Abstract
Bacillus anthracis, the anthrax agent, is a member of the Bacillus cereus sensu lato group, which includes invasive pathogens of mammals or insects as well as nonpathogenic environmental strains. The genes for anthrax pathogenesis are located on two large virulence plasmids. Similar virulence plasmids have been acquired by other B. cereus strains and enable the pathogenesis of anthrax-like diseases. Among the virulence factors of B. anthracis is the S-layer-associated protein BslA, which endows bacilli with invasive attributes for mammalian hosts. BslA surface display and function are dependent on the bacterial S-layer, whose constituents assemble by binding to the secondary cell wall polysaccharide (SCWP) via S-layer homology (SLH) domains. B. anthracis and other pathogenic B. cereus isolates harbor genes for the secretion of S-layer proteins, for S-layer assembly, and for synthesis of the SCWP. We review here recent insights into the assembly and function of the S-layer and the SCWP.
Keywords: Bacillus cereus sensu lato, virulence plasmid, Bacillus S-layer-associated proteins, BSLs, host invasion, S-layer homology domain, SLH domain, protein secretion, crystallization domain, secondary cell wall polysaccharide, SCWP
ORIGINS OF ANTHRAX AND ANTHRAX-LIKE DISEASE-CAUSING ISOLATES
Bacillus anthracis is an aerobic, spore-forming, gram-positive bacterium (139). Spores are the causative agent of anthrax, an infectious disease that is fatal to mammalian species (68). Herbivores are frequent hosts, ingesting spores that contaminate the environment (140). Spores germinate within the host, and their vegetative forms invade and replicate within all organ tissues, eventually with a lethal outcome (140). Vegetative bacilli sporulate in decaying carcasses so that dormant spores can again contaminate the environment, thereby completing the B. anthracis life cycle (68). Phylogenetic studies reveal that B. anthracis is a member of the Bacillus cereus sensu lato (s.l.) group, which includes the insect pathogen Bacillus thuringiensis and the soil microbe Bacillus cereus (53). Members of the B. cereus s.l. group are described as common inhabitants and symbionts of the invertebrate gut (56). Only occasionally do specific isolates of B. cereus s.l. enter into a pathogenic life cycle, infecting suitable hosts and multiplying without restraint (56). Multiple-locus sequence typing as well as 16S RNA and whole-genome sequencing of different B. cereus s.l. isolates demonstrate their relatedness: a core of 1,750 genes is found in every isolate, and an extended core of another 2,150 genes is present in almost every genome (24, 112, 114, 137, 152). Most members of B. cereus s.l. are classified into one of two clades yet carry few clade-specific genes (152). A third clade encompasses outliers including environmental isolates with significant gene decay (152). Three additional species, Bacillus mycoides, Bacillus pseudomycoides, and Bacillus weihenstephanensis, have been added to the B. cereus s.l. classification (112, 152). The pathogenic attributes of B. anthracis, B. thuringiensis, and B. cereus isolates are encoded on virulence plasmids, which are acquired via horizontal transfer (55, 66, 67, 115). Extensive genome analysis of B. anthracis isolates from many different geographic locations demonstrates the clonal relationship of this species (72). Keim & Wagner (60) proposed that members of the B. cereus s.l. group represent “hopeful monsters”: following acquisition of toxin-containing plasmids, successful clones rapidly expand and spread around the globe, aided by the mobility of humans.
VIRULENCE PLASMIDS OF B. ANTHRACIS
B. anthracis acquired two large plasmids, pXO1 and pXO2 (107). A key feature of the ∼182-kb pXO1 plasmid is the presence of three toxin genes (pag, lef, cya) whose secreted products [protective antigen (PA), lethal factor (LF), and edema factor (EF)] assemble into lethal toxin (LT) and edema toxin (ET) (76, 120, 144) (Figure 1). PA binds to the anthrax toxin receptors, TEM8 (ANTRX1) and CMG2 (ANTRX2), on the surface of mammalian host cells (14, 130). The host protease furin cleaves PA (49), generating mature 63-kDa PA, which then oligomerizes into an octameric structure (65, 97). Oligomerization provides for PA association with B. anthracis LF and EF (98). The assembled LT (PA+LF) and ET (PA+EF) are endocytosed by clathrin-coated vesicles; acidification of the endosomes promotes membrane insertion of PA and translocation of LF and EF across the endosomal membrane (73, 151). Once translocated into the cytoplasm of target cells, LF exerts zinc protease activity to cleave mitogen-activated protein (MAP) kinase and NLRP1 (31, 77). EF associates with calmodulin, a calcium-regulated host factor, to acquire adenylate cyclase activity, converting ATP into cyclic AMP messenger molecules (76). Both toxins make important contributions to the life cycle of B. anthracis, disabling innate and adaptive immune functions of the host and promoting the lethal outcome of anthrax infections (3, 100). The ∼95-kb pXO2 plasmid carries the capBCADE operon, which provides for the synthesis of the poly-D-γ-glutamic acid (PDGA) capsule (17, 18, 20, 50). The PDGA capsule endows vegetative bacilli with resistance to opsonization by host complement and opsonophagocytosis by granulocytes and macrophages; PDGA capsulation is an essential virulence determinant of B. anthracis (30, 111). Temperature shift to 37°C and increased environmental CO2, as occur following B. anthracis entry into mammalian tissues, induce the expression of toxin and capsule genes (45, 119). The transcriptional response to CO2 requires the trans-acting factor AtxA, whose gene (atxA) is located on pXO1 (9, 51, 69, 141, 142) (Figure 1). AtxA-mediated regulation of capsulation requires two additional transcription factors, AcpA and AcpB, whose structural genes are located on pXO2 (28, 29) (Figure 1). CO2-induced transcriptional activation also encompasses chromosomal genes, suggesting that AtxA is a global regulator of B. anthracis (12). Most B. cereus s.l. isolates regulate the expression of their secreted products via PlcR, a global transcriptional regulator and quorum sensor that is activated by binding to the PapR autoinducer (2, 11) (see sidebar titled “Virulence Plasmids of B. cereus Isolates Causing Anthrax-Like Disease”). Interference between PlcR and AtxA-mediated gene regulation is thought to have selected for the mutation in plcR that inactivates quorum sensing and PlcR-mediated gene regulation in B. anthracis (96).
Figure 1.
B. cereus s.l. isolates causing anthrax and anthrax-like diseases: (a) B. anthracis with its two virulence plasmids, pXO1 and pXO2; (b) B. cereus G9241, harboring pBCXO1 and pBC210; and (c) B. cereus biovar anthracis CI and plasmids pCI-XO1 and pCI-XO2. B. cereus G9241 and biovar anthracis CI have a third small plasmid not shown here. In addition to toxin genes (pag, lef, and cya), B. anthracis virulence plasmids pXO1 and pXO2 carry genes for poly-D-γ-glutamic acid (PDGA) capsular synthesis (capBCADE) and hyaluronic acid (HA) capsular synthesis (hasACB); the hasA gene, however, carries an inactivating mutation. The plasmids also carry three genes for B. anthracis S-layer-associated (BSL) proteins: bslA, bslB, and amiA. Virulence gene expression is regulated by the product of atxA, which activates toxin, bslA, and capBCADE gene expression in response to a host signal (increased CO2 concentration). The capBCADE operon is coregulated by acpA and acpB. B. cereus G9241 synthesizes HA (intact hasACB gene cluster). pBC210 encodes genes for three BSLs (bslO2, bslV, and bslW), the Certhrax toxin (pag2, lef 2), an atxA-like regulator (atxA2), and B. cereus exo-polysaccharide (BPS) (bpsXABCDEFG). B. cereus biovar anthracis capsulation includes both PDGA and HA.
S-LAYER AND S-LAYER PROTEINS OF B. ANTHRACIS
Two surface (S)-layer proteins, Sap and EA1, were identified in the extracellular medium and in the bacterial envelope of B. anthracis cultures (37, 38) (Figure 2). Expression of their structural genes, sap and eag (EA1), is regulated by CO2 (95). In response to increased CO2, the pXO1-encoded virulence regulator AtxA activates transcription of eag via PagR (95), the transcriptional regulator of pag expression, in addition to activating the expression of capsule genes (51, 94). As a result, encapsulated bacilli produce predominantly EA1 (94), while germinating bacilli or vegetative forms replicating at low CO2 produce large amounts of Sap and lesser quantities of EA1 (64). When analyzed by electron microscopy, B. anthracis variants lacking sap or eag each form two-dimensional crystalline arrays (23). Replicating at low CO2, B. anthracis forms Sap S-layers mid-cell and along the cylindrical axis of the cell, whereas EA1 S-layers are deposited at cell septa and near future division sites (64). PDGA polymers are anchored to the peptidoglycan of B. anthracis (118); electron microscopy studies revealed that PDGA polymers traverse the S-layer of bacilli (91) (see sidebar titled “Bacterial S-Layers”).
Figure 2.
The S-layer envelope of B. anthracis. (a) Model depicting different layers, including the plasma membrane, peptidoglycan, secondary cell wall polysaccharide (SCWP), and S-layer. The SCWP and poly-D-γ-glutamic acid strands (capsule) are tethered to the peptidoglycan of B. anthracis. (b) Secretion and assembly of S-layer proteins (SLPs), Sap and EA1, and B. anthracis S-layer-associated proteins (BSLs). Each protein is synthesized as a precursor with an N-terminal signal peptide (SP), three S-layer homology (SLH) domains and a C-terminal domain. SecA2 and SlaP contribute to the secretion of Sap and EA1, whereas SlaQ contributes to Sap and EA1 S-layer assembly. In Sap and EA1, the C-terminal domain is a crystallization domain (CD). (c) S-layer gene cluster (slc) in B. anthracis and other B. cereus s.l. isolates. The slc is flanked by sulfate permease and enoyl-CoA hydratase genes, shown in gray. The csaA and csaB (ketal pyruvyltransferase) and wpaA [Wzy-/WaaL (O-antigen ligase)-like] genes are absolutely conserved in all species and required for SCWP modifications that enable SLP or BSL assembly. Strains causing anthrax (B. anthracis) or anthrax-like disease (B. cereus G9241 and B. cereus biovar anthracis CI) encode genes for SLPs (Sap and EA1 with three SLH domains), SLP secretion and assembly (slaQ, slaP, and secA2), and SCWP acetylation (patA/patB). The slc of B. cereus ATCC 10987 harbors truncated eag and secA2 genes and lacks sap and slaP. The environmental isolate B. cereus ATCC 14579 lacks genes for SLPs, SLP secretion and assembly, and SCWP acetylation but encodes a murein hydrolase with three SLH domains in place of Sap and EA1. Gene clusters were obtained from GenBank using accession numbers AE017334.2 for B. anthracis str. Ames Ancestor, CP009590.1 for B. cereus G9241, CP001746.1 for B. cereus biovar anthracis CI, AE016877.1 for B. cereus ATCC 14579, and NC_00,3909.8 for B. cereus ATCC 10987. Colors of genes were assigned arbitrarily with the exception of flanking genes that are not part of the slc and which are shown in gray along with their respective locus number.
Sequence analysis of sap and eag indicates that both gene products are synthesized as precursors (Sap 814 residues, EA1 862 residues) with a cleavable N-terminal signal peptide followed by three tandem repeats of ∼55-residue S-layer homology (SLH) domains and a C-terminal crystallization domain (93). The 29-residue signal peptides differ by a single amino acid residue and are substrates for SecA2 (see below), whereas SLH domains display 66% identity, which is higher than the sequence identity between SLH domains of B. anthracis S-layer-associated proteins (BSLs) (63). However, the crystallization domains of Sap and EA1 contain only 22% amino acid identity (93). As reported for SLH domains in other bacteria, three tandem copies of the SLH domains of Sap and EA1 mediate attachment of fusion proteins to the envelope of B. anthracis (92). X-ray crystallography experiments show that SLH domains of Sap fold into a three-pronged spindle structure, generating binding sites for the secondary cell wall polysaccharide (SCWP) of B. anthracis (63). Binding of the SLH domains of Sap, EA1 and BSLs to the envelope of B. anthracis and B. cereus G9241 is dependent on ketal-pyruvylation of the terminal ManNAc residue at the nonreducing end of the SCWP (43, 61, 89, 147). Ketal-pyruvylation is thought to be catalyzed by the product of the csaB gene, which is located immediately adjacent to sap and eag on the chromosome of B. anthracis and of B. cereus isolates causing anthrax-like disease (89, 147) (Figure 2).
SECRETION AND ASSEMBLY OF S-LAYER PROTEINS
B. anthracis grows as chains of incompletely separated, rod-shaped cells, a trait contributing to its escape from phagocytic clearance (122). Mutations that abrogate expression of sap cause B. anthracis to form elongated chains owing to the mislocalization of the murein hydrolase BslO (64). Screening libraries of mutant bacilli for variants with increased chain length led to the identification of three genes: secA2, slaP, and slaQ (102, 103). These genes are located immediately adjacent to the S-layer gene cluster (csaA-csaB-sap-eag) (103) (Figure 2). Mutations in all three genes diminish the abundance of Sap and EA1 in the bacterial S-layer but do not affect the secretion of other products (102). Mutations in secA2 and slaP, but not in slaQ, diminish the secretion of mCherry hybrids fused to the N-terminal signal peptide and SLH domains of Sap (103). In contrast, mutations in slaQ affect only the secretion of mCherry hybrids fused to full-length Sap including the C-terminal crystallization domain (103). SecA2 and SlaP are thought to recognize the signal peptides and SLH domains of both Sap and EA1, respectively, whereas SlaQ acts on the crystallization domain of Sap (103) (Figure 2).
SecA, the canonical secretion ATPase of eubacteria, has been studied extensively in Escherichia coli (109, 138). E. coli SecA binds signal peptide–bearing precursors for delivery and translocation at the SecYEG translocon, which leads to precursor movement across the plasma membrane (34, 35, 78). Following signal peptide cleavage by signal peptidase, mature polypeptides are released on the trans side of the membrane (25). Auxiliary translocation factors (SecDF, YajC, and YidC) have been shown to increase membrane translocation of specific precursor proteins (32, 123, 129). The genome of B. anthracis harbors genes for the canonical SecA secretion ATPase and for components of the translocation machinery (116). Similar to other gram-positive bacterial pathogens, B. an-thracis encodes an accessory secretion gene, designated secA2 (15, 126). SecA2, a homolog of the canonical bacterial secretion ATPase SecA, is thought to recognize the signal peptide of Sap and EA1 and facilitate secretion of large amounts of S-layer proteins during vegetative growth (102). SlaP, a peripheral membrane protein of B. anthracis, also facilitates secretion of Sap and EA1 (102). Although this has not been tested experimentally, SlaP could play a role in the membrane translocation of SLH domains to prevent their premature association with SCWP precursors in the cytoplasm. Purified SlaQ assembles into high molecular weight structures and promotes Sap S-layer assembly in vitro (103). The secA2, slaP and slaQ genes are conserved in B. cereus s.l. isolates expressing S-layer proteins but are absent from isolates lacking S-layer proteins (SLPs) (103) (Figure 2).
VIRULENCE PLASMIDS ENCODE S-LAYER-ASSOCIATED PROTEINS
Vegetative forms of B. anthracis, when grown in the presence of increased CO2, display adherence to mammalian cells (62). This attribute is associated with the expression of bslA, a gene that is located on pXO1 (102) (Figure 1). The predicted 652-residue precursor product of bslA encompasses an N-terminal cleavable signal peptide, three SLH domains, and a C-terminal domain that, when purified as a recombinant protein, adheres to mammalian cells and functions as a competitive inhibitor for the binding of bacilli (102). BslA is deposited into the B. anthracis S-layer and displayed on the bacterial surface (102). B. anthracis mutants lacking bslA display a defect in adherence to mammalian cells. When analyzed in a guinea pig model of subcutaneous spore challenge and compared to wild-type B. anthracis Ames, the bslA mutants show delayed replication and dissemination into host tissues and an increase in the spore dose required for lethal disease (102). In a mouse model for disseminated anthrax meningitis, bslA mutant bacilli were defective in crossing the blood-brain barrier and in establishing anthrax (33). Thus, BslA represents an adhesin and virulence factor for B. anthracis. Biochemical experiments suggest that purified BslA binds to laminin, an extracellular matrix protein found in basal membranes (145). bslA homologs are also present on the pXO1-like plasmids of B. cereus G9241 and B. cereus biovar anthracis CI (147) (Figure 1). pXO1 and pXO1-like plasmids carry another gene for a secreted precursor with N-terminal SLH domains, designated bslB (147). The gene is absolutely conserved among B. anthracis and B. cereus isolates. However, a specific function has not yet been assigned to the bslB product.
pXO2 carries the amiA gene (pXO2–42), whose product is a secreted precursor of 503 amino acid residues with an N-terminal signal peptide, three SLH domains, and a C-terminal murein hydrolase domain (N-acetylmuramoyl-L-alanine amidase) (90) (Figure 1). When purified as a recombinant protein without the N-terminal signal peptide and SLH domains, the C-terminal amidase domain of AmiA cleaves B. anthracis peptidoglycan (90). The in vivo contributions of amiA to the life cycle of B. anthracis have, however, not yet been studied. amiA is conserved in the pXO2-like plasmids of B. cereus biovar anthracis CI but is absent from pBC210 of B. cereus G9241 (66) (Figure 1). The capsule plasmid (pBC210) of B. cereus G9241 encodes three genes for precursors bearing signal peptides and three SLH domains (54, 147). One of these, bslO2, is a predicted murein hydrolase with a C-terminal N-acetylglucosaminidase domain (147). Thus, genes for S-layer-associated murein hydrolases are a conserved feature of virulence plasmids with capsule genes, suggesting that these enzymes may contribute either to capsule assembly or to controlling the length of chains of capsulating bacterial cells with incompletely separated septal peptidoglycan (see sidebar titled “S-Layer Regulation of Chain Lengths in B. anthracis”).
B. ANTHRACIS AND B. CEREUS S-LAYER-ASSOCIATED PROTEINS
In addition to Sap and EA1, the chromosomes of B. anthracis and B. cereus G9241 encode 19 and 18 genes for precursor proteins with SLH domains, respectively (62, 147). Collectively these gene products have been designated BSLs (Bacillus S-layer-associated proteins) (62). Mutations in bslO cause mutant bacilli to form chains of vegetative forms with exaggerated lengths, a phenotype that is complemented when purified BslO protein is added in trans (5). In wild-type bacilli, the BslO protein is deposited near cell septa (5). BslO localization is dependent on S-layer assembly (64). Further, patA1 and patA2 (encoding two acetyltransferases that modify the SCWP) and wpaA [whose product belongs to the family of Wzy-/WaaL (also known as O-antigen ligase)-like enzymes] are also required for proper localization of BslO (83, 105) (Figure 2). Mutations in patA1 and patA2 affect the deposition of EA1 in the vicinity of cell septa, suggesting that SLP distribution is influenced by chemical modifications to the ligand of SLH domains (83). Computational simulation suggests that BslO effects a nonrandom distribution of B. anthracis chain lengths, implying that all septa are not equal candidates for BslO-mediated cell separation (5). BslS is an N-acetylmuramoyl-L-alanine amidase; its structural gene is located downstream of the wpaA gene and the S-layer locus (105). Deposition of BslS and BslT, a homolog of BslS, in the S-layer of B. anthracis is perturbed in wpaA mutant bacilli (105).
The bslK gene product carries a NEAT (heme-binding near-iron transporter) domain and binds heme-iron, as reported for other NEAT-domain-containing proteins (88, 136). Hal and IsdX2, secreted hemophores with NEAT domains, scavenge heme-iron from host hemoglobin and transfer the nutrient to BslK (7, 85). Further transport of heme-iron across the bacterial envelope involves other NEAT domain proteins, such as IsdX1 and IsdC, which is anchored by sortase B to the pep-tidoglycan of B. anthracis (84, 85). Heme-iron is imported into the bacterial cytoplasm by the IsdE ATP-binding protein and the IsdF membrane transporter (84). A hemoxygenase, IsdG, cleaves the tetrapyrrole of heme, and the released iron is incorporated into iron-containing proteins (117, 132).
STRUCTURE AND FUNCTION OF THE SECONDARY CELL WALL POLYSACCHARIDE
The ∼10- to 20-kDa SCWP released by hydrofluoric acid treatment from B. anthracis has the repeat structure [→4)-β-ManNAc-(1→4)-β-GlcNAc(O3-α-Gal)-(1→6)-α-GlcNAc(O3-α-Gal, O4-β-Gal)-(1→]6–12 (22) (Figure 3a). B. anthracis CDC684 is an avirulent isolate whose SCWP has the same trisaccharide repeat structure as wild-type B. anthracis yet lacks all galactosyl modifications (43, 108). NMR spectroscopy of CDC684 polysaccharide revealed both ketal pyruvyl (blue in Figure 3a) and acetyl modifications at the distal (nonreducing) end of the SCWP (43) (red in Figure 3a). Of note, only some molecules carry ketal pyruvyl at O4,O6 of ManNAc at the distal end of the SCWP and acetyl at O3 of the penultimate GlcNAc residue (43, 83). B. cereus G9241 and other B. cereus isolates causing anthrax-like disease synthesize SCWP with similar size and structure as B. anthracis, although wall polysaccharide from these isolates carries an additional α-Gal substitution at O3 of ManNAc (44) (Figure 3b). The environmental, nonpathogenic isolate B. cereus ATCC 10987 synthesizes SCWP with a distinct repeat structure, [→4)-β-ManNAc-(1→4)-β-GlcNAc-(1→6)-α-GalNAc-(1→]n, and β-Gal substitutions at the O3 of α-GalNAc as well as nonstoichiometrical acetylation at O3 of β-ManNAc (75) (Figure 3b). B. cereus ATCC 14579 SCWP has the same repeat structure and substitutions of β-GlcNAc at O3 of the β-GlcNAc in addition to β-Glc at O3 and α-ManNAc at O4 of the α-GalNAc residue (19) (Figure 3b). The SCWP of B. cereus ATCC 14579 is expressed constitutively, while another, structurally dissimilar polysaccharide is synthesized in biofilms but not during planktonic growth (19).
Figure 3.
Structure of the secondary cell wall polysaccharide (SCWP) in B. cereus s.l. isolates. (a) Structure of the terminal unit and the repeat unit of B. anthracis SCWP. The ketal pyruvyl modification of ManNAc at the nonreducing distal end of the SCWP is in blue. The acetyl group of the penultimate GlcNAc residue in the terminal unit is in red. The arrow identifies the glycosidic linkage to the next repeat unit. (b) Repeat unit structure of the SCWP from B. anthracis, B. cereus G9241, B. cereus ATCC 10987, and B. cereus ATCC 14579.
The pyruvylated SCWP of B. anthracis and B. cereus G9241 serves as the attachment site for the S-layer and its constituents, SLPs and BSLs with three SLH domains (61, 147). However, the SCWP is also a ligand for murein hydrolases of bacteriophages, for example, PlyG and PlyL (81, 127). PlyG, which is encoded by the γ-phage, binds to the SCWP from B. anthracis and B. cereus G9241, but not to the wall polysaccharides of B. cereus ATCC 10987 and ATCC 14597 (46, 81) (Figure 3b). Synthetic oligosaccharides variable for the galactosyl modifications of a single SCWP trisaccharide of B. anthracis revealed that high-affinity binding of PlyG and PlyL is dependent on the β-Gal modification at O4 of the α-GlcNAc residue (99). Thus, the SCWP of B. anthracis and pathogenic B. cereus strains functions as the structural determinant for bacteriophage-mediated lysis of bacteria by serving as ligand for the carbohydrate-binding domain of bacteriophage lysins (42). Furthermore, B. anthracis Sap functions as surface receptor for bacteriophage AP50c (110).
ASSEMBLY OF THE B. ANTHRACIS SECONDARY CELL WALL POLYSACCHARIDE
Unlike B. subtilis and Staphylococcus aureus, B. anthracis produces SCWP and lipoteichoic acid (LTA) but not wall teichoic acid (WTA) (47, 101). See Figure 4a for a model of SCWP assembly and synthesis reactions and Figure 4b for a summary of operons and genes involved in SCWP synthesis. B. subtilis and S. aureus synthesize polyribitol-phosphate teichoic acid on murein linkage units (GlcNAc-ManNAc) that are linked to undecaprenyl-pyrophosphate carrier [C55-(PO4)2-GlcNAc-ManNAc] (59, 70, 71). Murein linkage unit synthesis requires the products of two genes, tagO and tagA (74, 134). TagO transfers GlcNAc-1-phosphate from UDP-GlcNAc onto C55-(PO4)2 to generate C55-(PO4)2-GlcNAc (lipid III) (48, 121) (Figure 4a, Reaction 12); this reaction is inhibited by the antibiotic tunicamycin (52). TagA transfers N-acetylmannosamine from UDP-ManNAc onto O4 of GlcNAc within lipid III, releasing C55-(PO4)2-GlcNAc-ManNAc (26, 48) (Figure 4a, Reaction 13). S. aureus tagO mutants cannot synthesize WTA but also display subtle defects in peptidoglycan synthesis (40, 52, 148). The B. anthracis genome contains tagO and tagA homologs (116). When expressed in S. aureus tagO mutants, B. anthracis tagO restores WTA synthesis (61). Tunicamycin treatment or genetic repression of tagO inhibits vegetative growth, SCWP synthesis, and S-layer assembly of B. anthracis, causing expansions in size and spherical shapes of vegetative forms that can no longer divide (61, 82). These data suggest that the SCWP may be synthesized on murein linkage units that are tethered to an undecaprenyl-pyrophosphate carrier (Figure 4a). In agreement with this hypothesis, SCWP synthesis in B. anthracis also requires the LytR-CpsA-Psr (LCP) family of enzymes (79, 80) (Figure 4a). In B. subtilis and S. aureus, LCP enzymes tether teichoic acids to peptidoglycan by forming a phosphodiester bond between O6 of N-acetylmuramic acid (MurNAc) in glycan strands and O1 of GlcNAc in murein linkage units (21, 58). As expected, expression of LCPs is essential for B. anthracis growth and SCWP synthesis, and expression of some, but not all, B. anthracis LCP genes restores WTA synthesis in S. aureus mutants lacking endogenous LCPs (79). Genes involved in SCWP synthesis appear to be scattered among four gene clusters: slc (S-layer cluster), sps (surface polysaccharide), scwp1, and scwp2 (Figures 2 and 4b). Two of these clusters, sps (128) and scwp2, display variation between B. anthracis and various B. cereus isolates. We presume that some of these genes are responsible for the galactosylation patterns of B. anthracis and B. cereus SCWP (Figure 3b). Some of the genes predicted to be involved in SCWP synthesis are present in duplicate; their products often display functional redundancy (105, 106).
Figure 4.
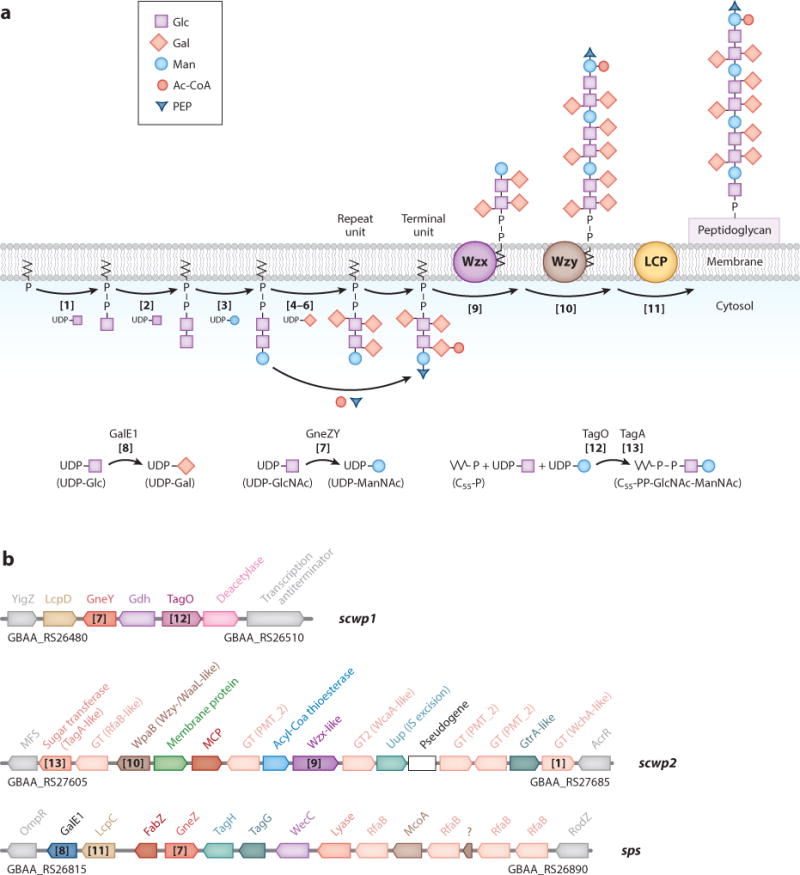
Model for the biosynthesis of the secondary cell wall polysaccharide (SCWP) in B. anthracis. (a) Putative synthesis of undecaprenyl-pyrophosphate-linked SCWP trisaccharide subunits that are modified with galactosyl, ketal pyruvyl, or acetyl groups, translocated across the membrane by Wzx-like and polymerized by Wzy-like catalysts. The SCWP is finally linked to peptidoglycan by LCP enzymes. (b) Gene clusters contributing to SCWP synthesis include slc (see Figure 2b) as well as scwp1, scwp2, and sps (surface polysaccharide). Genes in gray flank gene clusters but are not part of them. The GenBank identifiers of flanking genes are underneath gray gene symbols. Bracketed numbers in both panels indicate putative chemical reactions and enzymes for SCWP synthesis.
Bacteria evolved three biosynthetic pathways for polysaccharide synthesis: synthase, ABC transporter, and Wzy-polymerase-dependent assembly (150). Capsular polysaccharides and cell wall polysaccharides of gram-positive bacteria are frequently synthesized via the Wzy-polymerase-synthesis pathway. This has been extensively studied for the capsule genes of Streptococcus pneumoniae (150), and insights from this work informed our bioinformatic analysis of the B. anthracis SCWP synthesis pathway. Spratt and coworkers (10) characterized S. pneumoniae genes and putative enzymatic activities of their products involved in Wzy-dependent capsular polysaccharide (CPS) synthesis. cps genes are clustered in a single locus of 17–20 genes (10). S. pneumoniae evolved more than 90 serological variants with different CPS structures, and 1,999 genes in all of these variants are responsible for the synthesis of various carbohydrates (87). These genes have been classified into homology groups. Bioinformatic schemes have been employed to predict the function of these gene products, identifying proteins within the same homology group, Pfam family, or CAZy glycosyltransferase family. Correlating cps gene content with known CPS structures provides tentative assignments of function to the different homology groups: regulatory factors, enzymes for synthesis of polysaccharide constituents, polymerases, flippases, initial sugar trans-ferases, glycosyltransferases, phosphotransferases, acetyltransferases, and pyruvyltransferases (1).
Synthesis of B. anthracis SCWP Units
Wzy pathway synthesis begins with the addition of a UDP-linked sugar to undecaprenyl-PO4 (C55-PO4) (Figure 4a, Reaction 1). The reaction is catalyzed by the initiating glycosyltransferase WchA (WciI homology group), a member of the polyisoprenyl-phosphate hexose-1-P family (10). In S. pneumoniae 9L/9N CPS, the initial sugar is GlcNAc, similar to B. anthracis SCWP (10). Bioinformatic searches of B. anthracis genome sequences for WchA (WciI, CpsE9N/L) homologs identified the locus tag GBAA_RS27680 upstream of acrR in the scwp2 cluster (Figure 4b). B. an-thracis C55-(PO4)2-GlcNAc-specific glycosyltransferases are expected to extend the product of Reaction 1 first with β-GlcNAc (Reaction 2) and then with β-ManNAc (Reaction 3). Candidate glycosyltransferases for Reactions 2 and 3 are not readily obvious. The identity of the responsible enzymes may be guided from the S. pneumoniae CPS database for glycosyltransferases with retaining or inverting stereospecificity (α- or β-linkages) (87) and will need to be confirmed with biochemical tests. Nonetheless, it is likely that these enzymes are encoded within scwp2 given their genetic association with wchA- and wzx-like genes in S. pneumoniae CPS clusters (87).
UDP-GlcNAc, the substrate for Reactions 1 and 2, is also required for peptidoglycan synthesis and supplied by N-acetylglucosamine-1-PO4 uridyltransferase (GlmU). SCWP synthesis requires UDP-ManNAc, which is derived from UDP-GlcNAc by UDP-GlcNAc 2-epimerase. GneZ (encoded in sps) and GneY (encoded in scwp1) have been characterized as the corresponding enzymes in B. anthracis (Reaction 7) (128, 146). Galactosyl modifications of the SCWP require B. anthracis to synthesize UDP-Gal. The genome of B. anthracis encodes two functional UDP-Glc 4-epimerases. One of these genes, galE1, is located in the sps cluster (Figure 4b). The second gene is located elsewhere on the chromosome and has been shown to be expressed only during sporu-lation; it functions preferentially as UDP-GlcNAc 4-epimerase to synthesize UDP-GalNAc, a substrate for glycosyltransferases that modify BclA (27). BclA is the exosporium glycoprotein of B. anthracis spores (13, 135). galE1 is expressed during vegetative growth (128) and is the proposed candidate for UDP-GlcNAc 4-epimerase activity in support of SCWP synthesis. RfaB enzymes (COG0438) function as UDP-Gal-dependent galactosyltransferases (36). The B. anthracis genome harbors several genes for RfaB-type enzymes in sps and scwp2 (Figure 4b). These genes are candidate glycosyltransferases for galactosyl transfer to the SCWP trisaccharide repeat (Reactions 4–6).
Translocation and Polymerization of SCWP Units
Translocation of the SCWP precursor across the membrane requires Wzx flippase (RfbX) (Reaction 9); a candidate for the putative SCWP flippase activity is encoded within the scwp2 cluster (Figure 4b). Wzy polymerase/WaaL (Reaction 10) is responsible for generating SCWP polymers. WpaA (slc encoded) and WpaB (scwp2 encoded) belong to the same clan as members of the Wzy polymerase/WaaL family of proteins. Both genes are involved in the synthesis of the SCWP of B. anthracis (105). B. anthracis mutants blocking the expression of a single O-antigen ligase–like gene, wpaA or wpaB, cannot synthesize SCWP and form misshapen cells without S-layers (105). On the other hand, mutants expressing a single gene for Wzy-like/O-antigen ligase–like enzymes display either assembly defects for S-layer-associated murein hydrolases BslS and BslT (wpaA mutant) or defects in the length of B. anthracis cell chains and in S-layer assembly (wpaB mutant) (105). LCP enzymes are thought to transfer SCWP tethered to murein linkage units from C55-(PO4) to peptidoglycan, thereby generating phosphodiester bonds with the O6 of MurNAc that can be hydrolyzed with hydrofluoric acid and released from the bacterial envelope (61). B. anthracis expresses six genes whose products may catalyze this reaction: LcpB1, LcpB2, LcpB3, LcpB4 and LcpC, and LcpD (Reaction 11) (79, 80).
PERSPECTIVE
In addition to the secreted toxins and capsules of B. anthracis and pathogenic B. cereus, S-layer and S-layer-associated proteins are key virulence factors enabling host invasion and escape from phagocytic clearance during the pathogenesis of anthrax or anthrax-like diseases. However, only some of the S-layer-associated proteins have been studied. Further, the mechanisms whereby S-layer proteins and S-layer-associated proteins are deposited into the bacterial envelope to exert their functions are still poorly understood. The SCWP of B. anthracis and pathogenic B. cereus assumes strain-specific characteristics that affect the assembly of S-layers and the function of bacteriophage-encoded murein hydrolases. The genes and enzymatic mechanisms of SCWP synthesis and assembly are only beginning to be discovered. Additional insights into this field may explain the unique attributes of pathogenic B. cereus isolates, their envelope characteristics, the range of their mammalian hosts, and the tropism of bacteriophages.
VIRULENCE PLASMIDS OF B. CEREUS ISOLATES CAUSING ANTHRAX-LIKE DISEASE.
In addition to B. anthracis, many other B. cereus s.l. isolates, including B. cereus biovar anthracis CI and B. cereus G9241, have acquired pXO1-like plasmids and with them the ability to secrete anthrax toxins (54, 55, 57, 67, 149) (Figure 1). B. cereus G9241 and B. cereus biovar anthracis CI, along with B. anthracis, belong to clade 1 of B. cereus s.l. (152). B. cereus biovar anthracis CI was isolated from mammals (gorilla, chimpanzee, elephant, and goat) with anthrax-like disease in West and Central Africa (6, 67). B. cereus G9241 and its close relatives were isolated in North America from humans with severe pneumonia or cutaneous anthrax-like disease (55, 86). pXO1-like plasmids of B. cereus isolates provide for the expression of hyaluronic acid (HA) capsule via the hasACB operon, which is mutated and nonfunctional in B. anthracis (16, 104) (Figure 1). B. cereus biovar anthracis CI produces both HA and PDGA capsular material, as these strains acquired a pXO2-like plasmid (6, 16, 66). In contrast, B. cereus G9241 synthesizes HA in addition to another capsular polysaccharide designated BPS (B. cereus exo-polysaccharide), which requires the expression of the bpsXABCDEFGH genes located on the pBC210 virulence plasmid (104) (Figure 1). B. cereus G9241 and B. cereus biovar anthracis CI express atxA and carry an inactivating mutation in their chromosomal plcR gene that is distinct from that of B. anthracis (6, 16, 125). pBC210 carries an atxA-like gene (atxA2) that contributes to the pathogenesis of anthrax-like disease in B. cereus G9241 (125). pBC210 also carries pag2 (60% identity with pag) and lef 2 (36% identity with lef) (Figure 1); the lef 2 product shows the presence of a putative PA binding domain; however, the metalloprotease domain of LF is replaced with an ADP-ribosyltransferase domain (41, 143). The lef 2 product, which has been designated Certhrax, is translocated by PA2 into host cells, modifies the host factor vinculin, and disrupts focal adhesion complexes (131, 143). These features of B. anthracis and B. cereus may account for the observed variation in disease severity and geographic spread associated with spores causing anthrax or anthrax-like disease.
BACTERIAL S-LAYERS.
Bacterial S-layers are assembled from proteins that form monomolecular, two-dimensional crystalline arrays on the microbial envelope. S-layer proteins (SLPs) are endowed with a crystallization domain that, when purified and examined in isolation, promotes self-assembly of crystalline arrays (113, 124). A broad range of bacteria and archaea assemble S-layers as their outermost envelope component (4, 39, 133). S-layers form a porous mesh from unit sizes of 3–100 nm and pore sizes of 2–10 nm, thereby functioning as protective barriers with selective permeability (113). The thickness of S-layers varies from 5 to 20 nm in eubacteria but may reach 70 nm in archaea (113). In archaea, SLPs are tethered to the bacterial membrane via a hydrophobic peptide or lipid modification (4). In eubacteria, SLPs are retained on cell surfaces via noncovalent interactions with surface polymers, i.e., lipopolysaccharide (LPS) in gram-negative bacteria and wall polysaccharides in gram-positive bacteria (113). With nanobodies being used to block the formation of two-dimensional crystalline arrays, the structure of the crystallization domain of Geobacillus stearothermophilus SbsB was determined via X-ray crystallography (8). The crystallization domain comprises six subdomains with immunoglobulin-like structure (Pfam 02368) folding into a ϕ -shaped, disk-like monomeric unit (8). Interdomain interactions between units are stabilized by calcium ion coordination, which serves as a switch for the formation of a condensed quaternary structure (8). Pores approximately 30 Å in diameter are formed at the interface between three adjacent subunits (8). The crystallization domain of SbsB (residues 202–920) displays 22% amino acid identity with the crystallization domain of B. anthracis Sap (residues 211–814), suggesting that Sap may fold into a three-dimensional structure similar to SbsB.
S-LAYER REGULATION OF CHAIN LENGTHS IN B. ANTHRACIS.
Immediately following germination, B. anthracis emerges as a single vegetative cell that begins to elongate and divide, forming chains of 4–16 cells during mid-logarithmic growth (5). As B. anthracis vegetative forms approach stationary phase, chain length is decreased (2–4 cells) until eventually short chains and single cells predominate (5). Increased chain length is an impediment to B. anthracis host cell invasion and protects bacilli from phagocytosis by macrophages (147). In addition to SLPs, peptidoglycan hydrolases (BslO and BslS) function as determinants of B. anthracis chain length. Thus, SLPs and S-layer-associated proteins (BSLs) affect B. anthracis invasion of host cells (BslA) and evasion from innate defenses of the mammalian hosts.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Aanensen DM, Mavroidi A, Bentley SD, Reeves PR, Spratt BG. Predicted functions and linkage specificities of the products of the Streptococcus pneumoniae capsular biosynthetic loci. J Bacteriol. 2007;189:7856–76. doi: 10.1128/JB.00837-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agaisse H, Gominet M, Økstad OA, Kolstø AB, Lereclus D. PlcR is a pleiotropic regulator of extracellular virulence factor gene expression in Bacillus thuringiensis. Mol Microbiol. 1999;32:1043–53. doi: 10.1046/j.1365-2958.1999.01419.x. [DOI] [PubMed] [Google Scholar]
- 3.Agrawal A, Lingappa J, Leppla SH, Agrawal S, Jabbar A, et al. Impairment of dendritic cells and adaptive immunity by anthrax lethal toxin. Nature. 2003;424:329–34. doi: 10.1038/nature01794. [DOI] [PubMed] [Google Scholar]
- 4.Albers SV, Meyer BH. The archaeal cell envelope. Nat Rev Microbiol. 2011;9:414–26. doi: 10.1038/nrmicro2576. [DOI] [PubMed] [Google Scholar]
- 5.Anderson VJ, Kern JW, McCool JW, Schneewind O, Missiakas DM. The SLH domain protein BslO is a determinant of Bacillus anthracis chain length. Mol Microbiol. 2011;81:192–205. doi: 10.1111/j.1365-2958.2011.07688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Antonation KS, Grützmacher K, Dupke S, Mabon P, Zimmermann F, et al. Bacillus cereus biovar anthracis causing anthrax in sub-Saharan Africa—chromosomal monophyly and broad geographic distribution. PLOS Negl Trop Dis. 2016;10:e0004923. doi: 10.1371/journal.pntd.0004923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balderas MA, Nobles CL, Honsa ES, Alicki ER, Maresso AW. Hal is a Bacillus anthracis heme acquisition protein. J Bacteriol. 2012;194:5513–21. doi: 10.1128/JB.00685-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baranova E, Fronzes R, Garcia-Pino A, Van Gerven N, Papapostolou D, et al. SbsB structure and lattice reconstruction unveil Ca2+ triggered S-layer assembly. Nature. 2012;487:119–22. doi: 10.1038/nature11155. [DOI] [PubMed] [Google Scholar]
- 9.Bartkus JM, Leppla SH. Transcriptional regulation of the protective antigen gene of Bacillus anthracis. Infect Immun. 1989;57:2295–300. doi: 10.1128/iai.57.8.2295-2300.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bentley SD, Aanensen DM, Mavroidi A, Saunders D, Rabbinowitsch E, et al. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLOS Genet. 2006;2:e31. doi: 10.1371/journal.pgen.0020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bouillaut L, Perchat S, Arold S, Zorrilla S, Slamti L, et al. Molecular basis for group-specific activation of the virulence regulator PlcR by PapR heptapeptides. Nucleic Acids Res. 2008;36:3791–801. doi: 10.1093/nar/gkn149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bourgogne A, Drysdale M, Hilsenbeck SG, Peterson SN, Koehler TM. Global effects of virulence gene regulators in a Bacillus anthracis strain with both virulence plasmids. Infect Immun. 2003;71:2736–43. doi: 10.1128/IAI.71.5.2736-2743.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boydston JA, Chen P, Steichen CT, Turnbough CLJ. Orientation within the exosporium and structural stability of the collagen-like glycoprotein BclA of Bacillus anthracis. J Bacteriol. 2005;187:5310–17. doi: 10.1128/JB.187.15.5310-5317.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bradley KA, Mogridge J, Mourez M, Collier RJ, Young JA. Identification of the cellular receptor for anthrax toxin. Nature. 2001;414:225–29. doi: 10.1038/n35101999. [DOI] [PubMed] [Google Scholar]
- 15.Braunstein M, Brown AM, Kurtz S, Jacobs WRJ. Two nonredundant SecA homologues function in mycobacteria. J Bacteriol. 2001;183:6979–90. doi: 10.1128/JB.183.24.6979-6990.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brézillon C, Haustant M, Dupke S, Corre JP, Lander A, et al. Capsules, toxins and AtxA as virulence factors of emerging Bacillus cereus biovar anthracis. PLOS Negl Trop Dis. 2015;9:e0003455. doi: 10.1371/journal.pntd.0003455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bruckner V, Kovacs J, Denes G. Structure of poly-D-glutamic acid isolated from capsulated strains of B. anthracis. Nature. 1953;172:508. doi: 10.1038/172508a0. [DOI] [PubMed] [Google Scholar]
- 18.Candela T, Fouet A. Bacillus anthracis CapD, belonging to the γ-glutamyltranspeptidase family, is required for the covalent anchoring of capsule to peptidoglycan. Mol Microbiol. 2005;57:717–26. doi: 10.1111/j.1365-2958.2005.04718.x. [DOI] [PubMed] [Google Scholar]
- 19.Candela T, Maes E, Garenaux E, Rombouts Y, Krzewinski F, et al. Environmental and biofilm-dependent changes in a Bacillus cereus secondary cell wall polysaccharide. J Biol Chem. 2011;286:31250–62. doi: 10.1074/jbc.M111.249821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Candela T, Mock M, Fouet A. CapE, a 47-amino-acid peptide, is necessary for Bacillus anthracis polyglutamate capsule synthesis. J Bacteriol. 2005;187:7765–72. doi: 10.1128/JB.187.22.7765-7772.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan YGY, Frankel MB, Dengler V, Schneewind O, Missiakas DM. Staphylococcus aureus mutants lacking the LytR-CpsA-Psr (LCP) family of enzymes release wall teichoic acids into the extracellular medium. J Bacteriol. 2013;195:4650–59. doi: 10.1128/JB.00544-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choudhury B, Leoff C, Saile E, Wilkins P, Quinn CP, et al. The structure of the major cell wall polysaccharide of Bacillus anthracis is species specific. J Biol Chem. 2006;281:27932–41. doi: 10.1074/jbc.M605768200. [DOI] [PubMed] [Google Scholar]
- 23.Couture-Tosi E, Delacroix H, Mignot T, Mesnage S, Chami M, et al. Structural analysis and evidence for dynamic emergence of Bacillus anthracis S-layer networks. J Bacteriol. 2002;184:6448–56. doi: 10.1128/JB.184.23.6448-6456.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daffonchio D, Cherif A, Brusetti L, Rizzi A, Mora D, et al. Nature of polymorphisms in 16S-23S rRNA gene intergenic transcribed spacer fingerprinting of Bacillus and related genera. Appl Environ Microbiol. 2003;69:5128–37. doi: 10.1128/AEM.69.9.5128-5137.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dalbey RE, Wickner W. Leader peptidase catalyzes the release of exported proteins from the outer surface of the Escherichia coli plasma membrane. J Biol Chem. 1985;260:15925–31. [PubMed] [Google Scholar]
- 26.D’Elia MA, Henderson JA, Beveridge TJ, Heinrichs DE, Brown ED. The N-acetylmannosamine transferase catalyzes the first committed step of teichoic acid assembly in Bacillus subtilis and Staphylococcus aureus. J Bacteriol. 2009;191:4030–34. doi: 10.1128/JB.00611-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong S, Chesnokova ON, Turnbough CLJ, Pritchard DG. Identification of the UDP-N-acetylglucosamine 4-epimerase involved in exosporium protein glycosylation in Bacillus anthracis. J Bacteriol. 2009;191:7094–101. doi: 10.1128/JB.01050-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drysdale M, Bourgogne A, Hilsenbeck SG, Koehler TM. atxA controls Bacillus anthracis capsule synthesis via acpA and a newly discovered regulator, acpB. J Bacteriol. 2004;186:307–15. doi: 10.1128/JB.186.2.307-315.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drysdale M, Bourgogne A, Koehler TM. Transcriptional analysis of the Bacillus anthracis capsule regulators. J Bacteriol. 2005;187:5108–14. doi: 10.1128/JB.187.15.5108-5114.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drysdale M, Heninger S, Hutt J, Chen Y, Lyons CR, Koehler TM. Capsule synthesis by Bacillus anthracis is required for dissemination in murine inhalation anthrax. EMBO J. 2005;24:221–27. doi: 10.1038/sj.emboj.7600495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duesbery NS, Webb CP, Leppla SH, Gordon VM, Klimpel KR, et al. Proteolytic inactivation of Map-kinase-kinase by anthrax lethal factor. Science. 1998;280:734–37. doi: 10.1126/science.280.5364.734. [DOI] [PubMed] [Google Scholar]
- 32.Duong F, Wickner W. The SecDFyajC domain of preprotein translocase controls preprotein movement by regulating SecA membrane cycling. EMBO J. 1997;16:4871–79. doi: 10.1093/emboj/16.16.4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ebrahimi CM, Kern JW, Sheen TR, Ebrahimi-Fardooee MA, van Sorge NM, et al. Penetration of the blood-brain barrier by Bacillus anthracis requires the pXO1-encoded BslA protein. J Bacteriol. 2009;191:7165–73. doi: 10.1128/JB.00903-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Economou A, Pogliano JA, Beckwith J, Oliver DB, Wickner W. SecA membrane cycling at SecYEG is driven by distinct ATP binding and hydrolysis events and is regulated by SecD and SecF. Cell. 1995;83:1171–81. doi: 10.1016/0092-8674(95)90143-4. [DOI] [PubMed] [Google Scholar]
- 35.Economou A, Wickner W. SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell. 1994;78:835–43. doi: 10.1016/s0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 36.Endo A, Rothfield L. Studies of a phospholipid-requiring bacterial enzyme: I. Purification and properties of uridine diphosphate galactose; lipopolysaccharide alpha-3-galactosyl transferase. Biochemistry. 1969;8:3500–7. doi: 10.1021/bi00837a003. [DOI] [PubMed] [Google Scholar]
- 37.Etienne-Toumelin I, Sirard JC, Duflot E, Mock M, Fouet A. Characterization of the Bacillus anthracis S-layer: cloning and sequencing of the structural gene. J Bacteriol. 1995;177:614–20. doi: 10.1128/jb.177.3.614-620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ezzell JWJ, Abshire TG, Little SF, Lidgerding BC, Brown C. Identification of Bacillus anthracis by using monoclonal antibody to cell wall galactose-N-acetylglucosamine polysaccharide. J Clin Microbiol. 1990;28:223–31. doi: 10.1128/jcm.28.2.223-231.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fagan RP, Fairweather NF. Biogenesis and functions of bacterial S-layers. Nat Rev Microbiol. 2014;12:211–22. doi: 10.1038/nrmicro3213. [DOI] [PubMed] [Google Scholar]
- 40.Farha MA, Leung A, Sewell EW, D’Elia MA, Allison SE, et al. Inhibition of WTA synthesis blocks the cooperative action of PBPs and sensitizes MRSA to β-lactams. ACS Chem Biol. 2013;8:226–33. doi: 10.1021/cb300413m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fieldhouse RJ, Turgeon Z, White D, Merrill AR. Cholera- and anthrax-like toxins are among several new ADP-ribosyltransferases. PLOS Comput Biol. 2010;6:e1001029. doi: 10.1371/journal.pcbi.1001029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fischetti VA. Bacteriophage endolysins: a novel anti-infective to control gram-positive pathogens. Int J Med Microbiol. 2010;300:357–62. doi: 10.1016/j.ijmm.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Forsberg LS, Abshire TG, Friedlander A, Quinn CP, Kannenberg EL, Carlson RW. Localization and structural analysis of a conserved pyruvylated epitope in Bacillus anthracis secondary cell wall polysaccharides and characterization of the galactose deficient wall polysaccharide from avirulent B. anthracis CDC 684. Glycobiology. 2012;22:1103–17. doi: 10.1093/glycob/cws080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Forsberg LS, Choudhury B, Leoff C, Marston CK, Hoffmaster AR, et al. Secondary cell wall polysaccharides from Bacillus cereus strains G9241, 03BB87 and 03BB102 causing fatal pneumonia share similar glycosyl structures with the polysaccharides from Bacillus anthracis. Glycobiology. 2011;21:934–48. doi: 10.1093/glycob/cwr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fouet A. AtxA, a Bacillus anthracis global virulence regulator. Res Microbiol. 2010;161:735–42. doi: 10.1016/j.resmic.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 46.Ganguly J, Low LY, Kamal N, Saile E, Forsberg LS, et al. The secondary cell wall polysaccharide of Bacillus anthracis provides the specific binding ligand for the C-terminal cell wall-binding domain of two phage endolysins, PlyL and PlyG. Glycobiology. 2013;23:820–32. doi: 10.1093/glycob/cwt019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garufi G, Hendrickx AP, Beeri K, Kern JW, Sharma A, et al. Synthesis of lipoteichoic acids in Bacillus anthracis. J Bacteriol. 2012;194:4312–21. doi: 10.1128/JB.00626-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ginsberg C, Zhang YH, Yuan Y, Walker S. In vitro reconstitution of two essential steps in wall teichoic acid biosynthesis. ACS Chem Biol. 2006;1:25–28. doi: 10.1021/cb0500041. [DOI] [PubMed] [Google Scholar]
- 49.Gordon VM, Klimpel KR, Arora N, Henderson MA, Leppla SH. Proteolytic activation of bacterial toxins by eukaryotic cells is performed by furin and by additional cellular proteases. Infect Immun. 1995;63:82–87. doi: 10.1128/iai.63.1.82-87.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Green BD, Battisti L, Koehler TM, Thorne CB, Ivins BE. Demonstration of a capsule plasmid in Bacillus anthracis. Infect Immun. 1985;49:291–97. doi: 10.1128/iai.49.2.291-297.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guignot J, Mock M, Fouet A. AtxA activates the transcription of genes harbored by both Bacillus anthracis virulence plasmids. FEMS Microbiol Lett. 1997;147:203–7. doi: 10.1111/j.1574-6968.1997.tb10242.x. [DOI] [PubMed] [Google Scholar]
- 52.Hancock IC, Wiseman G, Baddiley J. Biosynthesis of the unit that links teichoic acid to the bacterial wall: inhibition by tunicamycin. FEBS Lett. 1976;69:75–80. doi: 10.1016/0014-5793(76)80657-6. [DOI] [PubMed] [Google Scholar]
- 53.Helgason E, Økstad OA, Caugant DA, Johansen HA, Fouet A, et al. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis—one species on the basis of genetic evidence. Appl Environ Microbiol. 2000;66:2627–30. doi: 10.1128/aem.66.6.2627-2630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoffmaster AR, Hill KK, Gee JE, Marston CK, De BK, et al. Characterization of Bacillus cereus isolates associated with fatal pneumonias: strains are closely related to Bacillus anthracis and harbor B. anthracis virulence genes. J Clin Microbiol. 2006;44:3352–60. doi: 10.1128/JCM.00561-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoffmaster AR, Ravel J, Rasko DA, Chapman GD, Chute MD, et al. Identification of anthrax toxin genes in a Bacillus cereus associated with an illness resembling inhalation anthrax. PNAS. 2004;101:8449–54. doi: 10.1073/pnas.0402414101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jensen GB, Hensen BM, Eilenberg J, Mahillon J. The hidden lifestyles of Bacillus cereus and relatives. Environ Microbiol. 2003;5:631–40. doi: 10.1046/j.1462-2920.2003.00461.x. [DOI] [PubMed] [Google Scholar]
- 57.Kaminska PS, Yernazarova A, Drewnowska JM, Zambrowski G, Swiecicka I. The worldwide distribution of genetically and phylogenetically diverse Bacillus cereus isolates harbouring Bacillus anthracis-like plasmids. Environ Microbiol Rep. 2015;7:738–45. doi: 10.1111/1758-2229.12305. [DOI] [PubMed] [Google Scholar]
- 58.Kawai Y, Marles-Wright J, Cleverley RM, Emmins R, Ishikawa S, et al. A widespread family of bacterial cell wall assembly proteins. EMBO J. 2011;30:4931–41. doi: 10.1038/emboj.2011.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaya S, Yokoyama K, Araki Y, Ito E. N-acetylmannosaminyl(1–4)N-acetylglucosamine, a linkage unit between glycerol teichoic acid and peptidoglycan in cell walls of several Bacillus strains. J Bacteriol. 1984;158:990–96. doi: 10.1128/jb.158.3.990-996.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Keim PS, Wagner DM. Humans and evolutionary and ecological forces shaped the phylogeography of recently emerged diseases. Nat Rev Microbiol. 2009;7:813–21. doi: 10.1038/nrmicro2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kern J, Ryan C, Faull K, Schneewind O. Bacillus anthracis surface-layer proteins assemble by binding to the secondary cell wall polysaccharide in a manner that requires csaB and tagO. J Mol Biol. 2010;401:757–75. doi: 10.1016/j.jmb.2010.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kern JW, Schneewind O. BslA, a pXO1-encoded adhesin of Bacillus anthracis. Mol Microbiol. 2008;68:504–15. doi: 10.1111/j.1365-2958.2008.06169.x. [DOI] [PubMed] [Google Scholar]
- 63.Kern JW, Wilton R, Zhang R, Binkowski A, Joachimiak A, Schneewind O. Structure of the SLH domains from Bacillus anthracis surface array protein. J Biol Chem. 2011;286:26042–49. doi: 10.1074/jbc.M111.248070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kern VJ, Kern JW, Theriot JA, Schneewind O, Missiakas DM. Surface (S)-layer proteins Sap and EA1 govern the binding of the S-layer associated protein BslO at the cell septa of Bacillus anthracis. J Bacteriol. 2012;194:3833–40. doi: 10.1128/JB.00402-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kintzer AF, Thoren KL, Sterling HJ, Dong KC, Feld GK, et al. The protective antigen component of anthrax toxin forms functional octameric complexes. J Mol Biol. 2009;392:614–29. doi: 10.1016/j.jmb.2009.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Klee SR, Brzuszkiewicz EB, Nattermann H, Brüggemann H, Dupke S, et al. The genome of a Bacillus isolate causing anthrax in chimpanzees combines chromosomal properties of B. cereus with B. anthracis virulence plasmids. PLOS ONE. 2010;5:e10986. doi: 10.1371/journal.pone.0010986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Klee SR, Ozel M, Appel B, Boesch C, Ellerbrok H, et al. Characterization of Bacillus anthracis-like bacteria isolated from wild great apes from Cote d’Ivoire and Cameroon. J Bacteriol. 2006;188:5333–44. doi: 10.1128/JB.00303-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Koch R. Die Ätiologie der Milzbrand-Krankheit, begründet auf die Entwicklungsgeschichte des Bacillus anthracis. Beitr Biol Pflanz. 1876;2:277–310. [Google Scholar]
- 69.Koehler TM, Dai Z, Kaufman-Yarbray M. Regulation of the Bacillus anthracis protective antigen gene: CO2 and a trans-acting element activate transcription from one of two promoters. J Bacteriol. 1994;176:586–95. doi: 10.1128/jb.176.3.586-595.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kojima N, Araki Y, Ito E. Structure of linkage region between ribitol teichoic acid and peptidoglycan in cell walls of Staphylococcus aureus H. J Biol Chem. 1983;258:9043–45. [PubMed] [Google Scholar]
- 71.Kojima N, Arakai Y, Ito E. Structure of the linkage units between ribitol teichoic acids and peptidoglycan. J Bacteriol. 1985;161:299–306. doi: 10.1128/jb.161.1.299-306.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kolstø AB, Tourasse NJ, Økstad OA. What sets Bacillus anthracis apart from other Bacillus species? Annu Rev Microbiol. 2009;63:451–76. doi: 10.1146/annurev.micro.091208.073255. [DOI] [PubMed] [Google Scholar]
- 73.Krantz BA, Melnyk RA, Zhang S, Juris SJ, Lacy DB, et al. A phenylalanine clamp catalyzes protein translocation through the anthrax toxin pore. Science. 2005;309:777–81. doi: 10.1126/science.1113380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lazarevic V, Abellan FX, Möller SB, Karamata D, Mauel C. Comparison of ribitol and glycerol teichoic acid genes in Bacillus subtilis W23 and 168: identical function, similar divergent organization, but different regulation. Microbiology. 2002;148:815–24. doi: 10.1099/00221287-148-3-815. [DOI] [PubMed] [Google Scholar]
- 75.Leoff C, Choudhury B, Saile E, Quinn CP, Carlson RW, Kannenberg EL. Structural elucidation of the non-classical secondary cell wall polysaccharide from Bacillus cereus ATCC 10987: Comparison with the polysaccharide from Bacillus anthracis and B. cereus type strain ATCC 14579 reveals both unique and common structural features. J Biol Chem. 2008;283:29812–21. doi: 10.1074/jbc.M803234200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Leppla SH. Anthrax toxin edema factor: a bacterial adenylate cyclase that increases cyclin AMP concentrations in eukaryotic cells. PNAS. 1982;79:3162–66. doi: 10.1073/pnas.79.10.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Levinsohn JL, Newman ZL, Hellmich KA, Fattah R, Getz MA, et al. Anthrax lethal factor cleavage of Nlrp1 is required for activation of the inflammasome. PLOS Pathog. 2012;8:e1002638. doi: 10.1371/journal.ppat.1002638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lill R, Dowhan W, Wickner W. The ATPase activity of SecA is regulated by acidic phospholipids, SecY, and the leader and mature domains of precursor proteins. Cell. 1990;60:271–80. doi: 10.1016/0092-8674(90)90742-w. [DOI] [PubMed] [Google Scholar]
- 79.Liszewski Zilla M, Chan YG, Lunderberg JM, Schneewind O, Missiakas D. LytR-CpsA-Psr enzymes as determinants of Bacillus anthracis secondary cell wall polysaccharide assembly. J Bacteriol. 2015;197:343–53. doi: 10.1128/JB.02364-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liszewski Zilla M, Lunderberg JM, Schneewind O, Missiakas D. Bacillus anthracis lcp genes support vegetative growth, envelope assembly and spore formation. J Bacteriol. 2015;197:3731–41. doi: 10.1128/JB.00656-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Low LY, Yang C, Perego M, Osterman A, Liddington RC. Structure and lytic activity of a Bacillus anthracis prophage endolysin. J Biol Chem. 2005;280:35433–39. doi: 10.1074/jbc.M502723200. [DOI] [PubMed] [Google Scholar]
- 82.Lunderberg JM, Liszewski Zilla M, Missiakas D, Schneewind O. Bacillus anthracis tagO is required for vegetative growth and secondary cell wall polysaccharide synthesis. J Bacteriol. 2015;197:3731–41. doi: 10.1128/JB.00494-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lunderberg JM, Nguyen-Mau SM, Richter GS, Wang YT, Dworkin J, et al. Bacillus anthracis acetyltransferases PatA1 and PatA2 modify the secondary cell wall polysaccharide and affect the assembly of S-layer proteins. J Bacteriol. 2013;195:977–89. doi: 10.1128/JB.01274-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Maresso AW, Chapa TJ, Schneewind O. Surface protein IsdC and sortase B are required for heme-iron scavenging of Bacillus anthracis. J Bacteriol. 2006;188:8145–52. doi: 10.1128/JB.01011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Maresso AW, Garufi G, Schneewind O. Bacillus anthracis secretes proteins that mediate heme acquisition from hemoglobin. PLOS Pathog. 2008;4:e1000132. doi: 10.1371/journal.ppat.1000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Marston CK, Ibrahim H, Lee P, Churchwell G, Gumke M, et al. Anthrax toxin-expressing Bacillus cereus isolated from an anthrax-like eschar. PLOS ONE. 2016;11:e0156987. doi: 10.1371/journal.pone.0156987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mavroidi A, Aanensen DM, Godoy D, Skovsted IC, Kaltoft MS, et al. Genetic relatedness of the Streptococcus pneumoniae capsular biosynthetic loci. J Bacteriol. 2007;189:7841–55. doi: 10.1128/JB.00836-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mazmanian SK, Skaar EP, Gaspar AH, Humayun M, Gornicki P, et al. Passage of heme-iron across the envelope of Staphylococcus aureus. Science. 2003;299:906–9. doi: 10.1126/science.1081147. [DOI] [PubMed] [Google Scholar]
- 89.Mesnage S, Fontaine T, Mignot T, Delepierre M, Mock M, Fouet A. Bacterial SLH domain proteins are non-covalently anchored to the cell surface via a conserved mechanism involving wall polysaccharide pyruvylation. EMBO J. 2000;19:4473–84. doi: 10.1093/emboj/19.17.4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mesnage S, Fouet A. Plasmid-encoded autolysin in Bacillus anthracis: modular structure and catalytic properties. J Bacteriol. 2002;184:331–34. doi: 10.1128/JB.184.1.331-334.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mesnage S, Tosi-Couture E, Gounon P, Mock M, Fouet A. The capsule and S-layer: two independent and yet compatible macromolecular structures in Bacillus anthracis. J Bacteriol. 1998;180:52–58. doi: 10.1128/jb.180.1.52-58.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mesnage S, Tosi-Couture E, Mock M, Fouet A. The S-layer homology domain as a means for anchoring heterologous proteins on the cell surface of Bacillus anthracis. J Appl Microbiol. 1999;87:256–60. doi: 10.1046/j.1365-2672.1999.00880.x. [DOI] [PubMed] [Google Scholar]
- 93.Mesnage S, Tosi-Couture E, Mock M, Gounon P, Fouet A. Molecular characterization of the Bacillus anthracis main S-layer component: evidence that it is the major cell-associated antigen. Mol Microbiol. 1997;23:1147–55. doi: 10.1046/j.1365-2958.1997.2941659.x. [DOI] [PubMed] [Google Scholar]
- 94.Mignot T, Couture-Tosi E, Mesnage S, Mock M, Fouet A. In vivo Bacillus anthracis gene expression requires PagR as an intermediate effector of the AtxA signalling cascade. Int J Med Microbiol. 2004;293:619–24. doi: 10.1078/1438-4221-00306. [DOI] [PubMed] [Google Scholar]
- 95.Mignot T, Mock M, Fouet A. A plasmid-encoded regulator couples the synthesis of toxins and surface structures in Bacillus anthracis. Mol Microbiol. 2003;47:917–27. doi: 10.1046/j.1365-2958.2003.03345.x. [DOI] [PubMed] [Google Scholar]
- 96.Mignot T, Mock M, Robichon D, Landier A, Lereclus D, Fouet A. The incompatibility between the PlcR- and AtxA-controlled regulons may have selected a nonsense mutation in Bacillus anthracis. Mol Microbiol. 2001;42:1189–98. doi: 10.1046/j.1365-2958.2001.02692.x. [DOI] [PubMed] [Google Scholar]
- 97.Milne J, Furlong D, Hanna PC, Wall JS, Collier RJ. Anthrax protective antigen forms oligomers during intoxication of mammalian cells. J Biol Chem. 1994;269:20607–12. [PubMed] [Google Scholar]
- 98.Milne JC, Blanke SR, Hanna PC, Collier RJ. Protective antigen-binding domain of anthrax lethal factor mediates translocation of a heterologous protein fused to its amino- or carboxy-terminus. Mol Microbiol. 1995;15:661–66. doi: 10.1111/j.1365-2958.1995.tb02375.x. [DOI] [PubMed] [Google Scholar]
- 99.Mo KF, Li X, Li H, Low LY, Quinn CP, Boons GJ. Endolysins of Bacillus anthracis bacteriophages recognize unique carbohydrate epitopes of vegetative cell wall polysaccharides with high affinity and selectivity. J Am Chem Soc. 2012;134:15556–62. doi: 10.1021/ja3069962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Moayeri M, Leppla SH, Vrentas C, Pomerantsev AP, Liu S. Anthrax pathogenesis. Annu Rev Microbiol. 2015;69:185–208. doi: 10.1146/annurev-micro-091014-104523. [DOI] [PubMed] [Google Scholar]
- 101.Molnár J, Prágai B. Attempts to detect the presence of teichoic acid in Bacillus anthracis. Acta Microbiol Acad Sci Hung. 1971;18:105–8. [PubMed] [Google Scholar]
- 102.Nguyen-Mau S-M, Oh S-Y, Kern V, Missiakas D, Schneewind O. Secretion genes as determinants of Bacillus anthracis chain length. J Bacteriol. 2012;194:3841–50. doi: 10.1128/JB.00384-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nguyen-Mau S-M, Oh S-Y, Schneewind DI, Missiakas D, Schneewind O. Bacillus anthracis SlaQ promotes S-layer protein assembly. J Bacteriol. 2015;197:3216–17. doi: 10.1128/JB.00492-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Oh S-Y, Budzik JM, Garufi G, Schneewind O. Two capsular polysaccharides enable Bacillus cereus G9241 to cause anthrax-like disease. Mol Microbiol. 2011;79:455–70. doi: 10.1111/j.1365-2958.2011.07582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Oh S-Y, Lunderberg JM, Chateau A, Schneewind O, Missiakas D. Genes required for Bacillus anthracis secondary cell wall polysaccharide synthesis. J Bacteriol. 2016;199:e00613–16. doi: 10.1128/JB.00613-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Oh S-Y, Richter SG, Missiakas DM, Schneewind O. Glutamate racemase mutants of Bacillus anthracis. J Bacteriol. 2015;197:1854–61. doi: 10.1128/JB.00070-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Okinaka R, Cloud K, Hampton O, Hoffmaster A, Hill K, et al. Sequence, assembly and analysis of pX01 and pX02. J Appl Microbiol. 1999;87:261–62. doi: 10.1046/j.1365-2672.1999.00883.x. [DOI] [PubMed] [Google Scholar]
- 108.Okinaka RT, Price EP, Wolken SR, Gruendike JM, Chung WK, et al. An attenuated strain of Bacillus anthracis (CDC 684) has a large chromosomal inversion and altered growth kinetics. BMC Genom. 2011;12:477–90. doi: 10.1186/1471-2164-12-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Oliver DB, Beckwith J. E. coli mutant pleiotropically defective in the export of secreted proteins. Cell. 1981;25:765–72. doi: 10.1016/0092-8674(81)90184-7. [DOI] [PubMed] [Google Scholar]
- 110.Plaut RD, Beaber JW, Zemansky J, Kaur AP, George M, et al. Genetic evidence for the involvement of the S-layer protein gene sap and the sporulation genes spo0A, spo0B, and spo0F in phage AP50c infection of Bacillus anthracis. J Bacteriol. 2014;196:1143–54. doi: 10.1128/JB.00739-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Preisz H. Experimentelle Studien über Virulenz, Empfänglichkeit und Immunität beim Milzbrand. Z Immunitäts-Forsch. 1909;5:341–452. [Google Scholar]
- 112.Priest FG, Barker M, Baillie LW, Holmes EC, Maiden MC. Population structure and evolution of the Bacillus cereus group. J Bacteriol. 2004;186:7959–70. doi: 10.1128/JB.186.23.7959-7970.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pum D, Toca-Herrera JL, Sleytr UB. S-layer protein self-assembly. Int J Mol Sci. 2013;14:2484–501. doi: 10.3390/ijms14022484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rasko DA, Altherr MR, Han CS, Ravel J. Genomics of the Bacillus cereus group of organisms. FEMS Microbiol Rev. 2005;29:303–29. doi: 10.1016/j.femsre.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 115.Rasko DA, Rosovitz MJ, Økstad OA, Fouts DE, Jiang L, et al. Complete sequence analysis of novel plasmids from emetic and periodontal Bacillus cereus isolates reveals a common evolutionary history among the B. cereus-group plasmids, including Bacillus anthracis pXO1. J Bacteriol. 2007;189:52–64. doi: 10.1128/JB.01313-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Read TD, Peterson SN, Tourasse N, Baille LW, Paulsen IT, et al. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature. 2003;423:81–86. doi: 10.1038/nature01586. [DOI] [PubMed] [Google Scholar]
- 117.Reniere ML, Ukpabi GN, Harry SR, Stec DF, Krull R, et al. The IsdG-family of haem oxygenases degrades haem to a novel chromophore. Mol Microbiol. 2010;75:1529–38. doi: 10.1111/j.1365-2958.2010.07076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Richter GS, Anderson VJ, Garufi G, Lu L, Joachimiak A, et al. Capsule anchoring in Bacillus anthracis occurs by a transpeptidation mechanism that is inhibited by capsidin. Mol Microbiol. 2009;71:404–20. doi: 10.1111/j.1365-2958.2008.06533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ristroph JD, Ivins BE. Elaboration of Bacillus anthracis antigens in a new, defined culture medium. Infect Immun. 1983;39:483–86. doi: 10.1128/iai.39.1.483-486.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Robertson DL, Leppla SH. Molecular cloning and expression in Escherichia coli of the lethal factor gene of Bacillus anthracis. Gene. 1986;44:71–78. doi: 10.1016/0378-1119(86)90044-2. [DOI] [PubMed] [Google Scholar]
- 121.Rubinchik E, Schneider T, Elliott M, Scott WR, Pan J, et al. Mechanism of action and limited cross-resistance of new lipopeptide MX-2401. Antimicrob Agents Chemother. 2011;55:2743–54. doi: 10.1128/AAC.00170-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ruthel G, Ribot WJ, Bavari S, Hoover T. Time-lapse confocal imaging of development of Bacillus anthracis in macrophages. J Infect Dis. 2004;189:1313–16. doi: 10.1086/382656. [DOI] [PubMed] [Google Scholar]
- 123.Samuelson JC, Chen M, Jiang F, Moller I, Wiedmann M, et al. YidC mediates membrane protein insertion in bacteria. Nature. 2000;406:637–41. doi: 10.1038/35020586. [DOI] [PubMed] [Google Scholar]
- 124.Sará M, Sleytr UB. S-layer proteins. J Bacteriol. 2000;182:859–68. doi: 10.1128/jb.182.4.859-868.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Scarff JM, Raynor MJ, Seldina YI, Ventura CL, Koehler TM, O’Brien AD. The roles of AtxA orthologs in virulence of anthrax-like Bacillus cereus G9241. Mol Microbiol. 2016;102:545–61. doi: 10.1111/mmi.13478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Schneewind O, Missiakas DM. Protein secretion and surface display in gram-positive bacteria. Philos Trans R Soc Lond B. 2012;367:1123–39. doi: 10.1098/rstb.2011.0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Schuch R, Nelson D, Fischetti VA. A bacteriolytic agent that detects and kills Bacillus anthracis. Nature. 2002;418:884–89. doi: 10.1038/nature01026. [DOI] [PubMed] [Google Scholar]
- 128.Schuch R, Pelzek AJ, Raz A, Euler CW, Ryan PA, et al. Use of a bacteriophage lysin to identify a novel target for antimicrobial development. PLOS ONE. 2013;8:e60754. doi: 10.1371/journal.pone.0060754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Schulze RJ, Komar J, Botte M, Allen WJ, Whitehouse S, et al. Membrane protein insertion and proton-motive-force-dependent secretion through the bacterial holo-translocon SecYEG-SecDF-YajC-YidC. PNAS. 2014;111:4844–99. doi: 10.1073/pnas.1315901111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Scobie HM, Rainey GJ, Bradley KA, Young JA. Human capillary morphogenesis protein 2 functions as an anthrax toxin receptor. PNAS. 2003;100:5170–74. doi: 10.1073/pnas.0431098100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Simon NC, Barbieri JT. Bacillus cereus Certhrax ADP-ribosylates vinculin to disrupt focal adhesion complexes and cell adhesion. J Biol Chem. 2014;289:10650–59. doi: 10.1074/jbc.M113.500710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Skaar EP, Gaspar AH, Schneewind O. Bacillus anthracis IsdG, a heme degrading monooxygenase. J Bacteriol. 2006;188:1071–80. doi: 10.1128/JB.188.3.1071-1080.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sleytr UB. Basic and applied S-layer research: an overview. FEMS Microbiol Rev. 1997;20:5–12. [Google Scholar]
- 134.Soldo B, Lazarevic V, Karamata D. tagO is involved in the synthesis of all anionic cell-wall polymers in Bacillus subtilis 168. Microbiology. 2002;148:2079–87. doi: 10.1099/00221287-148-7-2079. [DOI] [PubMed] [Google Scholar]
- 135.Steichen C, Chen P, Kearney JF, Turnbough CLJ. Identification of the immunodominant protein and other proteins of the Bacillus anthracis exosporium. J Bacteriol. 2003;185:1903–10. doi: 10.1128/JB.185.6.1903-1910.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tarlovsky Y, Fabian M, Solomaha E, Honsa E, Olson JS, Maresso AW. A Bacillus anthracis S-layer homology protein that binds heme and mediates heme delivery to IsdC. J Bacteriol. 2010;192:3503–11. doi: 10.1128/JB.00054-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tourasse NJ, Helgason E, Økstad OA, Hegna IK, Kolstø AB. The Bacillus cereus group: novel aspects of population structure and genome dynamics. J Appl Microbiol. 2006;101:579–93. doi: 10.1111/j.1365-2672.2006.03087.x. [DOI] [PubMed] [Google Scholar]
- 138.Tsirigotaki A, De Geyter J, Šoštaric N, Economou A, Karamanou S. Protein export through the bacterial Sec pathway. Nat Rev Microbiol. 2017;15:21–36. doi: 10.1038/nrmicro.2016.161. [DOI] [PubMed] [Google Scholar]
- 139.Turnbull PC. Definitive identification of Bacillus anthracis—a review. J Appl Microbiol. 1999;87:237–40. doi: 10.1046/j.1365-2672.1999.00876.x. [DOI] [PubMed] [Google Scholar]
- 140.Turnbull PC. Introduction: anthrax history, disease and ecology. Curr Top Microbiol Immunol. 2002;271:1–19. doi: 10.1007/978-3-662-05767-4_1. [DOI] [PubMed] [Google Scholar]
- 141.Uchida I, Hornung JM, Thorne CB, Klimpel KR, Leppla SH. Cloning and characterization of a gene whose product is a trans-activator of anthrax toxin synthesis. J Bacteriol. 1993;175:5329–38. doi: 10.1128/jb.175.17.5329-5338.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Uchida I, Makino S, Sekizaki T, Terakado N. Cross-talk to the genes for Bacillus anthracis capsule synthesis by atxA, the gene encoding the trans-activator of anthrax toxin synthesis. Mol Microbiol. 1997;23:1229–40. doi: 10.1046/j.1365-2958.1997.3041667.x. [DOI] [PubMed] [Google Scholar]
- 143.Visschedyk D, Rochon A, Tempel W, Dimov S, Park HW, Merrill AR. Certhrax toxin, an anthrax-related ADP-ribosyltransferase from Bacillus cereus. J Biol Chem. 2012;287:41089–102. doi: 10.1074/jbc.M112.412809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Vodkin MH, Leppla SH. Cloning of the protective antigen gene of Bacillus anthracis. Cell. 1983;34:693–97. doi: 10.1016/0092-8674(83)90402-6. [DOI] [PubMed] [Google Scholar]
- 145.Wang Y, Wei Y, Yuan S, Tao H, Dong J, et al. Bacillus anthracis S-layer protein BslA binds to extracellular matrix by interacting with laminin. BMC Microbiol. 2016;16:183. doi: 10.1186/s12866-016-0802-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wang YT, Missiakas D, Schneewind O. GneZ, a UDP-GlcNAc 2-epimerase, is required for S-layer assembly and vegetative growth of Bacillus anthracis. J Bacteriol. 2014;196:2969–78. doi: 10.1128/JB.01829-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wang YT, Oh SY, Hendrickx AP, Lunderberg JM, Schneewind O. Bacillus cereus G9241 S-layer assembly contributes to the pathogenesis of anthrax-like disease in mice. J Bacteriol. 2013;195:596–605. doi: 10.1128/JB.02005-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Weidenmaier C, Kokai-Kun JF, Kristian SA, Chanturiya T, Kalbacher H, et al. Role of teichoic acids in Staphylococcus aureus nasal colonization, a major risk factor in nosocomial infections. Nat Med. 2004;10:243–45. doi: 10.1038/nm991. [DOI] [PubMed] [Google Scholar]
- 149.Wright AM, Beres SB, Consamus EN, Long SW, Flores AR, et al. Rapidly progressive, fatal, inhalation anthrax-like infection in a human: case report, pathogen genome sequencing, pathology, and coordinated response. Arch Pathol Lab Med. 2011;135:1447–59. doi: 10.5858/2011-0362-SAIR.1. [DOI] [PubMed] [Google Scholar]
- 150.Yother J. Capsules of Streptococcus pneumoniae and other bacteria: paradigms for polysaccharide biosynthesis and regulation. Annu Rev Microbiol. 2011;65:563–81. doi: 10.1146/annurev.micro.62.081307.162944. [DOI] [PubMed] [Google Scholar]
- 151.Zhao J, Milne JC, Collier RJ. Effect of anthrax toxin’s lethal factor on ion channels formed by the protective antigen. J Biol Chem. 1995;270:18626–30. doi: 10.1074/jbc.270.31.18626. [DOI] [PubMed] [Google Scholar]
- 152.Zwick ME, Joseph SJ, Didelot X, Chen PE, Bishop-Lilly KA, et al. Genomic characterization of the Bacillus cereus sensu lato species: backdrop to the evolution of Bacillus anthracis. Genome Res. 2012;22:1512–24. doi: 10.1101/gr.134437.111. [DOI] [PMC free article] [PubMed] [Google Scholar]


