Abstract
Biomedical research in areas such as metabolic disorders, neuromodulatory, and immunomodulatory conditions involves lipid metabolism and demands a reliable and inexpensive method for quantification of short chain fatty acids (SCFAs). We report a GC-MS method for analysis of all straight-chain and branched-chain SCFAs using pentafluorobenzyl bromide (PFBBr) as derivatization reagent. We optimized the derivatization and GC-MS conditions using a mixture containing all eight SCFA standards, i.e., five straight-chain and three branched-chain SCFAs. The optimal derivatization conditions were derivatization time 90 min, temperature 60 °C, pH 7, and (CH3)2CO:H2O ratio 2:1 (v:v). Comparing the performance of different GC column configurations, a 30 m DB-225ms hyphenated with a 30 m DB-5ms column in tandem showed the best separation of SCFAs. Using the optimized experiment conditions, we simultaneously detected all SCFAs with much improved detection limit, 0.244 - 0.977 μM. We further applied the developed method to measure the SCFAs in mouse feces and all SCFAs were successfully quantified. The recovery rates of the eight SCFAs ranged from 55.7% to 97.9%.
Keywords: short chain fatty acids, GC-MS, PFBBr, metabolomics, hyphenated GC column
1. INTRODUCTION
Short chain fatty acids (SCFAs) are saturated aliphatic fatty acids with less than six carbon atoms. While five straight-chain SCFAs (formic acid, acetic acid, propionic acid, butyric acid, and valeric acid) are predominantly the end products of fermentation of dietary fibers by the anaerobic intestinal microbiota [1], the three branched-chain SCFAs (isobutyric acid, 2-methylbutyric acid, and isovaleric acid) are mainly derived from the catabolism of branched-chain amino acids such as valine, leucine, and isoleucine [2]. SCFAs play an important role in homeostasis due to their metabolic, neuromodulatory, and immunomodulatory actions. They can influence the growth and composition of gut microbiota, and thereby further affect the health of the host. While SCFAs are the main source of energy for the cells in colon, excess SCFAs can have other functions such as providing daily calorie needs and being involved in the metabolism of important nutrients such as carbohydrates and fats.
Emerging evidence indicates that SCFAs are associated with multiple metabolic diseases such as obesity, hypertension, and diabetes [3-5]. In fact, SCFAs stimulate leptin expression and inhibit lipolysis in adipocytes through G-coupled protein receptors. They also activate 5' adenosine monophosphate-activated protein kinase (AMPK) that acts as a major cellular fuel switch and a master regulator of metabolic homeostasis [6]. SCFAs also function in the synthesis of other metabolites. For instance, propionic acid may inhibit the synthesis of cholesterol in the liver [7]. Gastrointestinal disease could result in increased proteinous material in the colon and may increase the products of branched-chain amino acids [8]. Branched-chain amino acids are associated with the development of diabetes [9]. The oxidation of branched-chain amino acids provides energy for muscles, kidney, and other organs. As the derivatives of branched-chain amino acids, branched-chain fatty acids may become a signal for metabolic diseases.
Other than metabolic diseases, a reduction in SCFAs might induce alterations in the enteric nervous system and can contribute to gastrointestinal dysmotility in Parkinson's disease [10]. Butyric acid has a regulatory role in the skin immune system via increasing the gene expression of Treg-specific transcription factor foxp3 and IL-10 and expansion of Treg cells [11]. Butyric acid, as a gut product of dietary fiber by microflora, is also believed to play a critical role in cellular epigenetic function that promote the immune system by increasing Treg cells in the gut and extraenteric organs [12].
The measurement of SCFAs in biological samples receives considerable attention because of their important roles in physiological and pathological processes [13-15]. Different chemical derivatization reagents and extraction solvents were developed for analysis of SCFAs by gas chromatography (GC) coupled with a flame ionization detector (FID) or a mass spectrometer [16-21]. SCFAs were also analyzed by HPLC equipped with an electrochemical detector (ECD), a UV detector, or a mass spectrometer [22-24]. Some of the analytical methods for analysis SCFAs were summarized in a review paper [25].
While multiple analytical methods have been developed for analysis of SCFAs as described above, these methods are not able to simultaneously detect all SCFAs, especially formic acid and branched-chain SCFAs. However, formic acid and branched-chain SCFAs are pivotal in biological studies. For instance, formic acid has important regulatory role, and a low level of urinary formic acid correlates with increased blood pressure [26]. To analyze all these SCFAs, pentafluorobenzyl bromide (PFBBr) reagent was used as a derivatization reagent for GC-MS quantification of SCFAs in whole blood and urine from humans and mice [27, 28]. PFBBr was also used to derivatize the five straight-chain SCFAs in an isotopomer enrichment assay [29]. To this point, there has not been any report to simultaneously quantify all eight straight-chain and branched-chain SCFAs from biologic samples.
The objective of this work was to develop a method for simultaneous identification and quantification of all straight-chain and branched-chain SCFAs in a biological sample. We used PFBBr reagent to derivatize SCFAs and a GC-Ion Trap MS instrument to measure the derivatized SCFAs. To achieve high sensitivity in detecting the low abundance and branched-chain SCFAs, we optimized the experiment conditions of PFBBr derivatization and the selection of GC columns. The optimized experiment conditions were then used to simultaneously identify and quantify straight-chain and branched-chain SCFAs in mouse feces.
2. METHODS
2.1. Chemicals and reagents
Eight SCFA standards (sodium formate, sodium acetate, sodium propionate, sodium butyrate isobutyric acid, sodium pentanoate, 2-methylbutyric acid, and isovaleric acid) and 2, 3, 4, 5, 6-pentafluorobenzyl bromide (PFBBr) were purchased from Sigma-Aldrich Corp. (St. Louis, MO, USA).
2.2. Preparation of SCFA standards
Thirteen solutions were prepared for each of the eight SCFA standards in following concentrations: 0.977 μM, 1.95 μM, 3.91 μM, 7.81 μM, 15.63 μM, 31.3 μM, 62.5 μM, 125 μM, 250 μM, 500 μM, 1 mM, 2.5 mM, and 5 mM. These solutions were used for PFBBr derivatization. The derivatized standards were then used to optimize the experiment conditions and to construct calibration curves for SCFA quantification.
2.3. Animal and biological sample preparation
Eight-week-old male C57BL/6J mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Mice were housed in a pathogen-free barrier facility accredited by the Association for Assessment and Accreditation of Laboratory Animal Care. Food and tap water were allowed ad libitum. The procedures of animal care were approved by the University of Louisville Institutional Animal Care and Use Committee. Two groups of C57BL/6J mice fed different diets. One group fed an isocaloric maltose dextrin solution (Group1, n = 5) while the other group fed with Lieber DeCarli liquid diet containing 5% alcohol (Group 2, n = 9). After 24-day dietary intervention, mice were anesthetized with ketamine/xylazine (100/15 mg/kg i.m.) and feces were collected directly from the mouse colon. All fecal samples were frozen immediately in liquid nitrogen and stored in 1.5mL eppendorf tube at −80 °C freezer.
All fecal sample processing were performed at 4 °C to minimize the loss of volatile SCFAs, unless stated otherwise. For each sample, about 30 mg of mouse feces was weighed and ground in a 1.5 mL eppendorf tube. After adding water at a ratio of 100 mg feces/mL water, the mixture was sonicated for 20 min and then centrifuged at 4 °C and 12,000 rpm for 20 min. The supernatant was collected for SCFAs detection and recovery determination.
2.4. PFBBr derivatization
A 50 μL of standard solution or 150 μL of supernatant collected from a biological sample was used for derivatization. 100 mM PFBBr in acetone, 0.5 M phosphate buffer (PBS, pH 7), and a sample were mixed at a ratio of 14:2:5 (v:v:v) to make the acetone:water 14:7 (v:v) in a 2 mL glass tube. After 1 min of vigorous vortex mixing, the mixture was incubated in a water bath at 60 °C for 1.5 h. 200 μL or 150 μL of hexane was added after the mixture of standard or sample cooled down to room temperature. The sample was then vortexed for 3 min followed by centrifugation for 5 min in a speedvac. 100 μL of supernatant (hexane phase) was then transferred to a 200 μL GC vial for GC-MS analysis. A blank sample prepared using Distilled Milli-Q water was also derivatized as a reference for quality control purposes.
2.5. GC-MS analysis
A Thermo Scientific ITQ™ 1100 GC-Ion Trap MS instrument was coupled with a TRACE™ 1310 gas chromatography system and a 1310 autosampler (Thermo Fisher Scientific, Waltham, MS, USA). Two GC columns, DB-225ms (30 m × 0.25 mm 1dc × 0.25 μm 1dp, (50%-cyanopropylphenyl)-methylpolysiloxane) and DB-5ms (30 m × 0.25 mm 1dc × 0.25 μm 1dp, (5%-phenyl)-methylpolysiloxane), were used for SCFA separation. These columns were obtained from Agilent Technologies J&W (Santa Clara, CA, USA). The helium carrier gas (99.999% purity) flow rate was set to 1.5 mL/min for DB-225ms and DB-5ms columns, respectively. The flow rate was reduced to 1.0 mL/min when DB-225ms column and DB-5ms column were hyphenated together by a column connector purchased from Restek Corporation (Bellefonte, PA, USA), where DB-225ms was the first column and DB-5ms was the second column. The temperatures of inlet, ion source and transfer line were all set to 220 °C. The column temperature was programmed with an initial temperature of 80 °C for 0.5 min, then ramped to 158 °C at a rate of 10 °C/min, to 160 °C at 3 °C/min, to 220 °C at 20 °C/min, and then maintained at 220 °C for 8 min. The hyphenated column was ramped as follows: initial temperature, 80 °C for 0.5 min; 10 °C/min to 170 °C for 0.5 min; 5 °C/min to 220 °C, hold for 5 min. The energy of electron ionization (EI) was set to 70 eV.
One microliter of PFBBr derivatives was injected into GC-MS in splitless mode with a splitless time of 1.0 min. Solvent delay time was set to 6.1 min for the DB-225ms column, 5.38 min for the DB-5ms column, and 8.86 min for hyphenated columns, respectively. The mass spectral data were collected in a SIM mode (Table 1). All biological samples were analyzed on hyphenated columns. In addition, an aliquot of n-alkane series was also injected under each ramp condition for retention index calculation and quality control.
Table 1.
Ions monitored and corresponding retention time region for SCFA detection in SIM mode
| Compound Name | DB-225ms column | DB-5ms column | DB-225ms and DB-5ms column in tandem |
|||
|---|---|---|---|---|---|---|
| monitored ion m/z |
monitor region (min) |
monitored ion m/z |
monitor region (min) |
monitored ion m/z |
monitor region (min) |
|
| Formic acid | 226 | 6.10-6.60 | 226 | 5.38-5.91 | 181, 226 | 8.86-9.38 |
| Acetic acid | 240 | 6.60-7.40 | 181 | 5.91-18.00 | 181, 240 | 9.38-10.48 |
| Propionic acid | 254 | 7.40-7.64 | 181 | 5.91-18.00 | 181, 254 | 10.48-10.66 |
| Isobutyric acid | 268 | 7.64-7.95 | 181 | 5.91-18.00 | 181, 268 | 10.66-11.71 |
| Butyric acid | 181 | 7.95-22.00 | 181 | 5.91-18.00 | 181, 268 | 10.66-11.71 |
| 2-Methylbutyric acid | 181 | 7.95-22.00 | 181 | 5.91-18.00 | 181, 282 | 11.71-24.00 |
| Isovaleric acid | 181 | 7.95-22.00 | 181 | 5.91-18.00 | 181, 282 | 11.71-24.00 |
| Valeric acid | 181 | 7.95-22.00 | 181 | 5.91-18.00 | 181, 282 | 11.71-24.00 |
2.6. Data processing
Thermo Scientifc Xcalibur instrument control software Quan (2.2 SP1.48) was used to process the GC-MS data for peak picking, standard curve construction, and SCFA quantification. The signal-to-noise ratio (S/N) was set to S/N = 3. The concentration of each SCFA in a biological sample was calculated using the calibration curve constructed from the GC-MS data of a corresponding SCFA standard.
3. RESULTS AND DISCUSSION
We first optimized the derivatization and GC-MS experiment conditions using a mixture containing the eight SCFA standards, i.e., sodium formate, sodium acetate, sodium propionate, sodium butyrate, isobutyric acid, sodium pentanoate, 2-methylbutyric acid, and isovaleric acid. The derivatization time, pH, ratio of acetone to water (CH3)2CO:H2O, and temperature were individually optimized. Each optimization experiment was prepared in triplicate and the average peak area of the monitoring ion was used for comparison. To find the best GC column configuration, the SCFA standards were analyzed on DB-225ms, DB-5ms, and hyphenated DB-225ms and DB-5ms columns, respectively. The SCFAs extracted from mouse feces were then analyzed by GC-MS under the optimized experiment conditions.
3.1. Optimization of PFBBr derivatization
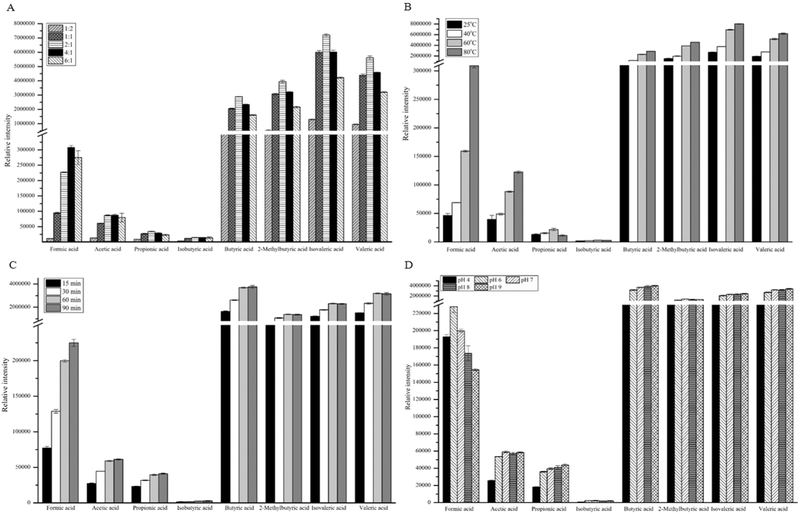
These experiments were executed on the DB-225ms column. The derivatization was carried out by mixing 100 mM PFBBr in acetone and water, where acetone served as a solvent for the derivatizing reagent PFBBr and aided in precipitation of proteins. To optimize the volume ratio of acetone and water, 100 mM PFBBr-acetone solution was mixed with 50 μL SCFA standards and 20 μL PBS buffer mixture in following (CH3)2CO:H2O ratios, 1:2, 1:1, 2:1, 4:1 and 6:1 (v:v). The concentration of each SCFA in the solution of SCFA standards was 1 mM. Figure 1 shows that the PFBBr derivatives of butyric acid, 2-methylbutyric acid, isovaleric acid, and valeric acid had much higher intensity than those of PFBBr derivatized formic acid, acetic acid, propionic acid, and isobutyric acid. This was mainly caused by the different types of ions used to quantify these compounds. Parents ions were used to quantify the PFBBr derivatives of formic acid, acetic acid, propionic acid, and isobutyric acid, while fragment ion m/z = 181 was used to quantify the PFBBr derivatives of the remaining SCFAs.
Figure 1.
PFBBr esters with short chain fatty acids formed under different reaction conditions, including (A) different acetone/water ratios, (B) different reaction temperatures, (C) different incubation times, and (D) different pH values of phosphate buffer.
Figure 1A shows that the intensity of PFBBr derivatives of all SCFAs increased with the increase of (CH3)2CO:H2O ratio. The PFBBr derivative of formic acid had its highest intensity at the (CH3)2CO:H2O ratio of 4:1, while the remaining SCFAs reached their maxima at 2:1 ratio. The intensity difference for the PFBBr derivative of formic acid was 35.7% between the selection of (CH3)2CO:H2O ratio 2:1 and 4:1, while the difference of intensity for the other SCFAs was less than 10%. For these reasons, we chose the (CH3)2CO:H2O ratio of 2:1 as the optimal ratio to make sure majority of SCFAs have the maximum intensity even though this selection was not the best for measurement of PFBBr derivatized formic acid. Choosing the (CH3)2CO:H2O ratio of 2:1 also avoided consuming additional PFBBr and acetone.
We also optimized the temperature of PFBBr derivatization reaction. Figure 1B shows that the intensity of PFBBr derivatized SCFAs increased with the increase of temperature. With the exception of the PFBBr derivatives of propionic acid and isobutyric acid, the PFBBr derivatives of other SCFAs reached their highest intensity at 80 °C. However, proteins could denature at 80 °C if biological samples were to be analyzed. The denatured proteins would affect the extraction of PFBBr derivatives. Therefore, 60 °C was chosen as the optimal derivatization temperature.
To investigate the effect of derivatization time, the PFBBr derivatization reaction was performed at 60 °C for 15, 30, 60, and 90 min, respectively. Figure 1C shows the relation of the intensity of the PFBBr derivatized SCFAs to the derivatization time. Except for PFBBr derivatized formic acid, the intensities of the other PFBBr derivatized SCFAs reached a plateau at 60 min and were stable up to 90 min. Therefore, 90 min was chosen as the optimal derivatization time.
The PFBBr derivatization reaction was also carried out at five pH values, including 4, 6, 7, 8, and 9. Figure 1D shows the relation between pH values and the intensity of the PFBBr derivatives. While the PFBBr derivative of formic acid had its highest intensity at pH 6, the PFBBr derivatives of other SCFAs had low intensity under acidic conditions. The PFBBr derivatized propionic acid, butyric acid, isovaleric acid, and valeric acid had their highest intensity in alkaline conditions. The PFBBr derivatized acetic acid, isobutyric acid, and 2-methylbutyric acid had their highest intensity in neutral condition. However, the intensity difference of all PFBBr derivatized SCFAs was less than 10% between neutral and alkaline conditions. For this reason, pH 7 was selected as the optimal condition for derivatization.
Collectively, we chose the optimal PFBBr derivatization condition as: the volume ratio of PFBBr acetone solution and SCFA solution 2:1, pH 7, derivatization time 90 mins, and temperature 60 °C. This optimal condition for analysis of all eight SCFAs agrees with the literature reported results, where PFBBr derivatization of five straight-chain fatty acids were optimized [29]. The differences are that the optimal derivatization time was 60 min and the optimal pH was 9.0 reported in reference 27.
3.2. Separation of SCFAs by GC-MS equipped with a DB-225ms column
To simultaneously quantify straight-chain and branched-chain SCFAs, the PFBBr derivatives of eight SCFA standards were first analyzed respectively by GC-MS to get the retention time and mass spectrum for each PFBBr derivatized SCFA. The PFBBr derivatives of propionic acid and isobutyric acid overlapped with a solvent peak eluted at tR = 7.67 min (Fig. S1A). It is impossible to quantify these two co-eluting SCFAs by integrating the total ion current (TIC) from the GC-MS data. Therefore, the SIM mode was used in GC-MS to measure the PFBBr derivatives of SCFAs.
The most abundant fragment ions in the EI mass spectra of the three overlapping compounds (PFBBr derivatized propionic acid and isobutyric acid, and the compound given rise the solvent peak) had the same m/z value (m/z =181), resulting that the most abundant fragment ion (m/z = 181) cannot be used to quantify propionic acid and isobutyric acid. For this reason, their parent ions were chosen for quantification (Table 1). The PFBBr derivatives of formic acid and acetic acid did not overlap with any chromatographic peaks, and the fragment ion with m/z = 181 was the most abundant ion in their EI mass spectra. Ideally, the most abundant fragment ion should be used for quantification of these two SCFAs to achieve high sensitivity. However, the baseline of selected ion chromatogram was dramatically decreased during the elution time of PFBBr derivatized propionic acid and isobutyric acid (7.20-7.92 min), resulting that the data processing software, Quan software, could not calculate the intensity of their fragment ion (Figure S2). For these reasons, the parent ions of PFBBr derivatized formic acid, acetic acid, propionic acid, and isobutyric acid were used to detect the abundance of these SCFAs eluted from DB-225ms column (Table 1). The PFBBr derivatives of the remaining SCFAs (butyric acid, 2-methylbutyric acid, isovaleric acid, and valeric acid) were separated from each other on the DB-225ms column and the fragment ion with m/z = 181 was the most intense ion in their EI mass spectra. Therefore, this m/z value was chosen to monitor the PFBBr derivatives of these SCFAs (Table 1).
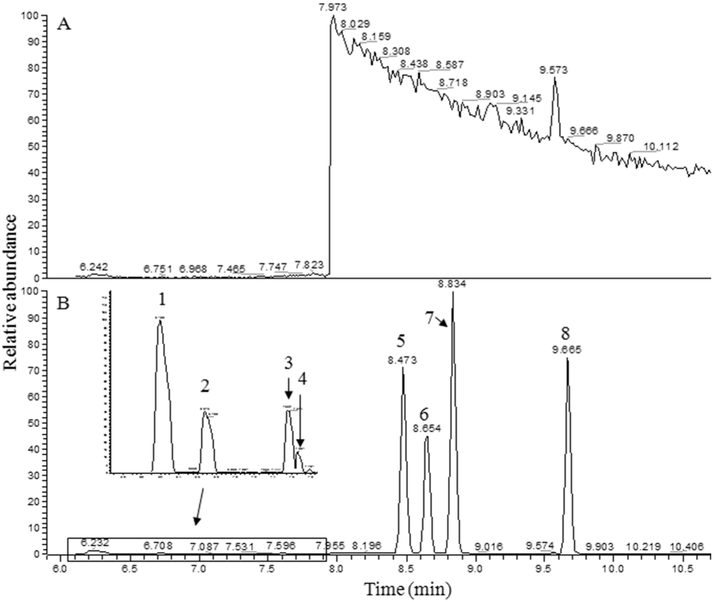
Figure 2 depicts two typical selected ion chromatograms acquired on GC-MS equipped with a DB-225ms column, one for a blank and other for PFBBr derivatives of a mixture of SCFAs. The significant chromatographic profile change at retention time tR = 7.95 min in Figure 2A was caused by monitoring different ions in the SIM mode. When blank samples were monitored for m/z = 181 in the retention time range between 7.95 and 22.0 min, one significant peak eluted at tR = 9.57 min (Figure 2A) was consistently present in the chromatogram. Figure 2B shows the selected ion chromatogram of PFBBr derivatized SCFAs. Though other SCFAs were separated from each other very well, the chromatographic peaks of propionic acid and isobutyric acid were overlapped, and affected the quantification accuracy of these two SCFAs.
Figure 2.
Selected ion chromatograms of blank and a mixture of SCFAs on DB-225 ms column under SIM mode. (A) is selected ion chromatogram of blank (i.e., 50 μL of distilled Milli-Q water, pH 7). (B) is selected ion chromatogram of a mixture of eight SCFAs, where 1 is formic acid; 2 is acetic acid; 3 is propionic acid; 4 is isobutyric acid; 5 is butyric acid; 6 is 2-methylbutyric acid; 7 is isovaleric acid; and 8 is valeric acid. The corresponding m/z for tracing those compounds were show in Table 1.
3.3. Detection of SCFAs by GC-MS equipped with a DB-5ms column
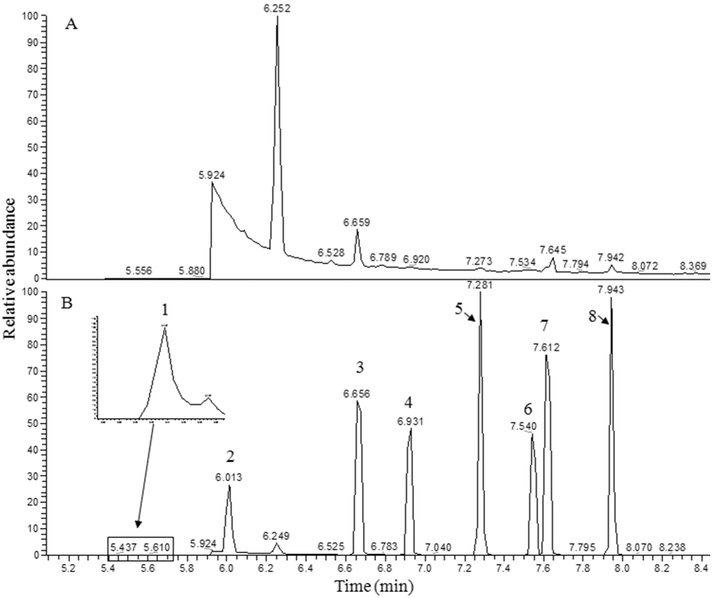
A DB-5ms column was used to explore the performance of a nonpolar column in separation of PFBBr derivatized SCFAs. The PFBBr derivatized formic acid co-eluted with the solvent peak at tR = 5.48 min, while the PFBBr derivatives of the other SCFAs were separated from each other (Fig. S1B). Therefore, the parent ion of PFBBr derivatized formic acid was used to monitor formic acid, and the fragment ion with m/z = 181 was used to monitor the PFBBr derivatives of other seven SCFAs. Figure 3 depicts the selected ion chromatograms of a blank and the PFBBr derivatized SCFAs. Comparing the separation of PFBBr derivatives on the DB-225ms column, the PFBBr derivatives of propionic acid and isobutyric acid achieved a better separation on the DB-5ms column. However, the intensity of PFBBr derivatized formic acid was decreased. The chromatographic peak of PFBBr derivatized formic acid on the DB-5ms column was also broader with significant right-side peak tailing compared to those on the DB-225ms column. Furthermore, there were four peaks in the selected ion chromatogram of the blank. Three of those co-eluted with the PFBBr derivatives of propionic acid, isovaleric acid and valeric acid at tR = 6.66 min, tR = 7.64 min and 7.94 min, respectively. The intensities of the fragment ion (m/z = 181) of these three peaks in the blank affect the quantification accuracy of the PFBBr derivatized propionic acid, isovaleric acid and valeric acid.
Figure 3.
Selected ion chromatograms of a blank and a mixture PFBBr derivatized SCFAs. The GC-MS was equipped with a DB-5 ms column and operated under SIM mode. (A) is the selected ion chromatogram of blank, i.e., 50 μL of distilled Milli-Q water with pH = 7. (B) is the selected ion chromatogram of PFBBr derivatized SCFAs where 1 is formic acid; 2 is acetic acid; 3 is propionic acid; 4 is isobutyric acid; 5 is butyric acid; 6 is 2-methylbutyric acid; 7 is isovaleric acid; and 8 is valeric acid. The corresponding m/z for tracing those compounds were show in Table 1.
3.4. Quantification of SCFAs by GC-MS equipped with two hyphenated columns
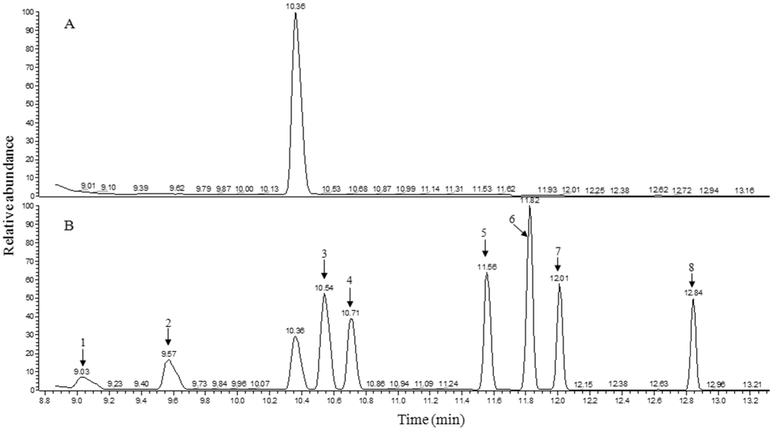
In order to separate the solvent peaks with propionic acid and isobutyric acid, the DB-225ms column was hyphenated with the DB-5ms column in GC-MS, where DB-225ms was the first column and DB-5ms was the second column. Figure S1C depicts the total ion chromatogram using the hyphenated columns. Three solvent peaks (SP1 at tR = 7.37 min, SP2 at tR = 8.60 min, and SP3 at tR = 10.38 min) were detected. The two major solvent peaks (SP1 and SP2) eluted much earlier than the eight SCFA peaks, while the solvent peak SP3 and the eight SCFAs peaks were completely separated from each other. Therefore, the two major solvent peaks can be cut by solvent delay and the most abundant fragment ion with m/z = 181 can be therefore used to quantify each SCFA to achieve the best sensitivity. Figure 4 shows two typical selected ion chromatograms of a blank and the PFBBr derivatized SCFAs. When the blank was monitored for m/z = 181 in the retention time range between 8.86 and 24.0 min, a peak was observed at tR = 10.36 min (Figure 4A), which was the solvent peak SP3 in Figure S1C. This solvent peak did not co-elute with any of the SCFA peaks (Figure 4B).
Figure 4.
Selected ion chromatograms of a blank and a mixture PFBBr derivatized SCFAs. The GC-MS was equipped with a combined column and operated under SIM mode. (A) is the selected ion chromatogram of blank, i.e., 50 μL of distilled Milli-Q water with pH = 7. (B) is the selected ion chromatogram of PFBBr derivatized SCFAs where 1 is formic acid; 2 is acetic acid; 3 is propionic acid; 4 is isobutyric acid; 5 is butyric acid; 6 is 2-methylbutyric acid; 7 is isovaleric acid; and 8 is valeric acid. The corresponding m/z for tracing those compounds were show in Table 1.
To determine the limit of detection (LOD) and the limit of quantification (LOQ) of GC-MS equipped with the hyphenated columns, 13 calibration solutions were prepared for each PFBBr derivatized SCFA standard, including 0.244 μM, 0.488 μM, 0.977 μM, 1.953 μM, 3.906 μM, 7.813 μM, 15.625 μM, 31.25 μM, 62.5 μM, 125 μM, 250 μM, 500 μM, and 1250 μM. Each calibrant was analyzed three times by GC-MS.
The criterion for determining the LOD of a SCFA was that the PFBBr derivative of that SCFA must be detected in at least two injections at a particular concentration. In other words, a detection of PFBBr derivative of a SCFA was considered as random if the PFBBr derivative of a SCFA was detected only in one injection. An iterative approach was used to determine the linear range of a calibration curve as follows. A narrow concentration range was initially selected for linear fitting. The linear range was then extended to both the high concentration and low concentration directions. To decide whether the linear range would be extended to a concentration, the difference between the experimentally measured intensity and the calculated intensity of the PFBBr derivatized SCFA at the concentration of interest was calculated. If the intensity difference was less than 20% of the experimentally measured value, the linear range was extended to that concentration. The intensity and concentration information of all data in the newly extended linear range were used to recalculate the linear fitting function. This process was repeated until the intensity difference was larger than 20% at a concentration.
Figure S3 depicts the calibration curves of the PFBBr derivatized SCFAs separated on the tandem columns. The solid line represents the calibration curve and the two dotted lines are the upper and low boundaries of 95% fitting confidence. Table 2 summarizes the information of the calibration curve of each SCFA including calibration equation, linear range, LOD, and LOQ. The calibration curve of formic acid was constructed using the data of its parent ion (m/z=226), while the calibration curves of the remaining SCFAs were respectively constructed using the data of their fragment ion with m/z = 181. The accuracy and reliability of these calibration curves were further evaluated by another person. Six mixtures of SCFA standards with known concentrations were prepared. Each of these mixtures was analyzed in the same manner using the optimized derivatization protocol and GC-MS method. The concentration of each SCFA was then determined using a corresponding calibration curve. The results showed that the calculated concentrations agreed with the true values within a 20% variation (Table S1).
Table 2.
Linearity, LOD, LOQ of PFBBr derivatized SCFAs detected on GC-MS with DB-225ms and DB-5ms column in tandem
| Compound Name | LODa (μM) |
Calibration equationb | LOQc (μM) |
Linear rage | R2 d | ne |
|---|---|---|---|---|---|---|
| Formic acid | 0.244 | Y=2152.4+727.75*x | 7.813 | LOQ-1.25mM | 0.9982 | 3 |
| Acetic acid | 0.977 | Y=4350.1+7528.6*x | 0.977 | LOQ-1.25mM | 0.9992 | 3 |
| Propionic acid | 0.977 | Y=−44946+17672*x | 7.813 | LOQ-0.625mM | 0.9998 | 3 |
| Isobutyric acid | 0.488 | Y=33959+17082*x | 3.906 | LOQ-0.625mM | 0.9989 | 3 |
| Butyric acid | 0.244 | Y=−9229.2+17967*x | 1.953 | LOQ-1.25mM | 0.9982 | 3 |
| 2-Methylbutyric acid | 0.244 | Y=23155+25256*x | 3.906 | LOQ-1.25mM | 0.9990 | 3 |
| Isovaleric acid | 0.244 | Y=−1946+14068*x | 3.906 | LOQ-1.25mM | 0.9996 | 3 |
| Valeric acid | 0.244 | Y=−995.52+11245*x | 0.977 | LOQ-1.25mM | 0.9974 | 3 |
LOD (μM) is the lowest calibration standard detected with a signal/noise ratio ≥ 3.
x: concentration (μM); Y: peak area.
LOQ (μM) is the limit of quantification determined by the linear range of a calibration curve, where the variation of calculated concentration and the true concentration was less than 20%.
R2: Regression coefficient.
The number of injections for each compound.
The linearity of calibration curves for all SCFAs were excellent among the validated concentration range (R2 ≥ 0.997). The LOD of low abundance SCFAs (such as formic acid, 2-methylbutyric acid, isovaleric acid, and valeric acid) reached 0.244 μM. The lowered LOD and wide range of these calibration curves allow for the analysis of SCFAs in samples with very different levels. The present method was more sensitive than the literature reported results. For instance, Zhao et al. showed the LOD for SCFAs was in a range of 10 μM, while the LOQ was below 31 μM [17]. Though Schwiertz et al. lowered the LOD about 10 times to about 1 μM in standard water solution [30]. However, these methods cannot simultaneously detect formic acid with other SCFAs. The method developed in this study not only offers better sensitivity but also can quantify formic acid together with other SCFAs in one injection.
3.5. Quantification of SCFAs in biological samples
To demonstrate the utility of this method for analysis of SCFAs in biological samples, feces were collected from C57BL/6J mice of two different feeding groups were analyzed to determinate the influence of alcohol on SCFAs in colon. The ratio of feces weight to water volume affects the numbers of metabolites extracted from feces (wf/vs), and it was reported that a maxima of metabolites was extracted from rat feces at the ratio of Wf/vs = ½ [31]. We decreased Wf/vs to 1/10 owing to the concentrations of acetic acid, propionic acid, and butyric acid beyond the linear range of these molecules.
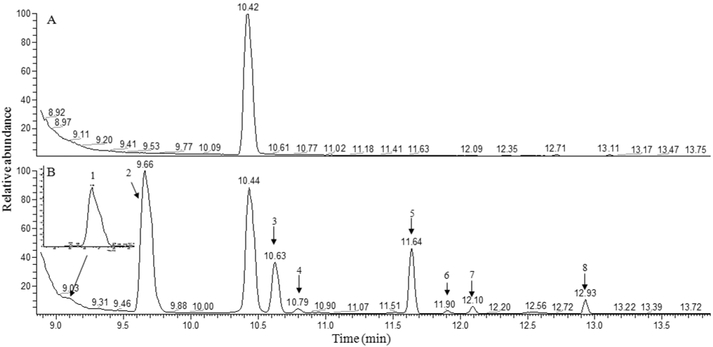
Figure 5 shows the selected ion chromatograms acquired from the blank and fecal samples. All eight SCFAs were identified and quantified in the fecal samples (Figure 5B). The concentrations of three SCFAs (acetic acid, propionic acid, and butyric acid) were predominant in feces samples. Acetic acid had the highest concentration (23.81 μmol/g feces), followed by butyric acid (8.93 μmol/g feces), and propionic acid (7.17 μmol/g feces). The concentrations of formic acid and valeric acid were 0.12 μmol/g feces and 0.48 μmol/g feces, respectively. The concentrations of three branched-chain SCFAs (isobutyric acid, 2-methylbutyric acid, and isovaleric acid) were 0.13, 0.18, and 0.12 μmol/g feces, respectively. However, the concentrations of all SCFAs in alcohol fed mice were significantly decreased except for 2-methylbutyric acid (Table 3). This result suggests that alcohol might significantly influence the function of the colon.
Figure 5.
Selected ion chromatograms of blank (A) and mouse fecal sample (B) obtained on GC-MS with the combined column, XIC for formic acid was made by its parent ion (m/z = 226). 1. Formic acid; 2. Acetic acid; 3 Propionic acid; 4. Isobutyric acid; 5. Butyric acid; 6. 2-Methylbutyric acid; 7. Isovaleric acid; 8. Valeric acid. The corresponding m/z for tracing those compounds were show in Table 1.
Table 3.
Quantitative analysis of SCFAs obtained from biological samples
| Compound Name | tR (min) | Ion used for quantitation (m/z) |
Group 1 (μmol/g feces) (n=5) |
Group 2 (μmol/g feces) (n=9) |
|---|---|---|---|---|
| Formic acid | 9.06 | 226 | 0.12±0.02 | 0.08±0.00 |
| Acetic acid | 9.63 | 181 | 23.81±1.26 | 6.40±0.13 |
| Propionic acid | 10.61 | 181 | 7.17±0.39 | 0.75±0.03 |
| Isobutyric acid | 10.78 | 181 | 0.13±0.02 | 0.03±0.01 |
| Butyric acid | 11.62 | 181 | 8.93±1.54 | 0.48±0.03 |
| 2-Methylbutyric acid | 11.88 | 181 | 0.18±0.02 | 0.21±0.02 |
| Isovaleric acid | 12.08 | 181 | 0.12±0.02 | 0.07±0.00 |
| Valeric acid | 12.91 | 181 | 0.48±0.03 | 0.08±0.01 |
For recovery studies, two standard mixtures (2.50 and 5 μmol/g feces) were spiked into fecal supernatant, respectively. The recovery rate was calculated as the ratio of the measured concentration of an added SCFA standard divided by the spiked concentration of that standard. Because the high concentration of acetic acid in fecal samples, the fecal supernatant was first diluted 10 times before the adding of standard mixtures. Table 4 lists the recovery rates for all SCFAs. The recovery rates were 55.7% and 57.3% for formic acid and isobutyric acid, while they were much higher for the remaining SCFAs ranging from 75.1% to 97.9%.
Table 4.
Recoveries of SCFAs in fecal samples
| Compound | Added SCFAs (μmol/g feces) |
n | Detected SCFAs (μmol/g feces) |
Recoveries (%) |
|
|---|---|---|---|---|---|
| Mean | SD | ||||
| Formic acid | 0 | 3 | 0.043 | 0.003 | |
| 2.5 | 3 | 1.525 | 0.019 | 59.27 | |
| 5.0 | 3 | 2.654 | 0.023 | 52.21 | |
| Acetic acid | 0 | 3 | 1.102 | 0.062 | |
| 2.5 | 3 | 2.991 | 0.078 | 75.58 | |
| 5.0 | 3 | 4.835 | 0.199 | 74.67 | |
| Propionic acid | 0 | 3 | 2.520 | 0.095 | |
| 2.5 | 3 | 4.967 | 0.144 | 97.9 | |
| 5.0 | 3 | - | - | ||
| Isobutyric acid | 0 | 3 | 0.064 | 0.004 | |
| 2.5 | 3 | 1.629 | 0.071 | 62.58 | |
| 5.0 | 3 | 2.666 | 0.068 | 54.96 | |
| Butyric acid | 0 | 3 | 1.835 | 0.083 | |
| 2.5 | 3 | 4.280 | 0.183 | 97.79 | |
| 5.0 | 3 | 5.477 | 0.097 | 72.84 | |
| 2-Methylbutyric acid | 0 | 3 | 0.040 | 0.009 | |
| 2.5 | 3 | 2.312 | 0.084 | 90.84 | |
| 5.0 | 3 | 3.916 | 0.084 | 77.52 | |
| Isovaleric acid | 0 | 3 | 0.195 | 0.045 | |
| 2.5 | 3 | 2.481 | 0.103 | 91.43 | |
| 5.0 | 3 | 3.989 | 0.101 | 75.87 | |
| Valeric acid | 0 | 3 | 0.185 | 0.010 | |
| 2.5 | 3 | 2.355 | 0.104 | 86.82 | |
| 5.0 | 3 | 3.940 | 0.082 | 75.12 | |
in table means the concentration of SCFAs is out of the liner range of this compound.
SD: standard deviation.
In this study, we coupled a 30 m DB-225ms column and a 30 m DB-5ms column together for quantification of SCFAs by SIM GC-MS. Such a column configuration enables not only the separation of the PFBBr derivatized SCFA peaks from each other but also the separation of very large solvent peaks from those peaks of PFBBr derivatized SCFAs. Lowering the start temperature of oven program to 40 °C can better focus the sample on the column head and therefore, results in further separation of formic acid from the solvent peak. Separating the large solvent peaks from the SCFA peaks made it possible to use the most abundant fragment ion (m/z = 181) for quantification of SCFAs (except formic acid) and therefore increased the sensitivity and accuracy of quantifying these SCFAs. It should be noted that we corrected the concentration of each SCFA after calculating them from the calibration curve to corrected the mismatch in the volume of standard and sample water volume. This approach may introduce a certain level of variations in the calculated concentration of each SCFA. Furthermore, adding internal standards to the sample before SCFA extraction and then use their results to normalize the results of SCFAs can further improve the accuracy of quantifying SCFAs.
To our knowledge, this work is the first study that could simultaneously detect and quantify all straight-chain and branch-chain SCFAs even though there were numerous reports in literature for SCFA quantification. For instance, Zheng et al. developed a method for quantification of SCFAs and branch-chain amino acids using propyl chloroformate (PCF) as derivatization reagent [19]. However, their method could not quantify formic acid. Most recently, Jiang and co-workers did an excellent job on fatty acid quantification by LC-MS [24]. But their method still could not detect and quantify formic acid. Furthermore, the linear quantification ranges for other SCFAs were smaller than those reported in this study. For instance, the linear quantification ranges for acetic acid, propionic acid, and butyric acid in reference 23 were 0.1 μM to 100 μM, 0.2 μM to 100 μM, and 0.1 μM to 40 μM, respectively. The corresponding linear quantification ranges for these three acids in this study were 0.977 μM to 1.25 mM, 7.8 μM to 0.625 mM, and 1.95 μM to 1.25 mM, which are large enough for the direct quantification of these three SCFAs in fecal samples. Direct quantification of these SCFAs in biological samples eliminated the dilution process and therefore, reduced the technical variations.
4. CONCLUSIONS
In order to simultaneously identify and quantify SCFAs, especially formic acid and branched-chain SCFAs, pentafluorobenzyl bromide (PFBBr) reagent was used to derivatize SCFAs and a GC-Ion Trap MS instrument with EI source was used to measure the derivatized SCFAs. Using rigorous optimization steps in sample preparation, we ensured maximum derivatization efficiency and reduced solvent consumption. Comparing the performance of different GC column configurations, a 30 m DB-225ms hyphenated with a 30 m DB-5ms column in tandem showed the best separation of SCFAs. This column configuration not only separated the three solvent peaks from the SCFAs but also allowed using the most abundant fragment ion for SCFA quantification. Using the optimized experiment conditions, the LOD of all SCFAs were improved to 0.244 - 0.977 μM. Application of the developed method to analyze SCFAs in mouse feces showed that all eight SCFAs were successfully quantified in the fecal samples, with the recovery rates of the eight SCFAs ranged from 55.7% to 97.9%. These analysis results demonstrated that the developed method can be used to analyze biological samples for simultaneous quantification of all eight SCFAs.
Supplementary Material
Highlights.
A method capable of simultaneous quantification of all SCFAs by GC-MS.
A DB225ms and a DB-5ms columns were connected in tandem for separation.
None of the solvent peaks overlaps with the SCFA peaks.
The best LOD and LOQ were achieved for detecting all SCFAs from biosamples.
ACKNOWLEDGEMENTS
The authors thank Mrs. Marion McClain for review of this manuscript.
FUNDINGS
This work was supported by NIH [grant nos. 1R01AA23190 (WF); 1P20GM113226 (CJM); 1P50AA024337 (CJM); 1U01AA021893-01 (CJM); 1U01AA021901-01 (CJM); 1U01AA022489-01A1 (CJM); and 1R01AA023681-01 (CJM)]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work was also supported by the Department of Veterans Affairs 1I01BX002996-01A2 (CJM).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Cummings JH, Short chain fatty acids in the human colon, Gut 22(1981) 763–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Macfarlane S, Macfarlane GT, Regulation of short-chain fatty acid production, Proc Nutr Soc 62(2003) 67–72. [DOI] [PubMed] [Google Scholar]
- 3.Roelofsen H, Priebe MG, Vonk RJ, The interaction of short-chain fatty acids with adipose tissue: relevance for prevention of type 2 diabetes, Benef Microbes. 1(2010) 433–7. [DOI] [PubMed] [Google Scholar]
- 4.Natarajan N, Hori D, Flavahan S, Steppan J, Flavahan NA, Berkowitz DE, Pluznick JL, Microbial short chain fatty acid metabolites lower blood pressure via endothelial G-protein coupled receptor 41, Physiol Genomics. physiolgenomics 00089 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pingitore A, Chambers ES, Hill T, M aldonado IR, Liu B, Bewick G, Morrison DJ, Preston T, Wallis GA, Tedford C, Castanera Gonzalez R, Huang GC, Choudhary P, Frost G, Persaud SJ, The diet-derived short chain fatty acid propionate improves beta-cell function in humans and stimulates insulin secretion from human islets in vitro, Diabetes Obes Metab 19(2017) 257–265. [DOI] [PubMed] [Google Scholar]
- 6.Samuel BS, Shaito A, Motoike T, Rey FE, Backhed F, Manchester JK, Hammer RE, Williams SC, Crowley J, Yanagisawa M, Gordon JI, Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41, Proc Natl Acad Sci U S A. 105(2008) 16767–16772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolever TM, Spadafora P, Eshuis H, Interaction between colonic acetate and propionate in humans, Am J Clin Nutr 53(1991) 681–687. [DOI] [PubMed] [Google Scholar]
- 8.Rasmussen HS, Holtug K, Mortensen PB, Degradation of amino acids to short-chain fatty acids in humans. An in vitro study, Scand J Gastroenterol 23(1988) 178–182. [DOI] [PubMed] [Google Scholar]
- 9.Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, Lewis GD, Fox CS, Jacques PF, Fernandez C, O'Donnell CJ, Carr SA, Mootha VK, Florez JC, Souza A, Melander O, Clish CB, Gerszten RE, Metabolite profiles and the risk of developing diabetes, Nat Med 17(2011) 448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Unger MM SJ, Dillmann KU, Grundmann D, Philippeit H, Bürmann J, Faßbender K, Schwiertz A, Schäfer KH, Short chain fatty acids and gut microbiota differ between patients with Parkinson's disease and age-matched controls, Parkinsonism Relat Disord 32(2016) 66–72. [DOI] [PubMed] [Google Scholar]
- 11.Schwarz A, Bruhs A, Schwarz T, The Short-Chain Fatty Acid Sodium Butyrate Functions as a Regulator of the Skin Immune System, J Invest Dermatol 137(2017) 855–864. [DOI] [PubMed] [Google Scholar]
- 12.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, Takahashi M, Fukuda NN, Murakami S, Miyauchi E, Hino S, Atarashi K, Onawa S, Fujimura Y, Lockett T, Clarke JM, Topping DL, Tomita M, Hori S, Ohara O, Morita T, Koseki H, Kikuchi J, Honda K, Hase K, Ohno H, Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells, Nature. 504(2013) 446–450. [DOI] [PubMed] [Google Scholar]
- 13.Hijova E, Chmelarova A, Short chain fatty acids and colonic health, Bratisl Lek Listy. 108(2007) 354–358. [PubMed] [Google Scholar]
- 14.Miller SJ, Cellular and physiological effects of short-chain fatty acids, Mini Rev Med Chem 4(2004) 839–845. [DOI] [PubMed] [Google Scholar]
- 15.Pluznick JL, Gut microbiota in renal physiology: focus on short-chain fatty acids and their receptors, Kidney Int 90(2016) 1191–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Villalba R, Gimenez-Bastida JA, Garcia-Conesa MT, Tomas-Barberan FA, Carlos Espin J, Larrosa M, Alternative method for gas chromatography-mass spectrometry analysis of short-chain fatty acids in faecal samples, J Sep Sci 35(2012) 1906–1913. [DOI] [PubMed] [Google Scholar]
- 17.Zhao G, Nyman M, Jonsson JA, Rapid determination of short-chain fatty acids in colonic contents and faeces of humans and rats by acidified water-extraction and direct-injection gas chromatography, Biomed Chromatogr 20(2006) 674–682. [DOI] [PubMed] [Google Scholar]
- 18.Sasaki R, Umezawa M, Tsukahara S, Ishiguro T, Sato S, Watanabe Y, Assignment of Milk Fat Fatty Acid Propyl Esters by GC-FID Analysis with the Aid of Ag-ion Solid-phase Extraction, J Oleo Sci 64(2015) 1251–1258. [DOI] [PubMed] [Google Scholar]
- 19.Zheng X, Qiu Y, Zhong W, Baxter S, Su M, Li Q, Xie G, Ore BM, Qiao S, Spencer MD, Zeisel SH, Zhou Z, Zhao A, Jia W, A targeted metabolomic protocol for short-chain fatty acids and branched-chain amino acids, Metabolomics. 9(2013) 818–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olivero SJ, Trujillo JP, A new method for the determination of short-chain fatty acids from the aliphatic series in wines by headspace solid-phase microextraction-gas chromatography-ion trap mass spectrometry, Anal Chim Acta 696(2011) 59–66. [DOI] [PubMed] [Google Scholar]
- 21.Hoving LR, Heijink M, van Harmelen V, van Dijk KW, Giera M, GC-MS Analysis of Short-Chain Fatty Acids in Feces, Cecum Content, and Blood Samples, Methods Mol Biol 1730(2018) 247–256. [DOI] [PubMed] [Google Scholar]
- 22.Kotani A, Miyaguchi Y, Kohama M, Ohtsuka T, Shiratori T, Kusu F, Determination of short-chain fatty acids in rat and human feces by high-performance liquid chromatography with electrochemical detection, Anal Sci 25(2009) 1007–1011. [DOI] [PubMed] [Google Scholar]
- 23.Schiffels J, Baumann ME, Selmer T, Facile analysis of short-chain fatty acids as 4-nitrophenyl esters in complex anaerobic fermentation samples by high performance liquid chromatography, J Chromatogr A 1218(2011) 5848–5851. [DOI] [PubMed] [Google Scholar]
- 24.Jiang R, Jiao Y, Zhang P, Liu Y, Wang X, Huang Y, Zhang Z, Xu F, Twin Derivatization Strategy for High-Coverage Quantification of Free Fatty Acids by Liquid Chromatography-Tandem Mass Spectrometry, Anal Chem 89(2017) 12223–12230. [DOI] [PubMed] [Google Scholar]
- 25.Primec M, Micetic-Turk D, Langerholc T, Analysis of short-chian fatty acids in human feces: A scoping review, Anaytical Biochemistry. 526(2017) 9–21. [DOI] [PubMed] [Google Scholar]
- 26.Holmes E, Loo RL, Stamler J, Bictash M, Yap IK, Chan Q, Ebbels T, De Iorio M, Brown IJ, Veselkov KA, Daviglus ML, Kesteloot H, Ueshima H, Zhao L, Nicholson JK, Elliott P, Human metabolic phenotype diversity and its association with diet and blood pressure, Nature. 453(2008) 396–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kage S, Kudo K, Ikeda H, Ikeda N, Simultaneous determination of formate and acetate in whole blood and urine from humans using gas chromatography-mass spectrometry, Journal of Chromatography B-Analytical Technologies in the Biomedical and Life Sciences. 805(2004) 113–117. [DOI] [PubMed] [Google Scholar]
- 28.Li Z, Yi C.x., Katiraei S, Kooijman S, Zhou E, Chung CK, Gao Y, Heuvel JK, Meijer OC, Berbee JFP, Giera M, Qillems van Dijk K, Groen AK, Rensen PCN, Wang Y, Butyrate reduces appetite and activates brown adipose tissue via the gut-brain neural circuit, Gut 0(2017) 1–11. [DOI] [PubMed] [Google Scholar]
- 29.Tomcik K, Ibarra RA, Sadhukhan S, Han Y, Tochtrop GP, Zhang GF, Isotopomer enrichment assay for very short chain fatty acids and its metabolic applications, Anal Biochem 410(2011) 110–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwiertz A, Taras D, Schafer K, Beijer S, Bos NA, Donus C, Hardt PD, Microbiota and SCFA in lean and overweight healthy subjects, Obesity (Silver Spring). 18(2010) 190–195. [DOI] [PubMed] [Google Scholar]
- 31.Deda O, Chatziioannou AC, Fasoula S, Palachanis D, Raikos Nu, Theodoridis GA, Gika HG, Sample preparation optimization in fecal metabolic profiling, J Chromatogr B Analyt Technol Biomed Life Sci 1047 (2017) 115–123. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.