Abstract
We have characterized the function of putative regulatory sequences upon the smooth muscle transcription of the SMGA gene, using promoter deletion analyses. We demonstrate that the SMGA promoter contains four domains: a basal promoter (−1 to −100), a smooth muscle specifier sequence (−100 to −400), a negative regulator (−400 to −1000), and a smooth muscle-specific modulator (−1000 to −2000). The basal or core promoter supports equivalent transcription in both smooth and skeletal muscle cells. Addition of sequences containing a CArG motif juxtaposed to an E-box element stimulates smooth muscle transcription by five- to sixfold compared to skeletal muscle. This smooth muscle-specific segment is maintained for about 200 bp, after which is a segment of DNA that appears to inhibit the transcriptional capacity of the SMGA promoter in smooth muscle cells. Within the boundary between the smooth muscle specifier and negative regulatory sequences (−400 to −500) are three E-box elements. The smooth muscle modulator domain contains two CArG elements and multiple Eboxes. When added to the SMGA promoter it causes an additional three- to fivefold increase in smooth musclespecific transcription over that stimulated by the smooth muscle specifier domain. Thus, our studies show that the appropriate cell-specific transcription of the SMGA gene involves complex interactions directed by multiple cis-acting elements. Moreover, our characterization of a cell culture system employing embryonic gizzard smooth muscle cells lays the foundation for further molecular analyses of factors that regulate or control SMGA and other smooth muscle genes during differentiation.
Keywords: Developmental gene regulation, Tissue-specific expression, Smooth muscle differentiation, Visceral smooth muscle, Transcriptional regulation, Cell culture
ACTIN represents a family of highly conserved proteins that are essential components of cellular organization and mobility. Six major isoforms of actin have been identified in vertebrates (64,65): classified as nonmuscle (β-cytoplasmic and γ-cytoplasmic), striated muscle (α-skeletal and α-cardiac), or smooth muscle (α-vascular and γ-enteric) isoforms. The individual actin isoforms are encoded by single, distinct genes, each of which demonstrates a discrete pattern of expression and tissue specificity throughout development (20,21,41,64–67). Thus, the actin family presents an excellent system for understanding mechanisms of differential gene expression. A key step in the developmental regulation of actin gene expression is the control of transcriptional initiation. The molecular mechanisms involved in skeletal and cardiac α-actin gene regulation have been the subject of extensive study (4,12,18,33,42,43,46,48,53). These analyses have revealed that the tissue-specific regulation of the skeletal and cardiac actins requires the interactions of trans-acting factors with DNA signal sequences found in cis with the actin coding regions.
The α-vascular smooth muscle actin gene contains a region of DNA adjacent to the gene that has multiple cis elements which confer either positive or negative transcriptional activity depending upon the cellular context (3,16). Many of the cis elements for the α-vascular smooth muscle gene have been identified, with Mhox, MCAT, TGF-β, and CArG/SRE elements playing a central role in its regulation (3,9,16,60). These studies have revealed that cis elements having the CArG/SRE [CC(A/T)6GG] sequence motif are vital for appropriate α-vascular actin gene transcription (3,9,16,60). This motif is found within the promoters of all vertebrate actin genes examined to date and has been shown to be a critical DNA element in the regulation of many muscle-specific genes, including the striated actins (33,43,46), skeletal and smooth myosin heavy chains (27,63), skeletal and cardiac myosin light chains (15,51), and the smooth muscle 22α gene (29,34,35,45). Although the mechanisms involved in the control of gene transcription in vascular smooth muscle cells have begun to be elucidated, the regulatory components involved in the smooth muscle α-actin (SMGA) gene transcriptional regulation in visceral or vascular cells are unknown. In humans (44) and mice (61), the sequence adjacent to the SMGA gene contains multiple CArG/SRE motifs. Although sequences 5′ to the SMGA gene and perhaps within the first intron are apparently needed for smooth muscle expression (54), the ability of specific cis element motifs to function in transcriptional activation of this gene has not been extensively investigated.
To study the regulation of visceral smooth muscle myogenesis, we have analyzed SMGA expression. In chicken, SMGA expression was found to be restricted to smooth muscle tissues (30). This highly tissue-restricted pattern of expression is consistent across species (41,44) and demonstrates its utility as a specific marker for smooth muscle differentiation. Further, this tissue-specific expression arises from the developmental regulation of the gene (30,41), indicating that the activation of SMGA expression is dependent upon factors unique to smooth muscle cells. In this study, we characterize the avian SMGA gene structure, and we devise a method to examine the function of specific cis elements of the SMGA promoter. DNA sequence analyses of the chicken SMGA gene revealed a high conservation of sequence not only within the gene coding region but also within the putative promoter DNA elements flanking the 5′ region of the gene. The results of gene transfer experiments presented here demonstrate that multiple exacting elements are required for the appropriate transcription of this gene and allow identification of four regions of the promoter that we refer to as core or basal promoter, smooth muscle specifier, negative regulator, and smooth muscle modulator DNA segments. Further, we demonstrate the ability to obtain embryonic gizzard visceral smooth muscle cells from embryos before they express overt smooth muscle phenotypic characteristics and induce them to undergo differentiation in vitro. As a result, we are able to begin to assess which factors regulate smooth muscle-specific gene expression during development.
MATERIALS AND METHODS
Isolation and DNA Sequencing of Chicken SMGA Genomic Clones
A chicken genomic library was constructed in EMBL-3 phage and was screened for SMGA clones using [α-32P]dCTP-labeled full-length cDNA, SMGA15-1, as a probe (30). Among the multiple potential positive clones two retained hybridization under conditions of high stringency and were purified to homogeneity. Restriction and hybridization analyses of both clones localized transcribed sequences within the genomic clones, named SMGA 6-1Z and SMGA 12-1Z, and revealed that the 3′ end of the gene was missing in the two clones. A DNA fragment spanning the missing genomic sequence was obtained by PCR amplification using purified high molecular weight chicken genomic DNA and oligonucleotide primers designed from sequences of the full-length cDNA. The 5′ PCR primer was constructed to a coding sequence in exon 7 of the gene (+1801 to +1822, 5′-GTGCGCGACATCAAGGAGAAG-3′), and the 3′ primer was constructed to include the reverse complement of sequences in the 3′ nontranslated region of the cDNA (5′-GGGAATTCCTGGAGAAAAGG-CTTTA-3′). Transcribed sequences contained within a ∼4.5-kb EcoRI-Sal I fragment derived from the phage insert of clone SMGA 6-1Z were subcloned into the pBluescript KS+ vector (Stratagene, La Jolla, CA). The 3′ genomic PCR product was initially subcloned using the TA Cloning System (Invitrogen, San Diego, CA), and was then ligated into the EcoRI site of pBluescript KS+ for ease of sequencing.
All sequencing reactions were performed by the Sanger-dideoxy chain termination method using the Sequenase Version 2.0 sequencing kit (U.S. Biochemicals, Cleveland, OH). DNA sequence data were determined from single-stranded templates derived from the subcloning of specific restriction fragments and the use of Exonuclease III to construct unidirectional deletions (40,68). DNA sequence for the entire chicken SMGA gene and 5′ flanking DNA was determined from multiple analyses of both DNA strands. Data from the multiple sequencing analyses were collated and analyzed using the University of Wisconsin Genetics Computer Group (GCG) package of programs. The composite sequence determined in this study has been deposited in the GenBank database and has been assigned the accession number AF012348.
Primer Extension and RNase Protection Analyses
Primer extension analysis was performed as described by Zimmer et al. (68). An 18-mer oligonucleotide probe that contained the reverse complement of nucleotides +770 to +787 of the SMGA genomic sequence (5′-GGTCTCCTCCTCGCACAT-3′) was 5′ end labeled with [α-32P]ATP (3000 Ci/mmol, New England Nuclear, Boston, MA) and T4 polynucleotide kinase. Ten micrograms of embryonic day 18 chicken gizzard total RNA isolated as described previously (30) and 2 × 106 cpm of labeled probe were hybridized at 60°C, followed by precipitation of the annealed products. Primer extension reactions were then performed at 45°C using the avian myeloblastosis virus (AMV) reverse transcriptase under conditions outlined by the supplier (Promega, Madison, WI).
RNase protection assays were performed as outlined previously (68). A genomic fragment spanning the putative transcriptional initiation site was generated by PCR using a primer adjacent to the single BamHI restriction site in the 5′ flanking DNA (nucleotides −98 to −80; 5′-CCATCACTTAGCCTATT-TAG-3′) and a primer containing the reverse complement of exon 1, which introduced a synthetic Xba I restriction site at nucleotide position +25. Following PCR, the fragment was digested with BamHI and Xba I, and the resultant 86 bp of DNA was cloned into like-digested pBluescript KS+. This clone, named pExl-2KS, was used to generate [32P]UTP radiolabeled sense and antisense probes by using T3 RNA polymerase and Xba I linearized pExl-2KS (sense) and T7 RNA polymerase in the presence of BamRI linearized pExl-2KS DNA (antisense). The 32P-labeled probes were purified on 5% acrylamide gels and coprecipitated with 10 μg of embryonic day 18 gizzard total RNA or 10 μg of yeast tRNA. The pellets were collected and RNase protection analyses were performed using the RPA II kit (Ambion, Austin, TX).
Both primer extension and RNase protected products were electrophoresed on 10% and 6% denaturing polyacrylamide gels and the results were visualized by autoradiography using Kodak AR5 or Biomax BMR1 X-ray film. A sequencing product of M13mp 18 single-strand DNA was included on the gels as a molecular size marker.
Reporter Gene Constructs
To facilitate the subcloning of truncated 5′ genomic sequences into the polylinker region of the pCAT-Basic vector (Promega), a unique Xba I restriction site at position +25 was introduced into exon 1 of the SMGA gene by PCR. The pExl-2KS clone containing the 86-bp genomic BamHI-synthetic Xba I fragment described above was the initial fragment generated for these experiments. This 86-bp fragment was extended upstream to position −2294 by the ligation of a 2.2-kb EcoRI-BamHI DNA fragment derived from clone 6-1Z (Fig. 1) containing sequences 5′ to the gene to yield the longest 2.3-kb EcoRI-Xba I promoter fragment (Fig. 6). To obtain other deletions in the SMGA promoter, unique restriction sites in the upstream 2.3-kb EcoRI-Xba I sequences were utilized: Nde I (−112), Sma I (−407), Xho I (−513), Apa I (−623), and Nar I (−1087). Following digestion with these enzymes, overhangs were filled in to create blunt ends using Klenow DNA polymerase (40), the DNA subsequently digested with Xba I, and the appropriate DNA fragment was isolated from agarose gels. The purified fragments were then ligated into the pCAT-Basic vector. The vector was prepared by digestion with Sal I, filled in with Klenow polymerase, and then digested with Xba I. Promoter fragments −236 and −296 were generated by PCR using oligonucleotides 5′-GTTGCCTCCTAAGCATAG-CCC-3′ (nts −236 to −213) and 5′-CTTGTGTCTCG-CCTGTTTATCG-3′ (nts −296 to −275) as forward primers in combination with the exon 1 synthetic Xba I primer. Constructs containing the sequences 5′ to the gene in addition to the first intervening sequence of the gene were derived by PCR, which placed a unique Xba I restriction site within exon 2 of the gene at nucleotide position +762. All constructs were verified by restriction mapping and DNA sequencing analyses prior to use in transfection experiments.
FIG. 1.
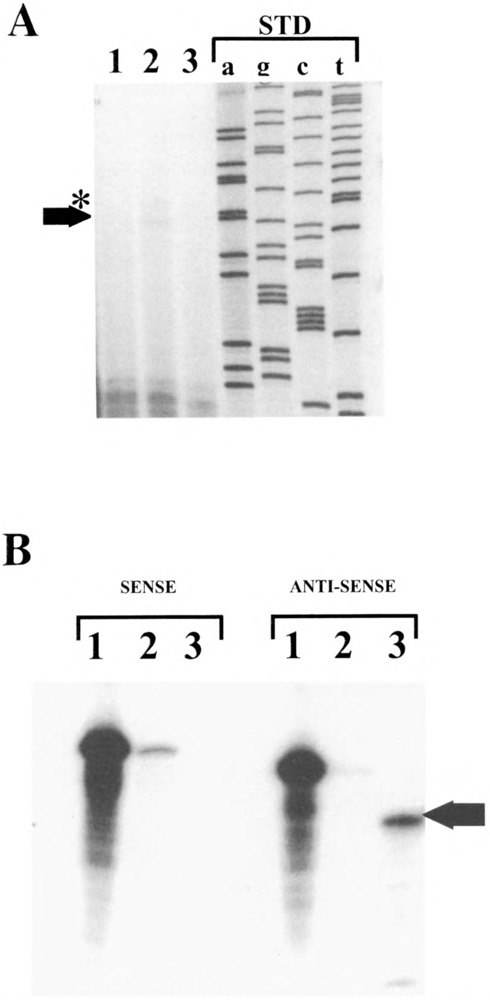
Localization of the SMGA transcriptional initiation site. The 5′ boundary of exon 1 was located using primer extension (A) and RNase protection analyses (B). (A) A primer made to complement sequences of exon 2 beginning at the ATG translation codon (nucleotides 770 to 787, Fig. 2) was labeled with 32P and annealed with 10 μg of yeast tRNA (lane 3) or 10 μg of total RNA isolated from embryonic day 18 gizzard tissue (lane 2). Primer extension was then performed as detailed in Materials and Methods. Lane 1 shows the results of primer extension reactions performed with no added RNA. The sequence ladder (shown a, g, c, t) was derived from single-stranded M13mpl8 and run on the same gel as a molecular weight standard. The arrow shows the position of the major extension product (∼92 bases) with the asterisk showing a minor product (∼95 bases). (B) Uniformly labeled sense and antisense RNA probes to the region spanning exon 1 (+25 to −68, Fig. 1) were annealed to 10 μg yeast tRNA (lanes 2) or 10 μg embryonic gizzard total RNA (lane 3) and the annealed products subjected to RNase protection analyses as outlined in Materials and Methods. Lanes 1 show the position of the probes (sense or anti-sense) that received no RNase treatment. The arrow denotes the position (∼27 bases) of the protected fragment corresponding to exon 1, which is only observed with the antisense probe in gizzard RNAs.
FIG. 6.
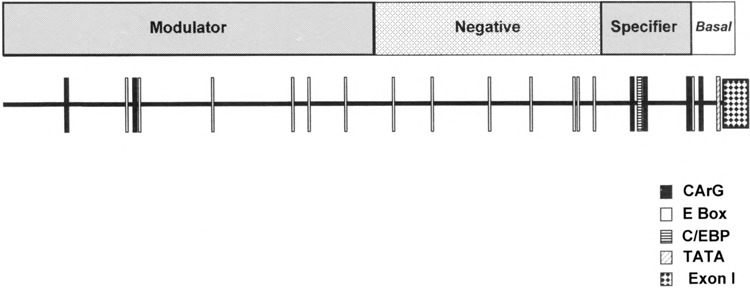
Summary of DNA transfection experiments. This is a model of the SMGA promoter activities in smooth muscle cells as determined by promoter deletions. The placement of potential, muscle-specific DNA elements is shown by the boxes along the ∼2.3 kb of DNA 5′ to the chicken SMGA gene. The promoter is divided into four segments or domains, with the boundaries shown above the promoter segment. The sequences extending to −100 bp 5′ to the gene confer basal promoter activity, exhibiting similar transcriptional capacity in smooth and skeletal muscle cells. The DNA segment between −100 and −400 has been termed a smooth muscle specifier because this DNA shows enhanced transcriptional capability in visceral smooth muscle compared to skeletal muscle myoblasts. The segment from −400 to −1087 imparts a diminished transcriptional activity and is referred to as a negative regulator. Finally, the sequences between −1087 and −2294 show enhanced smooth muscle transcriptional activation in visceral smooth muscle cells. We therefore have denoted this region as the smooth muscle modulator activity, as discussed in the text.
Primary Muscle Cell Cultures
Muscular tissue from Hamburger and Hamilton (23) stage 33–35 (day 8) embryonic gizzards were minced, transferred to medium 199 (Fisher Scientific, Norcross, GA), and dissociated into single cells by sequential incubation in 0.5% collagenase D (Boehringer Mannheim, Indianapolis, IN) at 37°C for 1 h followed by 0.125% trypsin (Gibco BRL, Gaithersburg, MD) treatment at 37°C for 15 min. Cells were collected by centrifugation and resuspended in medium 199 supplemented with 20% fetal bovine serum (Hyclone, Logan, UT), 10−6 M insulin, 2 mM L-glutamine, and 100 units/ml penicillin/ streptomycin. Cells were preplated in 75-cm2 tissue culture flasks by incubation for 20 min at 37°C, 5% CO2. Following initial preplating, the smooth muscle cells remaining in suspension were collected and plated onto collagen type IV-coated (Sigma, St. Louis, MO) 25-cm2 tissue culture flasks at a density of 2.5 × 104 cells/cm2. At a subconfluent stage of ∼80–85%, cells were collected by trypsinization and seeded onto collagen type IV-coated 100-mm tissue culture dishes at a density of 2 × 104 cells/cm2. After reaching 95–100% confluence, cells were induced to differentiate by switching the culture medium to DMEM/F12 supplemented with 2 mM l-glutamine, 10−6 units insulin, 100 units/ml penicillin/streptomycin, 5 μl/ml apo-transferrin, and 0.2 mM l-ascorbic acid. Cultures were maintained in this medium for 48 h prior to experimentation.
Primary skeletal myoblast and chicken embryonic fibroblast cultures were established based on the methodology of Hayward and Schwartz (24). To keep the skeletal myoblast cells in a replicative state, the medium was changed after 24 h to growth media supplemented with 10−5 M 5′-bromodeoxyuridine (BrdU). BrdU is a thymidine analogue and has been shown to block myogenic differentiation in primary embryonic skeletal muscle cultures (33,59). Cultures were used for transfections when the cell population reached ∼60% confluency.
DNA Transfections
SMGA 5′ deletion mutants and the control reporter plasmids were prepared using Qiagen columns (Chatsworth, CA) following an alkaline lysis procedure. The transient transfection of DNA into cultured cells was accomplished using DEAE-dextran in a protocol essentially as developed by Al-Molish and Dubes (1). Our procedure involved a limited pretreatment of cells with DEAE-dextran (1 mg/ml) for 9 min in order to minimize toxicity observed in primary cultures. The cells were rinsed with PBS, and the appropriate DNAs prepared in PBS were then added to the dishes and allowed to incubate at 37°C, 5% CO2 for 30 min. Following this incubation, 5–7 ml of differentiation media was added, and the cells were incubated at 37°C for 48 h.
Primary visceral smooth and skeletal muscle cells were cotransfected with 6 μg of CAT (chloramphenicol acetyl transferase) reporter plasmid construct, and 2 μg of a plasmid containing the β-galactosidase gene under the control of the cytomegalovirus promoter (kind gift of Dr. S. Kayes, University of South Alabama, College of Medicine, Mobile, AL) to normalize for transfection efficiency. Each experiment also included transfections of a CAT reporter plasmid under the control of the Rous sarcoma virus promoter (pCAT-Control vector, Promega), and a “promoter lacking” pCAT-Basic vector (Promega) to act as positive and negative controls, respectively. To ensure experimental reproducibility, all promoter constructs were evaluated in a minimum of three separate experiments, two culture dishes/plasmid construct in each experiment and a minimum of three separate plasmid preparations for each deletion mutant.
The level of CAT activity was determined using the Quant-T-CAT assay system developed by Amersham Corp. (Arlington Heights, IL). This system makes use of the biotin/streptavidin interaction to provide an efficient recovery method for the 3H-acetylated, biotinylated chloramphenicol produced by the CAT enzyme in cell extracts through binding to streptavidin-coated polystyrene beads. The radioactivity contained in the streptavidin-coated beads, indicative of the level of CAT activity for each sample, was counted in Ready Safe scintillation cocktail from Beckman Instruments (Palo Alto, CA). β-Galactosidase activity in the cell extracts was determined by the chemiluminescent Galacto-Light Plus Reporter Gene Assay system (Tropix, Inc., Bedford, MA) and measured in a Turner Model 20 Luminometer. Both β-galactosidase and CAT activity measurements were performed in duplicate for each sample. Normalized CAT activity (mean ± SEM) was expressed as a relative percentage of the maximal activity detected from both the pCAT-Control vector and the smooth muscle transfected (−2294 bp) promoter construct. An independent samples t-test was used to statistically analyze the generated CAT activity data. In instances where the variance was not equal between the populations tested, the Smith-Satterthwaite’s t-test was applied. The difference between populations was considered statistically significant when p < 0.05.
Immunofluorescence Microscopy
Murine monoclonal antibodies specific for SMGA (cat. No. 69-133, ICN Biochemical, Costa Mesa, CA) (37,38) and smooth muscle myosin (catt No. M-7786, Sigma) (17,32) were used in dilutions of 1:300 and 1:600, respectively. A general actin antibody (cat. No. A-2668, Sigma) developed in rabbit was used at a dilution of 1:40. Immunofluorescence microscopy was performed essentially as described by Balczon and West (2). Smooth muscle cells were grown on coverslips coated with type IV collagen, and the coverslips were rinsed in phosphate-buffered saline (PBS: 0.15 M NaCl, 10 mM Na2HPO4, 3 mM NaN3, pH 7.4) and fixed in −20°C MeOH for 6–8 min. The coverslips were rinsed for 5 min in PBS containing 0.1% Triton X-100, and then incubated in a humidified chamber at room temperature for 45 min with the appropriate primary antibody. Following rinsing in PBS, FITC-labeled antispecies IgG (1:20 dilution in PBS) was added to the coverslips and allowed to incubate at room temperature for 45 min. The cells were then rinsed and mounted in 50% glycerol/50% PBS (v/v) that also contained 25 μg/ml Hoechst 33258 to visualize cell nuclei.
Immunofluorescent staining was evaluated using a Zeiss 35 M Axiovert microscope equipped for epifluorescence microscopy. Cells were photographed using T-MAX 400 film (Kodak, Inc., Rochester, NY) and the film was developed in T-MAX developer.
RESULTS
Localization of the Chicken SMGA Transcription Initiation Site
The SMGA gene and 5′ flanking sequences were determined and the nucleotide sequence obtained for the SMGA gene has been deposited in the Genbank database (Accession number AF012348). The intron/ exon boundaries and the 94 nucleotides of 3′ non-translated region of the gene were identified by direct comparison to the chicken SMGA cDNA sequence (30). Furthermore, the intron/exon boundaries of the chicken gene were identical in placement to that of the human and mouse SMGA genes (44,61) and conform to consensus sequences (5). Although the deduced amino acid sequence of the gene was in complete agreement with that derived from the cDNA, minor variations between the nucleotide sequences of the chicken SMGA gene and SMGA cDNA were discernable in the form of silent nucleotide substitutions in the wobble position of codons encoding amino acid #13 (AAT), amino acid #57 (GAC), and amino acid #145 (GCG).
Primer extension and RNase protection analyses were performed to determine the chicken SMGA gene transcription initiation site. A primer extension product of 92 bases was obtained when a 5′ end labeled 18-base oligonucleotide primer complementary to the mRNA sequence coding for the N-terminal 6 amino acids (see Materials and Methods) was hybridized to chicken embryonic day 18 gizzard total RNA and extended using reverse transcriptase (Fig. 1A, lane 2). A faint band of 95 bases in size was observed above the major primer extension product, perhaps representing a minor secondary population of transcripts initiating 3 bases upstream from the primary start site. No extension product was obtained when yeast tRNA was used as a primer template (Fig. 1A, lane 3).
RNase protection analysis confirmed the location of the SMGA gene transcriptional start site. Uniformly labeled sense and antisense RNA probes (see Materials and Methods) were annealed to embryonic day 18 chicken gizzard total cellular RNA, and the hybrids were digested with RNases A and T1. The gizzard RNA protected a 27 nucleotide region of the antisense probe (Fig. 1B, antisense lane 3), whereas no protected fragment was observed with the sense RNA probe. A minor high molecular weight band was observed with yeast tRNA hybridized with the sense probe (Fig. 1B, sense lane 2), due to incomplete RNAase digestion. Although the protection experiments gave a slightly shorter exon 1 sequence (by 3–4 bp), this may be due to the breathing of the labled probe and the hybridized target mRNA; we therefore have chosen the +1 nucleotide as revealed by primer extension analysis. Collectively, these two assays revealed a single major transciptional start site for the chicken SMGA gene that is 74 bp 5′ from the ATG translation initiation codon.
In Vitro Tissue-Specific Expression of the SMGA Gene
To delimit DNA sequences in the SMGA gene that govern its cell-restricted expression, a primary culture system of low passage embryonic gizzard cells was established and evaluated for morphology and SMGA mRNA content (data not shown). The embryonic gizzard cell cultures were evaluated immunohistochemically to detect expression of smooth muscle differentiated products; namely SMGA and smooth muscle myosin heavy chain. As shown in Fig. 2C, when we used a monoclonal smooth muscle-specific γ-actin antibody on our cultures of embryonic smooth muscle cells, the replicating smooth muscle myoblast population exhibited only minor immunoreactivity to this antibody. This degree of staining was consistent with the presence of low levels of SMGA polypeptide in the gizzards of 8 day chicken embryos (26,31,56). After undergoing morphogenesis in culture, however, these same embryonic smooth muscle cells gave intense γ-actin staining (Fig. 2D) of the long, straight, noninterrupted fibrils of the contractile apparatus in the cytoplasm of the smooth muscle cells.
FIG. 2.
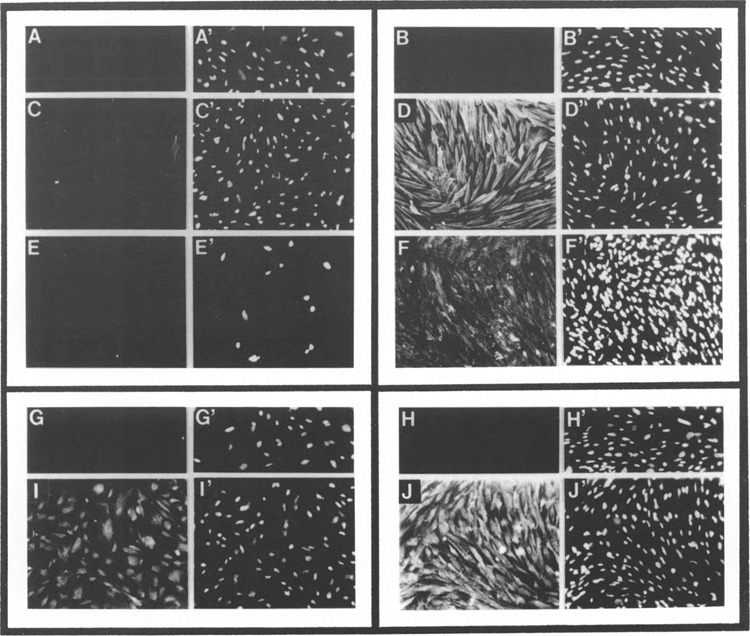
Characterization of embryonic gizzard smooth muscle cell primary cultures. Cultures of cells from 7–8 day embryonic gizzard tissue were established as outlined in Materials and Methods, and the cultured cells examined for the expression of SMGA (C, D) and smooth muscle myosin heavy chain (E, F) expression by immunofluorescence microscopy. (C) and (E) represent γ-actin and myosin heavy chain expression, respectively, in cells just prior to switching media to the differentiation media (DMEM:F12 plus 2 mM l-glutamine, 10−6 units insulin, 5 ml/ml Apo-transferrin, and 0.2 mM L-ascorbic acid), and (D) and (F) represent staining of cells after 48-h incubation in this media. (C), (D′), (E′), and (F) show the staining of nuclei in the same cultures using Hoechst 33258. (A)/(A′) and (B)/(B′) show staining obtained from replicating (A) and differentiated (B) cells using an unrelated, control monoclonal antibody (antispectrin, rat astrocyte-specific, monoclonal antibody, kind gift of Dr. S. Goodman, University of South Alabama). (I) and (J) show replicating and differentiated smooth muscle cells stained with a polyclonal actin antibody that is not isotype specific. (G) and (H) show staining in similar cultures using a preimmune rabbit control antibody.
These findings were confirmed by monoclonal antibodies against other smooth muscle specific proteins. Previous studies have demonstrated the utility of the myosin heavy chain as a marker for differentiated smooth muscle cells (8,10,17,19,32,58). As shown in Fig. 2, embryonic gizzard cells exhibited no staining for myosin heavy chain until they were induced to differentiate (Fig. 2E, F). All cells in the “replicating” (21) and “differentiated” (2J) cultures stained with an actin polyclonal antibody that reacts with all known actin isomers. No staining of myosin heavy chain in the replicating cultures is consistent with analyses of proteins in developing gizzard tissue demonstrating that actin (γ-actin) appeared to increase in content slightly before myosin heavy chain appearance (26,31,56). Importantly, these results showed that cells prepared from early embryonic gizzard tissue could be induced to express smooth muscle specific markers. Thus, the cultured cells provide an experimental environment to analyze the SMGA promoter for cis-acting elements that convey cell-specific transcription of this gene.
Deletion Analysis of the SMGA Promoter
A direct comparison between sequences flanking the 5′ end of the chicken, mouse, and human SMGA genes revealed an extensive homology (−70%) that the three promoters share within the first 130 proximal bases upstream from the transcriptional start site (Fig. 3). Furthermore, although the distal region is less conserved, the nucleotide sequence and spacing of four sequences similar to the CArG element and a single CCAAT/enhancer binding protein (C/EBP) motif (55) 5′ to the gene appear to be strictly maintained across species. No significant homologies were observed beyond the initial 400-bp segment adjacent to the SMGA genes. The level of evolutionary conservation in this ∼400-bp promoter segment from all three SMGA genes is suggestive of the functional importance of these 5′ upstream sequences in the transcriptional regulation of this actin isoform.
FIG. 3.
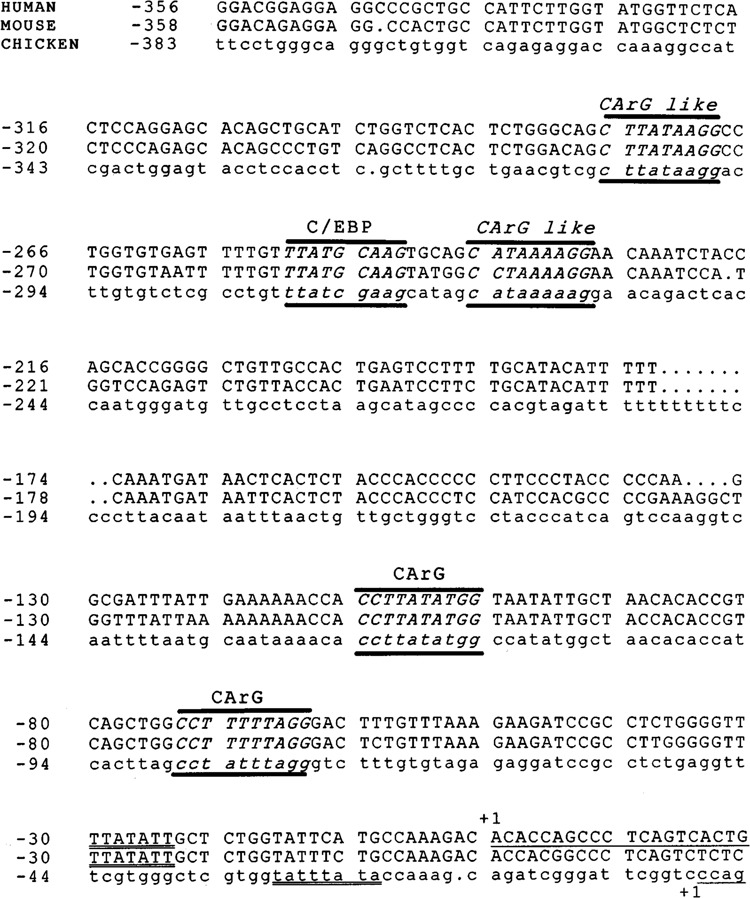
Comparison of SMGA gene proximal promoter segments. Sequences from the 5′ flanking regions of the human (44), mouse (61), and chicken SMGA genes were compared using the Bestfit and Pileup programs of the GCG package (University of Wisconsin). The top line shows the human sequence, the middle shows the mouse sequence, and the bottom line, in lowercase letters, represents the chicken sequence. The 5′ boundaries of the SMGA gene exon 1 segment are denoted by +1 and underlining. The sequences have been aligned to preserve maximal homology. Sequences conforming to CArG and C/EBP DNA elements that are conserved among the SMGA proximal promoters are illustrated with the motif identified above the sequence. The distal two CArG sequences deviate by one nucleotide from the CC(A/T)6GG motif. All the motifs were identified by the GCG program, patterns, with a threshold of 85%.
To initiate an analysis of DNA elements identified in sequences 5′ to the SMGA gene, nine deletion mutants were derived from the total ∼2.3-kb 5′ sequence of the gene (Fig. 4A). The nine chimeric deletion constructs were transfected into primary cultures of differentiated smooth muscle cells, and the data from these analyses were compared to those generated by transfections of identical DNAs into replicating skeletal myoblasts. All experiments included controls that consisted of a vector driven by the SV40 promoter/ enhancer (pCAT-Control), a promoterless CAT vector (pCAT-Basic), as well as examination of lysates from cells not transfected with DNA. In smooth muscle cells, the full-length γ-actin promoter construct (−2294) consistently stimulated transcription three- to fivefold above that obtained for pCAT Control. Thus, the −2294 construct in smooth muscle was given a value of 100% activity and the activities obtained with the other constructs were compared to this value (Fig. 4B). When the most 5′ ∼1 kb of DNA was removed from the −2294 SMGA gene promoter fragment, there was a strong diminution of transcriptional activity in smooth muscle cells (−1087, Fig. 4B). This reduction in transcriptional activity was present over −600 bp of DNA after which transcriptional activity of approximately 50% from that derived by the full-length promoter fragment was observed (−407, Fig. 4B). This 50% of maximal activity was maintained until sequences ∼100 bp 5′ to the SMGA gene were all that remained in the chimeric CAT gene constructs (deletion −112).
FIG. 4.
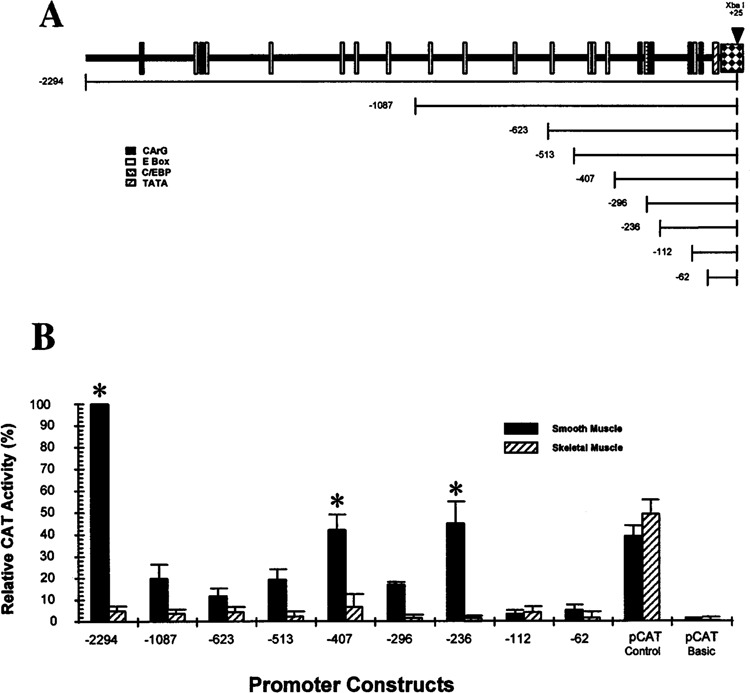
Analysis of the chicken SMGA promoter. Sequences flanking the 5′ region of the chicken SMGA gene were cloned in front of the CAT gene and the hybrid SMGA promoter/CAT genes then placed into cultured smooth muscle or skeletal muscle myoblast cells. (A) The top portion shows a diagram illustrating the position of CArG, E-box, C/EBP, and TATA consensus sequences in front of the chicken SMGA gene. The position of unique restriction sites (EcoRI: −2294, Nar I: −1087, Apa I: −623, Xho I: −513, Sma I: −407, Nde I: −112, and BamHI: −62) or synthetic oligonucleotides (−293 and −233) used to form SMGA promoter deletions are shown. The bottom segment shows the relative position of the promoter mutations tested, each having a common 3′ boundary within exon 1 of the SMGA gene. (B) The constructs shown in (A) were placed into either smooth muscle or skeletal muscle myoblast cells and the resultant CAT activity within the transfected cells assayed 48 h later as described in Materials and Methods. A constant amount of CMV-βgal plasmid was included in each experiment and the amount of CAT activity was evaluated relative to the activity measured from the CMV-βgal control in the same lysate. The data are reported as relative CAT activity, normalizing the −2294 construct to a value of 100%. Each construct was evaluated in a minimum of 20 separate assays (duplicate plates in three different cultures with three separate plasmid preparations) with the SEM shown by the error bars. *Statistical differences between the transcription observed in the smooth muscle and skeletal muscle cells (p < 0.05).
Comparison of the transcriptional activity of the various γ-actin promoter regions in smooth muscle to that in skeletal muscle myoblasts revealed two regions of DNA that appear to impart a significant difference (p < 0.05) in the cell type transcriptional activity: −2294 to −1087 and −407 to −112. Within the −407 to −112 segment of the SMGA promoter are multiple sequence motifs resembling the CArG DNA elements. In addition, this segment of DNA contains a C/EPB and a single E-box motif. The addition of 106 bp 5′ to the 407 deletion imparts a striking reduction in smooth muscle cell transcriptional activity.This DNA domain adds an additional three E-box motifs (−426, −479, −489), which may participate in diminishing the transcriptional capacity of the more proximal elements. Although the DNA segment between −1087 and −2294 is relatively large, when it is added to the SMGA promoter it provides maximal transcriptional activity (Fig. 4B). It is intriguing to note that this segment of the SMGA promoter contains two CArG/SRE motifs, spaced relatively closely together (−2098 and −1877), which may contribute to the smooth muscle-specific transcriptional activity of the SMGA promoter. There are also a number of E-box elements within the −1087 to −2294 as well as other DNA motifs that may participate in the observed enhanced transcriptional capacity in smooth muscle cells.
To further examine the specificity of the SMGA promoter, we placed selected deletions into differentiated smooth muscle cells, replicating smooth muscle myoblasts (analogous to cells shown in Fig. 2C), skeletal muscle cells, and embryonic fibroblasts (CEF). The data were evaluated by comparing of the values obtained with the SMGA promoter fragment to the transcriptional response derived from the reporter gene under the control of the SV40 promoter/ enhancer (pCAT-Control), in order to directly compare the SMGA response in the different cells (Fig. 5). There remained two segments of DNA demonstrating a smooth muscle-specific transcriptional response (−2294 and −407, −236) when compared with skeletal muscle or nonmuscle (CEF). The CEF cells did in most, but not all, cases exhibit a slightly higher response than that observed in the skeletal muscle myoblasts. There also were significant differences in the transcriptional responses obtained with the SMGA promoter constructs in replicating compared to differentiated smooth muscle cells. The −2294 and −236 promoter constructs demonstrated more transcriptional activity in differentiated compared to replicating cells; however, the −407 construct exhibited approximately equal transcription in both cell types, whereas the −513 promoter deletion demonstrated a higher transcriptional capacity in the replicating cells. The pattern of transcriptional capacity with the replicating smooth muscle cells appears different than in differentiated cells in that there were no changes in transcriptional ability observed with the SMGA promoter constructs past the −236 construct. Thus, the −236 SMGA promoter construct contained all the elements needed for transcriptional activation in replicating cells, and the more distal elements appeared to influence transcriptional capacity predominately in fully developed cells.
FIG. 5.
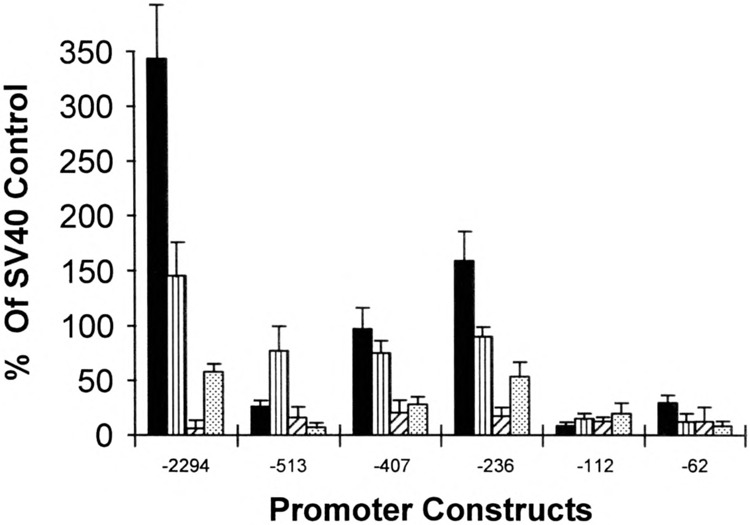
Cellular specificity of SMGA transcription. Selected SMGA promoter/reporter gene constructs were placed into parallel cultures of differentiated smooth muscle cells (filled bars), replicating smooth muscle mesenchyme (vertical striped bars), skeletal muscle myoblasts (striped bars), and chicken embryonic fibroblasts (dotted bars). The resultant reporter activity was assayed as described in Materials and Methods and this activity was normalized to the activity obtained by a constant amount of CMV-βgal plasmid as an internal control for transfection efficiency. Each cell type also recieved a reporter plasmid that contained the SV40 promoter and enhancer sequence (pCAT-Control), and the data are shown as the amount of activity obtained with the SMGA promoter fragment compared with the amount of activity derived for the SV40 control vector (% of SV40 control).
DISCUSSION
Using deletions to examine the cell-specific transcriptional capabilities of the SMGA promoter, we demonstrate that there are four district domains of activity within the DNA sequences flanking the 5′ boundary of the gene (Figs. 4, 5, 6). The initial ∼100 bp adjacent to the gene provide a basal transcriptional activity, regardless of cellular context. This portion of the proximal promoter contains a consensus TATA-box motif and a CArG element [CC(A/ T)6GG], located 80 bp from the transcriptional start site. Inclusion of sequences between -200 and −400 of the 5′ flanking DNA defines a domain that provides a smooth muscle-selective transcriptional capacity to the γ-actin gene. This domain of DNA is highly homologous among the y-genes elucidated to date (Fig. 3) and contains several potential regulatory elements found to be important in the regulation of muscle-specific genes, including multiple CArG-like motifs and E-box consensus sequences. Interestingly, the smooth muscle-selective transcription requires only ∼200–250 bp of DNA sequence adjacent to the gene that comprises the segment of the SMGA gene promoter that is most conserved across species. Although all the SMGA gene proximal promoter sequences elucidated to date [(44,61), this study] contain CArG and E-box sequences within this 200-bp region, only the CArG motifs are strictly conserved in structure and location (Fig. 3).
Flanking the distal boundary of the smooth muscle-selective domain is a segment of DNA that, when included in promoter constructs, appeared to inhibit or diminish transcription primarily in smooth muscle cells. Although we have not mapped the exact boundary of this inhibitory domain, it is clear from our work that the majority of activity is confined to the ∼100 bp between −513 and −407 (Fig. 4B). A region of negative or inhibitory DNA sequence is present in the proximal promoter of the chicken (3,9), rat (60), and mouse (16) smooth muscle α-actin genes. This is not a conserved structure between the α-smooth muscle genes and it appears to repress smooth muscle α-actin transcription in cells where it is not normally expressed (3,9,16,60). Although the −513 to −407 region of the SMGA promoter exhibits little homology across species, our results differ from that obtained with the α-smooth actin gene promoter in that this segment of the SMGA promoter diminished transcription in visceral smooth muscle cells, the cell type of maximal SMGA expression (30,41,44). Interestingly, a murine SMGA promoter fusion gene containing 571 bp of 5′ flanking sequence exhibited very reduced transcriptional capacity compared to longer 5′ constructs in transgenic mice (54). Thus, although not conserved in exact sequence, there may be a conservation of negative transcription domains within the SMGA gene promoters. There are three E-box motifs located within this 100 bp of the γ-actin promoter, each of which contains G/C in their internal two positions. Similar E-box motifs have been shown to bind various bHLH factors from muscle and nonmuscle cells (13,47). However, whether bHLH factors participate in the observed transcriptional inhibition of the SMGA gene is not known.
A key finding in the present study is the presence of a DNA domain (encompased in sequences from −1087 to −2294) that enhanced the cell-specific transcriptional capabilities of the SMGA promoter. The observation that CAT-reporter constructs that extended past the EcoRI −2294 border did not exhibit additional transcriptional activity (data not shown) supports the notion that the smooth muscle-specific enhancement of γ-actin transcription resides within the −2294 to −1087 domain. A similar result of enhanced transcriptional capacity for sequences flanking the murine SMGA gene (to ∼2.7 kb) was demonstrated using fusion genes in transgenic mice (54). From experiments presented here, this domain is inclusive of ∼1 kb of DNA. However, inspection of sequences within this domain (−2294 to −1087) draws attention to a subdomain of sequence from −2101 to −1859, which contains two CArG and two E-box sequence motifs. CArG sequences have been demonstrated to be important ds-acting regulators of skeletal, cardiac, and smooth muscle genes (3,12,22,33–35,45,57,60,62). Deletion of the −2294 to −1087 region containing multiple CArG elements caused a two- to fivefold reduction in smooth muscle specific transcriptional activity, indicating that these sequences may play a vital role in SMGA gene expression. However, it has been reported that the smooth muscle enhancer activity of the rabbit smooth muscle myosin heavy chain gene resides on a 107-bp DNA fragment that contains no apparent muscle-related cis-acting sequences (27). Therefore, it is possible that there are sequences other than the CArG boxes that provide the smooth muscle enhanced transcription to the γ-actin gene. Experiments to elucidate the role of the CArG boxes and/or other DNA elements within this distal segment of the SMGA promoter are currently being pursued in our laboratory.
We found a CArG motif located within intron 1 of the chicken SMGA gene. A CArG sequence structure is also located in the first intervening sequence of the human SMGA gene (44), which might imply that this motif plays a role in SMGA transcription. It has been reported that intron 1 of the human smooth muscle α-actin gene imparts an enhanced transcriptional capactity to the 5′ flanking sequences of the gene (49). Similar results have been demonstrated for other muscle (fast skeletal troponin I and slow cardiac tropomin C) and nonmuscle (human β-cytoplasmic actin) genes (28,52). Significantly, the recent work of Qian et al. (54) indicated that elements within intron 1 of the mouse SMGA gene are needed for high-level, smooth muscle-specific SMGA gene expression. Similar to the results using transgenic mice (54), we observed enhanced smooth muscle transcription when we included intron 1 of the chicken SMGA gene into our reporter gene constructs. However, this enhanced transcriptional capacity was not influenced by the presence of the CArG motif in intron 1, as there was no significant difference between the transcription induced by wild-type, unmodified intron 1 sequences and that observed with most of the intron, including the CArG motif, removed (data not shown). It is possible that the effect of intron 1 sequences upon transcription is a nonspecific enhancement due to RNA splicing upon the primary transcript of the reporter construct as has been reported previously (6,38,39). This concept is supported by the observation that removing most of intron 1, in a manner that maintains the splice donor/ acceptor sequences, of the mouse gene had little effect upon its tissue expression pattern, only causing a reduction in expression in transgenic mice (54). Our experiments differ from that obtained with the mouse gene in that removal of intron 1 sequences, maintaining splice sites, caused no reduction in transcription using our transient transfection assays. This difference may be due to species variation (avian compared to mammalian) or to in vitro verses in vivo analyses. Regardless, the role of specific sequences within intron 1 in smooth muscle-specific SMGA expression merits further investigation.
Significant progress has been made regarding the identification of specific transcription factors that program myogenic progenitor cells through skeletal muscle differentiation (7,50), and recent studies have begun to elucidate regulatory determinants that govern cardiac muscle development (11,37). However, the molecular mechanisms that regulate smooth muscle-specific gene expression are largely unknown. Initial studies of smooth muscle genes, including SM22α (34,35,45), α-vascular actin (3,9,16,60), my-osin heavy chain (27), and telokin (25), have implicated the CArG motif as an important cis-acting regulator. Although the CArG motif is bound by serum response factor (12,34–36,60), the factors that activate smooth muscle gene expression are largely unknown. The absence of an in vitro model of smooth muscle differentiation has limited the ability to employ strategies similar to that used for skeletal muscle (14) to uncover determinants of the smooth muscle lineage. We have developed methods by which cells isolated from embryonic gizzard tissue maintain an undifferentiated phenotype in culture, based upon morphological and biochemical criterion. These cells can be induced to acquire a morphology characteristic of smooth muscle and to express smooth muscle specific genes (Fig. 5). Importantly, our cultured visceral smooth muscle cells demonstrated transcriptional regulation of SMGA promoter constructs similar to that observed in experiments employing transgenic mice (54). This in vitro system affords us an attractive model system to initiate studies elucidating the molecular determinants of smooth muscle differentiation.
ACKNOWLEDGMENTS
The authors are grateful to Chan-Hee Porubcin, John Croushorn, and Ileana Aragon for excellent technical assistance. We thank Curtis Browning and Rebecca Repel for their help with the figures, writing, and reading, and to the rest of the Zimmer lab for critical discussions on this manuscript. Portions of this work were presented at the Society for Developmental Biology Annual meeting (Zimmer, W. E.; Browning, C; Kovacs, A. M. Dev. Biol. 175:399; 1996). This work represents partial fulfillment of the Ph.D. dissertation requirements for Ms. Kovacs. This work was supported by grants from the USDA (#9404341) and American Heart Association (#AL-G-940003) to Warren Zimmer.
REFERENCES
- 1. Al-Molish M. I.; Dubes G. R. Enhanced DNA transfer with dextrans. J. Gen. Virol. 18:189–193; 1973. [DOI] [PubMed] [Google Scholar]
- 2. Balczon R.; West K. The identification of mammalian centrosomal antigens using human autoimmune anti-centrosome antisera. Cell Motil. Cytoskeleton 20:121–135; 1991. [DOI] [PubMed] [Google Scholar]
- 3. Blank R. S.; McQuinn T. C.; Yin K. C.; Thompson M. M.; Takeyasu K.; Schwartz R. J.; Owens G. K. Elements of the smooth muscle α-actin promoter required in cis for transcriptional activation in smooth muscle. J. Biol. Chem. 267:984–989; 1992. [PubMed] [Google Scholar]
- 4. Boxer L. M.; Prywes R.; Roeder R. G.; Kedes L. The sarcomeric actin CArG-binding factor is indistinguishable from the c-fos serum response factor. Mol. Cell. Biol. 9:515–522; 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Breathnach R.; Chambon P. Organization and expression of eukaryotic split genes coding for proteins. Annu. Rev. Biochem. 50:349–383; 1981. [DOI] [PubMed] [Google Scholar]
- 6. Buchmann A.; Berg P. Comparison of intron dependent and intron independent gene expression. Mol. Cell. Biol. 8:4395–4405; 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Buckingham M. Molecular biology of muscle development. Cell 78:15–21; 1994. [DOI] [PubMed] [Google Scholar]
- 8. Cambell G. R.; Chamley J. H.; Burnstock G. Development of smooth muscle cells in tissue culture. J. Anat. 117:295–312; 1974. [PMC free article] [PubMed] [Google Scholar]
- 9. Carroll S. L.; Bergsma D. J.; Schwartz R. J. A 29-nucleotide DNA segment containing an evolutionarily conserved motif is required in cis for cell type-restricted repression of the chicken α-smooth muscle actin gene core promoter. Mol. Cell. Biol. 8:241–250; 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chamley J. H.; Groschel-Stewart U.; Campbell G. R.; Burnstock G. Distinction between smooth muscle fibroblasts and endothelial cells in culture by the use of fluoresceinated antibodies against smooth muscle actin. Cell Tissue Res. 177:445–457; 1977. [DOI] [PubMed] [Google Scholar]
- 11. Chen C. Y.; Croissant J.; Majesky M.; Topouzis S.; McQuinn, Tk.; Frankovsky M. J.; Schwartz R. J. Activation of the cardiac α-actin promoter depends upon serum response factor, tinman homologue, Nkx-2.5, and intact serum response elements. Dev. Genet. 19:119–130; 1996. [DOI] [PubMed] [Google Scholar]
- 12. Chow K. L.; Schwartz R. J. A combination of closely associated positive and negative cis-acting promoter elements regulates transcription of the skeletal α-actin gene. Mol. Cell. Biol. 10:528–538; 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cserjesi P.; Brown D.; Ligon K. L.; Lyons G. E.; Copeland N. G.; Gilbert D. J.; Jenkins N. A.; Olson E. N. Scleraxis: A bassc helix-loop-helix protein that prefigures skeletal formation during mouse embryogenesis. Development 121:1099–1110; 1995. [DOI] [PubMed] [Google Scholar]
- 14. Davis R. L.; Weintraub H.; Lassar A. B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51:987–1000; 1987. [DOI] [PubMed] [Google Scholar]
- 15. Ernt H.; Walsh K.; Harrison C. A.; Rosenthal N. The myosin light chain enhancer and the skeletal actin promoter share a binding site for factors involved in muscle-specific gene expression. Mol. Cell. Biol. 11:3735–3744; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Foster D. N.; Min B.; Foster L. K.; Stoflet E. S.; Sun S.; Getz M. J.; Straunch A. R. Positive and negative cis-acting regulatory elements mediate expression of the mouse vascular smooth muscle α-actin gene. J. Biol. Chem. 267:11995–12003; 1992. [PubMed] [Google Scholar]
- 17. Frid M. G.; Shekhonin B. V.; Koteliansky V. E.; Glukhova M. A. Phenotypic changes of human smooth muscle cells during development: Late expression of heavy caldesmon and calponin. Dev. Biol. 153:185–193; 1992. [DOI] [PubMed] [Google Scholar]
- 18. Grichnik J. M.; Bergsma D. J.; Schwartz R. J. Tissue restricted and stage specific transcription is maintained within 411 nucleotides flanking the 5′ end of the chicken α-actin skeletal actin gene. Nucleic Acids Res. 14:1683–1701; 1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Groschel-Stewart U.; Chamley J. H.; Campbell G. R.; Burnstock G. Changes in myosin distribution in redifferentiating and redifferentitating smooth muscle cells in tissue culture. Cell Tissue Res. 165:13–22; 1975. [DOI] [PubMed] [Google Scholar]
- 20. Gunning P.; Ponte P.; Blau H.; Kedes L. α-skeletal and α-cardiac actin genes are co-expressed in adult human skeletal muscle and heart. Mol. Cell. Biol. 3:1985–1995; 1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gunning P.; Mohun T.; Ng S. Y.; Ponte P.; Kedes L. Evolution of the human sarcomeric-actin: Evidence for units of selection within the 3′ untranslated regions of the mRNAs. J. Mol. Evol. 20:202–214; 1984. [DOI] [PubMed] [Google Scholar]
- 22. Gustafson T. A.; Miwa T.; Boxer L. M.; Kedes L. Interaction of nuclear proteins with muscle-specific regulatory sequences of the human cardiac α-actin promoter. Mol. Cell. Biol. 8:4110–4119; 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hamburger V.; Hamilton H. L. A series of normal stages in the development of the chick embryo. J. Morphol. 88:49–92; 1951. [PubMed] [Google Scholar]
- 24. Hayward L. J.; Schwartz R. J. Sequential expression of chicken actin genes during myogenesis. J. Cell Biol. 102:1485–1493; 1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Herring B. P.; Smith A. F. Telokin expression is mediated by a smooth muscle cell-specific promoter. Am. J. Physiol. 270:cl656–cl665; 1996. [DOI] [PubMed] [Google Scholar]
- 26. Hirai S.; Hirabayashi T. Developmental change of protein constituents in chicken gizzards. Dev. Biol. 97:483–493; 1983. [DOI] [PubMed] [Google Scholar]
- 27. Kallmeier R. C.; Samasundaram C.; Babij P. A novel smooth muscle-specific enhancer regulates transcription of the smooth muscle myosin heavy chain gene in vascular smooth muscle cells. J. Biol. Chem. 270:30949–30957; 1995. [DOI] [PubMed] [Google Scholar]
- 28. Kawamoto T.; Makino K.; Niwa H.; Sugiyama H.; Kimura S.; Amemusa M.; Nakata A.; Ka Kunagg T. Identification of the human β-actin enhancer and its binding factor. Mol. Cell. Biol. 8:267–272; 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim S.; Ip H.; Lu M.; Clendenin C.; Parmacek M. A serum response factor dependent transcriptional regulatory program identifies distinct smooth muscle cell sublineages. Mol. Cell. Biol. 17:2266–2278; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kovacs A. M.; Zimmer W. E. Molecular cloning and expression of the chicken smooth muscle γ-actin mRNA. Cell Motil. Cytoskeleton 24:67–81; 1993. [DOI] [PubMed] [Google Scholar]
- 31. Kuroda M. Changing of actin isomers during differentiation of smooth muscle. Biochim. Biophys. Acta 84:208–213; 1985. [DOI] [PubMed] [Google Scholar]
- 32. Lazard D.; Sastre X.; Frid M. G.; Glukhova M. A.; Thiery J.; Koteliansky V. E. Expression of smooth muscle-specific proteins in myoepithelium and stromal myofibroblasts of normal and malignant human breast tissue. Proc. Natl. Acad. Sci. USA 90:999–1003; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee T. C.; Chow K. L.; Fang P.; Schwartz R. J. Activation of skeletal α-actin gene transcription: The cooperative formation of serum response factor-binding complexes over positive cis-acting promoter serum response elements displaces a negative-acting nuclear factor enriched in replicating myoblasts and non-myogenic cells. Mol. Cell. Biol. 11:5090–5100; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li C.; Miano J. M.; Mercer B.; Olson E. N. Expression of the SM22α promoter in transgenic mice provides evidence for distinct transcriptional regulatory programs in vascular and visceral smooth muscle cells. J. Cell Biol. 132:849–859; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li L.; Liu Z.; Mercer B.; Overbeck P.; Olson E. Evidence for a serum response factor mediated regulatory networks governing SM22α transcription in smooth, skeletal, and cardiac muscle cells. Dev. Biol. 187:311–321; 1997. [DOI] [PubMed] [Google Scholar]
- 36. Lin Z.; Eshelman J. R.; Forry-Schaudres S.; Duran S.; Lessard J. L.; Holtzer H. Sequential disassembly of myofibrils induced by myristate acetate in cultured myotubes. J. Cell Biol. 105:1365–1376; 1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lints T. J.; Parsons L. M.; Hartley L.; Lyons I.; Harvey R. P. Nkx-2.5: A novel murine homeobox gene expressed in early heart progenitor cells and their myogenic decendants. Development 119:419–431; 1993. [DOI] [PubMed] [Google Scholar]
- 38. MacGregor G.; Caskey T. Construction of plasmids that express E. coliβ-galactosidase in mammalian cells. Nucleic Acids Res. 17:2365; 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. MacGregor G.; Nolan G.; Fiering S.; Roederer M.; Herzenberg L. Use of E. coli lacZ (β-galactosidase) as a reporter gene. In: Mwray E. S.; Walker J. M., eds. Gene transfer and expression protocols. Clifton, NJ: Humana Press; 1991:217–235. [DOI] [PubMed] [Google Scholar]
- 40. Maniatis T.; Fritsch E. F.; Sambrook J. Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 41. McHugh K. M.; Crawford K.; Lessard J. L. A comprehensive analysis of the developmental and tissue-specific expression of the isoactin multigene family in the rat. Dev. Biol. 148:442–458; 1991. [DOI] [PubMed] [Google Scholar]
- 42. Mitchell P. J.; Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science 245:371–378; 1989. [DOI] [PubMed] [Google Scholar]
- 43. Miwa T.; Kedes L. Duplicated CArG box domains have positive and mutually dependent regulatory role in expression of the human cardiac actin gene. Mol. Cell. Biol. 7:2803–2813; 1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Miwa T.; Manabe Y.; Kurokawa K.; Kamada S.; Kanda N.; Burns G.; Ueyama H.; Kakunaga T. Structure, chromosome location, and expression of the human smooth muscle (enteric type) γ-actin gene: Evolution of six human actin genes. Mol. Cell. Biol. 11:3296–3306; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Moessler H.; Mericskay M.; Li Z.; Nagl S.; Paulin D.; Small J. V. The SM 22 promoter directs tissue-specific expression in arterial but not in venous or viscerál smooth muscle cells in transgenic mice. Development 122:2415–2425; 1996. [DOI] [PubMed] [Google Scholar]
- 46. Moss J. B.; McQuinn T. C.; Schwartz R. J. The avian cardiac α-actin promoter is regulated through a pair of complex elements composed of E-boxes and serum response elements that bind both positive- and negative-acting factors. J. Biol. Chem. 269:12731–12740; 1994. [PubMed] [Google Scholar]
- 47. Murre C.; McCaw P. S.; Vassin H.; Caudy M.; Jan L. Y.; Jan Y. N.; Cabrera C. V.; Buskin J. M.; Hauschka S. D.; Lassar A. B.; Weintraub H.; Baltimore D. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell 58:537–544; 1989. [DOI] [PubMed] [Google Scholar]
- 48. Muscat G. E. O.; Gustafson T. A.; Kedes L. A common factor regulates skeletal and cardiac α-actin gene transcription in muscle. Mol. Cell. Biol. 8:4120–4133; 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nakano Y.; Nishihara T.; Sasayama S.; Miwa T.; Kamada S.; Kakunaga T. Transcriptional regulatory elements in the 5′ upstream and first intron regions of the human smooth muscle (aortic type) alpha-actin encoding gene. Gene 99:285–289; 1991. [DOI] [PubMed] [Google Scholar]
- 50. Olson E. N.; Klein W. H. bHLH factors in muscle develoment: Dead lines and commitments, what to leave in and what to leave out. Genes Dev. 8:1–8; 1994. [DOI] [PubMed] [Google Scholar]
- 51. Papadopoules N.; Crow M. T. Transcriptional control of the chicken cardiac myosin light chain gene is mediated by two AT-rich cis-acting DNA elements and binding of serum respones factor. Mol. Cell. Biol. 13:6907–6918; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Parmacek M.; Ip H.; Jung F.; Shen T.; Martin J.; Vora A.; Olson E.; Leiden J. A novel myogenic regulatory circuit controls slow/cardiac tropomine gene transcription in skeletal muscle. Mol. Cell. Biol. 14:1870–1885; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Petropoulous C. J.; Rosenberg M. P.; Jenkins N. A.; Copeland N. G.; Hughes S. H. The chicken skeletal muscle α-actin promoter is tissue-specific in transgenic mice. Mol. Cell. Biol. 9:3785–3792; 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Qian J.; Kumar A.; Szucsick J.; Lessard J. Tissue and developmental specific expression of murine smooth muscle γ-actin fusion genes in transgenic mice. Dev. Dynamics 207:135–144; 1996. [DOI] [PubMed] [Google Scholar]
- 55. Ryden T. A.; Beemont K. Avian retroviral long terminal repeats bind CCAAT/enhancer-binding protein. Mol. Cell. Biol. 9:1155–1164; 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Saborio J. L.; Segura M.; Flores M.; Garcia R.; Palmer E. Differential expression of gizzard actin genes during chick embryogenesis. J. Biol. Chem. 254:11119–11125; 1979. [PubMed] [Google Scholar]
- 57. Sartorelli V.; Webster K.; Kedes L. Muscle-specific expression of the cardiac alpha-actin gene requires MyoDl, CArG-box binding factor, and Spl. Genes Dev. 4:1811–1822; 1990. [DOI] [PubMed] [Google Scholar]
- 58. Schneider M. D.; Sellers J. R.; Vahey M.; Preston Y. A.; Adelstein R. S. Localization and topography of antigenic domains within the heavy chain of smooth muscle myosin. J. Cell Biol. 101:66–72; 1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Schwartz R. J.; Rothblum K. N. Gene switching in myogenesis: Differential expression of the chicken actin multigene family. Biochemistry 20:4122–4129; 1981. [DOI] [PubMed] [Google Scholar]
- 60. Shimizu R. T.; Blank R. S.; Jervis R.; Lawrenz-Smith S. C.; Owens G. K. The smooth muscle α-actin gene promoter is differentially regulated in smooth muscle versus non-smooth muscle cells. J. Biol. Chem. 270:7631–7643; 1995. [DOI] [PubMed] [Google Scholar]
- 61. Szucsik J. C.; Lessard J. L. Cloning and sequence analysis of the mouse smooth muscle γ-enteric actin gene. Genomics 28:154–162; 1995. [DOI] [PubMed] [Google Scholar]
- 62. Treisman R. The SRE: A growth factor responsive transcriptional regulator. Semin. Cancer Biol. 1:47–58; 1990. [PubMed] [Google Scholar]
- 63. Tuil D.; Soulez M.; Montarras D.; Pinset C.; Kahn A.; Phan-Dinh-Tuy F. Activation of gene expression via CArG boxes during myogenic differentiation. Exp. Cell Res. 205:32–38; 1993. [DOI] [PubMed] [Google Scholar]
- 64. Vandekerckhove J.; Weber K. At least six different actins are expressed in a higher mammal: An analysis based on the amino acid sequence of the amino-terminal tryptic peptide. J. Mol. Biol. 122:783–802; 1978. [DOI] [PubMed] [Google Scholar]
- 65. Vandekerckhove J.; Weber K. Actin typing on total cellular extracts. Eur. J. Biochem. 113:595–603; 1981. [DOI] [PubMed] [Google Scholar]
- 66. Wang D.; Greaser M. L.; Schultz E.; Bulinski J. C.; Lin J. J. C.; Lessard J. L. Studies on cardiac myofibrillogenesis with antibodies to titin, actin, tropomyosin, and myosin. J. Cell Biol. 107:1075–1083; 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zimmer W. E.; Chang K. S.; Bergsma D. J.; Schwartz R. J. Organization and expression of the chicken actin gene family. J. Cell Biol. 97:330a; 1983. [Google Scholar]
- 68. Zimmer W. E.; Schloss J. A.; Silflow C. D.; Youngblom J.; Watterson D. M. Structural organization, DNA sequence, and expression of the calmodulin gene. J. Biol. Chem. 263:19370–19383; 1988. [PubMed] [Google Scholar]


