Abstract
Introduction
The members of the family Flaviviridae, including West Nile virus, yellow fever virus, and dengue virus, are important human pathogens that are expanding their impact around the globe. The four serotypes of dengue infect 50 to 100 million people each year, yet the only clinical treatment is supportive care to reduce symptoms. Drugs that employ novel inhibition mechanisms and targets are urgently needed to combat the growing incidence of dengue worldwide.
Areas Covered
The authors discuss recently discovered flavivirus inhibitors with a focus on antivirals targeting non-enzymatic proteins of the dengue virus lifecycle. Specifically, the authors discuss the flaviviruses, the need for novel inhibitors, and the criteria for successful antiviral drug development. Current literature describing new advances in antiviral therapy at each stage of the flavivirus life cycle (entry, endosomal escape, viral RNA processing and replication, assembly, and immune evasion) are evaluated and summarized.
Expert Opinion
Overall, the prognosis of flavivirus antiviral drug development is positive: new effective compounds have been discovered and studied. However, repurposing existing compounds and a greater translation to the clinical setting are recommended in order to combat the growing threat of flaviviruses.
Keywords: antiviral, compound, dengue, inhibitor, peptide, small-molecule
1. Introduction
1.1 The flavivirus genus
The flavivirus genus is a member of the family Flaviviridae, which consists primarily of arthropod-borne human pathogens that infect millions of people each year. This genus includes dengue virus (DENV), West Nile virus (WNV), yellow fever virus (YFV), Japanese encephalitis virus (JEV), Kunjin virus (KUNV), and tick-borne encephalitis virus (TBEV) among others, and is closely related to the genus hepacivirus, whose sole member is hepatitis C virus (HCV). The four serotypes of DENV alone infect 50 to 100 million people each year, resulting in approximately 22,000 deaths annually. Upon DENV infection, patients first experience flu-like symptoms such as high fever, chills, nausea, joint pain, and dizziness. A few of the infections can progress to life-threatening dengue hemorrhagic fever and shock syndrome (DHF/DSS), characterized by rapid deterioration including rash, shock-like state, pinpoint dots of blood under the skin, drop in blood pressure, and amplification of primary symptoms (1). Over 100 countries are plagued by endemic DENV infection, mostly in the tropical and sub-tropical regions of the globe. The recent outbreaks of WNV in North America in particular, and the re-emergence of seemingly conquered YFV across the globe, have brought these viruses back to public awareness, if indeed they ever left (2).
Vaccines have been integral in combating YFV, JEV, and TBEV. However, even these strains have shown re-emergence in recent years most likely due to lax mosquito control, increased transportation of people and goods, unreachable populations, and the decline of vaccination in public opinion. For several members of the flavivirus genus, including DENV and WNV, vaccine development has stymied scientists despite many years of research. Although several clinical trials for vaccines are underway (1,3,4), the years that go into development require an interim substitute until such vaccines can be marketed.
Development of a vaccine for DENV presents a unique challenge. Not only would a vaccine need to address each of the four serotypes of DENV, but it would also have to overcome the potential for antibody-dependent enhancement (ADE). During a primary DENV infection, cross-reactive, non-neutralizing antibodies are produced. When an individual is infected with another DENV serotype, these antibodies can recognize, bind, but not neutralize the second infecting virus. The antibody-virus complexes can be internalized by Fc antibody receptors present on specific host cells. Thus these antibodies likely increase viremia by facilitating virus uptake through a novel mechanism and into a new subset of host cells (5,6).
Additionally, vaccination is only effective for those inoculated before infection with the virus. Those that lack such prevention and succumb to infection have no options for care beside treatment of symptoms. Countries that are already struggling to provide healthcare are stretched especially thin during DENV epidemics, and even such supportive treatment proves difficult (1). Clearly there is an urgent need for the development of antiviral drugs that will allow health professionals to cure, or at least diminish, these viruses after infection. Several studies have produced candidate drugs, such as ribavirin and its derivatives (7), that have passed the rigors of clinical trials to become at least rudimentary options for virus treatment. However, many of these treatments have serious limitations, and resistance is a constant concern (8). Not only are new drugs needed, but novel approaches to the development of effective antiviral drugs are crucial to control flavivirus infections.
1.2 The flavivirus proteins
Flaviviruses contain a single-stranded, positive-sense, 11KB RNA genome. The genomic RNA is translated into a single polyprotein that is cleaved by viral and cellular proteases to produce 10 viral proteins. Three proteins: capsid (C), pre-membrane (prM), and envelope (E) are structural proteins that facilitate assembly and budding. Together with cellular lipids and genomic RNA, these are the proteins that constitute the viral particle. The remaining C-terminal 7 proteins (NS1, NS2A/B, NS3, NS4A/B, NS5) are required for replication of the genomic RNA. The C protein interacts with the genomic RNA to promote packaging into immature virions. The prM and E proteins are embedded in the cellular membrane prior to budding and form the projections of the immature virion. The prM protein functions as an M protein precursor that primarily prevents premature rearrangement of the E protein under the mildly acidic conditions of the trans-Golgi prior to virion release. In the trans-Golgi network, the prM protein is cleaved by cellular furin to allow M/E rearrangement to produce the mature virion (9,10). The E protein of the released mature virion subsequently recognizes an unknown receptor on target host cells to induce viral uptake. After uptake, it mediates low pH-mediated membrane fusion to release the genomic RNA into the cytoplasm for replication. (2, 11, 12).
The nonstructural proteins are not found in the virion, but rather are found primarily in the cytoplasm and consist of the protease, helicase, polymerase, and other necessary proteins of RNA replication. Nonstructural protein 1 (NS1) is a current subject of study. It has primarily been implicated in host immune response evasion through modulation of complement activation although other activities are suspected (13, 14, 15). Likewise, the function of the NS2A protein is poorly defined. It has been implicated in a variety of roles, including immune response evasion, genomic replication, and even assembly (16, 17, 18). NS2B functions as a cofactor protein in the protease function of NS3, which also doubles as a helicase (19). The function of NS4A is a matter of debate, but studies suggest that it has a role in induction of membrane rearrangement and/or autophagy response to viral infection of host cells (20, 21). The NS4B membrane protein is thought to anchor and target the replication complex to the Endoplasmic Reticulum (ER) membrane and has been connected to immune response antagonism (22, 23). NS5 is the largest of the nonstructural proteins and it contains a classic RNA-dependent RNA polymerase (RdRp) domain as well as methyltransferase and guanylyltransferase domains for mRNA capping necessary for a virus that replicates its mRNA in the cytoplasm (2, 24, 25, 26).
1.3 The need for novel antiviral drugs
Traditionally, antiviral strategies attempt to target the enzymatic replication proteins such as helicase (NS3), protease (NS2B/3), and polymerase (NS5). This is largely because loss of these specific proteins has proven lethal for virus replication, and therefore drugs targeting these enzymes can be particularly effective at reducing viral load (17, 23, 27). Additionally, host cells do not express RdRp’s such as NS5, hence nonspecific detrimental effects of these drugs would theoretically be minimal. The same is not true for NS3, however, as molecules designed to target the WNV and DENV NS3 helicase would benefit from improved specificity (28). Furthermore, new viral targets are constantly in demand due to ready mutation of both NS3 and NS5 enzymes in the presence of inhibitors to regain function, as demonstrated in HCV (8, 24). It is of interest, therefore, to investigate new methods for targeting these infections, including pursuing novel targets in the viral lifecycle, such as entry, non-enzymatic replication proteins, membrane interaction, and others. Targeting non-enzymatic components of the virus may minimize both the appearance of resistance mutations and poor specificity, as well as allow synergistic use with anti-enzymatic drugs to address infection more efficiently in the clinical setting (28).
Effective development of candidate drugs takes several factors into account. First, a certain level of inhibition is desired; ideally a drug will eliminate virus replication. In practice, 50–90% inhibition is optimal during drug design. Even a 50% reduction in virus replication can reduce the viral load to a manageable level so that the host immune system can readily clear it. Second, the drug should ideally be target specific. Binding to, or otherwise inhibiting, an unintended target in the host cell can lead to detrimental side-effects, even causing cell death. The drug should therefore only inhibit the host or viral proteins for which it is intended. In order to consider this during drug development, cell cytotoxicity (cell death) is monitored in addition to overall potency of the drug. Also, efforts are made to target unique proteins, essential for the virus lifecycle, such as the RNA-dependent RNA polymerase (RdRp) that is not present in an uninfected host cell. Third, an ideal drug would be able to act effectively on multiple viruses in the family (broad spectrum inhibition). In practice, however, even the evolutionarily related flaviviruses prove broad spectrum drugs are difficult to generate. Fourth, an ideal drug is readily able to enter cells, whether though active transport, diffusion, or as a prodrug. Fifth, an ideal drug would be fast acting. Patients infected with DENV often do not show symptoms for 4–10 days, and therefore drugs need to prevent viral spread relatively late in the infection cycle.
1.4 The flavivirus life cycle
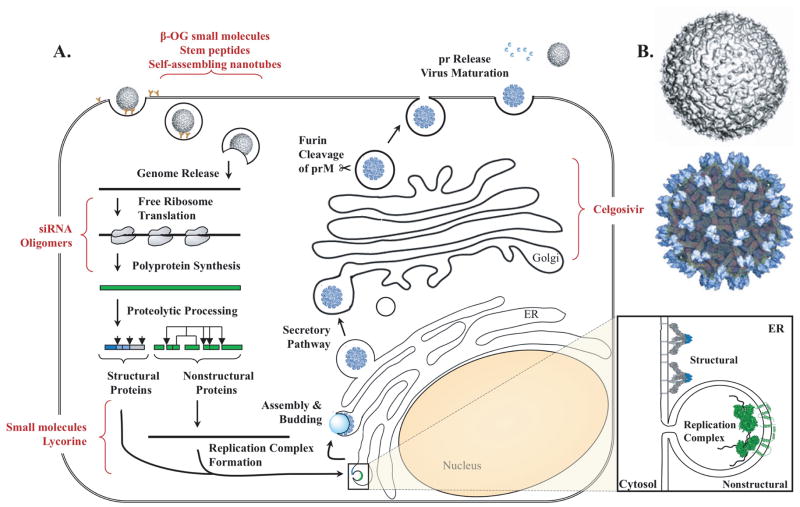
Flaviviruses begin uptake into host cells through the interaction of E protein with an unknown cellular receptor. Contact between E protein and the receptor induces clathrin-mediated endocytosis that transports the virion into the cytoplasm (29–32). The endocytic vesicle containing the virion undergoes a change in pH that causes a rearrangement of the E protein required to fuse the viral and cellular membranes, thereby releasing the RNA genome into the cytoplasm (2, 33, 34). The genomic RNA is subsequently translated into a long polyprotein which is auto-catalytically cleaved by the NS2B/3 viral protease and host proteases into its substituent proteins (2). The released nonstructural proteins are targeted to the site of replication on ER-derived vesicle packets to initiate transcription (35, 36). Meanwhile, the prM and E proteins are embedded into the ER membrane and enclose the newly formed nucleocapsid as it buds into the ER lumen to form the immature particle. The particle is trafficked to the plasma membrane via the secretory pathway. The low pH of the trans-Golgi network causes substantial rearrangement of the prM/E proteins and permits furin cleavage of prM to M. With the release of the pr peptide, the virion is released from the cell and is able to infect another naïve host cell (37–46). The inhibitors discussed subsequently target various stages of the flavivirus life cycle (Figure 1).
Figure 1.
DENV lifecycle and drug targets. A. DENV enters via receptor-mediated endocytosis. Low pH E rearrangement releases the viral genome. The viral RNA is translated into a polyprotein which is proteolytically processed into the nonstructural proteins after translocation into the Endoplasmic Reticulum (ER). The replication complex is formed and transcription begins. The newly synthesized RNA associates with C and collects prM and E as it buds into the lumen of the ER to form the immature particle. The particle enters the secretory pathway. In the trans-golgi network, prM is clipped by furin but remains associated due to the low pH. In the neutral pH of the extracellular milieu, pr is released, and the now mature virion is capable of infecting a naïve host cell. B. Structure of DENV mature virion (above) and immature particle (below). Figure adapted from Perera et al. (43) copyright 2002 with permission from Elsevier. Also adapted by permission from Macmillan Publishers Ltd: Nature Reviews Microbiology Mukhopadhyay et al. (90), copyright 2005, and partially derived from Miller and Krijnse-Locker (91), copyright 2008. Amended with permission from American Society for Microbiology from Quinkert et al. 2005 (92). PDB identifiers 2BHR, 2J7W, 1L9K, 3C6E.
2. Novel inhibitors of the flaviviruses
2.1 Entry
Entry of the virus into host cells is necessary for the virus to produce progeny since it has inadequate replication machinery on its own. An inhibitor that prevents entry into host cells is attractive because such a strategy potentially circumvents cytotoxicity and a variety of negative effects on the cell cycle, as well as leaving the virion vulnerable to immune system clearance. Furthermore, since the E protein is responsible for both entry and membrane fusion, targeting the E protein has additional potential to inhibit downstream steps in the lifecycle such as endosomal release, should the virion evade entry inhibition.
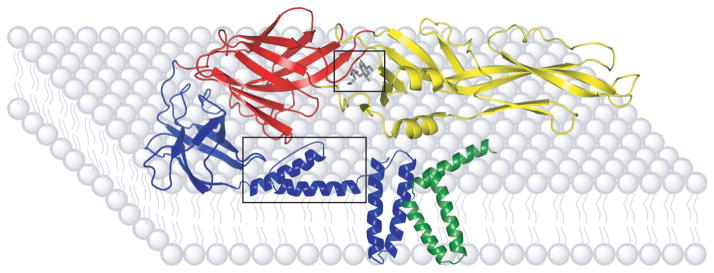
The E protein’s major conformational changes and well defined molecular structures, both pre- and post-fusion, present several targets amenable to inhibitor design (Figure 2). In particular, the crystal structure of the E protein displays a ligand-binding pocket that was occupied by a detergent molecule, n-octyl-β-D-glucoside (β-OG) (32). This discovery led to a veritable explosion of docking studies to identify and optimize potential inhibitors targeting this region of E (44-7).
Figure 2.
Sites of DENV E protein Inhibition. The E protein domains are indicated by color; domain I in red, domain II in yellow, and domain III in blue. The relative position of the M protein is indicated by green shading. The β-OG pocket and stem region (boxed) are common sites of E protein targeting. Stem and M protein structure and location determined from secondary structural predictions and cryo-electron microscopy densities. Figure adapted from Modis et al. 2003 (32) copyright National Academy of Sciences, U.S.A. Adapted by permission from Macmillan Publishers Ltd: Nature Structural Biology, Zhang et al. (42), copyright 2003, and Nature Reviews Microbiology, Mukhopadhyay et al. (90), copyright 2005.
One such computational study screened an NCI compound library to fit the β-OG pocket with an appropriate inhibitor. Using luciferase-tagged YFV constructs, the 23 top hits were narrowed to three top compounds that showed inhibition in the micromolar range (48). From these three compounds, several series of derivatives have been designed, including an inhibitor with a Selective Index (SI) of 256 (48–50). It is likely that these small molecule inhibitors prevent the E protein conformational change necessary for entry and uncoating and perhaps the late assembly maturation rearrangement. Further investigation is ongoing to confirm binding of these derivates to the β-OG pocket, and to define their mechanism of action (49).
A similar study was carried using a docking program to screen 135,000 compounds for inhibition of the E protein via the β-OG pocket (51). The resultant top hits were tested for biological efficacy and cytotoxicity against DENV, KUNV, and YFV, proving that several inhibitors displayed micromolar inhibition across multiple flaviviruses. Cell-based fusion assays were used to prove that at least one compound, A5, directly inhibits E protein-mediated fusion. Further study of this group of inhibitors is needed, as optimization of these compounds could yield even more potent inhibitors (51). However, the results suggest that virtual screening of compound libraries is an effective approach to discovery of novel inhibitors. Additionally, this study confirms that targeting conserved protein regions has the potential to develop broad-spectrum inhibitors useful for treating a variety of flavivirus infections.
2.2 Endosomal escape
In addition to targeting the β-OG pocket, small peptides have been designed against the E protein stem region. The stem is a conserved region at the C-terminus of the protein, adjacent to the membrane-bound anchor region, and is essential for membrane fusion (52, 42). The peptide inhibitors were designed based on conserved sequence-specific binding interactions of the postfusion DENV E. DENV lends itself well to this approach since the E protein sequence of this region is well conserved among DENV 1–4 and other flaviviruses. These peptides are unique in that they inhibit the virus following entry into the cell, rather than prevention of entry itself. It was hypothesized that the peptides are able to bind to the virion nonspecifically as it is taken up by the cell. The exposure of the virus to the low pH of the late endosome, results in conformational rearrangements, inducing tight binding of the peptides, and inhibiting membrane fusion (53–5).
A comparable study performed by Costin, et al. used the pre-fusion DENV-2 E protein for rational design of peptide inhibitors via biologically validated computer modeling techniques (46). Particles treated with the peptidic inhibitors displayed rough outer surface morphology, contrary to the smooth outer surface of mature DENV, thus indicating that the E proteins were likely rearranged. Subsequent attempts to generate a structure for the treated virions suggested that the virion was no longer icosahedral, confirming that the virion structure was grossly affected (55). Furthermore, comparable peptides added before or after attachment to cells yielded similar inhibition and even proved to inhibit ADE in vitro (56). It is likely that binding of these peptides inhibit the interaction of the transmembrane regions and the fusion loop, which has been proposed in other studies (54). These studies not only validate fusion inhibitors as powerful potential antiviral drugs, but also verify the effectiveness of rational de novo small molecule design (55, 56). However, most peptide-based antiviral compounds are not readily absorbed when administered orally, requiring intravenous delivery. This means of treatment is impractical for global use, especially in areas where DENV is most prevalent (44). Internalization of these peptides may be increased through the use of protective liposomes able to deliver the drug directly to the cell. Liposome-based drug delivery can be used to target inhibitors to specific cells as well as deliver the drug in high concentration (57). Furthermore, these peptides should require testing in an in vivo model to evaluate their efficacy during genuine DENV infection.
An exciting new possibility to circumvent peptide instability is presented by self-assembling nanotubes. Such an inhibitor was originally discovered to target bacterial membranes and adenovirus, but has now been applied to HCV (58–61). In the case of HCV, a cyclic D, L-α-peptide library was screened for anti-HCV activity and nine amphiphilic peptides with promise were identified. These peptides self-assemble into inhibitory nanotubes that act after entry but before protein synthesis, and also control spread of the virus in culture. It is likely that they interact with a “specialized cellular membrane” to inhibit either membrane fusion or pH control (62). Although these nanotubes inhibit a cellular membrane, further study could apply them specifically to the virion membrane. Additionally, these proteins are chemically and proteolytically stable, thus they may be amenable to in vivo application. Clearly, more investigation is needed to determine exactly how these peptides are inhibiting HCV, and how to apply them to DENV and related flaviviruses.
2.3 Viral RNA processing
Directly targeting the viral RNA is a tempting approach for antiviral development. However, the flavivirus genome is a positive-sense ssRNA that closely resembles cellular mRNA. Although convenient for the virus, this makes targeting viral RNA (vRNA) without collateral inhibition of cellular mRNA challenging. However, a unique study has been recently published that is able to specifically target the flavivirus vRNA.
Short antisense peptide-conjugated oligomers, called phosphorodiamidate morpholino oligomers (P-PMOs) were designed with short nucleotide sequences able to form Watson-Crick pairs with a complementary target sequence in the DENV and WNV genomes, conjugated with arginine-rich peptides that facilitate uptake in culture (63, 64). These P-PMOs can form short duplexes that are able to inhibit RNA-RNA or RNA-protein interactions in specific regions of the viral genome. Several P-PMOs were designed to target the initial 20 bases of the 5′ UTR of DENV-2, a 3′ cyclization sequence, and a 3′ terminal stem-loop. It was shown that a 5′ UTR targeted oligomer selectively inhibited translation of the viral transcripts, reducing virus production by 95 percent. Similarly, the 3′ cyclization sequence oligomer specifically reduced RNA synthesis by a similar amount. The 3′ stem-loop oligomer reduced both viral RNA synthesis and translation, resulting in an approximately 1000-fold reduction in virus replication. Furthermore, at low concentrations, all the P-PMOs were taken up into the cells and did not significantly affect cellular viability (63–5). These studies provide a novel mechanism of inhibition that neatly circumvents the non-specificity issues of targeting the viral RNA directly. However, these short oligomers are similar in design to siRNAs, and therefore may prove to have a short half-life in an in vivo model. A study investigating the long term effects of these P-PMOs needs to be conducted.
Another novel approach to inhibition of the vRNA involves small interfering RNA (siRNA) inhibition of flaviviruses. E protein targeted siRNAs proved to reduce TBEV particle production by 80 percent (66). Similarly, a study done in YFV targeted siRNAs to a variety of proteins including NS1, E, and NS5 (67). Cells treated with siRNA demonstrated up to 97 percent replication inhibition and even improved the infection outcome in a mouse model system. Although this study focused on YFV rather than DENV, targeting NS1 proves that the siRNA strategy can be applied to viral proteins not previously addressed by traditional approaches (67). Furthermore, the use of siRNA is an appealing strategy for antiviral drug design due to low cytotoxicity and sub-micromolar effective concentrations. However, these molecules have a short half-life in the host and often have difficulty entering the host cell (68). Although application of these powerful RNAs in DENV infection may prove fruitful, further development is needed.
2.4 Genomic Replication
2.4.1 NS4A/2K
Lycorine is a naturally occurring compound in daffodils (Narcissus pseuudonarcissus) and bush lilies (Clivia miniata). This compound has activity against poliovirus, Severe Acute Respiratory Syndrome-associated coronavirus, herpes simplex-1 virus, and enterovirus, primarily reducing protein and RNA synthesis (69–72). In recent studies, it was revealed that this drug reduced DENV, WNV, and YFV titers by 102 to 104 fold at 1.2 μM concentrations (73, 74). Further study selected for resistance in WNV and discovered a point mutation of Val9Met in the 2K peptide that confers resistance to the inhibitor (74). The 2K peptide is a 32 amino acid sequence at the C-terminus of NS4A that is important for targeting to the ER membrane. Upon insertion into the membrane, this 2K sequence is cleaved by virus and host proteases to produce mature NS4A and NS4B proteins (20, 75). It is unclear why interaction with 2K would confer such high susceptibility, especially since the function of 2K, beyond membrane targeting, is unknown. However, it has been hypothesized that 2K inhibition may interfere with membrane rearrangement or cleavage site recognition. Such a hypothesis implies that protease substrate targeting could also provide a wealth of new inhibitor strategies. Additionally, it seems logical that 2K could have another function in the replication complex besides protein targeting, since it localizes with the replication proteins. Therefore, inhibition of 2K requires further investigation for possible antiviral approaches.
2.4.2 NS4B
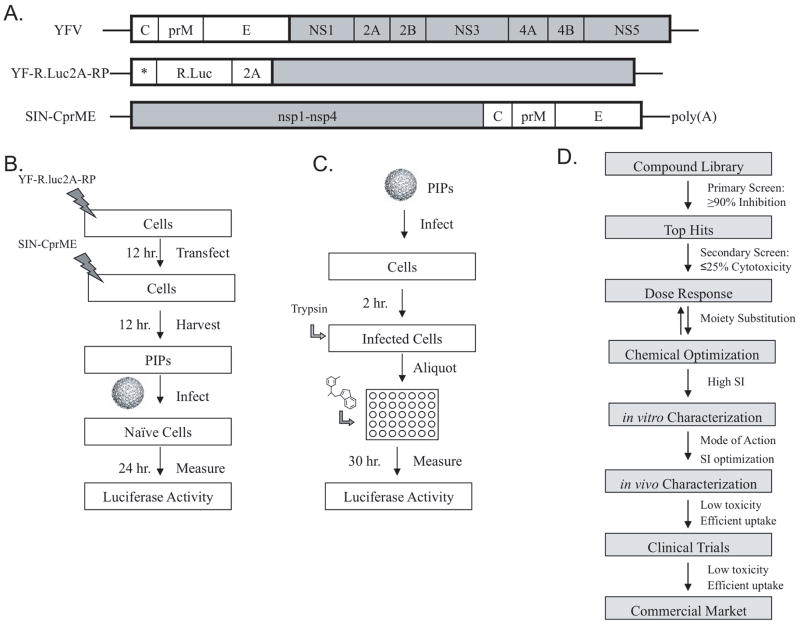
NS4B is an integral membrane protein that inserts into ER-derived membranes at the site of replication, and has proven vital for normal replication function (23, 76). Several inhibitors that interfere with NS4B’s function have been identified, however it is unclear how these molecules specifically inhibit the protein, or even why such inhibition would be adequate to reduce viral load (77, 78). A high-throughput screen (HTS) was performed on YFV using pseudo infectious particles (PIPs) (77). YFV PIPs are constructed by transfecting the YFV luciferase-tagged replicon into cells, later followed by Sindbis virus structural proteins (C, prM, E). Such transfection produces particles able to enter cells and replicate normally but unable to produce progeny virus. Thus, the PIP replicon provides a means to study viral transcription and translation specifically, and is a powerful tool for antiviral drug discovery (Figure 3). The PIP HTS identified twenty potent YFV inhibitors from a compound library that reduced luciferase activity by ≥ 90%, indicating that virus production was likewise affected. The top two compounds were used to propagate resistant virus stocks, and the mutations responsible for the resistance mapped to NS4B (77). A comparable HTS using a DENV-2 luciferase-tagged replicon cell line also discovered inhibitors targeting NS4B (78). A compound library screened against the cell line identified a compound that caused 85 percent reduction in viral replication with limited cytotoxicity, and specificity for DENV alone. Furthermore, the compound proved to inhibit RNA synthesis rather than translation, and did not directly inhibit protease, NTPase, methyltransferase, or RdRp function. Passaging and cell sorting methods were also used to isolate resistant strains to identify compensatory mutations in the NS4B protein. Based on the location of the selected resistance mutation, it was hypothesized that the compound interrupts the NS3-NS4B complex formation discovered previously (78, 79).
Figure 3.
PIP constructs and synthesis, drug design pipeline. A. YFV genome (top), YFV luciferase replicon construct (center), Sindbis virus replicon expressing YFV structural proteins for packaging (bottom). B. Schematic of pseudo-infectious particle production (PIPs) C. Schematic of PIP application for drug development D. Drug design pipeline. Figure adapted from Patkar et al. 2009 (77) with permission from American Society for Microbiology.
2.5 Assembly
To date, there are no antiviral compounds that target C protein and its function in virion assembly and uncoating. However, inhibition of the C protein has been shown to severely reduce viral production (80, 81). Thus, it may prove to be an effective target in the future and should be investigated. Short hairpin RNAs (shRNAs) targeting capsid of WNV reduced viral production significantly (80). Fusion of C protein with a nuclease also severely reduced virus production (81). Theoretically, it could be possible to design an inhibitor able to block inter-capsid interactions or “freeze” the nucleocapsid core in a conformation that is not amenable to release of the genomic RNA after entry. Development of an in vitro assembly assay would be instrumental in discovery of such anti-C inhibitors. Initial steps toward assembly assay development have resulted in the production of DENV nucleocapsid-like particles, which may prove foundational for further study of particle assembly. However, the exact mechanism of assembly is still elusive, and inhibitor addition has not yet been attempted (82).
2.6 Immune Response Evasion
2.6.1 NS1
Celgosivir is a clinically approved prodrug of castanospermine, a product of the Moreton Bay chestnut tree (Castanospermum australae), that is converted to Cast upon diffusion into the cell (83, 84). It has been shown to affect the folding of proteins by inhibiting the loss of the terminal glucose of N-linked glycans in HIV (85). This drug has also been shown to significantly inhibit the production of all four DENV serotypes both in vitro cell culture and in vivo mouse models (86). A combination of SDS-PAGE and Western blotting revealed that this drug prevents glycosylation of the NS1 protein, thus causing it to accumulate in the ER as shown through an immuno-fluorescence assay. Furthermore, it proved successful in vivo with an 80 percent survival rate for mice infected with DENV-2 and treated with Celgosivir within 1–2 days after infection (86). Although this inhibitor targets a cellular process, the efficiency of the inhibitor proves the vital role that NS1 plays in the replication of flavivirus infections, and also confirms that NS1 would be an effective novel target of antiviral design. More work is needed, however, with this particular natural compound. This study used DENV replicons to determine the efficacy and mechanism of the inhibitor. Yet both the prM and E proteins are also glycosylated during infection, and therefore this drug may be useful in combating various stages of the viral lifecycle (87). Since this compound is natural and has been previously studied, it could easily be used for new applications.
3. Conclusion
Collectively, these studies confirm that nontraditional protein targets have real promise for the development of new antiviral drugs. Additionally, such studies emphasize the need for further research into the functions of the individual nonstructural proteins. The greater the knowledge of viral protein functions the more opportunities arise to precisely target specific vital processes in the virus lifecycle. This development process is hampered by the fact that structures are often needed to guide the design of better inhibitors, however several replication proteins are integral membrane proteins and their protein structures have proven elusive. Thus it is difficult to design inhibitors based on structure-guided principles without known structures. Clearly, novel approaches are needed to address these proteins. HTS could be useful for further development of these proteins, not only to develop antiviral drugs for clinical use but also for determination of structure and function. Proteins such as NS2A and NS4A would greatly benefit from such studies as no direct antiviral approaches to these proteins have yet been developed.
3.1 Expert Opinion
Overall, there are many studies that have presented new options for DENV antiviral development and thus the field is clearly moving forward. It is especially interesting to note the studies involving new application of previously defined inhibitors. In particular, lycorine was developed to target other RNA viruses (69–72) and self-assembling nanotubes were originally discovered to inhibit bacteria (58), but these inhibitors have proved effective against Flaviviridae also (62, 74). Lycorine in particular demonstrates an exciting new application for a previously studied inhibitor, since 2K had not been previously targeted by antiviral drugs. Not only does lycorine present a new possible treatment option for flavivirus infection, it also encourages further investigation into the function of 2K. Furthermore, the efficacy of lycorine against a variety of viruses such as HSV-1 and SARS-CoV may even indicate shared function between 2K and viral proteins produced by viruses outside the flavivirus genus. Similarly, the transition of self-assembling nanotubes from bacterial to viral inhibition presents exciting new possibilities for antiviral treatment that may lead to further discovery about the nature of the membranes of the viral life cycle. Attention will not be lost on the rapidly expanding list of FDA-approved HCV inhibitors, which primarily target the NS3 proteinase and NS5B polymerase. These have been traditional drug targets, and are now validated by a member of the Flaviviridae family. Indeed, analogous molecules and targets are currently in development for DENV, and are likely to be the first drugs to enter clinical trials.
Although this review focuses on viral proteins, existing drug application also applies to host system targets. Additional studies should be conducted to encompass this area even as DENV host cell requirements, such as membrane fatty acid composition, are elucidated (88, 89). New insights into host cell requirements for viral replication present promising new targets for antiviral therapeutics.
These studies emphasize the need for constant re-evaluation of existing strategies in parallel with discovery of new inhibitors. Although discovery of novel inhibitors is clearly beneficial and should be encouraged, the journey from compound discovery to marketable drug is a long and expensive process (Figure 3D). Therefore, further studies should focus on development of novel uses for existing treatments, in addition to discovering new antiviral compounds.
Additionally, more in vivo studies would greatly benefit the drug design process. Many antiviral compounds demonstrate great success in vitro, but either fail, or are not tested, in vivo. In particular, many of the studies reviewed involved potent E protein inhibitors of the β-OG pocket. Only a few of the reviewed E protein inhibitors have been developed beyond preliminary cell culture luciferase assays, and none have been tested in vivo. Although mouse models are not always indicative of all facets of DENV progression, they are more representative of genuine DENV infection than cell culture assays. Since the goal of antiviral research is to control human infections, compounds are desperately needed to move beyond the bench and into the clinic.
Article Highlights.
The flaviviruses are a global health concern that lack effective treatment.
Novel antiviral approaches are needed to treat flavivirus infection.
Stem peptides, natural small molecules, peptide-conjugated oligomers, and self-assembling nanotubes are recent novel antiviral strategies.
Clear progress has been made, however existing inhibitors and in vivo trials should be further developed in addition to new drug discovery.
Acknowledgments
We thank T. Edwards, J. Grabowski, and R. Perera for helpful discussion and critical evaluation and A. Robinson for manuscript preparation.
Footnotes
Declaration of Interest
The authors thank the NIH/NIAID Regional Center of Excellence for Bio-defense and the Emerging Infectious Diseases Research (RCE) Program for financial support. We specifically recognize membership in and support from the Region V “Great Lakes” RECE (NIH award 2-U54-AI-057153).
4. Bibliography
- 1.Perry ST, Buck MD, Shresta S. Better late than never: antivirals for dengue. Expert Review of Anti-Infective Therapy. 2011 Jul;9(7):755–7. doi: 10.1586/eri.11.68. [DOI] [PubMed] [Google Scholar]
- 2*.Gubler D, Kuno G, Markoff L. Flaviviruses. In: Knipe DM, Howley PM, editors. Fields virology. 5. Vol. 1. Lippincott William & Wilkins; Philadelphia, PA: 2007. pp. 1153–1253. A comprehensive overview of the flaviviruses. [Google Scholar]
- 3.Watanaveeradej V, Simasathien S, Nisalak A, et al. Safety and Immunogenicity of a Tetravalent Live-Attenuated Dengue Vaccine in Flavivirus-Naive Infants. American Journal of Tropical Medicine and Hygiene. 2011 Aug;85(2):341–51. doi: 10.4269/ajtmh.2011.10-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rumyantsev AA, Giel-Moloney M, Liu YX, et al. Characterization of the RepliVax platform for replication-defective flavivirus vaccines. Vaccine. 2011 Jul;29(32):5184–94. doi: 10.1016/j.vaccine.2011.05.032. [DOI] [PubMed] [Google Scholar]
- 5.Gollins SW, Porterfield JS. Flavivirus infection enhancement in macrophages – radioactive and biological studies on the effect of antibody on viral fate. Journal of General Virology. 1984;65(AUG):1261–72. doi: 10.1099/0022-1317-65-8-1261. [DOI] [PubMed] [Google Scholar]
- 6.Gollins SW, Porterfield JS. Flavivirus infection enhancement in macrophages – an electron-microscopic study of viral cellular entry. Journal of General Virology. 1985;66(SEP):1969–82. doi: 10.1099/0022-1317-66-9-1969. [DOI] [PubMed] [Google Scholar]
- 7.Lin CC, Philips L, Xu C, et al. Pharmacokinetics and safety of viramidine, a prodrug of ribavirin, in healthy volunteers. Journal of Clinical Pharmacology. 2004 Mar;44(3):265–75. doi: 10.1177/0091270004262974. [DOI] [PubMed] [Google Scholar]
- 8.Manns MP, Foster GR, Rockstroh JK, et al. The way forward in HCV treatment - finding the right path. Nature Reviews Drug Discovery. 2007 Dec;6(12):991–1000. doi: 10.1038/nrd2411. [DOI] [PubMed] [Google Scholar]
- 9.Li L, Lok SM, Yu IM, et al. The flavivirus precursor membrane-envelope protein complex: Structure and maturation. Science. 2008 Mar;319(5871):1830–4. doi: 10.1126/science.1153263. [DOI] [PubMed] [Google Scholar]
- 10.Patkar CG, Jones CT, Chang YH, et al. Functional requirements of the yellow fever virus capsid protein. Journal of Virology. 2007 Jun;81(12):6471–81. doi: 10.1128/JVI.02120-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin YJ, Wu SC. Histidine at residue 99 and the transmembrane region of the precursor membrane prM protein are important for the prM-E heterodimeric complex formation of Japanese encephalitis virus. Journal of Virology. 2005 Jul;79(13):8535–44. doi: 10.1128/JVI.79.13.8535-8544.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smit JM, Moesker B, Rodenhuis-Zybert I, et al. Flavivirus Cell Entry and Membrane Fusion. Viruses-Basel. 2011 Feb;3(2):160–71. doi: 10.3390/v3020160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Avirutnan P, Hauhart RE, Somnuke P, et al. Binding of Flavivirus Nonstructural Protein NS1 to C4b Binding Protein Modulates Complement Activation. Journal of Immunology. 2011 Jul;187(1):424–33. doi: 10.4049/jimmunol.1100750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Avirutnan P, Fuchs A, Hauhart RE, et al. Antagonism of the complement component C4 by flavivirus nonstructural protein NS1. Journal of Experimental Medicine. 2010 Apr;207(4):793–806. doi: 10.1084/jem.20092545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung KM, Liszewski MK, Nybakken G, et al. West Nile virus nonstructural protein NS1 inhibits complement activation by binding the regulatory protein factor H. Proceedings of the National Academy of Sciences of the United States of America. 2006 Dec;103(50):19111–6. doi: 10.1073/pnas.0605668103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wen JL, Hua BC, Xiang JW, et al. Analysis of adaptive mutations in Kunjin virus replicon RNA reveals a novel role for the flavivirus nonstructural protein NS2A in inhibition of beta interferon promoter-driven transcription. Journal of Virology. 2004 Nov;78(22):12225–35. doi: 10.1128/JVI.78.22.12225-12235.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu WJ, Chen HB, Khromykh AA. Molecular and functional analyses of Kunjin virus infectious cDNA clones demonstrate the essential roles for NS2A in virus assembly and for a nonconservative residue in NS3 in RNA replication. Journal of Virology. 2003 Jul;77(14):7804–13. doi: 10.1128/JVI.77.14.7804-7813.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kummerer BM, Rice CM. Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. Journal of Virology. 2002 May;76(10):4773–84. doi: 10.1128/JVI.76.10.4773-4784.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chambers TJ, Weir RC, Grakoui A, et al. Evidence that the N-terminal domain of nonstructural protein NS3 from yellow fever virus is a serine protease responsible for site-specific cleavages in the viral polyprotein. Proceedings of the National Academy of Sciences of the United States of America. 1990 Nov;87(22):8898–902. doi: 10.1073/pnas.87.22.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roosendaal J, Westaway EG, Khromykh A, et al. Regulated cleavages at the West Nile virus NS4A-2K-NS4B junctions play a major role in rearranging cytoplasmic membranes and Golgi trafficking of the NS4A protein. Journal of Virology. 2006 May;80(9):4623–32. doi: 10.1128/JVI.80.9.4623-4632.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ambrose RL, Mackenzie JM. West Nile Virus Differentially Modulates the Unfolded Protein Response To Facilitate Replication and Immune Evasion. Journal of Virology. 2011 Mar;85(6):2723–32. doi: 10.1128/JVI.02050-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munoz-Jordan JL, Laurent-Rolle M, Ashour J, et al. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. Journal of Virology. 2005 Jul;79(13):8004–13. doi: 10.1128/JVI.79.13.8004-8013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grant D, Tan GK, Qing M, et al. A Single Amino Acid in Nonstructural Protein NS4B Confers Virulence to Dengue Virus in AG129 Mice through Enhancement of Viral RNA Synthesis. Journal of Virology. 2011 Aug;85(15):7775–87. doi: 10.1128/JVI.00665-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong HP, Zhang B, Shi PY. Flavivirus methyltransferase: A novel antiviral target. Antiviral Research. 2008 Oct;80(1):1–10. doi: 10.1016/j.antiviral.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iglesias NG, Filomatori CV, Gamarnik AV. The F1 Motif of Dengue Virus Polymerase NS5 Is Involved in Promoter-Dependent RNA Synthesis. Journal of Virology. 2011 Jun;85(12):5745–56. doi: 10.1128/JVI.02343-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Issur M, Geiss BJ, Bougie I, et al. The flavivirus NS5 protein is a true RNA guanylyltransferase that catalyzes a two-step reaction to form the RNA cap structure. RNA. 2009 Dec;15( 12):2340–50. doi: 10.1261/rna.1609709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang N, Chen HM, Koch V, et al. Ring-expanded (“fat”) nucleoside and nucleotide analogues exhibit potent in vitro activity against Flaviviridae NTPases/helicases, including those of the West Nile virus, hepatitis C virus, and Japanese encephalitis virus. Journal of Medicinal Chemistry. 2003 Sep 11;46(19):4149–64. doi: 10.1021/jm030842j. [DOI] [PubMed] [Google Scholar]
- 28.Ray D, Shi P-Y. Recent advances in flavivirus antiviral drug discovery and vaccine development. Recent patents on anti-infective drug discovery. 2006 Jan;1(1):45–55. doi: 10.2174/157489106775244055. [DOI] [PubMed] [Google Scholar]
- 29.Ishak R, Tovey DG, Howard CR. Morphogenesis of yellow fever virus 17D in infected cell cultures. Journal of General Virology. 1988 Feb;69:325–35. doi: 10.1099/0022-1317-69-2-325. [DOI] [PubMed] [Google Scholar]
- 30.van der Schaar HM, Rust MJ, Waarts B-L, et al. Characterization of the early events in dengue virus cell entry by biochemical assays and single-virus tracking. Journal of Virology. 2007 Nov;81(21):12019–28. doi: 10.1128/JVI.00300-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bressanelli S, Stiasny K, Allison SL, et al. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. Embo Journal. 2004 Feb 25;23(4):728–38. doi: 10.1038/sj.emboj.7600064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Modis Y, Ogata S, Clements D, et al. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proceedings of the National Academy of Sciences of the United States of America. 2003 Jun 10;100(12):6986–91. doi: 10.1073/pnas.0832193100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nemesio H, Palomares-Jerez F, Villalain J. The membrane-active regions of the dengue virus proteins C and E. Biochimica Et Biophysica Acta-Biomembranes. 2011 Oct;1808(10):2390–402. doi: 10.1016/j.bbamem.2011.06.019. [DOI] [PubMed] [Google Scholar]
- 34.Mackenzie JM, Khromykh AA, Westaway EG. Stable expression of noncytopathic Kunjin replicons simulates both ultrastructural and biochemical characteristics observed during replication of Kunjin virus. Virology. 2001 Jan 5;279(1):161–72. doi: 10.1006/viro.2000.0691. [DOI] [PubMed] [Google Scholar]
- 35.Elshuber S, Allison SL, Heinz FX, et al. Cleavage of protein prM is necessary for infection of BHK-21 cells by tick-borne encephalitis virus. Journal of General Virology. 2003 Jan;84:183–91. doi: 10.1099/vir.0.18723-0. [DOI] [PubMed] [Google Scholar]
- 36.Kuhn RJ, Zhang W, Rossmann MG, et al. Structure of dengue virus: Implications for flavivirus organization, maturation, and fusion. Cell. 2002 Mar 8;108(5):717–25. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li L, Lok SM, Yu IM, et al. The flavivirus precursor membrane-envelope protein complex: Structure and maturation. Science. 2008 Mar;319(5871):1830–4. doi: 10.1126/science.1153263. [DOI] [PubMed] [Google Scholar]
- 38.Modis Y, Ogata S, Clements D, et al. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004 Jan 22;427(6972):313–9. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- 39.Mukhopadhyay S, Kuhn RJ, Rossmann MG. A structural perspective of the Flavivirus life cycle. Nature Reviews Microbiology. 2005 Jan;3(1):13–22. doi: 10.1038/nrmicro1067. [DOI] [PubMed] [Google Scholar]
- 40.Stadler K, Allison SL, Schalich J, et al. Proteolytic activation of tick-borne encephalitis virus by furin. Journal of Virology. 1997 Nov;71(11):8475–81. doi: 10.1128/jvi.71.11.8475-8481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41**.Yu IM, Zhang W, Holdaway HA, et al. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science. 2008 Mar 28;319(5871):1834–7. doi: 10.1126/science.1153264. An excellent investigation of the rearrangement of the E protein during the flavivirus life cycle. [DOI] [PubMed] [Google Scholar]
- 42.Zhang W, Chipman PR, Corver J, et al. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nature Structural Biology. 2003 Nov;10(11):907–12. doi: 10.1038/nsb990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43*.Perera R, Khaliq M, Kuhn RJ. Closing the door on flaviviruses: Entry as a target for antiviral drug design. Antiviral Research. 2008 Oct;80(1):11–22. doi: 10.1016/j.antiviral.2008.05.004. Overview of flavivirus entry inhibitors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poh MK, Yip A, Zhang S, et al. A small molecule fusion inhibitor of dengue virus. Antiviral Research. 2009 Dec;84(3):260–6. doi: 10.1016/j.antiviral.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 45.Wang Q-Y, Patel SJ, Vangrevelinghe E, et al. A Small-Molecule Dengue Virus Entry Inhibitor. Antimicrobial Agents and Chemotherapy. 2009 May 1;53(5):1823–31. doi: 10.1128/AAC.01148-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Altmeyer R. Virus attachment and entry offer numerous targets for antiviral therapy. Current Pharmaceutical Design. 2004;10(30):3701–12. doi: 10.2174/1381612043382729. [DOI] [PubMed] [Google Scholar]
- 47.Yennamalli R, Subbarao N, Kampmann T, et al. Identification of novel target sites and an inhibitor of the dengue virus E protein. Journal of Computer-Aided Molecular Design. 2009 Jun;23(6):333–41. doi: 10.1007/s10822-009-9263-6. [DOI] [PubMed] [Google Scholar]
- 48.Li Z, Khaliq M, Zhou Z, et al. Design, synthesis, and biological evaluation of antiviral agents targeting flavivirus envelope proteins. Journal of Medicinal Chemistry. 2008 Aug 14;51(15):4660–71. doi: 10.1021/jm800412d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mayhoub AS, Khaliq M, Kuhn RJ, et al. Design, Synthesis, and Biological Evaluation of Thiazoles Targeting Flavivirus Envelope Proteins. Journal of Medicinal Chemistry. 2011 Mar 24;54(6):1704–14. doi: 10.1021/jm1013538. [DOI] [PubMed] [Google Scholar]
- 50.Zhou Z, Khaliq M, Suk J-E, et al. Antiviral Compounds Discovered by Virtual Screening of Small-Molecule Libraries against Dengue Virus E Protein. Acs Chemical Biology. 2008 Dec;3(12):765–75. doi: 10.1021/cb800176t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kampmann T, Yennamalli R, Campbell P, et al. In silico screening of small molecule libraries using the dengue virus envelope E protein has identified compounds with antiviral activity against multiple flaviviruses. Antiviral Research. 2009 Dec;84(3):234–41. doi: 10.1016/j.antiviral.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 52.Allison SL, Stiasny K, Stadler K, et al. Mapping of functional elements in the stem-anchor region of tick-borne encephalitis virus envelope protein E. Journal of Virology. 1999 Jul;73(7):5605–12. doi: 10.1128/jvi.73.7.5605-5612.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmidt AG, Yang PL, Harrison SC. Peptide Inhibitors of Dengue-Virus Entry Target a Late-Stage Fusion Intermediate. PLoS Pathogens. 2010 Apr;6(4):e1000851. doi: 10.1371/journal.ppat.1000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmidt AG, Yang PL, Harrison SC. Peptide Inhibitors of Flavivirus Entry Derived from the E Protein Stem. Journal of Virology. 2010 Dec;84(24):12549–54. doi: 10.1128/JVI.01440-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Costin JM, Jenwitheesuk E, Lok S-M, et al. Structural Optimization and De Novo Design of Dengue Virus Entry Inhibitory Peptides. Plos Neglected Tropical Diseases. 2010 Jun;4(6) doi: 10.1371/journal.pntd.0000721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nicholson CO, Costin JM, Rowe DK, et al. Viral entry inhibitors block dengue antibody-dependent enhancement in vitro. Antiviral Research. 2011 Jan;89(1):71–4. doi: 10.1016/j.antiviral.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 57.Tarallo R, Accardo A, Falanga A, et al. Clickable functionalization of liposomes with the gH625 peptide from herpes simplex virus type I for intracellular drug delivery. Chemistry. 2011 Nov;17(45):12659–68. doi: 10.1002/chem.201101425. [DOI] [PubMed] [Google Scholar]
- 58.Dartois V, Sanchez-Quesada J, Cabezas E, et al. Systemic antibacterial activity of novel synthetic cyclic peptides. Antimicrobial Agents and Chemotherapy. 2005 Aug;49(8):3302–10. doi: 10.1128/AAC.49.8.3302-3310.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fernandez-Lopez S, Kim HS, Choi EC, et al. Antibacterial agents based on the cyclic D,L-alpha-peptide architecture. Nature. 2001 Jul 26;412(6845):452–5. doi: 10.1038/35086601. [DOI] [PubMed] [Google Scholar]
- 60.Fletcher JT, Finlay JA, Callow ME, et al. A combinatorial approach to the discovery of biocidal six-residue cyclic D,L-alpha-peptides against the bacteria methicillin-resistant Staphylococcus aureus (MRSA) and E-coli and the biofouling algae Ulva linza and Navicula perminuta. Chemistry-a European Journal. 2007;13(14):4008–13. doi: 10.1002/chem.200((......)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Horne WS, Wiethoff CM, Cui CL, et al. Antiviral cyclic D,L-alpha-peptides: Targeting a general biochemical pathway in virus infections. Bioorganic & Medicinal Chemistry. 2005 Sep 1;13(17):5145–53. doi: 10.1016/j.bmc.2005.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Montero A, Gastaminza P, Law M, et al. Self-Assembling Peptide Nanotubes with Antiviral Activity against Hepatitis C Virus. Chemistry & biology 2011. 2011 Nov 23;18(11):1453–62. doi: 10.1016/j.chembiol.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Deas TS, Binduga-Gajewska I, Tilgner M, et al. Inhibition of flavivirus infections by antisense oligomers specifically suppressing viral translation and RNA replication. Journal of Virology. 2005 Apr;79(8):4599–609. doi: 10.1128/JVI.79.8.4599-4609.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kinney RM, Huang CYH, Rose BC, et al. Inhibition of dengue virus serotypes 1 to 4 in Vero cell cultures with morpholino oligomers. Journal of Virology. 2005 Apr;79(8):5116–28. doi: 10.1128/JVI.79.8.5116-5128.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Holden KL, Stein DA, Pierson TC, et al. Inhibition of dengue virus translation and RNA synthesis by a morpholino oligomer targeted to the top of the terminal 3 ′ stem-loop structure. Virology. 2006 Jan 20;344(2):439–52. doi: 10.1016/j.virol.2005.08.034. [DOI] [PubMed] [Google Scholar]
- 66.Achazi K, Patel P, Paliwal R, et al. RNA interference inhibits replication of tick-borne encephalitis virus in vitro. Antiviral Research. 2012 Jan;93(1):94–100. doi: 10.1016/j.antiviral.2011.10.023. [DOI] [PubMed] [Google Scholar]
- 67.Pacca CC, Severino AA, Mondini A, et al. RNA interference inhibits yellow fever virus replication in vitro and in vivo. Virus Genes. 2009 Apr;38(2):224–31. doi: 10.1007/s11262-009-0328-3. [DOI] [PubMed] [Google Scholar]
- 68.Ivacik D, Ely A, Arbuthnot P. Countering hepatitis B virus infection using RNAi: how far are we from the clinic? Reviews in Medical Virology. 2011 Nov;21(6):383–96. doi: 10.1002/rmv.705. [DOI] [PubMed] [Google Scholar]
- 69.Ieven M, Vlietinck AJ, Vandenberghe DA, et al. Plant antiviral agents.3. Isolation of alkaloids from clivia-minata regel (amaryllidaceae) Journal of Natural Products. 1982;45(5):564–73. doi: 10.1021/np50023a009. [DOI] [PubMed] [Google Scholar]
- 70.Li SY, Chen C, Zhang HQ, et al. Identification of natural compounds with antiviral activities against SARS-associated coronavirus. Antiviral Research. 2005 Jul;67(1):18–23. doi: 10.1016/j.antiviral.2005.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Renardnozaki J, Kim T, Imakura Y, et al. Effect of alkaloids isolated from amaryllidaceae on herpes-simplex virus. Research in Virology 1989. 1989;140(2):115–28. doi: 10.1016/s0923-2516(89)80089-5. [DOI] [PubMed] [Google Scholar]
- 72.Liu J, Yang Y, Xu Y, et al. Lycorine reduces mortality of human enterovirus 71-infected mice by inhibiting virus replication. Virology Journal. 2011 Oct;27:8. doi: 10.1186/1743-422X-8-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zou G, Puig-Basagoiti F, Zhang B, et al. A single-amino acid substitution in West Nile virus 2K peptide between NS4A and NS4B confers resistance to lycorine, a flavivirus inhibitor. Virology. 2009 Feb 5;384(1):242–52. doi: 10.1016/j.virol.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74*.Mertens E, Kajaste-Rudnitski A, Torres S, et al. Viral determinants in the NS3 helicase and 2K peptide that promote West Nile virus resistance to antiviral action of 2 ′,5 ′-oligoadenylate synthetase 1b. Virology. 2010 Mar 30;399(1):176–85. doi: 10.1016/j.virol.2009.12.036. Study integrates virus replication and host cell defenses as well as demonstrates the role of the Val9Met mutation. [DOI] [PubMed] [Google Scholar]
- 75.Miller S, Kastner S, Krijnse-Locker J, et al. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. Journal of Biological Chemistry. 2007 Mar;282(12):8873–82. doi: 10.1074/jbc.M609919200. [DOI] [PubMed] [Google Scholar]
- 76.Miller S, Sparacio S, Bartenschlager R. Subcellular localization and membrane topology of the dengue virus type 2 non-structural protein 4B. Journal of Biological Chemistry. 2006 Mar;281(13):8854–63. doi: 10.1074/jbc.M512697200. [DOI] [PubMed] [Google Scholar]
- 77*.Patkar CG, Larsen M, Owston M, et al. Identification of Inhibitors of Yellow Fever Virus Replication Using a Replicon-Based High-Throughput Assay. Antimicrobial Agents and Chemotherapy. 2009 Oct;53(10):4103–14. doi: 10.1128/AAC.00074-09. Effective application of YFV replicons; representative of general strategies employed for antiviral drug HTS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78*.Xie X, Wang Q-Y, Xu HY, et al. Inhibition of Dengue Virus by Targeting Viral NS4B Protein. Journal of Virology. 2011 Nov;85(21):11183–95. doi: 10.1128/JVI.05468-11. An excellent example of nonstructural protein targeting, HTS application, and resistance selection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Umareddy I, Chao A, Sampath A, et al. Dengue virus NS4B interacts with NS3 and dissociates it from single-stranded RNA. Journal of General Virology. 2006 Sep;87:2605–14. doi: 10.1099/vir.0.81844-0. [DOI] [PubMed] [Google Scholar]
- 80.Ong SP, Chu JJH, Ng ML. Inhibition of West Nile virus replication in cells stably transfected with vector-based shRNA expression system. Virus Research. 2008 Aug;135(2):292–7. doi: 10.1016/j.virusres.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 81.Qin CF, Qin ED. Capsid-targeted viral inactivation can destroy dengue 2 virus from within in vitro. Archives of Virology. 2006 Feb;151(2):379–85. doi: 10.1007/s00705-005-0631-9. [DOI] [PubMed] [Google Scholar]
- 82.Lopez C, Gil L, Lazo L, et al. In vitro assembly of nucleocapsid-like particles from purified recombinant capsid protein of dengue-2 virus. Archives of Virology. 2009 Apr;154(4):695–8. doi: 10.1007/s00705-009-0350-8. [DOI] [PubMed] [Google Scholar]
- 83.Molyneux RJ, Roitman JN, Dunnheim G, et al. 6-Epicatanospermine, a novel indolizidine alkaloid that inhibits alpha-glucosidase. Archives of Biochemistry and Biophysics. 1986 Dec;251(2):450–7. doi: 10.1016/0003-9861(86)90351-6. [DOI] [PubMed] [Google Scholar]
- 84.Kang MS. Uptake and metabolism of BuCast: A glycoprotein processing inhibitor and a potential anti-HIV drug. Glycobiology. 1996 Mar;6(2):209–16. doi: 10.1093/glycob/6.2.209. [DOI] [PubMed] [Google Scholar]
- 85.Taylor DL, Sunkara PS, Liu PS, et al. 6-0-Butanoylcastanospermine (MDL 28,574) inhibits glycoprotein processing and the growth of HIV. Aids. 1991 Jun;5(6):693–8. doi: 10.1097/00002030-199106000-00008. [DOI] [PubMed] [Google Scholar]
- 86.Rathore APS, Paradkar PN, Watanabe S, et al. Celgosivir treatment misfolds dengue virus NS1 protein, induces cellular pro-survival genes and protects against lethal challenge mouse model. Antiviral research. 2011 Dec;92(3):453–60. doi: 10.1016/j.antiviral.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 87.Somnuke P, Hauhart RE, Atkinson JP, et al. N-linked glycosylation of dengue virus NS1 protein modulates secretion, cell-surface expression, hexamer stability, and interactions with human complement. Virology. 2011 May;413(2):253–64. doi: 10.1016/j.virol.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Heaton NS, Perera R, Berger KL, et al. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proceedings of the National Academy of Sciences of the United States of America. 2010 Oct 5;107(40):17345–50. doi: 10.1073/pnas.1010811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Perera R, Riley C, Hopf-Jannasch AS, et al. Dengue virus perturbs lipid metabolism in infected mosquito cells. PLoS Pathogens. 2012 doi: 10.1371/journal.ppat.1002584. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mukhopadhyay S, Kuhn RJ, Rossmann MG. A structural perspective of the Flavivirus life cycle. Nature Reviews Microbiology. 2005 Jan;3(1):13–22. doi: 10.1038/nrmicro1067. [DOI] [PubMed] [Google Scholar]
- 91.Miller S, Krijnse-Locker J. Modification of intracellular membrane structures for virus replication. Nature Reviews Microbiology. 2008 May;6(5):363–74. doi: 10.1038/nrmicro1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Quinkert D, Bartenschlager R, Lohmann V. Quantitative analysis of the Hepatitis C Virus replication complex. Journal of Virology. 2005 Nov;79(21):13594–605. doi: 10.1128/JVI.79.21.13594-13605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]