Abstract
MicroRNAs and long noncoding RNAs have long been investigated due to their roles as diagnostic and prognostic biomarkers of cancers and regulators of tumorigenesis, and the potential regulatory roles of these molecules in anticancer therapies are attracting increasing interest as more in-depth studies are performed. The major clinical therapies for cancer include chemotherapy, immunotherapy, and targeted molecular therapy. MicroRNAs and long noncoding RNAs function through various mechanisms in these approaches, and the mechanisms involve direct targeting of immune checkpoints, cooperation with exosomes in the tumor microenvironment, and alteration of drug resistance through regulation of different signaling pathways. Herein we review the regulatory functions and significance of microRNAs and long noncoding RNAs in three anticancer therapies, especially in targeted molecular therapy, and their mechanisms.
Keywords: microRNAs, long noncoding RNAs, targeted therapy, chemoresistance, immune checkpoint
Main Text
MicroRNAs (miRNAs) and long noncoding RNAs (lncRNAs) are important noncoding RNAs (ncRNAs), which display a remarkable variety of biological functions.1 ncRNAs can be classified by length (small, 18–200 nt; long, >200 nt) or by function (housekeeping ncRNAs and regulatory ncRNAs), with research over the last two decades largely focusing on regulatory ncRNAs.2 miRNAs, which are ∼22 nt long, are the most widely studied class of regulatory ncRNAs, and these molecules mediate post-transcriptional gene silencing in animals by controlling the translation of mRNAs into proteins.3 lncRNAs, longer than 200 nt, are another subtype of regulatory ncRNAs that have a broad repertoire of functions in chromatin modification as well as in transcriptional, post-transcriptional, and translational regulation.2, 3
miRNAs and lncRNAs are expressed at different levels in multiple cell and tissue types; they are also involved in tumorigenesis and the progression of aggressive cancer phenotypes.4 These molecules are identified as either carcinogenetic or carcinostatic; are associated with cell growth, proliferation, migration, invasion, and apoptosis; and can even alter immune functions.5, 6, 7, 8, 9, 10 RNA sequencing has confirmed that miRNA and lncRNA profiles can serve as highly sensitive and specific diagnostic and prognostic biomarkers. Because these molecules can be detected in diverse tumor tissues compared to normal samples and are associated with different clinicopathologic characteristics, differentially expressed miRNAs and lncRNAs can be employed to assess the pathogenesis of diseases, including non-small-cell lung cancer (NSCLC), gastric cancer (GC), colorectal cancer (CRC), and melanoma, as well as clinical prognosis.11, 12, 13, 14, 15, 16
Recent studies of miRNAs and lncRNAs have indicated their latent therapeutic value for successful clinical translation. Results have confirmed that miRNAs and lncRNAs function as crucial regulators in different drug therapies, including chemotherapy, immunotherapy, and targeted molecular therapy, and the associated mechanisms have been investigated.
In this review, we discuss the ectopic expression of miRNAs and lncRNAs in multiple cancers and how they function in the three types of anticancer therapies, especially in targeted molecular therapy.
miRNAs and lncRNAs Participate in Chemotherapy
Although chemotherapy remains a mainstay of anticancer treatment, the multi-organ toxicity and chemoresistance associated with this treatment strategy continues to be problematic.17
Accumulating evidence shows that ncRNAs have an important role in cellular sensitivity to chemotherapy due to their specific regulatory features. The significance of miRNAs in anticancer chemotherapy has been demonstrated by multiple studies, and the associated mechanisms include regulation of different targets.18 For example, miR-197, miR-130b, and lncRNA MALAT1 confer cisplatin resistance in NSCLC by targeting the signal transducer and activator of transcription 3 (STAT3) and Wnt/β-catenin pathways, and lncRNA TP53TG1 enhances cellular sensitivity through the miR-18a/PTEN axis.19, 20, 21, 22 In contrast, miR-125a-5p and lncRNA TUSC7 are able to reverse cisplatin resistance in esophageal squamous cell carcinoma (ESCC) by reducing the levels of STAT3 and miR-224, respectively.23, 24 miR-503 and miR-623 inhibit resistance to different drugs by regulating cyclin D1-3 (CCND1-3),25, 26 and targeting Bcl-2, miR-374b-5p and miR-15 were found to enhance the chemosensitivity of cancer cells by modulating apoptotic pathways.27, 28
While investigating the role of lncRNAs involved in temozolomide (TMZ)-resistant glioma, Jia et al.29 and Cai and colleagues30 found that knockdown of lncRNAs H19 and MALAT1 reversed chemoresistance to TMZ by inhibiting or promoting their downstream targets. As a crucial regulator, lncRNA PVT1 directly acts on multiple drug resistance-associated molecules. Silencing of PVT1 downregulates the levels of multidrug resistance 1 (MDR1) and multidrug resistance protein 1 (MRP1) as well as the expression of antiapoptotic B cell lymphoma-2 (Bcl-2), but it upregulates levels of pro-apoptotic Bax and cleaved caspase-3.31 Mechanistically, the effects of lncRNAs TP53TG1, UCA1, MALAT1, and TUSC7 occur in an miRNA-dependent manner in which these molecules suppress expression of miRNAs, thus blocking relevant signaling pathways.22, 24, 30, 32 In summary, the regulatory roles of miRNAs and lncRNAs have been widely investigated (Table 1), and these functions are important for chemoresistance. The modulatory effects of these molecules mainly impact transcription and apoptosis, indicating that miRNAs and lncRNAs are potential targets that may improve drug efficacy.
Table 1.
miRNAs and lncRNAs Involved in Chemotherapy
| Cancer Type | ncRNA | Regulation of Chemoresistance | Target | Drug | Reference |
|---|---|---|---|---|---|
| NSCLC | miR-197 | promotion | CKS1B/STAT3 | DDP | 19 |
| miR-130b | promotion | Wnt/β-catenin pathway | DDP | 20 | |
| lncRNA MALAT1 | promotion | STAT3 | DDP | 21 | |
| lncRNA TP53TG1 | inhibition | miR-18a/PTEN | DDP | 22 | |
| PC | miR-455-3p | promotion | TAZ | GEM | 99 |
| miR-29c | inhibition | USP22 | GEM | 100 | |
| miR-374b-5p | inhibition | Bcl-2 | GEM | 27 | |
| BC | miR-503 | inhibition | CCND2, CCND3 | EPI, PTX | 25 |
| lncRNA LINP1 | promotion | – | ADM, 5-FU | 101 | |
| miR-17 | promotion | DEDD | DDP, 5-FU | 102 | |
| GC | miR-218 | inhibition | mTOR inhibitor | DDP | 103 |
| miR-623 | inhibition | CCND1 | 5-FU | 26 | |
| CRC | miR-191 | promotion | Wnt/β-catenin pathway | 5-FU | 104 |
| miR-519b-3p | inhibition | ARID4B mRNA | CAPE/OXA/5-FU | 105 | |
| miR-15 | inhibition | NF-κB, Bcl-2 | 5-FU/OXA | 28 | |
| lncRNA PVT1 | promotion | MDR1, MRP1, Bcl-2, Bax, cleaved caspase-3 | DDP | 31 | |
| lncRNA MALAT1 | promotion | EZH2 | OXA | 106 | |
| lncRNA UCA1 | promotion | miR-204-5p | 5-FU | 32 | |
| Glioma | lncRNA H19 | promotion | Wnt/β-catenin pathway | TMZ | 29 |
| lncRNA MALAT1 | promotion | MiR-101 | TMZ | 30 | |
| lncRNA DANCR | promotion | AXL/PI3K/Akt/ NF-κB | DDP | 107 | |
| ESCC | miR-125a-5p | inhibition | STAT3 | DDP | 23 |
| lncRNA TUSC7 | inhibition | MiR-224 | DDP, 5-FU, and ADM/PTX | 24 | |
| HCC | miR-16 | inhibition | NF-κB | PTX | 108 |
| OC | miR-630 | promotion | APAF-1 | PTX | 109 |
| miR-142-3p | inhibition | Sirtuin 1 | DDP | 110 |
NSCLC, non-small-cell lung cancer; PC, pancreatic cancer; BC, breast cancer; GC, gastric cancer; CRC, colorectal cancer; ESCC, esophageal squamous cell carcinoma; OC, ovarian cancer; HCC, hepatocellular carcinoma; APAF-1, apoptotic protease activating factor-1; CCND1-3, cyclin D1-3; DEDD, death effector domain-containing DNA-binding protein; EZH2, enhancer of zeste homolog 2; MDR1, multidrug resistance 1; MRP1, multidrug resistance protein 1; PTEN, phosphatase and tensin homolog deleted on chromosome 10; STAT3, signal transducer and activator of transcription 3; TAZ, transcriptional co-activator with PDZ-binding motif; TMZ, temozolomide; PI3K, phosphatidylinositol 3-kinase; USP22, ubiquitin-specific peptidase 22; Bcl-2, B cell lymphoma-2; DDP, cisplatin; GEM, gemcitabine; EPI, epirubicin; PTX, paclitaxel; 5-FU, 5-fluorouracil; CAPE, capecitabine; OXA, oxaliplatin; TMZ, temozolomide; ADM, adriamycin; PTX, paclitaxel.
By mediating cell-cell communication, exosomes have been suggested to exert profound effects on the development of drug resistance.33 Indeed, by transferring miR-503 from the endothelium to the tumor microenvironment, thus interfering with interaction between breast cancer (BC) cells and the microenvironment, endothelial exosomes contribute to chemotherapeutic response in BC.25 In addition, exosome-transferred miR-21 derived from M2-polarized macrophages confer cisplatin resistance in GC, suggesting a new therapeutic strategy for GC patients34, 35, 36 (Table 1).
miRNAs and lncRNAs Participate in Immunotherapy
The breakthrough of immune checkpoint therapy, which involves the use of monoclonal antibodies against the receptor cytotoxic T lymphocyte antigen-4 (CTLA-4), programmed cell death protein 1 (PD-1), and PD-1 ligand (PD-L1), represents a turning point in cancer immunotherapy.37 Notably, therapeutic success in clinical trials has been achieved with pembrolizumab, which targets the PD-1/PD-L1 pathway.38, 39
PD-L1 is a cell surface glycoprotein that maintains immunologic homeostasis; but, PD-L1 is overexpressed on tumor cells as well as immune cells in many cancers. Blockade of the PD-1/PD-L1 pathway reverses immune escape in tumors, and it provides strategies for cancer immunotherapy. As a biomarker of a response to immune checkpoint blockade, PD-L1 expression on tumor cells has been assessed in the prediction of therapeutic efficacy and chemoresistance.40
Altered expression of miRNAs in PD-1/PD-L1 immune checkpoint blockage and various cellular processes in cancer has recently gained attention (Table 2).41 miR-25-93-106b, miR-138-5p, miR-217, and miR-200 were found to suppress the expression of PD-L1, thus rescuing decreased tumor immunity and inhibiting multiple metastatic traits, such as cell migration, invasion, proliferation, apoptosis, and the epithelial-mesenchymal transition (EMT), as well as angiogenesis.42, 43, 44, 45
Table 2.
miRNAs and lncRNAs Involved in the PD-1/PD-L1 Immune Checkpoint
| Cancer Type | ncRNA | Expression | Regulation of PD-L1 (PD-1) | Reference |
|---|---|---|---|---|
| Bone marrow stromal niche | miR-25-93-106b | ↑ | ↓ | 42 |
| Colorectal cancer | miR-138-5p | ↓ | ↓ | 43 |
| Laryngeal cancer | miR-217 | ↓ | ↓ | 44 |
| Lung adenocarcinoma | miR-200 | ↓ | ↓ | 45 |
| Ovarian cancer | miR-424(322) | ↓ | ↓ | 46 |
| Oral squamous cell carcinoma | miR-197 | ↑ | ↓ | 47 |
| Melanoma | miR-17-5p | ↓ | ↑ | 50 |
| Glioma | miR-138 | ↓ | ↓ (PD-1) | 48 |
| Nasopharyngeal carcinoma | AFAP1-AS1 | ↑ | ↑ (PD-1) | 49 |
Additionally, miRNAs can enhance curative effects and restore immune functions indirectly through interaction with PD-L1. miR-424(322) regulates the PD-1/PD-L1 and CD80/CTLA-4 pathways in ovarian cancer by decreasing PD-L1 and CD80 expression, restoration of which enhances the drug sensitivity of ovarian cancer cells through PD-1/PD-L1 checkpoint blockage.46 Tumor-infiltrating lymphocytes (TILs) in oral squamous cell carcinoma (OSCC) are sites where immune escape arises, an effect that can be reversed by blocking the PD-1/PD-L1 pathway. miR-197 enhances anticancer immune responses by inhibiting PD-L1 expression, thus weakening the aggressive features of OSCC.47 In addition to PD-L1, PD-1 is also an effective target for PD-1/PD-L1 pathway blockade. miR-138 exhibits antiglioma efficacy by decreasing PD-1 expression, resulting in substantial tumor regression and a 43% increase in median survival time.48 In addition, co-expression of PD-1 and lncRNA AFAP1-AS1, which is associated with the poorest prognosis in nasopharyngeal carcinoma patients, suggests that this molecule is an ideal candidate for future clinical trials of anti-PD-1 immunotherapy.49
Most miRNAs play a positive role in anticancer immunology by targeting immune checkpoints; however, there are also miRNAs that carry out the opposite functions. For example, miR-17-5p post-transcriptionally upregulates PD-L1 in metastatic melanoma, leading to significantly enhanced invasive properties.50
Other immunologic mechanisms together with immune checkpoint blockade involve the anticancer functions of ncRNAs. For example, it has been demonstrated that ncRNAs drive exosome-mediated MAPK signaling by activating CD97 and proinflammatory cytokine production by activating cells of the mononuclear phagocytic system; because they are translated into short polypeptides, ncRNAs also present the best targets for immunotherapy51 (Table 2).
miRNAs and lncRNAs Are Involved in Targeted Molecular Therapy
Targeted therapy is personalized treatment that involves the application of agents targeted toward specific molecular features of cancer cells, thereby minimizing toxicity and decreasing the cost of cancer care. These unique molecular targets that recognize and eliminate cancer cells are genetic alterations that are primarily mutated versions of epidermal growth factor receptor (EGFR), epidermal growth factor receptor 2 (HER2), vascular endothelial growth factor (VEGF), vascular endothelial growth factor receptor 2 (VEGFR2), and v-Raf murine sarcoma viral oncogene homolog B (BRAF).52, 53, 54 In addition, the use of miRNAs and lncRNAs in targeted molecular therapy primarily involves the alteration of cellular sensitivity to drugs. Below we summarize the modulatory effects of miRNAs and lncRNAs on resistance to agents that have been approved in China.
EGFR and HER2 Mutations and Their Corresponding Agents
EGFR and HER2 are two common oncogenic mutations found in lung cancer and BC;52 they also occur in other types of malignancies.55, 56 Anticancer targeted molecular therapeutic drugs mainly include gefitinib, erlotinib, and cetuximab targeting EGFR;57 trastuzumab and pertuzumab targeting HER2;58 and afatinib and lapatinib targeting both EGFR and HER2.56, 57 Lapatinib, a tyrosine kinase inhibitor (TKI), was approved based on improvements in progression-free survival (PFS)58 and alleviation of side effects.56 In a survival analysis of HER2-positive BC, overall survival (OS) was significantly better in patients who were treated with the neoadjuvant lapatinib followed by the adjuvant trastuzumab than in those treated with trastuzumab alone (hazard ratio [HR], 0.32; p = 0.019).59 The addition of trastuzumab, a humanized monoclonal antibody, to carboplatin-paclitaxel was well tolerated by HER2-positive patients and increased PFS (12.6 months [experimental] versus 8.0 months [control], p = 0.005).55
However, numerous cases of acquired resistance reveal the limitation of targeted therapy. For example, acquired resistance to TKIs inevitably occurs in almost all NSCLC patients, and the major mechanisms include T790M, MET, and HER2/3 mutations as well as IGF1R and PI3K activation.60, 61 Additionally, emerging evidence highlights the master regulatory roles of miRNAs and lncRNAs in the acquisition of resistance, and it suggests potential targets for development in targeted therapy (Table 3).60, 62, 63, 64, 65, 66
Table 3.
miRNAs and lncRNAs Involved in the Resistance to Lapatinib, Gefitinib, Erlotinib, Pertuzumab, Cetuximab, and Trastuzumab
| Drug | Cancer Type | Regulation of Resistance | ncRNA | Target | Reference |
|---|---|---|---|---|---|
| Lapatinib | HER2(+) BC | inhibition | miR-630 | IGF1R | 111 |
| triple-negative BC | miR-7 | Raf-1/MAPK/IL-6 | 77 | ||
| HER2(-) BC | EGFR | 78 | |||
| Lapatinib + trastuzumab | HER2(+) BC+GC | inhibition | miR-16 | CCNJ, FUBP1 | 112 |
| Trastuzumab | HER2(+) BC | promotion | miR-7 | EGFR/Src | 76 |
| miR-21 | IL-6/STAT3/NF-κB, PTEN/PI3K | 71 | |||
| PTEN | 72 | ||||
| miR-221 | PTEN | 113 | |||
| lncRNA UCA1 | miR-18a/YAP1 | 80 | |||
| inhibition | miR-375 | IGF1R | 114 | ||
| miR-194 | TLN2 | 115 | |||
| miR-30b | CCNE2 | 116 | |||
| lncRNA GAS5 | miR-21/PTEN | 74 | |||
| HER2(+) GC | promotion | miR-125b | – | 117 | |
| Trastuzumab + gefitinib | melanoma | inhibition | miR-217 | CAGE | 118 |
| Gefitinib | NSCLC | promotion | miR-125b | – | 119 |
| miR-21 | PTEN, PDCD4, PI3K/Akt | 73 | |||
| lncRNA UCA1 | Akt/mTOR | 63 | |||
| miR-630 | YAP1/ERK | 120 | |||
| Erlotinib | NSCLC | promotion | miR-641 | NF1/ERK | 121 |
| Cetuximab | CRC | promotion | lncRNA MIR100HG, miR-100, miR-125b | Wnt/β-catenin pathway | 122 |
| miR-199a-5p, miR-375 | PHLPP1 | 123 | |||
| inhibition | miR-7 | EGFR | 79 | ||
| HCC | inhibition | let-7a | STAT3 | 124 | |
| miR-9 | eIF-5A-2 | 125 | |||
| Pertuzumab | OC | inhibition | miR-150 | Akt | 126 |
BC, breast cancer; GC, gastric cancer; NSCLC, non-small-cell lung cancer; CRC, colorectal cancer; OC, ovarian cancer; HCC, hepatocellular carcinoma; IGF1R, insulin growth factor receptor 1; MAPK, mitogen-activated protein kinase; IL-6, interleukin-6; CCNJ, cyclin J; FUBP1, far upstream element-binding protein 1; STAT3, signal transducer and activator of transcription 3; PI3K, phosphatidylinositol 3-kinase; PTEN, phosphatase and tensin homolog deleted on chromosome 10; TLN2, cytoskeleton protein talin2; CCNE2, cyclin E2; YAP1, Yes-associated protein 1; CAGE, cancer-associated gene; PDCD4, programmed cell death protein 4; NF1, neurofibromatosis 1; PHLPP1, PH domain and leucine-rich repeat protein phosphatase 1; eIF-5A-2, eukaryotic translation initiation factor 5A2.
miR-21, miR-7, and lncRNA UCA1 Regulate Drug Resistance
miR-21, which promotes cell proliferation and invasion and is upregulated in many cancers, is one of the most widely investigated miRNAs.67, 68, 69, 70 In HER2-positive BC, miR-21 was found to be inversely correlated with the expression of PTEN and PDCD4; by triggering an interleukin-6 (IL-6)/STAT3/nuclear factor κB (NF-κB)-mediated signaling loop and activating the PI3K pathway, it is also related to decreased trastuzumab sensitivity.71 Blocking the action of miR-21 with antisense oligonucleotides (ASOs) re-sensitized resistant cells to the therapeutic effects of trastuzumab.72 Similarly, miR-21 downregulates the expression of PTEN and PDCD4 and activates the PI3K/Akt pathway in gefitinib-resistant NSCLC cell lines, and inhibiting miR-21 with ASOs suppresses tumor growth in nude mice treated with gefitinib.73 Serving as a molecular sponge for miR-21, lncRNA GAS5 increases PTEN levels by competitively binding to miR-21 in a trastuzumab-resistant BC cell line (SKBR-3/Tr cell), thus inhibiting cell proliferation. In addition, GAS5 expression can be elevated by mTOR activation in lapatinib-treated SKBR-3/Tr cells, identifying GAS5 as candidate drug target for trastuzumab-resistant BC.74
miR-7 is another well-investigated miRNA that has been identified as both a tumor suppressor and promoter in a number of malignancies, such as BC, hepatocellular carcinoma (HCC), CRC, NSCLC, glioma, and melanoma.75 miR-7 also plays an indispensable role in drug resistance. Reestablished miR-7 expression abolishes HER2Δ16, the oncogenic isoform of HER2, and it induces cell proliferation and migration while sensitizing HER2Δ16-expressing cells to trastuzumab therapy.76 The off-target activity of lapatinib in inducing EGFR expression in BC was unexpectedly found to enhance metastasis, and this resistance-related phenotype was attributed to miR-7 downregulation.77, 78 Moreover, restoration of miR-7 expression inhibits Raf-1 signaling activation and EGFR expression, thereby restricting lapatinib-induced metastasis.77 By directly targeting EGFR and Raf-1, miR-7 also inhibits cell resistance to cetuximab in CRC.79
Previous studies have demonstrated the role of dysregulated miRNA expression in drug resistance, but, to date, few studies have examined lncRNAs. Nonetheless, Zhu et al.80 found that the lncRNA UCA1 desensitized BC cells to trastuzumab by impeding miR-18a repression of Yes-associated protein 1 (YAP1). In another study, UCA1 knockdown restored gefitinib sensitivity in cells with acquired resistance and no T790M mutations, and it inhibited activation of the Akt/mTOR pathway and EMT63 (Table 3).
miRNAs and lncRNAs Are Involved in the Effects of Sorafenib and Sunitinib
Sorafenib, the first systemic drug for patients with advanced HCC, inhibits the activity of multiple kinases, such as Raf kinase, VEGFR2, and platelet-derived growth factor receptor (PDGFR).53 This drug also increases the survival rate of renal cell carcinoma (RCC) patients. Regardless, poor primary response and acquired resistance remain the major obstacles for effective treatment with sorafenib.81 While assessing this urgent problem, researchers were able to identify the predictive and therapeutic functions of miRNAs and lncRNAs in sorafenib treatment (Table 4).82 By activating p53-dependent apoptosis, miR-27b was found to enhance the response to sorafenib in HCC and RCC, and the direct target of miR-27b was cyclin G1 (CCNG1), a negative regulator of p53.83
Table 4.
miRNAs and lncRNAs Involved in Sorafenib and Sunitinib Resistance
| Drug | Cancer Type | Regulation of Resistance | ncRNA | Target | Reference |
|---|---|---|---|---|---|
| Sorafenib | HCC | inhibition | miR-27b | CCNG1 | 83 |
| let-7 | Bcl-xL, Mcl-1 | 84 | |||
| miR-122 | ADAM10, SRF, IGF1R | 85, 86 | |||
| miR-338-3p | HIF-1α | 127 | |||
| miR-425-3p | – | 82 | |||
| miR-34a | Bcl-2, Mcl-1 | 128 | |||
| miR-193b | Mcl-1 | 129 | |||
| Ad5-AlncRNA | miR-21, miR-153, miR-216a, miR-217, miR-494, miR-10a-5p | 90 | |||
| promotion | miR-494 | PTEN, PI3K/Akt | 89 | ||
| miR-222 | PI3K/Akt | 130 | |||
| miR-21 | PTEN, PI3K/Akt | 88 | |||
| miR-181a | RASSF1 | 53 | |||
| lncTUC338 | RASAL1 | 131 | |||
| RCC | inhibition | miR-27b | CCNG1 | 83 | |
| miR-30a | Beclin-1 | 81 | |||
| miR-200c | HO-1 | 132 | |||
| promotion | lncRNA SRLR | NF-κB | 133 | ||
| lncRNA NEAT1 | miR-34a | 134 | |||
| Sunitinib | RCC | inhibition | lncRNA SARCC | AR/miR-143-3p | 93 |
| promotion | miR-144-3p | ARID1A | 92 | ||
| lncRNA ARSR | miR-34, miR-449 | 91 |
HCC, hepatocellular carcinoma; RCC, renal cell carcinoma; SRF, serum response factor; ADAM10, a distintegrin and metalloprotease family 10; IGF1R, insulin growth factor receptor 1; Bcl-2, B cell lymphoma-2; Bcl-xL, B cell lymphoma-extra large; Mcl-1, myeloid cell leukemia-1; HIF-1α, hypoxia-inducible factor-1; HO-1, heme oxygenase-1; RASAL1, RAS GTPase-activating protein (RasGAP) 1; RASAL1, RAS GTPase-activation protein (RasGAP) gene; CCNG1, cyclin G1; ARID1A, AT-rich interactive domain 1A; AR, androgen receptor.
Another miRNA that potentiates sorafenib-induced apoptosis in HCC is let-7, which reduces expression of the antiapoptotic Bcl-2 protein Bcl-xL and Mcl-1.84 miR-122 appears to sensitize HCC cells to sorafenib by targeting distintegrin and metalloprotease family 10 (ADAM10), serum response factor (SRF), and IGF1R.85, 86 Moreover, exosomes derived by adipose tissue-derived mesenchymal stem cells help to deliver miR-122 into HCC cells, further promoting the chemosensitivity of these cells.87 miR-494 and miR-21, which are both upregulated in HCC and reinforce sorafenib resistance, directly suppress the expression of PTEN but activate the PI3K/Akt-signaling pathway, thereby contributing to the promotion of proliferation, migration, and invasion.88, 89
Although these potential antiresistance targets have been identified, it is a challenge to restore sensitivity by regulating only one miRNA, because it may sequentially activate other compensatory pathways. Accordingly, Tang et al.90 generated an artificial lncRNA expressed by an adenoviral vector (Ad5-AlncRNA), which simultaneously targets multiple miRNAs, including miR-21, miR-153, miR-216a, miR-217, miR-494, and miR-10a-5p. As mentioned above, these miRNAs participate in the mechanisms underlying sorafenib resistance, and, thus, targeting multiple miRNAs may be a promising strategy for overcoming such resistance.
Sunitinib is the mainstay of therapeutic options for advanced RCC patients. This drug is a multitarget receptor TKI that mainly inhibits VEGFR and PDGFR. However, 10%–20% of advanced RCC patients are inherently resistant to sunitinib therapy, and most of the remaining patients exhibit drug resistance and tumor progression after 6–15 months of therapy.91 In studies of sunitinib resistance in RCC, miR-144-3p and lncRNAs ARSR and SARCC were found to affect malignancy via different targets91, 92, 93 (Table 4).
miRNAs and lncRNAs Are Involved in the Effects of Imatinib and Vemurafenib
Little is known about the effect of ncRNAs on imatinib and vemurafenib resistance in solid tumors. Sensitivity of melanoma to the BRAF(V600E) inhibitor vemurafenib is positively regulated by miR-579-3p, miR-216b, and miR-7, and it is negatively regulated by miR-204-5p and miR-211-5p.54, 94, 95, 96 Imatinib, a small-molecule inhibitor that targets several receptor tyrosine kinases, including KIT and PDGFR, is primarily applied in the treatment of chronic myelogenous leukemia (CML) and gastrointestinal stromal tumors (GISTs). One study on imatinib-resistant glioblastoma revealed that ectopic expression of miR-203 with miRNA mimics effectively sensitizes cells to chemotherapy by targeting SNAI2.97
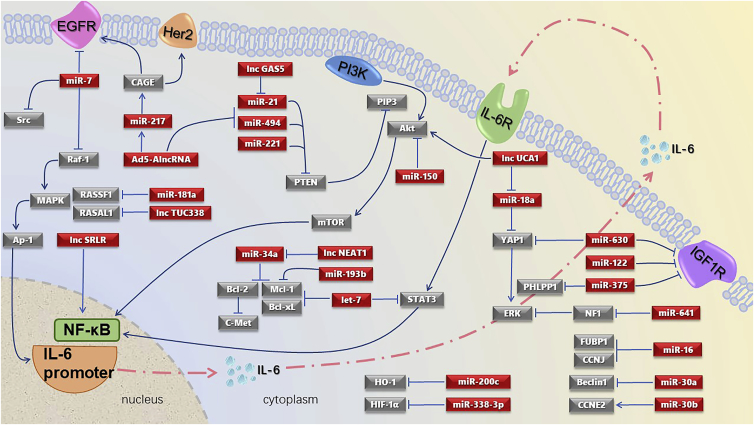
In summary, this review compiles the available literature on the miRNAs and lncRNAs involved in targeted therapy that have certain and explicit targets and pathways (Figure 1). All relevant publications were retrieved from the PubMed database, with keywords such as miRNA, lncRNA, exosome, PD-1/PD-L1, immunotherapy, chemoresistance, targeted therapy, lapatinib, gefitinib, trastuzumab, sorafenib, HER2, EGFR, and similar terms.
Figure 1.
miRNAs and lncRNAs in Targeted Therapy with Explicit Targets and Pathways
These pathways mainly comprise the Raf-1/MAPK/IL-6 axis, IL-6/STAT3/NF-κB axis, and PI3K/Akt/mTOR axis, and they center on targets of PTEN, IGF1R, and ERK. Among the ncRNAs involved, miR-7, miR-21, miR-630, and lncRNA UCA1 play important roles.
Conclusions
miRNAs and lncRNAs, subcategories of ncRNAs, have primarily been investigated as biomarkers for predicting the initiation and development of cancer, but they have recently been discovered to be involved in the curative process of three clinically adopted therapies. These molecules enhance or suppress cancer cell responses to chemotherapy drugs and targeted drugs indirectly by modulating relevant pathways, and they also affect immune checkpoint blockage therapy directly by altering the expression of PD-1/PD-L1. Overexpressing miRNAs and lncRNAs by mimics and silencing these molecules by small interfering RNAs (siRNAs) verify their therapeutic capacity in suppressing aggressive cell phenotypes and alleviating drug resistance.
Furthermore, rapid advances in elucidating the roles of miRNAs and lncRNAs in anticancer therapies have revealed several opportunities and challenges to address in the future. One opportunity is cooperation with extracellular vesicles, especially exosomes. As mentioned above, exosome-mediated miR-503 reduced chemoresistance after it was transferred from endothelial cells to tumor cells.25 Studies have demonstrated the communication shuttle function of exosomes between cells and that exosome-associated ncRNAs fulfill important jobs in regulating gene expression in cancer.3 However, more work on the therapeutic value of exosome-associated ncRNAs in cancer is needed. Second, miRNA-miRNA and miRNA-lncRNA networks reveal the complexity of ncRNA-mediated mechanisms in anticancer therapies, providing a better understanding of the ncRNA-mediated drug response and creative research approaches.98 One outstanding problem is whether ectopic miRNAs and lncRNAs actually function in vivo, and more research utilizing convenient in vivo model systems are needed. Future studies will likely focus on ncRNA-based drug development and integrated clinical trials, which may lead to a cure for cancer. Additionally, the investigation of circular RNAs, another ncRNA research hotspot, is needed to improve our understanding of the ncRNA therapeutic network.
All relevant publications were retrieved from the PubMed database, with key words such as miRNA, lncRNA, exosome, PD-1/PD-L1, immunotherapy, chemoresistance, targeted therapy, lapatinib, gefitinib, trastuzumab, sorafenib, HER2, EGFR and similar terms.
Author Contributions
M.X. designed the research and drafted the manuscript. L.M. and T.X. critically revised the manuscript. Y.P., Q.W., and Y.W. discussed and revised the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
The authors have no conflicts of interest.
Acknowledgments
This work was supported by grants from the National Key Research and Development Program: The key technology of palliative care and nursing for cancer patients (ZDZX2017ZL-01) and the Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX18_1482).
References
- 1.Cech T.R., Steitz J.A. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Glasgow A.M.A., De Santi C., Greene C.M. Non-coding RNA in cystic fibrosis. Biochem. Soc. Trans. 2018;46:619–630. doi: 10.1042/BST20170469. [DOI] [PubMed] [Google Scholar]
- 3.Ma P., Pan Y., Li W., Sun C., Liu J., Xu T., Shu Y. Extracellular vesicles-mediated noncoding RNAs transfer in cancer. J. Hematol. Oncol. 2017;10:57. doi: 10.1186/s13045-017-0426-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mohr A.M., Mott J.L. Overview of microRNA biology. Semin. Liver Dis. 2015;35:3–11. doi: 10.1055/s-0034-1397344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Z.H., Li L., Kang L.P., Wang Y. MicroRNA-92a promotes tumor growth and suppresses immune function through activation of MAPK/ERK signaling pathway by inhibiting PTEN in mice bearing U14 cervical cancer. Cancer Med. 2018;7:3118–3131. doi: 10.1002/cam4.1329. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Li C., Du X., Xia S., Chen L. MicroRNA-150 inhibits the proliferation and metastasis potential of colorectal cancer cells by targeting iASPP. Oncol. Rep. 2018;40:252–260. doi: 10.3892/or.2018.6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qiu M., Xu Y., Wang J., Zhang E., Sun M., Zheng Y., Li M., Xia W., Feng D., Yin R., Xu L. A novel lncRNA, LUADT1, promotes lung adenocarcinoma proliferation via the epigenetic suppression of p27. Cell Death Dis. 2015;6:e1858. doi: 10.1038/cddis.2015.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang L., Wang F., Na L., Yu J., Huang L., Meng Z.Q., Chen Z., Chen H., Ming L.L., Hua Y.Q. LncRNA AB209630 inhibits gemcitabine resistance cell proliferation by regulating PI3K/AKT signaling in pancreatic ductal adenocarcinoma. Cancer Biomark. 2018;22:169–174. doi: 10.3233/CBM-181182. [DOI] [PubMed] [Google Scholar]
- 9.Yang X., Song J.H., Cheng Y., Wu W., Bhagat T., Yu Y., Abraham J.M., Ibrahim S., Ravich W., Roland B.C. Long non-coding RNA HNF1A-AS1 regulates proliferation and migration in oesophageal adenocarcinoma cells. Gut. 2014;63:881–890. doi: 10.1136/gutjnl-2013-305266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang H., Wang S., Kang Y.J., Wang C., Xu Y., Zhang Y., Jiang Z. Long non-coding RNA SNHG1 predicts a poor prognosis and promotes colon cancer tumorigenesis. Oncol. Rep. 2018;40:261–271. doi: 10.3892/or.2018.6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding X., Zhang S., Li X., Feng C., Huang Q., Wang S., Wang S., Xia W., Yang F., Yin R. Profiling expression of coding genes, long noncoding RNA, and circular RNA in lung adenocarcinoma by ribosomal RNA-depleted RNA sequencing. FEBS Open Bio. 2018;8:544–555. doi: 10.1002/2211-5463.12397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tengda L., Shuping L., Mingli G., Jie G., Yun L., Weiwei Z., Anmei D. Serum exosomal microRNAs as potent circulating biomarkers for melanoma. Melanoma Res. 2018;28:295–303. doi: 10.1097/CMR.0000000000000450. [DOI] [PubMed] [Google Scholar]
- 13.Shukla S. Unravelling the Long Non-Coding RNA Profile of Undifferentiated Large Cell Lung Carcinoma. Noncoding RNA. 2018;4:E4. doi: 10.3390/ncrna4010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang M.H., Yang Y., Zhao Y., Wei H.B., Ma Y.Q., Yang C.J., Zhang X.J., Sun Y.L. LncRNA DQ786243 expression as a biomarker for assessing prognosis in patients with gastric cancer. Eur. Rev. Med. Pharmacol. Sci. 2018;22:2304–2309. doi: 10.26355/eurrev_201804_14819. [DOI] [PubMed] [Google Scholar]
- 15.Feng H., Xu M., Zhang Y., Han B., Wang J., Sun P. Identification of Differentially Expressed MicroRNAs involved in the Pathogenesis of Colorectal Cancer. Clin. Lab. 2018;64:797–804. doi: 10.7754/Clin.Lab.2017.171203. [DOI] [PubMed] [Google Scholar]
- 16.Shen Q.M., Wang H.Y., Xu S. LncRNA GHET1 predicts a poor prognosis of the patients with non-small cell lung cancer. Eur. Rev. Med. Pharmacol. Sci. 2018;22:2328–2333. doi: 10.26355/eurrev_201804_14823. [DOI] [PubMed] [Google Scholar]
- 17.Sonawane V.K., Mahajan U.B., Shinde S.D., Chatterjee S., Chaudhari S.S., Bhangale H.A., Ojha S., Goyal S.N., Kundu C.N., Patil C.R. A Chemosensitizer Drug: Disulfiram Prevents Doxorubicin-Induced Cardiac Dysfunction and Oxidative Stress in Rats. Cardiovasc. Toxicol. 2018;18:459–470. doi: 10.1007/s12012-018-9458-y. [DOI] [PubMed] [Google Scholar]
- 18.Riquelme I., Letelier P., Riffo-Campos A.L., Brebi P., Roa J.C. Emerging Role of miRNAs in the Drug Resistance of Gastric Cancer. Int. J. Mol. Sci. 2016;17:424. doi: 10.3390/ijms17030424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujita Y., Yagishita S., Hagiwara K., Yoshioka Y., Kosaka N., Takeshita F., Fujiwara T., Tsuta K., Nokihara H., Tamura T. The clinical relevance of the miR-197/CKS1B/STAT3-mediated PD-L1 network in chemoresistant non-small-cell lung cancer. Mol. Ther. 2015;23:717–727. doi: 10.1038/mt.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Q., Zhang B., Sun L., Yan Q., Zhang Y., Zhang Z., Su Y., Wang C. MicroRNA-130b targets PTEN to induce resistance to cisplatin in lung cancer cells by activating Wnt/β-catenin pathway. Cell Biochem. Funct. 2018;36:194–202. doi: 10.1002/cbf.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fang Z., Chen W., Yuan Z., Liu X., Jiang H. LncRNA-MALAT1 contributes to the cisplatin-resistance of lung cancer by upregulating MRP1 and MDR1 via STAT3 activation. Biomed. Pharmacother. 2018;101:536–542. doi: 10.1016/j.biopha.2018.02.130. [DOI] [PubMed] [Google Scholar]
- 22.Xiao H., Liu Y., Liang P., Wang B., Tan H., Zhang Y., Gao X., Gao J. TP53TG1 enhances cisplatin sensitivity of non-small cell lung cancer cells through regulating miR-18a/PTEN axis. Cell Biosci. 2018;8:23. doi: 10.1186/s13578-018-0221-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao Y., Ma K., Yang S., Zhang X., Wang F., Zhang X., Liu H., Fan Q. MicroRNA-125a-5p enhances the sensitivity of esophageal squamous cell carcinoma cells to cisplatin by suppressing the activation of the STAT3 signaling pathway. Int. J. Oncol. 2018;53:644–658. doi: 10.3892/ijo.2018.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang Z.W., Jia Y.X., Zhang W.J., Song L.J., Gao M., Li M.J., Zhao R.H., Li J., Zhong Y.L., Sun Q.Z., Qin Y.R. LncRNA-TUSC7/miR-224 affected chemotherapy resistance of esophageal squamous cell carcinoma by competitively regulating DESC1. J. Exp. Clin. Cancer Res. 2018;37:56. doi: 10.1186/s13046-018-0724-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bovy N., Blomme B., Frères P., Dederen S., Nivelles O., Lion M., Carnet O., Martial J.A., Noël A., Thiry M. Endothelial exosomes contribute to the antitumor response during breast cancer neoadjuvant chemotherapy via microRNA transfer. Oncotarget. 2015;6:10253–10266. doi: 10.18632/oncotarget.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang L., Yang W., Bian W., Yang H., Wu X., Li Y., Feng W., Liu X. microRNA-623 targets cyclin D1 to inhibit cell proliferation and enhance the chemosensitivity of cells to 5-fluorouracil in gastric cancer. Oncol. Res. 2018 doi: 10.3727/096504018X15193469240508. Published online March 1, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun D., Wang X., Sui G., Chen S., Yu M., Zhang P. Downregulation of miR-374b-5p promotes chemotherapeutic resistance in pancreatic cancer by upregulating multiple anti-apoptotic proteins. Int. J. Oncol. 2018;52:1491–1503. doi: 10.3892/ijo.2018.4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu L., Wang D., Qiu Y., Dong H., Zhan X. Overexpression of microRNA-15 increases the chemosensitivity of colon cancer cells to 5-fluorouracil and oxaliplatin by inhibiting the nuclear factor-κB signalling pathway and inducing apoptosis. Exp. Ther. Med. 2018;15:2655–2660. doi: 10.3892/etm.2017.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jia L., Tian Y., Chen Y., Zhang G. The silencing of LncRNA-H19 decreases chemoresistance of human glioma cells to temozolomide by suppressing epithelial-mesenchymal transition via the Wnt/β-Catenin pathway. OncoTargets Ther. 2018;11:313–321. doi: 10.2147/OTT.S154339. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Cai T., Liu Y., Xiao J. Long noncoding RNA MALAT1 knockdown reverses chemoresistance to temozolomide via promoting microRNA-101 in glioblastoma. Cancer Med. 2018;7:1404–1415. doi: 10.1002/cam4.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ping G., Xiong W., Zhang L., Li Y., Zhang Y., Zhao Y. Silencing long noncoding RNA PVT1 inhibits tumorigenesis and cisplatin resistance of colorectal cancer. Am. J. Transl. Res. 2018;10:138–149. [PMC free article] [PubMed] [Google Scholar]
- 32.Bian Z., Jin L., Zhang J., Yin Y., Quan C., Hu Y., Feng Y., Liu H., Fei B., Mao Y. LncRNA-UCA1 enhances cell proliferation and 5-fluorouracil resistance in colorectal cancer by inhibiting miR-204-5p. Sci. Rep. 2016;6:23892. doi: 10.1038/srep23892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ji R., Zhang B., Zhang X., Xue J., Yuan X., Yan Y., Wang M., Zhu W., Qian H., Xu W. Exosomes derived from human mesenchymal stem cells confer drug resistance in gastric cancer. Cell Cycle. 2015;14:2473–2483. doi: 10.1080/15384101.2015.1005530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanne A., Muniz L.R., Puzio-Kuter A., Leonova K.I., Gudkov A.V., Ting D.T., Monasson R., Cocco S., Levine A.J., Bhardwaj N., Greenbaum B.D. Distinguishing the immunostimulatory properties of noncoding RNAs expressed in cancer cells. Proc. Natl. Acad. Sci. USA. 2015;112:15154–15159. doi: 10.1073/pnas.1517584112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Charpentier M., Croyal M., Carbonnelle D., Fortun A., Florenceau L., Rabu C., Krempf M., Labarrière N., Lang F. IRES-dependent translation of the long non coding RNA meloe in melanoma cells produces the most immunogenic MELOE antigens. Oncotarget. 2016;7:59704–59713. doi: 10.18632/oncotarget.10923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng P., Chen L., Yuan X., Luo Q., Liu Y., Xie G., Ma Y., Shen L. Exosomal transfer of tumor-associated macrophage-derived miR-21 confers cisplatin resistance in gastric cancer cells. J. Exp. Clin. Cancer Res. 2017;36:53. doi: 10.1186/s13046-017-0528-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Y. Cancer immunotherapy: harnessing the immune system to battle cancer. J. Clin. Invest. 2015;125:3335–3337. doi: 10.1172/JCI83871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reck M., Rodríguez-Abreu D., Robinson A.G., Hui R., Csőszi T., Fülöp A., Gottfried M., Peled N., Tafreshi A., Cuffe S., KEYNOTE-024 Investigators Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2016;375:1823–1833. doi: 10.1056/NEJMoa1606774. [DOI] [PubMed] [Google Scholar]
- 39.Langer C.J., Gadgeel S.M., Borghaei H., Papadimitrakopoulou V.A., Patnaik A., Powell S.F., Gentzler R.D., Martins R.G., Stevenson J.P., Jalal S.I., KEYNOTE-021 investigators Carboplatin and pemetrexed with or without pembrolizumab for advanced, non-squamous non-small-cell lung cancer: a randomised, phase 2 cohort of the open-label KEYNOTE-021 study. Lancet Oncol. 2016;17:1497–1508. doi: 10.1016/S1470-2045(16)30498-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishino M., Ramaiya N.H., Hatabu H., Hodi F.S. Monitoring immune-checkpoint blockade: response evaluation and biomarker development. Nat. Rev. Clin. Oncol. 2017;14:655–668. doi: 10.1038/nrclinonc.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Q., Lin W., Tang X., Li S., Guo L., Lin Y., Kwok H.F. The Roles of microRNAs in Regulating the Expression of PD-1/PD-L1 Immune Checkpoint. Int. J. Mol. Sci. 2017;18:E2540. doi: 10.3390/ijms18122540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cioffi M., Trabulo S.M., Vallespinos M., Raj D., Kheir T.B., Lin M.L., Begum J., Baker A.M., Amgheib A., Saif J. The miR-25-93-106b cluster regulates tumor metastasis and immune evasion via modulation of CXCL12 and PD-L1. Oncotarget. 2017;8:21609–21625. doi: 10.18632/oncotarget.15450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao L., Yu H., Yi S., Peng X., Su P., Xiao Z., Liu R., Tang A., Li X., Liu F., Shen S. The tumor suppressor miR-138-5p targets PD-L1 in colorectal cancer. Oncotarget. 2016;7:45370–45384. doi: 10.18632/oncotarget.9659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miao S., Mao X., Zhao S., Song K., Xiang C., Lv Y., Jiang H., Wang L., Li B., Yang X. miR-217 inhibits laryngeal cancer metastasis by repressing AEG-1 and PD-L1 expression. Oncotarget. 2017;8:62143–62153. doi: 10.18632/oncotarget.19121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen L., Gibbons D.L., Goswami S., Cortez M.A., Ahn Y.H., Byers L.A., Zhang X., Yi X., Dwyer D., Lin W. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat. Commun. 2014;5:5241. doi: 10.1038/ncomms6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu S., Tao Z., Hai B., Liang H., Shi Y., Wang T., Song W., Chen Y., OuYang J., Chen J. miR-424(322) reverses chemoresistance via T-cell immune response activation by blocking the PD-L1 immune checkpoint. Nat. Commun. 2016;7:11406. doi: 10.1038/ncomms11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ahn H., Yang J.M., Kim H., Chung J.H., Ahn S.H., Jeong W.J., Paik J.H. Clinicopathologic implications of the miR-197/PD-L1 axis in oral squamous cell carcinoma. Oncotarget. 2017;8:66178–66194. doi: 10.18632/oncotarget.19842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wei J., Nduom E.K., Kong L.Y., Hashimoto Y., Xu S., Gabrusiewicz K., Ling X., Huang N., Qiao W., Zhou S. MiR-138 exerts anti-glioma efficacy by targeting immune checkpoints. Neuro-oncol. 2016;18:639–648. doi: 10.1093/neuonc/nov292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang Y., He Y., Shi L., Yang L., Wang J., Lian Y., Fan C., Zhang P., Guo C., Zhang S. Co-expression of AFAP1-AS1 and PD-1 predicts poor prognosis in nasopharyngeal carcinoma. Oncotarget. 2017;8:39001–39011. doi: 10.18632/oncotarget.16545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Audrito V., Serra S., Stingi A., Orso F., Gaudino F., Bologna C., Neri F., Garaffo G., Nassini R., Baroni G. PD-L1 up-regulation in melanoma increases disease aggressiveness and is mediated through miR-17-5p. Oncotarget. 2017;8:15894–15911. doi: 10.18632/oncotarget.15213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li C., Liu D.R., Li G.G., Wang H.H., Li X.W., Zhang W., Wu Y.L., Chen L. CD97 promotes gastric cancer cell proliferation and invasion through exosome-mediated MAPK signaling pathway. World J. Gastroenterol. 2015;21:6215–6228. doi: 10.3748/wjg.v21.i20.6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taus Á., Camacho L., Rocha P., Hardy-Werbin M., Pijuan L., Piquer G., López E., Dalmases A., Longarón R., Clavé S. Dynamics of EGFR Mutation Load in Plasma for Prediction of Treatment Response and Disease Progression in Patients With EGFR-Mutant Lung Adenocarcinoma. Clin. Lung Cancer. 2018;19:387–394.e2. doi: 10.1016/j.cllc.2018.03.015. [DOI] [PubMed] [Google Scholar]
- 53.Azumi J., Tsubota T., Sakabe T., Shiota G. miR-181a induces sorafenib resistance of hepatocellular carcinoma cells through downregulation of RASSF1 expression. Cancer Sci. 2016;107:1256–1262. doi: 10.1111/cas.13006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luo M., Wu L., Zhang K., Wang H., Wu S., O’Connell D., Gao T., Zhong H., Yang Y. miR-216b enhances the efficacy of vemurafenib by targeting Beclin-1, UVRAG and ATG5 in melanoma. Cell. Signal. 2018;42:30–43. doi: 10.1016/j.cellsig.2017.09.024. [DOI] [PubMed] [Google Scholar]
- 55.Fader A.N., Roque D.M., Siegel E., Buza N., Hui P., Abdelghany O., Chambers S.K., Secord A.A., Havrilesky L., O’Malley D.M. Randomized Phase II Trial of Carboplatin-Paclitaxel Versus Carboplatin-Paclitaxel-Trastuzumab in Uterine Serous Carcinomas That Overexpress Human Epidermal Growth Factor Receptor 2/neu. J. Clin. Oncol. 2018;36:2044–2051. doi: 10.1200/JCO.2017.76.5966. [DOI] [PubMed] [Google Scholar]
- 56.Jiao X.D., Ding C., Zang Y.S., Yu G. Rapid symptomatic relief of HER2-positive gastric cancer leptomeningeal carcinomatosis with lapatinib, trastuzumab and capecitabine: a case report. BMC Cancer. 2018;18:206. doi: 10.1186/s12885-018-4116-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heydt C., Michels S., Thress K.S., Bergner S., Wolf J., Buettner R. Novel approaches against epidermal growth factor receptor tyrosine kinase inhibitor resistance. Oncotarget. 2018;9:15418–15434. doi: 10.18632/oncotarget.24624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Escrivá-de-Romaní S., Arumí M., Bellet M., Saura C. HER2-positive breast cancer: Current and new therapeutic strategies. Breast. 2018;39:80–88. doi: 10.1016/j.breast.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 59.Untch M., von Minckwitz G., Gerber B., Schem C., Rezai M., Fasching P.A., Tesch H., Eggemann H., Hanusch C., Huober J., GBG and the AGO-B Study Group Survival Analysis After Neoadjuvant Chemotherapy With Trastuzumab or Lapatinib in Patients With Human Epidermal Growth Factor Receptor 2-Positive Breast Cancer in the GeparQuinto (G5) Study (GBG 44) J. Clin. Oncol. 2018;36:1308–1316. doi: 10.1200/JCO.2017.75.9175. [DOI] [PubMed] [Google Scholar]
- 60.Lin Y., Wang X., Jin H. EGFR-TKI resistance in NSCLC patients: mechanisms and strategies. Am. J. Cancer Res. 2014;4:411–435. [PMC free article] [PubMed] [Google Scholar]
- 61.Yang Z., Yang N., Ou Q., Xiang Y., Jiang T., Wu X., Bao H., Tong X., Wang X., Shao Y.W. Investigating novel resistance mechanisms to third-generation EGFR tyrosine kinase inhibitor osimertinib in non-small cell lung cancer patients. Clin. Cancer Res. 2018;24:3097–3107. doi: 10.1158/1078-0432.CCR-17-2310. [DOI] [PubMed] [Google Scholar]
- 62.Hannafon B.N., Ding W.Q. Intercellular communication by exosome-derived microRNAs in cancer. Int. J. Mol. Sci. 2013;14:14240–14269. doi: 10.3390/ijms140714240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cheng N., Cai W., Ren S., Li X., Wang Q., Pan H., Zhao M., Li J., Zhang Y., Zhao C. Long non-coding RNA UCA1 induces non-T790M acquired resistance to EGFR-TKIs by activating the AKT/mTOR pathway in EGFR-mutant non-small cell lung cancer. Oncotarget. 2015;6:23582–23593. doi: 10.18632/oncotarget.4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ricciuti B., Mecca C., Cenci M., Leonardi G.C., Perrone L., Mencaroni C., Crinò L., Grignani F., Baglivo S., Chiari R. miRNAs and resistance to EGFR-TKIs in EGFR-mutant non-small cell lung cancer: beyond ‘traditional mechanisms’ of resistance. Ecancermedicalscience. 2015;9:569. doi: 10.3332/ecancer.2015.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu X.H., Sun M., Nie F.Q., Ge Y.B., Zhang E.B., Yin D.D., Kong R., Xia R., Lu K.H., Li J.H. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol. Cancer. 2014;13:92. doi: 10.1186/1476-4598-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jung E.-J., Santarpia L., Kim J., Esteva F.J., Moretti E., Buzdar A.U., Di Leo A., Le X.F., Bast R.C., Jr., Park S.T. Plasma microRNA 210 levels correlate with sensitivity to trastuzumab and tumor presence in breast cancer patients. Cancer. 2012;118:2603–2614. doi: 10.1002/cncr.26565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yin C., Zhou X., Dang Y., Yan J., Zhang G. Potential Role of Circulating MiR-21 in the Diagnosis and Prognosis of Digestive System Cancer: A Systematic Review and Meta-Analysis. Medicine (Baltimore) 2015;94:e2123. doi: 10.1097/MD.0000000000002123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Melnik B.C. MiR-21: an environmental driver of malignant melanoma? J. Transl. Med. 2015;13:202. doi: 10.1186/s12967-015-0570-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang L.J., He C.C., Sui X., Cai M.J., Zhou C.Y., Ma J.L., Wu L., Wang H., Han S.X., Zhu Q. MiR-21 promotes intrahepatic cholangiocarcinoma proliferation and growth in vitro and in vivo by targeting PTPN14 and PTEN. Oncotarget. 2015;6:5932–5946. doi: 10.18632/oncotarget.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cissell K.A., Rahimi Y., Shrestha S., Hunt E.A., Deo S.K. Bioluminescence-based detection of microRNA, miR21 in breast cancer cells. Anal. Chem. 2008;80:2319–2325. doi: 10.1021/ac702577a. [DOI] [PubMed] [Google Scholar]
- 71.De Mattos-Arruda L., Bottai G., Nuciforo P.G., Di Tommaso L., Giovannetti E., Peg V., Losurdo A., Pérez-Garcia J., Masci G., Corsi F. MicroRNA-21 links epithelial-to-mesenchymal transition and inflammatory signals to confer resistance to neoadjuvant trastuzumab and chemotherapy in HER2-positive breast cancer patients. Oncotarget. 2015;6:37269–37280. doi: 10.18632/oncotarget.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gong C., Yao Y., Wang Y., Liu B., Wu W., Chen J., Su F., Yao H., Song E. Up-regulation of miR-21 mediates resistance to trastuzumab therapy for breast cancer. J. Biol. Chem. 2011;286:19127–19137. doi: 10.1074/jbc.M110.216887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li B., Ren S., Li X., Wang Y., Garfield D., Zhou S., Chen X., Su C., Chen M., Kuang P. MiR-21 overexpression is associated with acquired resistance of EGFR-TKI in non-small cell lung cancer. Lung Cancer. 2014;83:146–153. doi: 10.1016/j.lungcan.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 74.Li W., Zhai L., Wang H., Liu C., Zhang J., Chen W., Wei Q. Downregulation of LncRNA GAS5 causes trastuzumab resistance in breast cancer. Oncotarget. 2016;7:27778–27786. doi: 10.18632/oncotarget.8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Horsham J.L., Kalinowski F.C., Epis M.R., Ganda C., Brown R.A.M., Leedman P.J. Clinical Potential of microRNA-7 in Cancer. J. Clin. Med. 2015;4:1668–1687. doi: 10.3390/jcm4091668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huynh F.C., Jones F.E. MicroRNA-7 inhibits multiple oncogenic pathways to suppress HER2Δ16 mediated breast tumorigenesis and reverse trastuzumab resistance. PLoS ONE. 2014;9:e114419. doi: 10.1371/journal.pone.0114419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hsiao Y.C., Yeh M.H., Chen Y.J., Liu J.F., Tang C.H., Huang W.C. Lapatinib increases motility of triple-negative breast cancer cells by decreasing miRNA-7 and inducing Raf-1/MAPK-dependent interleukin-6. Oncotarget. 2015;6:37965–37978. doi: 10.18632/oncotarget.5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tu C.Y., Chen C.H., Hsia T.C., Hsu M.H., Wei Y.L., Yu M.C., Chen W.S., Hsu K.W., Yeh M.H., Liu L.C. Trichostatin A suppresses EGFR expression through induction of microRNA-7 in an HDAC-independent manner in lapatinib-treated cells. BioMed Res. Int. 2014;2014:168949. doi: 10.1155/2014/168949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Suto T., Yokobori T., Yajima R., Morita H., Fujii T., Yamaguchi S., Altan B., Tsutsumi S., Asao T., Kuwano H. MicroRNA-7 expression in colorectal cancer is associated with poor prognosis and regulates cetuximab sensitivity via EGFR regulation. Carcinogenesis. 2015;36:338–345. doi: 10.1093/carcin/bgu242. [DOI] [PubMed] [Google Scholar]
- 80.Zhu H.Y., Bai W.D., Ye X.M., Yang A.G., Jia L.T. Long non-coding RNA UCA1 desensitizes breast cancer cells to trastuzumab by impeding miR-18a repression of Yes-associated protein 1. Biochem. Biophys. Res. Commun. 2018;496:1308–1313. doi: 10.1016/j.bbrc.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 81.Zheng B., Zhu H., Gu D., Pan X., Qian L., Xue B., Yang D., Zhou J., Shan Y. MiRNA-30a-mediated autophagy inhibition sensitizes renal cell carcinoma cells to sorafenib. Biochem. Biophys. Res. Commun. 2015;459:234–239. doi: 10.1016/j.bbrc.2015.02.084. [DOI] [PubMed] [Google Scholar]
- 82.Vaira V., Roncalli M., Carnaghi C., Faversani A., Maggioni M., Augello C., Rimassa L., Pressiani T., Spagnuolo G., Di Tommaso L. MicroRNA-425-3p predicts response to sorafenib therapy in patients with hepatocellular carcinoma. Liver Int. 2015;35:1077–1086. doi: 10.1111/liv.12636. [DOI] [PubMed] [Google Scholar]
- 83.Mu W., Hu C., Zhang H., Qu Z., Cen J., Qiu Z., Li C., Ren H., Li Y., He X. miR-27b synergizes with anticancer drugs via p53 activation and CYP1B1 suppression. Cell Res. 2015;25:477–495. doi: 10.1038/cr.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shimizu S., Takehara T., Hikita H., Kodama T., Miyagi T., Hosui A., Tatsumi T., Ishida H., Noda T., Nagano H. The let-7 family of microRNAs inhibits Bcl-xL expression and potentiates sorafenib-induced apoptosis in human hepatocellular carcinoma. J. Hepatol. 2010;52:698–704. doi: 10.1016/j.jhep.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 85.Bai S., Nasser M.W., Wang B., Hsu S.-H., Datta J., Kutay H., Yadav A., Nuovo G., Kumar P., Ghoshal K. MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. J. Biol. Chem. 2009;284:32015–32027. doi: 10.1074/jbc.M109.016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xu Y., Huang J., Ma L., Shan J., Shen J., Yang Z., Liu L., Luo Y., Yao C., Qian C. MicroRNA-122 confers sorafenib resistance to hepatocellular carcinoma cells by targeting IGF-1R to regulate RAS/RAF/ERK signaling pathways. Cancer Lett. 2016;371:171–181. doi: 10.1016/j.canlet.2015.11.034. [DOI] [PubMed] [Google Scholar]
- 87.Lou G., Song X., Yang F., Wu S., Wang J., Chen Z., Liu Y. Exosomes derived from miR-122-modified adipose tissue-derived MSCs increase chemosensitivity of hepatocellular carcinoma. J. Hematol. Oncol. 2015;8:122. doi: 10.1186/s13045-015-0220-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.He C., Dong X., Zhai B., Jiang X., Dong D., Li B., Jiang H., Xu S., Sun X. MiR-21 mediates sorafenib resistance of hepatocellular carcinoma cells by inhibiting autophagy via the PTEN/Akt pathway. Oncotarget. 2015;6:28867–28881. doi: 10.18632/oncotarget.4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu K., Liu S., Zhang W., Jia B., Tan L., Jin Z., Liu Y. miR-494 promotes cell proliferation, migration and invasion, and increased sorafenib resistance in hepatocellular carcinoma by targeting PTEN. Oncol. Rep. 2015;34:1003–1010. doi: 10.3892/or.2015.4030. [DOI] [PubMed] [Google Scholar]
- 90.Tang S., Tan G., Jiang X., Han P., Zhai B., Dong X., Qiao H., Jiang H., Sun X. An artificial lncRNA targeting multiple miRNAs overcomes sorafenib resistance in hepatocellular carcinoma cells. Oncotarget. 2016;7:73257–73269. doi: 10.18632/oncotarget.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Qu L., Ding J., Chen C., Wu Z.J., Liu B., Gao Y., Chen W., Liu F., Sun W., Li X.F. Exosome-Transmitted lncARSR Promotes Sunitinib Resistance in Renal Cancer by Acting as a Competing Endogenous RNA. Cancer Cell. 2016;29:653–668. doi: 10.1016/j.ccell.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 92.Xiao W., Lou N., Ruan H., Bao L., Xiong Z., Yuan C., Tong J., Xu G., Zhou Y., Qu Y. Mir-144-3p Promotes Cell Proliferation, Metastasis, Sunitinib Resistance in Clear Cell Renal Cell Carcinoma by Downregulating ARID1A. Cell. Physiol. Biochem. 2017;43:2420–2433. doi: 10.1159/000484395. [DOI] [PubMed] [Google Scholar]
- 93.Zhai W., Sun Y., Guo C., Hu G., Wang M., Zheng J., Lin W., Huang Q., Li G., Zheng J., Chang C. LncRNA-SARCC suppresses renal cell carcinoma (RCC) progression via altering the androgen receptor(AR)/miRNA-143-3p signals. Cell Death Differ. 2017;24:1502–1517. doi: 10.1038/cdd.2017.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fattore L., Mancini R., Acunzo M., Romano G., Laganà A., Pisanu M.E., Malpicci D., Madonna G., Mallardo D., Capone M. miR-579-3p controls melanoma progression and resistance to target therapy. Proc. Natl. Acad. Sci. USA. 2016;113:E5005–E5013. doi: 10.1073/pnas.1607753113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sun X., Li J., Sun Y., Zhang Y., Dong L., Shen C., Yang L., Yang M., Li Y., Shen G. miR-7 reverses the resistance to BRAFi in melanoma by targeting EGFR/IGF-1R/CRAF and inhibiting the MAPK and PI3K/AKT signaling pathways. Oncotarget. 2016;7:53558–53570. doi: 10.18632/oncotarget.10669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Díaz-Martínez M., Benito-Jardón L., Alonso L., Koetz-Ploch L., Hernando E., Teixidó J. miR-204-5p and miR-211-5p Contribute to BRAF Inhibitor Resistance in Melanoma. Cancer Res. 2018;78:1017–1030. doi: 10.1158/0008-5472.CAN-17-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liao H., Bai Y., Qiu S., Zheng L., Huang L., Liu T., Wang X., Liu Y., Xu N., Yan X., Guo H. MiR-203 downregulation is responsible for chemoresistance in human glioblastoma by promoting epithelial-mesenchymal transition via SNAI2. Oncotarget. 2015;6:8914–8928. doi: 10.18632/oncotarget.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cilek E.E., Ozturk H., Gur Dedeoglu B. Construction of miRNA-miRNA networks revealing the complexity of miRNA-mediated mechanisms in trastuzumab treated breast cancer cell lines. PLoS ONE. 2017;12:e0185558. doi: 10.1371/journal.pone.0185558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhan T., Huang X., Tian X., Chen X., Ding Y., Luo H., Zhang Y. Downregulation of MicroRNA-455-3p Links to Proliferation and Drug Resistance of Pancreatic Cancer Cells via Targeting TAZ. Mol. Ther. Nucleic Acids. 2018;10:215–226. doi: 10.1016/j.omtn.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Huang L., Hu C., Cao H., Wu X., Wang R., Lu H., Li H., Chen H. MicroRNA-29c Increases the Chemosensitivity of Pancreatic Cancer Cells by Inhibiting USP22 Mediated Autophagy. Cell. Physiol. Biochem. 2018;47:747–758. doi: 10.1159/000490027. [DOI] [PubMed] [Google Scholar]
- 101.Liang Y., Li Y., Song X., Zhang N., Sang Y., Zhang H., Liu Y., Chen B., Zhao W., Wang L. Long noncoding RNA LINP1 acts as an oncogene and promotes chemoresistance in breast cancer. Cancer Biol. Ther. 2018;19:120–131. doi: 10.1080/15384047.2017.1394543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wu D.M., Hong X.W., Wang L.L., Cui X.F., Lu J., Chen G.Q., Zheng Y.L. MicroRNA-17 inhibition overcomes chemoresistance and suppresses epithelial-mesenchymal transition through a DEDD-dependent mechanism in gastric cancer. Int. J. Biochem. Cell Biol. 2018;102:59–70. doi: 10.1016/j.biocel.2018.06.007. [DOI] [PubMed] [Google Scholar]
- 103.Zhang X.L., Shi H.J., Wang J.P., Tang H.S., Wu Y.B., Fang Z.Y., Cui S.Z., Wang L.T. MicroRNA-218 is upregulated in gastric cancer after cytoreductive surgery and hyperthermic intraperitoneal chemotherapy and increases chemosensitivity to cisplatin. World J. Gastroenterol. 2014;20:11347–11355. doi: 10.3748/wjg.v20.i32.11347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Guo Z., Liu Z., Yue H., Wang J. Beta-elemene increases chemosensitivity to 5-fluorouracil through down-regulating microRNA-191 expression in colorectal carcinoma cells. J. Cell. Biochem. 2018;119:7032–7039. doi: 10.1002/jcb.26914. [DOI] [PubMed] [Google Scholar]
- 105.Luo J., Liu L., Zhou N., Shen J., Sun Q., Zhu Y., Chen M. miR-519b-3p promotes responsiveness to preoperative chemoradiotherapy in rectal cancer patients by targeting ARID4B. Gene. 2018;655:84–90. doi: 10.1016/j.gene.2018.02.056. [DOI] [PubMed] [Google Scholar]
- 106.Li P., Zhang X., Wang H., Wang L., Liu T., Du L., Yang Y., Wang C. MALAT1 Is Associated with Poor Response to Oxaliplatin-Based Chemotherapy in Colorectal Cancer Patients and Promotes Chemoresistance through EZH2. Mol. Cancer Ther. 2017;16:739–751. doi: 10.1158/1535-7163.MCT-16-0591. [DOI] [PubMed] [Google Scholar]
- 107.Ma Y., Zhou G., Li M., Hu D., Zhang L., Liu P., Lin K. Long noncoding RNA DANCR mediates cisplatin resistance in glioma cells via activating AXL/PI3K/Akt/NF-κB signaling pathway. Neurochem. Int. 2018;118:233–241. doi: 10.1016/j.neuint.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 108.Huang Y., Chen G., Wang Y., He R., Du J., Jiao X., Tai Q. Inhibition of microRNA-16 facilitates the paclitaxel resistance by targeting IKBKB via NF-κB signaling pathway in hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 2018;503:1035–1041. doi: 10.1016/j.bbrc.2018.06.113. [DOI] [PubMed] [Google Scholar]
- 109.Eoh K.J., Lee S.H., Kim H.J., Lee J.Y., Kim S., Kim S.W., Kim Y.T., Nam E.J. MicroRNA-630 inhibitor sensitizes chemoresistant ovarian cancer to chemotherapy by enhancing apoptosis. Biochem. Biophys. Res. Commun. 2018;497:513–520. doi: 10.1016/j.bbrc.2018.02.062. [DOI] [PubMed] [Google Scholar]
- 110.Gao J., Wu N., Liu X., Xia Y., Chen Y., Li S., Deng Z. MicroRNA-142-3p inhibits cell proliferation and chemoresistance in ovarian cancer via targeting sirtuin 1. Exp. Ther. Med. 2018;15:5205–5214. doi: 10.3892/etm.2018.6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Corcoran C., Rani S., Breslin S., Gogarty M., Ghobrial I.M., Crown J., O’Driscoll L. miR-630 targets IGF1R to regulate response to HER-targeting drugs and overall cancer cell progression in HER2 over-expressing breast cancer. Mol. Cancer. 2014;13:71. doi: 10.1186/1476-4598-13-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Venturutti L., Cordo Russo R.I., Rivas M.A., Mercogliano M.F., Izzo F., Oakley R.H., Pereyra M.G., De Martino M., Proietti C.J., Yankilevich P. MiR-16 mediates trastuzumab and lapatinib response in ErbB-2-positive breast and gastric cancer via its novel targets CCNJ and FUBP1. Oncogene. 2016;35:6189–6202. doi: 10.1038/onc.2016.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ye X., Bai W., Zhu H., Zhang X., Chen Y., Wang L., Yang A., Zhao J., Jia L. MiR-221 promotes trastuzumab-resistance and metastasis in HER2-positive breast cancers by targeting PTEN. BMB Rep. 2014;47:268–273. doi: 10.5483/BMBRep.2014.47.5.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ye X.M., Zhu H.Y., Bai W.D., Wang T., Wang L., Chen Y., Yang A.G., Jia L.T. Epigenetic silencing of miR-375 induces trastuzumab resistance in HER2-positive breast cancer by targeting IGF1R. BMC Cancer. 2014;14:134. doi: 10.1186/1471-2407-14-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Le X.F., Almeida M.I., Mao W., Spizzo R., Rossi S., Nicoloso M.S., Zhang S., Wu Y., Calin G.A., Bast R.C., Jr. Modulation of MicroRNA-194 and cell migration by HER2-targeting trastuzumab in breast cancer. PLoS ONE. 2012;7:e41170. doi: 10.1371/journal.pone.0041170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ichikawa T., Sato F., Terasawa K., Tsuchiya S., Toi M., Tsujimoto G., Shimizu K. Trastuzumab produces therapeutic actions by upregulating miR-26a and miR-30b in breast cancer cells. PLoS ONE. 2012;7:e31422. doi: 10.1371/journal.pone.0031422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sui M., Jiao A., Zhai H., Wang Y., Wang Y., Sun D., Li P. Upregulation of miR-125b is associated with poor prognosis and trastuzumab resistance in HER2-positive gastric cancer. Exp. Ther. Med. 2017;14:657–663. doi: 10.3892/etm.2017.4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kim Y., Kim H., Park D., Han M., Lee H., Lee Y.S., Choe J., Kim Y.M., Jeoung D. miR-217 and CAGE form feedback loop and regulates the response to anti-cancer drugs through EGFR and HER2. Oncotarget. 2016;7:10297–10321. doi: 10.18632/oncotarget.7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhao Q., Cao J., Wu Y.C., Liu X., Han J., Huang X.C., Jiang L.H., Hou X.X., Mao W.M., Ling Z.Q. Circulating miRNAs is a potential marker for gefitinib sensitivity and correlation with EGFR mutational status in human lung cancers. Am. J. Cancer Res. 2015;5:1692–1705. [PMC free article] [PubMed] [Google Scholar]
- 120.Wu D.W., Wang Y.C., Wang L., Chen C.Y., Lee H. A low microRNA-630 expression confers resistance to tyrosine kinase inhibitors in EGFR-mutated lung adenocarcinomas via miR-630/YAP1/ERK feedback loop. Theranostics. 2018;8:1256–1269. doi: 10.7150/thno.22048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen J., Cui J.D., Guo X.T., Cao X., Li Q. Increased expression of miR-641 contributes to erlotinib resistance in non-small-cell lung cancer cells by targeting NF1. Cancer Med. 2018;7:1394–1403. doi: 10.1002/cam4.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lu Y., Zhao X., Liu Q., Li C., Graves-Deal R., Cao Z., Singh B., Franklin J.L., Wang J., Hu H. lncRNA MIR100HG-derived miR-100 and miR-125b mediate cetuximab resistance via Wnt/β-catenin signaling. Nat. Med. 2017;23:1331–1341. doi: 10.1038/nm.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mussnich P., Rosa R., Bianco R., Fusco A., D’Angelo D. MiR-199a-5p and miR-375 affect colon cancer cell sensitivity to cetuximab by targeting PHLPP1. Expert Opin. Ther. Targets. 2015;19:1017–1026. doi: 10.1517/14728222.2015.1057569. [DOI] [PubMed] [Google Scholar]
- 124.Xue F., Liu Y., Zhang H., Wen Y., Yan L., Tang Q., Xiao E., Zhang D. Let-7a enhances the sensitivity of hepatocellular carcinoma cells to cetuximab by regulating STAT3 expression. OncoTargets Ther. 2016;9:7253–7261. doi: 10.2147/OTT.S116127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Xue F., Liang Y., Li Z., Liu Y., Zhang H., Wen Y., Yan L., Tang Q., Xiao E., Zhang D. MicroRNA-9 enhances sensitivity to cetuximab in epithelial phenotype hepatocellular carcinoma cells through regulation of the eukaryotic translation initiation factor 5A-2. Oncol. Lett. 2018;15:813–820. doi: 10.3892/ol.2017.7399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wuerkenbieke D., Wang J., Li Y., Ma C. miRNA-150 downregulation promotes pertuzumab resistance in ovarian cancer cells via AKT activation. Arch. Gynecol. Obstet. 2015;292:1109–1116. doi: 10.1007/s00404-015-3742-x. [DOI] [PubMed] [Google Scholar]
- 127.Xu H., Zhao L., Fang Q., Sun J., Zhang S., Zhan C., Liu S., Zhang Y. MiR-338-3p inhibits hepatocarcinoma cells and sensitizes these cells to sorafenib by targeting hypoxia-induced factor 1α. PLoS ONE. 2014;9:e115565. doi: 10.1371/journal.pone.0115565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yang F., Li Q.J., Gong Z.B., Zhou L., You N., Wang S., Li X.L., Li J.J., An J.Z., Wang D.S. MicroRNA-34a targets Bcl-2 and sensitizes human hepatocellular carcinoma cells to sorafenib treatment. Technol. Cancer Res. Treat. 2014;13:77–86. doi: 10.7785/tcrt.2012.500364. [DOI] [PubMed] [Google Scholar]
- 129.Mao K., Zhang J., He C., Xu K., Liu J., Sun J., Wu G., Tan C., Zeng Y., Wang J., Xiao Z. Restoration of miR-193b sensitizes Hepatitis B virus-associated hepatocellular carcinoma to sorafenib. Cancer Lett. 2014;352:245–252. doi: 10.1016/j.canlet.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 130.Liu K., Liu S., Zhang W., Ji B., Wang Y., Liu Y. miR-222 regulates sorafenib resistance and enhance tumorigenicity in hepatocellular carcinoma. Int. J. Oncol. 2014;45:1537–1546. doi: 10.3892/ijo.2014.2577. [DOI] [PubMed] [Google Scholar]
- 131.Jin W., Chen L., Cai X., Zhang Y., Zhang J., Ma D., Cai X., Fu T., Yu Z., Yu F., Chen G. Long non-coding RNA TUC338 is functionally involved in sorafenib-sensitized hepatocarcinoma cells by targeting RASAL1. Oncol. Rep. 2017;37:273–280. doi: 10.3892/or.2016.5248. [DOI] [PubMed] [Google Scholar]
- 132.Gao C., Peng F.H., Peng L.K. MiR-200c sensitizes clear-cell renal cell carcinoma cells to sorafenib and imatinib by targeting heme oxygenase-1. Neoplasma. 2014;61:680–689. doi: 10.4149/neo_2014_083. [DOI] [PubMed] [Google Scholar]
- 133.Xu Z., Yang F., Wei D., Liu B., Chen C., Bao Y., Wu Z., Wu D., Tan H., Li J. Long noncoding RNA-SRLR elicits intrinsic sorafenib resistance via evoking IL-6/STAT3 axis in renal cell carcinoma. Oncogene. 2017;36:1965–1977. doi: 10.1038/onc.2016.356. [DOI] [PubMed] [Google Scholar]
- 134.Liu F., Chen N., Gong Y., Xiao R., Wang W., Pan Z. The long non-coding RNA NEAT1 enhances epithelial-to-mesenchymal transition and chemoresistance via the miR-34a/c-Met axis in renal cell carcinoma. Oncotarget. 2017;8:62927–62938. doi: 10.18632/oncotarget.17757. [DOI] [PMC free article] [PubMed] [Google Scholar]