogo M. Archaic structure of the gene encoding transcription factor USF. J. Biol. Chem. 269:23894–23903; 1994. [PubMed] [Google Scholar]
- 27. Liu Z.; Thompson K. S.; Towle H. C. Carbohydrate regulation of the rat L-type pyruvate kinase gene requires two nuclear factors: LF-A1 and a member of the c-myc family. J. Biol. Chem. 268:12787–12795; 1993. [PubMed] [Google Scholar]
- 28. Liu Z.; Towle H. C. Functional synergism in the carbohydrate-induced activation of liver-type pyruvate kinase gene expression. Biochem. J. 308:105–111; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Luo X.; Sawadogo M. Functional domains of the transcription factor USF2: Atypical nuclear localization signals and context-dependent transcriptional activation domains. Mol. Cell. Biol. 16:1367–1375; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Miquerol L.; Cluzeaud F.; Porteu A.; Alexandre Y.; Vandewalle A.; Kahn A. Tissue specificity of L-pyruvate kinase transgenes results from the combinatorial effect of proximal promoter and distal activator regions. Gene Expr. 5:315–330; 1996. [PMC free article] [PubMed] [Google Scholar]
- 31. Mitanchez D.; Doiron B.; Chen R.; Kahn A. Glucose-stimulated genes and prospects of gene therapy for type I diabetes. Endocr. Rev. 18:520–540; 1997. [DOI] [PubMed] [Google Scholar]
- 32. Shih H. M.; Liu Z.; Towle H. C. Two CACGTG motifs with proper spacing dictate the carbohydrate regulation of hepatic gene transcription. J. Biol. Chem. 270:21991–21997; 1995. [DOI] [PubMed] [Google Scholar]
- 33. Shih H. M.; Towle H. C. Definition of the carbohydrate response element of the rat S14 gene. Evidence for a common factor required for carbohydrate regulation of hepatic genes. J. Biol. Chem. 267:13222–13228; 1992. [PubMed] [Google Scholar]
- 34. Sirito M.; Lin Q.; Maity T.; Sawadogo M. Ubiquitous expression of the 43- and 44-kDa forms of transcription factor USF in mammalian cells. Nucleic Acids Res. 22:427–133; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Thompson K. S.; Towle H. C. Localization of the carbohydrate response element of the rat L-type pyruvate kinase gene. J. Biol. Chem. 266:8679–8682; 1991. [PubMed] [Google Scholar]
- 36. Tobias K. E.; Shor J.; Kahana C. c-Myc and Max transregulate the mouse ornithine decarboxylase promoter through interaction with two downstream CAC-GTG motifs. Oncogene 11:1721–1727; 1995. [PubMed] [Google Scholar]
- 37. Tontonoz P.; Kim J. B.; Graves R. A.; Spiegelman B. M. ADD1: A novel helix-loop-helix transcription factor associated with adipocyte determination and differentiation. Mol. Cell. Biol. 13:4753–759; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Towle H. C. Metabolic regulation of gene transcription in mammals. J. Biol. Chem. 270:23235–23238; 1995. [DOI] [PubMed] [Google Scholar]
- 39. Vallet V.; Henrion A. A.; Bucchini D.; Casado M.; Raymondjean M.; Kahn A.; Vaulont S. Glucose-dependent liver gene expression in upstream stimulatory factor 2 -/- mice. J. Biol. Chem. 272:21944–21949; 1997. [DOI] [PubMed] [Google Scholar]
- 40. Vaulont S.; Kahn A. Transcriptional control of metabolic regulation genes by carbohydrates. FASEB J. 8:28–35; 1994. [DOI] [PubMed] [Google Scholar]
- 41. Vaulont S.; Puzenat N.; Levrat F.; Cognet M.; Kahn A.; Raymondjean M. Proteins binding to the liver-specific pyruvate kinase gene promoter. A unique combination of known factors. J. Mol. Biol. 209:205–219; 1989. [DOI] [PubMed] [Google Scholar]
- 42. Viollet B.; Kahn A.; Raymondjean M. Protein kinase A-dependent phosphorylation modulates DNA-binding activity of hepatocyte nuclear factor 4. Mol. Cell. Biol. 17:4208–4219; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Viollet B.; Lefrancois-Martinez A. M.; Henrion A.; Kahn A.; Raymondjean M.; Martinez A. Immunochemical characterization and transacting properties of upstream stimulatory factor isoforms. J. Biol. Chem. 271:1405–1415; 1996. [DOI] [PubMed] [Google Scholar]
- 44. Wang X.; Sato R.; Brown M. S.; Hua X.; Goldstein J. L. SREBP-1, a membrane-bound ttanscription factor released by sterol-regulated proteolysis. Cell 77:53–62; 1994. [DOI] [PubMed] [Google Scholar]
- 45. Yamada K.; Tanaka T.; Noguchi T. Members of the nuclear factor 1 family and hepatocyte nuclear factor 4 bind to overlapping sequences of the L-II element on the rat pyruvate kinase L gene promoter and regulate its expression. Biochem. J. 324:917–925; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yokoyama C.; Wang X.; Briggs M. R.; Admon M.; Wu J.; Hua X.; Goldstein J. L.; Brown M. S. SREBP-1, a basic-helix-loop-helix-leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell 75:187–197; 1993. [PubMed] [Google Scholar]


