Abstract
Since 1971, the Protein Data Bank (PDB) archive has served as the single, global repository for open access to atomic-level data for biological macromolecules. The archive currently holds >140,000 structures (>1 billion atoms). These structures are the molecules of life found in all organisms. Knowing the 3D structure of a biological macromolecule is essential for understanding the molecule’s function, providing insights in health and disease, food and energy production, and other topics of concern to prosperity and sustainability. PDB data are freely and publicly available, without restrictions on usage. Through bibliometric and usage studies, we sought to determine the impact of the PDB across disciplines and demographics. Our analysis shows that even though research areas such as molecular biology and biochemistry account for the most usage, other fields are increasingly using PDB resources. PDB usage is seen across 150 disciplines in applied sciences, humanities, and social sciences. Data are also re-used and integrated with >400 resources. Our study identifies trends in PDB usage and documents its utility across research disciplines.
Subject terms: Literature mining, Databases, Structural biology, Publishing
Introduction
The Protein Data Bank (PDB) is the single, global repository for structural data of the molecules of life. Understanding the 3D structure of a biological macromolecule is essential for understanding critical areas of science, including fundamental biology, medicine, energy, drug discovery, and education. As an established archive that continues to grow in size, the PDB provides an opportunity to study the impact of PDB data and resources in these different areas.
The PDB was established in 1971 to archive experimental data contributed by the new discipline of macromolecular crystallography, which was beginning to reveal three-dimensional (3D), atomic-level structures of biological macromolecules, including proteins, DNA, and RNA1,2. Today, the PDB also archives atomic coordinates and related experimental data from nuclear magnetic resonance spectroscopy and electron microscopy studies. Current archival holdings exceed 140,000 structures.
Since 2003, the Worldwide PDB (wwPDB) organization has managed the PDB archive and ensured that PDB data are freely and publicly available to Data Consumers around the globe3,4. Locally-funded, regional PDB Data Centers in the US5, Europe6, and Japan7 safeguard and disseminate PDB structures using a common data dictionary8 and a unified global system for data deposition-validation-biocuration9.
The Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB)5,10 has served as the US PDB Data Center since 1999. In 2017, RCSB PDB processed >6,200 new atomic level biomolecular structures plus experimental data and metadata contributed by PDB Data Depositors in the Americas and Oceania. wwPDB partners are together responsible for processing incoming data from elsewhere in the world. All data are available from the PDB archive; 11,124 new structures were released in 2017. The PDB archive and the RCSB PDB website (RCSB.org) are heavily used. During 2017, >680 million structure data files were downloaded from the archive by PDB Data Consumers worldwide. More than 1 million users from around the world benefited from open access to PDB data integrated with ~40 external resources at RCSB.org, providing rich structural views of fundamental biology, biomedicine, and energy sciences.
The most cited RCSB PDB publication, “The Protein Data Bank” by Berman et al.5, appeared in Nucleic Acids Research (NAR) in 2000. This inaugural article described the mission and vision of the resource and its operations, and provided data deposition and download instructions11. Berman et al. (2000) is routinely used to cite both the PDB data archive and the many services that the RCSB PDB provides to PDB Data Depositors submitting data (currently numbering >30,000) and PDB Data Consumers downloading data from the archive or using RCSB.org (currently >1 million). It has been heavily cited as noted by different reviews. A 2014 analysis12 ranked the inaugural article 92nd among the top 100 most-cited research publications of all time and a 2017 study13 placed it 5th among papers published since 2000.
By the end of 2016, Berman et al. (2000) had been cited by nearly 16,000 articles and other documents as recorded in the Web of Science database14. In 2017, RCSB PDB contracted with Clarivate Analytics to conduct an initial bibliometric analysis of citations to Berman et al. (2000) using in-house tools and proprietary data made available on a fee-for-service basis. Their report was made public by the RCSB PDB during the same year13.
The Clarivate analysis was per force limited to the activities of researchers who were referencing the Nucleic Acids Research publication in order to cite the RCSB PDB, specific PDB structures, and/or the PDB archive as a whole. Complicating matters further, many PDB Data Consumers do not cite Berman et al. (2000), electing instead to mention the RCSB PDB website (i.e., RCSB.org) or refer to individual PDB structures directly using a unique 4-character identifier (e.g., PDB ID 1vol)15. Some researchers have even grown so accustomed to using the RCSB PDB and the PDB archive that it is never mentioned in their publications, although close reading of their publications reveals reliance on the resource.
Herein, we report the results of systematic analyses of the types of published research citing the Berman et al. (2000) reference as well as PDB archival data reuse by >400 other online biodata resources. In addition, we relate these findings to outcomes from an online RCSB PDB user survey and the Clarivate study to provide a comprehensive picture of the breadth and depth of the impact on the scientific community of the RCSB PDB and the PDB archive.
Results
Exploration of Research Category
Between 2000 and 2016, the inaugural RCSB PDB publication, Berman et al. (2000), has garnered 15,711 citations in the Web of Science (Data set, Data Citation 1)14. Citing documents include articles, proceedings papers, reviews, book chapters, editorial material, and software reviews. The 2017 citation analysis13 by Clarivate Analytics focused on the >14,000 journal articles citing “The Protein Data Bank.” Web of Science assigns journals exclusively to 1 of 22 Essential Science Indicators (ESI) Subject Categories in science and social sciences. Citing articles were found in all ESI Subject Categories except Space Science; in particular, ESI categories Biology & Biochemistry, Chemistry, Molecular Biology & Genetics, and Computer Science contained the most citing articles. Clarivate determined that these citing articles were generally considered to be “high-quality” across fields. As citation rates vary between research areas and journal impact factors, Clarivate considers a variety of factors (e.g., world average citations per publication for the year, journal category, document type) to “normalize” citations in order to compare impact across disciplines. Using their Category-normalization, the citation impact of articles was at least the world-average except in the category of Psychiatry/Psychology. The greatest impact was seen in Computer Science, with a citation-based impact that was twice the world average.
In 2017, an online user survey was held to collect information about the research interests of RCSB PDB users (2017 RCSB.org Survey Data, Data Citation 1). 86% of respondents reported completing a Bachelor’s degree or higher. Subject area interests for all respondents mirrored the Web of Science analysis by Basner13. Most users predominantly identified their research interests as Biology & Biochemistry (72%), Molecular Biology & Genetics (31%), Chemistry (23%), Microbiology (8%), Computer Science (8%), and Immunology (6%). This finding strengthens the argument that the PDB is primarily used by individuals within the biological sciences and those working in fields with interdisciplinary applications. It is critical to note that 59% of survey respondents reported never having deposited a structure in the PDB, showing that the PDB plays a vital role in education and/or provides utility for researchers who are consumers of 3D structural information on biomolecules. In fact, 62% of all respondents report using the PDB-101 educational series Molecule of the Month.
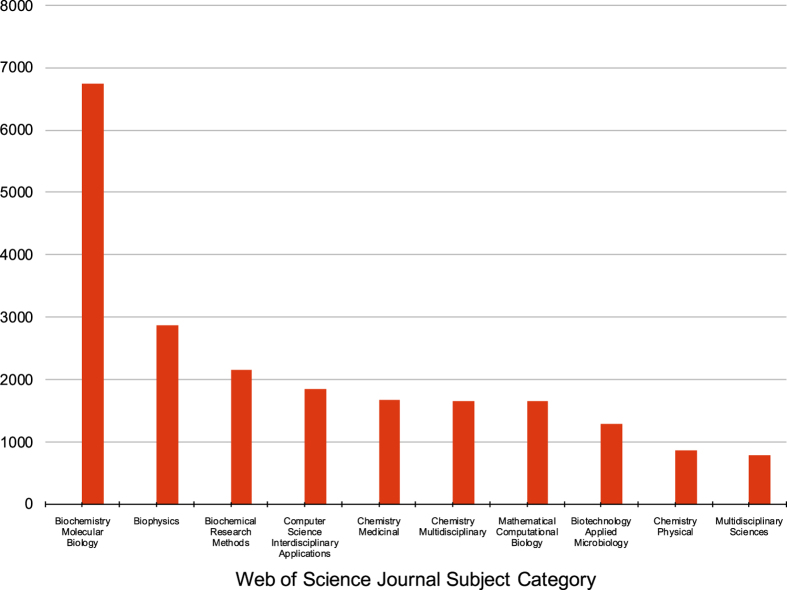
Web of Science also utilizes more granular Journal Subject Categories. To obtain a more detailed understanding of PDB usage, we used the online Web of Science interface14 to determine that the citing articles were themselves published in journals across 154 Journal Subject Categories (out of a possible 252). The top ten Journal Subject Categories (Fig. 1) revealed the expected large number of assignments associated with experimental structure determination: Biochemistry Molecular Biology (22%), Biophysics (9%), Biochemical Research Methods (7%), and Biotechnology Applied Microbiology (4%). Chemistry Medicinal (5%) is almost certainly prominent because of the importance of structural data in drug discovery. Chemistry Physical (3%) is also most likely common because it utilizes structural data to interpret thermodynamics and kinetics of macromolecules. However, the presence of Computer Science Interdisciplinary Applications (6%) and Mathematical Computational Biology (5%) among the top 10 underscore the widespread use of PDB data in conjunction with computational techniques.
Figure 1. Number of publications for the top-assigned Web of Science Journal Subject Category for all documents (2000–2016) citing the inaugural Berman et al. (2000) reference.
Biochemistry Molecular Biology is the largest category (6,735 publications), followed by Biophysics (2,872), Biochemical Research Methods (2,161), Computer Science Interdisciplinary Applications (1,852), Chemistry Medicinal (1,666), Chemistry Multidisciplinary (1,660), Mathematical Computational Biology (1,656), Biotechnology Applied Microbiology (1,297), Chemistry Physical (871), and Multidisciplinary Sciences (789).
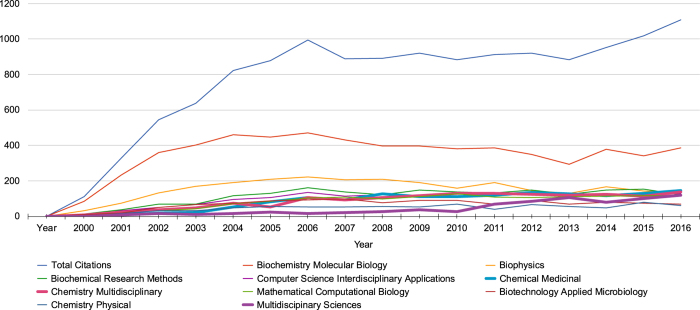
Since 2004, the annual number of citations of Berman et al. (2000) is consistently high, with an average of ~940 articles per year and no evidence that overall usage is on the verge of decline. Fig. 2 charts the total number of citing articles per year, with the top Journal Categories mapped underneath. Throughout 2004–2016, the majority of articles fall within the Biochemistry Molecular Biology category, while that number appears to be decreasing slowly in favor of other category topics.
Figure 2. Number of articles citing the inaugural Berman et al. (2000) reference each year.
Total number of articles is shown in blue; the top Journal Subject Categories are below. Growth in the areas of Chemical Medicinal, Chemistry Multidisciplinary, and Multidisciplinary Sciences is increasing (shown in bold); the number of articles in the areas of Biochemistry Molecular Biology, Biophysics, and Biotechnology Applied Microbiology do not have statistically significant growth.
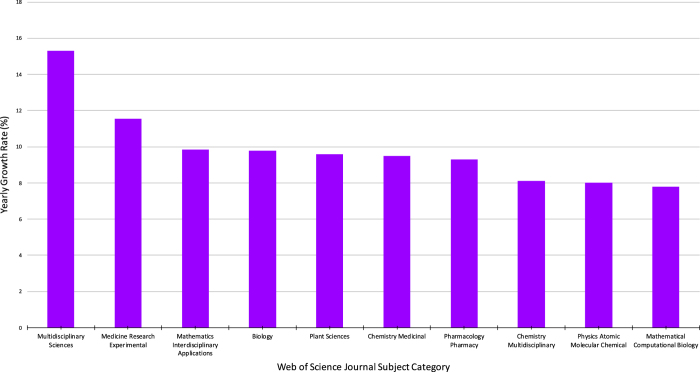
Analyses of the yearly growth rate of each Journal Subject Category provides information relating to the fastest growing research areas throughout 2000–2016. This perspective is important in understanding the future direction of PDB usage. Overall, the categories with the most citations were among categories with the smallest yearly growth rates (Biochemistry Molecular Biology, 1.1% and Biophysics, 1.5%). Other categories, with fewer total citations, exhibit much greater yearly growth rates (Multidisciplinary Sciences, 15.3% and Medicine Research Experimental, 11.6%). This trend suggests that the way the PDB data are being used is changing. It is remarkable that the categories Chemistry Medicinal, Chemistry Multidisciplinary, and Mathematical Computation Biology appear in the top ten Journal Subject Category by both count (Fig. 1) and growth rate (Fig. 3).
Figure 3. The top Web of Science Journal Subject Categories demonstrating the greatest yearly growth in all documents citing the Berman et al. (2000) reference (2000–2016).
The study compares 34 categories with at least 100 citations. Growth rate was calculated as the slope coefficient of the linear regression model between the number of citations in the category and year of publication, starting with the first year an article appeared, and expressed as a normalized percentage of the average yearly publication of that category. Multidisciplinary Sciences has grown at the greatest rate (15.3%), followed by Medicine Research Experimental (11.6%), Mathematics Interdisciplinary Applications (9.8%), Biology (9.8%), Plant Sciences (9.6%), Chemistry Medicinal (9.5%), Pharmacology Pharmacy (9.3%), Chemistry Multidisciplinary (8.1%), Physics Atomic Molecular Chemical (8.0%), and Mathematical Computational Biology (7.8%).
Articles citing Berman et al. (2000) are published in journals categorized in >100 additional subject categories, indicating the broad range of PDB data utilization. Examples drawn from categories that are not directly related to structural biology demonstrate both the unique applications and the breadth of influence of the PDB archive. Industrial-focused categories include Polymer Science, Agriculture Dairy Animal Science, Energy Fuels, and Forestry. An example from Agronomy studied flavonoid antifungal activity against Aspergillus ochraceus through in silico analysis of small molecule interactions with kinase structures in the PDB16. In Energy Fuels, one study refers to modeling protein structures using PDB data to design more efficient biocatalysts for industrial applications17. Other Medical categories that reference PDB include Oncology, Infectious Diseases, Nutrition Dietetics, and Veterinary Sciences. An example from Health Care Sciences Services calls for the establishment of a central repository of raw data for the human brain project, citing the PDB as an example of “highly successful bioinformatics efforts”18. Within the tail of the distribution of research areas citing, we found publications in unexpected categories, such as Optics, Telecommunications, and Social Issues. For example, a Management study exhibits the importance of management science tools to address issues in bioinformatics and cites the PDB as an important tool in disseminating macromolecular structural data19. Interestingly, publications were also noted in entirely unexpected disciplines and fields, such as Art20–24, Logic25–27, and Business Finance28.
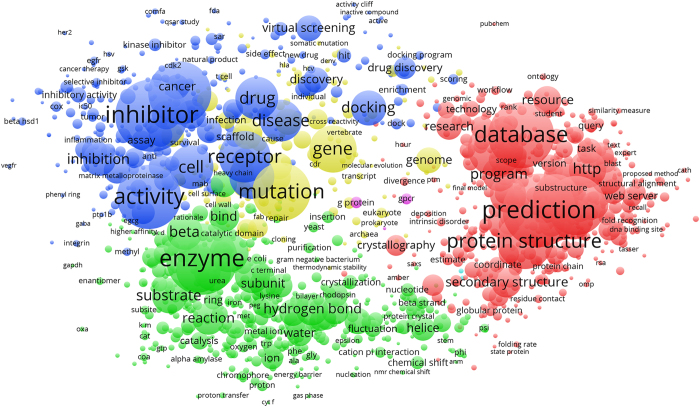
Creation of a co-occurrence network map reiterates the breadth of journal subject categories. The titles and abstracts of the ~15,000 articles published between 2000–2016 citing Berman et al. (2000) were used to generate a keyword map based on frequency of occurrence (Fig. 4; Network Visualization Map Data, Data Citation 1). Terms were concentrated in four main regions. On the right (red) are keywords linked to computational usage, including “prediction” (3,611 total occurrences), “database” (3,029), and “protein structure” (2,339). The bottom region (green) describes the 3D PDB structures themselves: “substrate” (1,127), “hydrogen bond” (978), “reaction” (952), and “subunit” (770). A dominating term in this region is “enzyme” (3,440), which falls between keywords linked to structure (green) and function (blue). Other terms in this region of the keyword plot include “inhibitor” (3,513), “activity” (2,901), “receptor” (2,046), and “drug” (1,536). The center of the map (yellow) reflects the impact of PDB data on overall understanding of genetics and genomics, with the terms “mutation” (2,504) and “gene” (1,780).
Figure 4. Network visualization of term occurrences extracted from abstracts and titles of 2000-2016 publications citing the inaugural Berman et al. (2000) reference.
Figure created using VOSviewer46. A threshold cutoff of 30 as number of term co-occurrence was used. The location of citation keywords is based on their overall position within the network; keywords located in more common regions of the map have higher network connectivity, i.e., they are more interconnected with surrounding keywords. Darker colors and font size represent keywords that appear more frequently among citations. Keywords are clustered in four main regions: red corresponds to keywords representing “computational” use of the data; green corresponds to 3D-structure and mechanism of action; blue corresponds to function; and yellow corresponds to keywords related to genetics and genomics.
PDB Usage in Other Databases
An important role of the PDB archive is to make structural data freely available for reuse by other resources29. The Nucleic Acids Research (NAR) Online Molecular Biology Database Collection30, a compilation of biological databases updated annually, was assessed to determine PDB usage by other resources. As of January 2018, the collection has 1,737 active databases organized into 15 NAR categories31. A review of this collection shows that 429 active bioinformatics resources utilize PDB data across 14 of the 15 categories (Table 1 (available online only); NAR Molecular Biology Database Collection Data, Data Citation 1). These databases encompass 44 subcategories, highlighting the variety of applications of PDB data. These databases add value to PDB data and function as resources for protein-protein interactions (e.g., AffinDB)32, enzymes (e.g., MultiTaskDB)33, metabolic pathways (e.g., MMCD)34, signaling pathways (e.g., REPAIRtoire)35, mapping cancer mutations in proteins (e.g., Cancer3D)36, antibody structure (e.g., SAbDab)37, viral capsid structures (e.g., VIPERdb)38, yeast genomics (e.g., SGD)39, drug-binding sites (e.g., sc-PDB)40, and more.
Table 1. Distribution of the 429 active resources in the NAR Molecular Biology Database Collection that utilize PDB archive data across major categories (bold) and subcategory (italics), and corresponding resources in the “golden set”41 of the NAR Molecular Biology Database Collection.
| NAR Category and Subcategory | Number that Utilize PDB Data | NAR Golden Set |
|---|---|---|
| wwPDB partners RCSB PDB, PDBe, and PDBj are included in the subcategory Protein Structure. | ||
| Structure Databases | 107 | |
| Protein Structure | 81 | MMDB47, PDBe,6 PDBsum48, RCSB PDB,5,10 SCOP249, SUPERFAMILY50, SWISS-MODEL Repository51 |
| Small Molecules | 11 | ChEBI52, PubChem53 |
| Nucleic Acid Structure | 8 | |
| Carbohydrates | 7 | |
| Protein Sequence Databases | 95 | |
| Databases of Individual Protein Families | 42 | GPCRDB54, MEROPS55 |
| Protein Domain Databases; Protein Classification | 14 | CDD56, CATH57, Pfam58, SMART59 |
| Protein Sequence Motifs and Active Sites | 15 | ELM60, PROSITE61 |
| Protein Properties | 10 | dbPTM62 |
| Protein Localization and Targeting | 8 | |
| General Sequence Databases | 6 | PIR63, TCDB64, UniProt65 |
| Metabolic and Signaling Pathways | 58 | |
| Protein-Protein Interactions | 30 | STITCH66, STRING67 |
| Enzymes and Enzyme Nomenclature | 14 | CAZy68 |
| Metabolic Pathways | 10 | BioCyc69, HMDB70, MODOMICS71, Reactome72 |
| Signaling Pathways | 4 | |
| Genomics Databases (Non-Vertebrate) | 47 | |
| Prokaryotic Genome Databases | 11 | EcoCyc73 |
| Viral Genome Databases | 9 | |
| Unicellular Eukaryotes Genome Databases | 8 | |
| Invertebrate Genome Databases | 7 | VectorBase74 |
| Fungal Genome Databases | 6 | CGD75, SGD76 |
| Genome Annotation Terms, Ontologies, and Nomenclature | 4 | Genenames77 |
| General Genomics Databases | 2 | KEGG78, MetaCyc69 |
| Human Genes and Diseases | 23 | |
| General Polymorphism Databases | 5 | |
| General Human Genetics Databases | 3 | |
| Gene-, System- or Disease-Specific Databases | 7 | |
| Cancer Gene Databases | 7 | CanSAR79 |
| Human Genes and Diseases | 1 | COSMIC80 |
| Other Molecular Biology Databases | 25 | |
| Drugs and Drug Design | 21 | ChEMBL81 |
| No Subcategory | 3 | |
| Molecular Probes and Primers | 1 | |
| Nucleotide Sequence Databases | 20 | |
| Transcriptional Regulator Sites and Transcription Factors | 11 | JASPAR82 |
| Gene Structure, Introns and Exons, Splice Sites | 5 | |
| International Nucleotide Sequence Database Collaboration | 3 | DDBJ83, ENA84 |
| Coding and Non-Coding DNA | 1 | |
| Immunological Databases | 12 | IEDB85, IMGT86 |
| RNA Sequence Databases | 12 | NONCODE87, Rfam88 |
| Human and Other Vertebrate Genomes | 11 | |
| Human ORFs | 5 | FANTOM89, neXtProt90 |
| Model Organisms, Comparative Genomics | 4 | Ensembl91, Mouse Genome Database92, Rat Genome Database93 |
| Human Genome Databases, Maps and Viewers | 2 | UCSC Genome Browser94 |
| Plant Databases | 7 | |
| General Plant Databases | 5 | |
| Arabidopsis thaliana | 1 | |
| Other Plants | 1 | |
| Proteomics Resources | 8 | dbPTM62 |
| Microarray Data and Other Gene Expression Databases | 2 | ArrayExpress95 |
| Organelle Databases | 2 |
From the 1,737 resources in the Online Molecular Biology Database Collection, the leading 108 databases used most intensively across the scientific community have been identified by NAR as the “golden set” of successful resources41. Our review revealed that 52 of these elite databases incorporate PDB data, demonstrating the importance of the PDB archive in resources widely used in the biological research community.
Discussion
While the PDB was initially established as an archive to serve structural biologists by hosting and providing access to their experimental data, bibliometric analyses of the heavily-cited Berman et al. (2000) reference5 demonstrate that the PDB has grown to serve a much more diverse community of users. While expected Web of Science journal categories, such as Biochemistry Molecular Biology and Biophysics, continue to generate the most citing publications per year (Fig. 1), disciplines such as Mathematical Computational Biology, Chemistry Medicinal, and Computer Science Interdisciplinary Applications are generating citations at much greater growth rates (Fig. 2). Moreover, keywords associated with structure determination and description were prevalent among citing publications in the early 2000s, but keywords pertinent to drug discovery, then genetics and genomics, and most recently protein structure prediction have come to the fore. Usage across disciplines is unmistakably confirmed by review of PDB usage within the NAR Online Molecular Biology Database Collection41.
Widespread usage of PDB data is also in line with survey results, which demonstrated that only a minority of RCSB PDB users have ever contributed a structure to the archive. The combination of these analyses indicates widespread use of the RCSB PDB and PDB archive in education and research, going well beyond studies of individual structures.
Methods
Publication data for articles and documents from 2000–2016 that cited the Berman et al. (2000) reference5 were exported from the Web of Science (Data set, Data Citation 1)14. Each publication can be assigned to more than one category and/or more than one country. Exported data were analyzed using Microsoft Excel in Fall 2017.
The data examined are limited to those citing this inaugural reference, and future work could show either expanded usage or usage in greater detail. Going beyond the scope of this work, additional analyses could be performed to include publication data for articles citing the wwPDB3, wwPDB data centers PDBe6 and PDBj7, wwPDB partner BioMagResBank42, as well as the NAR articles describing the wwPDB partners that are published regularly (for examples see recent articles for RCSB PDB10, PDBe43, PDBj7). Other analyses of PDB usage have examined citations and patents44 and usage of URLs in the literature15. The data set under consideration was limited to Web of Science data for articles citing the inaugural RCSB PDB publication to be consistent with the Clarivate Analytics report13.
The User Survey was hosted at SurveyMonkey during the month of October 2017, and promoted on RCSB.org, PDB101.rcsb.org, message boards, and social media. A total of 681 responses were received from high school students and teachers, undergraduates, graduate students, postdoctoral scholars, and faculty/staff (2017 RCSB.org Survey Data, Data Citation 1).
A co-occurrence network map of keywords from the same set of publication data was created using the VOSviewer server (Version 1.6.5; www.vosviewer.com)45,46. Approximately 227,000 keywords were extracted from citation titles and abstracts (Network Visualization Map Data, Data Citation 1). A network was computed for a total of 2,460 terms selected by the full-counting method and relevance scoring as implemented within VOSviewer. For analysis, we reviewed co-occurrence network maps for thresholds between 5 and 40. A total of 60% total number of terms with a default cutoff of 30 as the number of term co-occurrence is shown.
The online descriptions and related publication abstracts for the resources listed in the NAR Online Molecular Biology Database Collection42 as of January 2018 were text mined for the words “protein data bank,” “PDB,” and other terms related to protein structure. Only the 1688 databases with available abstracts were mined. Results were manually reviewed to confirm usage of PDB data and website availability (NAR Molecular Biology Database Collection Data, Data Citation 1).
Code and data availability
The following have been uploaded to FigShare: bibliographic data for articles citing the inaugural Berman et al. (2000) reference (with permission from Clarivate Analytics); anonymized user survey data; VOSviewer map file; and the current list of NAR Online Molecular Biology Database Collection resources that utilize PDB data.
The scripts used to obtain the initial data for the analysis of the NAR Online Molecular Biology Database Collection are available from GitHub: https://github.com/rcsb/review-NAR-Databases.
Additional information
How to cite this article: Markosian, C. et al. Analysis of impact metrics for the Protein Data Bank. Sci. Data. 5:180212 doi: 10.1038/sdata.2018.212 (2018).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Acknowledgments
The RCSB PDB is jointly funded by the National Science Foundation, the National Institutes of Health, and the Department of Energy (NSF-DBI 1338415). We gratefully acknowledge contributions from all members of the Research Collaboratory for Structural Bioinformatics PDB, our Worldwide Protein Data Bank partners, and the Rutgers Office of Research Analytics. We thank Laura Bowering Mullen and Laura Palumbo (Rutgers University), Anthony Molisani (Clarivate Analytics), and Ludo Waltman (Leiden University) for guidance.
Footnotes
C.M. declares no competing interests. L.D., M.S., C.S., S.K.B., and C.Z. receive research support from a grant to the RCSB PDB (DBI-1338415) from the National Science Foundation, the National Institutes of Health, and the US Department of Energy.
Data Citations
- Markosian C., et al. . 2018. Figshare. https://doi.org/10.6084/m9.figshare.c.4064561
References
- Protein Data Bank. Crystallography: Protein Data Bank. Nature New Biology 233, 223–223 ; DOI: 10.1038/newbio233223b0 (1971).20480989 [DOI] [Google Scholar]
- Berman H. The Protein Data Bank: a historical perspective. Acta Crystallogr A: Foundations of Crystallography 64, 88–95 ; DOI: 10.1107/S0108767307035623 (2008). [DOI] [PubMed] [Google Scholar]
- Berman H. M., Henrick K. & Nakamura H. Announcing the worldwide Protein Data Bank. Nat Struct Biol 10, 980 ; DOI: 10.1038/nsb1203-980 (2003). [DOI] [PubMed] [Google Scholar]
- Burley S. K. et al. in Methods In Molecular Biology: Protein Crystallography Methods and Protocols, Daughter Z., Wlodawer A. & Jaskolski M. eds 627–641 (Springer, 2017). [Google Scholar]
- Berman H. M. et al. The Protein Data Bank. Nucleic Acids Res 28, 235–242, ; DOI: 10.1093/nar/28.1.235 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velankar S. et al. PDBe: improved accessibility of macromolecular structure data from PDB and EMDB. Nucleic Acids Res 44, D385–D395 ; DOI: 10.1093/nar/gkv1047 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinjo A. R. et al. Protein Data Bank Japan (PDBj): updated user interfaces, resource description framework, analysis tools for large structures. Nucleic Acids Res 45, D282–D288, ; DOI: 10.1093/nar/gkw962 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westbrook J., Henrick K., Ulrich E. L. & Berman H. M. In International Tables for Crystallography Vol. G. Definition and exchange of crystallographic data Hall S. R. & McMahon B. 195–198 (Springer, 2005). [Google Scholar]
- Young, J. Y. et al. OneDep: Unified wwPDB System for Deposition, Biocuration, and Validation of Macromolecular Structures in the PDB Archive. Structure 25, 536–545, ; DOI: 10.1016/j.str.2017.01.004 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose P. W. et al. The RCSB protein data bank: integrative view of protein, gene and 3D structural information. Nucleic Acids Res 45, D271–D281 ; DOI: 10.1093/nar/gkw1000 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H. M., Burley S. K., Kleywegt G. J., Nakamura H. & Markley J. L. Response to On prompt update of literature references in the Protein Data Bank. Acta Crystallographica. Series D 70, 2780 ; DOI: 10.1107/S1399004714020513 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Noorden R., Maher B. & Nuzzo R. The top 100 papers. Nature 514, 550–553 ; DOI: 10.1038/514550a (2014). [DOI] [PubMed] [Google Scholar]
- Basner J. Impact Analysis of “Berman HM et al. (2000), The Protein Data Bank” ; DOI: 10.2210/rcsb_pdb/pdb-cit-anal-2017 (Clarivate Analytics 2017). [DOI]
- Web of Science (Clarivate Analytics, 2018).
- Huang Y. H., Rose P. W. & Hsu C. N. Citing a Data Repository: A Case Study of the Protein Data Bank. PLoS One 10, e0136631, ; DOI: 10.1371/journal.pone.0136631 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Júnior H. M. S. et al. Antifungal activity of flavonoids from Heteropterys byrsonimifolia and a commercial source against Aspergillus ochraceus: In silico interactions of these compounds with a protein kinase. Crop Protection 62, 107–114 (2014). [Google Scholar]
- Madhavan A., Sindhu R., Binod P., Sukumaran R. K. & Pandey A. Strategies for design of improved biocatalysts for industrial applications. Bioresour. Technol 245(Pt B), 1304–1313, ; DOI: 10.1016/j.biortech.2017.05.031 (2017). [DOI] [PubMed] [Google Scholar]
- Brinkley J. F. & Rosse C. Imaging and the Human Brain Project: a review. Methods Inf. Med. 41, 245–260 (2002). [PubMed] [Google Scholar]
- Abbas A. E. Bioinformatics and management science: Some common tools and techniques. Operations Research 52, 165–190 (2004). [Google Scholar]
- Balaji S. Biomorphic Presentation of Proteins: Artistic Science or Scientific Art? Leonardo 46, 226–231 (2013). [Google Scholar]
- Meyer E. F. Digital Design of Molecular Sculptures and Abstractions. Leonardo 44, 22-+ (2011). [Google Scholar]
- Shi X. J., Cai Y. Y. & Chan C. W. Electronic music for bio-molecules using short music phrases (Protein music). Leonardo 40, 137–141 (2007). [Google Scholar]
- Cai Y. Y. et al. Proteins, Immersive Games and Music. Leonardo 39, 135–137 (2006). [Google Scholar]
- Voss-Andreae J. Protein sculptures: Life’s building blocks inspire art. Leonardo 38, 41–45 (2005). [Google Scholar]
- Parpinelli R. S., Benitiez C. M., Cordeiro J. & Lopes H. S. Performance Analysis of Swarm Intelligence Algorithms for the 3D-AB off-lattice Protein Folding Problem. Multiple-Valued Logic and Soft Computing 22, 267–286 (2014). [Google Scholar]
- Alphonse E., Girschick T., Buchwald F., Kramer S. In 20th International Conference on Inductive Logic Programming (eds Frasconi P. & Lisi F. A.) 14–21 (Florence: Italy, 2011). [Google Scholar]
- Abraham M. R. & Wolfson H. J. Inexact Graph Matching by “Geodesic Hashing” for the Alignment of Pseudoknoted RNA Secondary Structures. In Proceedings of the Prague Stringology Conference 45–58 (Prague Stringology Club, 2011). [Google Scholar]
- Shi Y., Chen R., Wan J. & Zhang X. A Novel MCQP Approach for Predicting the Distance Range between Interface Residues in Antibody-Antigen Complex. In 20th International Conference on Multiple Criteria Decision Making 35, 643–648 (Springer, 2009). [Google Scholar]
- Wilkinson M. D. et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 3, 160018, ; DOI: 10.1038/sdata.2016.18 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxevanis A. D. The molecular biology database collection: an online compilation of relevant database resources. Nucleic Acids Res 28, 1–7, ; DOI: 10.1093/nar/28.1.1 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigden D. J. & Fernandez X. M. The 2018 Nucleic Acids Research database issue and the online molecular biology database collection. Nucleic Acids Res 46, D1–D7, ; DOI: 10.1093/nar/gkx1235 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block P., Sotriffer C. A., Dramburg I. & Klebe G. AffinDB: a freely accessible database of affinities for protein-ligand complexes from the PDB. Nucleic Acids Res 34, D522–D526, ; DOI: 10.1093/nar/gkj039 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez S. et al. MultitaskProtDB: a database of multitasking proteins. Nucleic Acids Res 42, D517–D520, ; DOI: 10.1093/nar/gkt1153 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Q. et al. Metabolite identification via the Madison Metabolomics Consortium Database. Nat Biotechnol 26, 162–164, ; DOI: 10.1038/nbt0208-162 (2008). [DOI] [PubMed] [Google Scholar]
- Milanowska K. et al. REPAIRtoire--a database of DNA repair pathways. Nucleic Acids Res 39, D788–D792, ; DOI: 10.1093/nar/gkq1087 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porta-Pardo E., Hrabe T. & Godzik A. Cancer3D: understanding cancer mutations through protein structures. Nucleic Acids Res 43, D968–D973, ; DOI: 10.1093/nar/gku1140 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar J. et al. SAbDab: the structural antibody database. Nucleic Acids Res 42, D1140–D1146, ; DOI: 10.1093/nar/gkt1043 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd C. M. et al. VIPERdb: a relational database for structural virology. Nucleic Acids Res 34, D386–D389 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry J. M. et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res 40, D700–D705, ; DOI: 10.1093/nar/gkr1029 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellenberger E. et al. sc-PDB: an annotated database of druggable binding sites from the Protein Data Bank. J Chem Inf Model 46, 717–727, ; DOI: 10.1021/ci050372x (2006). [DOI] [PubMed] [Google Scholar]
- Galperin M. Y., Fernández-Suárez X. M. & Rigden D. J. The 24th annual Nucleic Acids Research database issue: a look back and upcoming changes. Nucleic Acids Research 45, D1–D11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich E. L. et al. BioMagResBank. Nucleic Acids Res 36, D402–D408, ; DOI: 10.1093/nar/gkm957 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir S. et al. PDBe: towards reusable data delivery infrastructure at protein data bank in Europe. Nucleic Acids Res 46, D486–D492, ; DOI: 10.1093/nar/gkx1070 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousfield D. et al. Patterns of database citation in articles and patents indicate long-term scientific and industry value of biological data resources. F1000Res 5, ; DOI: 10.12688/f1000research.7911.1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eck N. J., Waltman L., Dekker R. & Van den Berg A comparison of two techniques for bibliometric mapping: Multidimensional scaling and VOS. J. Assoc. Inf. Sci. Technol 61, 2405–2416, ; DOI: 10.1002/asi.21421 (2010). [DOI] [Google Scholar]
- van Eck N. J. & Waltman L. Software survey: VOSviewer, a computer program for bibliometric mapping. Scientometrics 84, 523–538, ; DOI: 10.1007/s11192-009-0146-3 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madej T. et al. MMDB and VAST+: tracking structural similarities between macromolecular complexes. Nucleic Acids Res 42, D297–D303, ; DOI: 10.1093/nar/gkt1208 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Beer T. A., Berka K., Thornton J. M. & Laskowski R. A. PDBsum additions. Nucleic Acids Res 42, D292–D296, ; DOI: 10.1093/nar/gkt940 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva A., Howorth D., Chothia C., Kulesha E. & Murzin A. G. SCOP2 prototype: a new approach to protein structure mining. Nucleic Acids Res 42, D310–D314, ; DOI: 10.1093/nar/gkt1242 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oates M. E. et al. The SUPERFAMILY 1.75 database in 2014: a doubling of data. Nucleic Acids Res 43, D227–D233, ; DOI: 10.1093/nar/gku1041 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienert S. et al. The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res 45, D313–D319 ; DOI: 10.1093/nar/gkw1132 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings J. et al. ChEBI in 2016: Improved services and an expanding collection of metabolites. Nucleic Acids Res 44, D1214–D1219, ; DOI: 10.1093/nar/gkv1031 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. et al. PubChem Substance and Compound databases. Nucleic Acids Res 44, D1202–D1213, ; DOI: 10.1093/nar/gkv951 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isberg V. et al. GPCRdb: an information system for G protein-coupled receptors. Nucleic Acids Res 44, D356–D364, ; DOI: 10.1093/nar/gkv1178 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings N. D., Barrett A. J. & Finn R. Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res 44, D343–D350, ; DOI: 10.1093/nar/gkv1118 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A. et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res 43, D222–D226, ; DOI: 10.1093/nar/gku1221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillitoe I. et al. CATH: comprehensive structural and functional annotations for genome sequences. Nucleic Acids Res 43, D376–D381, ; DOI: 10.1093/nar/gku947 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D. et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44, D279–D285, ; DOI: 10.1093/nar/gkv1344 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Doerks T. & Bork P. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res 43, D257–D260, ; DOI: 10.1093/nar/gku949 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkel H. et al. ELM 2016--data update and new functionality of the eukaryotic linear motif resource. Nucleic Acids Res 44, D294–D300, ; DOI: 10.1093/nar/gkv1291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist C. J. et al. New and continuing developments at PROSITE. Nucleic Acids Res 41, D344–D347, ; DOI: 10.1093/nar/gks1067 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K. Y. et al. dbPTM 2016: 10-year anniversary of a resource for post-translational modification of proteins. Nucleic Acids Res 44, D435–D446, ; DOI: 10.1093/nar/gkv1240 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. H. et al. PIRSF: family classification system at the Protein Information Resource. Nucleic Acids Res 32, D112–D114, ; DOI: 10.1093/nar/gkh097 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier M. H. Jr. et al. The Transporter Classification Database (TCDB): recent advances. Nucleic Acids Res 44, D372–D379, ; DOI: 10.1093/nar/gkv1103 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium. UniProt: the universal protein knowledgebase. Nucleic Acids Res 45, D158–D169, ; DOI: 10.1093/nar/gkw1099 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D. et al. STITCH 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res 44, D380–D384, ; DOI: 10.1093/nar/gkv1277 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D. et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43, D447–452, ; DOI: 10.1093/nar/gku1003 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard V., Golaconda Ramulu H., Drula E., Coutinho P. M. & Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42, D490–D495, ; DOI: 10.1093/nar/gkt1178 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R. et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 44, D471–D480, ; DOI: 10.1093/nar/gkv1164 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D. S. et al. HMDB 3.0--The Human Metabolome Database in 2013. Nucleic Acids Res 41, D801–D807, ; DOI: 10.1093/nar/gks1065 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machnicka M. A. et al. MODOMICS: a database of RNA modification pathways--2013 update. Nucleic Acids Res 41, D262–D267, ; DOI: 10.1093/nar/gks1007 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabregat A. et al. The Reactome pathway Knowledgebase. Nucleic Acids Res 44, D481–D487, ; DOI: 10.1093/nar/gkv1351 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keseler I. M. et al. EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res 41, D605–D612, ; DOI: 10.1093/nar/gks1027 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo-Calderon G. I. et al. VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res 43, D707–D713, ; DOI: 10.1093/nar/gku1117 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binkley J. et al. The Candida Genome Database: the new homology information page highlights protein similarity and phylogeny. Nucleic Acids Res 42, D711–D716, ; DOI: 10.1093/nar/gkt1046 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard T. K. et al. The Saccharomyces Genome Database Variant Viewer. Nucleic Acids Res 44, D698–D702, ; DOI: 10.1093/nar/gkv1250 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray K. A., Yates B., Seal R. L., Wright M. W. & Bruford E. A. Genenames.org: the HGNC resources in 2015. Nucleic Acids Res 43, D1079–D1085, ; DOI: 10.1093/nar/gku1071 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Furumichi M., Tanabe M., Sato Y. & Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res, ; DOI: 10.1093/nar/gkw1092 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tym J. E. et al. canSAR: an updated cancer research and drug discovery knowledgebase. Nucleic Acids Res 44, D938–D943, ; DOI: 10.1093/nar/gkv1030 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes S. A. et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res 43, D805–D811, ; DOI: 10.1093/nar/gku1075 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bento A. P. et al. The ChEMBL bioactivity database: an update. Nucleic Acids Res 42, D1083–D1090, ; DOI: 10.1093/nar/gkt1031 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathelier A. et al. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res 44, D110–D115, ; DOI: 10.1093/nar/gkv1176 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashima J. et al. DNA data bank of Japan (DDBJ) progress report. Nucleic Acids Res 44, D51–D57, ; DOI: 10.1093/nar/gkv1105 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson R. et al. Biocuration of functional annotation at the European nucleotide archive. Nucleic Acids Res 44, D58–D66, ; DOI: 10.1093/nar/gkv1311 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vita R. et al. The immune epitope database (IEDB) 3.0. Nucleic Acids Res 43, D405–D412, ; DOI: 10.1093/nar/gku938 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefranc M. P. et al. IMGT(R), the international ImMunoGeneTics information system(R) 25 years on. Nucleic Acids Res 43, D413–D422, ; DOI: 10.1093/nar/gku1056 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. et al. NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res 44, D203–D208, ; DOI: 10.1093/nar/gkv1252 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki E. P. et al. Rfam 12.0: updates to the RNA families database. Nucleic Acids Res 43, D130–D137, ; DOI: 10.1093/nar/gku1063 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaji H. et al. Update of the FANTOM web resource: from mammalian transcriptional landscape to its dynamic regulation. Nucleic Acids Res 39, D856–D860, ; DOI: 10.1093/nar/gkq1112 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet P. et al. The neXtProt knowledgebase on human proteins: current status. Nucleic Acids Res 43, D764–D770, ; DOI: 10.1093/nar/gku1178 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey P. J. et al. Ensembl Genomes 2016: more genomes, more complexity. Nucleic Acids Res 44, D574–D580, ; DOI: 10.1093/nar/gkv1209 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bult C. J. et al. Mouse genome database 2016. Nucleic Acids Res 44, D840–D847, ; DOI: 10.1093/nar/gkv1211 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoyama M. et al. The Rat Genome Database 2015: genomic, phenotypic and environmental variations and disease. Nucleic Acids Res 43, D743–D750, ; DOI: 10.1093/nar/gku1026 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyner C. et al. The UCSC Genome Browser database: 2017 update. Nucleic Acids Res 45, D626–D634, ; DOI: 10.1093/nar/gkw1134 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnikov N. et al. ArrayExpress update–simplifying data submissions. Nucleic Acids Res 43, D1113–D1116, ; DOI: 10.1093/nar/gku1057 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Markosian C., et al. . 2018. Figshare. https://doi.org/10.6084/m9.figshare.c.4064561