Abstract
Although most in vitro (cell-free) synthetic biology projects are usually used for the purposes of fundamental research or the formation of high-value products, in vitro synthetic biology platform, which can implement complicated biochemical reactions by the in vitro assembly of numerous enzymes and coenzymes, has been proposed for low-cost biomanufacturing of bioenergy, food, biochemicals, and nutraceuticals. In addition to the most important advantage-high product yield, in vitro synthetic biology platform features several other biomanufacturing advantages, such as fast reaction rate, easy product separation, open process control, broad reaction condition, tolerance to toxic substrates or products, and so on. In this article, we present the basic bottom-up design principles of in vitro synthetic pathway from basic building blocks-BioBricks (thermoenzymes and/or immobilized enzymes) to building modules (e.g., enzyme complexes or multiple enzymes as a module) with specific functions. With development in thermostable building blocks-BioBricks and modules, the in vitro synthetic biology platform would open a new biomanufacturing age for the cost-competitive production of biocommodities.
Keywords: Biomanufacturing, Coenzyme regeneration, Pathway design, In vitro synthetic biology
1. Introduction
Synthetic biology is an interdisciplinary branch of biology, chemistry and engineering that combines the investigative nature of biology with engineering design principles [1]. Most efforts in synthetic biology have largely concentrated on the design and construction of artificial biological pathways in vivo, or on the redesign of existing natural biological systems for biological research [[2], [3], [4], [5]]. The ultimate engineering goal of synthetic biology is the cost-competitive production of new drugs, biochemical, nutraceuticals, and bioenergy via engineered bioentities to replace current manufacturing methods [[6], [7], [8]].
Synthetic biology can be roughly divided into two areas, in vivo and in vitro synthetic biology. In vivo synthetic biology focuses on living bioentities, which can duplicate themselves. There are numerous breakthroughs, especially in terms of fundamental researches and publications [2,9,10], However, some inherent constraints of living organisms (e.g., net ATP generation for cell growth and maintenance, intact cellular membrane for maintaining basic metabolism and achieving selective mass transfer and exchange) prevent them from implementing some important reactions. Whereas, in vitro synthetic biology focuses on the construction of synthetic enzymatic pathways outside cells to convert substrates to desired products. For example, 12H2 can be obtained from one glucose and water via ATP-free in vitro synthetic pathways [11,12]. This pathways cannot be applied to living organisms due to no bioenergetic benefits. It is for the reason that living microorganisms have their H2 yield limit (i.e., 4H2 per glucose), called the Thauer limit [13,14]. Another example is making starch from cellulose, whereas cellulose and starch, large-size polymers, cannot be transported across cellular membrane [15].
Although in vitro synthetic biology is largely ignored compared to in vivo synthetic biology, it has made great and rapid progress [7,[16], [17], [18], [19]]. These in vitro synthetic biology systems can be based on cell extracts [20] or purified enzymes [21,22] or their combinations. Their potential applications include cell-free protein synthesis (CFPS) [23,24], vaccines [[25], [26], [27]], and potentially low-cost production of bioenergy [12,[28], [29], [30], [31]], nutraceuticals [32] and biochemicals [33,34]. The in vitro synthetic biology platform has some distinctive advantages, such as high product yield, high volumetric productivity, high product titer, high tolerance in toxic environments, substrates, and/or products, easy product separation and easy process control and optimization [7], and so on. These features make it feasible to become a disruptive biomanufacturing platform [17].
The history of in vitro (cell-free) fundamental research and in vitro biomanufacturing accompanied with milestones is presented in Table 1. The development of in vitro (synthetic) biology originated from Eduard Buchner's paradigm-shifting discovery of “cell-free ethanol fermentation by non-living yeast lysate” (Nobel Chemistry Prize 1907). Later, whole-cell lysates were important scientific targets for understanding of biochemistry of natural organisms. Numerous scientists isolated and characterized individual enzymes, reconstituted metabolic pathways in vitro and in vivo, and understood natural organisms. For instance, Harden et al. discovered key enzymes in glycolysis (Nobel Chemistry Prize 1929), Krebs analyzed the citric acid cycle (Nobel Chemistry Prize 1952), and Calvin elucidated the CO2 assimilation in plants (Nobel Chemistry Prize 1961). Subsequently, Jacob et al. discovered concerning genetic control of enzyme and virus synthesis, and Nirenberg and Matthaei interpreted the genetic code and its function in protein synthesis (Nobel Physiology or Medicine Prize 1968). The next major technique breakthroughs were the invention of the PCR method and the establishment of site-directed mutagenesis in the 1990s. Fundamental studies and tools development of in vitro biology offer a versatile workforce for understanding the operation principle of nature and for enabling redesigned biosynthetic pathways for the biosynthesis of novel chemicals, sustainable fuel, and new tunable materials. For example, CFPS, used for decades as a fundamental research tool for understanding transcription and translation, has been suggested to be the fastest way to make recombinant proteins, especially for membrane or complicated proteins [23,24]. CFPS has been expanded to a 100-L scale recently, showing great potential in industrial biomanufacturing [35].
Table 1.
The history of cell-free fundamental research and in vitro biomanufacturing accompanied with milestones.
| Year | Leaders | Milestone (Award) | References |
|---|---|---|---|
| Milestones for cell-free fundamental research | |||
| 1907 | Arthur Harden, Hans Karl August Simon von Euler-Chelpin | Fermentation of sugar and fermentative enzymes (Nobel Chemistry 1929) | [109] |
| 1930s | James Batcheller Sumner, John Howard Northrop, Wendell Meredith Stanley | Preparation of enzymes and virus proteins in a pure form (Nobel Chemistry 1946) | [[110], [111], [112]] |
| 1940s | Hans Krebs | Discovery of the citric acid cycle (Nobel Physiology 1953) | [113] |
| 1940s | Melvin Calvin | Carbon dioxide assimilation in plants (Nobel Chemistry 1961) | [114] |
| 1960s | Francois Jacob, Jacques Monod, Andre Lwoff | Discoveries concerning genetic control of enzyme and virus synthesis (Nobel Physiology or Medicine 1965) | [115] |
| 1960s | Robert William Holley, Har Gobind Khorana, Marshall Warren Nirenbrg | Interpretation of the genetic code and its function in protein synthesis (Nobel Physiology or Medicine 1968) | [116,117] |
| 1970s | Werner Arber, Daniel Nathans, Hamilton Othanel Smith | Discovery of restriction enzymes and their application to problems of molecular genetics (Nobel Physiology or Medicine 1978) | [[118], [119], [120]] |
| 1990s | Kary Banks Mullis, Michael Smith | Invention of the polymerase chain reaction (PCR) method; Establishment of site-directed mutagenesis with application to protein studies (Nobel Chemistry 1993) | [121,122] |
| 2000s- | Natural/non-natural product synthesis used as pharmaceuticals, biochemicals and biofules (e.g., CFPS) | [24,35] | |
| Milestones for in vitro biomanufacturing | |||
| 1897 | Eduard Buchner | Cell-free ethanol fermentation by nonliving yeast lysate (Nobel Chemistry 1907) | [123] |
| 1960s-1970s | One-enzyme biotransformation for high fructose corn syrup production (i.e., more than 20 million tonnes yearly) and semi-synthetic antibiotics (e.g., cephalosporin) | [37] | |
| 1990s | Multi-enzyme biotransformation for fine chemicals and pharmaceuticals production | [36,40,41] | |
| 1990s | Cell-free protein synthesis | [35] | |
| 2000s | Hydrogen, artificial starch, and inositol production | [15,32,92] | |
The development of in vitro synthetic biology platform for biomanufacturing lags far behind fundamental research of in vitro biology-biochemistry [36]. Although Eduard Buchner discovered the phenomenon of cell-free ethanol fermentation in the 1890s, the use of one enzyme for industrial biomanufacturing came into being in the 1960s-1970s, for instance, high fructose corn syrup (i.e., more than 20 million tonnes yearly) and semi-synthetic antibiotics (e.g., cephalosporin) [37]. Such in vitro biosystems evolved to more complicated system containing two-three enzymes in one pot for enhancing volumetric productivity, decreasing product inhibition, shifting reaction equilibrium, and facilitating product/substrate separation [38,39]. For example, the pharmaceutical and fine chemistry industries adopted this platform to produce high-value chiral alcohols, α-hydroxy acids, and α-amino acids, such as, (S)-2-butanol, L-tert-leucine, (S)-ethyl-4-chloro-3-hydroxybutyrate, atorvastatin, and so on [36,40,41]. In the organic chemistry field, the synthesis of monosaccharides, activated monosaccharides, oligosaccharides, and glycopeptides by using two-three enzymes in one pot had been intensively investigated, such as, l-fructose, 5-deoxy-5-ethyl-d-xylulose, amylose, and so on [[42], [43], [44], [45], [46], [47]]. In this century, some researchers propose to put more than four biocatalytic components or even tens of ones in one vessel to implement very complicated reactions that is comparable to microbial cell factories [17,28,29,31,33,48,49]. This represents the emerging area-the in vitro synthetic biology platform, distinct from in vitro biocatalysis based on one or multiple enzymes. The first industrial biomanufacturing example is the production of myo-inositol (called inositol later) from starch, which has been demonstrated in China [32].
In this review, we are focused on the bottom-up design principles of in vitro enzymatic pathways including pathway design and reconstruction, enzyme selection, and coenzyme management, and we highlight three examples for industrial biomanufacturing.
2. Basic design principles for in vitro synthetic pathways
The basic bottom-up design principles of in vitro synthetic pathways include (i) pathway design and reconstruction, (ii) enzyme selection, and (iii) coenzyme management. For pathway design and reconstruction, several points, such as coenzyme balance and involvement, enzyme selection, thermodynamics, reaction equilibrium, product separation et al., need to be carefully considered [17,22]. For enzyme selection, the discovery and utilization of thermostable enzymes can greatly simplify numerous biotechnological processes and decrease potential biomanufacturing costs [7,17]. Furthermore, coenzyme regeneration and balancing in in vitro synthetic pathways is another important issue. Depletion or imbalance of specific coenzymes slows down the reaction and finally stops the entire cascade. To overcome this problem, modules for regenerating and balancing coenzymes have been proposed and developed [16].
2.1. Pathway design and reconstruction: coenzyme-free or coenzyme balancing
The design and reconstruction of an enzymatic pathway is the central point of the in vitro synthetic biology platform, which starts from basic building blocks-BioBricks (i.e., thermoenzymes, immobilized enzymes) to building modules, that is, enzyme complexes or several enzymes with defined functions (e.g., ATP regeneration, NAD(P)H regeneration) to a complicated synthetic pathway or system for the purpose of biomanufacturing [17]. The pathway design usually starts from natural metabolic pathways with necessary modifications. Because the same biochemical reactions can be conducted by several different pathways sometimes, the pathways need to be designed carefully by considering ATP and NAD(P)H balance, thermodynamics, reaction equilibrium, product separation, and so on [17,22,50]. Owing to the expensive and unstable characteristics of coenzymes, it is best to design an in vitro synthetic coenzyme-free enzymatic pathway. For example, an in vitro non-fermentative enzymatic pathway has been constructed to convert starch to inositol in one vessel, which is composed of four enzymes without ATP or NAD+ supplementation. Besides inositol, artificial starch [15,51] and fructose 1,6-diphosphate [33] have been produced by using similar in vitro synthetic enzymatic pathways without coenzyme involved.
However, most in vitro biocatalysts for biomanufacturing are restricted to coenzyme-independent enzymes such as hydrolases and isomerases. In comparison, coenzyme-dependent enzymes, such as oxidoreductases and transferases, are capable of performing more complex chemistry. As a result, by considering the increasing range of products, it is vital to maintain both ATP and reducing power carriers (NAD(P)H) recycling and balance for in vitro synthetic biosystems. As these coenzymes are too costly to be used as stoichiometric agents for preparative applications, the regeneration of coenzymes in situ are needed for low-cost production [52]. Abundant in vitro coenzyme regeneration methods have been developed to regenerate the required coenzymes, while simultaneously driving the reaction equilibrium toward desired products [[52], [53], [54]]. Besides, coenzyme regeneration can simplify product isolation and avoid the accumulation of inhibitory coenzymes [55]. Recently, a variety of in vitro synthetic pathways with coenzyme regeneration have been designed and implemented for the production of chiral alcohols [31,48,56], biopolymers [57], organic acids [49,58,59], hydrogen [12,60,61], and bioelectricity [28,62].
Moreover, phenomena of ATP hydrolysis or spontaneous NAD(P)H oxidation or undesired side-reactions may take place when the cell lysates containing undesired enzyme components were used or the system is exposed to the air [30,57,63]. Thus, the system would wind down when the levels of ATP or NAD(P)H dissipate over time. Coenzyme balancing must be maintained owing to the economic viability of in vitro synthetic biosystems, which run the systems in a self-sustaining manner for a long time. Several strategies can be implemented, such as, a molecular purge valve module for NADPH balance [29,48,57], a molecular ATP rheostat [63], and integration of an additional enzyme set of thermophilic adenylate kinase and polyphosphate kinase for the deceleration of ATP degradation [30].
2.2. Enzyme selection: stable enzymes as standardized building blocks
Stable enzymes used as BioBricks for in vitro synthetic biosystems are essentially significant to decrease production costs and increase the carbohydrate allocation to the desired products [7,17]. In general, three major strategies can be conducted for the selection of stable enzymes: enzyme mining and discovery from (hyper-) thermophilic hosts, protein engineering, and enzyme immobilization. The best and simplest starting point is mining and discovery of thermoenzymes from (hyper-) thermophilic hosts [64]. Several novel enzymes have been discovered from hyper-thermophilic microorganisms like Thermotoga maritima, Thermus thermophilus, Pyrococcus furiosus, Thermococcus kodakarensis, Sulfolobus tokodaii, and so on [17]. In vitro biosynthetic biosystems have been constructed by using numerous recombinant thermoenzymes produced in E. coli BL21(DE3), such as, artificial starch production [15], hydrogen generation [12,60], bioelectricity generation [28,62], fructose 1,6-diphosphate production [33], and inositol production [32]. Now several websites have provided valuable collections for putative enzyme sources, such as, the Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.jp/kegg/) and the comprehensive information of characterized enzymes-BRENDA (http://www.brenda-enzymes.org/).
Poor thermostability of enzymes is one of the main limiting factors preventing the industrial application of enzymes [65]. When thermoenzymes are not available in the database and literature, an enzyme from a mesophilic source needs to be modified to enhance the stability by enzyme engineering, which involves rational design and directed evolution or their combination. Rational design usually requires both the availability of the structure of the enzyme and knowledge about the relationships among sequence, structure, and mechanism/function. For example, the thermostability of Pseudoalteromonas carrageenovora arylsulfatase has been improved by using rational design [66]. Among the mutants, K253H/H260L is the best one with improved thermal stability, and structure modeling demonstrates that the additional hydrogen bonds, optimization of surface charge-charge interactions, and increasing of hydrophobic interaction could account for the improved thermostability imparted by K253H/H260L. On the other side, directed evolution is another potent protein engineering tool for improving enzyme performance without in-depth understanding of protein structure and enzyme-substrate interactions. By using error-prone PCR or other mutation strategies accompanied with thermal stress for screening, mutants of endoglucanase and cellobiohydrolases have been identified and characterized with improved thermostability [67,68]. Recently, new strategies have been developed for the improvement of enzyme thermostability. For example, Cornvik and his coworkers developed a new screening method for protein thermostability engineering, so called HotCoFi method [69]. Unlike the traditional screening methods based on activity, this method relies on the unfolding and aggregation quality of the protein above a critical temperature. Rather than playing off one approach against the others, future efforts should focus on how to combine these alternative approaches in order to improve the thermostability of the desired enzyme. A successful first study of this type has been reported by Cherry et al. [70]. In their endeavor to improve the stability of a haem peroxidase for laundry applications, four mutations have been rationally designed: one to increase the enzyme's thermostability and three to increase resistance to oxidative damage. The combination of these mutations with favorable amino acid exchanges identified in directed evolution experiments yields a final mutant with 174 times the thermal stability and 100 times the oxidative stability of the wild-type haem peroxidase.
Enzyme immobilization is a classic method to increase enzyme stability even before mining & discovery of thermoenzymes and protein engineering [7,17]. The underlying benefits for immobilization are improved stability, easy recyclability of immobilized enzymes, and easy separation of biocatalysts and products. Besides, low risk of production contamination and low allergenicity are further advantages of enzyme immobilization. Methods for enzyme immobilization can be classified into three principal types: adsorption, encapsulation and cross-linking [71]. Furthermore, combinations of two or more immobilization methods are designed to improve the performance of immobilized enzymes. For instance, Antrim and his coworkers immobilized glucose isomerase (GI) from Streptomyces rubiginosus to DEAE-cellulose–polystyrene–TiO2 resin using electrostatic binding, resulting in immobilized glucose isomerase (IGI) with catalytic densities of up to 1500 U g−1. IGI is very stable, with a half-life of over 1800 h under recommended operating conditions at a pH range of 7.2–8.2 with a preferred range of 7.6–7.8, and a temperature range of 54–62 °C with the temperature of optimum productivity being about 57 °C [72,73]. Novozyme 435 (a lipase) is utilized to synthesize specialty esters industrially. Lipase B from Candida antarctica (CaLB) is adsorbed on Lewatit VP OC 1600 (Lanxess, Germany), whose protein loading can be up to 1–10% and the thermal stability can be up to 110 °C in solvent-free systems [[74], [75], [76]]. As we known, the ideal immobilization should have no/little influence on enzyme activity. However, several essential trade-offs occur when considering the immobilization method, as immobilization procedures often inactivate a percentage of the enzymes prepared and mass transfer can become a limitation, slowing the reaction rate.
2.3. In vitro ATP regeneration or balancing
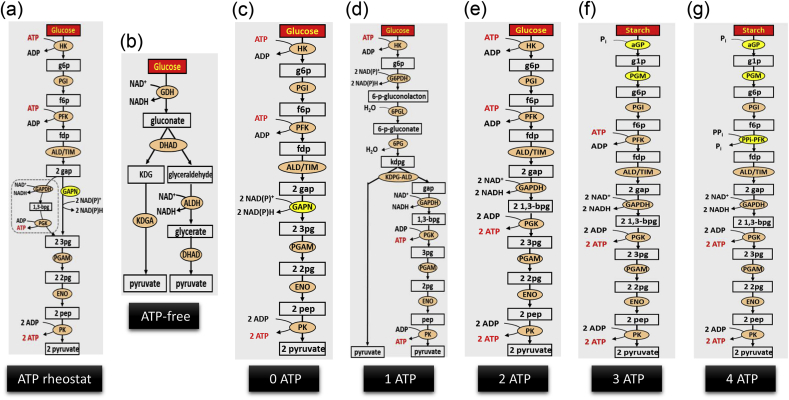
Adenosine triphosphate (ATP), the most influential energy currency for all living organisms, is essentially important for biosynthesis, mobility, signaling, and cell division [52]. Different from in vivo synthetic pathways where whole cells can obtain or depose extra ATP from or to cellular metabolism, in vitro synthetic enzymatic pathways must have a balance in ATP production and consumption although ATP may be needed for some enzymes. If net ATP is generated for the case of in vitro ethanol fermentation via the glycolytic pathway, the accumulation of ATP stops the cell-free system from running for a long time [77]. The best solution is cautious design of pathways without ATP involvement or with ATP balance. By contrast, unwanted ATP hydrolysis should be taken into account as a form of metabolite proofreading for maintaining high-energy coenzyme balance. A simple molecular ATP rheostat has been developed to regulate ATP levels by controlling the flow down either an ATP-generating or non-ATP-generating pathway in a function of free-phosphate concentration (Fig. 1a) [63]. This rheostat maintains adequate ATP concentrations even in the presence of ATPase contamination. Meanwhile, it is critical to use a low-cost sacrificial substrate for the regeneration of ATP due to high cost of ATP.
Fig. 1.
Pathway design for ATP regeneration or balancing. The enzymes are GDH, glucose dehydrogenase; DHAD, dihydroxy acid dehydratase; KDGA, 2-keto-3-desoxygluconate aldolase; ALDH glyceraldehyde dehydrogenase; HK, hexokinase; PGI, phosphoglucose isomerase; PFK, 6-phosphofructokinase; ALD, fructosebisphosphate aldolase; TIM, triosephosphate isomerase; GAPN, non-phosphorylating glyceraldehyde 3-phosphate dehydrogenase; PGAM, cofactor-independent phosphoglycerate mutase; ENO, enolase; PK, pyruvate kinase; G6PDH, glucose 6-phosphate dehydrogenase; 6PGL, 6-phosphogluconolactonase; 6PG, 6-phosphogluconate dehydratase; KDPG-ALD, 2-keto-3-deoxy-phosphogluconate aldolase; GAPDH glyceraldehyde 3-phosphate dehydrogenase; PGK phosphoglycerate dehydrogenase; aGP, alpha-glucan phosphorylase; PGM, phosphoglucomutase; PPi-PFK, pyrophosphate-dependent fructose-6-phosphate 1-phosphotransferase. Metabolites are KDG, 2-keto-3-deoxygluconate; g6p, glucose 6-phosphate; f6p, fructose 6-phosphate; fdp, fructose 1,6-diphosphate; gap, glyceraldehyde 3-phosphate; 3pg, 3-phosphoglycerate; 2pg, 2-phosphoglycerate; pep, phosphoenolpyruvate; kdpg, 2-keto-3-deoxy-6-phosphogluconate; 1,3-bpg, 1,3-diphosphoglycerate; dhap, dihydroxacetone phosphate.
In vitro ATP regeneration technologies are performed through glycolysis or by using different phosphate donors based on substrate-level phosphorylation. Various metabolic pathway modules from glucose or anhydroglucose from starch to pyruvate can be implemented in a function of different numbers of ATP generated, from zero to four (Fig. 1b–g). The in vitro ATP-free pathway has been shown to produce two pyruvate from glucose (Fig. 1b) [56], but this pathway suffer from very slow reaction rates. Alternatively, another in vitro ATP-balanced pathway has been designed by modification of the glycolytic pathway, generating two pyruvate and two NADH from one glucose with zero net ATP produced (Fig. 1c) [31]. Furthermore, if a small amount of ATP is necessary for the synthesis of desired products, several pathways can be selected as below. The Entner-Doudoroff pathway (ED pathway) can produce a net yield of one ATP per glucose (Fig. 1d) and the Embden-Meyerhof-Parnas pathway can generate two net ATP per glucose (Fig. 1e). The use of alpha-glucan phosphorylase and phosphoglucomutase to phosphorylate starch to generate glucose 6-phosphate, following the glycolytic pathway can generate three net ATP per glucose (Fig. 1f). When the introduction of a pyrophosphate-dependent fructose 6-phosphate kinase to replace ATP-dependent fructose 6-phosphate kinase enables the generation of four net ATP for a glucose unit of starch (Fig. 1g).
For phosphorylation with phosphate donors, low-cost polyphosphate and pyrophosphate will be economically feasible for biocommodity production. Meanwhile, numerous enzymes are found to be able to accept polyphosphate as phosphate donor for ATP regeneration. For instance, a thermophilic polyphosphate-dependent glucokinase from Thermobifida fusca YX has been applied into hydrogen production from glucose [60,78]; A new polyphosphate-dependent xylulokinase from T. maritima has been used to convert xylose to xylulose 5-phosphate along with xylose isomerase by using polyphosphate instead of ATP [79]; Pyrophosphate-dependent phosphofructokinase from T. maritima has been used along with three thermopilic enzymes to produce a high-energy phosphate metabolite fructose 1,6-diphosphate from starch and pyrophosphate [33].
2.4. In vitro NAD(P)+/NAD(P)H regeneration and balance
While living organisms can adjust NAD(P)+/NAD(P)H balance through anabolism and catabolism, in vitro synthetic biosystem must have NAD(P)+/NAD(P)H balanced in its pathway design at the beginning [16,17]. It means that the amount of reduced NAD(P)H generated from substrates should match that of NAD(P)H consumption for the production of desired products. The accumulation of the reduced NAD(P)H leads to depletion of the corresponding oxidized NAD(P)+, which is necessary for continuous utilization of substrate. For example, a molecular purge valve module for balancing the availability of NAD(P)+/NADPH has been designed, which is useful for the reaction module where NADPH production upstream in the reaction is in excess over its consumption downstream [57].
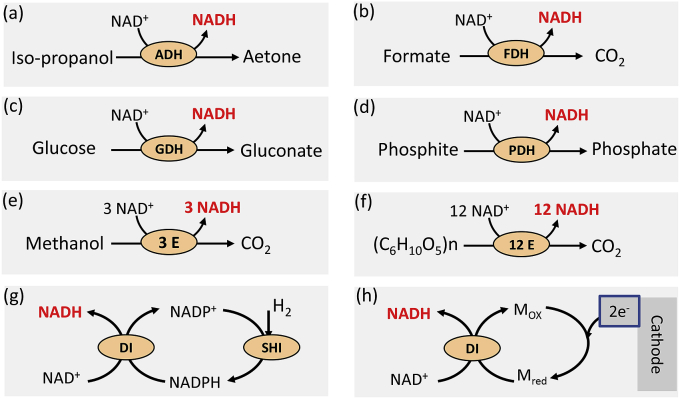
Most in vitro NAD(P)H regeneration methods can be implemented by using another substrate and its respective enzymes (Fig. 2). NAD(P)H can be generated by using a hydrogen-donor substrate and one of the followings: a single enzyme, cascade enzymes, and bioelectrochemistry. Single-enzyme systems include alcohol/alcohol dehydrogenase [80], formate/formate dehydrogenase [81], glucose/glucose dehydrogenase [82], glucose 6-phosphate (G6P)/G6P dehydrogenase [83], dihydrogen/hydrogenase [84,85], and phosphite/phosphite dehydrogenase [86]. Single-enzyme NAD(P)H regeneration systems have been widely used in the synthesis of high-value chiral compounds in the pharmaceutical industry. Four representative single-enzyme substrates to regenerate NADH are the dehydrogenation of isopropanol, formate, glucose, and phosphite (Fig. 2a–d). As an example of cascade enzymes for NADH regeneration, three enzymes-formate dehydrogenase, formaldehyde dehydrogenase, and alcohol dehydrogenase-can completely oxidize methanol to carbon dioxide, generating three NADH (Fig. 2e) [87]. A 12-enzyme system is utilized to produce nearly 12 NADPH from one glucose unit of cellobiose (Fig. 2f) [88]. In addition, our group has designed an NAD+- based electron transport chain (ETC) for in vitro NADH regeneration, where diaphorase as a transhydrogenase was used to convert NADPH and NAD+ to NADH and NADP+, matching with NADP+-preferred hydrogenase (submitted for publication) (Fig. 2g). NADH can also be regenerated by electrochemistry based on the mediator-conjugated diaphorase system (Fig. 2h) [89]. Among all hydrogen-donor compounds, renewable sugars have the lowest substrate costs, but they require more enzymes and increase system complexity. Utilization of electrochemistry to generate reduced cofactors is low-cost and clean, but the instability of NADH under high over-potential must be solved before this technique becomes industrially feasible.
Fig. 2.
In vitro NAD(P)H generation catalyzed by one enzyme or synthetic enzymatic pathways. NAD(P)H can be generated by using a hydrogen-donor substrate and one of the followings: a single enzyme (a–d), cascade enzymes (e–f), and bioelectrochemistry (g–h). The enzymes are ADH, alcohol dehydrogenase; FDH, formate dehydrogenase; GDH, glucose dehydrogenase; PDH, phosphite dehydrogenase; DI, diaphorase; SHI, soluble [NiFe] hydrogenase I. Mox and Mred are oxidized and reduced mediator.
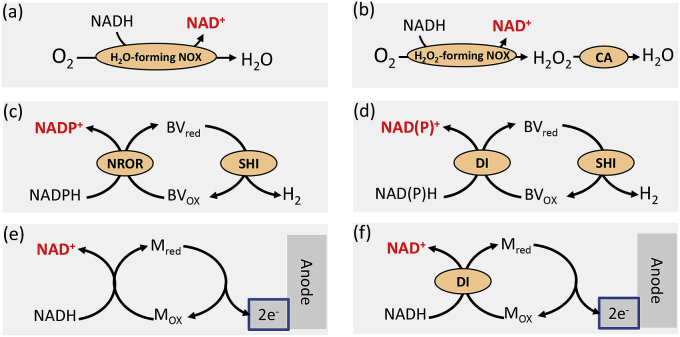
Sometimes designed products have a lower degree of reduction than those of substrates, for example, the production of 1,3-butanediol or fatty acid ethyl esters from glucose [90], that is, extra NAD(P)H is generated in in vitro pathways. Unlike microbial fermentation that can consume NAD(P)H through oxidation or cell mass synthesis, it is vital to remove extra NAD(P)H from in vitro synthetic biosystems. Fig. 3 presents four different ways to remove extra NAD(P)H: enzymatic (Fig. 3a–d) and electrochemical (Fig. 3e–f). NADH can be converted to NAD+ by using a water-forming NADH oxidase (Fig. 3a) or a hydrogen peroxide-forming NADH oxidase combined with catalase (Fig. 3b). For example, a water-forming NADH oxidase from Lactobacillus pentosus has been used for the regeneration of NAD+ from NADH during the conversion of glucuronate to α-ketoglutarate [34] and the cell-free production of monoterpenes from glucose [29]. Furthermore, extra NADPH can also be removed by hydrogenase to produce H2 via the biomimetic ETC, which has been designed by the introduction of an electron mediator benzyl viologen (BV) and an enzyme NADPH rubredoxin oxidoreductase (NROR) (Fig. 3c) [91] or BV-conjugated diaphorase system (Fig. 3d) [92]. Lastly, another way to remove extra NADH occurs in enzymatic fuel cells through an electron mediator (Fig. 3e–f) [28,93].
Fig. 3.
In vitro NAD(P) oxidative regeneration. NAD(P) oxidative regeneration by a water-forming NADH oxidase (a), a hydrogen peroxide-forming NADH oxidase combined with catalase (b), cascade enzymes combined with an electron mediator (c–d), and electrochemistry through a mediator with or without enzyme (e–f). The enzymes are NOX, NADH oxidase; CA, catalase; NROR, NADPH rubredoxin oxidoreductase; DI, diaphorase; SHI, soluble [NiFe] hydrogenase I. BVox and BVred are oxidized and reduced benzyl viologen; Mox and Mred are oxidized and reduced electron mediator.
NAD(P)+ and NAD(P)H are known to have relatively low thermal stability. Thermal instability of NAD(P)H is problematic, especially at high temperature [31,59]. To overcome this obstacle, the NAD+ salvage module has been designed to re-synthesize NAD+ from its thermal decomposition products of nicotinamide and ADP-ribose using eight thermophilic enzymes [94]. NAD+ concentration remains nearly constant for 15 h at 60 °C with the NAD+ salvage module, while the concentration decreased by a half in 6 h without the module [94].
3. Representative examples of biomanufacturing
It is highly likely that more biocommodities with huge-market sizes could be preferentially produced by the in vitro synthetic biology platform if synergetic efforts are taken for the design of enzymatic pathway, production of low-cost stable enzyme, enzyme immobilization and recycle, utilization of biomimetic coenzymes, coenzyme recycle, and product separation. Although the in vitro synthetic biology is just an emerging frontier, many high-value biochemicals and biofuels have been produced via this platform, such as 1,3-propanediol [95], poly-3-hydroxylbutyrate [96], amylose [15], n-butanol [31], isobutanol [56,63], terpenoids [29], and so on. Here three representative examples are highlighted for the ingeniousness of pathway design.
3.1. Pathway design for inositol production
Inositol is important in the cosmetics, pharmaceutical and functional food industries, which is predominately obtained by acid hydrolysis of inositol hexakisphosphate (IP6). However, this production method suffers from costly feedstock, serious phosphorous pollution, and complicated feedstock and product separation, resulting in relatively high price and limited supply.
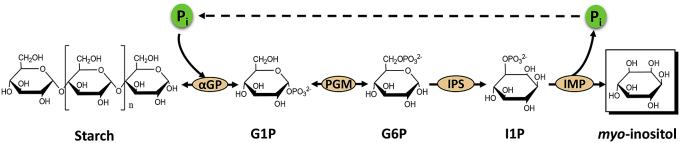
You and his coworkers have constructed an in vitro synthetic enzymatic pathway that can convert starch to inositol without external coenzyme supplement [32]. This pathway is comprised of four steps (Fig. 4): (i) glucose 1-phosphate (G1P) generation from starch and phosphate; (ii) G6P generation from G1P; (iii) inositol 1-phosphate (I1P) generation from G6P; and (iv) inositol generation accompanied by phosphate generation from I1P. Phosphate generated from the fourth module is recycled in the first step. The consolidation of four step reactions has an overall Gibbs energy of −80.1 kJ/mol, that is, this pathway has a very high equilibrium constant to push the overall reaction toward completeness with very high product yield. Later, Atomi and his coworkers also demonstrate the synthesis of inositol from starch [97]; Tao and his coworkers demonstrate the synthesis of inositol from glucose with ATP regeneration from polyphosphate [98]; and Zhang and his coworkers demonstrate inositol production from sucrose [99].
Fig. 4.
Scheme of the in vitro synthetic enzymatic pathway for the production of inositol from starch. The enzymes are aGP, alpha-glucan phosphorylase; PGM, phosphoglucomutase; IPS, inositol 1-phosphate synthase; IMP inositol monophosphatase. Metabolites are G1P, glucose 1-phosphate; G6P, glucose 6-phosphate; I1P, inositol 1-phosphate; Pi, inorganic phosphate.
This new synthesis of inositol from starch is a disruptive method for green production of inositol compared to the acid hydrolysis of IP6. It has many biomanufacturing advantages: (i) less costly substrate with starch; (ii) decreased phosphorous pollution and COD emission; (iii) easy product separation; (iv) scalable low-cost production of all thermoenzymes; and (v) nearly no odds for microbial contamination.
3.2. High-yield production of hydrogen
Hydrogen (H2) as a future transportation fuel offers enhanced energy conversion efficiency and tremendous potential to reduce greenhouse gas emissions [100]. In spite of intensive efforts in metabolic engineering and synthetic biology, none of natural or engineered microorganisms can produce H2 beyond the Thauer limit (4H2/glucose) [[101], [102], [103]]. Moreover, in vitro hydrogen production from low-cost biomass and water is an excellent solution for producing low-cost H2 without net carbon emissions [12,92,104].
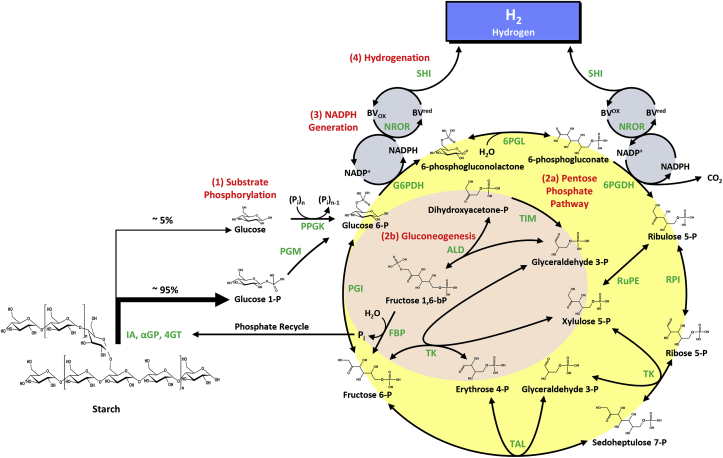
Starch has been proposed as a new high-density hydrogen storage carrier with its gravimetric density of up to 14% H2 mass. Zhang and his coworkers have carried out a proof of-concept experiment for H2 production from glycogen (animal starch) using an in vitro enzymatic pathway [105] with the maximum H2 production yield (43% of the theoretical yield of 12H2 per glucose) exceeding the Thauer limit. Later they have redesigned and demonstrated several in vitro pathways for H2 production from various carbohydrates, including cellulosic materials [106], xylose [79], sucrose [61], a mixture of biomass monosaccharides [60], and xylooligosaccharides [104]. Recently, Kim and his coworkers have constructed an in vitro synthetic pathway for generating H2 at theoretical yield from starch with the maximum volumetric productivity of 90.2 mmol/L/h [12]. This reconstituted ATP-free and cofactor-balanced enzymatic pathway composed of 17 enzymes and it can be grouped into four modules (Fig. 5): (i) ATP-free phosphorylation of starch generating G6P; (ii) NADPH generation via the oxidative pentose phosphate pathway (PPP); (iii) hydrogen generation catalyzed by soluble [NiFe]-hydrogenase I from a hyperthermophilic archaeon P. furiosus (SHI) from NADPH via a biomimetic ETC comprised of NROR and BV as an abiotic electron mediator [91]; and (iv) G6P regeneration via the non-oxidative PPP (iv-a) and partial gluconeogenesis pathway (iv-b). Phosphate generated from the fourth module is recycled by αGP for starch phosphorolysis in the first module.
Fig. 5.
Scheme of the in vitro synthetic pathway for the production of hydrogen through complete utilization of starch. The enzymes are IA, isoamylase; 4GT, 4-alpha-glucanotransferase; αGP, alpha-glucan phosphorylase; PPGK, polyphosphate glucokinase; PGM, phosphoglucomutase; G6PDH, glucose 6-phosphate dehydrogenase; 6PGDH, 6-phosphogluconate dehydrogenase; NROR, NADPH rubredoxin oxidoreductase; SHI, soluble [NiFe] hydrogenase I; 6PGL, 6-phosphogluconolactonase; RPI, ribose 5-phosphate isomerase; RuPE, ribulose 5-phosphate 3-epimerase; TK, transketolase; TAL, transaldolase; TIM, triose phosphate isomerase; ALD, aldolase; FBP, fructose 1,6-biphosphatase; PGI, phosphoglucose isomerase. Pi and (Pi)n are inorganic phosphate and polyphosphate with a degree of polymerization of n. BVox and BVred are oxidized and reduced benzyl viologen.
Thermodynamic analysis indicates that the overall reaction is spontaneous with an overall Gibbs free energy change of −48.9 kJ/mol. Meanwhile, due to the gaseous products (H2 and CO2) are simultaneously removed from the liquid reaction solution, the real Gibbs free energy change is much less than −48.9 kJ/mol to drive the overall reaction toward completeness.
3.3. N-butanol production
N-butanol, a primary 4-carbon alcohol, is regarded as the advanced liquid biofuel with an energy density (27 MJ/L) comparable to gasoline (32 MJ/L). It is traditionally produced by acetone-butanol-ethanol (ABE) fermentation using Clostridium acetobutylicum [7,107]. However, its fermentation involves a complicated transition from acidogenesis to solvogenesis and suffers from low product yields and severe product inhibition, resulting in low product titers and yields [108].
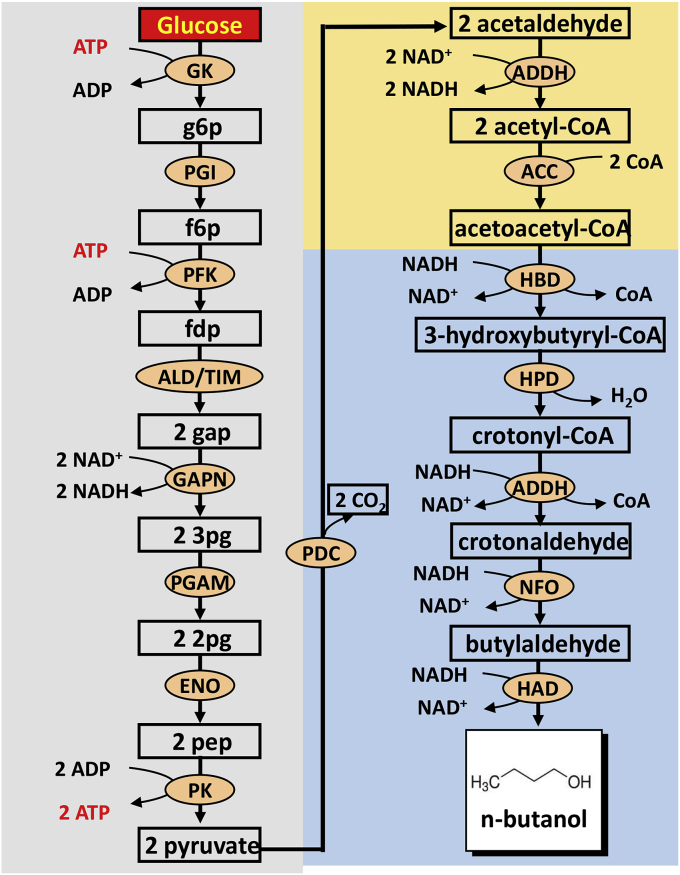
Honda and his coworkers have constructed a non-natural, cofactor-balanced, and oxygen-insensitive pathway for the direct conversion of glucose to n-butanol using 16 thermostable enzymes [31]. This pathway comprises three modules (Fig. 6): (i) generation of two pyruvate and two NADH from one glucose without ATP accumulation, (ii) generation of acetyl-CoA from pyruvate; and (iii) production of one n-butanol from acetyl-CoA consuming two NADH. As a consequence, one glucose can produce one n-butanol, two CO2 and one water. This synthetic pathway has three key features pertaining to the regenerations of ATP and redox cofactors (i.e., NADH and CoA): (i) ATP balance, where the ATP consumption during the conversion of glucose to fructose-1,6-diphosphate matches the ATP regeneration from phosphoenolpyruvate to pyruvate mediated by pyruvate kinase; (ii) NADH balance, where NADH regeneration by non-phosphorylating GAP dehydrogenase (GAPN) and CoA-acylating aldehyde dehydrogenase (ADDH) matches its consumption by hydroxybutyryl-CoA dehydrogenase (HBD), ADDH, NADH dependent flavinoxidoreductase (NFO), and 3-hydroxyacyl-CoA dehydrogenase (HAD); (iii) CoA balance, where CoA is needed by ADDH and is released by acetyl-CoA acetyltransferase (ACC) and ADDH. The overall reaction is an enthalpy-driven reaction with an overall Gibbs free energy change of −265.9 kJ/mol and a slight loss of chemical energy exists in the reaction. By optimizing enzyme loading and replenishing of redox cofactors NAD+ and NADH, n-butanol could be produced from glucose with a molar yield of 82% at a rate of 8.2 μmol l-1 min-1, comparable with the best product yield in ABE fermentation [108].
Fig. 6.
Scheme of the in vitro synthetic enzymatic pathway for the production of n-butanol from glucose. The enzymes are HK, hexokinase; PGI, phosphoglucose isomerase; PFK, 6-phosphofructokinase; FBA, fructosebisphosphate aldolase; TIM, triosephosphate isomerase; GAPN, non-phosphorylating GAP dehydrogenase; PGAM, cofactor-independent phosphoglycerate mutase; ENO, enolase; PK, pyruvate kinase; PDC, pyruvate decarboxylase; ADDH, CoA-acylating aldehyde dehydrogenase; HBD, hydroxybutyryl-CoA dehydrogenase; HPD, 3-hydroxypropionyl-CoA dehydratase; NFO, NADH-dependent flavinoxidoreductase; and HAD, 3-hydroxyacyl-CoA dehydrogenase. Metabolites are g6p, glucose 6-phosphate; f6p, fructose 6-phosphate; fdp, fructose 1,6-diphosphate; gap, glyceraldehyde 3-phosphate; dhap, dihydroxacetone phosphate; 3pg, 3-phosphoglycerate; 2pg, 2-phosphoglycerate; and pep, phosphoenolpyruvate.
4. Conclusions
The appealing advantages, such as high product yield, fast reaction rate, broad reaction condition, as well as easy process control and regulation, are motivating the in vitro synthetic biology platform to be a novel biomanufacturing platform compared to the predominant fermentation. Here, in vitro synthetic pathways are comprised of stable BioBricks (e.g., thermoenzymes, immobilized enzymes) and modules (e.g., enzyme complexes or multiple enzymes as a module) with specific functions (e.g., coenzyme regeneration). Mining and discovery of thermoenzymes, protein engineering, and enzyme immobilization would result in ultra-stable enzymes as basic BioBricks. Many efficient coenzyme regeneration systems, including ATP and NAD(P)H, have been developed as building modules. The assembly of BioBricks and modules would make cost-competitive production of biocommodities. The value-added inositol from starch is the first manufactured on an industrial scale [32]. In a word, the in vitro synthetic biology platform would open a new biomanufacturing age for the cost-competitive manufacturing of bioenergy, food, biochemicals, and nutraceuticals [7].
Declaration of interest
None.
Acknowledgement
This work was supported by the National Natural Science Foundation of China (Grant No. 31700033) and the Key Research Program of the Chinese Academy of Sciences (Grant No. ZDRW-ZS-2016-3).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Contributor Information
Chun You, Email: you_c@tib.cas.cn.
Yi-Heng P. Job Zhang, Email: yhjob_zhang@outlook.com.
References
- 1.Khalil A.S., Collins J.J. Synthetic biology: applications come of age. Nat Rev Genet. 2010;11:367–379. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang M.C., Keasling J.D. Production of isoprenoid pharmaceuticals by engineered microbes. Nat Chem Biol. 2006;2:674–681. doi: 10.1038/nchembio836. [DOI] [PubMed] [Google Scholar]
- 3.Stephanopoulos G. Synthetic biology and metabolic engineering. ACS Synth Biol. 2012;1:514–525. doi: 10.1021/sb300094q. [DOI] [PubMed] [Google Scholar]
- 4.Lee S.Y. Metabolic engineering and synthetic biology in strain development. ACS Synth Biol. 2012;1:491–492. doi: 10.1021/sb300109d. [DOI] [PubMed] [Google Scholar]
- 5.Keasling J.D. Synthetic biology and the development of tools for metabolic engineering. Metab Eng. 2012;14:189–195. doi: 10.1016/j.ymben.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 6.Clomburg J.M., Crumbley A.M., Gonzalez R. Industrial biomanufacturing: the future of chemical production. Science. 2017;355 doi: 10.1126/science.aag0804. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y.P., Sun J., Ma Y. Biomanufacturing: history and perspective. J Ind Microbiol Biotechnol. 2017;44:773–784. doi: 10.1007/s10295-016-1863-2. [DOI] [PubMed] [Google Scholar]
- 8.Dudley Q.M., Karim A.S., Jewett M.C. Cell-free metabolic engineering: biomanufacturing beyond the cell. Biotechnol J. 2015;10:69–82. doi: 10.1002/biot.201400330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paddon C.J., Westfall P.J., Pitera D.J., Benjamin K., Fisher K., McPhee D., Leavell M.D., Tai A., Main A., Eng D. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature. 2013;496:528–532. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]
- 10.Yim H., Haselbeck R., Niu W., Pujol-Baxley C., Burgard A., Boldt J., Khandurina J., Trawick J.D., Osterhout R.E., Stephen R. Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nat Chem Biol. 2011;7:445–452. doi: 10.1038/nchembio.580. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y.H.P. Renewable carbohydrates are a potential high-density hydrogen carrier. Int J Hydrogen Energy. 2010;35:10334–10342. [Google Scholar]
- 12.Kim J.E., Kim E.J., Chen H., Wu C.H., Adams M.W.W., Zhang Y.P. Advanced water splitting for green hydrogen gas production through complete oxidation of starch by in vitro metabolic engineering. Metab Eng. 2017;44:246–252. doi: 10.1016/j.ymben.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 13.Thauer R.K., Kaster A.K., Seedorf H., Buckel W., Hedderich R. Methanogenic archaea: ecologically relevant differences in energy conservation. Nat Rev Microbiol. 2008;6:579–591. doi: 10.1038/nrmicro1931. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y.H.P. Simpler is better: high-yield and potential low-cost biofuels production through cell-free synthetic pathway biotransformation (SyPaB) ACS Catal. 2011;1:998–1009. [Google Scholar]
- 15.You C., Chen H., Myung S., Sathitsuksanoh N., Ma H., Zhang X.Z., Li J., Zhang Y.H. Enzymatic transformation of nonfood biomass to starch. P Natl Acad Sci USA. 2013;110:7182–7187. doi: 10.1073/pnas.1302420110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taniguchi H., Okano K., Honda K. Modules for in vitro metabolic engineering: pathway assembly for bio-based production of value-added chemicals. Synth Syst Biotechnol. 2017;2:65–74. doi: 10.1016/j.synbio.2017.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y.H. Production of biofuels and biochemicals by in vitro synthetic biosystems: opportunities and challenges. Biotechnol Adv. 2015;33:1467–1483. doi: 10.1016/j.biotechadv.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 18.Guo W., Sheng J., Feng X. Mini-review: in vitro metabolic engineering for biomanufacturing of high-value products. Comput Struct Biotechnol J. 2017;15:161–167. doi: 10.1016/j.csbj.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan G.Y., Zhu F., Deng Z., Liu T. In vitro reconstitution guide for targeted synthetic metabolism of chemicals, nutraceuticals and drug precursors. Synth Syst Biotechnol. 2016;1:25–33. doi: 10.1016/j.synbio.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dudley Q.M., Anderson K.C., Jewett M.C. Cell-Free Mixing of Escherichia coli crude extracts to prototype and rationally engineer high-titer mevalonate synthesis. ACS Synth Biol. 2016;5:1578–1588. doi: 10.1021/acssynbio.6b00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang H.H., Huang P.Y., Xu G., Haas W., Marblestone A., Li J., Gygi S.P., Forster A.C., Jewett M.C., Church G.M. Multiplexed in vivo his-tagging of enzyme pathways for in vitro single-pot multienzyme catalysis. ACS Synth Biol. 2012;1:43–52. doi: 10.1021/sb3000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rollin J.A., Tam T.K., Zhang Y.H.P. New biotechnology paradigm: cell-free biosystems for biomanufacturing. Green Chem. 2013;15:1708–1719. [Google Scholar]
- 23.Carlson E.D., Gan R., Hodgman C.E., Jewett M.C. Cell-free protein synthesis: applications come of age. Biotechnol Adv. 2012;30:1185–1194. doi: 10.1016/j.biotechadv.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogonah O.W., Polizzi K.M., Bracewell D.G. Cell free protein synthesis: a viable option for stratified medicines manufacturing? Curr Opin Chem Eng. 2017;18:77–83. [Google Scholar]
- 25.Kanter G., Yang J., Voloshin A., Levy S., Swartz J.R., Levy R. Cell-free production of scFv fusion proteins: an efficient approach for personalized lymphoma vaccines. Blood. 2007;109:3393–3399. doi: 10.1182/blood-2006-07-030593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeltins A. Construction and characterization of virus-like particles: a review. Mol Biotechnol. 2013;53:92–107. doi: 10.1007/s12033-012-9598-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jennings G.T., Bachmann M.F. The coming of age of virus-like particle vaccines. Biol Chem. 2008;389:521–536. doi: 10.1515/bc.2008.064. [DOI] [PubMed] [Google Scholar]
- 28.Zhu Z., Zhang Y.P. In vitro metabolic engineering of bioelectricity generation by the complete oxidation of glucose. Metab Eng. 2017;39:110–116. doi: 10.1016/j.ymben.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 29.Korman T.P., Opgenorth P.H., Bowie J.U. A synthetic biochemistry platform for cell free production of monoterpenes from glucose. Nat Commun. 2017;8 doi: 10.1038/ncomms15526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Honda K., Kimura K., Ninh P.H., Taniguchi H., Okano K., Ohtake H. In vitro bioconversion of chitin to pyruvate with thermophilic enzymes. J Biosci Bioeng. 2017;124:296–301. doi: 10.1016/j.jbiosc.2017.04.013. [DOI] [PubMed] [Google Scholar]
- 31.Krutsakorn B., Honda K., Ye X., Imagawa T., Bei X., Okano K., Ohtake H. In vitro production of n-butanol from glucose. Metab Eng. 2013;20:84–91. doi: 10.1016/j.ymben.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 32.You C., Shi T., Li Y., Han P., Zhou X., Zhang Y.P. An in vitro synthetic biology platform for the industrial biomanufacturing of myo-inositol from starch. Biotechnol Bioeng. 2017;114:1855–1864. doi: 10.1002/bit.26314. [DOI] [PubMed] [Google Scholar]
- 33.Wang W., Liu M., You C., Li Z., Zhang Y.P. ATP-free biosynthesis of a high-energy phosphate metabolite fructose 1,6-diphosphate by in vitro metabolic engineering. Metab Eng. 2017;42:168–174. doi: 10.1016/j.ymben.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 34.Beer B., Pick A., Sieber V. In vitro metabolic engineering for the production of alpha-ketoglutarate. Metab Eng. 2017;40:5–13. doi: 10.1016/j.ymben.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 35.Zawada J.F., Yin G., Steiner A.R., Yang J.H., Naresh A., Roy S.M., Gold D.S., Heinsohn H.G., Murray C.J. Microscale to manufacturing scale-up of cell-free cytokine production-a new approach for shortening protein production development timelines. Biotechnol Bioeng. 2011;108:1570–1578. doi: 10.1002/bit.23103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bornscheuer U.T., Huisman G.W., Kazlauskas R.J., Lutz S., Moore J.C., Robins K. Engineering the third wave of biocatalysis. Nature. 2012;485:185–194. doi: 10.1038/nature11117. [DOI] [PubMed] [Google Scholar]
- 37.Demain A.L. Pickles, pectin, and penicillin. Annu Rev Microbiol. 2004;58:1–42. doi: 10.1146/annurev.micro.58.030603.123757. [DOI] [PubMed] [Google Scholar]
- 38.Santacoloma P.A., Sin G., Gernaey K.V., Woodley J.M. Multienzyme-catalyzed processes: next-generation biocatalysis. Org Process Res Dev. 2011;15:203–212. [Google Scholar]
- 39.Schoffelen S., van Hest J.C.M. Multi-enzyme systems: bringing enzymes together in vitro. Soft Matter. 2012;8:1736–1746. [Google Scholar]
- 40.De Wildeman S.M.A., Sonke T., Schoemaker H.E., May O. Biocatalytic reductions: from lab curiosity to “first choice”. Accounts Chem Res. 2007;40:1260–1266. doi: 10.1021/ar7001073. [DOI] [PubMed] [Google Scholar]
- 41.Huisman G.W., Liang J., Krebber A. Practical chiral alcohol manufacture using ketoreductases. Curr Opin Chem Biol. 2010;14:122–129. doi: 10.1016/j.cbpa.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 42.Endo T., Koizumi S. Large-scale production of oligosaccharides using engineered bacteria. Curr Opin Struct Biol. 2000;10:536–541. doi: 10.1016/s0959-440x(00)00127-5. [DOI] [PubMed] [Google Scholar]
- 43.Fessner W.D. Enzyme mediated C-C bond formation. Curr Opin Chem Biol. 1998;2:85–97. doi: 10.1016/s1367-5931(98)80040-9. [DOI] [PubMed] [Google Scholar]
- 44.Fessner W.D., Helaine V. Biocatalytic synthesis of hydroxylated natural products using aldolases and related enzymes. Curr Opin Biotechnol. 2001;12:574–586. doi: 10.1016/s0958-1669(01)00265-8. [DOI] [PubMed] [Google Scholar]
- 45.Huang K.T., Wu B.C., Lin C.C., Luo S.C., Chen C.P., Wong C.H., Lin C.C. Multi-enzyme one-pot strategy for the synthesis of sialyl Lewis X-containing PSGL-1 glycopeptide. Carbohydr Res. 2006;341:2151–2155. doi: 10.1016/j.carres.2006.04.047. [DOI] [PubMed] [Google Scholar]
- 46.Schoevaart R., van Rantwijk F., Sheldon R.A. A four-step enzymatic cascade for the one-pot synthesis of non-natural carbohydrates from glycerol. J Org Chem. 2000;65:6940–6943. doi: 10.1021/jo000492y. [DOI] [PubMed] [Google Scholar]
- 47.Franke D., Machajewski T., Hsu C.C., Wong C.H. One-pot synthesis of L-fructose using coupled multienzyme systems based on rhamnulose 1-phosphate aldolase. J Org Chem. 2003;68:6828–6831. doi: 10.1021/jo030021m. [DOI] [PubMed] [Google Scholar]
- 48.Opgenorth P.H., Korman T.P., Bowie J.U. A synthetic biochemistry module for production of bio-based chemicals from glucose. Nat Chem Biol. 2016;12:393–395. doi: 10.1038/nchembio.2062. [DOI] [PubMed] [Google Scholar]
- 49.Ninh P.H., Honda K., Sakai T., Okano K., Ohtake H. Assembly and multiple gene expression of thermophilic enzymes in Escherichia coli for in vitro metabolic engineering. Biotechnol Bioeng. 2015;112:189–196. doi: 10.1002/bit.25338. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Y.H.P. Production of biocommodities and bioelectricity by cell-free synthetic enzymatic pathway biotransformations: challenges and opportunities. Biotechnol Bioeng. 2010;105:663–677. doi: 10.1002/bit.22630. [DOI] [PubMed] [Google Scholar]
- 51.Qi P., You C., Zhang Y.H.P. One-pot enzymatic conversion of sucrose to synthetic amylose by using enzyme cascades. ACS Catal. 2014;4:1311–1317. [Google Scholar]
- 52.Andexer J.N., Richter M. Emerging enzymes for ATP regeneration in biocatalytic processes. Chembiochem. 2015;16:380–386. doi: 10.1002/cbic.201402550. [DOI] [PubMed] [Google Scholar]
- 53.Hummel W., Groger H. Strategies for regeneration of nicotinamide coenzymes emphasizing self-sufficient closed-loop recycling systems. J Biotechnol. 2014;191:22–31. doi: 10.1016/j.jbiotec.2014.07.449. [DOI] [PubMed] [Google Scholar]
- 54.Weckbecker A., Groger H., Hummel W. Regeneration of nicotinamide coenzymes: principles and applications for the synthesis of chiral compounds. Adv Biochem Eng Biot. 2010;120:195–242. doi: 10.1007/10_2009_55. [DOI] [PubMed] [Google Scholar]
- 55.Koeller K.M., Wong C.H. Enzymes for chemical synthesis. Nature. 2001;409:232–240. doi: 10.1038/35051706. [DOI] [PubMed] [Google Scholar]
- 56.Guterl J.K., Garbe D., Carsten J., Steffler F., Sommer B., Reisse S., Philipp A., Haack M., Ruhmann B., Koltermann A. Cell-free metabolic engineering: production of chemicals by minimized reaction cascades. ChemSusChem. 2012;5:2165–2172. doi: 10.1002/cssc.201200365. [DOI] [PubMed] [Google Scholar]
- 57.Opgenorth P.H., Korman T.P., Bowie J.U. A synthetic biochemistry molecular purge valve module that maintains redox balance. Nat Commun. 2014;5 doi: 10.1038/ncomms5113. [DOI] [PubMed] [Google Scholar]
- 58.Ye X.T., Honda K., Morimoto Y., Okano K., Ohtake H. Direct conversion of glucose to malate by synthetic metabolic engineering. J Biotechnol. 2013;164:34–40. doi: 10.1016/j.jbiotec.2012.11.011. [DOI] [PubMed] [Google Scholar]
- 59.Ye X., Honda K., Sakai T., Okano K., Omasa T., Hirota R., Kuroda A., Ohtake H. Synthetic metabolic engineering-a novel, simple technology for designing a chimeric metabolic pathway. Microb Cell Factories. 2012;11:120. doi: 10.1186/1475-2859-11-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rollin J.A., del Campo J.M., Myung S., Sun F.F., You C., Bakovic A., Castro R., Chandrayan S.K., Wu C.H., Adams M.W.W. High-yield hydrogen production from biomass by in vitro metabolic engineering: mixed sugars coutilization and kinetic modeling. P Natl Acad Sci USA. 2015;112:4964–4969. doi: 10.1073/pnas.1417719112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Myung S., Rollin J., You C., Sun F.F., Chandrayan S., Adams M.W.W., Zhang Y.H.P. In vitro metabolic engineering of hydrogen production at theoretical yield from sucrose. Metab Eng. 2014;24:70–77. doi: 10.1016/j.ymben.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 62.Zhu Z., Kin Tam T., Sun F., You C., Percival Zhang Y.H. A high-energy-density sugar biobattery based on a synthetic enzymatic pathway. Nat Commun. 2014;5:3026. doi: 10.1038/ncomms4026. [DOI] [PubMed] [Google Scholar]
- 63.Opgenorth P.H., Korman T.P., Iancu L., Bowie J.U. A molecular rheostat maintains ATP levels to drive a synthetic biochemistry system. Nat Chem Biol. 2017;13 doi: 10.1038/nchembio.2418. 938-+ [DOI] [PubMed] [Google Scholar]
- 64.You C., Zhang Y.H. Cell-free biosystems for biomanufacturing. Adv Biochem Eng Biot. 2013;131:89–119. doi: 10.1007/10_2012_159. [DOI] [PubMed] [Google Scholar]
- 65.Denard C., Ren H., Zhao H. Improving and repurposing biocatalysts via directed evolution. Curr Opin Chem Biol. 2015;25:55–64. doi: 10.1016/j.cbpa.2014.12.036. [DOI] [PubMed] [Google Scholar]
- 66.Zhu Y., Qiao C., Li H., Li L., Xiao A., Ni H., Jiang Z. Improvement thermostability of Pseudoalteromonas carrageenovora arylsulfatase by rational design. Int J Biol Macromol. 2018;108:953–959. doi: 10.1016/j.ijbiomac.2017.11.014. [DOI] [PubMed] [Google Scholar]
- 67.Wu I., Arnold F.H. Engineered thermostable fungal Cel6A and Cel7A cellobiohydrolases hydrolyze cellulose efficiently at elevated temperatures. Biotechnol Bioeng. 2013;110:1874–1883. doi: 10.1002/bit.24864. [DOI] [PubMed] [Google Scholar]
- 68.Liu W.J., Zhang X.Z., Zhang Z.M., Zhang Y.H.P. Engineering of Clostridium phytofermentans endoglucanase Cel5A for improved thermostability. Appl Environ Microbiol. 2010;76:4914–4917. doi: 10.1128/AEM.00958-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Asial I., Cheng Y.X., Engman H., Dollhopf M., Wu B.H., Nordlund P., Cornvik T. Engineering protein thermostability using a generic activity-independent biophysical screen inside the cell. Nat Commun. 2013;4 doi: 10.1038/ncomms3901. [DOI] [PubMed] [Google Scholar]
- 70.Cherry J.R., Lamsa M.H., Schneider P., Vind J., Svendsen A., Jones A., Pedersen A.H. Directed evolution of a fungal peroxidase. Nat Biotechnol. 1999;17:379–384. doi: 10.1038/7939. [DOI] [PubMed] [Google Scholar]
- 71.Sheldon R.A., van Pelt S. Enzyme immobilisation in biocatalysis: why, what and how. Chem Soc Rev. 2013;42:6223–6235. doi: 10.1039/c3cs60075k. [DOI] [PubMed] [Google Scholar]
- 72.Antrim R.L., Lloyd N.E., Auterinen A.L. New isomerization technology for high fructose syrup production. Starch Staerke. 1989;41:155–159. [Google Scholar]
- 73.Antrim R.L., Auterinen A.L. A new regenerable immobilized glucose isomerase. Starch Staerke. 1986;38:132–137. [Google Scholar]
- 74.Tufvesson P., Annerling A., Hatti-Kaul R., Adlercreutz D. Solvent-free enzymatic synthesis of fatty alkanolamides. Biotechnol Bioeng. 2007;97:447–453. doi: 10.1002/bit.21258. [DOI] [PubMed] [Google Scholar]
- 75.Mei Y., Miller L., Gao W., Gross R.A. Imaging the distribution and secondary structure of immobilized enzymes using infrared microspectroscopy. Biomacromolecules. 2003;4:70–74. doi: 10.1021/bm025611t. [DOI] [PubMed] [Google Scholar]
- 76.Kirk O., Christensen M.W. Lipases from Candida Antarctica: unique biocatalysts from a unique origin. Org Process Res Dev. 2002;6:446–451. [Google Scholar]
- 77.Welch P., Scopes R.K. Studies on cell-free metabolism: ethanol production by a yeast glycolytic system reconstituted from purified enzymes. J Biotechnol. 1985;2:257–273. [Google Scholar]
- 78.Liao H., Myung S., Zhang Y.H. One-step purification and immobilization of thermophilic polyphosphate glucokinase from Thermobifida fusca YX: glucose 6-phosphate generation without ATP. Appl Microbiol Biotechnol. 2012;93:1109–1117. doi: 10.1007/s00253-011-3458-1. [DOI] [PubMed] [Google Scholar]
- 79.Martin del Campo J.S., Rollin J., Myung S., Chun Y., Chandrayan S., Patino R., Adams M.W.W., Zhang Y.H.P. High-yield production of dihydrogen from xylose by using a synthetic enzyme cascade in a cell-free system. Angew Chem Int Ed. 2013;52:4587–4590. doi: 10.1002/anie.201300766. [DOI] [PubMed] [Google Scholar]
- 80.Wichmann R., Vasic-Racki D. Cofactor regeneration at the lab scale. Adv Biochem Eng Biot. 2005;92:225–260. doi: 10.1007/b98911. [DOI] [PubMed] [Google Scholar]
- 81.Bozic M., Pricelius S., Guebitz G.M., Kokol V. Enzymatic reduction of complex redox dyes using NADH-dependent reductase from Bacillus subtilis coupled with cofactor regeneration. Appl Microbiol Biotechnol. 2010;85:563–571. doi: 10.1007/s00253-009-2164-8. [DOI] [PubMed] [Google Scholar]
- 82.Xu Z.N., Jing K.J., Liu Y., Cen P.L. High-level expression of recombinant glucose dehydrogenase and its application in NADPH regeneration. J Ind Microbiol Biotechnol. 2007;34:83–90. doi: 10.1007/s10295-006-0168-2. [DOI] [PubMed] [Google Scholar]
- 83.Wong C.H., Whitesides G.M. Enzyme-catalyzed organic synthesis: NAD(P)H cofactor regeneration by using glucose 6-phosphate and the glucose 6-phosphate dehydrogenase from Leuconostoc mesenteroides. J Am Chem Soc. 1981;103:4890–4899. [Google Scholar]
- 84.Mertens R., Liese A. Biotechnological applications of hydrogenases. Curr Opin Biotechnol. 2004;15:343–348. doi: 10.1016/j.copbio.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 85.Wong C.H., Daniels L., Ormejohnson W.H., Whitesides G.M. Enzyme-catalyzed organic synthesis: NAD(P)H regeneration using dihydrogen and the hydrogenase from methanobacterium Thermoautotrophicum. J Am Chem Soc. 1981;103:6227–6228. [Google Scholar]
- 86.Johannes T.W., Woodyer R.D., Zhao H.M. Efficient regeneration of NADPH using an engineered phosphite dehydrogenase. Biotechnol Bioeng. 2007;96:18–26. doi: 10.1002/bit.21168. [DOI] [PubMed] [Google Scholar]
- 87.Palmore G.T.R., Bertschy H., Bergens S.H., Whitesides G.M. A methanol/dioxygen biofuel cell that uses NAD(+)-dependent dehydrogenases as catalysts: application of an electro-enzymatic method to regenerate nicotinamide adenine dinucleotide at low overpotentials. J Electroanal Chem. 1998;443:155–161. [Google Scholar]
- 88.Wang Y.R., Huang W.D., Sathitsuksanoh N., Zhu Z.G., Zhang Y.H.P. Biohydrogenation from biomass sugar mediated by in vitro synthetic enzymatic pathways. Chem Biol. 2011;18:372–380. doi: 10.1016/j.chembiol.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 89.Dinh T.H., Lee S.C., Hou C.Y., Won K. Diaphorase-viologen conjugates as bioelectrocatalysts for NADH regeneration. J Electrochem Soc. 2016;163:H440–H444. [Google Scholar]
- 90.Huang W.D., Zhang Y.H.P. Energy efficiency analysis: biomass-to-wheel efficiency related with biofuels production, fuel distribution, and powertrain systems. PLoS One. 2011;6 doi: 10.1371/journal.pone.0022113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kim E.J., Wu C.H., Adams M.W., Zhang Y.P. Exceptionally high rates of biological hydrogen production by biomimetic in vitro synthetic enzymatic pathways. Chem Eur J. 2016;22:16047–16051. doi: 10.1002/chem.201604197. [DOI] [PubMed] [Google Scholar]
- 92.Kim E.J., Kim J.E., Zhang Y.H.P.J. Ultra-rapid rates of water splitting for biohydrogen gas production through in vitro artificial enzymatic pathways. Energy Environ Sci. 2018 [Google Scholar]
- 93.Sokic-Lazic D., Minteer S.D. Citric acid cycle biomimic on a carbon electrode. Biosens Bioelectron. 2008;24:945–950. doi: 10.1016/j.bios.2008.07.043. [DOI] [PubMed] [Google Scholar]
- 94.Honda K., Hara N., Cheng M., Nakamura A., Mandai K., Okano K., Ohtake H. In vitro metabolic engineering for the salvage synthesis of NAD+ Metab Eng. 2016;35:114–120. doi: 10.1016/j.ymben.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 95.Rieckenberg F., Ardao I., Rujananon R., Zeng A.-P. Cell-free synthesis of 1,3-propanediol from glycerol with a high yield. Eng Life Sci. 2014;14:380–386. [Google Scholar]
- 96.Satoh Y., Tajima K., Tannai H., Munekata M. Enzyme-catalyzed poly(3-hydroxybutyrate) synthesis from acetate with CoA recycling and NADPH regeneration in vitro. J Biosci Bioeng. 2003;95:335–341. doi: 10.1016/s1389-1723(03)80064-6. [DOI] [PubMed] [Google Scholar]
- 97.Fujisawa T., Fujinaga S., Atomi H. An in vitro enzyme system for the production of myo-inositol from starch. Appl Environ Microbiol. 2017;83 doi: 10.1128/AEM.00550-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lu Y., Wang L., Teng F., Zhang J., Hu M., Tao Y. Production of myo-inositol from glucose by a novel trienzymatic cascade of polyphosphate glucokinase, inositol 1-phosphate synthase and inositol monophosphatase. Enzym Microb Technol. 2018;112:1–5. doi: 10.1016/j.enzmictec.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 99.Zhong C., You C., Wei P., Zhang Y.H.P. Thermal cycling cascade biocatalysis of myo-inositol synthesis from sucrose. ACS Catal. 2017;7:5992–5999. [Google Scholar]
- 100.Armaroli N., Balzani V. The hydrogen issue. ChemSusChem. 2011;4:21–36. doi: 10.1002/cssc.201000182. [DOI] [PubMed] [Google Scholar]
- 101.Chou C.J., Jenney F.E., Jr., Adams M.W., Kelly R.M. Hydrogenesis in hyperthermophilic microorganisms: implications for biofuels. Metab Eng. 2008;10:394–404. doi: 10.1016/j.ymben.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 102.Veit A., Akhtar M.K., Mizutani T., Jones P.R. Constructing and testing the thermodynamic limits of synthetic NAD(P)H: H2 pathways. Microb Biotechnol. 2008;1:382–394. doi: 10.1111/j.1751-7915.2008.00033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wu Y.R., Zhang M., Zhong M., Hu Z. Synergistic enzymatic saccharification and fermentation of agar for biohydrogen production. Bioresour Technol. 2017;241:369–373. doi: 10.1016/j.biortech.2017.05.117. [DOI] [PubMed] [Google Scholar]
- 104.Moustafa H.M.A., Kim E.J., Zhu Z.G., Wu C.H., Zaghloul T.I., Adams M.W.W., Zhang Y.H.P. Water splitting for high-yield hydrogen production energized by biomass xylooligosaccharides catalyzed by an enzyme cocktail. ChemCatChem. 2016;8:2898–2902. [Google Scholar]
- 105.Zhang Y.H.P., Evans B.R., Mielenz J.R., Hopkins R.C., Adams M.W.W. High-yield hydrogen production from starch and water by a synthetic enzymatic pathway. PLoS One. 2007;2 doi: 10.1371/journal.pone.0000456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ye X.H., Wang Y.R., Hopkins R.C., Adams M.W.W., Evans B.R., Mielenz J.R., Zhang Y.H.P. Spontaneous high-yield production of hydrogen from cellulosic materials and water catalyzed by enzyme cocktails. Chemsuschem. 2009;2:149–152. doi: 10.1002/cssc.200900017. [DOI] [PubMed] [Google Scholar]
- 107.Dusseaux S., Croux C., Soucaille P., Meynial-Salles I. Metabolic engineering of Clostridium acetobutylicum ATCC 824 for the high-yield production of a biofuel composed of an isopropanol/butanol/ethanol mixture. Metab Eng. 2013;18:1–8. doi: 10.1016/j.ymben.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 108.Ezeji T.C., Qureshi N., Blaschek H.P. Bioproduction of butanol from biomass: from genes to bioreactors. Curr Opin Biotechnol. 2007;18:220–227. doi: 10.1016/j.copbio.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 109.Harden A., Young W.J. The alcoholic ferment of yeast-juice. P R Soc Lond B Conta. 1906;77:405–420. [Google Scholar]
- 110.Sumner J.B. The isolation and crystallization of the enzyme urease. Preliminary paper. J Biol Chem. 1926;69:435–441. [Google Scholar]
- 111.Northrop J.H. Crystalline pepsin III. Preparation of active crystalline pepsin from inactive denatured pepsin. J Gen Physiol. 1931;14:713–724. doi: 10.1085/jgp.14.6.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Stanley W.M. Chemical studies on the virus of tobacco mosaic VI. The isolation from diseased Turkish tobacco plants of a crystalline potein possessing the properties of tobacco-mosaic virus. Phytopathology. 1936;26:305–320. [Google Scholar]
- 113.Krebs H.A., Eggleston L.V. Metabolism of acetoacetic acid in animal tissues. Nature. 1944;154:209–210. [Google Scholar]
- 114.Calvin M., Benson A.A. The path of carbon in photosynthesis. Science. 1948;107:476–480. doi: 10.1126/science.107.2784.476. [DOI] [PubMed] [Google Scholar]
- 115.Jacob F., Sanchez D.P.C., Monod J. The operon: a group of genes with expression coordinated by an operator. Cr Biol. 2005;328:518–520. [Google Scholar]
- 116.Eiserlin F., Levin J.G., Byrne R., Karlsson U., Nirenber Mw, Sjostran Fs. Polyribosomes and DNA-dependent amino acid incorporation in Escherichia coli extracts. J Mol Biol. 1964;10 doi: 10.1016/s0022-2836(64)80073-5. 536-&. [DOI] [PubMed] [Google Scholar]
- 117.Holley R.W., Apgar J., Everett G.A., Madison J.T., Marquisee M., Merrill S.H., Penswick J.R., Zamir A. Structure of a ribonucleic acid. Science. 1965;147 doi: 10.1126/science.147.3664.1462. 1462-+ [DOI] [PubMed] [Google Scholar]
- 118.Kelly T.J., Smith H.O. A restriction enzyme from Hemophilus influenzae .2. Base sequence of recognition site. J Mol Biol. 1970;51 doi: 10.1016/0022-2836(70)90150-6. 393-+ [DOI] [PubMed] [Google Scholar]
- 119.Smith H.O., Wilcox K.W. A restriction enzyme from Hemophilus influenzae .1. Purification and general properties. J Mol Biol. 1970;51 doi: 10.1016/0022-2836(70)90149-x. 379-+ [DOI] [PubMed] [Google Scholar]
- 120.Danna K., Nathans D. Studies of Sv40 DNA .1. Specific cleavage of simian virus 40 DNA by restriction endonuclease of Hemophilus influenzae. P Natl Acad Sci USA. 1971;68 doi: 10.1073/pnas.68.12.2913. 2913-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hutchison C.A., Phillips S., Edgell M.H., Gillam S., Jahnke P., Smith M. Mutagenesis at a specific position in a DNA-sequence. J Biol Chem. 1978;253:6551–6560. [PubMed] [Google Scholar]
- 122.Saiki R.K., Gelfand D.H., Stoffel S., Scharf S.J., Higuchi R., Horn G.T., Mullis K.B., Erlich H.A. Primer-directed enzymatic amplification of DNA with a thermostable DNA-polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 123.Buchner E. Alkoholische Gärung ohne Hefezellen (Vorläufige Mitteilung) Ber Dtsch Chem Ges. 1897;30:117–124. [Google Scholar]