CONSPECTUS
Chemical tools are transforming our understanding of biomolecules and living systems. Included in this group are bioorthogonal reagents – functional groups that are inert to most biological species, but can be selectively ligated with complementary probes, even in live cells and whole organisms. Applications of these tools have revealed fundamental new insights into biomolecule structure and function—information often beyond the reach of genetic approaches. In many cases, the knowledge gained from bioorthogonal probes has enabled new questions to be asked and innovative research to be pursued. Thus, the continued development and application of these tools promises to both refine our view of biological systems and facilitate new discoveries.
Despite decades of achievements in bioorthogonal chemistry, limitations remain. Several reagents are too large or insufficiently stable for use in cellular environments. Many bioorthogonal groups also cross-react with one another, restricting them to singular tasks. In this Account, we describe our work to address some of the voids in the bioorthogonal toolbox. Our efforts to date have focused on small reagents with a high degree of tunability: cyclopropenes, triazines, and cyclopropenones. These motifs react selectively with complementary reagents, and their unique features are enabling new pursuits in biology.
The Account is organized by common themes that emerged in our development of novel bioorthogonal reagents and reactions. First, natural product structures can serve as valuable starting points for probe design. Cyclopropene, triazine, and cyclopropenone motifs are all found in natural products, suggesting that they would be metabolically stable and compatible with a variety of living systems. Second, fine-tuning bioorthogonal reagents is essential for their successful translation to biological systems. Different applications demand different types of probes; thus, generating a collection of tools that span a continuum of reactivities and stabilities remains an important goal. We have used both computational analyses and mechanistic studies to guide the optimization of various cyclopropene and triazine probes. Along the way, we identified reagents that are chemoselective, but best suited for in vitro work. Others are selective and robust enough for use in living organisms.
The last section of this Account highlights the need for the continued pursuit of new reagents and reactions. Challenges exist when bioorthogonal chemistries must be used in concert, given that many exploit similar mechanisms and cannot be used simultaneously. Such limitations have precluded many multi-component labeling studies and other biological applications. We have relied on mechanistic and computational insights to identify mutually orthogonal sets of reactions, in addition to exploring unique genres of reactivity. The continued development of mechanistically distinct, biocompatible reactions will further diversify the bioorthogonal reaction portfolio for examining biomolecules.
Graphical Abstract
Introduction
Over the past twenty years, bioorthogonal chemistries have become a mainstay of chemical biology. These transformations are routinely used to target individual biomolecules with imaging agents or other probes, even in live cells and organisms. Key to their success are functional groups that react reliably and selectively in complex settings (Figure 1). Considering the breadth of functionality present in cells and tissues, the demands on bioorthogonal reactions are enormous.1,2 The reagents must tolerate aqueous environments and large concentrations of cellular nucleophiles. While remaining inert to their surroundings, the functional groups must also react robustly with one another to provide stable adducts. Despite these stringent criteria, transformations have been identified that are well recognized as being bioorthogonal. Many have been used for decades to tag proteins and other biomolecules,3 profile active enzymes,4,5 and identify drug targets.6,7 More recent advances in bioorthogonal reactions are enabling new pursuits in drug delivery,8,9 genetic code expansion,10,11 and protein activation.12–16
Figure 1.
Bioorthogonal chemistries comprise reagents (blue circle and arc) that react reliably and with high specificity in complex environments. These reactions enable target biomolecules to be covalently ligated with fluorophores, affinity tags, or other probes (red star).
Even with its impressive résumé, the field of bioorthogonal chemistry is not without limitation. Few reagents can be used in the harshest cellular confines and in conjunction with the smallest biomolecules.17 Furthermore, many of the most popular bioorthogonal reagents cross-react with one another and thus cannot be employed concurrently.18 These and other drawbacks continue to inspire new explorations for chemistries with potential biological utility. In our own group, we have focused on constructing reagents that are small in size, highly stable and tunable—features that can facilitate their widespread adoption. This Account highlights the development of three such bioorthogonal functional groups: cyclopropenes, triazines, and cyclopropenones. These motifs can also be used together in live cells, enabling multicomponent imaging and other multiplexed analyses. Our work is presented against a backdrop of design considerations for new bioorthogonal reactions. These principles can guide the continued pursuit of useful chemistries.
How many bioorthogonal reactions are ultimately necessary? A strong case for hundreds can be made, based on analogy to other classes of synthetic organic transformations. Dozens of distinct methods exist to construct amides and other classic bonds, with each having its pros and cons. The approach selected is dictated by the individual experiment. Similar parallels exist in bioorthogonal chemistry. Each transformation has its strengths and weaknesses, with no single reaction type being suitable for all applications. In some cases, the fastest-reacting probes are necessary; in others, slower, but more selective reagents should be employed. Some reagents exhibit background reactivities with cellular nucleophiles and other instabilities, hindering in vivo applications. Such liabilities can often be mitigated via steric and electronic tuning, and dozens of examples exist among the major classes of bioorthogonal ligations. The time invested to fine-tune and optimize bioorthogonal scaffolds is often critical to their long-term success. Thus, it is important to appreciate the bioorthogonal “workbench” just as much as the tools in the toolbox.
Laying the foundation
At the outset of new reaction development, it is important to tackle a fundamental question: what does it mean to be bioorthogonal? Quite literally, bioorthogonal reagents and transformations are orthogonal to (i.e., independent of) biology. Thus, in the strictest sense, the functional groups must not be present in living systems. This type of “bioorthogonality” is rarely (if ever) achieved, as many of the classic motifs have precedence in microbial natural products and other metabolites. “Bioorthogonal”, in practice, tends to be more loosely applied to reagents that minimally interfere with the system under study. Even with this broader definition, perfect compatibility can be difficult to achieve. Nearly all bioorthogonal reagents have liabilities in certain cellular environments.1 Such drawbacks do not necessarily lessen the impact of a given probe, but can provide motivation to optimize and develop new reactions that address the inherent shortcomings.
Our work to build better bioorthogonal reactions (via the latter definition) has often been inspired by nature. The diversity of chemical functionality present in natural products and other metabolites is immense (Figure 2). Many of the motifs are not found in higher eukaryotes, making them “bioorthogonal” in a heterologous context. The groups also possess some degree of stability in cellular environments, owing to their presence in living systems. Thus, they can be ideal starting points for new reaction development and eventual translation in vivo. One of the best examples of a naturally occurring bioorthogonal functional group is the terminal alkyne.19,20 Alkynes comprise numerous small molecule metabolites (often as part of di-yne or poly-yne scaffolds) in sponges and other organisms. The stability and unique reactivities of this functional group have rendered it one of the most widely used labels in all of bioorthogonal chemistry.
Figure 2.
Many popular bioorthogonal motifs (blue) are found in natural products. The functional groups highlighted here have been re-purposed for selective tagging reactions in heterologous environments.
We took additional cues from nature in developing bioorthogonal chemistries based on cyclopropenes,21–26 triazines,27–29 and cyclopropenones.30–33 All three motifs are found in natural product structures, suggesting that they were suitable starting points for probe development. Indeed, the native scaffolds inspired our initial choices of model probes and analogs to prepare. Further optimization to tune stabilities and reactivities was guided by physical organic principles and computation (described below). We anticipate that continued mining of natural product structures will uncover new functionality and reaction platforms to pursue.
Filling the voids
Many applications of bioorthogonal chemistry involve profiling or visualizing small targets, including metabolites and drugs. In these cases, there is a clear need for functional groups that minimally perturb the systems under study. Even with proteins and larger targets, it is often desirable to use small tags to preserve native functions and interactions. When steric considerations are important, the azide and terminal alkyne have long dominated as the bioorthogonal reagents of choice (Figure 3). Comprising just a few atoms, these groups have been ubiquitously employed in chemical biology. Both are well tolerated in biological systems and can be readily detected via copper-catalyzed azide-alkyne cycloaddition (CuAAC).34 The azide and alkyne are also quite “user friendly”. Many popular azido- and alkynyl-functionalized probes are commercially available or otherwise easily constructed. Consequently, they have found widespread application in bioconjugation, materials chemistry, drug discovery, and many other areas.34 Limitations of CuAAC in cellular environments also drove numerous efforts to improve the scope and biocompatibility of azide-alkyne cycloadditions.34,35
Figure 3.
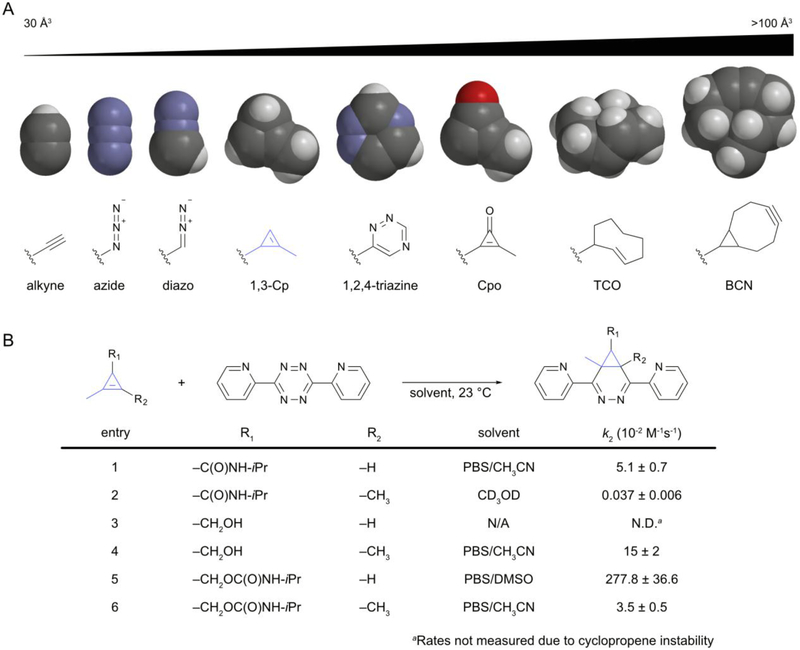
Bioorthogonal reagents come in all shapes and sizes. A) Sample probes are shown, arranged by approximate size. Volumes were estimated using atomic radii measurements in Spartan. B) Cyclopropenes can be tuned for reaction with tetrazines. Minimally substituted cyclopropenes exhibit the fastest rates.
Inspired by the versatility of small and stable reagents, we set our sights initially on cyclopropenes as candidate bioorthogonal reagents. As noted earlier, cyclopropenes are found in some natural products, suggesting compatibility with living systems. While slightly larger than both azides and alkynes, cyclopropenes are the smallest of the isolable strained alkenes. This class of reactants includes the venerable trans−cyclooctene (TCO), a motif that has emerged as a powerful and versatile addition to the bioorthogonal toolkit. Strained alkenes react with tetrazines via inverse electron-demand Diels–Alder (IED-DA) reactions, exhibiting remarkably fast kinetics.36–39 The TCO-tetrazine ligation is unrivaled in its reaction speed, and such rapid reactivity has enabled applications in rodent models and other large organisms—settings where only minimal reaction times and reagent concentrations are tolerated.
While TCO is routinely used for IED-DA reactions, it is roughly double the size of a standard cyclopropene (Figure 3B), making it less attractive for some applications. We were motivated to examine the utility of the smaller strained alkene for bioorthogonal labeling. In early work, we synthesized a small panel of substituted cyclopropenes.21 Methyl-substituted scaffolds were found to be stable in a variety of biological environments, even in the presence of common nucleophiles. The reactivities of the probes with various tetrazines were also measured, and the fastest reactions were observed with the least sterically congested cyclopropenes. Complementary studies were performed by the Devaraj group.40,41 Rapid reactions were also observed in more polar solvents and with cyclopropenes with reduced electron-withdrawing character at C-3, consistent with the inverse electron-demand of the cycloaddition.
The cyclopropene-tetrazine reaction rates are markedly slower than the corresponding ligations with TCO. Nonetheless, the reaction has been applied in numerous biomolecule tagging experiments, including protein and nucleic acid visualization,42–45 cell surface labeling,46,47 and in vivo proteomics.48,49 Importantly, the small size of the cyclopropene has enabled experiments that would be difficult to achieve with larger motifs. One poignant example includes metabolic targeting of cellular glycans with functionalized monosaccharides. The enzymes involved in glycan biosyntheses can be quite stringent, allowing only minimally perturbed scaffolds to be processed. The fact that cyclopropene sugars were metabolized on par with analogous azido substrates is a testament to the potential broad utility of the new bioorthogonal reagent.21,46,50
The small size of the cyclopropenes has also been a boon to genetic code expansion efforts. This powerful technology enables noncanonical amino acids (ncAAs) to be site-specifically installed into target proteins in response to a stop codon (Figure 4).3 The key step involves an orthogonal aminoacyl-tRNA synthetase (AARS) that charges the ncAA onto a cognate tRNA. The tRNA is similarly orthogonal to the cell’s endogenous machinery. To identify an appropriate AARS/tRNA pair, large libraries of mutants must typically be screened and extensive optimization for use in mammalian cells must be performed.51,52 Interestingly, ncAAs bearing cyclopropene motifs can be efficiently processed by a native pyrrolysine AARS from archaebacteria, without the need for additional mutagenesis.48,53,54 This feature has enabled cyclopropenes to be immediately applied in a variety of contexts, including cell-specific proteome labeling in flies and mouse brain tissue.48,55 More recent work has capitalized on cyclopropene ncAAs for dual protein labeling experiments.56,57
Figure 4.
Genetic code expansion with bioorthogonal functional groups. Using an orthogonal AARS/tRNA pair, ncAAs can be site-specifically incorporated into proteins of interest. A variety of small bioorthogonal motifs (including cyclopropenes) have been installed via the pyrrolysine synthetase (PylRS) machinery.
Hammering out the details
Understanding the stability and reactivity profiles of bioorthogonal reagents is critical to their successful application in vitro, in cells, and in vivo. Such analyses are often aided by detailed investigations of reaction mechanism and substituent effects. Ideally, these studies provide insight on how to tune scaffolds for desired reaction speeds, biocompatibilities, or other parameters. In our work, a deep dive into reaction mechanism has been best exemplified in studies of the other half of the IED-DA reaction: the electron-deficient diene. To date, tetrazines have dominated in this role. Tetrazines react robustly with cyclopropenes, TCO, and a variety of other strained dienophiles. Like other families of bioorthogonal reagents, tetrazines have been tuned to achieve desired levels of reactivity (Figure 5). Electron-withdrawing substituents at the 3 or 6 positions lower tetrazine LUMO energies and increase reaction rates.58 Steric effects can also play a role in tetrazine reactivity, with less encumbered scaffolds displaying the fastest rates. Improvements in speed, though, often come at the expense of stability. For many applications this is a fair trade, but for others it is desirable to minimize background labeling.
Figure 5.
Bioorthogonal reagents can be tuned for downstream applications. A panel of tetrazine and triazine motifs have been developed that exhibit a wide range of stabilities and reactivities.
The need for additional tuning inspired our search for new dienes. In particular, we were drawn to triazine scaffolds. 1,2,4-Triazines have been identified in microbial natural products and pigments, suggesting that they were stable in physiological environments. Density functional theory (DFT) calculations performed by the Houk group further suggested that 1,2,4-triazines would exhibit enhanced stability relative to tetrazines, yet retain reactivity with TCO (Figure 5). Based on these observations, we hypothesized that triazines would be good candidates for bioorthogonal reaction development. We synthesized a small panel of triazine probes, and found them to be highly stable to aqueous conditions and cellular thiols. Importantly, they were still reactive with dienophiles such as TCO and some strained alkynes.27,59 Because triazines require fewer substituents than analogous tetrazines for long-term stability, they can leave a smaller footprint. We capitalized on these features to showcase the utility of triazines for recombinant protein production.27 The long incubation times required in this process are not compatible with many tetrazines and other bioorthogonal scaffolds. Triazines have also found recent application in labeling nucleic acids,60 and further tuning has identified fluorogenic analogs.61
The increase in stability gained with triazines comes at the expense of decreased reaction rates. Modifications to the triazine core (e.g., electron-withdrawing substituents) can recover some of the reactivity and even small modifications were found to have dramatic impacts.27 Further calculations predicted that 1,2,4-triazines would be nonreactive with other strained alkenes, including cyclopropenes and norbornene. These results were verified experimentally and set the stage for mutually orthogonal reaction development.27,62
Mechanistic studies and computational analyses further enabled efficient reagent tuning in the case of the cyclopropenes. Our initial investigations revealed that cyclopropene-tetrazine reactions occur from the least hindered face of the cyclopropene. The adducts can undergo additional rearrangements to produce mixtures of diastereomers.21 Computational analyses predicted that the addition of a single methyl group at C-3 would be sufficient to impede tetrazine reactivity (Figure 6A).63 The methyl substituent was predicted to engender a steric clash that could control cycloaddition preference. Indeed, 3,3-disubstituted cyclopropenes (3,3-Cp) were refractory to tetrazine ligation, while their 1,3-disubstitued counterparts (1,3-Cp) reacted robustly. Both scaffolds were readily ligated with less sterically encumbered 1,3-dipoles (e.g., nitriles imines). The unique reactivity profiles of the isomeric cyclopropenes enabled tandem labeling of biomolecules in a single pot (Figure 6B). Recent work by the Lin group has uncovered a more strained 3,3-disubstituted cyclopropene that can be effectively ligated with tetrazines, suggesting that even more finely tuned reagent pairs will be uncovered.53
Figure 6.
Cyclopropene isomers exhibit unique reactivity profiles. A) Predicted transition state geometries indicated a steric clash in the reaction of a 3,3-disubstituted cyclopropenes (3,3-Cp) with tetrazine.63 The unfavorable interaction was predicted to markedly the diminish reaction rate compared to the 1,3-disubstituted isomer (1,3-Cp). B) The unique reactivities of 1,3-Cp and 3,3-Cp enabled tandem labeling of distinct biomolecules.
Thinking outside the (tool)box
As the number of bioorthogonal tools continues to expand, their application to multi-component labeling becomes feasible. Such studies require reactions that are not only bioorthogonal, but also orthogonal to one another. The challenges in this context are immense, considering the number of potential side products (Figure 7). Additionally, most bioorthogonal reactions, and nearly all reported in recent years, comprise cycloadditions.1 Many of the underlying reagents are incompatible with one another and cannot be used concurrently.18,64 In some cases, differences in rate can be exploited for sequential labeling. However, such strategies often require the removal of excess reagents from the first reaction, before the second can be initiated. Our initial work with isomeric cyclopropenes63 (described above) enabled in-tandem labeling, but only with sequential reagent addition.
Figure 7.
Mutually orthogonal reactions enable simultaneous tagging of multiple biomolecules. The reagents and products in such transformations must exhibit no off-target or cross-reactivities. Even in the simplest case (two targets, A and B) the large number of undesirable reactions that must be avoided is large.
A potentially more general approach to developing mutually orthogonal reactions involves focusing on transformations that operate via distinct mechanisms (Figure 8). In early work, we showed that the cyclopropene-tetrazine ligation is compatible with azide-alkyne cycloadditions. We used these reactions to simultaneously tag cell surface glycans for downstream imaging applications.46 The compatible ligations have also enabled concurrent labeling of multiple bacterial targets.65 In recent years, additional orthogonal reactions have been reported that promise to bolster multi-parameter imaging and other applications.18,62,66–69
Figure 8.
Examples of bioorthogonal chemistries that exploit distinct reaction mechanisms.
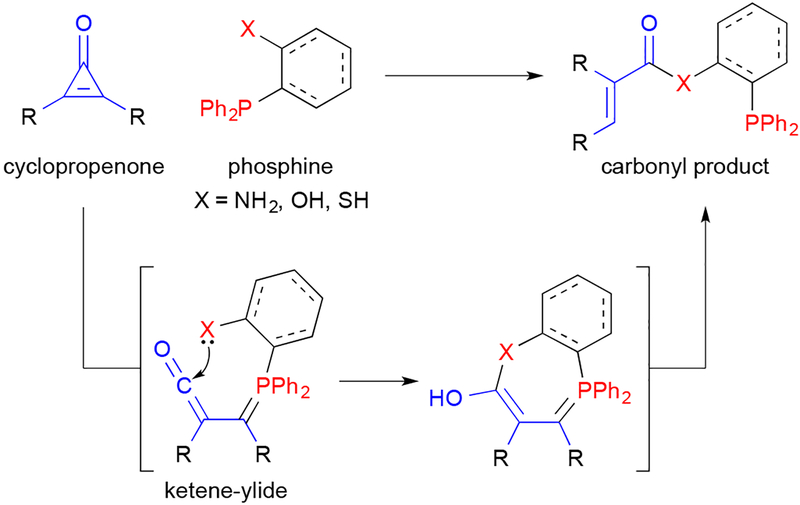
In our own group, we have begun examining polar reactions—chemistries that are likely compatible with existing bioorthogonal ligations based on their unique mechanisms. In one area, we have been exploring the reaction between cyclopropenones and bioorthogonal phosphines.30,70 Mono-substituted cyclopropenones are found in some natural products (Figure 2), suggesting that they were suitable candidates for biocompatible reaction development.31,33 We synthesized a panel of cyclopropenones, and analyzed their stabilities in aqueous solution and in the presence of cellular nucleophiles.70 The most stable scaffolds were structurally similar to bioorthogonal cyclopropenes, yet exhibited unique manifolds of reactivity. Cyclopropenones react with phosphines to generate ketene-ylides; these intermediates can be trapped by pendant nucleophiles on the phosphine probe (Figure 9). We demonstrated that this ligation can be performed on model proteins in vitro and in cell lysate. The cyclopropenone-phosphine reaction also holds promise for multi-component labeling, as the reagents are not expected to interfere in common cycloadditions. Indeed, preliminary work in our lab has shown that cyclopropenones and phosphines do not interfere with tetrazines, cyclopropenes, or TCO.
Figure 9.
The cyclopropenone-phosphine ligation. Cyclopropenones react with phosphines to generate ketene-ylide intermediates. The ketenes are subject to intramolecular trapping by pendant nucleophiles. Subsequent protonation and elimination steps provide carbonyl products.
Ongoing work in the field has also revealed new platforms for “orthogonal bioorthogonal” labeling. Davis and others have demonstrated that radical reactions can be employed for bioconjugation reactions.71,72 Other transformations that are garnering attention include metal-catalyzed ligations73–76 and bioorthogonal cleavage reactions.12 It should also be noted that many bioorthogonal chemistries can be triggered with light or other exogenous stimuli, enabling spatiotemporal control of reactivity.45,77
Paving the way for new discoveries
The application of bioorthogonal reactions to diverse problems has not only revealed new and often unanticipated discoveries, but also driven the development of new tools and synthetic methods.78 For example, bioorthogonal transformations have been used to expediently prepare diverse libraries of complex molecules.79 Mild methods to affix azides and other bioorthogonal motifs to complex drugs and natural products have also been reported.80,81 Systematic efforts to tune and optimize biocompatible reactions will continue to provide probes with novel capabilities; these tools, in turn, will spur new advances. This iterative cycle of tool development and discovery will continue as the field expands. We are still in the midst of establishing a fleet of sensitive, selective reagents that will bolster chemistries in living systems and thus enable new research directions.
As the number of bioorthogonal reagents and their spectrum of reactivities grows, new challenges are emerging. The sheer number of possible reagents and conditions can be daunting to the non-specialist. Unlike other areas of organic chemistry, where organized catalogs of reaction conditions exist, there is no comparable “Larock book” of bioorthogonal transformations. Tool users must wade through an ever expanding and complex body of literature to identify reagents best suited for their applications. Thus, knowing which probes to select for a given experiment remains difficult. Additionally, while numerous chemical tools have been developed in recent years, their transition to the broader scientific commmunity—and widespread adoption—have often been quite sluggish. This is due, in part, to limited probe accessibilities. Many of the best reagents for a given application are not commercially available or require complex syntheses. Renewed efforts to develop more accessible probes will ease the transition of these reactions from the hands of the toolmaker to the tool user.
There is also an ongoing need to fill gaps along the continuums of reactivity and stability. Few reagents meet the strict requirements for use in living animals. Such probes must often be both exquisitely stable and potently reactive with complementary functionality. Further advances in reagent design will likely address this challenge and broaden the scope of possible applications. Along the way, probes that fall short of the bar for in vivo will likely be useful in other contexts. Already, many reagents that are insufficiently stable for intracellular application have found utility in antibody-drug conjugate formation and materials research.82 Recent developments in methionine labeling,83 cysteine conjugations,84 and carbonyl ligations85–87 are also addressing the need for diverse chemical transformations.
Conclusions
Bioorthogonal chemistries have become indispensible tools for modern chemical biology research. These reactions enable biomolecules to be studied in real time and in their native environments. Despite decades of achievements in crafting biocompatible reagents and reactions, though, limitations remain. Only a few probes are suitable for use in intracellular labeling experiments. Even fewer are small enough to traverse native biosynthetic pathways. Many popular probes also cross-react with one another, limiting applications in multi-component studies. Others and we have been addressing these challenges in recent years, by focusing on small and stable reagents that react via unique mechanisms.
Crafting new bioorthogonal reactions is not trivial, but successful examples can illuminate some guiding principles. In many cases, new transformations have been inspired by natural product structures. Such scaffolds often comprise unique functional groups, that when used in heterologous hosts, exhibit bioorthogonal character. These naturally occurring motifs thus offer advantageous starting points for developing new reactions Further optimization can be achieved via classic physical organic studies or computational analyses. Together, these approaches can reveal unanticipated modes of reactivity and guide new reaction development. Embracing a spectrum of reactivity will encourage the development and application of new reactions without the unrealistic constraints of a one-size-fits-all transformation. Reactions that operate via distinct mechanistic pathways will further enable simultaneous labeling of multiple targets. As new biological questions continue to emerge, the demand for new probes will grow. We anticipate that efforts to expand the collection of bioorthogonal tools will not only enable new pursuits in biology, but also push the frontiers of chemistry.
ACKNOWLEDGMENTS
R.D.R. is a National Science Foundation Graduate Research Fellow. J.A.P. is a Cottrell Scholar, Alfred P. Sloan Fellow, and Dreyfus Scholar. Our work on bioorthogonal reaction development is funded by the National Institutes of Health (R01 GM126226). We thank members of the Prescher laboratory for helpful discussions during the manuscript preparation.
Biographies
R. David Row obtained his B.S. in Biochemistry from Western Washington University in 2014. He is currently a Ph.D. candidate at UC Irvine, developing new bioorthogonal reactions in the Prescher laboratory.
Jennifer A. Prescher is a Professor of Chemistry, Molecular Biology and Biochemistry, and Pharmaceutical Sciences at UC Irvine. She earned her B.S. degree at the University of Wisconsin–La Crosse in 2001 and obtained her Ph.D. in Chemistry at UC Berkeley in 2006 with Prof. Carolyn Bertozzi. She conducted postdoctoral research with Prof. Christopher Contag at Stanford University before joining the UC Irvine faculty in 2010.
REFERENCES
- (1). Patterson DM; Nazarova LA; Prescher JA Finding the right (bioorthogonal) chemistry. ACS Chem. Biol 2014, 9, 592–605. [DOI] [PubMed] [Google Scholar]
- (2). Prescher JA; Bertozzi CR Chemistry in living systems. Nat. Chem. Biol 2005, 1, 13–21. [DOI] [PubMed] [Google Scholar]
- (3). Lang K; Chin JW Cellular incorporation of unnatural amino acids and bioorthogonal labeling of proteins. Chem. Rev 2014, 114, 4764–4806. [DOI] [PubMed] [Google Scholar]
- (4). Willems LI; van der Linden WA; Li N; Li K-Y; Liu N; Hoogendoorn S; van der Marel GA; Florea BI; Overkleeft HS Bioorthogonal chemistry: Applications in activity-based protein profiling. Acc. Chem. Res 2011, 44, 718–729. [DOI] [PubMed] [Google Scholar]
- (5). Nomura DK; Dix MM; Cravatt BF Activity-based protein profiling for biochemical pathway discovery in cancer. Nat. Rev. Cancer 2010, 10, 630–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6). Yang KS; Budin G; Tassa C; Kister O; Weissleder R Bioorthogonal approach to identify unsuspected drug targets in live cells. Angew. Chem. Int. Ed 2013, 52, 10593–10597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7). Rutkowska A; Thomson DW; Vappiani J; Werner T; Mueller KM; Dittus L; Krause J; Muelbaier M; Bergamini G; Bantscheff M A modular probe strategy for drug localization, target identification and target occupancy measurement on single cell level. ACS Chem. Biol 2016, 11, 2541–2550. [DOI] [PubMed] [Google Scholar]
- (8). Versteegen RM; Rossin R; ten Hoeve W; Janssen HM; Robillard MS Click to release: Instantaneous doxorubicin elimination upon tetrazine ligation. Angew. Chem. Int. Ed 2013, 52, 14112–14116. [DOI] [PubMed] [Google Scholar]
- (9). Oneto JMM; Khan I; Seebald L; Royzen M In vivo bioorthogonal chemistry enables local hydrogel and systemic pro-drug to treat soft tissue sarcoma. ACS Cent. Sci 2016, 2, 476–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10). Stone SE; Glenn WS; Hamblin GD; Tirrell DA Cell-selective proteomics for biological discovery. Curr. Opin. Chem. Biol 2017, 36, 50–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11). Alvarez-Castelao B; Schanzenbächer CT; Hanus C; Glock C; Dieck ST; Doörrbaum AR; Bartnik I; Nassim-Assir B; Ciirdaeva E; Mueller A; Dieterich DC; Tirrell DA; Langer JD; Schuman EM Cell-type-specific metabolic labeling of nascent proteomes in vivo. Nat. Biotechnol 2017, 35, 1196–1201. [DOI] [PubMed] [Google Scholar]
- (12). Li J; Chen PR Development and application of bond cleavage reactions in bioorthogonal chemistry. Nat. Chem. Biol 2016, 12, 129–137. [DOI] [PubMed] [Google Scholar]
- (13). Fan X; Yun Ge FL; Yang Y; Zhang G; Ngai WSC; Zhi Lin SZ; Wang J; Zhao J; Li J; Chen PR Optimized tetrazine derivatives for rapid bioorthogonal decaging in living cells. Angew. Chem. Int. Ed 2016, 55, 14046–14050. [DOI] [PubMed] [Google Scholar]
- (14). Zhang G; Li J; Xie R; Fan X; Liu Y; Zheng S; Ge Y; Chen PR Bioorthogonal chemical activation of kinases in living systems. ACS Cent. Sci 2016, 2, 325–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15). Völker T; Meggers E Transition-metal-mediated uncaging in living human cells — an emerging alternative to photolabile protecting groups. Curr. Opin. Chem. Biol 2015, 25, 48–54. [DOI] [PubMed] [Google Scholar]
- (16). Li J; Yu J; Zhao J; Wang J; Zheng S; Lin S; Chen L; Yang M; Jia S; Zhang X; Chen PR Palladium-triggered deprotection chemistry for protein activation in living cells. Nat. Chem 2014, 6, 352–361. [DOI] [PubMed] [Google Scholar]
- (17). Murrey HE; Judkins JC; am Ende CW; Ballard TE; Fang Y; Riccardi K; Di L; Guilmette ER; Schwartz JW; Fox JM; Johnson DS Systematic evaluation of bioorthogonal reactions in live cells with clickable HaloTag ligands: Implications for intracellular imaging. J. Am. Chem. Soc 2015, 137, 11461–11475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18). Patterson DM; Prescher JA Orthogonal bioorthogonal chemistries. Curr. Opin. Chem. Biol 2015, 28, 141–149. [DOI] [PubMed] [Google Scholar]
- (19). Chai Q-Y; Yang Z; Lin H-W; Han B-N Alkynyl-containing peptides of marine origin: A review. Mar. Drugs 2016, 14, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20). Cimino G; De Giulio A; De Rosa S; Di Marzo V High molecular weight polyacetylenes from Petrosia ficiformis: Further structural analysis and biological activity. Tetrahedron Lett 1989, 30, 3563–3566. [Google Scholar]
- (21). Patterson DM; Nazarova LA; Xie B; Kamber DN; Prescher JA Functionalized cyclopropenes as bioorthogonal chemical reporters. J. Am. Chem. Soc 2012, 134, 18638–18643. [DOI] [PubMed] [Google Scholar]
- (22). Matsuura M; Saikawa Y; Inui K; Nakae K; Igarashi M; Hashimoto K; Nakata M Identification of the toxic trigger in mushroom poisoning. Nat. Chem. Biol 2009, 5, 465–467. [DOI] [PubMed] [Google Scholar]
- (23). Fattorusso E; Magno S; Mayol L; Satacroce C; Sica D Calysterol: A C29 cyclopropene-containing marine sterol from the sponge Calyx nicaensis. Tetrahedron 1975, 31, 1715–1716. [Google Scholar]
- (24). Doss GA; Djerassi C Sterols in marine invertebrates. 60. Isolation and structure elucidation of four new steroidal cyclopropenes from the sponge Calyx podatypa. J. Am. Chem. Soc 1988, 110, 8124–8128. [Google Scholar]
- (25). Pasha MK; Ahmad F Analysis of triacylglycerols containing cyclopropene fatty acids in Sterculia foetida (Linn.) seed lipids. J. Agric. Food Chem 1992, 40, 626–629. [Google Scholar]
- (26). Zoeller RA; Wood R Effects of cyclopropene fatty acids on the lipid composition of the Morris hepatoma 7288C. Lipids 1984, 19, 529–538. [DOI] [PubMed] [Google Scholar]
- (27). Kamber DN; Liang Y; Blizzard RJ; Liu F; Mehl RA; Houk KN; Prescher JA 1,2,4-Triazines are versatile bioorthogonal reagents. J. Am. Chem. Soc 2015, 137, 8388–8391. [DOI] [PubMed] [Google Scholar]
- (28). Smirnov VV; Kiprianova EA; Garagulya AD; Esipov SE; Dovjenko SA Fluviols, bicyclic nitrogen-rich antibiotics produced by Pseudomonas fluorescens. FEMS Microbiol. Lett 1997, 153, 357–361. [DOI] [PubMed] [Google Scholar]
- (29). Lindner HJ; Schaden G Pyrazolo[4.3-e]as-triazin, ein neues heterocyclisches system aus Pseudomonas fluorescens var. pseudoiodinum. Chem. Ber 1972, 105, 1949–1955. [DOI] [PubMed] [Google Scholar]
- (30). Shih H-W; Prescher JA A bioorthogonal ligation of cyclopropenones mediated by triarylphosphines. J. Am. Chem. Soc 2015, 137, 10036–10039. [DOI] [PubMed] [Google Scholar]
- (31). Kogen H; Kiho T; Tago K; Miyamoto S; Fujioka T; Otsuka N; Suzuki-Konagai K; Ogita T Alutacenoic acids A and B, rare naturally occurring cyclopropenone derivatives isolated from fungi: Potent non-peptide Factor XIIIa inhibitors. J. Am. Chem. Soc 2000, 122, 1842–1843. [Google Scholar]
- (32). Okuda T; Yokose K; Furumai T; Maruyama HB Penitricin, a new class of antibiotic produced by Penicillium aculearum I. Isolation and characterization. J. Antibiot 1984, 37, 718–722. [DOI] [PubMed] [Google Scholar]
- (33). Bohlmann F; Jakupovic J; Müller L; Schusrer A Naturally occurring cyclopropenone derivatives. Angew. Chem. Int. Ed 1981, 3, 292–293. [Google Scholar]
- (34). Meldal M; Tornøe CW Cu-catalyzed azide-alkyne cycloaddition. Chem. Rev 2008, 108, 2952–3015. [DOI] [PubMed] [Google Scholar]
- (35). Jewett JC; Bertozzi CR Cu-free click cycloaddition reactions in chemical biology. Chem. Soc. Rev 2010, 39, 1272–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36). Blackman ML; Royzen M; Fox JM Tetrazine ligation: Fast bioconjugation based on inverse-electron-demand Diels-Alder reactivity. J. Am. Chem. Soc 2008, 130, 13518–13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37). Taylor MT; Blackman ML; Dmitrenko O; Fox JM Design and synthesis of highly reactive dienophiles for the tetrazine trans-cyclooctene ligation. J. Am. Chem. Soc 2011, 133, 9646–9649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38). Lambert WD; Scinto SL; Dmitrenko O; Boyd SJ; Magboo R; Mehl RA; Chin JW; Fox JM; Wallace S Computationally guided discovery of a reactive, hydrophilic trans-5-oxocene dienophile for bioorthogonal labeling. Org. Biomol. Chem 2017, 15, 6640–6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39). Blizzard RJ; Backus DR; Brown W; Bazewicz CG; Li Y; Mehl RA Ideal bioorthogonal reactions using a site-specifically encoded tetrazine amino acid. J. Am. Chem. Soc 2015, 137, 10044–10047. [DOI] [PubMed] [Google Scholar]
- (40). Yang J; Šecǩute J; Cole CM; Devaraj NK Live-cell imaging of cyclopropene tags with fluorogenic tetrazine cycloadditions. Angew. Chem. Int. Ed 2012, 51, 7476–7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41). Yang J; Liang Y; Šečkutė J; Houk KN; Devaraj NK Synthesis and reactivity comparisons of 1-methyl-3-substituted cyclopropene mini-tags for tetrazine bioorthogonal reactions. Chem. Eur. J 2014, 20, 3365–3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42). Sachdeva A; Wang K; Elliott T; Chin JW Concerted, rapid, quantitative, and site-specific dual labeling of proteins. J. Am. Chem. Soc 2014, 136, 7785–7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43). Wu H; Cisneros BT; Cole CM; Devaraj NK Bioorthogonal tetrazine-mediated transfer reactions facilitate reaction turnover in nucleic acid-templated detection of microRNA. J. Am. Chem. Soc 2014, 136, 17942–17945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44). Eggert F; Kath-Schorr S A cyclopropene-modified nucleotide for site-specific RNA labeling using genetic alphabet expansion transcription. Chem. Commun 2016, 52, 7284–7287. [DOI] [PubMed] [Google Scholar]
- (45). Yu Z; Pan Y; Wang Z; Wang J; Lin Q Genetically encoded cyclopropene directs rapid, photoclick-chemistry-mediated protein labeling in mammalian cells. Angew. Chem. Int. Ed 2012, 51, 10600–10604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46). Patterson DM; Jones KA; Prescher JA Improved cyclopropene reporters for probing protein glycosylation. Mol. BioSyst 2014, 10, 1693–1697. [DOI] [PubMed] [Google Scholar]
- (47). Spate A-K; Bußkamp H; Niederwieser A; Schart VF; Marx A; Wittmann V Rapid labeling of metabolically engineered cell-surface glycoconjugates with a carbamate-linked cyclopropene reporter. Bioconjugate Chem 2014, 25, 147–154. [DOI] [PubMed] [Google Scholar]
- (48). Elliott TS; Townsley FM; Bianco A; Ernst RJ; Sachdeva A; Elsässer SJEA; Davis L; Lang K; Pisa R; Greiss S; Lilley KS; Chin JW Proteome labeling and protein identification in specific tissues and at specific developmental stages in an animal. Nat. Biotechnol 2014, 32, 465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49). Elliott TS; Bianco A; Townsley FM; Fried SD; Chin JW Tagging and enriching proteins enables cell-specific proteomics. Cell Chemical Biology 2016, 23, 805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50). Ravasco JMJM; Monteiro CM; Trindade AF Cyclopropenes: A new tool for the study of biological systems. Org. Chem. Front 2017, 4, 1167–1198. [Google Scholar]
- (51). Chin JW Expanding and reprogramming the genetic code of cells and animals. Annu. Rev. Biochem 2014, 83, 379–408. [DOI] [PubMed] [Google Scholar]
- (52). Xie J; Schultz PG An expanding genetic code. Methods 2005, 36, 227–238. [DOI] [PubMed] [Google Scholar]
- (53). Ramil CP; Dong M; An P; Lewandowski TM; Yu Z; Miller LJ; Lin Q Spirohexene-tetrazine ligation enables bioorthogonal labeling of class B G protein-coupled receptors in live cells. J. Am. Chem. Soc 2017, 139, 13376–13386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54). Peng T; Hang HC Site-specific bioorthogonal labeling for fluorescence imaging of intracellular proteins in living cells. J. Am. Chem. Soc 2016, 138, 14423–14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55). Ernst RJ; Krogager TP; Maywood ES; Zanchi R; Beránek V; Elliott TS; Barry NP; Hastings MH; Chin JW Genetic code expansion in the mouse brain. Nat. Chem. Biol 2016, 12, 776–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56). Zheng Y; Addy PS; Mukherjee R; Chatterjee A Defining the current scope and limitations of dual noncanonical amino acid mutagenesis in mammalian cells. Chem. Sci 2017, 8, 7211–7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57). Zheng Y; Mukherjee R; Chin MA; Igo P; Gilgenast MJ; Chatterjee A Expanding the scope of single and dual noncanonical amino acid mutagenesis in mammalian cells using orthogonal polyspecific leucyl-tRNA synthetases. Biochemistry 2017, 57, 441–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58). Karver MR; Weissleder R; Hilderbrand SA Synthesis and evaluation of a series of 1,2,4,5-tetrazines for bioorthogonal conjugation. Bioconjugate Chem 2011, 22, 2263–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59). Horner KA; Valette NM; Webb ME Strain-promoted reaction of 1,2,4-triazines with bicyclononynes. Chem. Eur. J 2015, 21, 14376–14381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60). Peewasan K; Wagenknecht H-A 1,2,4-Triazine-modified 2’-deoxyuridine triphosphate for efficient bioorthogonal fluorescent labeling of DNA. ChemBioChem 2017, 18, 1473–1476. [DOI] [PubMed] [Google Scholar]
- (61). Siegl SJ; Dzijak R; Vázquez A; Pohl R; Vrabel M The discovery of pyridinium 1,2,4-triazines with enhanced performance in bioconjugation reactions. Chem. Sci 2017, 8, 3593–3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62). Liu F; Liang Y; Houk KN Bioorthogonal cycloadditions: Computational analysis with the distortion/interaction model and predictions of reactivities. Acc. Chem. Res 2017, 50, 2297–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63). Kamber DN; Nazarova LA; Liang Y; Lopez SA; Patterson DM; Shih H-W; Houk KN; Prescher JA Isomeric cyclopropenes exhibit unique bioorthogonal reactivities. J. Am. Chem. Soc 2013, 135, 13680–13683. [DOI] [PubMed] [Google Scholar]
- (64). Shih H-W; Kamber DN; Prescher JA Building better bioorthogonal reactions. Curr. Opin. Chem. Biol 2014, 21, 103–111. [DOI] [PubMed] [Google Scholar]
- (65). Hudak JE; Alvarez D; Skelly A; von Andrian UH; Kasper DL Illuminating vital surface molecules of symbionts in health and disease. Nat. Microbiol 2017, 2, 17099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66). Sherratt AR; Chigrinova M; MacKenzie DA; Rastogi NK; Ouattara MTM; Pezacki AT; Pezacki JP Dual strain-promoted alkyne−nitrone cycloadditions for simultaneous labeling of bacterial peptidoglycans. Bioconjugate Chem 2016, 27, 1222–1226. [DOI] [PubMed] [Google Scholar]
- (67). Narayanam MK; Liang Y; Houk KN; Murphy JM Discovery of new mutually orthogonal bioorthogonal cycloaddition pairs through computational screening. Chem. Sci 2016, 7, 1257–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68). Aronoff MR; Gold B; Raines RT 1,3-Dipolar cycloadditions of diazo compounds in the presence of azides. Org. Lett 2016, 18, 1538–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69). Eising S; Lelivelt F; Bonger KM Vinylboronic acids as fast reacting, synthetically accessible, and stable bioorthogonal reactants in the Carboni–Lindsey reaction. Angew. Chem. Int. Ed 2016, 55, 12243–12247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70). Row RD; Shih H-W; Alexander AT; Mehl RA; Prescher JA Cyclopropenones for metabolic targeting and sequential bioorthogonal labeling. J. Am. Chem. Soc 2017, 139, 7370–7375. [DOI] [PubMed] [Google Scholar]
- (71). Wright TH; Bower BJ; Chalker JM; Bernardes GJL; Wiewiora R; Ng W-L; Raj R; Faulkner S; Vallée MRJ; Phanumartwiwath A; Coleman OD; Thézénas M-L; Khan M; Galan SRG; Lercher L; Schombs MW; Gerstberger S; Palm-Espling ME; Baldwin AJ; Kessler BM; Claridge TDW; Mohammed S; Davis BG Posttranslational mutagenesis: A chemical strategy for exploring protein side-chain diversity. Science 2016, 354, aag1465. [DOI] [PubMed] [Google Scholar]
- (72). Yang A; Ha S; Ahn J; Kim R; Kim S; Lee Y; Kim J; Söll D; Lee H-Y; Park H-S A chemical biology route to site-specific authentic protein modifications. Science 2016, 354, 623–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73). Chalker JM; Wood CSC; Davis BG A convenient catalyst for aqueous and protein Suzuki-Miyaura cross-coupling. J. Am. Chem. Soc 2009, 131, 16346–16347. [DOI] [PubMed] [Google Scholar]
- (74). Willwacher J; Raj R; Mohammed S; Davis BG Selective metal-site-guided arylation of proteins. J. Am. Chem. Soc 2016, 138, 8678–8681. [DOI] [PubMed] [Google Scholar]
- (75). Li N; Ramil CP; Lim RKV; Lin Q A genetically encoded alkyne directs palladium-mediated protein labeling on live mammalian cell surface. ACS Chem. Biol 2015, 10, 379–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76). Vinogradova EV; Zhang C; Spokoyny AM; Pentelute BL; Buchwald SL Organometallic palladium reagents for cysteine bioconjugation. Nature 2015, 526, 687–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77). Poloukhtine AA; Mbua NE; Wolfert MA; Boons G-J; Popik VV Selective labeling of living cells by a photo-triggered click reaction. J. Am. Chem. Soc 2009, 131, 15769–15776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78). Lopchuk JM; Fjelbye K; Kawamata Y; Malins LR; Pan C-M; Gianatassio R; Wang J; Prieto L; Bradow J; Brandt TA; Collins MR; Elleraas J; Ewanicki J; Farrell W; Fadeyi OO; Gallego GM; Mousseau JJ; Oliver R; Sach NW; Smith JK; Spangler JE; Zhu H; Zhu J; Baran PS Strain-Release Heteroatom Functionalization: Development, Scope, and Stereospecificity. J. Am. Chem. Soc 2017, 139, 3209–3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79). Thirumurugan P; Matosiuk D; Jozwiak K Click chemistry for drug development and diverse chemical−biology applications. Chem. Rev 2013, 113, 4905–4979. [DOI] [PubMed] [Google Scholar]
- (80). Sharma A; Hartwig JF Metal-catalysed azidation of tertiary C–H bonds suitable for late-stage functionalization. Nature 2015, 517, 600–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81). Karimov RR; Sharma A; Hartwig JF Late stage azidation of complex molecules. ACS Cent. Sci 2016, 2, 715–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82). Beck A; Goetsch L; Dumontet C; Corvaïa N Strategies and challenges for the next generation of antibody-drug conjugates. Nat. Rev. Drug. Discov 2017, 16, 315–337. [DOI] [PubMed] [Google Scholar]
- (83). Lin S; Yang X; Jia S; Weeks AM; Hornsby M; Lee PS; Nichiporuk RV; Lavarone AT; Wells JA; Toste FD; Chang CJ Redox-based reagents for chemoselective methionine bioconjugation. Science 2017, 355, 597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84). Zhang C; Welborn M; Zhu T; Yang NJ; Santos MS; Voorhis TV; Pentelute BL π-Clamp-mediated cysteine conjugation. Nat. Chem 2016, 8, 120–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85). Kölmel DK; Kool ET Oximes and hydrazones in bioconjugation: Mechanism and catalysis. Chem. Rev 2017, 117, 10358–10376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86). Schmidt P; Stress C; Gillingham D Boronic acids facilitate rapid oxime condensations at neutral pH. Chem. Sci 2015, 6, 3329–3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87). Bode JW Chemical protein synthesis with the α‐ketoacid−hydroxylamine ligation. Acc. Chem. Res 2017, 50, 2104–2115. [DOI] [PubMed] [Google Scholar]