Abstract
Initially, pilE transcription in Neisseria gonorrhoeae appeared to be complicated, yet it was eventually simplified into a model where integration host factor activates a single − 35/ − 10 promoter. However, with the advent of high-throughput RNA sequencing, numerous small pil-specific RNAs (sense as well as antisense) have been identified at the pilE locus as well as at various pilS loci. Using a combination of in vitro transcription, site-directed mutagenesis, Northern analysis and quantitative reverse transcriptase PCR (qRT-PCR) analysis, we have identified three additional non-canonical promoter elements within the pilE gene; two are located within the midgene region (one sense and one antisense), with the third, an antisense promoter, located immediately downstream of the pilE ORF. Using strand-specific qRT-PCR analysis, an inverse correlation exists between the level of antisense expression and the amount of sense message. By their nature, promoter sequences tend to be AT-rich. In Escherichia coli, the small DNA-binding protein H-NS binds to AT-rich sequences and inhibits intragenic transcription. In N. gonorrhoeae hns mutants, pilE antisense transcription was increased twofold, with a concomitant decrease in sense transcript levels. However, most noticeably in these mutants, the absence of H-NS protein caused pilE/pilS recombination to increase dramatically when compared with WT values. Consequently, H-NS protein suppresses pilE intragenic transcription as well as antigenic variation through the pilE/pilS recombination system.
Introduction
Neisseria gonorrhoeae (gonococcus) is a Gram-negative diplococcus and the aetiological agent of the human sexually transmitted disease gonorrhoea. Gonorrhoea is generally characterized by a pustular discharge due to neutrophil infiltration. However, if left untreated, the infection can lead to more severe sequellae such as bacteraemia, sterility and pelvic inflammatory disease. Pathogenesis associated with a gonococcal infection is best characterized by the sophistication of its major virulence factors that are employed to survive and reproduce within the human host. The elaboration of type IV pili on the cell surface appears to be particularly important as the infectivity of pilus-minus bacteria is severely reduced (Hill & Davies, 2009).
The major protein component of the pilus organelle is PilE polypeptide, which is encoded by the pilE gene. Initially, regulation of pilE gene expression appeared complex, as there are three functional promoters (P1, P2 and P3), with the P1 and P2 promoters utilizing the σ70 sigma factor, in contrast to P3, which is recognized by the σ54 sigma factor (Fyfe et al., 1995). However, in the gonococcus, only the P1 promoter is actively transcribed. In addition, there appeared to be an absence of transcription factors that provided either positive or negative regulation. However, recently CrgA (contact-regulated gene A) has been implicated in maintaining pilE transcript levels in the gonococcus in both a contact-dependent and -independent manner (Matthias & Rest, 2014). However, these observations are at odds with a study involving Neisseria meningitidis, where CrgA protein did not appear to regulate pilus expression, despite the fact that it had been shown to be an active transcriptional regulator in this organism (Ieva et al., 2005).
What is firmly established in the gonococcus is the role played by integration host factor (IHF) on pilE expression. Upstream of the pilE gene resides an IHF-binding site, and, in gonococcal mutants where the IHF-binding site has been deleted, pilE transcription is reduced approximately 10-fold (Hill et al., 1997). These observations were further embellished through the identification of two AT-rich segments of DNA (UP-like elements) that flank the IHF-binding site and are both required for optimum transcription (Fyfe & Davies, 1998). IHF binds to a consensus DNA sequence, and, when bound, causes a bend in the surrounding DNA, which is believed to facilitate contact of the UP-like elements with RNA polymerase, thus potentiating transcription. Recently, small RNA (sRNA) regulation has also been considered as a means by which pilE expression is modulated. At the time, no sRNA had been identified that theoretically could hybridize with the pilE transcript, yet PilE polypeptide was found to be absent in N. gonorrhoeae hfq mutants, suggesting that the Hfq RNA chaperone is required for efficient PilE translation, and, by implication, such regulation involves a potential sRNA molecule (Dietrich et al., 2009). Subsequently, pilE-specific antisense RNA has been identified in the gonococcus (Wachter & Hill, 2015). Consequently, this antisense RNA may mediate translation efficiency; however, this has not yet been formally tested.
With the advent of high-throughput RNA sequencing, transcriptome analysis, principally using Artemis and RNA-Seq technologies, has allowed the identification of many previously unknown sRNAs within a cell. Recently, gonococcal small transcriptome analysis revealed that the majority of genes within the chromosome were associated with sense and/or antisense intragenic sRNAs (Wachter & Hill, 2015). Moreover, many other sRNAs were also found within the chromosomal intergenic regions (Wachter & Hill, 2015). Consequently, gonococci exhibit the recently described phenomenon known as pervasive transcription (Sorek & Cossart, 2010; Wade & Grainger, 2014; Wachter & Hill, 2015; Wachter et al., 2015). An unusual aspect of this type of transcription is the use of non-canonical promoters that deviate from the accepted rules governing transcription. An in-depth analysis also showed that the majority of the pil genes in the chromosome also produced sense and antisense sRNAs, with sRNAs also observed at the pilE locus (Wachter et al., 2015). Indeed, antisense sRNA appears to be the predominant sRNA species that is observed at pilE. Therefore, given these observations, the purpose of this study was to further examine transcription at pilE with the view of identifying these new putative pilE promoter elements and determining the biological consequence(s), if any, for sense and antisense pilE transcription. In this report we have identified three new promoter elements within pilE (one yielding a sense sRNA and two yielding antisense sRNAs) and have shown through mutational analysis in the gonococcus that transcription from these novel promoters appears to modulate the level of the pilE sense transcript. Furthermore, we also observed that the small DNA-binding protein histone-like nucleoid structuring protein (H-NS) changed the level of pilE intragenic transcription within the cell, and, more dramatically, influenced the level of pilE/pilS recombination.
Methods
N. gonorrhoeae strain MS11 pilE7 : 30 : 2 (Bergström et al., 1986) was grown at 37 °C in a 5 % CO2 atmosphere on a previously described growth medium (Swanson, 1982). When appropriate, antibiotics were added to the growth medium: erythromycin at a final concentration of 5 μg ml− 1, chloramphenicol at a final concentration of 10 μg ml− 1 and kanamycin at a final concentration of 80 μg ml− 1.
All recombinant work utilized Escherichia coli DH5α, which was grown in LB medium at 37 °C. Antibiotics were added to the medium when appropriate: carbenicillin at a final concentration of 100 μg ml− 1 and kanamycin at a final concentration of 50 μg ml− 1.
In vitro transcription
In vitro transcription was performed as follows: 3 μg purified template DNA was resuspended in 8 μl 5 × RNA polymerase transcription buffer (0.2 M Tris/HCl, 0.75 M KCl, 50 mM MgCl2 and 0.05 % Triton X-100; Epicentre), 2 μl 100 mM DTT, 4 μl rNTPs (1 μl each 100 mM rATP, rUTP, rCTP and rGTP), 1 μl RNase inhibitor (20 U μl− 1; Promega), 4 μl E. coli RNA polymerase holoenzyme (1 U μl− 1; Epicentre) and sufficient diethyl pyrocarbonate (DEPC)-treated water to bring the total volume of the reaction to 40 μl. The reaction was then incubated at 37 °C for 2 h. Template DNA was removed by an overnight HaeIII enzymic digestion at 37 °C. The restriction-digested in vitro transcription reaction was then treated with 6 μl 10 × RQ1 DNase buffer (Promega) and 4 μl RQ1 DNase (1 U μl− 1; Promega) and subsequently incubated for 3 h at 37 °C. This was followed by the addition of 4 μl RQ1 DNase Stop Solution (Promega) and incubation for 10 min in a 65 °C water bath, followed by addition of 36 μl DEPC-treated water. The RNA was then purified as described below. Quantification of in vitro-transcribed RNA utilized a NanoDrop ND-2000 1-Position Spectrophotometer (Fisher Scientific).
RNA analysis
RNA isolation, size fractionation, standard quantitative reverse transcriptase PCR (qRT-PCR) analysis, and RNA circularization experiments have been described (Wachter & Hill, 2015; Wachter et al., 2015). Oligonucleotide sequences for primers that were used in the qRT-PCR analysis are presented in Tables 1 and 2. For strand-specific qRT-PCR analysis, the initial cDNA reaction utilized a single specific oligonucleotide primer that was complementary to either sense or antisense RNA (the various oligonucleotide sequences are in Table 3). The cDNA sample was then denatured followed by the addition of RNaseA to remove the RNA from the DNA/RNA hybrid. The remaining single-stranded cDNA product was then amplified with pilE-specific primer pairs. In control experiments with gonococcal RNA extracts, a potential RNA secondary structure inhibitor, betaine, was tested. However, with these studies self-priming became evident in the strand-specific scores (data not shown). Therefore, an important consideration in strand-specific qRT-PCR was assessing self-priming of the RNA transcripts at the 5′ or 3′ end, as further control experiments indicated that reverse transcriptase utilized the looped-back segment of RNA as a primer in cDNA synthesis to a significant degree. Consequently, for the final cycle threshold (Ct) values from the strand-specific reactions, no primer controls (which reflect self-priming) were subtracted from the actual strand-specific Ct score. The viability of this approach was initially tested on in vitro transcription reactions, where it was hypothesized that subtraction of the no-primer expression value from the overall strand-specific cDNA values obtained utilizing a specific primer should yield approximate expression levels comparable with cDNA synthesized using a random oligonucleotide. The results (data not shown) of such a comparison from randomly primed, specifically primed and non-primed cDNAs from an in vitro-transcribed RNA template confirmed that removing the expression value from a no-primer control allows strand-specific cDNA synthesis to be a viable method of quantifying RNA from either strand at any given location along pilE. The Northern blotting protocol that was used has been published previously (Hill et al., 1990, 1997). The sequence of the oligonucleotide primer designed to target pilE-specific antisense RNA is 5′-GUCGGCAUUUUGGCGGCAGUCG-3′.
Table 1. Oligonucleotides used in reverse transcription of pilE RNAs.
| Oligonucleotide | Description | Sequence |
|---|---|---|
| 14728 | Random decamer | 5′-NNNNNNNNNN-3′ |
| 07053 | RNA3 reverse primer | 5′-GATGTAGCCCATGAGAGCCAAGCAGATACGTGTG-3′ |
| 09085 | pilE 3′ end primer | 5′-CGGCAGGTTGACGGCAGGTGCTTGGTG-3′ |
| RC3 | pilE 5′ end primer | 5′-GTCGGCATTTTGGCGGCAGTCG-3′ |
Table 3. Oligonucleotides used in pilE strand-specific qRT-PCR.
| Nucleotide | Sequence |
|---|---|
| 09085 | 5′-CGGCAGGTTGACGGCAGGTGCTTGGTG-3′ |
| RC3 | 5′-GTCGGCATTTTGGCGGCAGTCG-3′ |
| 09173 | 5′-TTGACCTTCGGCCAAAAGGATGGCTT CGGAAAC-3′ |
| 09263 | 5′-CATTGTGGCGGTAACGACGCCGTT-3′ |
| 10685-tsp4 | 5′-CGCCGGCAGAAGTGTTGTTTTC-3′ |
Table 2. Control primers used in qRT-PCR.
| Primer | Description | Nucleotide sequence |
|---|---|---|
| 07052 | RNA3 forward primer | 5′-GGCAAGCAAGCGGGAGGTACTAGCGATGAGAAGC-3′ |
| 07053 | RNA3 reverse primer | 5′-TGGATGTAGCCCATGAGAGCCAAGCAGATACGTGTG-3′ |
| 06971 | E. coli rrsA (16S rRNA) forward | 5′-CGGTGGAGCATGTGGTTTAA-3′ |
| 06972 | E. coli rrsA (16S rRNA) reverse | 5′-GAAAACTTCCGTGGATGTCAAGA-3′ |
| 09484 | GC 16S rRNA forward | 5′-GTTATCCCAGGAGGCTGCCTTCGCCATCGGTATTCC-3′ |
| 09485 | GC 16S rRNA reverse | 5′-GTAATACGTAGGGTGCGAGCGTTAATCGGAATTACTGGG-3′ |
| 09170 | GC recA forward | 5′-GCCATCATGAAAATGGACGGCAGCCAGCAGGAAG-3′ |
| 09171 | GC recA reverse | 5′-CTGGCATTGGGCGACGGCTTCGAGGCAGAGTGTGG-3′ |
Site-directed mutagenesis protocol
Site-directed mutagenesis protocols have been described previously (Wachter et al., 2015). The oligonucleotide sequences that were used are presented in Table 4. The mutant sequence was designed so that it corresponded to a restriction endonuclease cleavage site for ease of analysis.
Table 4. Oligonucleotides used to generate mutations in pilE promoter regions.
| Oligonucleotide | Description | Sequence |
|---|---|---|
| 12561-MutC2F | Midgene mutation | 5′-CCCCCCTCCGACATCAAAGGCAAATATGTGATCAAGGTTGAAGTTAAAAAC-3′ |
| 12562-MutC2R | Midgene mutation | 5′-GTTTTTAACTTCAACCTTGATCACATATTTGCCTTTGATGTCGGAGGGGGG-3′ |
| 12490-P2F | 3′ end mutation | 5′-AAGGCATCTGATGCCAAATGAGGCAAATTAGGCGGATCCTTTTAAATAAATCAAGCGGTAAGTGATTTTCCA-3′ |
| 12491-P2R | 3′ end mutation | 5′-TGGAAAATCACTTACCGCTTGATTTATTTAAAAGGATCCGCCTAATTTGCCTCATTTGGCATCAGATGCCTT-3′ |
Construction of an N. gonorrhoeae hns deletion strain
In order to construct an N. gonorrhoeae hns insertional mutation, segments of the hns gene were PCR-amplified using the following primer pairs: 5′-GTGAGGGAATACAGTGTTTAATGGTTCGG-3′ and 5′-AAGGTACCATACATTTAAAAATACCTCGGGGCATCC-3′; and 5′-AATCTAGAGCCTATGCGCTCAAAATCCTGC-3′ and 5′-AAGATATCTTTTGCCGCCTTTTTCAATATTCCGCC-3′. The DNA amplicons were then sequentially ligated to a kanamycin antibiotic resistance marker. The final construct was then used to transform gonococci to kanamycin drug resistance using standard protocols to generate an N. gonorrhoeae hns : : kan strain.
Construction of gonococcal hns complementation strain
Previously, no correlation has been demonstrated between pilE/pilS recombination and opa expression (Swanson & Barrera, 1983). Therefore, a WT copy of the hns gene was placed into the opaE locus of the hns− strain, which resides immediately downstream of the pilE gene. The entire hns segment was PCR-amplified using the appropriate primers described above and was then ligated to a chloramphenicol drug-resistance marker that carried a transformation uptake sequence in plasmid pCR2.1. The resulting hns : cat construct was then amplified utilizing the M13 forward and reverse primers, with the resulting amplicon being phosphorylated by T4 polynucleotide kinase (New England Biolabs), which allowed for blunt-end ligation into a unique SalI site (blunt-ended treatment with Klenow; New England Biolabs). The resulting hns : cat : opaE construct was then used to transform the N. gonorrhoeae hns− strain to chloramphenicol/kanamycin drug resistance.
pilE/pilS recombination assay
pilE/pilS recombination was monitored at both the DNA and RNA levels in various gonococcal strains that carried the pilE promoter mutations, in conjunction with either a WT or hns mutant genetic background. For the RNA-based recombination assay, total RNA was purified from the various gonococcal strains and quantified, followed by DNase treatment and cDNA synthesis utilizing a random decamer. The newly synthesized cDNAs were then used as templates for either end-point PCRs or qRT-PCR using a conserved pilE primer (RC3) and a pilS-specific opposing primer (either pilS5c1 or pilS6c1). Recombination efficiency between pilE and pilS was determined by comparing the expression level of the pilE/pilS recombinant cDNAs with the expression of a recA internal control. For the recombination assay at the DNA level, recombination PCRs and qRT-PCRs were performed on isolated chromosomal DNAs. Each reaction utilized 120 ng DNA template and the above pilE/pilS primer pairs. Amplified recombinant pilE products were again quantified against a recA control signal. In addition, reactions using conserved 5′ and 3′ pilE primers (RC3 and 09085, respectively) were also included as a positive control in the assays.
Bioinformatics analysis
All algorithms that were used for promoter predictions, RNA-Seq analysis and Artemis GC genome analysis have been described previously (Wachter & Hill, 2015; Wachter et al., 2015). The NCBI GenBank accession number for the N. gonorrhoeae strain MS11 genome sequence that was used is CP003909.1.
Results
Antisense RNA identified at the pilE locus
In a previous study, RNA-Seq analysis revealed the presence of cis-encoded sense and antisense sRNAs at the majority of loci within the gonococcal chromosome (Wachter & Hill, 2015). When similar analysis focused on the pil genes within the chromosome, sense and antisense sRNAs were detected at the pilE locus (Wachter et al., 2015), with antisense RNA being observed throughout the entire pilE gene (Fig. S1). The presence of pilE-specific antisense RNA was confirmed by Northern blotting of total RNA, as well as size-fractionated sRNA, using an oligonucleotide probe designed to hybridize with pilE-derived antisense RNA (Fig. 1; the probe targets a unique 5′ pilE gene sequence). As can be seen in the Northern blot, pilE antisense RNA varied in size from approximately 250 to 1000 bp in length. The presence of a 250 bp signal in the size-fractionated sRNA suggests a promoter yielding antisense RNA may be present internally within the pilE gene. As qualitatively similar results were obtained with RNA extracted from an N. gonorrhoeae rppH mutant, this suggests that the smaller RNA species observed in Fig. 1 represent true transcripts and are not a consequence of RNA degradation (S. A. Hill & J. Wachter unpublished observations; Wachter & Hill, 2015). RNA circularization experiments then identified the 3′ end points of pilE-specific RNAs. The pilE sense transcript from the cognate pilE promoter ends within the downstream Sma/Cla repeat at a rho-independent transcription terminator (data not shown). A potential pilE antisense transcript was also identified with the 5′ end point located within the pilE midgene region, with the 3′ end point located 20 bp downstream from the AUG start codon at the base of a predicted stem–loop structure (Fig. S2).
Fig. 1.
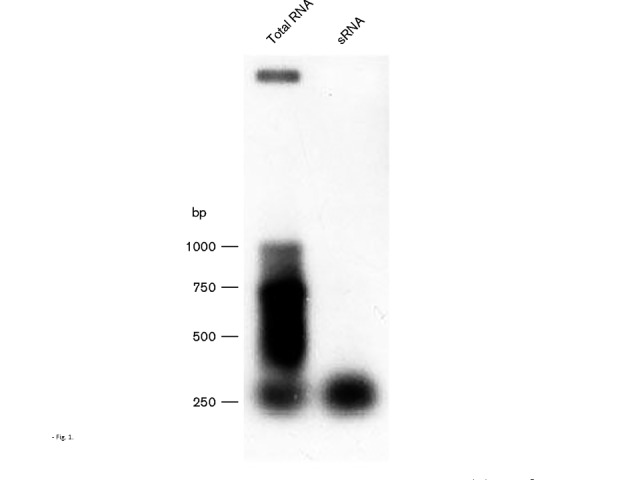
Northern analysis of pilE antisense RNA. Total RNA and size-fractionated sRNA were isolated from N. gonorrhoeae strain MS11. The Northern blot was then probed with an oligonucleotide designed to hybridize with pilE-specific antisense RNA.
Identification of intragenic antisense promoters within pilE
In order to identify the pilE antisense promoters, we combined observations from in vitro transcription experiments, qRT-PCR analysis, RNA circularization and Northern blotting. From these initial studies, an antisense promoter was believed to be located within the pilE midgene region, with an additional antisense promoter being located towards the 3′ end of the pilE gene.
Several potential non-canonical midgene promoters were identified using bprom and Neural Network online prediction tools. In order to further determine whether any of these putative promoters could be transcribed, in vitro transcription profiling was utilized, with a sequential series of DNA fragments as transcription templates (Fig. 2a). Each putative promoter was then changed through site-directed mutagenesis (Fig. 2b) and was reassessed for the ability to engage in transcription (Fig. 2c). The analysis was further refined by utilizing a strand-specific qRT-PCR protocol. From such analysis, it was determined that the mutated promoter designated mut2 lost the ability to transcribe both sense and antisense RNA, whereas the mutated promoter designated mut5 lost the ability to transcribe sense RNA (Fig. 2c; P < 0.01, n = 4). These observations were then confirmed through Northern blotting with strand-specific oligonucleotides in conjunction with deletion analysis (data not shown). Therefore, the pilE gene appears to possess two non-canonical midgene promoters that yield both sense (5′-GTTAAAAA-3′) and antisense (5′-TAACATAT-3′) pil-specific RNA, with the midgene antisense promoter being appropriately located to account for the approximate 250 bp antisense RNA species in the Northern blot (Fig. 1).
Fig. 2.
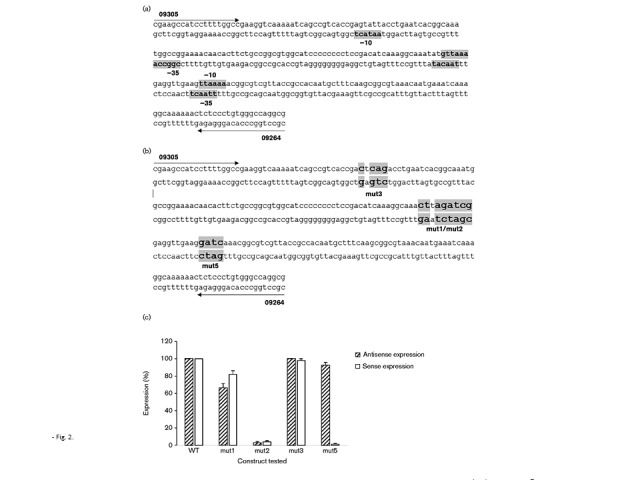
Identification of the pilE midgene sense and antisense promoters. (a) Putative promoter elements were initially identified by the bprom and Neural Network online prediction tools. (b) Mutated promoter sequences. (c) Expression levels of WT and mutated constructs through strand-specific qRT-PCR analysis using in vitro-transcribed RNA. Strand-specific cDNA was made utilizing primers 09305 and 09264 for antisense and sense, respectively. WT expression for both antisense and sense expression was normalized to 100 %. Data represent mean ± sd; n = 4.
The two pilE midgene promoters are located within the mc5 variable segment of the pil genes (Hagblom et al., 1985). Consequently, as pilE engages in extensive recombination with the various pilS genes, it is possible that the midgene promoter elements could be lost as the pilE gene varies. The clustal analysis presented in Fig. 3 indicates that there is strong conservation of the midgene antisense promoter (promoter 2) across the various Neisseria pil gene copies within the database, whereas the sense promoter (promoter 5) appears to be less conserved and may be lost following pilE/pilS recombination (Fig. 3).
Fig. 3.

clustal analysis of Neisseria pil gene sequences. The accession numbers for each strain are as follows: Neisseria gonorrhoeae MS11 GenBank accession CP003909.1 (pilE = NGFG_01821; pilS1 copy 1 = NGFG_02253 (2122534–2123035), copy 2 = NGFG_02253 (2123072–2123479), copy 3 = NGFG_02487, copy 4 = NGFG_00014 (2124309–2124659), copy 5 = NGFG_00014 (2124696–2125043); pilS2 copy 1 = NGFG_02484, copy 2 = NGFG_02485; pilS5 = NGFG_02405; pilS6 copy 1 = NGFG_02481, copy 3 = NGFG_02482; pilS7 = NGFG_02431); Neisseria gonorrhoeae FA1090 GenBank accession NC_002946.2 (pilE = 2038021–2037383*; pilS1 copy 1 = 2043862–2044354*, copy 2 = 2044392–2044736*, copy 3 = 2044773–2045127*, copy 4 = 2045506–2045842*, copy 5 = 2045843–2046256*; pilS2 copy 1 = 2040723–2041203*, copy 2 = 2041256–2041603*, copy 3 = 2041646–2042026*, copy 4 = 2042470–2042800*, copy 5 = 2042801–2043147*, copy 6 = 2043148–2043522*; pilS3 copy 1 = 2013715–2014201*, copy2 = 2014238–2014567*, copy 3 = 2014626–2015018*; pilS6 NGO2041a copy 1 = 2015343–2015868, copy 2 = 2016302–2016669, copy 3 = 2016707–2017060; pilS7 = 1480693–1481248*); Neisseria gonorrhoeae NCCP 11945 GenBank accession NC_011035.1 [pilE = NGK_RS08820, pilS1 = NGK_RS09875 (1818155–1818627), pilS2 = NGK_RS09875 (1818628–1819032), pilS3 = NGK_RS09875 (1819033–1819327)]; Neisseria meningitidis FAM18 GenBank accession NC_008767.1 (pilE = NMC_RS01135, pilS1 = NMC_RS00015, pilS2 = NMC_RS00020); Neisseria meningitidis 053442 GenBank accession NC_010120.1 (pilE = NMCC_RS00970; pilS1 = NMCC_RS00130; pilS2 = NMCC_RS00125 copy 1 = 19974–20401, copy 2 = 19532–19973 ; pilS3 = NMCC_RS03380; pilS4 = NMCC_RS07650; pilS5 = NMCC_RS07655; pilS6 = NMCC_RS08075; pilS7 = NMCC_RS08080); Neisseria meningitidis MC58 GenBank accession NC_003112.2 (pilE = NMB0018; pilS1 = NMB0019; pilS2 = NMB0020; pilS3 = NMB0021; pilS4 = NMB0022; pilS5 = NMB0023; pilS6 = NMB0024; pilS7 = NMB0025 copy 1 = 23169–23502, copy 2 = 22825–23168; pilS8 = NMB0026); *Not annotated in final assembly. The midgene antisense promoter is indicated by the left arrow; the midgene sense promoter is indicated by the right arrow. The colour-shaded regions highlight the locations of the antisense and sense promoters within the sequences.
A similar strategy as outlined above was also employed to identify the 3′ antisense promoter that was predicted by RNA-Seq analysis to reside between the end of the pilE gene and the downstream Sma/Cla repeat. Several putative promoters were predicted by bprom analysis, with a single non-canonical promoter ( − 10 motif, 5′-TTTAAG-3′; Fig. 4a) found to be transcriptionally active (Fig. 4b, c). When mutated via site-directed mutagenesis, a 90 % reduction in transcription from the mutated construct was observed compared with the non-mutated promoter (Fig. 4c; P < 0.001, n = 10).
Fig. 4.
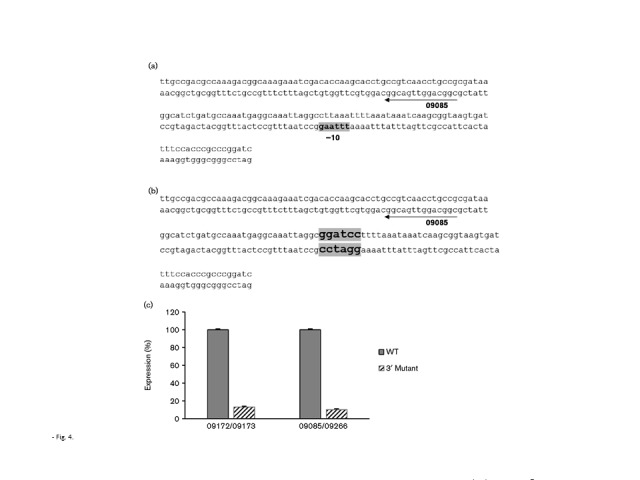
Identification of the antisense promoter at the 3′ end of pilE. (a) Location of the predicted antisense promoter at the 3′ end of pilE. (b) 3′ Mutated promoter. (c) Expression of pilE antisense RNAs from recombinant WT and the 3′ mutated constructs in E. coli using strand-specific qRT-PCRs. WT expression is normalized to 100 %. Error bars represent mean ± sd; n = 10.
Collectively, these observations indicate that the pilE gene contains two antisense promoters, one with the potential to yield full-length antisense RNA, and a second antisense promoter capable of producing an approximately 250 bp antisense RNA, with both antisense RNAs being theoretically able to pair with pilE sense RNA arising from the cognate sense promoter.
pilE antisense transcription in gonococci
The mutated midgene and 3′ antisense promoters were crossed individually, or together, into the MS11 pilE7/30 : 2 gene utilizing an antibiotic resistance marker located downstream of pilE for the initial selection of transformants. Owing to the presence of − 35/ − 10 elements, promoter motifs deviated from the mean chromosomal nucleotide content, which was especially evident for both the midgene and 3′ antisense promoters (AT content: midgene promoter 72.5 %, 3′ promoter 75 % versus cognate 65 %; Fig. 5a). The small DNA-binding protein H-NS binds to AT-rich segments of DNA and in doing so suppresses intragenic transcription (Navarre et al., 2006; Singh & Grainger, 2013). In light of this, the various promoter mutations were crossed into both a WT genetic background and an N. gonorrhoeae hns mutant background.
Fig. 5.
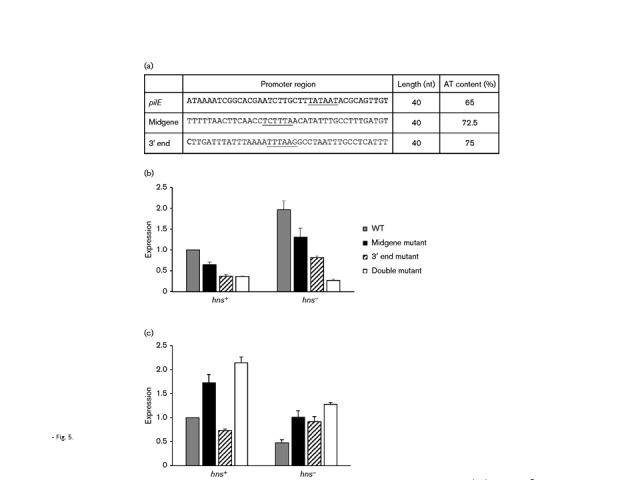
Strand-specific qRT-PCR analyses of in vivo-transcribed gonococcal RNAs. Total RNA was isolated from gonococci carrying either a non-mutated pilE gene, a pilE midgene antisense promoter mutation, a pilE 3′ antisense mutation, or a pilE midgene antisense promoter mutation as well as a pilE 3′ antisense mutation. Strand-specific cDNAs were synthesized using primer 09172 for antisense expression and 09085 for sense expression. (a) AT-richness of the various promoter sequences and surrounding DNA. The promoter is underlined. (b, c) Expression of pilE antisense (b) and sense (c) RNAs in all four GC constructs. The left side of each panel reflects RNAs extracted from WT bacteria; the right side of each panel reflects RNAs extracted from an isogenic hns mutant. Expression is normalized relative to the WT value, which is set to 1. Error bars represent mean ± sd; n = 10.
The data in Fig. 5(b, c) present strand-specific qRT-PCR analyses. In a WT genetic background, mutation of the midgene antisense promoter and the 3′ antisense promoter, as well as both the midgene antisense and the 3′ antisense promoters together, resulted in a reduction of pilE antisense RNA relative to the non-mutated promoter (Fig. 5b, left panel; midgene mutated promoter, P = 0.001; 3′ mutated promoter, P = 0.001; both promoters mutated, P < 0.001, n = 10). In accordance with this reduction of antisense RNA, pilE sense RNA inversely increased with respect to the non-mutated promoter (Fig. 5c, left panel; midgene mutated promoter, P < 0.001; 3′ mutated promoter, P < 0.001; both promoters mutated, P < 0.001, n = 10). When both antisense promoters were mutated, approximately 40 % of pilE antisense RNA remained compared with transcript levels from the non-mutated gene, which may reflect antisense transcription that originates from the downstream opaE locus (Figs 5b & S1).
In the absence of H-NS protein, a more dramatic reduction in antisense RNA is observed between the non-mutated pilE locus and the various pilE promoter mutations (Fig. 5b, right panel; midgene mutated promoter, P < 0.001; 3′ mutated promoter, P < 0.001; both promoters mutated, P < 0.001, n = 10). Again, approximately 40 % pilE antisense RNA remains when both antisense promoters are mutated. Regarding sense expression, a perfect inverse correlation is observed with a non-mutated pilE gene, with a reduced inverse correlation for the various pilE promoter mutation combinations (Fig. 5c, right panel; midgene mutated promoter, P = 0.001; 3′ mutated promoter, P = 0.02; both promoters mutated, P < 0.001, n = 10). Therefore, these data indicate an imperfect inverse correlation between pilE sense and antisense RNAs that manifests most evidently in a WT genetic background. The data also indicate that H-NS protein suppresses transcription from the pilE intragenic antisense promoters.
Temporal changes of pilE sense and antisense RNA
Northern blotting analysis has previously shown that the pilE transcript is not present following growth for 24 h in liquid culture (Wachter et al., 2015). A similar experiment is presented in Fig. 6(a), where pilE transcription was monitored over several time points, and the pilE transcript appeared to accumulate after approximately 12 h of growth with a subsequent decline. In Fig. 6(b), we determined the temporal ratio of antisense : sense RNA levels using strand-specific qRT-PCR analysis. As Ct values inversely reflect the amount of RNA that is present within a sample, then an antisense : sense RNA ratio that increases reflects more sense RNA than antisense RNA within the sample. The results indicate that the antisense : sense RNA ratio varies over time, with sense RNA accumulating within the 6 and 12 h cultures, at which point antisense RNA begins to increase, causing the antisense : sense ratio to drop thereafter. However, a final 1 : 1 ratio is not observed, which may reflect the fact that antisense transcription from the midgene promoter may also be occurring; the design of the experiment would not identify the contribution of this pilE antisense RNA to the sense levels.
Fig. 6.
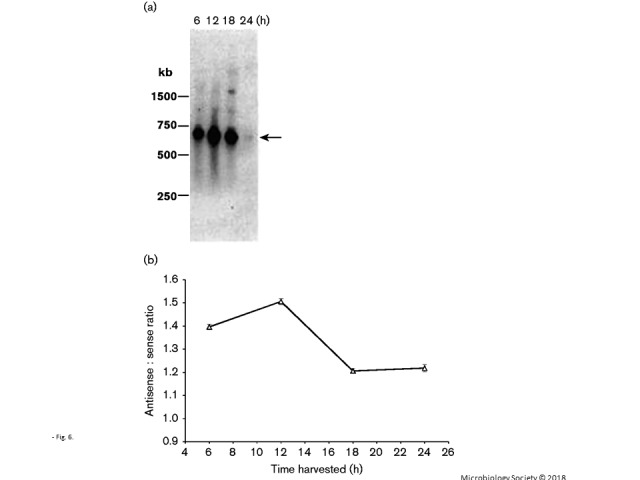
pilE RNA expression over time. (a) Northern blot of total RNA samples isolated from liquid-grown cells at the indicated time points. The total RNAs from each time point were quantified by spectrophotometric analysis and equal amounts of RNA were applied to the gel. The blot was probed with oligonucleotide probe 245, which recognizes pilE sense RNA at the 5′ end (Wachter et al., 2015). The arrow indicates the full-length pilE message. (b) Antisense : sense ratio of pilE RNA. The strand-specific Ct values were determined through qRT-PCR. The initial strand-specific cDNA was made using primer 09172 for antisense expression and primer 09085 for sense expression. As a Ct value is inversely related to the extent of expression, the higher the ratio that is observed, the more sense expression is apparent when compared with antisense expression. Error bars represent mean ± sd; n = 4.
H-NS protein suppresses pilE/pilS recombination
In a WT genetic background, recombinational repair of pilE genes carrying the antisense promoter mutations (which yielded pilus-minus phenotype colonies) appeared to be impeded when examined microscopically on plates, as they exhibited a lower frequency of reversion to the pilus-plus colony phenotype (data not shown), with the apparent phase variation defect being exacerbated in an hns− background (Fig. S3). To further examine this apparent H-NS effect on pilE recombination, a previously used pilE/pilS recombination assay was modified so that it could be quantified by qRT-PCR analysis (Hill et al., 2007; Hill & Davies, 2009). For this analysis, one qRT-PCR primer is specific for pilE, and the other, opposing primer, is specific for a pilS locus. Therefore, in order to obtain a signal, a pilE/pilS recombination event needs to occur. The starting templates that were assessed were either chromosomal DNA extracts (Fig. 7a) or, alternatively, total RNA extracts (Fig. 7b), which were derived from either WT cells or N. gonorrhoeae hns mutant cells. When nucleic acid was isolated from WT cells, little difference was observed for recombination between a non-mutated pilE gene and one that carried the promoter mutations with pilS5 copy 1 (Fig. 7a, b, left panels). In contrast, when using a different pilS-specific primer that targeted pilS6 copy 1, a slight decrease in recombination was observed when the pilE genes carried the antisense promoter mutations (Fig. S4a, b, left panels). For all pilE/pilS recombination experiments, the pilE G4 quartet sRNA (Cahoon & Seifert, 2013) was monitored and was found to be equivalent in each sample (data not shown).
Fig. 7.
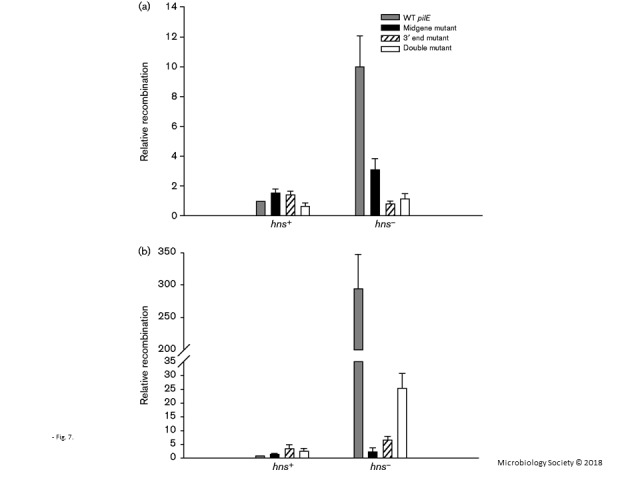
pilE/pilS recombination assay. (a) pilE/pilS recombination assessed at the DNA level. Identical quantities of chromosomal DNA were used to assess pilE/pilS5 recombinants using qRT-PCR. (b) pilE/pilS recombination assessed at the RNA level. Identical amounts of total RNA were used to assess pilE/pilS5 recombinant cDNAs levels using qRT-PCR. The left grouping reflects a WT genetic background (hns+); the right grouping reflects an hns− genetic background. The primers that were used for the recombination assay were the pilE-specific primer (RC3) and the pilS5-specific primer (pilS5c1). The relative level of recombinant RNAs/DNAs was initially standardized against a recA control. Each group was then standardized against WT pilE (hns+), which was set to 1. The data reflect mean ± sd; n = 3.
In contrast to these observations in a WT genetic background, when nucleic acid was prepared from N. gonorrhoeae hns mutant cells, a dramatic increase in recombination between an intact pilE gene and pilS5 copy 1 (Fig. 7a, b, right panels) as well as recombination with pilS6 copy 1 (Fig. 4Sa, b, right panels) was observed. For recombination with pilS5 copy 1, a 10-fold increase in recombination was observed using DNA templates; a 200–300-fold increase was observed in recombination using total RNA extracts (qualitatively similar increases can be seen for recombination with pilS6 copy 1; Fig. S4a, b). When the hns mutation was complemented with a WT hns gene, recombination decreased to levels approaching those observed with WT cells (Fig. S5). In the hns mutants, when recombination was monitored between pilE genes carrying the various antisense promoter mutations and pilS, recombination declined significantly, in contrast to the observations with WT cells [Figs 7a, b (compare the right and left panels) and S4a, b (compare the right and left panels)]. From these results, we conclude; (i) H-NS protein suppresses recombination between pilE and pilS; (ii) in the absence of pilE antisense transcription, pilE/pilS recombination is significantly reduced in cells lacking H-NS protein; and (iii), in a WT background, the absence of pilE antisense transcription may slightly influence pilE/pilS recombination but only in a pilS-dependent fashion.
Discussion
In this study, three additional non-canonical promoters were identified in the gonococcal pilE gene; one yielding sense sRNA, with the other two yielding antisense sRNA. Thus, the pilE transcription profile conforms to the recently described phenomenon known as pervasive transcription, which was also observed at the multiple pilS loci in the gonococcus (Sorek & Cossart, 2010; Wade & Grainger, 2014; Wachter & Hill, 2015; Wachter et al., 2015). Moreover, through mutational analysis of pilE in the gonococcus, an inverse correlation is evident between pilE sense and antisense RNA levels, most noticeably in a WT genetic background. Therefore, pilE antisense RNA may be pairing with the pilE sense transcript, thus allowing double-stranded RNA templates to form, which then may serve as substrates for RNase III degradation. Alternatively, transcriptional interference may also be occurring when the respective RNA polymerase enzymes arising from the converging promoters either collide or pause, depending upon the relative strength of the converging promoters (Callen et al., 2004; Shearwin et al., 2005). Consequently, an added layer of complexity in pilE transcriptional control may have been revealed, where the levels of pilE sense RNA are moderated by concurrent pilE antisense transcription. However, in the absence of a perfect inverse correlation between the two RNA species, other unidentified factors/features may also be involved in modulating pilE transcript levels. A possibility could be that protective stem–loop structures may form at the 5′ ends of both the pilE sense and antisense RNAs, as such structures have been shown to stabilize mRNAs in other organisms (Chen et al., 1991). Indeed, RNA secondary structural algorithms do predict the formation of such loop structures, and, if these 5′ stem–loops form in vivo, then this may also provide additional protection from degradation (T. L. Masters, J. Wachter, J. Mason and S. A. Hill, in preparation).
In a recent study with N. meningitidis, pilE antisense RNA was also detected during the stationary growth phase (as well as under salt stress conditions) from a promoter that is located at the 3′ end of the pilE gene (Tan et al., 2015). However, in contrast to the observations reported herein, this antisense RNA species did not appear to play a role in modulating pilE expression, yet antisense expression was shown to negatively impact PilE antigenic variation. The identified meningococcal antisense promoter was located 6 nt downstream of the pilE stop codon (which corresponds to promoter P1; Fig. S6). When the equivalent nucleotide sequence was mutated in the MS11 pilE7 : 30 : 2 gene as well as another potential promoter sequence (designated P3; located further downstream), no significant changes in the amount of pilE antisense transcripts were observed (data not shown). Instead, it was only by mutating the 3′ P2 promoter (Fig. S6), which is located 12 nt downstream from the stop codon, that significant changes in 3′ antisense transcription were observed compared with WT. Consequently, the 3′ P2 promoter appears to function in the gonococcus and is responsible for transcription of the antisense RNA originating at the 3′ end of pilE, which perhaps indicates either a species-specific difference in promoter usage, or possibly reflects other effects due to variations in the respective pilE DNA sequences.
In the meningococcal study, no correlation was observed between antisense transcription and pilE sense transcript levels (Tan et al., 2015), which contrasts with the analysis with the gonococcal pilE RNAs, as an inverse correlation is evident. The observed difference between the two studies may reflect the contribution that self-priming causes in the initial strand-specific reverse transcription reaction for the qRT-PCR analysis as this was factored out in the current study. Reverse transcription due to self-priming has been extensively reported in studies involving RNA viruses, where it significantly impacts the analysis of strand-specific RNA templates (Haddad et al., 2007; Tuiskunen et al., 2010). Whether reverse transcription as a consequence of self-priming of pilE sense and antisense RNAs impacts gonococcal pilE transcript levels in vivo remains unknown. However, the possibility exists as N. gonorrhoeae strain MS11 has been shown to engage in reverse transcription, even though a reverse transcriptase enzyme has not been identified within the gonococcal genome (Barten & Meyer, 1997). In the absence of such an enzyme, it was postulated that reverse transcription may have arisen through DNA polymerase I operating on RNA templates (Barten & Meyer, 1997).
As H-NS protein has been shown to suppress intragenic transcription in E. coli (Singh et al., 2014), we next explored whether gonococcal H-NS protein also played a similar role. H-NS protein is widely found in many bacterial species (Dillon & Dorman, 2010), where it has been suggested that the presence of the protein impacts genome evolution through its ability to silence transcription from horizontally acquired DNAs (Navarre et al., 2006; Singh et al., 2014). H-NS appears to be a universal transcriptional repressor and has been shown to regulate many genes that encode virulence factors (Navarre et al., 2006). In the current study, gonococcal H-NS protein modulated the pilE antisense transcriptional profiles, with more pilE antisense RNA being apparent in hns mutants than in WT. Also, with the hns mutant, an ordered decrease in the amount of antisense RNA was observed when each of the pilE antisense promoters was mutated. Therefore, these observations indicate that H-NS protein may bind to the pilE intragenic promoters (presumably owing to the AT-richness of these sequences; Fig. 5a) and suppress pilE intragenic antisense transcription.
In WT gonococci, pilE antisense transcription did not appear to significantly impact pilE/pilS recombination, with negative effects not being universally apparent and seemingly dependent on which pilS locus pilE was engaged with during recombination (cf. Figs 7 and S4, WT, left panels). H-NS protein has also been shown to influence homologous recombination (Shiraishi et al., 2007; Sharadamma et al., 2010). For example, in Mycobacterium tuberculosis, H-NS protein was shown to bind with high affinity to Holliday junctions (an important recombination intermediate), and, in doing so, restrained RecA-mediated strand exchange (Sharadamma et al., 2010). The data presented herein show that the hns mutation dramatically influenced the extent of pilE/pilS recombination (Figs 7 and S4), indicating that H-NS protein also influences intragenic recombination in the gonococcus. Therefore, gonococcal H-NS protein can be regarded as a suppressor of pilE/pilS recombination, which complements the positive effect of the G4-associated sRNA regulation of pilE/pilS recombination (Cahoon & Seifert, 2013).
The extent of pilE/pilS recombination was monitored using an assay that utilized either total RNA extracts or isolated chromosomal DNA. When pilE/pilS recombination was assessed using chromosomal DNA, there was an approximate 10-fold increase in recombination when the hns gene was mutated, with the extent of pilE/pilS recombination decreasing when the pilE antisense promoters were individually mutated (Figs 7a and S4a). However, when total RNA extracts of hns mutants were used for the recombination assay, a 200–300-fold increase in recombination was observed (Figs 7b and S4b), suggesting a differential suppressive effect by H-NS protein on transcripts derived from newly recombined pilE genes. A possible explanation for this effect could be that Holliday junctions will naturally form in pilE genes engaged in pilE/pilS recombination, which would allow targeting of recombining pilEs by H-NS protein; if H-NS protein remains bound to the rearranged pilE gene, then transcription of those recombinant genes could be preferentially suppressed, which could account for the dramatic recombination effect that was observed when total RNAs were used for the recombination assay (Figs 7 and S4). Indeed, when the extent of transcription from the pilS5/pilE recombinant genes was measured against overall pilE transcription, H-NS clearly suppressed transcription from the recombinant pilE genes in contrast to WT (Fig. 8; P < 0.001, n = 3). Therefore, besides silencing transcription from horizontally acquired DNA, H-NS protein may preferentially silence transcription from newly rearranged genes that arise by intracellular recombination.
Fig. 8.
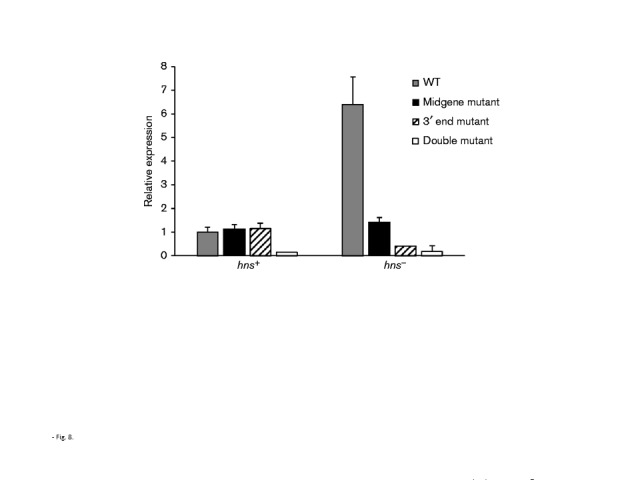
Preferential inhibition of transcription of recombinant genes by H-NS protein. The overall extent of pilE transcription was determined by qRT-PCR analysis using primers RC3/173, which specifically amplify all pilE transcripts in a total RNA sample. The pilE recombinant scores were determined using the primers that were used for the recombination assay, which were the pilE-specific primer (RC3) and the pilS5-specific primer (pilS5c1). The data represent the ratios of the recombinant pilE Ct scores to the overall pilE Ct score. The left grouping reflects a WT genetic background (hns+); the right grouping reflects an hns− genetic background. Each group was also standardized against WT pilE (hns+), which was set to 1. The data reflect mean ± sd; n = 3.
Acknowledgements
The authors thank Scott Grayburn for technical help. The work was supported by NIH grant 1R15 AI072720-01A1 to S. A. H. and the Northern Illinois University's graduate student grant programme.
Supplementary Data
Supplementary Data
Footnotes
Abbreviations: H-NS, histone-like nucleoid structuring protein; IHF, integration host factor; qRT-PCR, quantitative reverse transcriptase PCR; sRNA, small RNA.
Six supplementary figures are available with the online Supplementary Material.
Edited by: R. Lan
References
- Barten R., Meyer T. F. (1997). Neisseria gonorrhoeae reverse transcriptase activity does not mediate pilin gene conversion Mol Microbiol 24665–670 [DOI] [PubMed] [Google Scholar]
- Bergström S., Robbins K., Koomey J. M., Swanson J. (1986). Piliation control mechanisms in Neisseria gonorrhoeae Proc Natl Acad Sci U S A 833890–3894 10.1073/pnas.83.11.3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon L. A., Seifert H. S. (2013). Transcription of a cis-acting, noncoding, small RNA is required for pilin antigenic variation in Neisseria gonorrhoeae PLoS Pathog 9e1003074. 10.1371/journal.ppat.1003074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callen B. P., Shearwin K. E., Egan J. B. (2004). Transcriptional interference between convergent promoters caused by elongation over the promoter Mol Cell 14647–656 10.1016/j.molcel.2004.05.010. [DOI] [PubMed] [Google Scholar]
- Chen L. H., Emory S. A., Bricker A. L., Bouvet P., Belasco J. G. (1991). Structure and function of a bacterial mRNA stabilizer: analysis of the 5′ untranslated region of ompA mRNA J Bacteriol 1734578–4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich M., Munke R., Gottschald M., Ziska E., Boettcher J. P., Mollenkopf H., Friedrich A. (2009). The effect of hfq on global gene expression and virulence in Neisseria gonorrhoeae FEBS J 2765507–5520 10.1111/j.1742-4658.2009.07234.x. [DOI] [PubMed] [Google Scholar]
- Dillon S. C., Dorman C. J. (2010). Bacterial nucleoid-associated proteins, nucleoid structure and gene expression Nat Rev Microbiol 8185–195 10.1038/nrmicro2261. [DOI] [PubMed] [Google Scholar]
- Fyfe J.A.M., Davies J. K. (1998). An AT-rich tract containing an integration host factor-binding domain and two UP-like elements enhances transcription from the pilEp1 promoter of Neisseria gonorrhoeae J Bacteriol 1802152–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyfe J.A.M., Carrick C. S., Davies J. K. (1995). ThepilE gene of Neisseria gonorrhoeae MS11 is transcribed from a σ70 promoter during growth in vitro J Bacteriol 1773781–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad F., Qin A. X., Giger J. M., Guo H., Baldwin K. M. (2007). Potential pitfalls in the accuracy of analysis of natural sense-antisense RNA pairs by reverse transcription-PCR BMC Biotechnol 721. 10.1186/1472-6750-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagblom P., Segal E., Billyard E., So M. (1985). Intragenic recombination leads to pilus antigenic variation in Neisseria gonorrhoeae Nature 315156–158 10.1038/315156a0. [DOI] [PubMed] [Google Scholar]
- Hill S. A., Davies J. K. (2009). Pilin gene variation in Neisseria gonorrhoeae: reassessing the old paradigms FEMS Microbiol Rev 33521–530 10.1111/j.1574-6976.2009.00171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill S. A., Morrison S. G., Swanson J. (1990). The role of direct oligonucleotide repeats in gonococcal pilin gene variation Mol Microbiol 41341–1352 10.1111/j.1365-2958.1990.tb00713.x. [DOI] [PubMed] [Google Scholar]
- Hill S. A., Samuels D. S., Carlson J. H., Wilson J., Hogan D., Lubke L., Belland R. J. (1997). Integration host factor is a transcriptional cofactor of pilE in Neisseria gonorrhoeae Mol Microbiol 23649–656 10.1046/j.1365-2958.1997.2321612.x. [DOI] [PubMed] [Google Scholar]
- Hill S. A., Woodward T., Reger A., Baker R., Dinse T. (2007). Role for the RecBCD recombination pathway for pilE gene variation in repair-proficient Neisseria gonorrhoeae J Bacteriol 1897983–7990 10.1128/JB.00980-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieva R., Alaimo C., Delany I., Spohn G., Rappuoli R., Scarlato V. (2005). CrgA is an inducible LysR-type regulator of Neisseria meningitidis, acting both as a repressor and as an activator of gene transcription J Bacteriol 1873421–3430 10.1128/JB.187.10.3421-3430.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthias K. A., Rest R. F. (2014). Control of pili and sialyltransferase expression in Neisseria gonorrhoeae is mediated by the transcriptional regulator CrgA Mol Microbiol 911120–1135 10.1111/mmi.12522. [DOI] [PubMed] [Google Scholar]
- Sharadamma N., Harshavardhana Y., Singh P., Muniyappa K. (2010). Mycobacterium tuberculosis nucleoid-associated DNA-binding protein H-NS binds with high-affinity to the Holliday junction and inhibits strand exchange promoted by RecA protein Nucleic Acids Res 383555–3569 10.1093/nar/gkq064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearwin K. E., Callen B. P., Egan J. B. (2005). Transcriptional interference – a crash course Trends Genet 21339–345 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi K., Ogata Y., Hanada K., Kano Y., Ikeda H. (2007). Roles of the DNA binding proteins H-NS and StpA in homologous recombination and repair of bleomycin-induced damage in Escherichia coli Genes Genet Syst 82433–439 10.1266/ggs.82.433. [DOI] [PubMed] [Google Scholar]
- Singh S. S., Grainger D. C. (2013). H-NS can facilitate specific DNA-binding by RNA polymerase in AT-rich gene regulatory regions PLoS Genet 9e1003589. 10.1371/journal.pgen.1003589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S. S., Singh N., Bonocora R. P., Fitzgerald D. M., Wade J. T., Grainger D. C. (2014). Widespread suppression of intragenic transcription initiation by H-NS Genes Dev 28214–219 10.1101/gad.234336.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R., Cossart P. (2010). Prokaryotic transcriptomics: a new view on regulation, physiology and pathogenicity Nat Rev Genet 119–16 10.1038/nrg2695. [DOI] [PubMed] [Google Scholar]
- Swanson J. (1982). Colony opacity and protein II compositions of gonococci Infect Immun 37359–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson J., Barrera O. (1983). Gonococcal pilus subunit size heterogeneity correlates with transitions in colony piliation phenotype, not with changes in colony opacity J Exp Med 1581459–1472 10.1084/jem.158.5.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan F.Y.Y., Wörmann M. E., Loh E., Tang C. M., Exley R. M. (2015). Characterization of a novel antisense RNA in the major pilin locus of Neisseria meningitidis influencing antigenic variation J Bacteriol 1971757–1768 10.1128/JB.00082-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuiskunen A., Leparc-Goffart I., Boubis L., Monteil V., Klingström J., Tolou H. J., Lundkvist A., Plumet S. (2010). Self-priming of reverse transcriptase impairs strand-specific detection of dengue virus RNA J Gen Virol 911019–1027 10.1099/vir.0.016667-0. [DOI] [PubMed] [Google Scholar]
- Wachter J., Hill S. A. (2015). Small transcriptome analysis indicates that the enzyme RppH influences both the quality and quantity of sRNAs in Neisseria gonorrhoeae FEMS Microbiol Lett 3621–7 10.1093/femsle/fnu059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachter J., Masters T. L., Wachter S., Mason J., Hill S. A. (2015). pilS loci in Neisseria gonorrhoeae are transcriptionally active Microbiology 1611124–1135 10.1099/mic.0.000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade J. T., Grainger D. C. (2014). Pervasive transcription: illuminating the dark matter of bacterial transcriptomes Nat Rev Microbiol 12647–653 10.1038/nrmicro3316. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data


