Abstract
Aim:
This study aims to evaluate the genetic and population structure of Vibrio parahaemolyticus in the major coastal regions of China.
Materials & methods:
Multilocus sequence typing was performed.
Results:
Insertion of large sequence into recA happened in nearly 30 strains, which were untypeable by multilocus sequence typing. A collection of 307 V. parahaemolyticus isolates were typed into 160 sequence types, including 117 novel ones. eBURST analysis revealed five clonal complexes, 11 doublets, and 108 singletons. The 160 sequence types formed two main lineages in the phylogenetic analysis.
Conclusion:
V. parahaemolyticus along the Chinese coastal regions exhibits high levels of genetic diversity and has undergone significant purifying selection and frequent recombination. A deeper understanding of V. parahaemolyticus genetic diversity could be obtained at the level of genome sequences.
Keywords: : China, genetic diversity, MLST, population structure, Vibrio parahaemolyticus
Vibrio parahaemolyticus is a major food-borne pathogen that widely inhabits estuarine and marine environments [1,2]. It is one of the major causes of food poisoning in China [3–6]. Outbreaks are mainly caused by the consumption of V. parahaemolyticus- contaminated raw or undercooked seafood [7], which causes gastroenteritis, and even wound infection or septicemia [2,8]. Virulence factors of this species are associated with thermostable direct hemolysin, TDH-related hemolysin and two type 3 secreation systems [9,10]. A high level of genetic diversity of V. parahaemolyticus has been identified by various molecular typing methods, such as pulsed-field Gel electrophoresis and multilocus sequence typing (MLST) [11]. Pulsed-field Gel electrophoresis is a highly discriminative molecular technique to type V. parahaemolyticus strains from food-poisoning outbreaks [12] and nosocomial outbreaks [13], and also implied to differentiate pandemic and nonpandemic V. parahaemolyticus strains. However, the highly discriminatory nature of this method may cover the relationship in closely related clusters, and it is likely to yield untypeable results as a result of DNA degradation [14]. MLST is a typing method based on several housekeeping genes that can provide insights into epidemiology and population structure, and allows a continuous understanding of the molecular epidemiology and evolution of the typed bacteria because the comparison of results from different laboratories and the exchange via public databases are feasible [15].
The present work evaluated a collection of 307 V. parahaemolyticus strains isolated from the coastal regions of the Liaoning, Zhejiang and Guangdong provinces of China, and depicted the distribution of MLST sequence types (STs) in these three regions and their population structure.
Materials & methods
Bacterial strains
A total of 307 V. parahaemolyticus strains (Supplementary table 1) included 57 (isolated between 2006 and 2016) from Liaoning Entry-Exit Inspection and Quarantine Bureau, China, 152 (isolated between 2009 and 2012) from Zhejiang Center for Disease Control and Prevention (Zhejiang CDC), China, and 97 (isolated between 2013 and 2014) from Guangzhou Center for Disease Control and Prevention (Guangzhou CDC), China. ATCC17802 was provided by Fujian Entry-Exit Inspection and Quarantine Bureau. These 307 strains were composed of 89 clinical isolates, 103 nonclinical strains (45 from seawater, 43 from seafood and 15 from other environment samples) and 115 strains of unknown origins. All isolates were inoculated on heart infusion agar overnight. Chromosomal DNA was extracted using the phenol/chloroform method plus methoxyethanol to remove polysaccharides. Some of the DNA was prepared using Blood & Cell Culture DNA Maxi Kit (Qiagen, Germany). The genomic DNA was further arrayed in 96-well plates and used for PCR templates.
PCR amplification & sequencing
Four of seven housekeeping genes were amplified using the primers presented on the V. parahaemolyticus MLST website (www.pubmlst.org/vparahaemolyticus). For the other three loci, we designed new primers with better amplification than suggested ones (Supplementary table 2). The PCR mixture with a volume of 50 μl contained 50 mM KCl, 10 mM Tris-HCl (pH8.0), 2.5 mM MgCl2, 0.001% gelatin, 0.1% BSA, 100 μl of each of dATP, dTTP, dCTP and dGTP, 0.1 μM of each primer, two units of ExTaq polymerase (TaKaRa), and 5 ng of template DNA. PCR conditions were as follows: predenaturation at 95°C for 5 min; 30 cycles of 94°C for 50 s, an appropriate annealing temperature for 50 s, and 72°C for 80 s; with a final extension step of 72°C for 7 min. The PCR products were tested by 1.5% agarose gel electrophoresis with Gold-view staining. Products were bidirectionally sequenced with PCR primers on an ABI-3700 sequencer. DNA sequences were aligned using Contig-Express, bioedit, and MUSCLE 3.8.31.
Sequence diversity
The G+C content, single-nucleotide polymorphic sites, and difference per site (π) for individual loci and concatenated sequences were calculated using DnaSP 5.10 [16]. The ratio of nonsynonymous/synonymous rates (dN/dS) was calculated using KaKs Calculator Version 2.0 [17], and the first base of open reading frame for each allele was designated using V. parahaemolyticus RIMD2210633 whole-genome sequences (accession number NC_004603 and NC_004605 for chromosome I and II, respectively) [9] as the reference.
Allelic diversity analysis
The concatenated sequence for each strain with a final sequence of 3682 bp (in the following order: dnaE, gyrB, recA, dtdS, pntA, pyrC and tnaA) was used as a query against the V. parahaemolyticus PubMLST database, to determine its allelic profile (AP) and ST (Supplementary table 1). Novel alleles were submitted to PubMLST. Each distinct allele sequence within a locus was assigned a different allele number, and different STs were composed of distinct AP. Cluster analysis of APs was carried out using eBURST version 3 (http://eburst.mlst.net/) with the most stringent definition, where a member of a group must share six of seven loci with at least one other member of the group. A clonal complex (CC) comprised at least three STs with a single-locus variant.
The phylogenetic analysis
The concatenated sequences of each of the STs were aligned using MUSCLE 3.8.31, and the unrooted neighbor-joining trees of STs were generated from the indicative aligned sequences using MEGA 7 by the bootstrapping method (1000 replicates). The Bayesian Markov chain Monte Carlo (MCMC) method in STRUCTURE software version 2.3 was implemented to predict the ancestry of 160 STs. K stood for the number of genotypes of sampled isolates. In this study, K was set from 2 to 10 with 20,000 burn-in and 100,000 MCMC updates. The value of 3 was chosen as the appropriate ancestry number. The split network of 160 STs was generated using SplitsTree version 4.0 with the neighbor-net method.
Recombination analysis
The phi test for recombination was performed with SplitsTree version 4.0, and a p-value <0.05 indicates the occurrence of recombination. The ‘standardized’ index of association (st. I A) for the AP was calculated using The Linkage Analysis version 3.6 with 10,000 iterations in Monte Carlo mode. The LDhat program of the RDP4 package was used to calculate the mean per site recombination (rho)/mutation (theta) ratios based on concatenated sequences of STs with 1,000,000 MCMC updates, and the parameter of rho/theta ratio was used as an index of the probability that a given site was altered by recombination or mutation.
Results
Insufficient amplification of recA
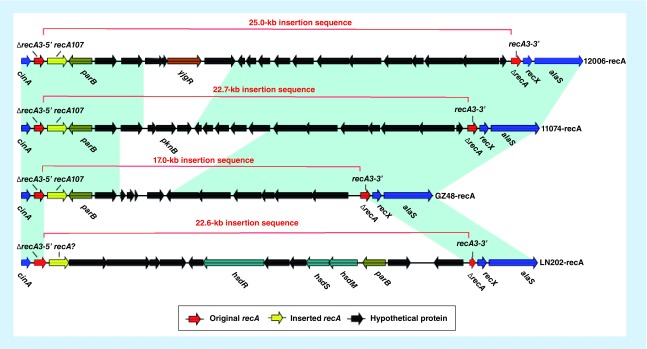
Amplification of recA using the primers described on the V. parahaemolyticus MLST website resulted in no amplification or products larger than expected 773 bp for a collection 30 V. parahaemolyticus strains in this work. We chose four of these 30 strains to obtain their draft genome sequences (data not shown). It was found that all these four selected strains had large DNA fragments (17–25 kb in length), which contained a non-V. parahaemolyticus recA gene (recA107 and recA? in Figure 1) and inserted into the original V. parahaemolyticus recA gene (recA3 in Figure 1), interrupting the original recA into two separate parts (recA3-5′ and recA3-3′ in Figure 1). These large inserts were mostly the same, with the exception of strain LN202. These 30 strains were not included in the MLST analysis.
Figure 1. . A linear comparison of inserted sequences in four V. parahaemolyticus strains.
Genes are denoted by arrows. Genes are colored based on function classification. The original V. parahaemolyticus recA genes are shown in red. Shading denotes regions of homology (>95% nucleotide identity).
Genetic diversity & purifying selection at each locus
A total of 307 strains of V. parahaemolyticus were typed into 160 STs, including 117 STs newly found in this study (Supplementary table 1). There were 24, 23 and 71 new STs among the 47, 36 and 79 STs of the analyzed isolates in Liaoning, Zhejiang and Guangzhou provinces, respectively (Table 1). Out of the 160 STs, ST3 contained the largest number of strains (104 of 307), followed by ST189 containing ten strains, while there are 134 STs contained only one strain.
Table 1. . Properties of each locus and sequence type.
| Regions | Number and proportion of alleles/STs | Number and proportion of new alleles/STs | Whole | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Liaoning (%) | Zhejiang (%) | Guangzhou (%) | Liaoning (%) | Zhejiang (%) | Guangzhou (%) | Total | New | |||||||
| dnaE | 38 | (51.4) | 20 | (27.0) | 39 | (52.7) | 4 | (33.3) | 1 | (8.3) | 7 | (58.3) | 74 | 12 |
| gyrB | 38 | (43.7) | 26 | (29.9) | 40 | (46.0) | 4 | (26.7) | 2 | (13.3) | 9 | (60) | 87 | 15 |
| recA | 38 | (50.0) | 23 | (30.3) | 35 | (46.1) | 2 | (20) | 3 | (30) | 5 | (50) | 76 | 10 |
| dtdS | 37 | (48.1) | 29 | (37.7) | 38 | (49.4) | 7 | (43.8) | 3 | (18.8) | 5 | (31.3) | 77 | 15 |
| pntA | 31 | (58.5) | 21 | (39.6) | 25 | (47.2) | 1 | (25) | 0 | (0) | 3 | (75) | 53 | 4 |
| pyrC | 36 | (47.4) | 29 | (38.2) | 39 | (51.3) | 2 | (22.2) | 4 | (44.4) | 3 | (33.4) | 76 | 9 |
| tnaA | 28 | (53.8) | 23 | (44.2) | 29 | (55.8) | 2 | (40) | 1 | (20) | 2 | (40) | 52 | 5 |
| STs | 47 | (29.4) | 36 | (22.5) | 79 | (49.4) | 24 | (20.5) | 23 | (19.7) | 71 | (60.7) | 160 | 117 |
ST: Sequence type.
The gyrB locus had the largest number of alleles (n = 87), while tnaA had the smallest (n = 52). The dtdS and gyrB alleles had the most new alleles (n = 16). Among the three regional subsets, Guangzhou had the largest number of STs and new STs, although Zhejiang had the most isolates. Regarding the number and proportion of alleles of dnaE, gyrB, tnaA, dtdS and pyrC, the value for Guangzhou was higher than those for Liaoning and Zhejiang, while Liaoning had higher value than the other two regarding the number and proportion for recA and pntA. However, Guangzhou still contributed the majority of new alleles (Table 1).
Table 2 presented the nucleotide and allelic diversity of seven housekeeping genes. There were 374 SNPs in concatenated sequences, ranging in number from 40 (pntA) to 70 (recA) for seven loci. The nucleotide diversity per site of these seven genes ranged from 0.01143 (pntA) to 0.02483 (dtdS). The dN/dS ratios (ratios of nonsynonymous to synonymous evolutionary changes) for all of them were <1. Therefore, all of the loci seemed to have been under purifying selection.
Table 2. . Nocleotide and allelic sequences diversity.
| Locus | Length (bp) | Number of alleles/STs | Number of new alleles/STs (%) | Average G+C content % | Number of SNPs (%) | Average dN/dS ratio | π |
|---|---|---|---|---|---|---|---|
| dnaE | 557 | 74 | 12 (16.22) | 48.6 | 55 (9.87) | 0.022 | 0.01191 |
| gyrB | 592 | 87 | 15 (17.24) | 47.6 | 63 (10.64) | 0.008 | 0.01500 |
| recA | 729 | 76 | 10 (13.16) | 45.2 | 70 (9.60) | 0.007 | 0.02252 |
| dtdS | 458 | 77 | 16 (20.78) | 50.1 | 51 (11.14) | 0.004 | 0.02483 |
| pntA | 430 | 53 | 4 (7.55) | 43.9 | 40 (9.30) | 0.064 | 0.01143 |
| pyrC | 493 | 76 | 9 (11.84) | 48.3 | 54 (10.95) | 0.119 | 0.01357 |
| tnaA | 423 | 52 | 5 (9.62) | 48.8 | 41 (9.69) | 0.049 | 0.01212 |
| Concatenated | 3682 | 160 | 117 (73.13) | 47.4 | 374 (10.16) | 0.039 | 0.01492 |
SNP: Single nucleotide polymorphism; ST: Sequence type.
Population of snapshots
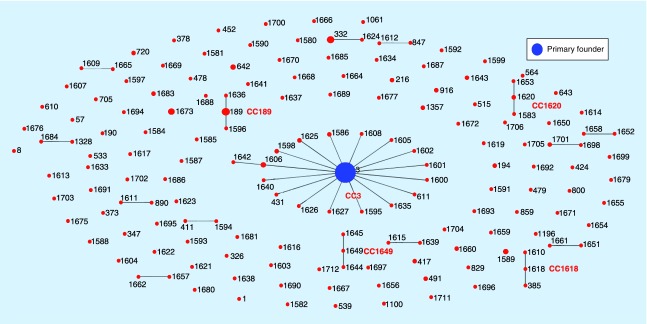
The result of the CC analysis with eBURST v3.0 software was shown in Figure 2. Overall, The 307 strains (160 STs) were clustered into five CCs (CC3, C189, CC1618, CC1649 and CC1620), 11 doublets and 108 singletons. The predominant CC was CC3, which contained 18 STs; moreover, ST3 was defined as the founder of this CC. CC3 had the largest number of isolates (126/307) and ST3 was the predominant ST among all the isolates (104/307).
Figure 2. . V. parahaemolyticus population snapshot obtained using eBURST v3 with stringent criteria (6/7 shared alleles).
STs that are SLVs of each other are shown connected by black lines. The sizes of the circles are related to the number of strains within each ST. The primary founder is shown in blue.
SLV: Single Locus variant; ST: Sequence type.
CC189 was composed of three STs (12 strains), namely, ST189, ST1636 and ST1596. ST1636 and ST1596 were newly classified in our study and ST189 was the predominant ST, with 10 of 12 strains in this CC. ST385, ST1610 and ST1618 were clustered together in CC1618, while ST1583, ST1620, and ST1653 were included in CC1620. For CC1649, it was composed of ST1644, ST1645 and ST1649.
Predominant recombination & linkage disequilibrium
The p-values of the phi test for comparisons within and across the three geographic regions were <0.0001, indicating that recombinant events had occurred within and across those regions. The probabilities of rho/theta values of the strains within and across the three geographic regions were all above 1, suggesting that recombination was more likely to have occurred than mutation during the evolution of the strains. The rho/theta values of each geographic region were 35.31, 50.88 and 27.45 for Liaoning, Zhejiang, and Guangzhou provinces, respectively, all of which are higher than the value for the whole population. The p-values of st. I A of all STs and STs within the three geographic regions were all <0.0001, indicating the linkage disequilibrium of these alleles (Table 3).
Table 3. . Recombination test and estimation.
| Population (n) | ST (New ST) | phi | Recombination | Linkage disequilibrium | |||||
|---|---|---|---|---|---|---|---|---|---|
| theta/site | rho/site | LB 95% | UP 95% | rho/theta | IA | p-value | |||
| Whole | 160 (117) | <0.001 | 1.63E-02 | 2.89E-01 | 1.49E-01 | 5.57E-01 | 17.69 | 0.2327 | <0.0001 |
| Liaoning | 47 (24) | <0.001 | 1.48E-02 | 5.21E-01 | 2.07E-01 | 11.62E-01 | 35.31 | 0.0915 | <0.0001 |
| Zhejiang | 36 (23) | <0.001 | 1.34E-02 | 1.98E-01 | 1.19E-01 | 3.20E-01 | 50.88 | 0.1239 | <0.0001 |
| Guangzhou | 79 (71) | <0.001 | 1.36E-02 | 3.73E-01 | 1.21E-01 | 8.45E-01 | 27.45 | 0.4133 | <0.0001 |
ST: Sequence type.
Two distinct lineages in the whole population
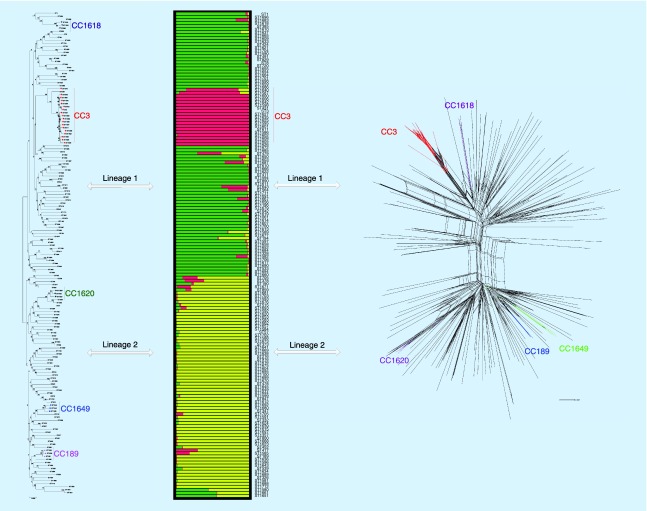
Phylogenetic analysis of 160 STs was conducted using concatenated sequences, and an Neighbor Joining (NJ) tree was built with no root (Figure 3A).The 160 STs were clustered into two lineages, the bootstrap values were low. Each branch showed a mixture in terms of the geographical origins of its constituents. An NJ tree of all STs (1712 STs) submitted to the MLST database of V. parahaemolyticus was also constructed (Supplementary figure 1) (last accessed on 5 March 2017), and the 160 STs in this study were well distributed. Therefore, the 160 STs in this study collected from three regions of China could reflect the main population structure of the studied species.
Figure 3. . Diagrams denoting the population structure.
(A) The unrooted NJ tree of the 160 STs based on the concatenated sequences of seven loci formed two lineages (Lineage 1, 2) with low bootstrap support. (B) Two main populations of the 160 STs and different colors represent distinct populations corresponding to Lineage 1 and 2. (C) Splits network of the 128 STs generated by the neighbor-net method using SplitsTree4 based on the concatenated sequences of seven loci. Five clonal complexes defined by eBURST v3 are highlighted in the three diagrams in different colors.
NJ: Neighbor Joining; ST: Sequence type;.
Phylogenetic analysis was applied to the dataset of the 160 STs’ sequences. The number of genotypes K was set from 2 to 10, and K = 3 was chosen for its maximal posterior probability (Figure 3B). The 160 STs were separated into two main groups, which corresponded to Lineages 1 and 2 observed in the phylogenetic analysis, which were distinct from each other with low admixture. Notably, ST1587 and all STs in CC3 (in red in the figure) formed a semiclonal structure existed within Lineage 1 conservatively (in green).
A splits network of the 160 STs was created using SplitsTree4 by the neighbor-net method. The 160 STs of the 307 strains were divided into the above two lineages (Figure 3C). STs between the two lineages showed strong interaction and each lineage displayed a very complex network. CC3 and CC1618 were included in Lineage 1, while CC189, CC1620 and CC1649 were in Lineage 2.
Discussion
Previous studies already used MLST analysis to characterize V. parahaemolyticus strains, and some researchers also evaluated the epidemiology of this pathogen in China [18]. The strains used in some studies had a global distribution [19,20], with a focus being placed on both clinical and environmental isolates [21]. Considering the high prevalence of V. parahaemolyticus in China, our study evaluated this species in three main coastal regions from north to south in China, with sufficient numbers of isolates to achieve a representative overall assessment. Moreover, the isolates in this study were mainly collected from 2011 to 2016, making them suitable to assess the recent conditions of this species. The 117 new STs, which have been submitted to the MLST database, will enable further analyses to keep pace with new evolutionary trends of this species in China.
Frequent horizontal recombination occurred in recA
Previous studies identified frequent interspecies horizontal gene transfer and intragenetic recombination at the recA locus [21], which play a great role in the apparent evolutionary classification of V. parahaemolyticus [20]. Narjol González-Escalona [22] also identified that an event involving the insertion of a large fragment occurred in V. parahaemolyticus, which disrupted the original recA gene and substituted it with a non-V. parahaemolyticus recA. Some of the strains in our study were untypable due to similar large sequence insertions, indicating that horizontal gene transfer of V. parahaemolyitcus is frequent at a global scale. MLST may not be suitable to track the routes of dispersion of V. parahaemolyticus strains with large fragment insertion into recA. By contrast, whole genome analysis represents a better method to delineate the routes and mechanisms of dispersion of these strains.
High genetic diversity of V. parahaemolyticus
The collection of 307 strains in this study was typed into 160 STs, 117 of which were newly found, indicating the high genetic diversity of this species. The majority of the identified STs were recovered once, matching the results obtained in other studies [19,21]. The high proportion of new STs can be explained by frequent recombination, especially in environmental strains [23,24]. The p-value of st. IA was significantly different from zero, indicating that the alleles were not generally randomly distributed, but that recombination could occur within different subpopulations [11]. The gyrB locus was the most diverse, with 87 alleles, while possessing only 63 (10.6%) variable sites. This is consistent with the findings of previous studies [18,19]. The dN/dS values of each locus were all lower than 1.0, revealing that these loci had been subjected to purifying selection [24].
ST distribution & epidemic population structure
All of the 160 STs analyzed in this study were independently distributed among the three geographic regions, except that ST3 and ST1586 were recovered in two regions: Zhejiang and Guangdong. ST3 is the most common ST, which matches the findings of previous studies performed on a global scale [20]. The distinct distribution of STs between those regions suggests that some environmental factors are associated with the prevalence and distribution of V. parahaemolyticus, such as water temperature and salinity [25].
The population structure of this pathogen indicates that it is an epidemic population [24,26,27], in which recombination has occurred much more frequently than mutation. The trees produced by the phylogenetic analysis of V. parahaemolyticus were usually star-like, matching the result obtained in previous studies, and the distribution of STs was found to be independent of the geographic origin [19]. However, for the population analysis, 307 strains of 160 STs were divided into two main lineages, indicating that there are some differences between the two lineages that are unrelated to their regions.
Both clinical and environmental populations were illustrated to have a semiclonal structure, which clustered together with high homogeneity [28]. In this study, ST1587 and all of the STs in CC3 existed independently within one lineage and formed a semiclonal population structure, as determined by STRUCTURE software. The large proportion of the clinical isolates in this semiclone suggests that environmental isolates are more heterogeneous than clinical ones. Some environmental isolates could potentially have become virulent by acquiring virulence genes, which may explain their heterogeneity [29].
Conclusion & future perspective
A collection of 307 V. parahaemolyticus strains isolated from three major coastal regions (Liaoning, Zhejiang and Guangzhou) in China were typed into 160 STs, and notably, 117 of them were newly found in this study. All of 160 STs showed high genetic diversity and had undergone purifying selection and significant recombination. STs were distributed distinctly in these three geographic regions, and formed two main lineages, which were unrelated to their geographical origins, in the phylogenetic analysis. Horizontal recombination and large sequence insertion frequently occurred in recA, which usually resulted in limited application of MLST in the epidemiological analysis of V. parahaemolyticus. High genetic diversity of V. parahaemolyticus required more suitable and highly discriminative methods, such as genome sequencing, to obtain a comprehensive understanding of them and, moreover, continuous studies and routine surveillance of the genetic diversity of this species are needed in global scale.
Summary points.
This study presented the genetic diversity and population structure of V. parahaemolyticus in three major coastal regions of China using multilocus sequence typing (MLST).
In total, 160 sequence types (STs) were identified, and 117 of them were newly typed in this study, and the predominant ST was ST3.
The 160 STs could be assigned into five clonal complex (CCs) (CC3, C189, CC1618, CC1649 and CC1620), 11 doublets and 108 singletons. CC3 was the predominant Clonal Complex.
The 307 strains (160 STs) formed two distinct lineages, which were unrelated to geographical origins, in the phylogenetic analysis.
All STs in CC3 and ST1587 showed higher homogeneity than other STs in Lineage 1, because they formed a semiclone in this lineage.
V. parahaemolyticus showed high levels of genetic diversity and had undergone purifying selection and significant recombination.
MLST acts as a quick and feasible method to assess the epidemiological relatedness of V. parahaemolyticus isolates. However, as for the strains containing insertion of large sequence into anyone of the MLST loci, genome sequence analysis is more suitable than MLST.
Supplementary Material
Acknowledgements
The authors would like to thank J Cao from Liaoning Entry-Exit Inspection and Quarantive Bureau, X Wu from Guangzhou Centre for Disease Control and Prevention and D Jin from Zhejiang Province Center for Disease Control and Prevention for supplying a large number of strains of V. parahaemolyticus. We thank L Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Footnotes
Supplementary data
To view the supplementary data that accompany this paper please visit the journal website at: www.futuremedicine.com/doi/full/10.2217/fmb-2018-0060
Author's Contributions
W Yang and J Wang conceived and designed the experiments; P Li, W Xin and L Kang performed the experiments; P Li, W Xin, Z Chen, C Guo analyzed the data; S Gao, H Yang, B Ji, D Zhou, Y Yan and H Wang contributed reagents/materials/analysis tools; P Li and W Xin wrote the paper. All authors have read and approved the final version of the manuscript.
Financial & competing interests disclosure
This work was supported by the National Hi-Tech Research and Development (863) Program of China (grant number 2014AA021402). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Writing assistance was utilized in theproduction of this manuscript. Writing assistance was provided by Edanz Group China.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Open access
This work is licensed under the Attribution-NonCommercial-NoDerivatives 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
References
- 1.Su YC, Liu C. Vibrio parahaemolyticus: a concern of seafood safety. Food Microbiol. 2007;24(6):549–558. doi: 10.1016/j.fm.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Zhang L, Orth K. Virulence determinants for Vibrio parahaemolyticus infection. Curr. Opin. Microbiol. 2013;16(1):70–77. doi: 10.1016/j.mib.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Chen Y, Liu XM, Yan JW, et al. Foodborne pathogens in retail oysters in south China. Biomed. Environ. Sci. 2010;23(1):32–36. doi: 10.1016/s0895-3988(10)60028-1. [DOI] [PubMed] [Google Scholar]
- 4.Ma C, Deng X, Ke C, et al. Epidemiology and etiology characteristics of foodborne outbreaks caused by Vibrio parahaemolyticus during 2008-2010 in Guangdong province, China. Foodborne Pathog. Dis. 2014;11(1):21–29. doi: 10.1089/fpd.2013.1522. [DOI] [PubMed] [Google Scholar]
- 5.Zhang C, Hu X, Luo J, et al. Degradation dynamics of glyphosate in different types of citrus orchard soils in China. Molecules. 2015;20(1):1161–1175. doi: 10.3390/molecules20011161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qi XL, Wang HX, Bu SR, Xu XG, Wu XY, Lin DF. Incidence rates and clinical symptoms of Salmonella, Vibrio parahaemolyticus, and Shigella infections in China, 1998-2013. J. Infect. Dev. Ctries. 2016;10(2):127–133. doi: 10.3855/jidc.6835. [DOI] [PubMed] [Google Scholar]
- 7.Pal D, Das N. Isolation, identification and molecular characterization of Vibrio parahaemolyticus from fish samples in Kolkata. Eur. Rev. Med. Pharmacol. Sci. 2010;14(6):545–549. [PubMed] [Google Scholar]
- 8.Daniels NA, Mackinnon L, Bishop R, et al. Vibrio parahaemolyticus infections in the United States, 1973-1998. J. Infect. Dis. 2000;181(5):1661–1666. doi: 10.1086/315459. [DOI] [PubMed] [Google Scholar]
- 9.Makino K, Oshima K, Kurokawa K, et al. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V cholerae. Lancet. 2003;361(9359):743–749. doi: 10.1016/S0140-6736(03)12659-1. [DOI] [PubMed] [Google Scholar]
- 10.Zhang L, Krachler AM, Broberg CA, et al. Type III effector VopC mediates invasion for Vibrio species. Cell Rep. 2012;1(5):453–460. doi: 10.1016/j.celrep.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzalez-Escalona N, Martinez-Urtaza J, Romero J, Espejo RT, Jaykus LA, Depaola A. Determination of molecular phylogenetics of Vibrio parahaemolyticus strains by multilocus sequence typing. J. Bacteriol. 2008;190(8):2831–2840. doi: 10.1128/JB.01808-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong HC, Liu SH, Ku LW, et al. Characterization of Vibrio parahaemolyticus isolates obtained from foodborne illness outbreaks during 1992 through 1995 in Taiwan. J. Food Prot. 2000;63(7):900–906. doi: 10.4315/0362-028x-63.7.900. [DOI] [PubMed] [Google Scholar]
- 13.Lu PL, Chang SC, Pan HJ, Chen ML, Luh KT. Application of pulsed-field gel electrophoresis to the investigation of a nosocomial outbreak of Vibrio parahaemolyticus. J. Microbiol. Immunol. Infect. 2000;33(1):29–33. [PubMed] [Google Scholar]
- 14.Marshall S, Clark CG, Wang G, Mulvey M, Kelly MT, Johnson WM. Comparison of molecular methods for typing Vibrio parahaemolyticus. J. Clin. Microbiol. 1999;37(8):2473–2478. doi: 10.1128/jcm.37.8.2473-2478.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maiden MC, Bygraves JA, Feil E, et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl Acad. Sci. USA. 1998;95(6):3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11):1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 17.Wang D, Zhang Y, Zhang Z, Zhu J, Yu J. KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinformatics. 2010;8(1):77–80. doi: 10.1016/S1672-0229(10)60008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han D, Tang H, Ren C, Wang G, Zhou L, Han C. Prevalence and genetic diversity of clinical Vibrio parahaemolyticus isolates from China, revealed by multilocus sequence typing scheme. Front. Microbiol. 2015;6:291. doi: 10.3389/fmicb.2015.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Urmersbach S, Alter T, Koralage MS, et al. Population analysis of Vibrio parahaemolyticus originating from different geographical regions demonstrates a high genetic diversity. BMC Microbiol. 2014;14:59. doi: 10.1186/1471-2180-14-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han D, Tang H, Lu J, et al. Population structure of clinical Vibrio parahaemolyticus from 17 coastal countries, determined through multilocus sequence analysis. PLoS ONE. 2014;9(9):e107371. doi: 10.1371/journal.pone.0107371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Theethakaew C, Feil EJ, Castillo-Ramirez S, et al. Genetic relationships of Vibrio parahaemolyticus isolates from clinical, human carrier and environmental sources in Thailand, determined by multilocus sequence analysis. Appl. Environ. Microbiol. 2013;79(7):2358–2370. doi: 10.1128/AEM.03067-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalez-Escalona N, Gavilan RG, Brown EW, Martinez-Urtaza J. Transoceanic spreading of pathogenic strains of Vibrio parahaemolyticus with distinctive genetic signatures in the recA gene. PLoS ONE. 2015;10(2):e0117485. doi: 10.1371/journal.pone.0117485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vos M, Didelot X. A comparison of homologous recombination rates in bacteria and archaea. Isme J. 2009;3(2):199–208. doi: 10.1038/ismej.2008.93. [DOI] [PubMed] [Google Scholar]
- 24.Yan Y, Cui Y, Han H, et al. Extended MLST-based population genetics and phylogeny of Vibrio parahaemolyticus with high levels of recombination. Int. J. Food Microbiol. 2011;145(1):106–112. doi: 10.1016/j.ijfoodmicro.2010.11.038. [DOI] [PubMed] [Google Scholar]
- 25.Cabrera-Garcia ME, Vazquez-Salinas C, Quinones-Ramirez EI. Serologic and molecular characterization of Vibrio parahaemolyticus strains isolated from seawater and fish products of the Gulf of Mexico. Appl. Environ. Microbiol. 2004;70(11):6401–6406. doi: 10.1128/AEM.70.11.6401-6406.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feil EJ. Small change: keeping pace with microevolution. Nat. Rev. Microbiol. 2004;2(6):483–495. doi: 10.1038/nrmicro904. [DOI] [PubMed] [Google Scholar]
- 27.Chao G, Wang F, Zhou X, et al. Origin of Vibrio parahaemolyticus O3:K6 pandemic clone. Int. J. Food Microbiol. 2011;145(2-3):459–463. doi: 10.1016/j.ijfoodmicro.2011.01.022. [DOI] [PubMed] [Google Scholar]
- 28.Turner JW, Paranjpye RN, Landis ED, et al. Population structure of clinical and environmental Vibrio parahaemolyticus from the Pacific Northwest Coast of the United States. PLoS ONE. 2013;8(2):e55726. doi: 10.1371/journal.pone.0055726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson CN, Flowers AR, Young VC, et al. Genetic relatedness among tdh+ and trh+ Vibrio parahaemolyticus cultured from Gulf of Mexico oysters (Crassostrea virginica) and surrounding water and sediment. Microb. Ecol. 2009;57(3):437–443. doi: 10.1007/s00248-008-9418-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.