Abstract
Microbial natural products are a tremendous source of new bioactive chemical entities for drug discovery. Next generation sequencing has revealed an unprecedented genomic potential for production of secondary metabolites by diverse micro-organisms found in the environment and in the microbiota. Genome mining has further led to the discovery of numerous uncharacterized ‘cryptic’ metabolic pathways in the classical producers of natural products such as Actinobacteria and fungi. These biosynthetic gene clusters may code for improved biologically active metabolites, but harnessing the full genetic potential has been hindered by the observation that many of the pathways are ‘silent’ under laboratory conditions. Here we provide an overview of the various biotechnological methodologies, which can be divided to pleiotropic, biosynthetic gene cluster specific, and targeted genome-wide approaches that have been developed for the awakening of microbial secondary metabolic pathways.
Keywords: Biosynthesis, Chemical diversity, Genome mining, Metabolic engineering, Synthetic biology
1. Microbes: promising and prolific source of new drug leads
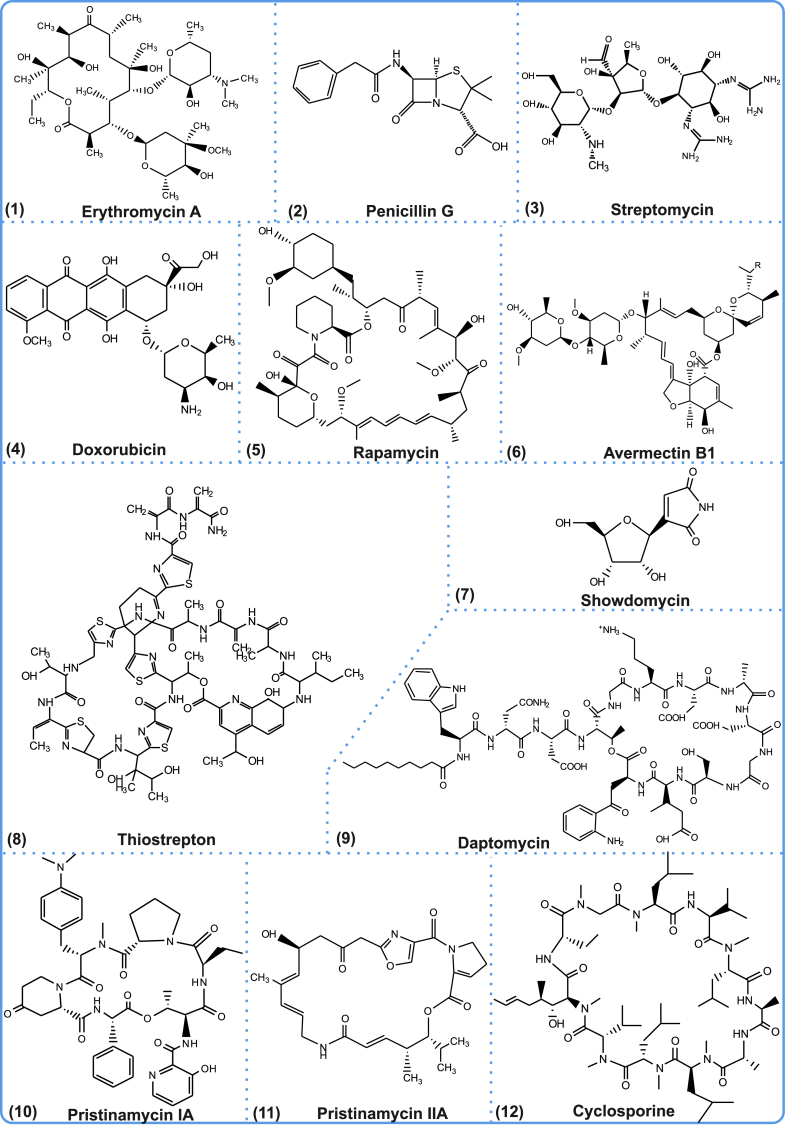
Natural products (also referred to as secondary metabolites or specialized metabolites; SMs) represent a group of low-molecular weight structurally diverse and complex bioactive compounds occupying an unusual chemical property space [1]. Microbes in particular are prolific antibiotic factories [2], and have proven to be a bountiful source of SMs that have been successfully developed as crucial drug leads [3,4]. To date, more than 5000 antibiotics have been identified from the genus Actinobacteria, while 500 natural products have been isolated from Myxobacteria [[5], [6], [7]]. Soil-dwelling Streptomyces bacteria are particularly proficient producers with 7600 SMs identified until 2005 [8], while computational predictions have estimated that these bacteria may have the capability to produce 150,000 chemically distinct antimicrobial agents [[5], [6], [7]]. In addition to classical antibiotics, microbial cultures have been a source for immune-suppressive agents [9], lantibiotics, anti-proliferative [10], cytotoxic [11], anti-hypertensive [12], antiviral compounds [13,14] and various enzyme inhibitors [15]. Circa 35% of drugs approved by the FDA/EMA are estimated to be either natural products or their derivatives [16], and >50% of clinical antibiotics are of Actinomycetes origin [8]. Widely used antibiotics include erythromycin A, penicillin G and streptomycin, while examples of microbial metabolites that have been successfully launched for other therapeutic areas comprise the anticancer agent doxorubicin, the immunosuppressants rapamycin and cyclosporine and the anthelmintic drug avermectin B1 (Fig. 1).
Fig. 1.
Chemical structures of selected microbial natural products.
The chemical entities described above (Fig. 1) were discovered several decades ago, most during the “Golden Era of Antibiotics” in the 1950s and 1960s. Starting from the 1980s and 1990s, traditional bioactivity-based screening of microbial culture extracts became severely affected by diminishing returns due to high probability of discovering previously known metabolites [17]. Structure elucidation of hit molecules is time consuming and ultimately the high rediscovery rate of known molecules led to decreased interest of pharmaceutical companies in natural products [2], which turned their attention to structure-based drug design and combinatorial chemistry instead [18,19]. However, despite very strong investments into this field in the last 15 years, these efforts have not provided a single novel synthetic antimicrobial agent that has proceeded beyond preliminary clinical trials [20]. One key problem has been that while many synthetic compounds with high efficacies towards their molecular targets have been successfully developed, it has become apparent that these compounds have difficulties in reaching their target site in vivo and in penetrating bacterial cell membranes [17].
However, recent technological advancements have created a renaissance of interest in natural products and new chemical entities with novel mode of action [21,22]. The foundation was laid in the 2000s by the diligent work of the Academic sector in elucidating the biosynthetic logic of the main classes of microbial natural products made by polyketide synthase (PKS) [23,24], non-ribosomal peptide synthetase (NRPS) [25] and ribosomally synthesized and post-translationally modified peptide (RiPP) pathways [26]. The classical pre-genomics view was that an individual microbial strain could produce only a limited number of SMs, but the first genome sequences of Streptomyces [27] and Aspergillus [28] published in the 2000s and the subsequent explosion of genome sequencing data in the 2010s has demonstrated that these micro-organisms have the genetic capability to produce a significantly larger number of compounds with great strain-to-strain variations. These unknown metabolic pathways are likely to encode numerous bioactive molecules, which could be used to solve the current problems with drug resistant pathogens and to obtain improved chemotherapy agents with reduced side effects. The key question remaining to answer in the field is how to access this hidden potential, since it would appear that the most of these cryptic pathways are silent or poorly expressed under laboratory conditions. In our current discourse, we attempt to provide crucial insights into methodologies that have been developed in an attempt to harness this tremendous chemical diversity.
2. Genomics-driven natural products discovery
The paradigm shift to genomics-based discovery of microbial SMs may be considered to be initiated by the sequencing of the genomes of S. coelicolor and S. avermitilis, which led to the discovery of 22 [27] and 25 [29] putative biosynthetic gene clusters (BGC), respectively, that could code for secondary metabolic pathways. The genome of the filamentous fungi Aspergillus nidulans harbored an even greater potential with 56 putative pathways, as observed by genome mining [30]. However, recent next generation sequencing efforts have vastly expanded this phenomenon in an unparalleled scale to diverse microbial genus that have not typically been associated with natural products such as Burkholderia, Clostridium and Pseudomonas [31,32]. In addition, probing microbial communities in various ecological niches such as the human microbiota [33] and environmental samples [34] have revealed exceptional metabolic diversity. Many lactic acid bacteria, including species of Enterococcus, Lactobacillus and Streptococcus associated with the gastrointestinal tract have been reported to produce bioactive modified peptides [33], in particular those synthesized via RiPP pathways [35,36]. The marine sponge Theonella swinhoei has been shown to host many uncultivated bacterial symbionts, which harbor pathways for production of exotic SMs [37]. It should also be noted that the potential of soil microbes for production of SMs remains elusive, since ca. 99% of environmental microbes are un-cultured under laboratory settings. In order to circumvent this barrier, environmental DNA isolated directly from soil samples may be used for cloning and expression of BGCs, as in the case of the malacidins pathway [38]. In another approach, the broad spectrum antibiotic teixobactin was discovered from an uncultured soil bacteria ‘Eleftheria terrae’ by growing the microbe directly in situ in the soil using a bacterial iChip [39].
The development of bioinformatics software for analysis of sequencing data for the presence of BGCs has been instrumental in genomics-driven natural products discovery. Programs such as antiSMASH [40,41], SMURF [42], BAGEL3 [43] and PRISM [44,45] are able to detect and decipher all of the most common biosynthetic types. In some instances, such as type I polyketides and NRPSs and RiPPs, these programs are able to even provide predictions for the chemical scaffolds of the metabolites [45], but for other compound classes such as type II PKSs and glycosylated natural product, the information acquired through computational studies is limited. Linking genomic data to chemistry has also been aided by community standards such as MIBiG (Minimum Information about a Biosynthetic Gene cluster) [46], which define standardized annotation and metadata procedures for characterization of pathways thus facilitating future research efforts.
3. Metabolic gene clusters in microbes
The genome mapping approaches are facilitated by the fact that all genes required for the biosynthesis of the metabolite, its regulation, resistance and transport are typically located in within the BGCs [[47], [48], [49]]. The size of metabolic gene clusters varies greatly depending on the complexity of the end product of the pathway. As an example, the gene cluster responsible for biosynthesis of the glycosylated anthracycline nogalamycin encodes 32 enzymes [50]. In addition, the type of biosynthetic machinery has a significant influence to the size of the BGC with large differences noted between the 30-kb thiostrepton and 128-kb daptomycin gene clusters [51,52], even though both are assembled from amino acid building blocks and are of similar complexity. The difference can be explained with the fact that thiostrepton is RiPP [51], whereas daptomycin is made through a NRPS pathway. Wide variation may also be found within the same classes of biosynthetic machineries, since the chemically simple NRP showdomycin is encoded within a 12-kb fragment of DNA, while the pristinamycin supercluster, which encodes two complex hybrid PK-NRP molecules, spans as large as 210-kb [53].
The onset of SMs production typically occurs during the early stationary phase, and involves complex metabolic changes within the organism [54]. Pleiotropic (or global) regulators are localized distantly from the biosynthetic cluster, and control the expression of secondary metabolism by responding to diverse signals [55]. These pleiotropic regulators have wider coverage, simultaneously controlling the expression of several BGCs. Events of nitrogen or phosphate starvation or distinct signaling compounds like N-acetylglucosamine (GlcNAc), stressors like heat, pH and damage to the cell wall provide proper stimuli for triggering the expression of these regulatory genes [56,57]. The production is also guided by regulatory genes present in the biosynthetic clusters, which are pathway-specific and may either be activators or repressors [58].
The genes responsible for providing resistance and for transporting SMs outside of the cells are also typically clustered within BGCs. In particular, if the end product of the pathway has biological activity against the producing host, then the resistance genes for that particular metabolite may be encoded in the genomic locus. Similarly, genes encoding efflux pumps (for instance, ATP-binding cassette (ABC) and Major Facilitator Superfamily proteins) have also been traced along the BGCs [59] and are responsible for the reduction of both toxicity [60] and feedback inhibition effects. For instance, in S. avermitilis, the ABC transporter AvtAB responsible for the exporting avermectin is found within the biosynthesis gene cluster [61].
4. Strategies for the activation of unknown metabolic pathways
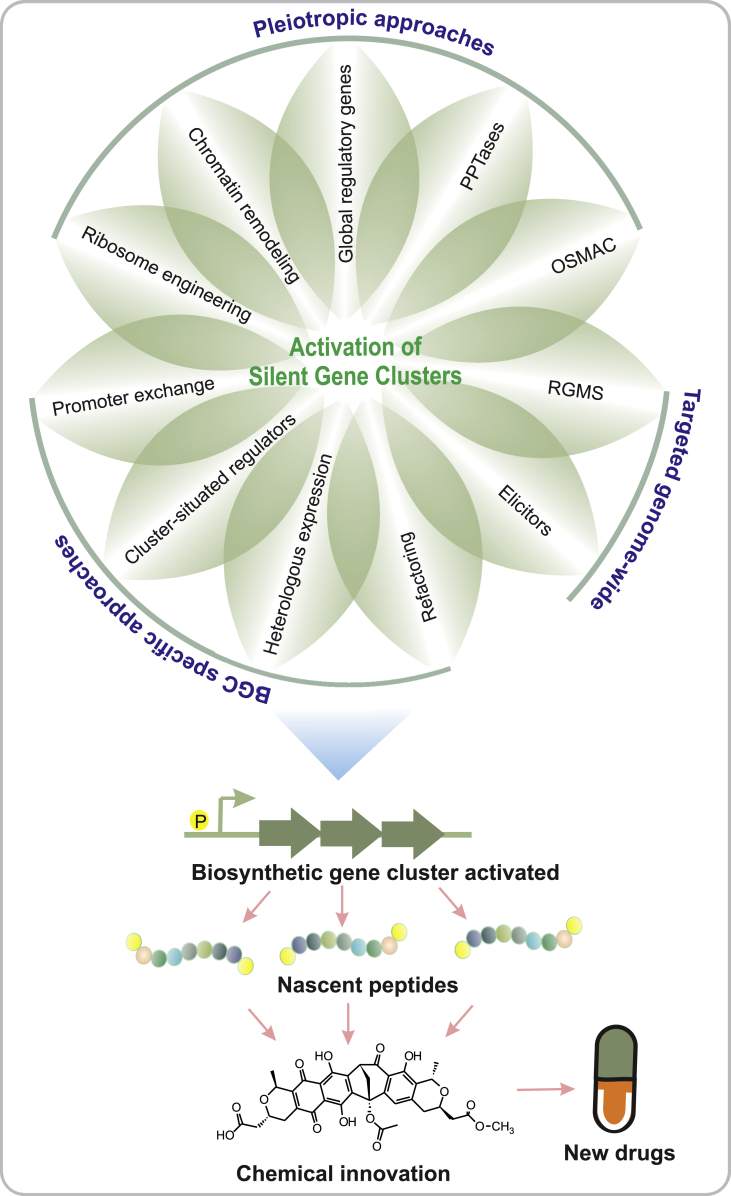
A growing body of evidence has indicated that the activation of gene clusters has the potential to greatly facilitate discovery of new natural products of high-therapeutic leads. Several methods have been developed for the activation of silent or poorly expressed cryptic gene clusters, which can be broadly classified to three major categories (Fig. 2). In one group, there are methodologies that aim to modify the whole metabolome of the target strain and generate pleiotropic effects to activate randomly any pathway residing in the strain. These methods are generally technically simple and are therefore suitable for scaling up to high-throughput systems. Unfortunately, these techniques generally forfeit the advantages provided by next-generation sequencing and genome mining, since they are not able to target the most interesting gene clusters for activation. They also suffer from the same metabolite rediscovery issues as traditional natural products discovery. In contrast, the methodologies lying under the second category are able to focus on desired pathways, but are technically challenging and suffer from low throughput. Finally, the techniques in the third group aim to gain the benefits of both of the approaches mentioned above, usually by combining pleiotropic activation methods to gene cluster specific reporter systems. The diversity of these methodologies offer many tools for the research community, but to date no single superior method for activation of biosynthetic pathways has been presented.
Fig. 2.
Methodologies for the activation of silent gene clusters to obtain new natural products. Petals within the circle represents different techniques that have been developed with three major categories, viz., pleiotropic, BGC-specific and targeted-genome wide approaches highlighted. Abbreviations: OSMAC, One strain many compounds; P, Promoters; PPtases, Phosphopantetheinyl transferases; RGMS, Reporter-Guided Mutant Selection.
4.1. Genome-wide pleiotropic methods
4.1.1. Ribosome engineering and its applicability
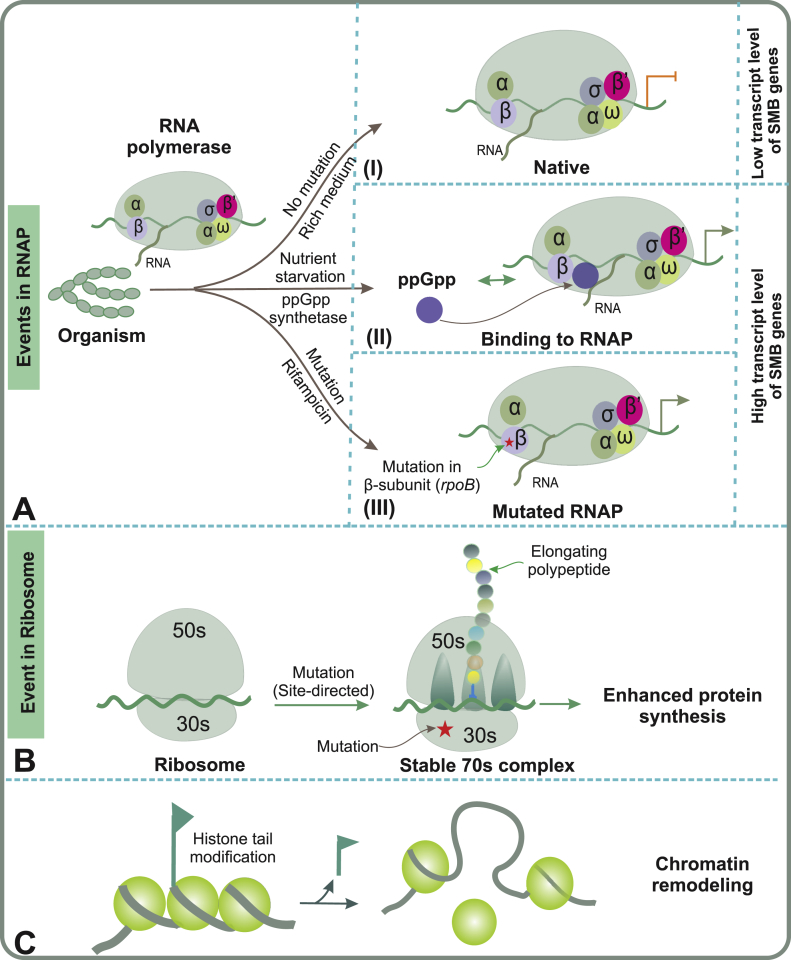
One of the most important adaptations in bacteria to environmental stresses, e.g. depletion of amino acids, is the ‘‘stringent response’’ governed by an intracellular transient accumulation of guanosine tetraphosphate (ppGpp) [62]. The bacterial alarmone ppGpp is synthesized by the ribosomal enzyme ppGpp synthetase (relA) and is initiated by the binding of an uncharged tRNA molecule to the aminoacyl binding site (A-site) on the ribosome [63]. Subsequent binding of ppGpp to the RNA polymerase (RNAP) modulates transcription by upregulating and downregulating various promoters [64,65] through mechanisms that can vary between species [66]. Investigations into the RNA polymerase/ppGpp complex in a thermophilic bacterium Thermus thermophilus revealed three modes of transcriptional regulation by ppGpp depending on the nucleic acid composition of the promoter, which influenced the conformation of the RNAP [64]. Therefore it is unsurprising that ppGpp has also been linked to secondary metabolism and initiation of antibiotic production [67]. In Streptomyces, relA/relC mutants with significantly decreased ppGpp contents display lowered production of SMs when transferred to a media with nutritional shift-down [[68], [69], [70]] (Fig. 3A and B). It has been revealed that mutations conferring resistance to rifampicin in the rpoB gene, which encodes the RNA polymerase β-subunit, alter the conformation of the RNAP to resemble ppGpp bound status. Therefore, the requirement for ppGpp for the expression of silent or poorly expressed gene clusters is bypassed in these mutants in various actinomycetes [[71], [72], [73]].
Fig. 3.
Pleiotropic approaches for activation of silent gene clusters: A) Events that occur in the RNAP when an organism is grown in alternative conditions: (i) Nutrient rich-medium, where the conditions suppress secondary metabolism and transcription levels from BGCs are low; (ii) Microbe subjected to nutritional down-shift (depletion of amino-acids) conditions, which leads to the production of ppGpp (Guainosine tetraphosphate), that ultimately binds to RNAP, giving rise to high transcription levels from the BGC; (iii) Addition of antibiotics like rifampicin induces a mutation in the β-subunit of the polymerase, which in turn increases the transcription level of the BGC. B) A mutation in the 30S subunit of ribosome stabilizes the 70S ribosomal complex and causes upregulation of protein synthesis in the stationary phase. C) Packaging of DNA around histones prevents gene transcription and is regulated through post-translational modifications of histone tails. In chromatin remodeling, enzymes responsible for histone modifications are manipulated to allow histone release, which enables expression of BGCs from the uncovered stretch of DNA.
In an analogous manner, the acquisition of mutations in ribosomal proteins conferring resistance to antibiotics targeting the ribosome has also been reported to promote the expression of silent genes. However, the molecular mechanism of this phenomena remains obscure. The introduction of streptomycin-resistance mutations in rel mutants of S. coelicolor and S. griseus reinstated actinorhodin production without the requirement of ppGpp [74,75]. On the contrary, elevated concentrations of ppGpp (30-fold) and actinorhodin (180-fold) has been detected in S. coelicolor, when the strain was exposed to eight antibiotics in a successive manner, where all of the antibiotics were targeting the translational machinery with the exception of rifampicin [76]. In addition, relA disruption resulted in the total loss of ppGpp accumulation as well as a significant decrease in actinorhodin production, indicating the key role of ppGpp in the activation of biosynthesis. Cluster-activated strains resulting from mutations in the ribosome have been reported in Streptomycetes and other bacteria including Bacillus spp., Mycobacterium spp. and Pseudomonas spp. [74,77,78]. Strikingly, two single rpoB mutants, an rpoB+rpsL double mutant, and a single gentamicin-resistant mutant induced the production of piperidamycins in S. mauvecolor, whereas the wild-type does not produce these metabolites at any detectable level in various media [79].
4.1.2. Chromatin remodeling
Chromatin remodeling is a vital mechanism for the modulation of transcription of eukaryotic genomes [80]. Chromatin is composed of repeating nucleosome units, which consist of histones (2 copies each of H2A, and H2B, H3 and H4 [80]) wrapped around chromosomal DNA and linked together through a linker histone (H1) (Fig. 3C) [81,82]. Histones cause dynamic variations to packaging and accessibility of DNA in response to physiological and developmental cues [83,84]. These are mediated via post-translational modifications such as deacetylation and methylation of histone tails, which ultimately regulate gene expression [85]. Gene clusters for secondary metabolism are clustered at subtelomeric regions and are co-regulated [86], where histone acetylation and methylation largely impact transcription [87].
For activation of SMs, the state of chromatin packaging has been influenced by artificial expression of histone modification genes and by utilizing small molecule inhibitors against histone deacetylase enzymes. In A. nidulans, inactivation of the histone deacetylase (HDAC) hdaA down-regulated secondary metabolic pathways [88], while loss-in-function of cclA (an ortholog of bre2 involved in histone H3 lysine 4 methylation) activated the expression of the cryptic secondary metabolic clusters generating monodictyphenone, emodin and its derivatives [89]. The use of the HDAC inhibitor suberoylanilide hydroxamic acid (SAHA) led to stimulation of production of new cladochromes and calphostin B in Cladosporium cladosporioides [90] and histone deacetylase (HDAC) inhibitors have also caused over-expression of SM genes in A. nidulans [88,91].
Unlike in fungi and other eukaryotes, prokaryotes are entirely deprived of histones. However, the bacterial genome, for instance, Streptomyces sp. comprises their own versions of HDAC [92] and the structure of the nucleoid may have a role in the global regulation of metabolism [93]. Access to chromosomal DNA may be restricted by nucleoid-associated proteins, various RNAs and differential supercoiling in the nucleoid, which may prevent aberrant transcription of many gene clusters [94]. In Streptomyces, a bifunctional nucleoid-associated protein (DdbA) has also been characterized that comprises of an N-terminal DNA-binding histone H1-like domain and a C-terminal DksA-like domain, which could modulate the RNA polymerase activity along with ppGpp to induce transcription [95].
4.1.3. OSMAC approach and environmental cues
One strain many compounds “OSMAC” is a relatively simple and versatile approach that allows activation of diverse-metabolic pathways [96]. The biosynthesis of SMs is stimulated through alterations in cultivation parameters [97,98], e.g. media composition, aeration rate, type of culturing vessel or a combination of these factors. In some cases, even subtle changes may result in drastic changes, as demonstrated in fermentations of Paraphaeosphaeria quadriseptata, where switching from the use of tap water to distilled water lead to the isolation of six new SMs [99]. These experiments aim to mimic more precisely the natural growth conditions found in the environment to trigger the onset of secondary metabolism, and has been successfully applied to both bacteria and fungi, as reviewed elsewhere [97,98].
The classical OSMAC approach may be extended to more drastic measures by modulating the culture conditions with stress or chemicals. Heat shock has been shown to induce jadomycin production [100] and increase the yields of validamycin [101], whereas limitation of nutrients such as alanine and/or an acidic pH shock led to methylenomycin production in Streptomyces coelicolor A3(2) [102]. In particular, numerous SMs have been shown to be regulated negatively by inorganic phosphate concentrations [57], while GlcNAc appears to function as an important sensory molecule for the onset of secondary metabolism [103]. Both biotic (microbial lysates or soil extracts) and abiotic (antibiotics, synthetic compounds) chemical elicitors have been proved to be effective strategies for the induction of silent gene clusters. Two small molecule elicitors have been particularly successful; ARC2, a synthetic compound discovered through large scale screening [104], and goadsporin [105,106], a natural product that promotes secondary metabolism in Streptomyces. Sub-inhibitory concentrations of other xenobiotic antibiotics, such as jadomycin B [107] and monensin [108] have been shown to induce endogenous production of SMs. Interestingly, it has been shown that the addition of rare earth elements, e.g. scandium and lanthanum, at low concentrations resulted in the activation of the cryptic secondary metabolite biosynthetic gene clusters [78,109].
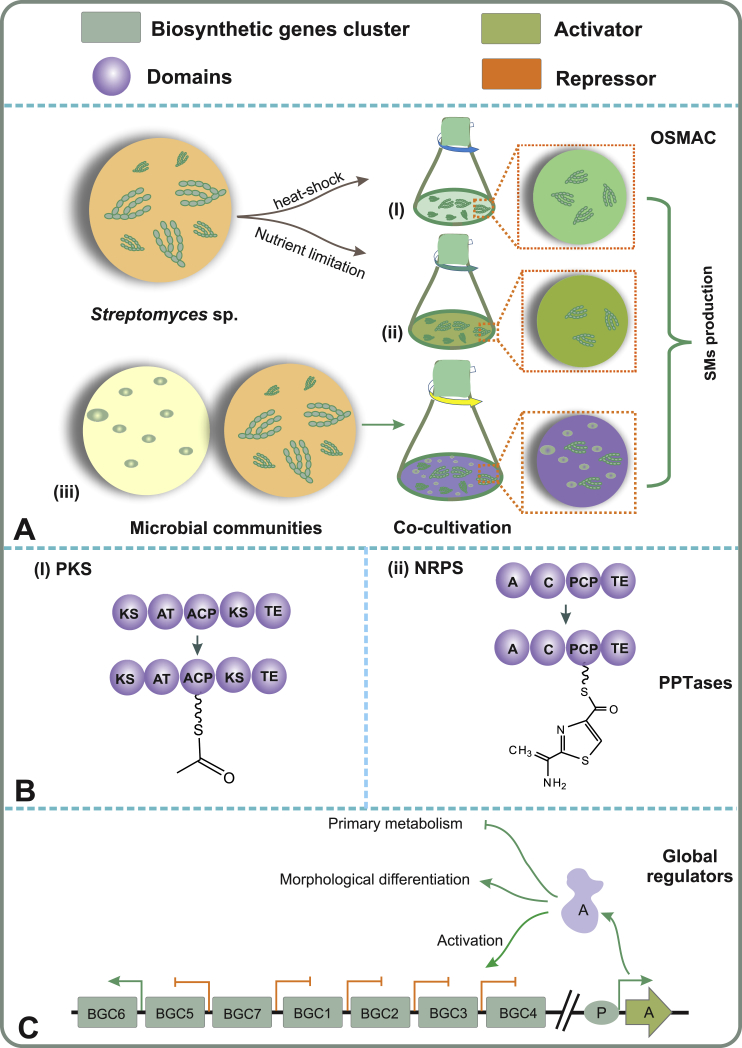
Co-cultivation, mimicking the ecological habitats of micro-organisms, has been emerged as an effective strategy to increase the chemical diversity of secondary metabolites (Fig. 4A). It is believed that the social networking in ecological environment is mediated by diffusible low-molecular-weight signaling molecules that are secreted from and received by cells [110,111]. For example, co-cultivation of the marine-derived fungal isolate A. fumigatus MR2012 with S. leeuwenhoekii strain C34 led to induction of metabolite production in both organisms, while co-cultivation of the fungus with bacterial strain C58 resulted in the detection of pentalenic acid from the bacteria [112]. Another marine-derived fungus Emericella sp. was induced to produce emericellamides A and B upon co-culture with Salinispora arenicola [113], whereas promomycin produced by one Streptomyces strain led to initiation of antibiotic production in another strain by cross-feeding [114]. Interestingly, dialysis experiments and electron microscopy have revealed that not only diffusible signals, but also intimate physical interactions contribute to the networking and induction of silent clusters during co-cultures of A. nidulans with S. hygroscopicus [115] or A. fumigatus with S. rapamycinicus [116].
Fig. 4.
Pleiotropic approaches for the activation of the silent gene clusters: Global regulators, PPTases and the OSMAC approach. A) In the OSMAC approach, cultivation conditions are manipulated to uncover the environmental signal required for activation of biosynthesis. As an example, co-cultivation of two or more microbes may induce microbial chemical communication, where one or even both participants produce novel compounds. Abbreviations: A, Activator; BGC, Biosynthetic gene cluster; P, Promoter. B) PPTases catalyze the post-translational modification of carrier proteins, which is essential for the biosynthesis of polyketides and nonribosomal peptides. Overexpression of PPTases may influence the production of SMs under conditions where the native PPTase may be downregulated. Abbreviations: A, Adenylation domain; ACP, Acyl-carrier protein; AT, Acyltransferase; C, Condensation domain; KS, Ketosynthase domain; NRPS, Non-ribosomal peptide synthetase; PCP, Peptidyl-carrier protein; PKS, Polyketide synthases; TE, Thioesterase domain. C) Pleiotropic regulators control many aspects in the life cycle of microbes. Manipulation of these regulatory networks may lead to the activation of BGCs.
4.1.4. Phosphopantheteine transferases
The biosynthesis of polyketides and nonribosomal peptides only occurs if the essential acyl - and peptidyl carrier proteins, respectively, are post-translationally modified into active holo-form [117]. This reaction is catalyzed by phosphopantetheinyl transferases (PPtases) through covalent attachment of a 4′-phosphopantetheine group to a highly conserved serine residue (Fig. 4B). Microbial genomes may contain up to three PPtases, which harbor specificity to various biosynthetic pathways [118], but these genes are typically not found within BGCs. Since PPtases regulate primary metabolism that leads to physiological changes and metabolic flux alterations, overexpression of PPtases may also influence secondary metabolism [119]. Indeed, expression of two PPtase genes sfp and svp resulted in activation of silent pathways with a success rate of 70% in a study using 33 Actinomycetes strains [120]. The sfp and svp gene products, from Bacillus subtilis and Streptomyces verticillus, respectively, exhibit broad substrate specificity to guarantee sufficient phosphopantetheinylation of diverse biosynthetic enzymes. Recently, PPtase-based activation also led to the identification of three nucleosides including puromycin A and two new congeners in S. alboniger NRRL B-1832 [121]. Interestingly, the biosynthetic pathways of nucleosides do not contain carrier proteins, suggesting that the result was achieved indirectly via modulation of cellular regulatory networks by the PPtase.
4.1.5. Manipulation of global regulatory systems
The engineering of global regulatory genes aims to influence the same regulatory networks as the OSMAC approach, but instead of trying to find the environmental cues, it does it at a more refined molecular level through over-expression or inactivation of the genes controlling the cellular response to these phenomena (Fig. 4C). Investigations into pleiotropic regulatory networks in bacteria and fungi have revealed that production of SMs is linked to morphological development and nutrient availability (carbon and nitrogen sources, phosphate starvation, amino acid and iron availability) [55]. Other examples include response to pH stress in fungi mediated by the PacC zinc finger transcription factor [122] and the master regulator DasR in Streptomyces, which responds to the presence of N-acetylglucosamine that may be considered as a signal for autolytic degradation of the vegetative mycelium [103]. An important distinction to cluster situated regulatory genes is that the global regulatory systems also control the transcription of many other genes not related to secondary metabolism. In fungi, pathway-specific regulatory genes have been discovered only in about 60% BGCs [123], which indicates that the production of many SMs is modulated strictly by global regulatory networks. Similarly, the moenomycin biosynthetic gene cluster does not harbor a pathway-specific regulator and is under the control of pleiotropic regulators in Streptomyces ghanaensis [124].
In Streptomyces, manipulation of regulatory elements that respond to butyrolactone hormones such as adpA [125] have been particularly successful with cryptic oviedomycin [126] and germicidin [127] gene clusters recently activated. Disruption of whiB-like regulatory genes, which are associated with control of morphological development [128], have also led to the discovery novel violapyrones [129] and antimycin-type depsipeptides [130].
4.2. Biosynthetic gene cluster specific approaches
4.2.1. Transcription factors and promoter exchange experiments
In eukaryotes, each gene is transcribed from its own promoter, whereas in prokaryotes (e.g., Streptomyces sp.) several genes are often clustered together to form operons (Fig. 5A). In all bacterial species, the promoter sequences residing within BGCs are typically associated with specific sigma factors (σ-factors) that are vital for binding of the RNA polymerase (RNAP) in the formation of the translation initiation complex [131]. The various σ-factors confer promoter selectivity and are thereby responsible for switching on specific regulons [132]. For instance, the filamentous prokaryotes, S. albus J1074, S. coelicolor and S. avermitilis comprise 35, 65 and 60 σ-factors, respectively [27,133]. On the other hand, promoter recognition in fungi relies on more diverse transcription factors. Eighty families of transcription factors were revealed through a whole-genome analysis of more than 200 fungal genomes [134]. For example, the filamentous fungi A. nidulans and N. crassa contain 81 and 58 transcription factors, respectively [135]. These aforementioned mechanisms enable alterations in transcription patterns in response to environmental cues.
Fig. 5.
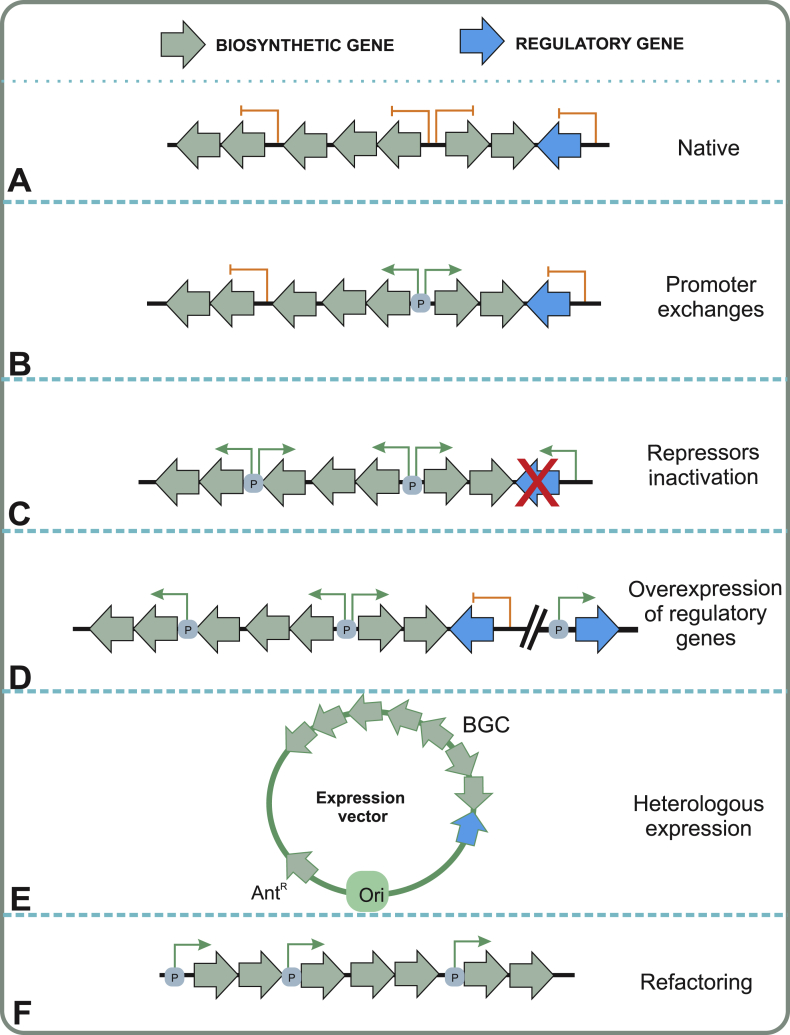
Gene cluster specific approaches for activating silent biosynthetic pathways. A) The target gene cluster is silent under laboratory conditions and no transcription or product formation can be observed. B) The gene cluster may be activated by exchanging the promoters from the silent gene cluster to constitutively active promoters. C) In instances where the gene cluster is controlled by a repressor, inactivation of the regulatory element may lead to expression of the biosynthetic genes. D) Overexpression of cluster situated activator regulatory genes may also lead to activation of biosynthesis. E) Cloning of the whole biosynthetic gene cluster and heterologous expression in a surrogate host can be used for artificial activation of the BGC. Abbreviations: AntR, Antibiotic selection marker; BGC, Biosynthetic gene cluster; Ori, Origin of replication; P, Promoters. F) Synthetic approaches include refactoring of the gene cluster and the use of constitutive or strong promoters to drive expression of the silent gene cluster.
The σ-factors influence the expression of larger groups of genes, therefore σ-factor-based engineering methods have been utilized to activate gene expressions associated with secondary metabolite biosynthesis. For example, overexpression of the σ-factor ‘Orf21’ in Streptomyces clavuligerus NRRL3585 resulted in a slight increase of clavulanic acid production through the induction of early biosynthetic genes (ceas2, cas2) and the activator gene ccaR [136]. Moreover, the overexpression of σ-factors led to the accumulation of higher level of FK506, a hybrid polyketide-nonribosomal peptide and carotenoid productions in Streptomyces sp. KCCM 11116P [137] and Corynebacterium glutamicum, respectively [138]. Successful expression of a polyketide biosynthetic pathway (oxytetracycline) was achieved for the first time in a surrogate host E. coli by overexpressing the host σ54-factor [139]. The deletion of σ-factor ‘Sig6’ caused over-production of avermectin by 2–2.7 fold in S. avermitilis, while overexpression of an extra copy lead to significant decrease in yields [140]. In addition, S. avermitilis hrdB mutants lacking σhrdB exhibited higher levels of avermectin production than the parental strain [141].
The promoter sequences residing in BGCs do not typically promote transcription in all growth conditions and are highly influenced by intracellular regulation networks. Manipulation of promoter sequences by exchanging native silent promoters with functionally active ones directly in the producing organism has proven to be a useful strategy to drive expression of the silent genes by circumventing the complexity of the regulatory genes (Fig. 5B). The methodology requires advanced genome editing tools that might not be available for the target organisms and is best suited for small BGCs that are composed of very few operons. The activation of a silent polycyclic tetramate macrolactam (PTM) gene cluster in Streptomyces albus has been achieved by the independent introduction of ermEp* upstream of ftdA and ftdB encoding putative desaturase and hybrid polyketide synthase-nonribosomal peptide synthetase, respectively, and led to the identification of 6-epi-alteramides A and B. Furthermore, the biosynthesis of a blue pigment ‘indigoidine’, which is not typically produced in standard laboratory conditions, has been induced in S. albus by expressing the NRPS gene bpsA under ermEp* promoter [142]. The introduction of strong engineered promoter kasOp* also resulted in successful activation of silent lycopene biosynthetic cluster in S. avermitilis [143]. Recently, replacement of the native promoters of the silent actinorhodin, undecylprodigiosin and PTM clusters with kasOp* produced targeted compounds in S. albus, S. lividans and S. roseosporus, respectively [144].
In Aspergillus nidulans, replacing the native promoter of the non-ribosomalpeptide synthetase, acvA, with a promoter from the ethanol dehydrogenase, alcAp, from the same host led to a 30-fold increase in penicillin production [145]. In addition, sequential exchange of promoters for six genes in the silent fellutamide B gene cluster (inp) with alcAp enabled the activation of the cluster and led to the production of the target compound in A. nidulans [146].
4.2.2. Manipulation of pathway-specific regulatory genes
Pathway-specific transcriptional regulatory genes are typically found within the BGCs and they may either repress or activate the biosynthesis of the corresponding SM. These genes represent the lowest organizational level in the cellular regulatory networks and are typically tightly regulated by environmental sensors and pleiotropic regulators. However, in reality the regulatory circuits are likely to be even more complex with cross-talk between distinct pathways [147] and even between global and BGC specific regulatory genes reported [148]. The co-localization of pathway-specific regulatory genes offers a possibility for targeted activation of the desired BGC either by inactivation of the repressor or over-expression of the activator genes (Fig. 5C and D). The caveat is that for many families of transcriptional regulators it is not possible to identify whether the gene is a repressor or an activator based on sequence information alone. The diversity of microbial regulatory proteins is expansive [149], and only two systems are described here to demonstrate the functional differences.
The TetR family repressors consist of a helix-turn-helix DNA binding domain and a receptor domain that responds to the presence of a small molecule ligand [150]. In the absence of the specific signaling molecule, TetR dimers bind to DNA and prevent transcription of downstream genes. Binding of the ligand induces a conformational shift that repositions the helix-turn-helix motifs of the dimer far apart in such a way that the proteins lose their ability to bind DNA [151]. Interestingly, some TetR proteins have been proven to respond to γ-butyrolactone autoinducers during the onset of secondary metabolism [152], while others bind directly to both endogenous and exogenous SMs [107,153]. Inactivation of TetR family repressors have led to the activation of jadomycin [153,154], kinamycin [155], auricin [156] and ceoelimycin biosynthesis [157,158].
Another predominant class of regulators is homologous to LuxR from Vibrio fischeri, which is a transcription factor involved in quorum sensing [159]. The protein consists of an α/β/α sandwich domain that binds the activator ligand and a DNA binding response domain. In contrast to the TerR family, most characterized LuxR proteins are transcriptional activators, where ligand binding induces homodimerization, which enables binding of the protein to DNA in so-called tra box sequences and initiate transcription [160]. An important subfamily of LuxR regulators in Actinobacteria are LAL–proteins (Large ATP-binding regulators of the LuxR family) [161], originally discovered in the regulon for maltose utilization, that contains an additional ATP-binding domain at the N-terminus responsible for the interaction with the inducers, maltotriose and ATP [162]. In a recent example, LuxR-type activators were over-expressed and TetR-type repressors were inactivated to initiate production of totopotensamides [163].
4.2.3. Heterologous expression in surrogate hosts
Many microbes that have the capability to produce natural products, such as strains of Cyanobacteria and filamentous fungi, are genetically intractable [164]. Heterologous expression provides an opportunity to access the genomic potential these organisms harbor in a genetically compliant host. The capture of target BGCs was classically done through screening of cosmid gene libraries, which was both time-consuming and could not accurately clone the desired DNA fragment, but advances in direct cloning strategies and DNA assembly tools have provided attractive alternatives. These methodologies are described in more detail in Section 4.2.4., since changing the expression host may require manipulation of control elements such as promoter sequences to ensure sufficient transcription levels (Fig. 5E).
The choice of expression host is far from trivial and many factors need to be considered. The genetically malleable Escherichia coli and Saccharomyces cerevisiae provide attractive hosts for production of bacterial and fungal SMs, respectively, but since neither is naturally equipped for production of complex chemicals, issues with metabolic flux, codon usage, self-resistance and activation of biosynthetic acyl/peptidyl carrier proteins need to be resolved. E. coli seems to be well-suited for expression of pathways originating from Cyanobacteria that are challenging to modify genetically, with production of microcystin [165] and lyngbyatoxin reported [166]. Several fungal terpene synthases have also been expressed in E. coli, but multi-gene clusters are much more challenging due to the requirement to remove introns and modify promoter regions. E. coli has had less success with BGCs originating from high-GC Actinobacteria, even though the entire erythromycin pathways has been expressed in this host [[167], [168], [169]]. The titres of SMs produced in E. coli have been generally low. Other promising heterologous hosts for production of prokaryotic SMs include Myxococcus xanthus, Pseudomonas putida [170] and Bacillus subtilis [171].
In many instances, it would appear that hosts that are evolutionarily closely linked to the organism harbouring the target pathway may be best suited for heterologous expression. Several well-characterized Streptomyces sp. are easily amenable and are efficient surrogate hosts for the heterologous characterization of different genes from Actinobacteria. In particular, S. albus J1074 [172], S. lividans TK24 [173], S. venezuelae ATCC10712 and S. coelicolor A3(2) have been widely used for the expression of cryptic pathways. S. coelicolor is genetically the most studied actinomycete and a large array of tools are available for manipulation of the organism [174], whereas the closely related S. lividans TK24 has particularly excelled at expression of proteins [175]. S. venezuelae, is well-suited for the heterologous expression of the PKs and NRPS due to the presence of large number of innate PPTase genes [176]. Finally, cloning genes in S. albus is facilitated by a defective restriction modification system and the naturally minimized genome may help increasing product titres [177]. In filamentous fungi, Aspergillus species have been widely used as heterologous hosts. An early example includes the transfer of the entire penicillin gene cluster to A. nidulans, while more recently geodin has been heterologously produced in this host [[178], [179], [180]]. A. oryzae has gained interest for heterologous expression, as it has traditionally been used in the food industry and has been given the status of GRAS (generally regarded as safe) organism. Selected recent examples of SMs produced in this host include paxilline [181], alfatrem [182] and pleuromutilin [183]. The heterologous expression of A. terreus asperfuranone cluster in the model ascomycete, A. nidulans, resulted in awakening of the asperfuranone biosynthetic pathway when several non-reducing polyketide synthase genes were placed under the control of alcAp [184].
An ideal host for heterologous expression would provide sufficient metabolic building blocks for the synthesis of SMs, but would not have native pathways that could interfere with the biosynthesis or generate background. To this end, several strains have been genome minimized to function better as expression chasses. Four gene clusters have been deleted from the genomes of S. coelicolor M1152 and M1154, which have been further improved for metabolite production by ribosome engineering [185]. Extensive manipulation, including deletion of a 1.5 Mb subtelomeric region and 78% of putative transposase genes of wild type S. avermitilis led to high success rates and product yields in the expression of exogenous biosynthetic pathways [186]. These ongoing efforts are likely to result in a handful of “superhosts” for the heterologous expression of diverse metabolic pathways.
4.2.4. Refactoring of gene clusters
The emergence of new metabolic engineering methods and the continually decreasing costs of DNA synthesis have enabled complete refactoring of entire BGCs, which typically includes three distinct stages. The first step involves the architectural design of the synthetic gene cluster from biological part libraries to obtain desired promoter, gene and terminator sequences, which may be ordered synthetically or amplified using PCR. In the next step, the pathway is assembled together with advanced cloning methods such as recombineering or TAR-cloning typically in E. coli or Sacc. cerevisiae. TAR cloning takes advantage of the high efficiency natural homologous recombination in yeast that allows for both direct capture of BGCs and simultaneous cloning of multiple DNA fragments [187]. In contrast, recombineering utilizes recombination systems from phages to enable homologous recombination to occur in E. coli [188]. In addition, the combination of long-amplicon PCR and DNA recombination (DiPaC) has recently been utilized for the direct cloning of the anabaenopeptins and erythromycin pathways [189]. The ultimate step involves the transfer of the artificial BGC to the expression host selected for the production of SMs (Fig. 5F).
Reports on fully synthetic refactoring of BGCs have still been scarce due to the significant costs associated with the method. Even though this approach would allow for unprecedented freedom in pathway design, the complexity of metabolic pathways required for the production of even simple SMs makes the task of designing functional BGC in silico far from trivial. For these reasons, the methodology appears to be best suited for improvements in the production titres of known value-added metabolites and not for the activation of unknown pathways. Examples of fully synthetic refactored gene clusters include rebeccamycin and pyrrolnitrin [190].
A more widely used approach in refactoring has been to modify regulatory elements or promoter regions of natural gene clusters prior to heterologous expression. Expression of the unmodified taromycin gene cluster directly in S. coelicolor M1146 did not lead to the production of the lipopeptide, but activation of the pathway was achieved by deletion of pathway –specific repressors using in vivo yeast recombination-mediated PCR targeting [191]. Conversely, refactoring of a strong constitutive promoter in front of a cluster situated activator enabled the heterologous production of pacidamycin D [190]. In a more arduous approach, individual genes or short operons from BGCs may also be amplified by PCR and placed in front of active promoters. After reassembly of full length pathways, polycyclic tetramate macrolactams [192,193] and spectinabilin [194] have been produced in heterologous hosts. The PCR –amplification step was circumvented in the refactoring of the novobiocin gene cluster, where a cosmid harbouring the BGC was modified using recombineering to facilitate the generation of plug-and-play expression modules [195]. Through a plug-and-play promoter insertion strategy, introduction of three novel constitutive promoters of S. coelicolor, upstream of jadJ in the cryptic jadomycin B gene cluster in S. venezuelae, led to cluster activation. Importantly, expression of jadJ from these three promoters led to higher transcription levels than when the widely used ermEp* was utilized [196]. The benefit of strong constitutive promoters is that they may lead to increased transcription levels and product yields, as has been shown in the engineering of polycyclic tetramate macrolactams [193].
4.3. Targeted genome-wide methodologies
To date, only few approaches have been developed that are able to focus on the desired cryptic BGC detected by genome mining, which are able to utilize pleiotropic methods to widely probe conditions that lead to activation of the pathway. These targeted genome-wide methodologies (Fig. 6) typically take advantage of a biosensor to report increased transcription or other activity from the desired BGC, but differ in the ways of generating alterations to the global transcriptome.
Fig. 6.
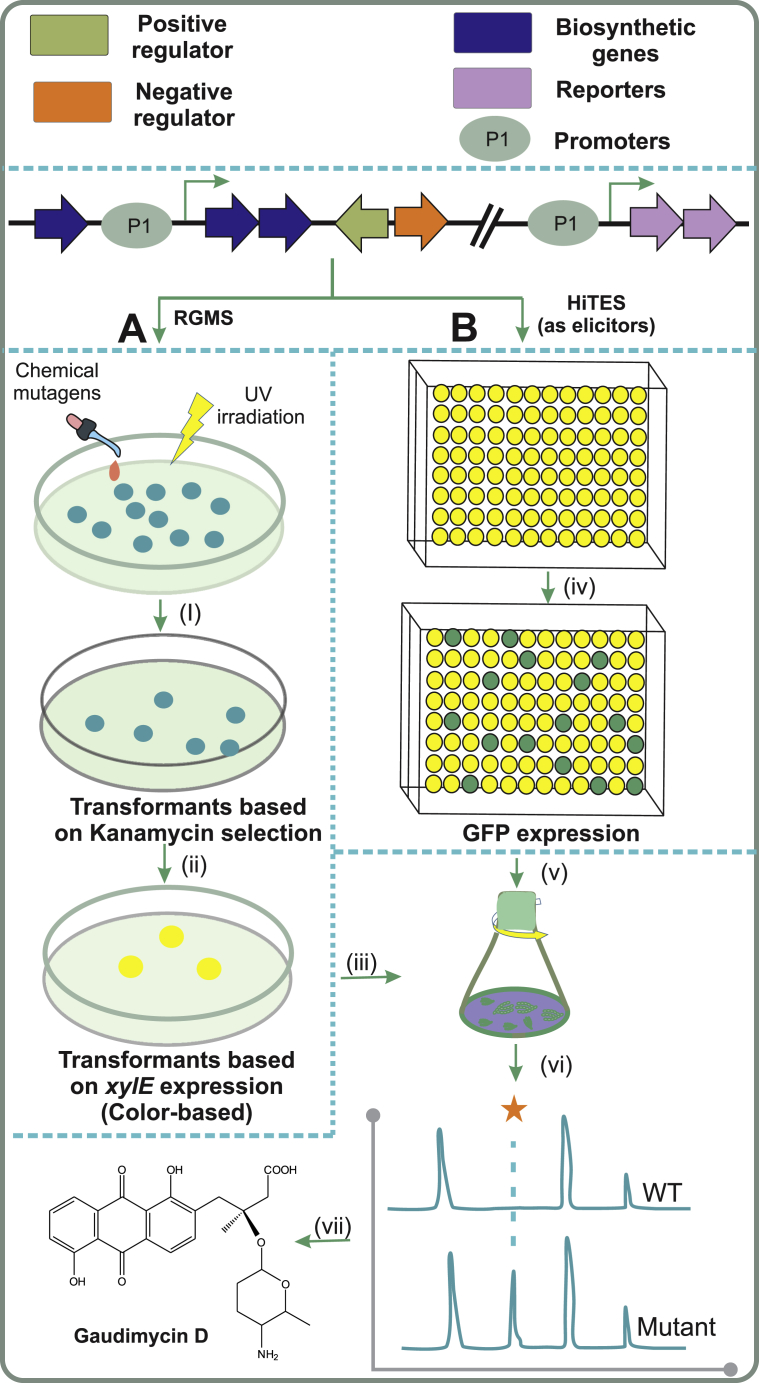
Targeted genome-wide approaches for the activation of the silent gene clusters. A) In Reporter Guided Mutant selection, promoters identified from the target pathway are utilized to report increased transcription from the BGC. The selected BGC is activated (i) either by random chemical mutagens or UV irradiation to generate mutants where the pathway has been upregulated. (ii) The double-reporter system (xylE-neo) allows for selection of mutants in the presence of kanamycin and screening of the mutant library for highest expression levels of xylE. Abbreviation: RGMS, Reporter-guided mutant selection. B) In high-throughput elicitor screening, activation of the gene clusters is achieved by finding a small-molecular weight compound that triggers expression. (iv) Elicitors from a chemical library are added to individual wells for performing the elicitation response, which is (v) followed by monitoring transcription levels from a promoter probe construct expressing GFP. (vi) Metabolic profiling can be used to detect novel SMs from cultivations of wild type and biosynthesis activated strains, followed by (vii) structural elucidation of the target metabolite. Abbreviations: GFP, Green fluorescence protein; HiTES, High-throughput elicitor screening; WT, wild-type strain.
4.3.1. Reporter guided systems
A pioneering Reporter-Guided Mutant Selection (RGMS) method developed in the group of Prof. Keqian Yang provides a widely applicable approach for targeted activation of silent gene clusters in the native organism [197]. RGMS combines two effective yet simple techniques: cloning of key promoter sequences from the gene cluster of interest in front of a double xylE-neo reporter system and the use of genome-wide mutagenesis to generate a library of mutants that contain tremendous genomic diversity. The core rationale relies on randomly modulating the complex regulatory cascades that are controlling secondary metabolism and converting expression of the target pathway into a signal that enables selection of a mutant with the desired phenotype. The reporter cassette allows initial selection by kanamycin resistance (neo), followed by colour-based detection of catechol oxidase (xylE) activity, which allows for distinction between spontaneous kanamycin-resistant and genuine target-activated mutants (Fig. 6A).
RGMS was initially developed for industrial strain improvement purposes by taking advantage of the correlation between increased transcription levels from the target BGC and product yields. The method has been used to improve production of lovastatin [198], clavulanic acid [199] and natamycin [200] in Aspergillus terreus, S. clavuligerus and S. gilvosporeus, respectively. In addition, the methodology has also been successfully carried out in combination with ribosome engineering, through exposure to streptomycin, to achieve over-production of daptomycin in S. roseosporus NRRL11379 [201]. Recently, BGC situated TetR –family transcriptional repressors have been developed as alternative biosensors to aid screening of mutant libraries. These reporter proteins bind end products of the pathways and are therefore able to report increased production of the target metabolites directly, which reduces the number of false positives encountered with reporters that only monitor transcription levels. The method was utilized for increasing the production of panamycins in S. alboniger [202].
Many of the cryptic gene clusters denoted “silent” may, in effect, be transcribed at a low basal level, but in such low quantities that the corresponding metabolite remains undetected. Therefore RGMS is also suitable for the activation of BGCs and has been successfully utilized on the jadomycin and gaudimycin pathways in S. venezuelae ISP5230 and Streptomyces sp. PGA64, respectively [197]. The jadomycin BGC is silent under typical culture conditions unless special environmental cues such as heat-shock are applied [203]. To establish proof of concept, jadomycin biosynthesis could be re-activated by using RGMS strategy, whereas work on the cryptic gaudimycin pathway led to the isolation of two novel anthraquinone aminoglycosides [197]. The indigoidine synthetase gene bpsA, which is responsible for synthesis of a blue pigment ‘indigoidine’, has also been utilized as a reporter in the activation of the cryptic coelimycin gene cluster in S. lividans, where the activation was achieved through over-expression of a pathway-specific regulatory gene [204].
4.3.2. High-throughput elicitor screening
Many SMs are produced in response to various chemical signals, which may either be communication molecules or chemical weapons made by other organisms cohabiting the same environment. If the environmental signal can be identified in the laboratory using small molecule libraries, expression of silent BGCs can be achieved (Fig. 6B). The chemogenetic HiTES (high-throughput elicitor screening) method does this by coupling the screening to a promoter-probe system, where either lacZ [205] or eGFP [206] is utilized to report increased transcription from the target BGC. The approach has been successfully applied to two gene clusters in the Gram-negative pathogenic model bacterium Burkholderia thailandensis E264, where a silent virulence factor, namely malleilactone (mal) and a poorly expressed burkholdac (bhc) cluster were targeted [205]. The strategy has also been implemented in S. albus J1074, where the nonribosomal peptide surugamide (sur) cluster was awakened [207]. Interestingly, the cytotoxins etoposide and ivermectin were found to be effective inducers resulting the occurrence of 14 novel compounds in the culture media.
5. Concluding thoughts and future perspectives
The abundance of silent and cryptic pathways in microbial genomes provides an exciting opportunity for the discovery of new chemical entities with high therapeutic potential. The diverse strategies developed for awakening these pathways offer an excellent starting point, but to date no single superior methodology has been developed. This is not unexpected, since the BGCs reside in vastly diverse micro-organisms and in many cases are strictly regulated to be activated only under highly specific conditions. The availability of the numerous methods described above, which all have their strengths and weaknesses, can therefore be considered as a significant advantage and it will remain the responsibility of the researcher to choose the most appropriate one for any given project.
Acknowledgements
The authors would like to dedicate this article to the memory of Prof. Keqian Yang, who made significant contributions to our understanding of secondary metabolic pathways in Streptomyces. The authors would like to thank the Jane and Aatos Erkko Foundation for their financial support.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
References
- 1.Singh S.B. Confronting the challenges of discovery of novel antibacterial agents. Bioorg Med Chem Lett. 2014;24:3683–3689. doi: 10.1016/j.bmcl.2014.06.053. [DOI] [PubMed] [Google Scholar]
- 2.Awan A.R., Shaw W.M., Ellis T. Biosynthesis of therapeutic natural products using synthetic biology. Adv Drug Deliv Rev. 2016;105:96–106. doi: 10.1016/j.addr.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 3.Rutledge P.J., Challis G.L. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat Rev Microbiol. 2015;13:509–523. doi: 10.1038/nrmicro3496. [DOI] [PubMed] [Google Scholar]
- 4.Newman D.J., Cragg G.M. Natural products as sources of new drugs over the last 25 years. J Nat Prod. 2007;70:461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 5.Challinor V.L., Bode H.B. Bioactive natural products from novel microbial sources. Ann N Y Acad Sci. 2015;1354:82–97. doi: 10.1111/nyas.12954. [DOI] [PubMed] [Google Scholar]
- 6.Schäberle T.F., Lohr F., Schmitz A., König G.M. Antibiotics from myxobacteria. Nat Prod Rep. 2014;31:953. doi: 10.1039/c4np00011k. [DOI] [PubMed] [Google Scholar]
- 7.Watve M.G., Tickoo R., Jog M.M., Bhole B.D. How many antibiotics are produced by the genus Streptomyces? Arch Microbiol. 2001;176:386–390. doi: 10.1007/s002030100345. [DOI] [PubMed] [Google Scholar]
- 8.Bérdy J. Bioactive microbial metabolites. J Antibiot (Tokyo) 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 9.Hatanaka H., Kino T., Miyata S., Inamura N., Kuroda A., Goto T. FR-900520 and FR-900523, novel immunosuppressants isolated from a Streptomyces. II. Fermentaion, isolation and physio-chemical and biological characteristics. J Antibiot (Tokyo) 1988;41:1592–1601. doi: 10.7164/antibiotics.41.1592. [DOI] [PubMed] [Google Scholar]
- 10.Hu J., Wunderlich D., Thiericke R., Dahse H.-M., Grabley S., Feng X.-Z. Jenamidines A to C: unusual alkaloids from Streptomyces sp. with specific antiproliferative properties obtained by chemical screening. J Antibiot (Tokyo) 2003;56:747–754. doi: 10.7164/antibiotics.56.747. [DOI] [PubMed] [Google Scholar]
- 11.Tapiolas D.M., Roman M., Fenical W., Stout T.J., Clardy J. Octalactins A and B: cytotoxic eight-membered-ring lactones from a marine bacterium, Streptomyces sp. J Am Chem Soc. 1991;113:4682–4683. [Google Scholar]
- 12.Patel R.N. Biocatalytic synthesis of chiral intermediates for antiviral and antihypertensive drugs. J Am Oil Chem Soc. 1999;76:1275–1281. [Google Scholar]
- 13.Maullu C., Lampis G., Deidda D., Petruzzelli S., Pompei R. A rapid method for screening large numbers of environmental microorganisms for antiviral activity. Appl Environ Microbiol. 1998;64:1161–1162. doi: 10.1128/aem.64.3.1161-1162.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tong J., Trapido-Rosenthal H., Wang J., Wang Y., Li Q.X., Lu Y. Antiviral activities and putative identification of compounds in microbial extracts from the Hawaiian coastal waters. Mar Drugs. 2012;10:521–538. doi: 10.3390/md10030521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imada C. Enzyme inhibitors and other bioactive compounds from marine actinomycetes. Antonie Leeuwenhoek. 2005;87:59–63. doi: 10.1007/s10482-004-6544-x. [DOI] [PubMed] [Google Scholar]
- 16.Newman D.J. Developing natural product drugs: supply problems and how they have been overcome. Pharmacol Ther. 2016;162:1–9. doi: 10.1016/j.pharmthera.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 17.Lewis K. Platforms for antibiotic discovery. Nat Rev Drug Discov. 2013;12:371–387. doi: 10.1038/nrd3975. [DOI] [PubMed] [Google Scholar]
- 18.Anderson A.C. The process of structure-based drug design. Chem Biol. 2003;10:787–797. doi: 10.1016/j.chembiol.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 19.Antel J. Integration of combinatorial chemistry and structure-based drug design. Curr Opin Drug Discov Dev. 1999;2:224–233. [PubMed] [Google Scholar]
- 20.Piddock L.J.V. The crisis of no new antibiotics-what is the way forward? Lancet Infect Dis. 2012;12:249–253. doi: 10.1016/S1473-3099(11)70316-4. [DOI] [PubMed] [Google Scholar]
- 21.Wright G.D. Something old, something new: revisiting natural products in antibiotic drug discovery. Can J Microbiol. 2014;60:147–154. doi: 10.1139/cjm-2014-0063. [DOI] [PubMed] [Google Scholar]
- 22.Georgiev M.I. Natural products utilization. Phytochem Rev. 2014;13:339–341. [Google Scholar]
- 23.Zhang Z., Pan H.-X., Tang G.-L. New insights into bacterial type II polyketide biosynthesis. F1000Research. 2017;6:172. doi: 10.12688/f1000research.10466.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robbins T., Liu Y.C., Cane D.E., Khosla C. Structure and mechanism of assembly line polyketide synthases. Curr Opin Struct Biol. 2016;41:10–18. doi: 10.1016/j.sbi.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Payne J.A.E., Schoppet M., Hansen M.H., Cryle M.J. Diversity of nature's assembly lines-recent discoveries in non-ribosomal peptide synthesis. Mol Biosyst. 2017;13:9–22. doi: 10.1039/c6mb00675b. [DOI] [PubMed] [Google Scholar]
- 26.Hudson G.A., Mitchell D.A. RiPP antibiotics: biosynthesis and engineering potential. Curr Opin Microbiol. 2018;45:61–69. doi: 10.1016/j.mib.2019.06.001. [DOI] [PubMed] [Google Scholar]
- 27.Bentley S.D., Chater K.F., Cerdeño-Tárraga A.-M., Challis G.L., Thomson N.R., James K.D. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 28.Machida M., Asai K., Sano M., Tanaka T., Kumagai T., Terai G. Genome sequencing and analysis of Aspergillus oryzae. Nature. 2005;438:1157–1161. doi: 10.1038/nature04300. [DOI] [PubMed] [Google Scholar]
- 29.Omura S., Ikeda H., Ishikawa J., Hanamoto A., Takahashi C., Shinose M. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc Natl Acad Sci. 2001;98:12215–12220. doi: 10.1073/pnas.211433198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yaegashi J., Oakley B.R., Wang C.C.C. Recent advances in genome mining of secondary metabolite biosynthetic gene clusters and the development of heterologous expression systems in Aspergillus nidulans. J Ind Microbiol Biotechnol. 2014;41:433–442. doi: 10.1007/s10295-013-1386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pidot S.J., Coyne S., Kloss F., Hertweck C. Antibiotics from neglected bacterial sources. Int J Med Microbiol. 2014;304:14–22. doi: 10.1016/j.ijmm.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 32.Gross H., Loper J.E. Genomics of secondary metabolite production by Pseudomonas spp. Nat Prod Rep. 2009;26:1408–1446. doi: 10.1039/b817075b. [DOI] [PubMed] [Google Scholar]
- 33.Mousa W.K., Athar B., Merwin N.J., Magarvey N.A. Antibiotics and specialized metabolites from the human microbiota. Nat Prod Rep. 2017;34:1302–1331. doi: 10.1039/c7np00021a. [DOI] [PubMed] [Google Scholar]
- 34.Milshteyn A., Schneider J.S., Brady S.F. Mining the metabiome: identifying novel natural products from microbial communities. Chem Biol. 2014;21 doi: 10.1016/j.chembiol.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burkhart B.J., Hudson G.A., Dunbar K.L., Mitchell D.A. A prevalent peptide-binding domain guides ribosomal natural product biosynthesis. Nat Chem Biol. 2015;11:564–570. doi: 10.1038/nchembio.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sardar D., Lin Z., Schmidt E.W. Modularity of RiPP enzymes enables designed synthesis of decorated peptides. Chem Biol. 2015;22:907–916. doi: 10.1016/j.chembiol.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piel J., Hui D., Wen G., Butzke D., Platzer M., Fusetani N. Antitumor polyketide biosynthesis by an uncultivated bacterial symbiont of the marine sponge Theonella swinhoei. Proc Natl Acad Sci U S A. 2004;101:16222–16227. doi: 10.1073/pnas.0405976101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hover B.M., Kim S.H., Katz M., Charlop-Powers Z., Owen J.G., Ternei M.A. Culture-independent discovery of the malacidins as calcium-dependent antibiotics with activity against multidrug-resistant Gram-positive pathogens. Nat Microbiol. 2018;3:415–422. doi: 10.1038/s41564-018-0110-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ling L.L., Schneider T., Peoples A.J., Spoering A.L., Engels I., Conlon B.P. A new antibiotic kills pathogens without detectable resistance. Nature. 2015;517:455–459. doi: 10.1038/nature14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weber T., Blin K., Duddela S., Krug D., Kim H.U., Bruccoleri R. antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015;43:W237–W243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Medema M.H., Blin K., Cimermancic P., de Jager V., Zakrzewski P., Fischbach M.A. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011;39:W339–W346. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khaldi N., Seifuddin F.T., Turner G., Haft D., Nierman W.C., Wolfe K.H. SMURF: genomic mapping of fungal secondary metabolite clusters. Fungal Genet Biol. 2010;47:736–741. doi: 10.1016/j.fgb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Heel A.J., de Jong A., Montalbán-López M., Kok J., Kuipers O.P. BAGEL3: automated identification of genes encoding bacteriocins and (non-) bactericidal posttranslationally modified peptides. Nucleic Acids Res. 2013;41:448–453. doi: 10.1093/nar/gkt391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skinnider M.A., Johnston C.W., Edgar R.E., Dejong C.A., Merwin N.J., Rees P.N. Genomic charting of ribosomally synthesized natural product chemical space facilitates targeted mining. Proc Natl Acad Sci U S A. 2016;113:E6343–E6351. doi: 10.1073/pnas.1609014113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skinnider M.A., Dejong C.A., Rees P.N., Johnston C.W., Li H., Webster A.L.H. Genomes to natural products PRediction informatics for secondary metabolomes (PRISM) Nucleic Acids Res. 2015;43:9645–9662. doi: 10.1093/nar/gkv1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Medema M.H., Kottmann R., Yilmaz P., Cummings M., Biggins J.B., Blin K. Minimum information about a biosynthetic gene cluster. Nat Chem Biol. 2015;11:625–631. doi: 10.1038/nchembio.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bibb M.J. Regulation of secondary metabolism in streptomycetes. Curr Opin Microbiol. 2005;8:208–215. doi: 10.1016/j.mib.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 48.Cimermancic P., Medema M.H., Claesen J., Kurita K., Wieland Brown L.C., Mavrommatis K. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell. 2014;158:412–421. doi: 10.1016/j.cell.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wisecaver J.H., Slot J.C., Rokas A. The evolution of fungal metabolic pathways. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Siitonen V., Claesson M., Patrikainen P., Aromaa M., Mäntsälä P., Schneider G. Identification of late-stage glycosylation steps in the biosynthetic pathway of the anthracycline nogalamycin. Chembiochem. 2012;13:120–128. doi: 10.1002/cbic.201100637. [DOI] [PubMed] [Google Scholar]
- 51.Kelly W.L., Pan L., Li C. Thiostrepton biosynthesis: prototype for a new family of bacteriocins. J Am Chem Soc. 2009;131:4327–4334. doi: 10.1021/ja807890a. [DOI] [PubMed] [Google Scholar]
- 52.Miao V., Coëffet-LeGal M.F., Brian P., Brost R., Penn J., Whiting A. Daptomycin biosynthesis in Streptomyces roseosporus: cloning and analysis of the gene cluster and revision of peptide stereochemistry. Microbiology. 2005;151:1507–1523. doi: 10.1099/mic.0.27757-0. [DOI] [PubMed] [Google Scholar]
- 53.Mast Y., Weber T., Gölz M., Ort-Winklbauer R., Gondran A., Wohlleben W. Characterization of the “pristinamycin supercluster” of Streptomyces pristinaespiralis. Microb Biotechnol. 2011;4:192–206. doi: 10.1111/j.1751-7915.2010.00213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nieselt K., Battke F., Herbig A., Bruheim P., Wentzel A., Jakobsen Ø.M. The dynamic architecture of the metabolic switch in Streptomyces coelicolor. BMC Genom. 2010;11:1–9. doi: 10.1186/1471-2164-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van der Heul H.U., Bilyk B.L., McDowall K.J., Seipke R.F., van Wezel G.P. Regulation of antibiotic production in Actinobacteria: new perspectives from the post-genomic era. Nat Prod Rep. 2018;00:1–30. doi: 10.1039/c8np00012c. [DOI] [PubMed] [Google Scholar]
- 56.Rigali S., Titgemeyer F., Barends S., Mulder S., Thomae A.W., Hopwood D.A. Feast or famine: the global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep. 2008;9:670–675. doi: 10.1038/embor.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martín J.F. Phosphate control of the biosynthesis of antibiotics and other secondary metabolites is mediated by the PhoR-PhoP system: an unfinished story. J Bacteriol. 2004;186:5197–5201. doi: 10.1128/JB.186.16.5197-5201.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martín J.F., Liras P. Engineering of regulatory cascades and networks controlling antibiotic biosynthesis in Streptomyces. Curr Opin Microbiol. 2010;13:263–273. doi: 10.1016/j.mib.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 59.Martín J.F., Casqueiro J., Liras P. Secretion systems for secondary metabolites: how producer cells send out messages of intercellular communication. Curr Opin Microbiol. 2005;8:282–293. doi: 10.1016/j.mib.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 60.Ikeno S., Yamane Y., Ohishi Y., Kinoshita N., Hamada M., Tsuchiya K.S. ABC transporter genes, kasKLM, responsible for self-resistance of a kasugamycin producer strain. J Antibiot (Tokyo) 2000;53:373–384. doi: 10.7164/antibiotics.53.373. [DOI] [PubMed] [Google Scholar]
- 61.Chen J., Liu M., Liu X., Miao J., Fu C., Gao H. Interrogation of Streptomyces avermitilis for efficient production of avermectins. Synth Syst Biotechnol. 2016;1:7–16. doi: 10.1016/j.synbio.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ochi K. Metabolic initiation of differentiation and secondary metabolism by Streptomyces griseus: significance of the stringent response (ppGpp) and GTP content in relation to A factor. J Bacteriol. 1987;169:3608–3616. doi: 10.1128/jb.169.8.3608-3616.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ochi K. From microbial differentiation to ribosome engineering. Biosci Biotechnol Biochem. 2007;71:1373–1386. doi: 10.1271/bbb.70007. [DOI] [PubMed] [Google Scholar]
- 64.Artsimovitch I., Patlan V., Sekine S.-I., Vassylyeva M.N., Hosaka T., Ochi K. Structural basis for transcription regulation by alarmone ppGpp. Cell. 2004;117:299–310. doi: 10.1016/s0092-8674(04)00401-5. [DOI] [PubMed] [Google Scholar]
- 65.Ross W., Vrentas C.E., Sanchez-Vazquez P., Gaal T., Gourse R.L. The magic spot: a ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol Cell. 2013;50:420–429. doi: 10.1016/j.molcel.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu K., Bittner A.N., Wang J.D. Diversity in (p)ppGpp metabolism and effectors. Curr Opin Microbiol. 2015;24:72–79. doi: 10.1016/j.mib.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ochi K. Microb. Resour., Elsevier; 2017. Cryptic pathways and implications for novel drug discovery; pp. 189–203. [Google Scholar]
- 68.Martinez-Costa O.H., Arias P., Romero N.M., Parro V., Mellado R.P., Malpartida F. A relA/spoT homologous gene from Streptomyces coelicolor A3(2) controls antibiotic biosynthetic genes. J Biochem Chem. 1996;271:10627–10634. doi: 10.1074/jbc.271.18.10627. [DOI] [PubMed] [Google Scholar]
- 69.Chakraburtty R., Bibb M. The ppGpp synthetase gene (relA) of Streptomyces coelicolor A3(2) plays a conditional role in antibiotic production and morphological differentiation. J Bacteriol. 1997;179:5854–5861. doi: 10.1128/jb.179.18.5854-5861.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gomez-Escribano J.P., Liras P., Pisabarro A., Martín J.F. An rplKΔ29-PALG-32 mutation leads to reduced expression of the regulatory genes ccaR and claR and very low transcription of the ceaS2 gene for clavulanic acid biosynthesis in Streptomyces clavuligerus. Mol Microbiol. 2006;61:758–770. doi: 10.1111/j.1365-2958.2006.05266.x. [DOI] [PubMed] [Google Scholar]
- 71.Lai C., Xu J., Tozawa Y., Okamoto-Hosoya Y., Yao X., Ochi K. Genetic and physiological characterization of rpoB mutations that activate antibiotic production in Streptomyces lividans. Microbiology. 2002;148:3365–3373. doi: 10.1099/00221287-148-11-3365. [DOI] [PubMed] [Google Scholar]
- 72.Xu J., Tozawa Y., Lai C., Hayashi H., Ochi K. A rifampicin resistance mutation in the rpoB gene confers ppGpp-independent antibiotic production in Streptomyces coelicolor A3(2) Mol Genet Genom. 2002;268:179–189. doi: 10.1007/s00438-002-0730-1. [DOI] [PubMed] [Google Scholar]
- 73.Tanaka Y., Kasahara K., Hirose Y., Murakami K., Kugimiya R., Ochi K. Activation and products of the cryptic secondary metabolite biosynthetic gene clusters by rifampin resistance (rpoB) mutations in actinomycetes. J Bacteriol. 2013;195:2959–2970. doi: 10.1128/JB.00147-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shima J., Hesketh A., Okamoto S., Kawamoto S., Ochi K. Induction of actinorhodin production by rpsL (encoding ribosomal protein S12) mutations that confer streptomycin resistance in Streptomyces lividans and Streptomyces coelicolor A3(2) J Bacteriol. 1996;178:7276–7284. doi: 10.1128/jb.178.24.7276-7284.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ochi K., Zhang D., Kawamoto S., Hesketh A. Molecular and functional analysis of the ribosomal L11 and S12 protein genes (rplK and rpsL) of Streptomyces coelicolor A3(2) Mol Gen Genet. 1997;256:488–498. doi: 10.1007/pl00008614. [DOI] [PubMed] [Google Scholar]
- 76.Wang G., Hosaka T., Ochi K. Dramatic activation of antibiotic production in Streptomyces coelicolor by cumulative drug resistance mutations. Appl Environ Microbiol. 2008;74:2834–2840. doi: 10.1128/AEM.02800-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hosoya Y., Okamoto S., Muramatsu H., Ochi K. Acquisition of certain streptomycin-resistant (str) mutations enhances antibiotic production in bacteria. Antimicrob Agents Chemother. 1998;42:2041–2047. doi: 10.1128/aac.42.8.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tanaka Y., Hosaka T., Ochi K. Rare earth elements activate the secondary metabolite-biosynthetic gene clusters in Streptomyces coelicolor A3(2) J Antibiot (Tokyo) 2010;63:477–481. doi: 10.1038/ja.2010.53. [DOI] [PubMed] [Google Scholar]
- 79.Hosaka T., Ohnishi-Kameyama M., Muramatsu H., Murakami K., Tsurumi Y., Kodani S. Antibacterial discovery in actinomycetes strains with mutations in RNA polymerase or ribosomal protein S12. Nat Biotechnol. 2009;27:462–464. doi: 10.1038/nbt.1538. [DOI] [PubMed] [Google Scholar]
- 80.Whitehouse I., Rando O.J., Delrow J., Tsukiyama T. Chromatin remodelling at promoters suppresses antisense transcription. Nature. 2007;450:1031–1035. doi: 10.1038/nature06391. [DOI] [PubMed] [Google Scholar]
- 81.Bednar J., Horowitz R.A., Grigoryev S.A., Carruthers L.M., Hansen J.C., Koster A.J. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc Natl Acad Sci U S A. 1998;95:14173–14178. doi: 10.1073/pnas.95.24.14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Saha A., Wittmeyer J., Cairns B.R. Chromatin remodelling: the industrial revolution of DNA around histones. Nat Rev Mol Cell Biol. 2006;7:437–447. doi: 10.1038/nrm1945. [DOI] [PubMed] [Google Scholar]
- 83.Cedar H., Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 84.Jenuwein T., Allis C. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 85.Marks P.A., Richon V.M., Breslow R., Rifkind R.A. Histone deacetylase inhibitors as new cancer drugs. Curr Opin Oncol. 2001;13:477–483. doi: 10.1097/00001622-200111000-00010. [DOI] [PubMed] [Google Scholar]
- 86.Yu J.-H., Keller N. Regulation of secondary metabolism in filamentous fungi. Annu Rev Phytopathol. 2005;43:437–458. doi: 10.1146/annurev.phyto.43.040204.140214. [DOI] [PubMed] [Google Scholar]
- 87.Gacek A., Strauss J. The chromatin code of fungal secondary metabolite gene clusters. Appl Microbiol Biotechnol. 2012;95:1389–1404. doi: 10.1007/s00253-012-4208-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shwab E.K., Jin W.B., Tribus M., Galehr J., Graessle S., Keller N.P. Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryot Cell. 2007;6:1656–1664. doi: 10.1128/EC.00186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bok J.W., Chiang Y.M., Szewczyk E., Reyes-Domingez Y., Davidson A.D., Sanchez J.F. Chromatin-level regulation of biosynthetic gene clusters. Nat Chem Biol. 2009;5:462–464. doi: 10.1038/nchembio.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Williams R.B., Henrikson J.C., Hoover A.R., Lee A.E., Cichewicz R.H. Epigenetic remodeling of the fungal secondary metabolome. Org Biomol Chem. 2008;6:1895. doi: 10.1039/b804701d. [DOI] [PubMed] [Google Scholar]
- 91.Reyes-Dominguez Y., Bok J.W., Berger H., Shwab E.K., Basheer A., Gallmetzer A. Heterochromatic marks are associated with the repression of secondary metabolism clusters in Aspergillus nidulans. Mol Microbiol. 2010;76:1376–1386. doi: 10.1111/j.1365-2958.2010.07051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lombardi P.M., Angell H.D., Whittington D.A., Flynn E.F., Rajashankar K.R., Christianson D.W. Structure of prokaryotic polyamine deacetylase reveals evolutionary functional relationships with eukaryotic histone deacetylases. Biochemistry. 2011;50:1808–1817. doi: 10.1021/bi101859k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Williams R.M., Rimsky S. Molecular aspects of the E. coli nucleoid protein, H-NS: a central controller of gene regulatory networks. FEMS Microbiol Lett. 1997;156:175–185. doi: 10.1111/j.1574-6968.1997.tb12724.x. [DOI] [PubMed] [Google Scholar]
- 94.Moore J.M., Bradshaw E., Seipke R.F., Hutchings M.I., McArthur M. Use and discovery of chemical elicitors that stimulate biosynthetic gene clusters in Streptomyces bacteria. In: Hopwood D.A., editor. vol. 517. Elsevier Inc.; 2012. pp. 367–385. (Methods enzymol). [DOI] [PubMed] [Google Scholar]
- 95.Aldridge M., Facey P., Francis L., Bayliss S., Del Sol R., Dyson P. A novel bifunctional histone protein in Streptomyces: a candidate for structural coupling between DNA conformation and transcription during development and stress? Nucleic Acids Res. 2013;41:4813–4824. doi: 10.1093/nar/gkt180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Scherlach K., Hertweck C. Triggering cryptic natural product biosynthesis in microorganisms. Org Biomol Chem. 2009;7:1753. doi: 10.1039/b821578b. [DOI] [PubMed] [Google Scholar]
- 97.Bode H.B., Bethe B., Höfs R., Zeeck A. Big effects from small changes: possible ways to explore nature's chemical diversity. Chembiochem. 2002;3:619. doi: 10.1002/1439-7633(20020703)3:7<619::AID-CBIC619>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 98.Reen F.J., Romano S., Dobson A.D.W., O'Gara F. The sound of silence: activating silent biosynthetic gene clusters in marine microorganisms. Mar Drugs. 2015;13:4754–4783. doi: 10.3390/md13084754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Paranagama P.A., Wijeratne E.M.K., Gunatilaka A.A.L. Uncovering biosynthetic potential of plant-associated fungi: effect of culture conditions on metabolite production by Paraphaeosphaeria quadriseptata and Chaetomium chiversii. J Nat Prod. 2007;70:1939–1945. doi: 10.1021/np070504b. [DOI] [PubMed] [Google Scholar]
- 100.Doull J.L., Singh A.K., Hoare M., Ayer S.W. Conditions for the production of jadomycin B by Streptomyces venezuelae ISP5230: effects of heat shock, ethanol treatment and phage infection. J Ind Microbiol. 1994;13:120–125. doi: 10.1007/BF01584109. [DOI] [PubMed] [Google Scholar]
- 101.Liao Y., Wei Z.H., Bai L., Deng Z., Zhong J.J. Effect of fermentation temperature on validamycin A production by Streptomyces hygroscopicus 5008. J Biotechnol. 2009;142:271–274. doi: 10.1016/j.jbiotec.2009.04.015. [DOI] [PubMed] [Google Scholar]
- 102.Hayes A., Hobbs G., Smith C.P., Oliver S.G., Butler P.R. Environmental signals triggering methylenomycin production by Streptomyces coelicolor A3(2) J Bacteriol. 1997;179:5511–5515. doi: 10.1128/jb.179.17.5511-5515.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rigali S., Titgemeyer F., Barends S., Mulder S., Thomae A.W., Hopwood D.A. Feast or famine: the global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep. 2008;9:670–675. doi: 10.1038/embor.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Craney A., Ozimok C., Pimentel-Elardo S.M., Capretta A., Nodwell J.R. Chemical perturbation of secondary metabolism demonstrates important links to primary metabolism. Chem Biol. 2012;19:1020–1027. doi: 10.1016/j.chembiol.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 105.Onaka H., Tabata H., Igarashi Y., Sato Y., Furumai T. Goadsporin, a chemical substance which promotes secondary metabolism and morphogenesis in Streptomycetes. I. Purification and Characterization. J Antibiot (Tokyo) 2001;54:1036–1044. doi: 10.7164/antibiotics.54.1036. [DOI] [PubMed] [Google Scholar]
- 106.Onaka H. Novel antibiotic screening methods to awaken silent or cryptic secondary metabolic pathways in actinomycetes. J Antibiot (Tokyo) 2017:1–6. doi: 10.1038/ja.2017.51. [DOI] [PubMed] [Google Scholar]
- 107.Wang W., Ji J., Li X., Wang J., Li S., Pan G. Angucyclines as signals modulate the behaviors of Streptomyces coelicolor. Proc Natl Acad Sci U S A. 2014;111:5688–5693. doi: 10.1073/pnas.1324253111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Amano S.I., Sakurai T., Endo K., Takano H., Beppu T., Furihata K. A cryptic antibiotic triggered by monensin. J Antibiot (Tokyo) 2011;64:703. doi: 10.1038/ja.2011.69. [DOI] [PubMed] [Google Scholar]
- 109.Kawai K., Wang G., Okamoto S., Ochi K. The rare earth, scandium, causes antibiotic overproduction in Streptomyces spp. FEMS Microbiol Lett. 2007;274:311–315. doi: 10.1111/j.1574-6968.2007.00846.x. [DOI] [PubMed] [Google Scholar]
- 110.Davies J., Spiegelman G.B., Yim G. The world of subinhibitory antibiotic concentrations. Curr Opin Microbiol. 2006;9:445–453. doi: 10.1016/j.mib.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 111.Davies J. Are antibiotics naturally antibiotics? J Ind Microbiol Biotechnol. 2006;33:496–499. doi: 10.1007/s10295-006-0112-5. [DOI] [PubMed] [Google Scholar]
- 112.Wakefield J., Hassan H.M., Jaspars M., Ebel R., Rateb M.E. Dual induction of new microbial secondary metabolites by fungal bacterial co-cultivation. Front Microbiol. 2017;8:1–10. doi: 10.3389/fmicb.2017.01284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Oh D.C., Kauffman C.A., Jensen P.R., Fenical W. Induced production of emericellamides A and B from the marine-derived fungus Emericella sp. in competing co-culture. J Nat Prod. 2007;70:515–520. doi: 10.1021/np060381f. [DOI] [PubMed] [Google Scholar]
- 114.Amano S.I., Morota T., Kano Y.K., Narita H., Hashidzume T., Yamamoto S. Promomycin, a polyether promoting antibiotic production in Streptomyces spp. J Antibiot (Tokyo) 2010;63:486–491. doi: 10.1038/ja.2010.68. [DOI] [PubMed] [Google Scholar]
- 115.Schroeckh V., Scherlach K., Nutzmann H.-W., Shelest E., Schmidt-Heck W., Schuemann J. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc Natl Acad Sci U S A. 2009;106:14558–14563. doi: 10.1073/pnas.0901870106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.König C.C., Scherlach K., Schroeckh V., Horn F., Nietzsche S., Brakhage A.A. Bacterium induces cryptic meroterpenoid pathway in the pathogenic fungus Aspergillus fumigatus. Chembiochem. 2013;14:938–942. doi: 10.1002/cbic.201300070. [DOI] [PubMed] [Google Scholar]
- 117.Beld J., Sonnenschein E.C., Vickery C.R., Noel J.P., Burkart M.D. The phosphopantetheinyl transferases: catalysis of a post-translational modification crucial for life. Nat Prod Rep. 2014;31:61–108. doi: 10.1039/c3np70054b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bunet R., Riclea R., Laureti L., Hôtel L., Paris C., Girardet J.M. A single Sfp-type phosphopantetheinyl transferase plays a major role in the biosynthesis of PKS and NRPS derived metabolites in Streptomyces ambofaciens ATCC23877. PLoS One. 2014;9:1–12. doi: 10.1371/journal.pone.0087607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gunnarsson N., Eliasson A., Nielsen J. Control of fluxes towards antibiotics and the role of primary metabolism in production of antibiotics. Adv Biochem Eng Biotechnol. 2004;88:137–178. doi: 10.1007/b99260. [DOI] [PubMed] [Google Scholar]
- 120.Zhang B., Tian W., Wang S., Yan X., Jia X., Pierens G.K. Activation of natural products biosynthetic pathways via a protein modification level regulation. ACS Chem Biol. 2017;12:1732–1736. doi: 10.1021/acschembio.7b00225. [DOI] [PubMed] [Google Scholar]
- 121.Yan X., Zhang B., Tian W., Dai Q., Zheng X., Hu K. Puromycin A, B and C, cryptic nucleosides identified from Streptomyces alboniger NRRL B-1832 by PPtase-based activation. Synth Syst Biotechnol. 2018;3:76–80. doi: 10.1016/j.synbio.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tilburn J., Sarkar S., Widdick D.A., Espeso E.A., Orejas M., Mungroo J. The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline-expressed genes by ambient pH. EMBO J. 1995;14:779–790. doi: 10.1002/j.1460-2075.1995.tb07056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Brakhage A.A. Regulation of fungal secondary metabolism. Nat Rev Microbiol. 2013;11:21–32. doi: 10.1038/nrmicro2916. [DOI] [PubMed] [Google Scholar]
- 124.Makitrynskyy R., Ostash B., Tsypik O., Rebets Y., Doud E., Meredith T. Pleiotropic regulatory genes bldA, adpA and absB are implicated in production of phosphoglycolipid antibiotic moenomycin. Open Biol. 2013;3 doi: 10.1098/rsob.130121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ohnishi Y., Yamazaki H., Kato J., Tomono A., Horinouchi S. AdpA, a central transcriptional regulator in the A-Factor regulatory cascade that leads to morphological development and secondary metabolism in Streptomyces griseus. Biosci Biotechnol Biochem. 2005;69:431–439. doi: 10.1271/bbb.69.431. [DOI] [PubMed] [Google Scholar]
- 126.Xu J., Zhang J., Zhuo J., Li Y., Tian Y., Tan H. Activation and mechanism of a cryptic oviedomycin gene cluster via the disruption of a global regulatory gene, adpA, in Streptomyces ansochromogenes. J Biol Chem. 2017;292:19708–19720. doi: 10.1074/jbc.M117.809145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Becerril A., Álvarez S., Braña A.F., Rico S., Díaz M., Santamaría R.I. Uncovering production of specialized metabolites by Streptomyces argillaceus: activation of cryptic biosynthesis gene clusters using nutritional and genetic approaches. PLoS One. 2018;13 doi: 10.1371/journal.pone.0198145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fowler-Goldsworthy K., Gust B., Mouz S., Chandra G., Findlay K.C., Chater K.F. The actinobacteria-specific gene wblA controls major developmental transitions in Streptomyces coelicolor A3(2) Microbiology. 2011;157:1312–1328. doi: 10.1099/mic.0.047555-0. [DOI] [PubMed] [Google Scholar]
- 129.Huang H., Hou L., Li H., Qiu Y., Ju J., Li W. Activation of a plasmid-situated type III PKS gene cluster by deletion of a wbl gene in deepsea-derived Streptomyces somaliensis SCSIO ZH66. Microb Cell Factories. 2016;15:116. doi: 10.1186/s12934-016-0515-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Li H., Huang H., Hou L., Ju J., Li W. Discovery of antimycin-type depsipeptides from a wbl gene mutant strain of deepsea-derived Streptomyces somaliensis SCSIO ZH66 and their effects on pro-inflammatory cytokine production. Front Microbiol. 2017;8:1–8. doi: 10.3389/fmicb.2017.00678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gruber T.M., Gross C.A. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu Rev Microbiol. 2003;57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- 132.Paget M.S., Helmann J.D. The σ70 family of sigma factors. Genome Biol. 2003;4:203. doi: 10.1186/gb-2003-4-1-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jiang C., Wang H., Kang Q., Liu J., Bai L. Cloning and characterization of the polyether salinomycin biosynthesis gene cluster of Streptomyces albus XM211. Appl Environ Microbiol. 2012;78:994–1003. doi: 10.1128/AEM.06701-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Shelest E. Transcription factors in fungi: TFome dynamics, three major families, and dual-specificity TFs. Front Genet. 2017;8:53. doi: 10.3389/fgene.2017.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hu Y., Qin Y., Liu G. Collection and curation of transcriptional regulatory interactions in Aspergillus nidulans and Neurospora crassa reveal structural and evolutionary features of the regulatory networks. Front Microbiol. 2018;9:1–8. doi: 10.3389/fmicb.2018.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Jnawali H.N., Liou K., Sohng J.K. Role of σ-factor (orf21) in clavulanic acid production in Streptomyces clavuligerus NRRL3585. Microbiol Res. 2011;166:369–379. doi: 10.1016/j.micres.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 137.Lee S., Yang S.H., Kang C., Mo S., Suh J. Overexpression of the putative extracytoplasmic function sigma (σ) factor FujE enhances FK506 production in Streptomyces sp. strain KCCM 11116P. Can J Microbiol. 2014;60:363–369. doi: 10.1139/cjm-2014-0166. [DOI] [PubMed] [Google Scholar]
- 138.Taniguchi H., Henke N.A., Heider S.A.E., Wendisch V.F. Overexpression of the primary sigma factor gene sigA improved carotenoid production by Corynebacterium glutamicum: application to production of β-carotene and the non-native linear C50 carotenoid bisanhydrobacterioruberin. Metab Eng Commun. 2017;4:1–11. doi: 10.1016/j.meteno.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Stevens D.C., Conway K.R., Pearce N., Villegas-Peñaranda L.R., Garza A.G., Boddy C.N. Alternative sigma factor over-expression enables heterologous expression of a Type II polyketide biosynthetic pathway in Escherichia coli. PLoS One. 2013;8 doi: 10.1371/journal.pone.0064858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jiang L., Liu Y., Wang P., Wen Y., Song Y., Chen Z. Inactivation of the extracytoplasmic function sigma factor Sig6 stimulates avermectin production in Streptomyces avermitilis. Biotechnol Lett. 2011;33:1955–1961. doi: 10.1007/s10529-011-0673-x. [DOI] [PubMed] [Google Scholar]
- 141.Zhuo Y., Zhang W., Chen D., Gao H., Tao J., Liu M. Reverse biological engineering of hrdB to enhance the production of avermectins in an industrial strain of Streptomyces avermitilis. Proc Natl Acad Sci U S A. 2010;107:11250–11254. doi: 10.1073/pnas.1006085107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Olano C., García I., González A., Rodriguez M., Rozas D., Rubio J. Activation and identification of five clusters for secondary metabolites in Streptomyces albus J1074. Microb Biotechnol. 2014;7 doi: 10.1111/1751-7915.12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bai C., Zhang Y., Zhao X., Hu Y., Xiang S., Miao J. Exploiting a precise design of universal synthetic modular regulatory elements to unlock the microbial natural products in Streptomyces. Proc Natl Acad Sci U S A. 2015;112:12181–12186. doi: 10.1073/pnas.1511027112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhang M.M., Wong F.T., Wang Y., Luo S., Lim Y.H., Heng E. CRISPR–Cas9 strategy for activation of silent Streptomyces biosynthetic gene clusters. Nat Chem Biol. 2017;13:607–609. doi: 10.1038/nchembio.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kennedy J., Turner G. δ-(L-α-Aminoadipyl)-L-cysteinyl-D-valine synthetase is a rate limiting enzyme for penicillin production in Aspergillus nidulans. Mol Gen Genet MGG. 1996;253:189–197. doi: 10.1007/s004380050312. [DOI] [PubMed] [Google Scholar]
- 146.Yeh H.-H., Ahuja M., Chiang Y.-M., Oakley C.E., Moore S., Yoon O. Resistance gene-guided genome mining: serial promoter exchanges in Aspergillus nidulans reveal the biosynthetic pathway for Fellutamide B, a proteasome inhibitor. ACS Chem Biol. 2016;11:2275–2284. doi: 10.1021/acschembio.6b00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Huang J., Shi J., Molle V., Sohlberg B., Weaver D., Bibb M.J. Cross-regulation among disparate antibiotic biosynthetic pathways of Streptomyces coelicolor. Mol Microbiol. 2005;58:1276–1287. doi: 10.1111/j.1365-2958.2005.04879.x. [DOI] [PubMed] [Google Scholar]
- 148.Santos-Beneit F., Rodríguez-García A., Sola-Landa A., Martín J.F. Cross-talk between two global regulators in Streptomyces: PhoP and AfsR interact in the control of afsS, pstS and phoRP transcription. Mol Microbiol. 2009;72:53–68. doi: 10.1111/j.1365-2958.2009.06624.x. [DOI] [PubMed] [Google Scholar]
- 149.Romero-Rodríguez A., Robledo-Casados I., Sánchez S. An overview on transcriptional regulators in Streptomyces. Biochim Biophys Acta Gene Regul Mech. 2015;1849:1017–1039. doi: 10.1016/j.bbagrm.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 150.Bhukya H., Anand R. TetR regulators: a structural and functional perspective. J Indian Inst Sci. 2017;97:245–259. [Google Scholar]
- 151.Orth P., Schnappinger D., Hillen W., Saenger W., Hinrichs W. Structural basis of gene regulation by the tetracycline inducible Tet repressor-operator system. Nat Struct Biol. 2000;7:215–219. doi: 10.1038/73324. [DOI] [PubMed] [Google Scholar]
- 152.Onaka H., Horinouchi S. DNA-binding activity of the A-factor receptor protein and its recognition DNA sequences. Mol Microbiol. 1997;24:991–1000. doi: 10.1046/j.1365-2958.1997.4081772.x. [DOI] [PubMed] [Google Scholar]
- 153.Xu G., Wang J., Wang L., Tian X., Yang H., Fan K. “Pseudo” γ-butyrolactone receptors respond to antibiotic signals to coordinate antibiotic biosynthesis. J Biol Chem. 2010;285:27440–27448. doi: 10.1074/jbc.M110.143081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhang Y.Y., Zou Z.Z., Niu G.Q., Tan H.R. jadR* and jadR2 act synergistically to repress jadomycin biosynthesis. Sci China Life Sci. 2013;56:584–590. doi: 10.1007/s11427-013-4508-y. [DOI] [PubMed] [Google Scholar]
- 155.Bunet R., Song L., Mendes M.V., Corre C., Hotel L., Rouhier N. Characterization and manipulation of the pathway-specific late regulator AlpW reveals Streptomyces ambofaciens as a new producer of kinamycins. J Bacteriol. 2011;193:1142–1153. doi: 10.1128/JB.01269-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lee H.N., Huang J., Im J.H., Kim S.H., Noh J.H., Cohen S.N. Putative TetR family transcriptional regulator SCO1712 encodes an antibiotic downregulator in Streptomyces coelicolor. Appl Environ Microbiol. 2010;76:3039–3043. doi: 10.1128/AEM.02426-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Takano E., Kinoshita H., Mersinias V., Bucca G., Hotchkiss G., Nihira T. A bacterial hormone (the SCB1) directly controls the expression of a pathway-specific regulatory gene in the cryptic type I polyketide biosynthetic gene cluster of Streptomyces coelicolor. Mol Microbiol. 2005;56:465–479. doi: 10.1111/j.1365-2958.2005.04543.x. [DOI] [PubMed] [Google Scholar]
- 158.Gomez-Escribano J.P., Song L., Fox D.J., Yeo V., Bibb M.J., Challis G.L. Structure and biosynthesis of the unusual polyketide alkaloid coelimycin P1, a metabolic product of the cpk gene cluster of Streptomyces coelicolor M145. Chem Sci. 2012;3:2716. [Google Scholar]
- 159.Zhang R., Pappas K.M., Brace J.L., Miller P.C., Oulmassov T., Molyneaux J.M. Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA. Nature. 2002;417:971–974. doi: 10.1038/nature00833. [DOI] [PubMed] [Google Scholar]
- 160.White C.E., Winans S.C. Identification of amino acid residues of the Agrobacterium tumefaciens quorum-sensing regulator TraR that are critical for positive control of transcription. Mol Microbiol. 2005;55:1473–1486. doi: 10.1111/j.1365-2958.2004.04482.x. [DOI] [PubMed] [Google Scholar]
- 161.Schrijver A.D., Mot R.D. A subfamily of MalT-related ATP-dependent regulators in the LuxR family. Microbiology. 1999;145:1287–1288. doi: 10.1099/13500872-145-6-1287. [DOI] [PubMed] [Google Scholar]
- 162.Danot O. A complex signaling module governs the activity of MalT, the prototype of an emerging transactivator family. Proc Natl Acad Sci U S A. 2001;98:435–440. doi: 10.1073/pnas.98.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chen R., Zhang Q., Tan B., Zheng L., Li H., Zhu Y. Genome mining and activation of a silent PKS/NRPS gene cluster direct the production of Totopotensamides. Org Lett. 2017;19:5697–5700. doi: 10.1021/acs.orglett.7b02878. [DOI] [PubMed] [Google Scholar]
- 164.Wilkinson B., Micklefield J. Mining and engineering natural-product biosynthetic pathways. Nat Chem Biol. 2007;3:379–386. doi: 10.1038/nchembio.2007.7. [DOI] [PubMed] [Google Scholar]
- 165.Liu T., Mazmouz R., Ongley S.E., Chau R., Pickford R., Woodhouse J.N. Directing the heterologous production of specific cyanobacterial toxin variants. ACS Chem Biol. 2017;12:2021–2029. doi: 10.1021/acschembio.7b00181. [DOI] [PubMed] [Google Scholar]
- 166.Ongley S.E., Bian X., Zhang Y., Chau R., Gerwick W.H., Müller R. High-titer heterologous production in E. coli of lyngbyatoxin, a protein kinase C activator from an uncultured marine cyanobacterium. ACS Chem Biol. 2013;8:1888–1893. doi: 10.1021/cb400189j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Gustafsson C., Govindarajan S., Minshull J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004;22:346–353. doi: 10.1016/j.tibtech.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 168.Zhang Z., Smart T.J., Choi H., Hardy F., Lohans C.T., Abboud M.I. Structural and stereoelectronic insights into oxygenase-catalyzed formation of ethylene from 2-oxoglutarate. Proc Natl Acad Sci. 2017;114:4667–4672. doi: 10.1073/pnas.1617760114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Peiru S., Menzella H.G., Rodriguez E., Carney J., Gramajo H. Production of the potent antibacterial polyketide Erythromycin C in Escherichia coli. Appl Environ Microbiol. 2005;71:2539–2547. doi: 10.1128/AEM.71.5.2539-2547.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Choi K.R., Cho J.S., Cho I.J., Park D., Lee S.Y. Markerless gene knockout and integration to express heterologous biosynthetic gene clusters in Pseudomonas putida. Metab Eng. 2018;47:463–474. doi: 10.1016/j.ymben.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 171.Liu Q., Shen Q., Bian X., Chen H., Fu J., Wang H. Simple and rapid direct cloning and heterologous expression of natural product biosynthetic gene cluster in Bacillus subtilis via Red/ET recombineering. Sci Rep. 2016;6:1–10. doi: 10.1038/srep34623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wendt-Pienkowski E., Huang Y., Zhang J., Li B., Jiang H., Kwon H. Cloning, sequencing, analysis, and heterologous expression of the fredericamycin biosynthetic gene cluster from Streptomyces griseus. J Am Chem Soc. 2005;127:16442–16452. doi: 10.1021/ja054376u. [DOI] [PubMed] [Google Scholar]
- 173.Young T.S., Walsh C.T. Identification of the thiazolyl peptide GE37468 gene cluster from Streptomyces ATCC 55365 and heterologous expression in Streptomyces lividans. Proc Natl Acad Sci U S A. 2011;108:13053–13058. doi: 10.1073/pnas.1110435108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Gomez-Escribano J.P., Bibb M.J. Heterologous expression of natural product biosynthetic gene clusters in Streptomyces coelicolor: from genome mining to manipulation of biosynthetic pathways. J Ind Microbiol Biotechnol. 2014;41:425–431. doi: 10.1007/s10295-013-1348-5. [DOI] [PubMed] [Google Scholar]
- 175.Rückert C., Albersmeier A., Busche T., Jaenicke S., Winkler A., Fridjónsson Ó.H. Complete genome sequence of Streptomyces lividans TK24. J Biotechnol. 2015;199:21–22. doi: 10.1016/j.jbiotec.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 176.Baltz R.H. Genetic manipulation of secondary metabolite biosynthesis for improved production in Streptomyces and other actinomycetes. J Ind Microbiol Biotechnol. 2016;43:343–370. doi: 10.1007/s10295-015-1682-x. [DOI] [PubMed] [Google Scholar]
- 177.Zaburannyi N., Rabyk M., Ostash B., Fedorenko V., Luzhetskyy A. Insights into naturally minimised Streptomyces albus J1074 genome. BMC Genom. 2014;15:1–11. doi: 10.1186/1471-2164-15-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Nielsen M.T., Nielsen J.B., Anyaogu D.C., Holm D.K., Nielsen K.F., Larsen T.O. Heterologous reconstitution of the intact Geodin gene cluster in Aspergillus nidulans through a simple and versatile PCR based approach. PLoS One. 2013;8 doi: 10.1371/journal.pone.0072871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Alberti F., Foster G.D., Bailey A.M. Natural products from filamentous fungi and production by heterologous expression. Appl Microbiol Biotechnol. 2017;101:493–500. doi: 10.1007/s00253-016-8034-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Smith D.J., Burnham M.K.R., Edwards J., J EA, Turner G. Cloning and heterologous expression of the Penicillin biosynthetic gene cluster from Penicillium chrysogenum. Biotechnology. 1990;8:39–41. doi: 10.1038/nbt0190-39. [DOI] [PubMed] [Google Scholar]
- 181.Tagami K., Liu C., Minami A., Noike M., Isaka T., Fueki S. Reconstitution of biosynthetic machinery for indole-diterpene paxilline in Aspergillus oryzae. J Am Chem Soc. 2013;135:1260–1263. doi: 10.1021/ja3116636. [DOI] [PubMed] [Google Scholar]
- 182.Tagami K., Minami A., Fujii R., Liu C., Tanaka M., Gomi K. Rapid reconstitution of biosynthetic machinery for fungal metabolites in Aspergillus oryzae: total biosynthesis of aflatrem. Chembiochem. 2014;16:2076–2080. doi: 10.1002/cbic.201402195. [DOI] [PubMed] [Google Scholar]
- 183.Alberti F., Khairudin K., Venegas E.R., Davies J.A., Hayes P.M., Willis C.L. Heterologous expression reveals the biosynthesis of the antibiotic pleuromutilin and generates bioactive semi-synthetic derivatives. Nat Commun. 2017;8:1–9. doi: 10.1038/s41467-017-01659-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Chiang Y.-M., Oakley C.E., Ahuja M., Entwistle R., Schultz A., Chang S.-L. An efficient system for heterologous expression of secondary metabolite genes in Aspergillus nidulans. J Am Chem Soc. 2013;135:7720–7731. doi: 10.1021/ja401945a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Gomez-Escribano J.P., Bibb M.J. Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb Biotechnol. 2011;4:207–215. doi: 10.1111/j.1751-7915.2010.00219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Komatsu M., Komatsu K., Koiwai H., Yamada Y., Kozone I., Izumikawa M. Engineered Streptomyces avermitilis host for heterologous expression of biosynthetic gene cluster for secondary metabolites. ACS Synth Biol. 2013;2:384–396. doi: 10.1021/sb3001003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kouprina N., Larionov V. Transformation-associated recombination (TAR) cloning for genomics studies and synthetic biology. Chromosoma. 2016;125:621–632. doi: 10.1007/s00412-016-0588-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Wang H., Li Z., Jia R., Hou Y., Yin J., Bian X. RecET direct cloning and Redαβ recombineering of biosynthetic gene clusters, large operons or single genes for heterologous expression. Nat Protoc. 2016;11:1175–1190. doi: 10.1038/nprot.2016.054. [DOI] [PubMed] [Google Scholar]
- 189.Greunke C., Duell E.R., D'Agostino P.M., Glöckle A., Lamm K., Gulder T.A.M. Direct Pathway Cloning (DiPaC) to unlock natural product biosynthetic potential. Metab Eng. 2018;47:334–345. doi: 10.1016/j.ymben.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 190.Casini A., Chang F.Y., Eluere R., King A.M., Young E.M., Dudley Q.M. A pressure test to make 10 molecules in 90 days: external evaluation of methods to engineer biology. J Am Chem Soc. 2018;140:4302–4316. doi: 10.1021/jacs.7b13292. [DOI] [PubMed] [Google Scholar]
- 191.Yamanaka K., Reynolds K.A., Kersten R.D., Ryan K.S., Gonzalez D.J., Nizet V. Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. Proc Natl Acad Sci U S A. 2014;111:1957–1962. doi: 10.1073/pnas.1319584111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Luo Y., Zhang L., Barton K.W., Zhao H. Systematic identification of a panel of strong constitutive promoters from Streptomyces albus. ACS Synth Biol. 2015;4:1001–1010. doi: 10.1021/acssynbio.5b00016. [DOI] [PubMed] [Google Scholar]
- 193.Luo Y., Huang H., Liang J., Wang M., Lu L., Shao Z. Activation and characterization of a cryptic polycyclic tetramate macrolactam biosynthetic gene cluster. Nat Commun. 2013;4:2894. doi: 10.1038/ncomms3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Shao Z., Rao G., Li C., Abil Z., Luo Y., Zhao H. Refactoring the silent spectinabilin gene cluster using a plug-and-play scaffold. ACS Synth Biol. 2013;2:662–669. doi: 10.1021/sb400058n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Basitta P., Westrich L., Rösch M., Kulik A., Gust B., Apel A.K. AGOS: a plug-and-play method for the assembly of artificial gene operons into functional biosynthetic gene clusters. ACS Synth Biol. 2017;6:817–825. doi: 10.1021/acssynbio.6b00319. [DOI] [PubMed] [Google Scholar]
- 196.Li S., Wang J., Li X., Yin S., Wang W., Yang K. Genome-wide identification and evaluation of constitutive promoters in streptomycetes. Microb Cell Factories. 2015;14:172. doi: 10.1186/s12934-015-0351-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Guo F., Xiang S., Li L., Wang B., Rajasärkkä J., Gröndahl-Yli-Hannuksela K. Targeted activation of silent natural product biosynthesis pathways by reporter-guided mutant selection. Metab Eng. 2015;28:134–142. doi: 10.1016/j.ymben.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 198.Askenazi M., Driggers E.M., Holtzman D.A., Norman T.C., Iverson S., Zimmer D.P. Integrating transcriptional and metabolite profiles to direct the engineering of lovastatin-producing fungal strains. Nat Biotechnol. 2003;21:150–156. doi: 10.1038/nbt781. [DOI] [PubMed] [Google Scholar]
- 199.Xiang S.-H., Li J., Yin H., Zheng J.-T., Yang X., Wang H.-B. Application of a double-reporter-guided mutant selection method to improve clavulanic acid production in Streptomyces clavuligerus. Metab Eng. 2009;11:310–318. doi: 10.1016/j.ymben.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 200.Wang Y., Tao Z., Zheng H., Zhang F., Long Q., Deng Z. Iteratively improving natamycin production in Streptomyces gilvosporeus by a large operon-reporter based strategy. Metab Eng. 2016;38:418–426. doi: 10.1016/j.ymben.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 201.Wang L., Zhao Y., Liu Q., Huang Y., Hu C., Liao G. Improvement of A21978C production in Streptomyces roseosporus by reporter-guided rpsL mutation selection. J Appl Microbiol. 2012;112:1095–1101. doi: 10.1111/j.1365-2672.2012.05302.x. [DOI] [PubMed] [Google Scholar]
- 202.Rebets Y., Schmelz S., Gromyko O., Tistechok S., Petzke L., Scrima A. Design, development and application of whole-cell based antibiotic-specific biosensor. Metab Eng. 2018;47:263–270. doi: 10.1016/j.ymben.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 203.Yang K., Han L., He J., Wang L., Vining L.C. A repressor-response regulator gene pair controlling jadomycin B production in Streptomyces venezuelae ISP5230. Gene. 2001;279:165–173. doi: 10.1016/s0378-1119(01)00723-5. [DOI] [PubMed] [Google Scholar]
- 204.Sun Y.-Q., Busche T., Rückert C., Paulus C., Rebets Y., Novakova R. Development of a biosensor concept to detect the production of cluster-specific secondary metabolites. ACS Synth Biol. 2017;6:1026–1033. doi: 10.1021/acssynbio.6b00353. [DOI] [PubMed] [Google Scholar]
- 205.Seyedsayamdost M.R. High-throughput platform for the discovery of elicitors of silent bacterial gene clusters. Proc Natl Acad Sci U S A. 2014;111 doi: 10.1073/pnas.1400019111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Rosen P.C., Seyedsayamdost M.R. Though much is taken, much abides: finding new antibiotics using old ones. Biochemistry. 2017;56:4925–4926. doi: 10.1021/acs.biochem.7b00782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Xu F., Nazari B., Moon K., Bushin L.B., Seyedsayamdost M.R. Discovery of a cryptic antifungal compound from Streptomyces albus J1074 using high-throughput elicitor screens. J Am Chem Soc. 2017;139:9203–9212. doi: 10.1021/jacs.7b02716. [DOI] [PMC free article] [PubMed] [Google Scholar]