Abstract
Background
Breast cancer is the most common tumor among women. miR-638 has been demonstrated to play an important role in various cancers.
Purpose
In this study, we aimed to investigate the function and prognostic value of miR-638 in breast cancer.
Methods
Quantitative real-time polymerase chain reaction analysis was used to evaluate the expression of miR-638 in breast cancer tissues and cell lines. The correlation of miR-638 with clinicopathological features was analyzed using the chi-squared test. Kaplan–Meier survival analysis and Cox regression assay were performed to investigate the prognostic value of miR-638 in breast cancer patients. The effects of miR-638 on the biological behavior of breast cancer cells were evaluated using functional assays.
Results
The expression of miR-638 was downregulated in breast cancer tissues and cell lines (all P<0.05). Decreased expression of miR-638 was significantly correlated with lymph node metastasis (P=0.015) and TNM stage (P=0.021). Patients with low miR-638 expression had shorter overall survival compared with those with high levels (Log-rank P=0.025). The miR-638 could be considered as an independent prognostic factor for the patients (HR =0.321, 95% CI =0.117–0.882, P=0.027). Downregulation of miR-638 was capable of promoting cell proliferation, migration, and invasion in MDA-MB-231 and MCF-7 cells.
Conclusion
All the results indicate that miR-638 is a tumor suppressor in breast cancer and is involved in the progression of breast cancer. Thus, it may serve as a prognostic biomarker for breast cancer.
Keywords: miR-638, cell proliferation, migration, invasion, prognosis, breast cancer
Introduction
Breast cancer is a common malignancy and a leading cause of cancer-related mortality in women.1 It is a heterogeneous disease that can be categorized into three major subtypes, of which triple-negative breast cancer (TNBC), accounting for 10%–20% of all breast cancer cases, has the worst prognosis because of high recurrence rates and early metastasis.2–4 Although advances in both detection and therapy have led to the improvement of prognosis, recurrence, and metastasis remain to be major challenges in breast cancer management that are related to the poor prognosis.5,6 Current biological markers used in the management of breast cancer are mainly estrogen receptor (ER), progesterone receptor (PR), and HER2.7,8 Rising evidence demonstrated that the prognosis could be improved by using some molecular biomarkers. Therefore, identification of novel precise biomarkers is important and needed for the development of prognosis in breast cancer patients.
MicroRNAs (miRNAs) are small endogenous noncoding RNAs, which function in the transcriptional and posttranscriptional regulation of gene expression.9–12 Evidence has shown that miRNAs play a crucial role in different biological processes, such as inflammation, cell proliferation, migration, invasion, and differentiation.13–16 Furthermore, a large number of miRNAs have been identified to function as oncogenes or tumor suppressor genes in the tumorigenesis process.17–19 Recently, dysregulation of miR-638 has been confirmed in several types of cancer, such as gastric cancer, basal cell carcinoma, and non-small-cell lung cancer (NSCLC).20–22 A recent study by Tan et al23 showed the expression of miR-638 was decreased in most non-TNBC and played an important role in TNBC progression via BRCA1 deregulation. However, the role of miR-638 in breast cancer is still limited.
In this study, we investigated the expression levels of miR-638 in breast cancer tissues and cell lines. The effects of miR-638 on biological behaviors of cancer cells were also assessed. This study aimed to identify the prognostic significance of miR-638 in breast cancer.
Methods and materials
Patients and tissue samples
Breast cancer tissues and adjacent normal tissues (located >3 cm away from the tumors) were collected from 125 breast cancer patients between January 2007 and December 2012 in Yidu Central Hospital of Weifang (Weifang, People’s Republic of China). None of the patients had received radiotherapy or chemotherapy or any other treatment before surgery. The tissues were verified by experienced pathologists. All the surgical specimens of the tumor resection were snap-frozen in liquid nitrogen after surgery until use. Written informed consent was obtained from all patients that agreed to participate in our study. This study was approved by the Ethics Committee of Yidu Central Hospital of Weifang (Weifang, People’s Republic of China). Detailed clinical characteristics of the patients are summarized in Table 1. A 5-year follow-up information was collected for the subsequent analysis.
Table 1.
Relationship between miR-638 expression and clinical characteristics of breast cancer patients
| Characteristics | Total number (N=125) |
miR-638 expression
|
P-values | |
|---|---|---|---|---|
| High (n=55) |
Low (n=70) |
|||
|
| ||||
| Age (years) | 0.363 | |||
| <50 | 58 | 23 | 35 | |
| ≥50 | 67 | 32 | 35 | |
| Tumor size (cm) | 0.315 | |||
| <2 | 54 | 21 | 33 | |
| ≥2 | 71 | 34 | 37 | |
| ER status | 0.676 | |||
| Negative | 61 | 28 | 33 | |
| Positive | 64 | 27 | 37 | |
| PR status | 0.245 | |||
| Negative | 55 | 21 | 34 | |
| Positive | 70 | 34 | 36 | |
| HER2 status | 0.696 | |||
| Negative | 57 | 24 | 33 | |
| Positive | 68 | 31 | 37 | |
| Lymph node metastasis | 0.015 | |||
| Negative | 53 | 30 | 23 | |
| Positive | 72 | 25 | 47 | |
| TNM stage | 0.021 | |||
| I–II | 65 | 35 | 30 | |
| III–IV | 60 | 20 | 40 | |
Abbreviations: ER, estrogen receptor; PR, progesterone receptor.
Cell lines and transfection
The breast cancer cell lines (MDA-MB-231, MCF-7) and normal breast cell line MCF10A were obtained from the American Type Culture Collection (Manassas, VA, USA). These breast cell lines were maintained in Roswell Park Memorial Institute 1640 (RPMI 1640) with 10% fetal bovine serum and 1% antibiotics (Invitrogen, Carlsbad, CA, USA). MCF-10A cells were cultured in mammary epithelial cell growth medium (MEGM) (CC-3150, Lonza, Basel, Switzerland) containing 100 ng/mL of cholera toxin to make a complete growth culture medium. All cells were cultured in a humidified incubator at 37°C with 5% CO2. A total of 1×105 cells were seeded into a 6-well plate. Then, the cells were transfected with the miR-638 mimic or mimic negative control (mimic NC), miR-638 inhibitor or negative inhibitor control (inhibitor NC) using the Lipofectamine 2000 regent (Invitrogen) according to the manufacturer’s instructions. Cells transfected with only transfection reagent were used as mock. The transfection efficiency was detected by quantitative real-time PCR (qRT-PCR) analysis.
RNA extraction and qRT-PCR analysis
Total RNA was isolated from tissues or cell lines using TRIzol reagent (Invitrogen) in accordance with the manufacturer’s instructions. Subsequently, purified total RNA was reverse transcribed to cDNA by using M-MLV reverse transcriptase (Promega, Madison, WI, USA) following the manufacturer’s instructions. The qRT-PCR experiment was performed using SYBR Green (Takara, Kusatsu, Japan) on an ABI 7500 system (Applied Biosystems, Foster City, CA, USA). The relative miR-638 levels were detected from the threshold cycle with the 2−ΔΔCt method using the U6 snRNA as an endogenous control. All the experiments were performed in triplicate.
MTT assay
The colorimetric MTT analysis was performed to examine the effects of miR-638 on cell proliferation of breast cancer cells. In brief, the cells were grown in 96-well plates and transfected. After transfection and incubation for 5 days, to each well was added 10 µL MTT (5 mg/mL, Sigma-Aldrich, St Louis, MO, USA) and incubated at 37°C. After 4 hours incubation, the supernatant was removed and 150 µL DMSO (Sigma-Aldrich) was added to each well to dissolve formazan crystals. The absorbance was measured at 490 nm using a spectrophotometer (Multiskan MK3, Thermo Fisher Scientific, Waltham, MA, USA). The experiment was repeated at least three times.
Transwell migration and invasion assay
To measure the effects of miR-638 on breast cancer cell migration and invasion ability, a transwell kit (Corning, Tewksbury, MA, USA, 8.0 µm pores) was used with a 24-well transwell chamber according to the manufacturer’s instruction. A total of 5×104 cells transfected with miR-638 mimic or NC, miR-638 inhibitor or NC, or mock were placed in the upper chamber in RPMI 1640 with no serum. The bottom chamber contained RPMI 1640 with 10% fetal bovine serum, using as the chemotactic factor. For the invasion assay, the upper chamber was first coated with Matrigel (BD Company, Bedford, MA, USA). After incubating for 24 hours, the cells that had migrated or invaded to the bottom chamber were fixed in 3.7% formaldehyde for 5 minutes and stained with 0.1% crystal violet for 15 minutes. The number of cells on the bottom of the chamber was counted under a microscope (Olympus, Tokyo, Japan). The process of the transwell migration assay was similar to the transwell invasion assay except for the Matrigel-coated membrane.
Statistical analysis
Statistical analysis of data was performed using the SPSS 21.0 software (IBM Corporation, Armonk, NY, USA), and the data were expressed as the mean ± SD at least three times. Student’s t-test was performed to analyze the significance of differences between the two groups. The association between miR-638 expression and clinicopathological parameters of patients was analyzed by chi-squared test. Kaplan–Meier method was used to analyze the association between overall survival and miR-638 expression in breast cancer patients according to the log-rank test. Cox regression analysis was used to assess the prognostic factors. Differences were considered statistically significant when P<0.05.
Results
Relative expression of miR-638 in breast cancer tissues and cell lines
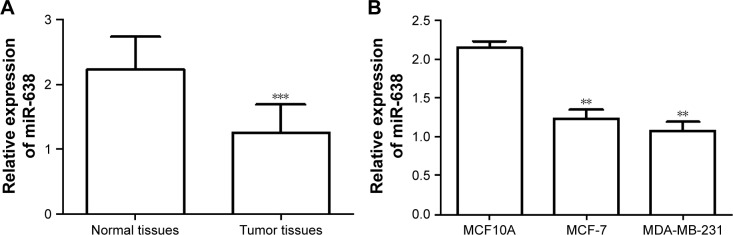
We used qRT-PCR analysis to measure the miR-638 expression levels in breast cancer tissues. As shown in Figure 1A, the expression level of miR-638 was significantly lower in breast cancer tissues than in normal breast tissues (P<0.001).
Figure 1.
Expression of miR-638 in breast cancer tissues and cell lines was measured by qRT-PCR.
Notes: (A) The expression levels of miR-638 were lower in breast cancer tissues than in normal tissues. (B) The level of miR-638 in breast cancer cell lines (MDA-MB-231 and MCF-7) was downregulated compared with normal breast cell line MCF10A. **P<0.01, ***P<0.001.
Abbreviation: qRT-PCR, quantitative real-time polymerase chain reaction.
The expression of miR-638 was also evaluated in two breast cancer cell lines, MDA-MB-231 and MCF-7, compared to a normal breast cell line MCF10A. The result showed that the expression level of miR-638 was decreased significantly in breast cancer cell lines than in normal cell line (all P<0.01, Figure 1B).
Correlation of miR-638 expression and clinical characteristics
According to the mean value of miR-638 expression levels in breast cancer tissues, which was used as the cutoff value, we classified the patients into low miR-638 expression group (n=70) and high expression group (n=55). As shown in Table 1, the expression of miR-638 was significantly associated with lymph node metastasis (P=0.015) and TNM stage (P=0.021). There was no dramatical association between miR-638 expression and other clinical parameters, such as age, tumor size, ER status, PR status, and HER2 status (all P>0.05, Table 1).
Prognostic significance of miR-638 in breast cancer
We used the Kaplan–Meier analysis to evaluate the association of miR-638 expression with overall survival of breast cancer patients. As shown in Figure 2, the breast cancer patients with lower miR-638 expression in tumor tissues suffered from unfavorable overall survival compared with those with higher miR-638 expression (Log-rank P=0.025, Figure 2). Furthermore, multivariate Cox regression analysis results indicated that miR-638 expression (HR =0.321, 95% CI =0.117–0.882, P=0.027, Table 2) was an independent prognostic factor for breast cancer patients.
Figure 2.
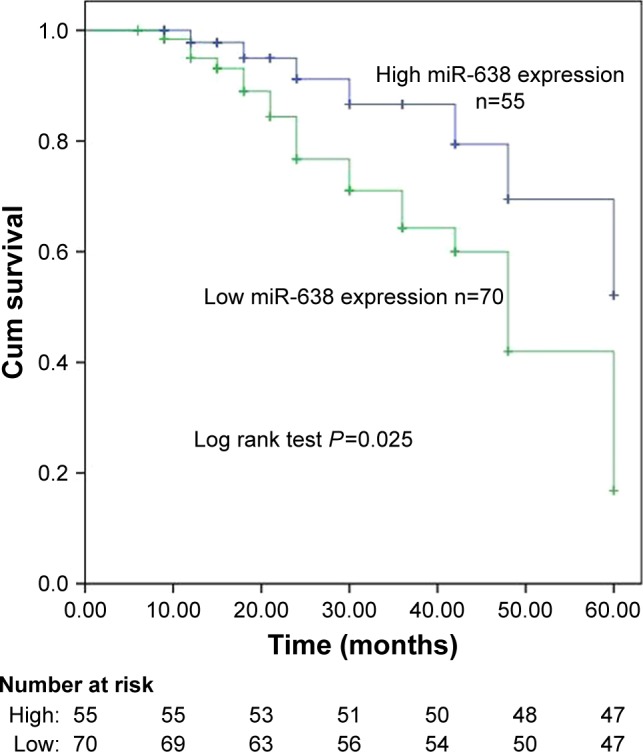
The prognosis of miR-638 in breast cancer.
Note: Patients with low miR-638 expression showed shorter overall survival than those with high expression levels.
Table 2.
Univariate and multivariate Cox analyses for miR-638 in breast cancer patients
| Characteristics | Univariate analyses
|
Multivariate analysis
|
||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value | |
|
| ||||||
| miR-638 | 0.325 | 0.119–0.886 | 0.028 | 0.325 | 0.119–0.886 | 0.028 |
| Age | 1.828 | 0.857–3.900 | 0.119 | – | – | – |
| Tumor size | 1.067 | 0.464–2.454 | 0.879 | – | – | – |
| ER status | 1.221 | 0.547–2.728 | 0.626 | – | – | – |
| PR status | 0.798 | 0.323–1.970 | 0.625 | – | – | – |
| HER-2 status | 1.634 | 0.743–3.592 | 0.222 | – | – | – |
| Lymph node metastasis | 0.799 | 0.337–1.894 | 0.611 | – | – | – |
| TNM stage | 1.521 | 0.721–3.210 | 0.271 | – | – | – |
Note: –, indicated no related data.
Abbreviations: ER, estrogen receptor; HR, hazard ratio; PR, progesterone receptor.
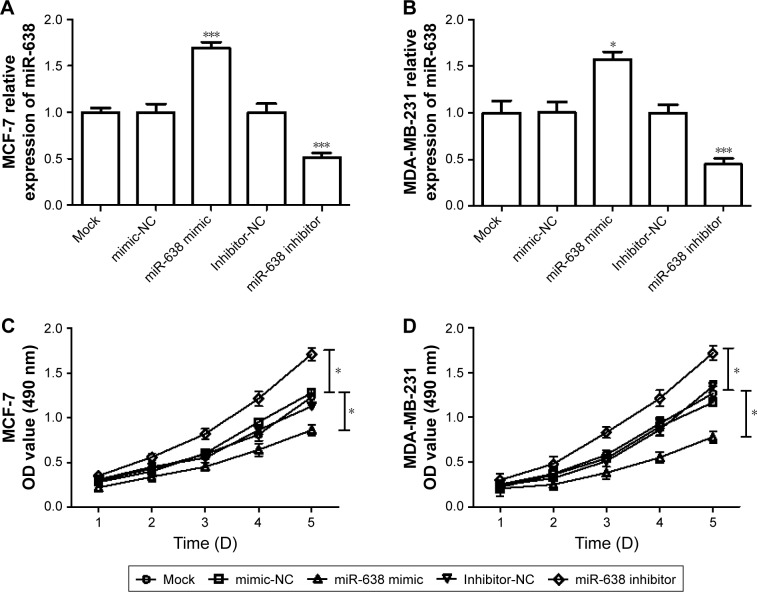
Overexpression of miR-638 can suppress breast cancer cell proliferation, migration, and invasion
To investigate the function of miR-638 in breast cancer cell lines, two breast cancer cell lines, MDA-MB-231 and MCF-7, were transfected with miR-638 mimic or mimic NC, miR-638 inhibitor or inhibitor NC. The qRT-PCR analysis results indicated that miR-638 mimic could increase the expression of miR-638 in MDA-MB-231 and MCF-7 cells, and miR-638 inhibitor can decrease the expression of miR-638 in MDA-MB-231 and MCF-7 cells, respectively (P<0.05, Figure 3A and B). MTT assays were then performed to investigate the effects of miR-638 on cell growth in vitro. We observed that the proliferation of cells was inhibited in miR-638 mimic-transfected cells and promoted in miR-638 inhibitor-transfected cells (P<0.05, Figure 3C and D).
Figure 3.
Expression of miR-638 expression on proliferation of breast cancer cells.
Notes: (A, B) qRT-PCR was used to detect the transfection efficiency of miR-638 mimic or inhibitor in MDA-MB-231 and MCF-7 cells. (C, D) MTT assay was used to determine the cell proliferation. *P<0.05, ***P<0.001.
Abbreviation: qRT-PCR, quantitative real-time polymerase chain reaction.
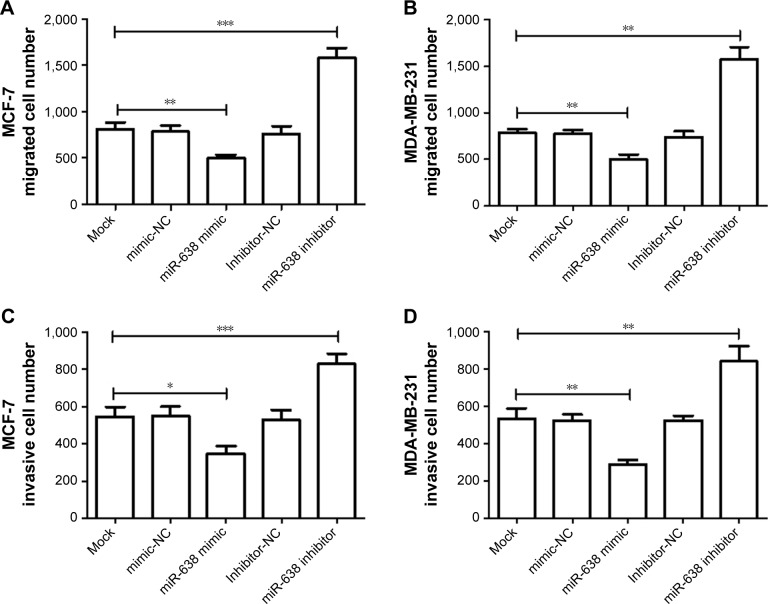
To further investigate whether miR-638 affects cell metastasis, transwell assay was used in this study. The transwell migration assay results showed that cell migration was inhibited in cells transfected with miR-638 mimic, and promoted in the cells transfected with miR-638 inhibitor (P<0.05, Figure 4A and B). The transwell invasion assay with Matrigel results demonstrated that miR-638 mimic could inhibit the cell invasion, and miR-638 inhibitor could promote the cell invasion (P<0.05, Figure 4C and D).
Figure 4.
Expression of miR-638 expression on migration and invasion of breast cancer cells.
Notes: (A, B) Transwell migration assay were performed in breast cancer cell lines (MDA-MB-231 and MCF-7) transfected with miR-638 mimic or mimic NC, inhibitor or inhibitor NC. (C, D) Transwell invasion assay were performed in breast cancer cell lines (MDA-MB-231 and MCF-7). *P<0.05, **P<0.01, ***P<0.001.
Abbreviation: NC, negative control.
Discussion
The incidence of breast cancer has increased rapidly with economic development in People’s Republic of China in recent years.24 Breast cancer patients who are diagnosed and received treatment at an early stage usually have longer overall survival, while those at advanced stage usually relapse or recur, leading to poor prognosis.25 Nowadays, the incidence of breast cancer in young women is more aggressive and invasive than that in older patients, leading to an adverse prognosis.26,27 Despite the advances of both detection and treatment, the rates of breast cancer remain high in most countries. Therefore, more accurate cancer-related biomarkers are needed to help predict patient prognosis and treatment strategies.
An increasing number of molecular markers, including miRNAs, have been identified play crucial roles in tumor pathogenesis and development, acting as biomarkers for diagnosis, prognosis, or treatment of cancers.28,29 Increasing miRNAs have been reported as new biological markers in cancers. For instance, Shidfar et al30 found that miR-18a and miR-210 were significantly highly expressed in breast cancer, and could be used as a candidate for breast cancer risk biomarkers. Marques et al31 evaluated miR-195 and let-7a expression in invasive breast cancer and showed miR-195 and let-7a could be used as a noninvasive biomarker for breast cancer detection. The study by He et al32 indicated that miR-638 might act as a sensitizer in cancer chemotherapy and accompany chemotherapy drugs to enhance chemotherapeutic efficacy. To better understand the role of miR-638 in breast cancer, its expression patterns, and prognostic significance, this study was carried out in breast cancer patients.
The study by Zavala et al33 investigated the expression of miR-638 in TNBC and found that miR-638 was overexpressed in most TNBC tumors, consistent with the result in the TNBC samples of Tan et al.23 In our study, we mainly measured the expression of miR-638 in the breast cancer tissues and cell lines. Similar to the non-TNBC result of Tan et al,23 expression of miR-638 was significantly downregulated in breast cancer tissues, as well as in cell lines, compared to normal control. Moreover, the decreased expression of miR-638 was correlated with lymph node metastasis and TNM stage. Taken together, we considered that miR-638 might be a suppressor gene in breast cancer and be involved in the progression of this malignancy. Furthermore, we investigated the clinical significance of miR-638 in breast cancer prognosis using the Kaplan–Meier analysis and Cox regression analysis. The Kaplan–Meier result with the data from follow-up showed that patients with low expression of miR-638 had a shorter overall survival rate than those with high miR-638 expression, which suggested that miR-638 expression was related to the prognostic value for patients with breast cancer. Multivariate Cox regression analysis results showed that miR-638 was an independent biomarker for indicating the overall survival of breast cancer patients.
To investigate the function of miR-638 in breast cancer progression, the effects of miR-638 on biological behaviors of breast cancer were also analyzed. The analysis results demonstrated that cell proliferation, migration, and invasion were suppressed by the overexpression of miR-638, which showed the potential functional role of miR-638 in breast cancer progression. Many of the studies have indicated the effects of miR-638 on biological behaviors during cancer progression.22,34 For instance, the study by Xia et al22 showed that downregulated miR-638 could promote cell invasion and proliferation in NSCLC, indicating that miR-638 may play a pivotal role in the development of NSCLC. Zhang et al34 revealed that miR-638 inhibited colorectal carcinoma cell growth, invasion, and functioned as a tumor suppressor in human colorectal cancer by inhibiting TSPAN1. The study by Zhao et al35 found that miR-638 was downregulated in gastric cancer, and molecular mechanistic investigation results demonstrated that MeCP2, a target of miR-638, facilitated gastric cancer cell proliferation through activation of the MEK1/2-ERK1/2 signaling pathway by upregulating GIT1. Besides the cell experiments, the precise molecular mechanistic investigations underlying role of miR-638 in breast cancer need to be assessed in the further studies. Also, due to the limitation of sample size in the present study, further studies are necessary with a larger research cohort.
In conclusion, we identified that expression of miR-638 was downregulated in breast cancer tissue and was correlated with the progression of breast cancer. This study results showed that miR-638 might be an independent prognostic biomarker for breast cancer.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.di Cosimo S, Baselga J. Management of breast cancer with targeted agents: importance of heterogeneity. [corrected] Nat Rev Clin Oncol. 2010;7(3):139–147. doi: 10.1038/nrclinonc.2009.234. [DOI] [PubMed] [Google Scholar]
- 3.Metzger-Filho O, Tutt A, de Azambuja E, et al. Dissecting the heterogeneity of triple-negative breast cancer. J Clin Oncol. 2012;30(15):1879–1887. doi: 10.1200/JCO.2011.38.2010. [DOI] [PubMed] [Google Scholar]
- 4.Yang Q, Liu HY, Liu D, Song YQ. Ultrasonographic features of triple-negative breast cancer: a comparison with other breast cancer subtypes. Asian Pac J Cancer Prev. 2015;16(8):3229–3232. doi: 10.7314/apjcp.2015.16.8.3229. [DOI] [PubMed] [Google Scholar]
- 5.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331(6024):1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 6.Scully OJ, Bay BH, Yip G, Yu Y. Breast cancer metastasis. Cancer Genomics Proteomics. 2012;9(5):311–320. [PubMed] [Google Scholar]
- 7.Sørlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu RZ, Garcia E, Glubrecht DD, Poon HY, Mackey JR, Godbout R. CRABP1 is associated with a poor prognosis in breast cancer: adding to the complexity of breast cancer cell response to retinoic acid. Mol Cancer. 2015;14:129. doi: 10.1186/s12943-015-0380-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10.Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8(2):93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- 11.Li P, Dong J, Zhou X, et al. Expression patterns of microRNA-329 and its clinical performance in diagnosis and prognosis of breast cancer. Onco Targets Ther. 2017;10:5711–5718. doi: 10.2147/OTT.S147974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang Y, Jian W, Zhao J, Wang G. Overexpression of HDAC9 is associated with poor prognosis and tumor progression of breast cancer in Chinese females. Onco Targets Ther. 2018;11:2177–2184. doi: 10.2147/OTT.S164583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh RP, Massachi I, Manickavel S, et al. The role of miRNA in inflammation and autoimmunity. Autoimmun Rev. 2013;12(12):1160–1165. doi: 10.1016/j.autrev.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 14.Chen L, Song J, Cui J, et al. microRNAs regulate adipocyte differentiation. Cell Biol Int. 2013;37(6):533–546. doi: 10.1002/cbin.10063. [DOI] [PubMed] [Google Scholar]
- 15.Lu GJ, Dong YQ, Zhang QM, et al. miRNA-221 promotes proliferation, migration and invasion by targeting TIMP2 in renal cell carcinoma. Int J Clin Exp Pathol. 2015;8(5):5224–5229. [PMC free article] [PubMed] [Google Scholar]
- 16.Viñas JL, Ventayol M, Brüne B, et al. miRNA let-7e modulates the Wnt pathway and early nephrogenic markers in mouse embryonic stem cell differentiation. PLoS ONE. 2013;8(4):e60937. doi: 10.1371/journal.pone.0060937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farazi TA, Hoell JI, Morozov P, Tuschl T. MicroRNAs in human cancer. Adv Exp Med Biol. 2013;774:1–20. doi: 10.1007/978-94-007-5590-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang YX, Gao WQ. Roles of microRNAs during prostatic tumorigenesis and tumor progression. Oncogene. 2014;33(2):135–147. doi: 10.1038/onc.2013.54. [DOI] [PubMed] [Google Scholar]
- 19.Jiang C, Chen X, Alattar M, Wei J, Liu H. MicroRNAs in tumorigenesis, metastasis, diagnosis and prognosis of gastric cancer. Cancer Gene Ther. 2015;22(6):291–301. doi: 10.1038/cgt.2015.19. [DOI] [PubMed] [Google Scholar]
- 20.Yao Y, Suo AL, Li ZF, et al. MicroRNA profiling of human gastric cancer. Mol Med Rep. 2009;2(6):963–970. doi: 10.3892/mmr_00000199. [DOI] [PubMed] [Google Scholar]
- 21.Sand M, Skrygan M, Sand D, et al. Expression of microRNAs in basal cell carcinoma. Br J Dermatol. 2012;167(4):847–855. doi: 10.1111/j.1365-2133.2012.11022.x. [DOI] [PubMed] [Google Scholar]
- 22.Xia Y, Wu Y, Liu B, Wang P, Chen Y. Downregulation of miR-638 promotes invasion and proliferation by regulating SOX2 and induces EMT in NSCLC. FEBS Lett. 2014;588(14):2238–2245. doi: 10.1016/j.febslet.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 23.Tan X, Peng J, Fu Y, et al. miR-638 mediated regulation of BRCA1 affects DNA repair and sensitivity to UV and cisplatin in triple-negative breast cancer. Breast Cancer Res. 2014;16(5):435. doi: 10.1186/s13058-014-0435-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong W, Dong E. The past, present and future of breast cancer research in China. Cancer Lett. 2014;351(1):1–5. doi: 10.1016/j.canlet.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 25.Naumov GN, Townson JL, Macdonald IC, et al. Ineffectiveness of doxorubicin treatment on solitary dormant mammary carcinoma cells or late-developing metastases. Breast Cancer Res Treat. 2003;82(3):199–206. doi: 10.1023/B:BREA.0000004377.12288.3c. [DOI] [PubMed] [Google Scholar]
- 26.Darwish AD, Helal AM, Aly El-Din NH, Solaiman LL, Amin A. Breast cancer in women aging 35 years old and younger: the Egyptian National Cancer Institute (NCI) experience. Breast. 2017;31:1–8. doi: 10.1016/j.breast.2016.09.018. [DOI] [PubMed] [Google Scholar]
- 27.Slaoui M, Mouh FZ, Ghanname I, Razine R, El Mzibri M, Amrani M. Outcome of breast ZC cancer in moroccan young women correlated to clinic-pathological features, risk factors and treatment: a comparative study of 716 cases in a single institution. PLoS ONE. 2016;11(10):e0164841. doi: 10.1371/journal.pone.0164841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jagarlamudi KK, Shaw M. Thymidine kinase 1 as a tumor biomarker: technical advances offer new potential to an old biomarker. Biomark Med. 2018;12(9):1035–1048. doi: 10.2217/bmm-2018-0157. [DOI] [PubMed] [Google Scholar]
- 29.Givechian KB, Garner C, Garban H, Rabizadeh S, Soon-Shiong P. CAD/POLD2 gene expression is associated with poor overall survival and chemoresistance in bladder urothelial carcinoma. Oncotarget. 2018;9(51):29743–29752. doi: 10.18632/oncotarget.25701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shidfar A, Costa FF, Scholtens D, et al. Expression of miR-18a and miR-210 in normal breast tissue as candidate biomarkers of breast cancer risk. Cancer Prev Res. 2017;10(1):89–97. doi: 10.1158/1940-6207.CAPR-16-0177. [DOI] [PubMed] [Google Scholar]
- 31.Marques MM, Evangelista AF, Macedo T, et al. Expression of tumor suppressors miR-195 and let-7a as potential biomarkers of invasive breast cancer. Clinics. 2018;73:e184. doi: 10.6061/clinics/2018/e184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He M, Lin Y, Tang Y, et al. miR-638 suppresses DNA damage repair by targeting SMC1A expression in terminally differentiated cells. Aging. 2016;8(7):1442–1456. doi: 10.18632/aging.100998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zavala V, Pérez-Moreno E, Tapia T, Camus M, Carvallo P. miR-146a and miR-638 in BRCA1-deficient triple negative breast cancer tumors, as potential biomarkers for improved overall survival. Cancer Biomark. 2016;16(1):99–107. doi: 10.3233/CBM-150545. [DOI] [PubMed] [Google Scholar]
- 34.Zhang J, Fei B, Wang Q, et al. MicroRNA-638 inhibits cell proliferation, invasion and regulates cell cycle by targeting tetraspanin 1 in human colorectal carcinoma. Oncotarget. 2014;5(23):12083–12096. doi: 10.18632/oncotarget.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao LY, Tong DD, Xue M, et al. MeCP2, a target of miR-638, facilitates gastric cancer cell proliferation through activation of the MEK1/2-ERK1/2 signaling pathway by upregulating GIT1. Oncogenesis. 2017;6(7):e368. doi: 10.1038/oncsis.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]