Abstract
In nature, a limited, conservative set of amino acids are utilized to synthesize proteins. Genetic code expansion technique reassigns codons and incorporates noncanonical amino acids (ncAAs) through orthogonal aminoacyl-tRNA synthetase (aaRS)/tRNA pairs. The past decade has witnessed the rapid growth in diversity and scope for therapeutic applications of this technology. Here, we provided an update on the recent progress using genetic code expansion in the following areas: antibody-drug conjugates (ADCs), bispecific antibodies (BsAb), immunotherapies, long-lasting protein therapeutics, biosynthesized peptides, engineered viruses and cells, as well as other therapeutic related applications, where the technique was used to elucidate the mechanisms of biotherapeutics and drug targets.
Keywords: Genetic code expansion, Noncanonical amino acids, Biotherapeutics, Antibody-drug conjugates, Live-attenuated vaccines
1. Introduction
While DNA and RNA are two ways how creatures on Earth store hereditary information, it is protein that constitutes most of cells’ dry mass and carry out most information-bearing functions. Natural proteins are synthesized by 20 canonical amino acids. The different chemical properties of these building blocks equip proteins with multifarious structures and functions, however, at the same time, restrict the development of protein properties. Post-translational modifications (PTM) such as methylation, phosphorylation and glycosylation, as well as the binding of co-enzyme factors are different strategies natural proteins take to overcome these limitations at a certain extent. To unleash the full power of amino acid diversities and meet the challenges for protein related applications, scientists have devised a variety of ways to incorporate noncanonical amino acids (ncAAs) into proteins, over the past few decades, greatly expanding protein properties.
Genetic code expansion is one of these techniques, which allows site-specific incorporation of ncAAs into any protein of interest in cells. This is achieved with engineered aminoacyl-tRNA synthetase/tRNA (aaRS/tRNA) pairs which can specifically aminoacylates its cognate tRNA with a given ncAA, devoted to decoding reassigned codons, which are typically nonsense codons (UAG/UGA/UAA). The aminoacylated suppressor tRNA charged with the provided ncAA is then recognized by ribosome allowing the ncAA to be site-specifically incorporated into the synthesizing peptides during translation in response to assigned codons. This system must be orthogonal with the native translation system, including endogenous tRNA, canonical amino acids as well as endogenous aaRS (Fig. 1(a)).
Fig. 1.
(a) Principles of genetic code expansion technique. aaRS: aminoacyl-tRNA synthetase. (b) Applications of ncAAs in studying protein structures and functions.
Several aaRS/tRNA pairs have been developed to incorporate more than 200 different ncAAs in different species. For recombinant expression therapeutic proteins in E. coli, the most widely used aaRS/tRNA pairs are the Archean TyrRS/tRNATyr pair from Methanocaldococcus jannaschii (MjTyrRS/tRNATyr) [[1], [2], [3], [4]] and the Archean pyrrolysine aaRS/tRNAPyl pair (Mb/MmPylRS/tRNAPyl) [[5], [6], [7], [8], [9]]. The MjTyrRS/tRNATyr pair was the first genetic code expansion system developed for site-specific ncAAs incorporation in living cells in 2000 [1] and has now been widely used in prokaryotic genetic code expansion. This system is highly efficient in incorporation of aromatic ncAAs with a wide range of novel chemical, physical and biological properties. While the MjTyrRS/tRNATyr pair is commonly used in E. coli, it is not orthogonal in higher organisms. Unlike other systems, Mb/MmPylRS/tRNAPyl pair was a gift from nature. The discovery of this naturally existing orthogonal system was first described in 2002 and it allows the genetic incorporation of a natural ncAA, pyrrolysine, which was then referred as the 22nd amino acid [10,11], site-specifically into the proteome of living cells in response to amber stop codon. The pyrrolysine system has shown a great orthogonality in both prokaryotes and eukaryotic cells. Such orthogonality enables the evolution of PylRS/tRNAPyl pairs in E. coli and the evolved PylRSs can be directly applied in mammalian cells. In addition, the evolved PylRS mutants can incorporate not only pyrrolysine analogs but also a diverse side chain structures [12].
Systems most widely used in eukaryotes nowadays are E. coli TyrRS/tRNATyr (EcTyrRS/tRNATyr) [[13], [14], [15], [16], [17]], E. coli LeuRS/tRNALeu (EcLeuRS/tRNALeu) [16,18,19] and Mb/MmPylRS/tRNAPyl pair [17,[20], [21], [22], [23], [24], [25], [26], [27], [28]]. The incorporation of ncAAs in Saccharomyces cerevisiae was first achieved with E. coli TyrRS/tRNATyr pair by Chin, J. W. et al. [14]. The E. coli TyrRS has also been cooperated with the Bacillus stearothermophilus tRNATyr [13,29] to achieve more efficient suppression. These pairs successfully introduced phenylalanine derivatives containing chemical structures like benzophenone [15], ketone [30], iodide [30] and azide [30] groups. E. coli LeuRS/tRNALeu pair was first developed in 2004 [18]. A variety of structures can be incorporated by this pair through mult-site mutations for its tolerance for structural diversity. Mb/MmPylRS/tRNAPyl pair has also been extensively applied in mammalian cells as previously described.
Other platforms like TrpRS/tRNATrp [7,[31], [32], [33], [34]], PheRS/tRNAPhe [35,36], LysRS/tRNALys [37], SepRS/tRNACys [38,39] have also been developed and were further explained elsewhere [40]. The orthogonality feature of these pairs determines the disadvantage that two distinct platforms need to be developed for applications of one single ncAA in both bacterial and eukaryotic cells. Italia, J. S. et al. recently reported a novel strategy to expand the genetic code of E. coli by replacing its endogenous pair with an eukaryotic-archaeal counterpart, achieving a general way to develop new aaRS/tRNA pairs for both E. coli and eukaryotes [34,41].
Genetic code expansion has been commonly used to study protein structures and functions both in vitro and in vivo (Fig. 1(b)). Chemical cross-linkable ncAAs were used to catch proximity-based protein-protein interactions in live cells [42,43]. Fluorescent ncAAs [44] or fluorophores that react with ncAAs through bioorthogonal ligation reactions [45,46] were used in live cell imaging studies. Through incorporation of photo-switchable [47,48] or chemical-switchable ncAAs [49], a new field in controllable protein function has also been opened. In addition to the above applications, expanding the genetic code has shown great advantages in the study of PTM, like serine phosphorylation [39,50], threonine phosphorylation [51], tyrosine phosphorylation [52], lysine acetylation [6], lysine crotonylation [53] and lysine hydroxyisobutyrylation [54].
The site-specific modification characteristic of genetic code expansion technique makes it a powerful tool in the improvement of pharmacological properties of protein therapeutics as well as in developing novel therapeutic methods. In this review, we will be focused on the therapeutic applications of genetic code expansion in recent years.
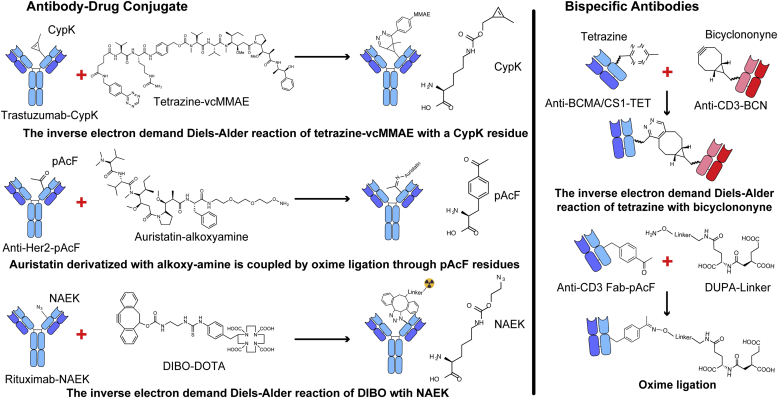
2. Antibody-drug conjugate (ADC)
Antibody-drug conjugate (ADC) is a class of novel biopharmaceutical drugs with a significantly enhanced therapeutic window against cancer. Four ADCs have been approved by FDA and EMA in clinical use [55]. Conventional method of ADC production is through nonselectively coupling to lysine or cysteine residues, yielding heterogeneous products, which decreases ADC potency, circulation half-life, stability, tolerability as well as antigen binding [56]. In 2010, the first approved ADC, gemtuzumab ozogamicin (Mylotarg™) was voluntarily withdrawn from the market due to toxicity and a lack of efficacy [57]. To avoid the problem of heterogeneity, different ways have been exploited to engineer ADCs. Reactive cysteine residues were introduced into antibody-Fab, which is called THIOMAB (the antibodies with engineered reactive cysteine residues), exhibiting an improved therapeutic window [58]. However, in the process of producing THIOMABs, other side products, like triple light antibodies (3LC) may form [59]. The inherent cysteine-maleimide instability and the multistep process precluded it from clinical applications as well. Other site-specific methods utilizing enzymatic modifications like β1,4-galactosyltransferase (Gal T), α2,6-sialyltransferase (Sial T) [60], transglutaminase (TG) [61], formylglycine-generating enzyme (FGE) or anaerobic sulfatase-maturating enzyme (anSME) [62] have also been demonstrated to successfully introduce specific residues into antibodies. Nevertheless, the positions that can be targeted on antibodies are limited. The incorporation of ncAAs using genetic expression systems like auxotrophic strain or selenocysteine have provided new insights [63,64].
Genetic code expansion owns the advantage of synthesizing homogenous ADCs through incorporating ncAAs with different moieties at one or more specific sites, such as ketone, azide, alkyne and alkene groups [57,65]. To produce antibodies with excellent potency, pharmacokinetics and safety, p-acetylphenylalanine (pAcF) was genetically incorporated into two sites on an anti-Her2 antibody Fab fragment and coupled with the microtubule toxin auristatin F (AF) (Fig. 2) [66]. The synthesis of chemical defined CXCR4-auristatin ADC, anti-CD11a IgG conjugated with liver X receptor (LXR) agonists or phosphodiesterase 4 (PDE4) embraced the same strategy [[67], [68], [69]]. These ADCs not only showed significant effects in in vivo disease models but also reduced unwanted side-effects due to the improvement of drug specificity. To produce more complex proteins like full-length antibodies, Tian, F et al. utilized CHO cells to develop a stable expression system and promoted the titer of antibodies containing ncAAs to over 1 g/L [70]. The genetic code expansion has also been used in radioimmunotherapy. Through incorporation of an azido group-bearing amino acid, Nε-2-azideoethyloxycarbonyl-l-lysine (NAEK), a newly synthesized bifunctional linker DIBO-DOTA was site-specifically conjugated to the anti-CD20 antibody rituximab, followed by radiolabeling (Fig. 2) [71]. To improve conjugation chemistry, Koehler, C et al. utilized MutiBac system in insect cells to introduce large, hydrophobic, carbocyclic reactive ncAAs into antibodies [27]. Later, Oller-Salvia, B et al. engineered mammalian expression systems that efficiently incorporated a cyclopropene derivative of lysine (CypK) into trastuzumab through genetic code expansion [72]. Above all else, this technique has also been applied to immunosuppressive drugs. This was exemplified by the site-specific conjugation of dasatinib with humanized antibody HLCX, selectively delivering this Lck inhibitor to human T lymphocytes [73].
Fig. 2.
Coupling reactions used in genetic code expansion. CypK: cyclopropene derivative of lysine; pAcF: p-acetylphenylalanine; NAEK: Nε-2-azideoethyloxycarbonyl-l-lysine; MMAE: monomethyl auristatin E; DIBO: 4-dibenzocyclooctynol; DOTA: 1,4,7,10-tetraazacyclote-tradecane-1,4,7,10-tetraacetic acid; DUPA: 2-[3-(1, 3-dicarboxy propyl)-ureido] pentanedioic acid.
3. Bispecific antibody (BsAb)
Bispecific antibody is a type of bioengineered antibodies with the ability to simultaneously recognize two different types of epitopes. In 2009, catumaxomab (Removab) was approved in the European Union for the treatment of malignant ascites [74]. The FDA granted accelerated approval of blinatumomab (Blincyto) in December, 2014 for the treatment of Philadelphia chromosome-negative relapsed or refractory precursor B-cell acute lymphoblastic leukemia (R/R ALL) [75]. However, the development of bispecific antibodies is hampered by the production of correctly assembled structures (i.e., correct assembly of heavy and light chains). One way to produce bispecific antibodies is through chemical conjugations using lysine or cysteine residues within the antibody, which often yields heterogenous products unsuitable for therapeutic applications.
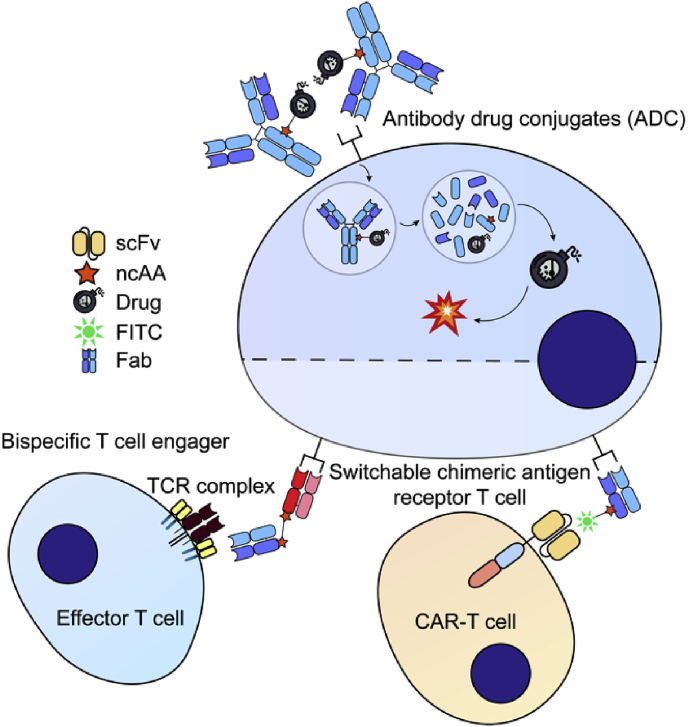
Through site-specific encoding pAcF into anti-Her2 and anti-CD3 Fabs and subsequently coupling the Fabs to bifunctional ethylene glycol linkers with an alkoxy-amine on one end and an azide or cyclooctyne group on the other, an anti-Her2/anti-CD3 bispecific antibody was synthesized by a copper-free click chemistry, effectively crosslinking HER2+ cells and CD3+ cells (Fig. 3) [76,77]. Similar strategy has also been applied to the synthesis of a novel bispecific antibody, αCLL1-αCD3, which completely eliminated established tumors in a cell line xenograft model, showing potential as a treatment for acute myeloid leukemia (AML) [78]. A general and straightforward approach using Watson-Crick base pairing interactions has been demonstrated for producing homodimeric, heterodimeric and multimeric Fabs with defined composition, valency and geometry. The chemical reactive ncAA in this platform was used to site-specifically couple oligonucleotides or peptide nucleic acids (PNAs) to antibodies, with the assemblies showing enhanced anti-tumor activity [79]. Different types of chemical reactions have been constantly exploited to develop novel bispecific antibodies. In 2015, Ramadoss, N. S et al. utilized the reaction between tetrazine (TET) and bicyclononyne (BCN) to develop anti-BCMA and anti-CS1 bispecific antibodies (Fig. 2). The resulting BiFab-BCMA robustly activated T cells and mediated rapid tumor regression [80].
Fig. 3.
Antibody-related applications of an expanded genetic code. scFv: single-chain variable fragment; ncAA: noncanonical amino acid; FITC: fluorescein isothiocyanate; Fab: antigen-binding fragment; TCR: T cell receptor; CAR: chimeric antigen receptor.
Apart from bispecific antibodies, bispecific targeting therapeutics consisting of a small molecule 2-[3-(1, 3-dicarboxy propyl)-ureido] pentanedioic acid (DUPA), that selectively targets a tumor-associated antigen, prostate-specific membrane antigen (PSMA) were also developed (Fig. 2). With DUPA conjugating to pAcF site-specifically expressed on anti-CD3 Fab, the EC50 of the conjugate has been reduced to 100pM, and serum half-life has been improved to 5–6 h, resulting effective in a treatment in prostate cancer xenograft mouse models [81]. Same strategy was also employed to synthesize an anti-CD3 Fab-folate conjugate [82] and a switchable αGCN4-Fab conjugate [83].
4. Chimeric antigen receptor T (CAR-T) cell immunotherapy
Chimeric antigen receptor T (CAR-T) cell therapy is a promising immunotherapy for cancer treatment. CARs endow the engineered T cells derived from patients to recognize cancer cells through equipment of an anti-tumor single-chain variable fragment (scFv), overcoming the limitations of the need for MHC expression. In August 2017, a CAR-T therapy targeting CD19 was approved by FDA for the treatment of refractory pre-B cell acute lymphoblastic leukemia and diffuse large B cell lymphoma [84]. The conventional CAR-T cell therapy is facing a number of challenges. One of the most important obstacles is the severe cytokine release syndrome (CRS). Hence, it is important to control T cell activities when necessary. In 2012, Tamada, K et al. developed an anti-tag CAR technology which was adaptable and capable of regulating CAR T-cell functions. T cells were genetically engineered to express a CAR that recognizes fluorescein isothiocyanate (FITC) molecules and various antibodies conjugated with FITC served as an intermediary [85]. However, antibodies in this study were conjugated with FITC using nonspecific labeling methods, with an average of 3 FITC molecules conjugating per one molecule of antibody. FITC-labelled Abs-induced in vivo anti-FITC immune responses might shorten the half-life of these intermediary molecules and thus weakened curative effects. In this case, a site specifically modified “switch” molecule is needed to minimize the potential for immunogenicity. Ma, J. S. Y et al. and Cao, Y et al. later developed switchable CAR-T (sCAR-T) systems based on genetic code expansion, in which anti-CD19, anti-CD22 and anti-Her2 antibody fragments were site-specifically modified with FITC through para-azidophenylalanine (pAzF) and mediated distinct spatial interactions between sCAR-T cells and cancer cells (Fig. 3) [86,87]. The strictly switch dose-dependency of this sCAR-T system enabled CAR-T cell activity to become potent and controllable, thus improving efficacy and safety. Furthermore, this system is able to target multiple antigens without further genetic engineering of a patient's T cells, making it a versatile immunotherapy.
5. Increasing therapeutic agent serum half-lives
PEGylation is a well-established technique for improving the pharmacokinetics and biocompatibility of therapeutic agent with short in vivo half-lives. The first unspecific multi-PEGylated drug, Adagen, was approved by FDA in 1990 [88]. PEGylation is usually achieved through chemical conjugation to reactive side chains [89]. The acquired heterogeneous mixture gave rise to difficulties in batch-to-batch reproducibility and quality control, making site-specific PEGylation attractive.
In spite of the efficacy of human growth hormone (hGH) therapy for treatment for pathological short stature, it requires daily subcutaneous injection due to its short circulating half-life [90]. Through site-specific conjugation of hGH containing pAcF with PEG, the homogeneous hGH variant was demonstrated to be as effective and safe as the native hGH. Meanwhile, the injection frequency was reduced to once a week [90]. In addition, site-specific multi-PEGylation of hGH was achieved by Wu, L et al., which showed further reduced immunogenicity and improved pharmacokinetic profiles [91]. Apart from hGH, site-specific PEGylation using genetic code expansion has also been expanded to the optimization of IFN-α2b [92], fibroblast growth factor 21 (FGF21) [93] as well as AAV vectors [94]. Similar strategy was developed to achieve site-specific POxylation of Interleukin-4 (IL-4), either [95].
In addition to site-selective conjugation, ncAAs themselves are able to enhance protein stability through formation of more stable bonds. ncAAs containing long side-chain thiols were incorporated into β-lactamase, forming unnatural disulfide bond and exhibiting significantly enhanced thermostability both in vitro and in vivo [96]. Genetically incorporation of p‐fluorophenylalanine (pFF) [97] or para‐isothiocyanate phenylalanine (pNCSF) [98] was also capable of enhancing protein stability at elevated temperature.
6. Biosynthesized therapeutic peptides
Genetic code expansion was used in combination with split intein catalyzed ligation of proteins and peptides (SICLOPPS) to construct a large cyclic peptide library, which was subsequently harnessed to evolve an HIV protease inhibitor [99]. More recently, co-translational and site-specific incorporation of ncAAs has been applied to the biosynthesis of ribosomally synthesized and post-translationally modified peptide (RiPP) natural products to expand their chemistry and structure repertoire. For example, incorporation of ncAAs at discrete positions on Nisin expanded building blocks that can be biosynthetically incorporated into lanthipeptide and diversified macrocyclic topologies of this antibacterial peptide [100]. Likewise ncAAs have been successfully introduced into other lanthipeptides [101,102], lasso peptides [103,104], macrocyclic peptides [105], thiopeptides [106] as well as in the natural product cinnamycin, recently [107].
7. Site specifically modified viruses
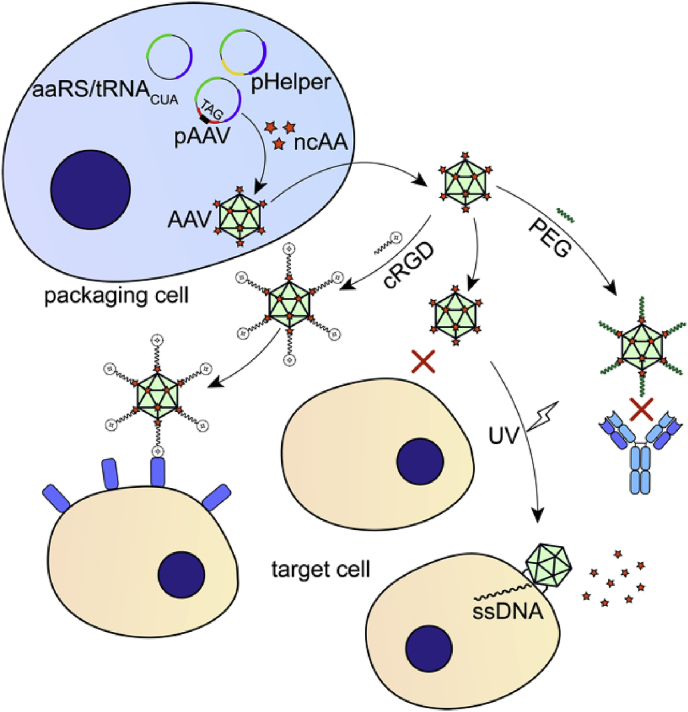
Adeno-associated virus (AAV) is a small non-enveloped human virus that has been proven as a viral vector for gene therapy with its attractive feature of lacking apparent pathogenicity, replication deficiency, persistent transgene expression as well as the capability to infect nondividing and dividing cells [108]. Its icosahedral capsid is made up of three capsid proteins, VP1, VP2, and VP3 [109]. However, the serum instability, immunogenicity and nonspecific cell infectivity remain major obstacles for effective gene transfer. Although reengineering of altered capsid proteins [110,111], fusion of high-affinity ligands to capsid proteins [112] and chemical conjugation of small molecule effector functionality [113] were applied to overcome these challenges, few method can produce heterogeneous vectors for specific targets.
To alleviate the immunogenicity and instability, polyethylene glycols (PEG) was covalently conjugated to AAV-2 site-selectively through genetic code expansion (Fig. 4) [94]. In this study, NAEK was introduced to a series of chosen sites on the capsid protein VP1. PEGylation at these sites showed protective effect and improvement in stability. Selective tropism towards αvβ3 integrin receptors overexpressing tumor cells was enhanced through bioorthogonally and stoichiometrically attachment of AAV2 with a cyclic-RGD peptide precisely, constructing chimeric viruses as simple and viable targeting delivery vehicles (Fig. 4) [114,115]. This method greatly expanded the versatility of AAV vectors for it could be easily applied to different ligands and to other types of viral vectors for gene therapy. Recently, Erickson, S. B et al. combined AAV with photocaged derivatives of lysine to precisely perturb virus-host interaction with high in vivo spatial and temporal control [116]. A photocaged amino acid NBK was site-specifically incorporated to a functionally important site on the virus capsid protein VP1, abrogating the positive charge on the lysine residue and subsequently undergoing efficient decaging to non-toxic products upon irradiation with 365 nm light (Fig. 4). With this methodology, it is able to synchronize the invading viruses at a specific stage. Fluorescent molecules were also lately labelled to AAV2 for visualization and tracking of virus vectors [72].
Fig. 4.
Applications of genetic code expansion related to viral vectors. aaRS: aminoacyl-tRNA synthetase; ncAA: noncanonical amino acid; AAV: adeno-associated virus; cRGD: cyclic Arg-Gly-Asp; PEG: polyethylene glycols.
8. Therapeutic vaccines
8.1. Protein-based vaccines
Immunization of patients with self-proteins or specific epitopes like tumor antigens is difficult due to weakly immunogenicity and immunotolerance. A number of strategies including adjuvants, the introduction of foreign immunodominant T-helper (TH) epitopes into chimeric antigens [117,118], chemical derivatization of self-antigens [119], and DNA vaccines [120] have been exploited to break immunological self-tolerance. Autologous tumor cells modified with a hapten, dinitrophenyl (DNP), was demonstrated to elicit patients’ immune responses cross-reacting with unmodified, native tumor-cell antigens, likely due to the formation of strong stacking and van der Waals interaction [121]. Therefore, genetic code expansion provided a way to introduce immunogenic ncAAs into autologous proteins and induce immune responses to these self-tolerated epitopes. Site-specific incorporation of p-nitrophenylalanine (pNO2Phe) to murine tumor necrosis factor-α (mTNF-α) successfully generated a sustained high-titer antibody response to both parental and antigenically distinct mTNF-α variants without the need for strong adjuvants [122]. Another DNP-containing ncAA, N6‐(2‐(2,4‐dinitrophenyl)acetyl)lysine (DnpK) was later incorporated into HEK 293T cells, although unstable in Escherichia coli [123]. Sulfotyrosine (SO3Tyr) and 3-nitrotyrosine (3NO2Tyr), two PTM-related ncAAs were introduced to specific sites in mTNF-α and EGF, indicating that PTM might play a role in autoimmune disorders [124]. This methodology could also be applied to a self-protein unrelated to immune function, murine retinol-binding protein 4 (RBP4) [125] and might as well be applied to other weakly immunogenic antigens.
8.2. Organism-based vaccines
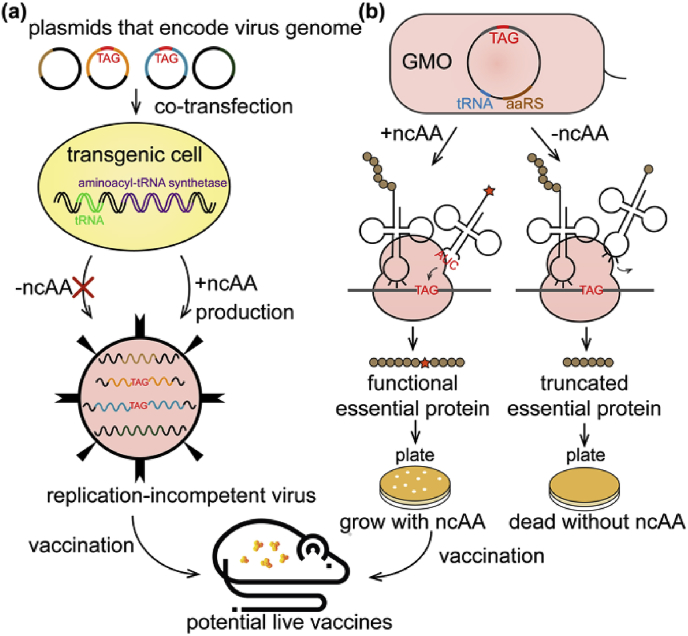
Although traditional live-attenuated vaccines (LAVs) are potent, the potential pathogenic consequences have aroused safety concerns [126]. Genetic code expansion strategy was first coupled with intact and infectious virus engineering using hepatitis D virus (HDV) as a model system [127]. The resultant HDV virions maintained near wild-type viability and infectivity. Then this strategy was expanded to influenza A viruses and human immunodeficiency virus type 1 (HIV-1) virus for the development of LAVs and could be potentially adapted to almost any viral vaccines. Live but replication-incompetent influenza A virus vaccine was generated in a special transgenic cell line harboring MbPylRS/tRNAPylCUA, eliciting robust immunity against both wild type and ncAA-containing strains (Fig. 5(a)) [128]. By means of introducing blank codons on HIV-1 essential proteins within the genome, all-in-one live-attenuated HIV-1 mutants were constructed, and HIV-1 viability was precisely controlled by ncAAs [129,130]. This live-attenuated HIV-1 vaccine constructed through genetic code expansion marked an important step towards combating AIDS pandemic worldwide.
Fig. 5.
(a) Principles of live-attenuated vaccines (LAVs) using genetic code expansion. ncAA: noncanonical amino acid. (b) Principles of genetically modified organisms (GMOs).
Genetic code expansion showed its enormous potential in developing organisms strictly depending on ncAA for growth. After ncAA incorporation had been achieved in the intracellular pathogen Mycobacterium tuberculosis (Mtb), which facilitated studies of tuberculosis (TB) vaccine development [131], increasing number of genetically modified organisms (GMOs) were developed. Unlike viruses, evolutionary pressure compels cellular organisms to impose mutagenic drift, environmental supplementation and horizontal gene transfer (HGT), resulting in high escape frequencies [132]. Computationally redesign of essential enzymes and the combination of multiple redesigned enzymes resulted in remarkable reduced escape rate, alleviating this problem to some degree [132]. Using multiplex automated genome engineering, TAG codons were introduced into permissive locations of 22 essential genes in an Escherichia coli strain that lacked all TAG codons and release factor 1, creating a notable strain with robust growth and undetectable escape frequencies (Fig. 5(b)) [133]. To addict the modified organisms to ncAAs, a selection method was developed using TEM-1 β-lactamase as a model system [134].
Inspired by PTM, scientists developed a general strategy to create genetically modified organisms with stringent dependency on the presence of N‐ε‐acetyl‐L‐Lys (AcK), which blocked a functionally essential lysine residue required for cell growth and can be rescued by endogenous deacetylases. The combination of this approach with a barnase-based suicide switch finally gave a low escape frequency [135]. Other ncAAs with specific chemical properties have also been utilized for biological containment. The hydrophobic amino acid p-benzoyl-l-phenylalanine (pBzF) was introduced to a homodimeric chorismate mutase (CM) to disrupt protein-protein interfaces for its large nonpolar side chain which cannot be replaced by 20 canonical amino acids in theory. Through screening of ncAA surrounding residues, CM was evolved to an enzyme that could only retain active in presence of pBzF, thus developing Escherichia coli strain dependent on ncAAs [136]. However, this deficiency might be complemented by aromatic amino acids in the host. Afterwards, replacing key histidine (His) residues in essential metalloproteins with 3-methyl-l-histidine (MeH) provided a straightforward strategy to solve this problem [137]. Due to the requirement of two single nucleotide mutations to substitute UAG codon with native codons for His (CAU and CAC), this strategy showed extremely low reversion rate and was feasible to be applied to ncAA conditional live vaccines.
9. Therapeutic related applications
Genetic code expansion has also been applied to elucidate the mechanisms of drug targets and biotherapeutics. For example, the genetic code expansion system has been used to explore interactions between drug targets like G protein-coupled receptors (GPCRs) and their binding partners (antibodies or ligands). Back in 2008, the noncanonical amino acids with reactive keto groups, pAcF and pBzF, were incorporated into two different GPCRs, CCR5 and rhodopsin at different sites [29]. Later, this strategy was further applied to map the antibody epitopes on another GPCR, CXC chemokine receptor 4 (CXCR4) [138]. Similar strategy was also applied to map the binding path of neuropeptide Urocortin-I (Ucn1) to the corticotropin releasing factor receptor type 1 (CRF1R) [139], the binding sites between antidepressant drugs and human serotonin transporter (hSERT) [140] and the interactions between exendin-4 peptide and the glucagon-like peptide-1 receptor (GLP-1R) [141], which may contribute to the pursuit of structure-based drug design.
Apart from these applications, it is worth mentioning that clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system have also been combined with a genetically installed photocaged lysine (PCK), achieving opto-chemical controllable gene editing [142]. Although the CRISPR system has not yet been applied in clinic, it has shown a great potential in future gene therapy and cell therapy. We think that the genetic code expansion technology has not yet shown its full potential in the gene editing field.
10. Challenges and prospects
Since the first ncAA, O-methyl-l-tyrosine was incorporated into proteins in Escherichia coli, expansion of the genetic code has allowed over 200 ncAAs to be site-specifically incorporated into proteins in bacteria, yeast [14,16,18,20,[142], [143], [144], [145], [146], [147]], plant [23], worm [21,148,149], insect [22,25,27,150], fish [151] and mammal [13,15,17,19,[24], [25], [26],28,31,34,144,[151], [152], [153]], including stem cells [154,155], thus affording novel avenues for biotherapeutic applications both in vitro and in vivo. Drug candidates produced with genetic code expansion like αHER2 ADC (Ambrx, ARX788) and PEG-FGF21 (Ambrx, ARX618) are in phase 1 and phase 2 clinical trial respectively.
Although the competence of precise and highly tailored structural perturbations to proteins provides abundant advantages, an expanded genetic code is far from reaching its full potential. While ncAAs were incorporated at high levels in E. coli and several cell lines (e.g. HEK293T cells), one of the major challenges of this technique is the low suppression efficiency in more challenging cell lines and species due to its competition with endogenous termination system. This was partly solved by engineering components in the expression system like eRF1 [[156], [157], [158]], the elongation factor Tu (EF-Tu) [159], ribosome [160] and the nonsense-mediated mRNA decay (NMD) pathway [16,28], codon reassignment [161,162] or by improving expression vectors [163]. Up to 30 ncAA residues incorporated with high yields and accuracy into one recombinant protein can now be achieved in E. coli [4]. Optimization of tRNA structure have also been applied in bacteria. However, the evolution of the tRNA sequence in mammalian cells is still challenging [164]. This might also arouse the suspicion that suppressor tRNAs would have toxic effects to cell fitness due to uncontrolled proteome-wide ncAA incorporation. Quadruplet codons offer an optional route to avoiding this problem [3,37]. Another issue is the limited ncAA core structure motif owing to the limited orthogonal pairs. Many natural PTMs still cannot be encoded either in natural or mimic forms. This can only be resolved by exploiting novel aaRS/tRNA system and higher throughput screening methods. Recently, a phage-assisted continuous evolution (PACE) [165] and a phage-assisted noncontinuous evolution (PANCE) [166] have been developed, achieving rapid production of orthogonal aaRS variants that are of highly activity and amino acid specificity. Although PylRS does not recognize the tRNAPyl anticodon, the fact that many aaRS use the anticodon as an identity element [167] and tRNAPylUCA along with tRNAPylUUA display much lower suppression efficiency than tRNAPylCUA [168] largely limited the incorporation of different ncAAs at the same time. Willis, J. C. et al. recently created new PylRS/tRNAPyl pairs that are orthogonal with MmPylRS/tRNAPyl pairs [169].
Future directions, in terms of biotherapeutic applications, include genetic encoding of more building blocks with multifarious chemical, physical and biological characteristics, enhancing suppression efficiencies in therapeutic related species, and to apply this system to relevant therapeutic agents including proteins, cells and even organisms. Merging between genetic code expansion with other disciplines like synthetic biology may force it to a low-cost and available technique to a great extent. Currently, the genetic code expansion technology can allow a maximum of two additional ncAAs to be site-specifically incorporated into one protein [9,24,170,171]. Recent advances in cell engineering [132,133], codon reassignment [172] and expansion of the genetic alphabet [173,174] would theoretically allow the creation of organisms with multiple ncAAs in the proteome. On the other hand, more mutually orthogonal systems and structurally diverse ncAAs need to be developed to accommodate these engineered organisms with free codons or expanded codons. And more importantly, more researchers need to apply the genetic code expansion technique in their research to exploit new applications and functions of ncAAs, which will eventually benefit human therapy. As this technique develops, we are sure that new generations of biotherapeutics will emerge.
Acknowledgments
We thank the following agencies for financial support on related ongoing projects in our lab: National Key Research and Development Program of China (No. 2016YFA0201400), the National Natural Science Foundation of China (21778005), Peking University Health Science Center (BMU20160537 and BMU2017QQ006) and Youth Thousand-Talents Program of China.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
References
- 1.Wang L., Magliery T.J., Liu D.R., Schultz P.G. A new functional suppressor tRNA/aminoacyl-tRNA synthetase pair for the in vivo incorporation of unnatural amino acids into proteins. J Am Chem Soc. 2000;122:5010–5011. [Google Scholar]
- 2.Wang L., Brock A., Herberich B., Schultz P.G. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 3.Neumann H., Wang K., Davis L., Garcia-Alai M., Chin J.W. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature. 2010;464:441–444. doi: 10.1038/nature08817. [DOI] [PubMed] [Google Scholar]
- 4.Amiram M., Haimovich A.D., Fan C.G., Wang Y.S., Aerni H.R., Ntai I. Evolution of translation machinery in recoded bacteria enables multi-site incorporation of nonstandard amino acids. Nat Biotechnol. 2015;33:1272–1279. doi: 10.1038/nbt.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blight S.K., Larue R.C., Mahapatra A., Longstaff D.G., Chang E., Zhao G. Direct charging of tRNA(CUA) with pyrrolysine in vitro and in vivo. Nature. 2004;431:333–335. doi: 10.1038/nature02895. [DOI] [PubMed] [Google Scholar]
- 6.Neumann H., Peak-Chew S.Y., Chin J.W. Genetically encoding N(epsilon)-acetyllysine in recombinant proteins. Nat Chem Biol. 2008;4:232–234. doi: 10.1038/nchembio.73. [DOI] [PubMed] [Google Scholar]
- 7.Ellefson J.W., Meyer A.J., Hughes R.A., Cannon J.R., Brodbelt J.S., Ellington A.D. Directed evolution of genetic parts and circuits by compartmentalized partnered replication. Nat Biotechnol. 2014;32:97–101. doi: 10.1038/nbt.2714. [DOI] [PubMed] [Google Scholar]
- 8.Sachdeva A., Wang K.H., Elliott T., Chin J.W. Concerted, rapid, quantitative, and site-specific dual labeling of proteins. J Am Chem Soc. 2014;136:7785–7788. doi: 10.1021/ja4129789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang K., Sachdeva A., Cox D.J., Wilf N.M., Lang K., Wallace S. Optimized orthogonal translation of unnatural amino acids enables spontaneous protein double-labelling and FRET. Nat Chem. 2014;6:393–403. doi: 10.1038/nchem.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hao B., Gong W., Ferguson T.K., James C.M., Krzycki J.A., Chan M.K. A new UAG-encoded residue in the structure of a methanogen methyltransferase. Science. 2002;296:1462–1466. doi: 10.1126/science.1069556. [DOI] [PubMed] [Google Scholar]
- 11.Srinivasan G., James C.M., Krzycki J.A. Pyrrolysine encoded by UAG in Archaea: charging of a UAG-decoding specialized tRNA. Science. 2002;296:1459–1462. doi: 10.1126/science.1069588. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y.S., Fang X., Wallace A.L., Wu B., Liu W.R. A rationally designed pyrrolysyl-tRNA synthetase mutant with a broad substrate spectrum. J Am Chem Soc. 2012;134:2950–2953. doi: 10.1021/ja211972x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakamoto K., Hayashi A., Sakamoto A., Kiga D., Nakayama H., Soma A. Site-specific incorporation of an unnatural amino acid into proteins in mammalian cells. Nucleic Acids Res. 2002;30:4692–4699. doi: 10.1093/nar/gkf589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chin J.W., Cropp T.A., Anderson J.C., Mukherji M., Zhang Z., Schultz P.G. An expanded eukaryotic genetic code. Science. 2003;301:964–967. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- 15.Hino N., Okazaki Y., Kobayashi T., Hayashi A., Sakamoto K., Yokoyama S. Protein photo-cross-linking in mammalian cells by site-specific incorporation of a photoreactive amino acid. Nat Methods. 2005;2:201–206. doi: 10.1038/nmeth739. [DOI] [PubMed] [Google Scholar]
- 16.Wang Q., Wang L. New methods enabling efficient incorporation of unnatural amino acids in yeast. J Am Chem Soc. 2008;130:6066–6067. doi: 10.1021/ja800894n. [DOI] [PubMed] [Google Scholar]
- 17.Chatterjee A., Xiao H., Bollong M., Ai H.W., Schultz P.G. Efficient viral delivery system for unnatural amino acid mutagenesis in mammalian cells. Proc Natl Acad Sci USA. 2013;110:11803–11808. doi: 10.1073/pnas.1309584110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu N., Deiters A., Cropp T.A., King D., Schultz P.G. A genetically encoded photocaged amino acid. J Am Chem Soc. 2004;126:14306–14307. doi: 10.1021/ja040175z. [DOI] [PubMed] [Google Scholar]
- 19.Wang W., Takimoto J.K., Louie G.V., Baiga T.J., Noel J.P., Lee K.F. Genetically encoding unnatural amino acids for cellular and neuronal studies. Nat Neurosci. 2007;10:1063–1072. doi: 10.1038/nn1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hancock S.M., Uprety R., Deiters A., Chin J.W. Expanding the genetic code of yeast for incorporation of diverse unnatural amino acids via a pyrrolysyl-tRNA synthetase/tRNA pair. J Am Chem Soc. 2010;132:14819–14824. doi: 10.1021/ja104609m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greiss S., Chin J.W. Expanding the genetic code of an animal. J Am Chem Soc. 2011;133:14196–14199. doi: 10.1021/ja2054034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bianco A., Townsley F.M., Greiss S., Lang K., Chin J.W. Expanding the genetic code of Drosophila melanogaster. Nat Chem Biol. 2012;8:748–750. doi: 10.1038/nchembio.1043. [DOI] [PubMed] [Google Scholar]
- 23.Li F.H., Zhang H., Sun Y., Pan Y.C., Zhou J.Z., Wang J.Y. Expanding the genetic code for photoclick chemistry in E.coli, mammalian cells, and A.thaliana. Angew Chem Int Ed Engl. 2013;52:9700–9704. doi: 10.1002/anie.201303477. [DOI] [PubMed] [Google Scholar]
- 24.Xiao H., Chatterjee A., Choi S.H., Bajjuri K.M., Sinha S.C., Schultz P.G. Genetic incorporation of multiple unnatural amino acids into proteins in mammalian cells. Angew Chem Int Ed Engl. 2013;52:14080–14083. doi: 10.1002/anie.201308137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elliott T.S., Townsley F.M., Bianco A., Ernst R.J., Sachdeva A., Elsasser S.J. Proteome labeling and protein identification in specific tissues and at specific developmental stages in an animal. Nat Biotechnol. 2014;32:465–472. doi: 10.1038/nbt.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ernst R.J., Krogager T.P., Maywood E.S., Zanchi R., Beranek V., Elliott T.S. Genetic code expansion in the mouse brain. Nat Chem Biol. 2016;12:776–778. doi: 10.1038/nchembio.2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koehler C., Sauter P.F., Wawryszyn M., Girona G.E., Gupta K., Landry J.J.M. Genetic code expansion for multiprotein complex engineering. Nat Methods. 2016;13:997–1000. doi: 10.1038/nmeth.4032. [DOI] [PubMed] [Google Scholar]
- 28.Han S., Yang A., Lee S., Lee H.-W., Park C.B., Park H.-S. Expanding the genetic code of Mus musculus. Nat Commun. 2017;8:14568. doi: 10.1038/ncomms14568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye S., Kohrer C., Huber T., Kazmi M., Sachdev P., Yan E.C. Site-specific incorporation of keto amino acids into functional G protein-coupled receptors using unnatural amino acid mutagenesis. J Biol Chem. 2008;283:1525–1533. doi: 10.1074/jbc.M707355200. [DOI] [PubMed] [Google Scholar]
- 30.Liu W.S., Brock A., Chen S., Chen S.B., Schultz P.G. Genetic incorporation of unnatural amino acids into proteins in mammalian cells. Nat Methods. 2007;4:239–244. doi: 10.1038/nmeth1016. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Z., Alfonta L., Tian F., Bursulaya B., Uryu S., King D.S. Selective incorporation of 5-hydroxytryptophan into proteins in mammalian cells. Proc Natl Acad Sci U S A. 2004;101:8882–8887. doi: 10.1073/pnas.0307029101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hughes R.A., Ellington A.D. Rational design of an orthogonal tryptophanyl nonsense suppressor tRNA. Nucleic Acids Res. 2010;38:6813–6830. doi: 10.1093/nar/gkq521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chatterjee A., Xiao H., Yang P.Y., Soundararajan G., Schultz P.G. A tryptophanyl-tRNA synthetase/tRNA pair for unnatural amino acid mutagenesis in E.coli. Angew Chem Int Ed Engl. 2013;52:5106–5109. doi: 10.1002/anie.201301094. [DOI] [PubMed] [Google Scholar]
- 34.Italia J.S., Addy P.S., Wrobel C.J., Crawford L.A., Lajoie M.J., Zheng Y. An orthogonalized platform for genetic code expansion in both bacteria and eukaryotes. Nat Chem Biol. 2017;13:446–450. doi: 10.1038/nchembio.2312. [DOI] [PubMed] [Google Scholar]
- 35.Kwon I., Kirshenbaum K., Tirrell D.A. Breaking the degeneracy of the genetic code. J Am Chem Soc. 2003;125:7512–7513. doi: 10.1021/ja0350076. [DOI] [PubMed] [Google Scholar]
- 36.Kwon I., Wang P., Tirrell D.A. Design of a bacterial host for site-specific incorporation of p-bromophenylalanine into recombinant proteins. J Am Chem Soc. 2006;128:11778–11783. doi: 10.1021/ja0626281. [DOI] [PubMed] [Google Scholar]
- 37.Anderson J.C., Wu N., Santoro S.W., Lakshman V., King D.S., Schultz P.G. An expanded genetic code with a functional quadruplet codon. Proc Natl Acad Sci USA. 2004;101:7566–7571. doi: 10.1073/pnas.0401517101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park H.S., Hohn M.J., Umehara T., Guo L.T., Osborne E.M., Benner J. Expanding the genetic code of Escherichia coli with phosphoserine. Science. 2011;333:1151–1154. doi: 10.1126/science.1207203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rogerson D.T., Sachdeva A., Wang K., Haq T., Kazlauskaite A., Hancock S.M. Efficient genetic encoding of phosphoserine and its nonhydrolyzable analog. Nat Chem Biol. 2015;11:496–503. doi: 10.1038/nchembio.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mukai T., Lajoie M.J., Englert M., Soll D. Rewriting the genetic code. Annu Rev Microbiol. 2017;71:557–577. doi: 10.1146/annurev-micro-090816-093247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Italia J.S., Latour C., Wrobel C.J.J., Chatterjee A. Resurrecting the bacterial tyrosyl-tRNA synthetase/tRNA pair for expanding the genetic code of both E. coli and eukaryotes. Cell Chem Biol. 2018 doi: 10.1016/j.chembiol.2018.07.002. pii: S2451-9456(18)30229-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Y., Song H.P., He D., Zhang S., Dai S.Z., Lin S.X. Genetically encoded protein photocrosslinker with a transferable mass spectrometry-identifiable label. Nat Commun. 2016;7 doi: 10.1038/ncomms12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang B., Tang S.B., Ma C., Li S.T., Shao G.C., Dang B.B. Spontaneous and specific chemical cross-linking in live cells to capture and identify protein interactions. Nat Commun. 2017;8 doi: 10.1038/s41467-017-02409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chatterjee A., Guo J., Lee H.S., Schultz P.G. A genetically encoded fluorescent probe in mammalian cells. J Am Chem Soc. 2013;135:12540–12543. doi: 10.1021/ja4059553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uttamapinant C., Howe J.D., Lang K., Beranek V., Davis L., Mahesh M. Genetic code expansion enables live-cell and super-resolution imaging of site-specifically labeled cellular proteins. J Am Chem Soc. 2015;137:4602–4605. doi: 10.1021/ja512838z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peng T., Hang H.C. Site-specific bioorthogonal labeling for fluorescence imaging of intracellular proteins in living cells. J Am Chem Soc. 2016;138:14423–14433. doi: 10.1021/jacs.6b08733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsai Y.H., Essig S., James J.R., Lang K., Chin J.W. Selective, rapid and optically switchable regulation of protein function in live mammalian cells. Nat Chem. 2015;7:554–561. doi: 10.1038/nchem.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walker O.S., Elsasser S.J., Mahesh M., Bachman M., Balasubramanian S., Chin J.W. Photoactivation of mutant isocitrate dehydrogenase 2 reveals rapid cancer-associated metabolic and epigenetic changes. J Am Chem Soc. 2016;138:718–721. doi: 10.1021/jacs.5b07627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang G., Li J., Xie R., Fan X.Y., Liu Y.J., Zheng S.Q. Bioorthogonal chemical activation of kinases in living systems. Acs Central Sci. 2016;2:325–331. doi: 10.1021/acscentsci.6b00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ambrogelly A., Palioura S., Soll D. Natural expansion of the genetic code. Nat Chem Biol. 2007;3:29–35. doi: 10.1038/nchembio847. [DOI] [PubMed] [Google Scholar]
- 51.Zhang M.S., Brunner S.F., Huguenin-Dezot N., Liang A.D., Schmied W.H., Rogerson D.T. Biosynthesis and genetic encoding of phosphothreonine through parallel selection and deep sequencing. Nat Methods. 2017;14:729–736. doi: 10.1038/nmeth.4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luo X., Fu G., Wang R.E., Zhu X., Zambaldo C., Liu R. Genetically encoding phosphotyrosine and its nonhydrolyzable analog in bacteria. Nat Chem Biol. 2017;13:845–849. doi: 10.1038/nchembio.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim C.H., Kang M., Kim H.J., Chatterjee A., Schultz P.G. Site-specific incorporation of epsilon-N-crotonyllysine into histones. Angew Chem Int Ed Engl. 2012;51:7246–7249. doi: 10.1002/anie.201203349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiao H., Xuan W., Shao S., Liu T., Schultz P.G. Genetic incorporation of epsilon-N-2-Hydroxyisobutyryl-lysine into recombinant histones. ACS Chem Biol. 2015;10:1599–1603. doi: 10.1021/cb501055h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoffmann R.M., Coumbe B.G.T., Josephs D.H., Mele S., Ilieva K.M., Cheung A. Antibody structure and engineering considerations for the design and function of Antibody Drug Conjugates (ADCs) OncoImmunology. 2018;7 doi: 10.1080/2162402X.2017.1395127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sievers E.L., Senter P.D. Antibody-drug conjugates in cancer therapy. Annu Rev Med. 2013;64:15–29. doi: 10.1146/annurev-med-050311-201823. [DOI] [PubMed] [Google Scholar]
- 57.Kim C.H., Axup J.Y., Schultz P.G. Protein conjugation with genetically encoded unnatural amino acids. Curr Opin Chem Biol. 2013;17:412–419. doi: 10.1016/j.cbpa.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Panowski S., Bhakta S., Raab H., Polakis P., Junutula J.R. Site-specific antibody drug conjugates for cancer therapy. Mabs-Austin. 2014;6:34–45. doi: 10.4161/mabs.27022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gomez N., Vinson A.R., Ouyang J., Nguyen M.D.H., Chen X.N., Sharma V.K. Triple light chain antibodies: factors that influence its formation in cell culture. Biotechnol Bioeng. 2010;105:748–760. doi: 10.1002/bit.22580. [DOI] [PubMed] [Google Scholar]
- 60.Zhou Q., Stefano J.E., Manning C., Kyazike J., Chen B., Gianolio D.A. Site-specific antibody-drug conjugation through glycoengineering. Bioconjugate Chem. 2014;25:510–520. doi: 10.1021/bc400505q. [DOI] [PubMed] [Google Scholar]
- 61.Farias S.E., Strop P., Delaria K., Casas M.G., Dorywalska M., Shelton D.L. Mass spectrometric characterization of transglutaminase based site-specific antibody-drug conjugates. Bioconjugate Chem. 2014;25:240–250. doi: 10.1021/bc4003794. [DOI] [PubMed] [Google Scholar]
- 62.Appel M.J., Bertozzi C.R. Formylglycine, a post-translationally generated residue with unique catalytic capabilities and biotechnology applications. ACS Chem Biol. 2015;10:72–84. doi: 10.1021/cb500897w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hofer T., Skeffington L.R., Chapman C.M., Rader C. Molecularly defined antibody conjugation through a selenocysteine interface. Biochemistry. 2009;48:12047–12057. doi: 10.1021/bi901744t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thyer R., Shroff R., Klein D.R., d'Oelsnitz S., Cotham V.C., Byrom M. Custom selenoprotein production enabled by laboratory evolution of recoded bacterial strains. Nat Biotechnol. 2018;36(7):624–631. doi: 10.1038/nbt.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang L., Zhang Z.W., Brock A., Schultz P.G. Addition of the keto functional group to the genetic code of Escherichia coli. Proc Natl Acad Sci USA. 2003;100:56–61. doi: 10.1073/pnas.0234824100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Axup J.Y., Bajjuri K.M., Ritland M., Hutchins B.M., Kim C.H., Kazane S.A. Synthesis of site-specific antibody-drug conjugates using unnatural amino acids. Proc Natl Acad Sci USA. 2012;109:16101–16106. doi: 10.1073/pnas.1211023109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kularatne S.A., Deshmukh V., Ma J., Tardif V., Lim R.K., Pugh H.M. A CXCR4-targeted site-specific antibody-drug conjugate. Angew Chem Int Ed Engl. 2014;53:11863–11867. doi: 10.1002/anie.201408103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lim R.K.V., Yu S., Cheng B., Li S.J., Kim N.J., Cao Y. Targeted delivery of LXR agonist using a site-specific antibody drug conjugate. Bioconjugate Chem. 2015;26:2216–2222. doi: 10.1021/acs.bioconjchem.5b00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu S., Pearson A.D., Lim R.K.V., Rodgers D.T., Li S., Parker H.B. Targeted delivery of an anti-inflammatory PDE4 inhibitor to immune cells via an antibody-drug conjugate. Mol Ther. 2016;24:2078–2089. doi: 10.1038/mt.2016.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tian F., Lu Y.C., Manibusan A., Sellers A., Tran H., Sun Y. A general approach to site-specific antibody drug conjugates. Proc Natl Acad Sci USA. 2014;111:1766–1771. doi: 10.1073/pnas.1321237111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu Y.M., Zhu H., Zhang B., Liu F., Chen J.X., Wang Y.F. Synthesis of site-specific radiolabeled antibodies for radioimmunotherapy via genetic code expansion. Bioconjugate Chem. 2016;27:2460–2468. doi: 10.1021/acs.bioconjchem.6b00412. [DOI] [PubMed] [Google Scholar]
- 72.Zhang C., Zhou X., Yao T., Tian Z., Zhou D. Precision fluorescent labeling of an adeno-associated virus vector to monitor the viral infection pathway. Biotechnol J. 2018;13:1700374. doi: 10.1002/biot.201700374. [DOI] [PubMed] [Google Scholar]
- 73.Wang R.S.E., Liu T., Wang Y., Cao Y., Du J.T., Luo X.Z. An immunosuppressive antibody-drug conjugate. J Am Chem Soc. 2015;137:3229–3232. doi: 10.1021/jacs.5b00620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Holmes D. Buy buy bispecific antibodies. Nat Rev Drug Discov. 2011;10:798–800. doi: 10.1038/nrd3581. [DOI] [PubMed] [Google Scholar]
- 75.Przepiorka D., Ko C.W., Deisseroth A., Yancey C.L., Candau-Chacon R., Chiu H.J. FDA approval: blinatumomab. Clin Cancer Res. 2015;21:4035–4039. doi: 10.1158/1078-0432.CCR-15-0612. [DOI] [PubMed] [Google Scholar]
- 76.Kim C.H., Axup J.Y., Dubrovska A., Kazane S.A., Hutchins B.A., Wold E.D. Synthesis of bispecific antibodies using genetically encoded unnatural amino acids. J Am Chem Soc. 2012;134:9918–9921. doi: 10.1021/ja303904e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cao Y., Axup J.Y., Ma J.S., Wang R.E., Choi S., Tardif V. Multiformat T-cell-engaging bispecific antibodies targeting human breast cancers. Angew Chem Int Ed Engl. 2015;54:7022–7027. doi: 10.1002/anie.201500799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lu H., Zhou Q., Deshmukh V., Phull H., Ma J., Tardif V. Targeting human C-type lectin-like molecule-1 (CLL1) with a bispecific antibody for immunotherapy of acute myeloid leukemia. Angew Chem Int Ed Engl. 2014;53:9841–9845. doi: 10.1002/anie.201405353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kazane S.A., Axup J.Y., Kim C.H., Ciobanu M., Wold E.D., Barluenga S. Self-assembled antibody multimers through peptide nucleic acid conjugation. J Am Chem Soc. 2013;135:340–346. doi: 10.1021/ja309505c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ramadoss N.S., Schulman A.D., Choi S.H., Rodgers D.T., Kazane S.A., Kim C.H. An anti-B cell maturation antigen bispecific antibody for multiple myeloma. J Am Chem Soc. 2015;137:5288–5291. doi: 10.1021/jacs.5b01876. [DOI] [PubMed] [Google Scholar]
- 81.Kim C.H., Axup J.Y., Lawson B.R., Yun H., Tardif V., Choi S.H. Bispecific small molecule-antibody conjugate targeting prostate cancer. Proc Natl Acad Sci USA. 2013;110:17796–17801. doi: 10.1073/pnas.1316026110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kularatne S.A., Deshmukh V., Gymnopoulos M., Biroc S.L., Xia J.M., Srinagesh S. Recruiting cytotoxic T cells to folate-receptor-positive cancer cells. Angew Chem Int Ed Engl. 2013;52:12101–12104. doi: 10.1002/anie.201306866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lyu Z., Kang L., Buuh Z.Y., Jiang D., McGuth J.C., Du J. A switchable site-specific antibody conjugate. ACS Chem Biol. 2018;13:958–964. doi: 10.1021/acschembio.8b00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen Y.Y. Increasing T cell versatility with SUPRA CARs. Cell. 2018;173:1316–1317. doi: 10.1016/j.cell.2018.05.030. [DOI] [PubMed] [Google Scholar]
- 85.Tamada K., Geng D.G., Sakoda Y., Bansal N., Srivastava R., Li Z.Y. Redirecting gene-modified T cells toward various cancer types using tagged antibodies. Clin Cancer Res. 2012;18:6436–6445. doi: 10.1158/1078-0432.CCR-12-1449. [DOI] [PubMed] [Google Scholar]
- 86.Ma J.S.Y., Kim J.Y., Kazane S.A., Choi S.H., Yun H.Y., Kim M.S. Versatile strategy for controlling the specificity and activity of engineered T cells. Proc Natl Acad Sci USA. 2016;113:E450–E458. doi: 10.1073/pnas.1524193113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cao Y., Rodgers D.T., Du J., Ahmad I., Hampton E.N., Ma J.S. Design of switchable chimeric antigen receptor T cells targeting breast cancer. Angew Chem Int Ed Engl. 2016;55:7520–7524. doi: 10.1002/anie.201601902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Swierczewska M., Lee K.C., Lee S. What is the future of PEGylated therapies? Expet Opin Emerg Drugs. 2015;20:531–536. doi: 10.1517/14728214.2015.1113254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pfister D., Morbidelli M. Process for protein PEGylation. J Contr Release. 2014;180:134–149. doi: 10.1016/j.jconrel.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 90.Cho H., Daniel T., Buechler Y.J., Litzinger D.C., Maio Z.W., Putnam A.M.H. Optimized clinical performance of growth hormone with an expanded genetic code. Proc Natl Acad Sci USA. 2011;108:9060–9065. doi: 10.1073/pnas.1100387108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wu L., Chen J.X., Wu Y.M., Zhang B., Cai X.C., Zhang Z.W. Precise and combinatorial PEGylation generates a low-immunogenic and stable form of human growth hormone. J Contr Release. 2017;249:84–93. doi: 10.1016/j.jconrel.2017.01.029. [DOI] [PubMed] [Google Scholar]
- 92.Zhang B., Xu H., Chen J.X., Zheng Y.X., Wu Y.M., Si L.L.L. Development of next generation of therapeutic IFN-alpha 2b via genetic code expansion. Acta Biomater. 2015;19:100–111. doi: 10.1016/j.actbio.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 93.Mu J., Pinkstaff J., Li Z.H., Skidmore L., Li N.N., Myler H. FGF21 analogs of sustained action enabled by orthogonal biosynthesis demonstrate enhanced antidiabetic pharmacology in rodents. Diabetes. 2012;61:505–512. doi: 10.2337/db11-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yao T.Z., Zhou X.Y., Zhang C.L., Yu X.J., Tian Z.Y., Zhang L.H. Site-specific PEGylated adeno-associated viruses with increased serum stability and reduced immunogenicity. Molecules. 2017;22 doi: 10.3390/molecules22071155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Luhmann T., Schmidt M., Leiske M.N., Spieler V., Majdanski T.C., Grube M. Site-specific POxylation of Interleukin-4. ACS Biomater Sci Eng. 2017;3:304–312. doi: 10.1021/acsbiomaterials.6b00578. [DOI] [PubMed] [Google Scholar]
- 96.Liu T., Wang Y., Luo X.Z., Li J., Reed S.A., Xiao H. Enhancing protein stability with extended disulfide bonds. Proc Natl Acad Sci USA. 2016;113:5910–5915. doi: 10.1073/pnas.1605363113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang C.Y., Renfrew P.D., Olsen A.J., Zhang M., Yuvienco C., Bonneau R. Improved stability and half-life of fluorinated phosphotriesterase using rosetta. Chembiochem. 2014;15:1761–1764. doi: 10.1002/cbic.201402062. [DOI] [PubMed] [Google Scholar]
- 98.Xuan W.M., Li J., Luo X.Z., Schultz P.G. Genetic incorporation of a reactive isothiocyanate group into proteins. Angew Chem Int Ed Engl. 2016;55:10065–10068. doi: 10.1002/anie.201604891. [DOI] [PubMed] [Google Scholar]
- 99.Young T.S., Young D.D., Ahmad I., Louis J.M., Benkovic S.J., Schultz P.G. Evolution of cyclic peptide protease inhibitors. Proc Natl Acad Sci USA. 2011;108:11052–11056. doi: 10.1073/pnas.1108045108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zambaldo C., Luo X., Mehta A.P., Schultz P.G. Recombinant macrocyclic lanthipeptides incorporating non-canonical amino acids. J Am Chem Soc. 2017;139:11646–11649. doi: 10.1021/jacs.7b04159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shi Y.X., Yang X.A., Garg N., van der Donk W.A. Production of lantipeptides in Escherichia coli. J Am Chem Soc. 2011;133:2338–2341. doi: 10.1021/ja109044r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bindman N.A., Bobeica S.C., Liu W.S.R., van der Donk W.A. Facile removal of leader peptides from lanthipeptides by incorporation of a hydroxy acid. J Am Chem Soc. 2015;137:6975–6978. doi: 10.1021/jacs.5b04681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Al Toma R.S., Kuthning A., Exner M.P., Denisiuk A., Ziegler J., Budisa N. Site-Directed and global incorporation of orthogonal and isostructural noncanonical amino acids into the ribosomal lasso peptide capistruin. Chembiochem. 2015;16:503–509. doi: 10.1002/cbic.201402558. [DOI] [PubMed] [Google Scholar]
- 104.Piscotta F.J., Tharp J.M., Liu W.R., Link A.J. Expanding the chemical diversity of lasso peptide MccJ25 with genetically encoded noncanonical amino acids. Chem Commun. 2015;51:409–412. doi: 10.1039/c4cc07778d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Frost J.R., Jacob N.T., Papa L.J., Owens A.E., Fasan R. Ribosomal synthesis of macrocyclic peptides in vitro and in vivo mediated by genetically encoded aminothiol unnatural amino acids. ACS Chem Biol. 2015;10:1805–1816. doi: 10.1021/acschembio.5b00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Luo X.Z., Zambaldo C., Liu T., Zhang Y.H., Xuan W.M., Wang C. Recombinant thiopeptides containing noncanonical amino acids. Proc Natl Acad Sci USA. 2016;113:3615–3620. doi: 10.1073/pnas.1602733113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lopatniuk M., Myronovskyi M., Luzhetskyy A. Streptomyces albus: a new cell factory for non-canonical amino acids incorporation into ribosomally synthesized natural products. ACS Chem Biol. 2017;12:2362–2370. doi: 10.1021/acschembio.7b00359. [DOI] [PubMed] [Google Scholar]
- 108.Daya S., Berns K.I. Gene therapy using adeno-associated virus vectors. Clin Microbiol Rev. 2008;21:583–593. doi: 10.1128/CMR.00008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Agbandje-McKenna M., Kleinschmidt J. AAV capsid structure and cell interactions. Meth Mol Biol. 2011;807:47–92. doi: 10.1007/978-1-61779-370-7_3. [DOI] [PubMed] [Google Scholar]
- 110.Li W., Asokan A., Wu Z., Van Dyke T., DiPrimio N., Johnson J.S. Engineering and selection of shuffled AAV genomes: a new strategy for producing targeted biological nanoparticles. Mol Ther. 2008;16:1252–1260. doi: 10.1038/mt.2008.100. [DOI] [PubMed] [Google Scholar]
- 111.Asokan A., Conway J.C., Phillips J.L., Li C., Hegge J., Sinnott R. Reengineering a receptor footprint of adeno-associated virus enables selective and systemic gene transfer to muscle. Nat Biotechnol. 2010;28:79–82. doi: 10.1038/nbt.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Munch R.C., Janicki H., Volker I., Rasbach A., Hallek M., Buning H. Displaying high-affinity ligands on adeno-associated viral vectors enables tumor cell-specific and safe gene transfer. Mol Ther. 2013;21:109–118. doi: 10.1038/mt.2012.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Banerjee P.S., Ostapchuk P., Hearing P., Carrico I. Chemoselective attachment of small molecule effector functionality to human adenoviruses facilitates gene delivery to cancer cells. J Am Chem Soc. 2010;132:13615–13617. doi: 10.1021/ja104547x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang C.L., Yao T.Z., Zheng Y.X., Li Z.J., Zhang Q., Zhang L.H. Development of next generation adeno-associated viral vectors capable of selective tropism and efficient gene delivery. Biomaterials. 2016;80:134–145. doi: 10.1016/j.biomaterials.2015.11.066. [DOI] [PubMed] [Google Scholar]
- 115.Kelemen R.E., Mukherjee R., Cao X.F., Erickson S.B., Zheng Y.A., Chatterjee A. A precise chemical strategy to alter the receptor specificity of the adeno-associated virus. Angew Chem Int Ed Engl. 2016;55:10645–10649. doi: 10.1002/anie.201604067. [DOI] [PubMed] [Google Scholar]
- 116.Erickson S.B., Mukherjee R., Kelemen R.E., Wrobel C.J.J., Cao X.F., Chatterjee A. Precise photoremovable perturbation of a virus-host interaction. Angew Chem Int Ed Engl. 2017;56:4234–4237. doi: 10.1002/anie.201700683. [DOI] [PubMed] [Google Scholar]
- 117.Dalum I., Butler D.M., Jensen M.R., Hindersson P., Steinaa L., Waterston A.M. Therapeutic antibodies elicited by immunization against TNF-alpha. Nat Biotechnol. 1999;17:666–669. doi: 10.1038/10878. [DOI] [PubMed] [Google Scholar]
- 118.Zuany-Amorim C., Manlius C., Dalum I., Jensen M.R., Gautam A., Pay G. Induction of TNF-alpha autoantibody production by AutoVac TNF106: a novel therapeutic approach for the treatment of allergic diseases. Int Arch Allergy Immunol. 2004;133:154–163. doi: 10.1159/000076441. [DOI] [PubMed] [Google Scholar]
- 119.Weigle W.O. The induction of autoimmunity in rabbits following injection of heterologous or altered homologous thyroglobulin. J Exp Med. 1965;121:289–308. doi: 10.1084/jem.121.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Leitner W.W., Hwang L.N., deVeer M.J., Zhou A., Silverman R.H., Williams B.R. Alphavirus-based DNA vaccine breaks immunological tolerance by activating innate antiviral pathways. Nat Med. 2003;9:33–39. doi: 10.1038/nmxx. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Berd D. M-Vax: an autologous, hapten-modified vaccine for human cancer. Expert Rev Vaccines. 2004;3:521–527. doi: 10.1586/14760584.3.5.521. [DOI] [PubMed] [Google Scholar]
- 122.Grunewald J., Tsao M.L., Perera R., Dong L., Niessen F., Wen B.G. Immunochemical termination of self-tolerance. Proc Natl Acad Sci USA. 2008;105:11276–11280. doi: 10.1073/pnas.0804157105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ren W., Ji A., Wang M.X., Ai H.W. Expanding the genetic code for a dinitrophenyl hapten. Chembiochem. 2015;16:2007–2010. doi: 10.1002/cbic.201500204. [DOI] [PubMed] [Google Scholar]
- 124.Gauba V., Grunewald J., Gorney V., Deaton L.M., Kang M., Bursulaya B. Loss of CD4 T-cell-dependent tolerance to proteins with modified amino acids. Proc Natl Acad Sci USA. 2011;108:12821–12826. doi: 10.1073/pnas.1110042108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Grunewald J., Hunt G.S., Dong L., Niessen F., Wen B.G., Tsao M.L. Mechanistic studies of the immunochemical termination of self-tolerance with unnatural amino acids. Proc Natl Acad Sci USA. 2009;106:4337–4342. doi: 10.1073/pnas.0900507106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang N.X., Yuan Z., Niu W., Li Q.S., Guo J.T. Synthetic biology approach for the development of conditionally replicating HIV-1 vaccine. J Chem Technol Biotechnol. 2017;92:445–452. doi: 10.1002/jctb.5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lin S., Yan H., Li L., Yang M., Peng B., Chen S. Site-specific engineering of chemical functionalities on the surface of live hepatitis D virus. Angew Chem Int Ed Engl. 2013;52:13970–13974. doi: 10.1002/anie.201305787. [DOI] [PubMed] [Google Scholar]
- 128.Si L.L., Xu H., Zhou X.Y., Zhang Z.W., Tian Z.Y., Wang Y. Generation of influenza A viruses as live but replication-incompetent virus vaccines. Science. 2016;354:1170–1173. doi: 10.1126/science.aah5869. [DOI] [PubMed] [Google Scholar]
- 129.Wang N.X., Li Y., Niu W., Sun M., Cerny R., Li Q.S. Construction of a live-attenuated HIV-1 vaccine through genetic code expansion. Angew Chem Int Ed Engl. 2014;53:4867–4871. doi: 10.1002/anie.201402092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yuan Z., Wang N.X., Kang G.B., Niu W., Li Q.S., Guo J.T. Controlling multicycle replication of live-attenuated HIV-1 using an unnatural genetic switch. ACS Synth Biol. 2017;6:721–731. doi: 10.1021/acssynbio.6b00373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wang F., Robbins S., Guo J., Shen W., Schultz P.G. Genetic incorporation of unnatural amino acids into proteins in Mycobacterium tuberculosis. PLoS One. 2010;5 doi: 10.1371/journal.pone.0009354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mandell D.J., Lajoie M.J., Mee M.T., Takeuchi R., Kuznetsov G., Norville J.E. Biocontainment of genetically modified organisms by synthetic protein design. Nature. 2015;518:55–60. doi: 10.1038/nature14121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rovner A.J., Haimovich A.D., Katz S.R., Li Z., Grome M.W., Gassaway B.M. Recoded organisms engineered to depend on synthetic amino acids. Nature. 2015;518:89–93. doi: 10.1038/nature14095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tack D.S., Ellefson J.W., Thyer R., Wang B., Gollihar J., Forster M.T. Addicting diverse bacteria to a noncanonical amino acid. Nat Chem Biol. 2016;12:138–140. doi: 10.1038/nchembio.2002. [DOI] [PubMed] [Google Scholar]
- 135.Xuan W.M., Schultz P.G. A strategy for creating organisms dependent on noncanonical amino acids. Angew Chem Int Ed Engl. 2017;56:9170–9173. doi: 10.1002/anie.201703553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Koh M., Nasertorabi F., Han G.W., Stevens R.C., Schultz P.G. Generation of an orthogonal protein-protein interface with a noncanonical amino acid. J Am Chem Soc. 2017;139:5728–5731. doi: 10.1021/jacs.7b02273. [DOI] [PubMed] [Google Scholar]
- 137.Gan F., Liu R., Wang F., Schultz P.G. Functional replacement of histidine in proteins to generate noncanonical amino acid dependent organisms. J Am Chem Soc. 2018;140:3829–3832. doi: 10.1021/jacs.7b13452. [DOI] [PubMed] [Google Scholar]
- 138.Ray-Saha S., Huber T., Sakmar T.P. Antibody epitopes on g protein-coupled receptors mapped with genetically encoded photoactivatable cross-linkers. Biochemistry. 2014;53:1302–1310. doi: 10.1021/bi401289p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Coin I., Katritch V., Sun T., Xiang Z., Siu F.Y., Beyermann M. Genetically encoded chemical probes in cells reveal the binding path of urocortin-I to CRF class B GPCR. Cell. 2013;155:1258–1269. doi: 10.1016/j.cell.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rannversson H., Andersen J., Sorensen L., Bang-Andersen B., Park M., Huber T. Genetically encoded photocrosslinkers locate the high-affinity binding site of antidepressant drugs in the human serotonin transporter. Nat Commun. 2016;7 doi: 10.1038/ncomms11261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Koole C., Reynolds C.A., Mobarec J.C., Hick C., Sexton P.M., Sakmar T.P. Genetically encoded photocross-linkers determine the biological binding site of exendin-4 peptide in the N-terminal domain of the intact human glucagon-like peptide-1 receptor (GLP-1R) J Biol Chem. 2017;292:7131–7144. doi: 10.1074/jbc.M117.779496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hemphill J., Borchardt EK, Brown K, Asokan A, Deiters A. Optical control of CRISPR/Cas9 gene editing. J Am Chem Soc. 2015;137:5642–5645. doi: 10.1021/ja512664v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chen S., Schultz P.G., Brock A. An improved system for the generation and analysis of mutant proteins containing unnatural amino acids in Saccharomyces cerevisiae. J Mol Biol. 2007;371:112–122. doi: 10.1016/j.jmb.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 144.Mukai T., Kobayashi T., Hino N., Yanagisawa T., Sakamoto K., Yokoyama S. Adding l-lysine derivatives to the genetic code of mammalian cells with engineered pyrrolysyl-tRNA synthetases. Biochem Biophys Res Commun. 2008;371:818–822. doi: 10.1016/j.bbrc.2008.04.164. [DOI] [PubMed] [Google Scholar]
- 145.Young T.S., Ahmad I., Brock A., Schultz P.G. Expanding the genetic repertoire of the methylotrophic yeast Pichia pastoris. Biochemistry. 2009;48:2643–2653. doi: 10.1021/bi802178k. [DOI] [PubMed] [Google Scholar]
- 146.Palzer S., Bantel Y., Kazenwadel F., Berg M., Rupp S., Sohn K. An expanded genetic code in Candida albicans to study protein-protein interactions in vivo. Eukaryot Cell. 2013;12:816–827. doi: 10.1128/EC.00075-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Shao N., Singh N.S., Slade S.E., Jones A.M., Balasubramanian M.K. Site specific genetic incorporation of azidophenylalanine in Schizosaccharomyces pombe. Sci Rep. 2015;5:17196. doi: 10.1038/srep17196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Parrish A.R., She X.Y., Xiang Z., Coin I., Shen Z.X., Briggs S.P. Expanding the genetic code of Caenorhabditis elegans using bacterial aminoacyl-tRNA synthetase/tRNA pairs. ACS Chem Biol. 2012;7:1292–1302. doi: 10.1021/cb200542j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Chang H., Han M., Huang W., Wei G., Chen J., Chen P.R. Light-induced protein translocation by genetically encoded unnatural amino acid in Caenorhabditis elegans. Protein Cell. 2013;4:883–886. doi: 10.1007/s13238-013-3118-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mukai T., Wakiyama M., Sakamoto K., Yokoyama S. Genetic encoding of non-natural amino acids in Drosophila melanogaster Schneider 2 cells. Protein Sci. 2010;19:440–448. doi: 10.1002/pro.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Chen Y.T., Ma J., Lu W.Q., Tian M.L., Thauvin M., Yuan C.G. Heritable expansion of the genetic code in mouse and zebrafish. Cell Res. 2017;27:294–297. doi: 10.1038/cr.2016.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kang J.Y., Kawaguchi D., Coin I., Xiang Z., O'Leary D.D.M., Slesinger P.A. In vivo expression of a light-activatable potassium channel using unnatural amino acids. Neuron. 2013;80:358–370. doi: 10.1016/j.neuron.2013.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Niu W., Schultz P.G., Guo J. An expanded genetic code in mammalian cells with a functional quadruplet codon. ACS Chem Biol. 2013;8:1640–1645. doi: 10.1021/cb4001662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Shen B., Xiang Z., Miller B., Louie G., Wang W.Y., Noel J.P. Genetically encoding unnatural amino acids in neural stem cells and optically reporting voltage-sensitive domain changes in differentiated neurons. Stem Cell. 2011;29:1231–1240. doi: 10.1002/stem.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Elsasser S.J., Ernst R.J., Walker O.S., Chin J.W. Genetic code expansion in stable cell lines enables encoded chromatin modification. Nat Methods. 2016;13:158–164. doi: 10.1038/nmeth.3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Schmied W.H., Elsasser S.J., Uttamapinant C., Chin J.W. Efficient multisite unnatural amino acid incorporation in mammalian cells via optimized pyrrolysyl tRNA synthetase/tRNA expression and engineered eRF1. J Am Chem Soc. 2014;136:15577–15583. doi: 10.1021/ja5069728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Johnson D.B.F., Xu J.F., Shen Z.X., Takimoto J.K., Schultz M.D., Schmitz R.J. RF1 knockout allows ribosomal incorporation of unnatural amino acids at multiple sites. Nat Chem Biol. 2011;7:779–786. doi: 10.1038/nchembio.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Johnson D.B.F., Wang C., Xu J.F., Schultz M.D., Schmitz R.J., Ecker J.R. Release factor one is nonessential in Escherichia coli. ACS Chem Biol. 2012;7:1337–1344. doi: 10.1021/cb300229q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Schrader J.M., Chapman S.J., Uhlenbeck O.C. Tuning the affinity of aminoacyl-tRNA to elongation factor Tu for optimal decoding. Proc Natl Acad Sci USA. 2011;108:5215–5220. doi: 10.1073/pnas.1102128108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wang K., Neumann H., Peak-Chew S.Y., Chin J.W. Evolved orthogonal ribosomes enhance the efficiency of synthetic genetic code expansion. Nat Biotechnol. 2007;25:770–777. doi: 10.1038/nbt1314. [DOI] [PubMed] [Google Scholar]
- 161.Lajoie M.J., Rovner A.J., Goodman D.B., Aerni H.R., Haimovich A.D., Kuznetsov G. Genomically recoded organisms expand biological functions. Science. 2013;342:357–360. doi: 10.1126/science.1241459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ostrov N., Landon M., Guell M., Kuznetsov G., Teramoto J., Cervantes N. Design, synthesis, and testing toward a 57-codon genome. Science. 2016;353:819–822. doi: 10.1126/science.aaf3639. [DOI] [PubMed] [Google Scholar]
- 163.Zheng Y., Lewis T.L., Jr., Igo P., Polleux F., Chatterjee A. Virus-enabled optimization and delivery of the genetic machinery for efficient unnatural amino acid mutagenesis in mammalian cells and tissues. ACS Synth Biol. 2017;6:13–18. doi: 10.1021/acssynbio.6b00092. [DOI] [PubMed] [Google Scholar]
- 164.Guo J., Melancon C.E., 3rd, Lee H.S., Groff D., Schultz P.G. Evolution of amber suppressor tRNAs for efficient bacterial production of proteins containing nonnatural amino acids. Angew Chem Int Ed Engl. 2009;48:9148–9151. doi: 10.1002/anie.200904035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Bryson D.I., Fan C., Guo L.T., Miller C., Soll D., Liu D.R. Continuous directed evolution of aminoacyl-tRNA synthetases. Nat Chem Biol. 2017;13:1253–1260. doi: 10.1038/nchembio.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Suzuki T., Miller C., Guo L.T., Ho J.M.L., Bryson D.I., Wang Y.S. Crystal structures reveal an elusive functional domain of pyrrolysyl-tRNA synthetase. Nat Chem Biol. 2017;13:1261–1266. doi: 10.1038/nchembio.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Chin J.W. Expanding and reprogramming the genetic code. Nature. 2017;550:53–60. doi: 10.1038/nature24031. [DOI] [PubMed] [Google Scholar]
- 168.Odoi K.A., Huang Y., Rezenom Y.H., Liu W.R. Nonsense and sense suppression abilities of original and derivative Methanosarcina mazei pyrrolysyl-tRNA synthetase-tRNA(Pyl) pairs in the Escherichia coli BL21(DE3) cell strain. PLoS One. 2013;8 doi: 10.1371/journal.pone.0057035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Willis J.C.W., Chin J.W. Mutually orthogonal pyrrolysyl-tRNA synthetase/tRNA pairs. Nat Chem. 2018;10:831–837. doi: 10.1038/s41557-018-0052-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wan W., Huang Y., Wang Z.Y., Russell W.K., Pai P.J., Russell D.H. A facile system for genetic incorporation of two different noncanonical amino acids into one protein in Escherichia coli. Angew Chem Int Ed Engl. 2010;49:3211–3214. doi: 10.1002/anie.201000465. [DOI] [PubMed] [Google Scholar]
- 171.Zheng Y., Addy P.S., Mukherjee R., Chatterjee A. Defining the current scope and limitations of dual noncanonical amino acid mutagenesis in mammalian cells. Chem Sci. 2017;8:7211–7217. doi: 10.1039/c7sc02560b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wang K., Fredens J., Brunner S.F., Kim S.H., Chia T., Chin J.W. Defining synonymous codon compression schemes by genome recoding. Nature. 2016;539:59–64. doi: 10.1038/nature20124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zhang Y., Ptacin J.L., Fischer E.C., Aerni H.R., Caffaro C.E., Jose K.S. A semi-synthetic organism that stores and retrieves increased genetic information. Nature. 2017;551 doi: 10.1038/nature24659. 644-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zhang Y., Lamb B.M., Feldman A.W., Zhou A.X., Lavergne T., Li L. A semisynthetic organism engineered for the stable expansion of the genetic alphabet. Proc Natl Acad Sci U S A. 2017;114:1317–1322. doi: 10.1073/pnas.1616443114. [DOI] [PMC free article] [PubMed] [Google Scholar]