Abstract
A central question in neuroscience is how developmental programs instruct the formation of complex neural circuits with temporal, spatial, and numerical precision. Pinto-Teixeira et al. (2018) reveal simple developmental rules that govern sequential neurogenesis to concurrently establish highly organized retinotopic maps in the Drosophila visual system.
The human brain contains more than 100 billion neurons assembled into intricate neural networks to support numerous functions. While the daunting complexity of the mammalian brain has been a major obstacle to understanding its organizing principles, the stereotypical neural circuit wiring of compound eyes in the Drosophila brain provides a simple and tractable model system to tackle this question. In contrast to a wealth of information on the generation of diverse types of neurons following a precise spatiotemporal pattern during Drosophila brain development (Lin and Lee, 2012), how genetic programs drive developmental algorithms to construct complicated neural circuits is just starting to be explored. It has been a common practice to divide the neurodevelopment process into multiple sequential stages, from neurogenesis, neuronal migration, and axon guidance to synapse and circuit formation, and focus on each stage for investigation. In this issue of Cell, Pinto-Teixeira et al. examine the whole developmental process and reveal that specific modes of neurogenesis not only control fates of sibling daughter neurons but are also coupled with the spatial formation of concurrent retinotopic maps in the fly motion detection circuit (Pinto-Teixeira et al., 2018).
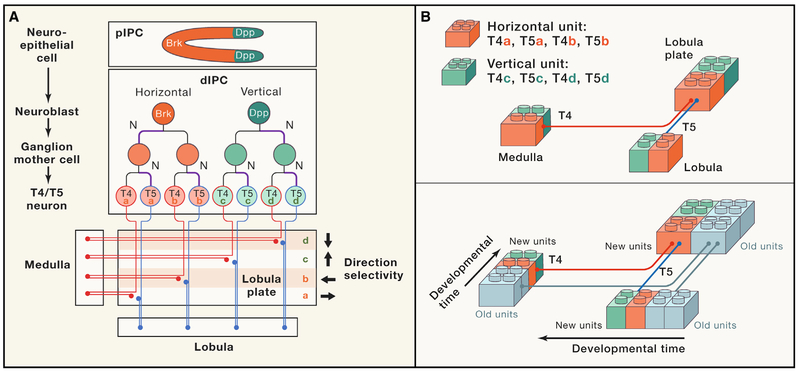
In Drosophila, visual information from 800 retinal ommatidia is processed into 800 matching retinotopic columns. During optic-lobe development, numerous neuronal subtypes are concurrently generated from different neuroblasts. Therefore, it is a daunting task to coordinate fate determination, migration, and projection pattern of neuronal progenies into a well-organized retinotopic column with exquisite precision. Neurons in four optic lobes originate from two crescent-shaped neuroepithelial domains, the outer (OPC) and inner (IPC) proliferation centers. The IPC crescent is separated into the proximal IPC (pIPC) and the distal IPC (dIPC) (Figure 1A). Neuroblasts in the dIPC originate from the pIPC and undergo a temporal transition of their competence. dIPC neuroblasts first express Dichaete and generate C/T neurons and later express Ato and Dac and produce T4/T5 neurons (Apitz and Salecker, 2015). The T4 and T5 cells are the elementary motion-detector neurons, responding preferentially to moving bright edges (ON, T4) or to moving dark edges (OFF, T5). Both T4 and T5 neurons exist in four subtypes (a, b, c, d) and respond to one of the four cardinal directions. The dIPC is formed by migrating progenitors from two distinct Decapentaplegic (Dpp)-expressing or Brinker (Brk)-expressing pIPC neuroepithelial domains. Using various lineage-tracing tools, Pinto-Teix-eira et al. show that vertical and horizontal T4/T5 motion detectors are produced by distinct neuroblast populations distinguished by Dpp-signaling activity. dIPC neuroblasts from the Brk domain exclusively generate T4/T5 neurons of the horizontal system (subtypes a and b), while the Dpp domain generates all T4/T5 neurons of the vertical system (subtypes c and d). Therefore, directionality of motion-detector neurons is pre-determined before the onset of neuroblast division by spatial patterning at pIPC (Figure 1A).
Figure 1. Modes of T4/T5 Neurogenesis Establish Retinotopic Organization in the Visual System of Drosophila.
(A) Specification of eight T4/T5 neuronal subtypes corresponding to eight concurrent retinotopic maps in motion-detection circuits. dIPC neuroblasts originate from pIPC neuroepithelium spatially patterned with Brk or Dpp expression. In dIPC, T4/T5 neurons of the horizontal system (orange, subtypes a and b) are produced by Brk+ neuroblasts, while those of the vertical system (green, subtypes c and d) are from Dpp+ neuroblasts. Notch signaling (N, violet lines) mediates binary cell-fate specification during two consecutive divisions. Axons of eight T4 and T5 neurons project across the lobula plate into the medulla (T4) or the lobula (T5) then return to the lobula plate and innervate to the specific retinotopic location (a, b, c, d).
(B) Schematic illustrating the establishment of the retinotopy of T4/T5 neurons. A horizontal unit represents four T4/T5 neurons (subtype a and b) from a single Brk+ neuroblast, and a vertical unit represents four T4/T5 neurons (subtype c and d) from a single Dpp+ neuroblast. Newborn units incorporate into existing units (gray) according to their birth time, establishing the temporal axis of axonal projection.
How do dIPC neuroblasts divide to generate the set of T4/T5 neurons with specific connectivity? Pinto-Teixeira et al. examine the fate of four sibling neurons from a single T4/T5 neuroblast using Twin-Spot MARCM technology for clonal lineage-tracing. They uncover a previously unknown mode of amplifying neuroblast division to produce two ganglion mother cells (GMCs), which in turn produce a unit of two T4 and two T5 neurons detecting motion in opposite directions (Figure 1A). To address the molecular mechanism underlying fate determination of progenies from dIPC neuroblasts, they focus on Notch signaling, a well-known binary cell-fate regulator. MARCM analysis of Notch mutant clones suggests that Notch signaling is critical for the identity of both T4/T5 neurons and the two sister GMCs (Figure 1A). Therefore, a simple molecular switch of Notch signaling, together with Dpp signaling, specifies the four T4 and four T5 neuron subtypes. How can the eight subtypes of T4 and T5 neurons project retinotopically to the proper target locations? They find that the temporal axis of axonal projections is progressively established according to the birth time of neuroblasts. Since each T4/T5 neuroblast produces whole components of either the horizontal or the vertical system as a unit, sequential projection of four sister neurons according to birth time ensures the synchronous connection to newly produced target columns with proper stoichiometry to build precise retinotopic maps (Figure 1B).
In this study, Pinto-Teixeira et al. reveal simple developmental rules that can establish the complex neural-circuit organization of the fly optic lobes. First, the pIPC neuroepithelium is spatially patterned with differential Dpp signaling to generate distinct neuroblast populations in the dIPC, which exclusively generate either horizontal or vertical motion-selective T4/T5 neurons. Second, neuroblasts undergo sequential temporal patterning by a series of transcription factors to produce district progeny types, namely from Dichaete+ neuroblasts to C/T neurons and from Ato+Dac+ neuroblasts to T4/T5 neurons. Third, two consecutive Notch-mediated binary fate decisions during the division of a single neuroblast give rise to two T4 and two T5 neurons with opposite motion-direction selectivity. This elegant study raises a number of questions. Why does Notch signaling instruct different lineage fates at each round of consecutive divisions? What are the molecular mechanisms to guide axons of sibling neurons born in close proximity to distinct targets? Single-cell transcriptome analyses can provide a holistic view of molecular cascades underlying sequential stem/progenitor states to answer these questions (Shin et al., 2015; Yang et al., 2016). The step-by-step cell-fate-specification program also implies progressive changes of cell-type-specific transcriptomes in every round of cell division. Considering the rapid cell division of neuroblasts, well-designed neurogenic programs, such as sequential expression of temporal transcription factors (Doe, 2017), might be pre-encoded before the onset of neurogenesis. Future studies are needed to address potential epigenetic and epitranscriptomic pre-patterning mechanisms before cell division (Yoon et al., 2017). Functionally, do four sister T4/T5 neurons derived from the same neuroblast cooperate for motion sensing? Single-cell lineage-tracing can be combined with neural-activity monitoring, such as calcium imaging, to resolve the relationship between the development of connectivity and functional properties of neural circuits.
The study by Pinto-Teixeira et al. exemplifies the power of genetic tools in fly models, such as lineage-tracing and genetic manipulations at the single-cell level, to greatly facilitate our understanding of neural-circuit development (del Valle Rodríguez et al., 2011). Columnar organization is frequently found in the nervous system of higher organisms, including mammalian neocortex (Rakic, 1988). During adult hippocampal neurogenesis in mice, primary axonal projections of sibling newborn neurons organize into laminar patterns with staggered terminations that stack along the septo-temporal hippocampal axis (Sun et al., 2013). It will be fascinating to investigate whether the remarkably simple developmental rules revealed by this study in flies may also instruct the complex wiring of mammalian brains, including human.
REFERENCES
- Apitz H, and Salecker I (2015). A region-specific neurogenesis mode requires migratory progenitors in the Drosophila visual system. Nat. Neurosci 18, 46–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Valle Rodríguez A, Didiano D, and Desplan C (2011). Power tools for gene expression and clonal analysis in Drosophila. Nat. Methods 9, 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CQ (2017). Temporal Patterning in the Drosophila CNS. Annu. Rev. Cell Dev. Biol 33, 219–240. [DOI] [PubMed] [Google Scholar]
- Lin S, and Lee T (2012). Generating neuronal diversity in the Drosophila central nervous system. Dev. Dyn 241, 57–68. [DOI] [PubMed] [Google Scholar]
- Pinto-Teixeira F, Koo C, Rossi AM, Neriec N, Bertet C, Li X, Del-Valle-Rodriguez A, and Desplan C (2018). Development of concurrent retinotopic maps in the fly motion detection circuit. Cell 173, this issue, 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P (1988). Specification of cerebral cortical areas. Science 241, 170–176. [DOI] [PubMed] [Google Scholar]
- Shin J, Berg DA, Zhu Y, Shin JY, Song J, Bonaguidi MA, Enikolopov G, Nauen DW, Christian KM, Ming GL, and Song H (2015). Single-Cell RNA-Seq with Waterfall Reveals Molecular Cascades underlying Adult Neurogenesis. Cell Stem Cell 17, 360–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun GJ, Sailor KA, Mahmood QA, Chavali N, Christian KM, Song H, and Ming GL (2013). Seamless reconstruction of intact adultborn neurons by serial end-block imaging reveals complex axonal guidance and development in the adult hippocampus. J. Neurosci 33, 11400–11411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CP, Fu CC, Sugino K, Liu Z, Ren Q, Liu LY, Yao X, Lee LP, and Lee T (2016). Transcriptomes of lineage-specific Drosophila neuroblasts profiled by genetic targeting and robotic sorting. Development 143, 411–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon KJ, Ringeling FR, Vissers C, Jacob F, Pokrass M, Jimenez-Cyrus D, Su Y, Kim NS, Zhu Y, Zheng L, et al. (2017). Temporal control of mammalian cortical neurogenesis by m6A methylation. Cell 171, 877–889.e817. [DOI] [PMC free article] [PubMed] [Google Scholar]