Abstract
Developing bioactive hydrogels with potential to guide the differentiation behavior of stem cells has become increasingly important in the biomaterials field. Here, silk hydrogels with tunable mechanical properties were developed by introducing inert silk fibroin nanofibers (SNF) within an enzyme crosslinked system of regenerated silk fibroin (RSF). After the crosslinking reaction of RSF, the inert SNF was embedded into the RSF hydrogel matrix, resulting in improved mechanical properties. Tunable stiffness in the range of 9–60 KPa was achieved by adjusting the amount of the added NSF, significantly higher than SNF-free hydrogels formed under same conditions (about 1 KPa). In addition, the proliferation of rat bone marrow derived mesenchymal stem cells cultured on the composite hydrogels and differentiated into endothelial cells, myoblast and osteoblast cells was improved, putatively due to the control of stiffness of the hydrogels. Bioactive and tunable silk-based hydrogels were prepared via a composite SNF and crosslinked RSF system, providing a new strategy to design silk biomaterials with tunable mechanical and biological performance.
Keywords: Silk, Nanofiber, Hydrogel, Stem cells, Differentiation
Table of contents
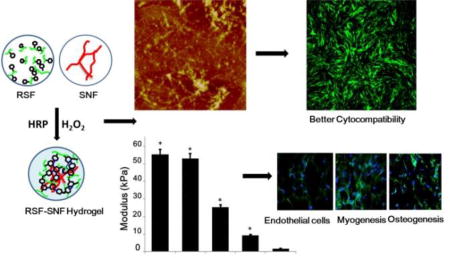
Hydrogels with tunable stiffness and differentiation capacity were achieved through introducing inert SF nanofibers into HRP crosslinking system of SF.
Introduction
Cell-matrix interactions play critical roles in various cell biological processes such as adhesion, growth, differentiation and apoptosis.1–2 Many studies confirmed that mechanical properties of the matrix can guide the differentiation of bone marrow mesenchymal stem cells (MSCs) into various anchorage-dependent cell types, including neurons, myoblasts, and osteoblasts, suggesting a feasible strategy for developing bioactive biomaterials by modulation of stiffness.3–5 Considering the wide range of stiffness, from kilopascal (KPa) to several hundred megapascals (MPa) in various tissues, different strategies such as blending and crosslinking different polymers and components were developed to match the stiffness of various tissues.6–7 However, it remains a challenge to finely tune the stiffness of biomaterials over a wide range related to the needs above.
Silk fibroin (SF) shows promising applications as cell-support matrices for stem cells, fibroblasts, nerve cells, and osteoblasts,8–10 and as scaffolds for bone, cartilage, skin, nerve and blood vessel tissue regeneration due to the biocompatibility, minimal inflammatory reactions and tunable biodegradability.11–15 Because of their simulation with the extracellular cell matrix (ECM), SF hydrogel is a choice for these applications. Although several processes were developed to regulate the stiffness of the SF hydrogels to improve the biocompatibility and applicability,16–18 SF hydrogels with innate bioactivity to induce the differentiation of MSCs into specific anchorage-dependent cells has been relatively unexplored. In addition, SF-based composite hydrogels with other natural or synthetic biomaterials were prepared to achieve preferable mechanical properties for specific applications, but this approach can compromise biocompatibility.19
Enzyme catalyzed crosslinking of SF by reactions with horseradish peroxidase (HRP) and hydrogen peroxide was used to form hydrogels with highly elastic and tunable stiffness.20–22 The stiffness could be controlled in the range of 200–10,000 Pa through changing molecular weight and solvent composition, but which falls short of the mechanical match with many tissues, especially bone and cartilage.23–25 Limited options are also available for hydrogels that are fine tuned in terms of stiffness for specific tissues. The introduction of additional biomaterials and beta-sheet domains within silk materials can improve the stiffness to about several MPa, but provided limited fine tuning.26 Therefore, new strategies are required to finely regulate the stiffness of the SF hydrogels to optimize applications in various tissue regeneration studies.
Recently, it was found that beta-sheet rich SF nanofibers (SNF) developed in our group remained inert in the presence of HRP due to their special conformational composition.27 Considering that the original high stiffness of the nanofibers stemmed from beta-sheet structure, the NSFs are suitable additives for tuning the stiffness of the SF hydrogels without the influence on the HRP crosslinking in traditional SF solution. The nanofibers also introduce ECM-like nano-morphology, which can improve biocompatibility.28,29 Therefore, as proof-of-concept, different ratios of SNF were blended with traditional SF solutions and formed into hydrogels with tunable stiffness. The delicate adjustment of stiffness, improved cytocompatibility as well as controlled differentiation of MSC by changing the content of the SNFs suggested a simple way to develop bioactive SF hydrogels with tunable mechanical properties.
Experimental
Preparation of Aqueous Regenerated Silk Fibroin Solutions
Regenerated SF solution (RSF) was prepared based on the protocol described previously.30 Bombyx mori cocoons dissolved in 0.02 M Na2CO3 solution were boiled for 20 min at room temperature, and then rinsed thoroughly with distilled water to extract the sericin proteins. The extracted silk was dissolved in 9.3 M LiBr solution (Sigma-Aldrich, St. Louis, MO) at 60°C, yielding a 20 wt% solution. This solution was dialyzed against distilled water to remove the salt. Then this solution was centrifuged at 9,000 rpm for 20 min at 4°C to remove silk aggregates formed, achieving aqueous RSF solution with concentration of about 6 wt%.
SNF Formation
To prepare SNFs, the RSF solution was treated according to the protocol described previously.31 The solution (6 wt%) was slowly concentrated to about 20 wt% at 60°C for 24 h to form metastable nanoparticles, and then diluted to below 2 wt% with distilled water. This diluted silk solution was incubated at 60°C for about 24h to induce nanofiber formation.
Preparation of Enzymatically Crosslinked SF Hydrogels
Enzymatically cross-linked hydrogels were prepared through a modified HRP crosslinking process.20 Different ratios of RSF (6 wt%) and SNF (2 wt%) solutions were mixed directly to form homogeneous aqueous solutions with various ratios of RSF and SNF at 3:1, 4:1, 6:1, 12:1, 100:0, respectively, and then adjusted to a final RSF concentration of 3 wt% with distilled water. According to the ratios of RSF and SNF, the hydrogels were termed R3-SN1, R4-SN1, R6-SN1, R12-SN1, respectively, while the NSF-free hydrogels were termed R100. HRP solution (type VI lyophilized powder, Sigma-Aldrich, St Louis, MO) was dissolved in deionized water and added to the RSF-SNF solutions with a ratio of 10U HRP to 1 mL silk solution. Then 10 µL of hydrogen peroxide solution (165 mM, Sigma Aldrich, St. Louis, MO) was added to 1 mL of silk solution containing HRP to initiate gelation. The RSF and SNF-RSF hydrogels formed within 1 h. Gels were cultured at room temperature for 4 h to complete the crosskinking. All the hydrogels were sterilized with 60Co γ-irradiation at dose of 50 kGy before co-culture with cells.
Scanning Electron Microscopy (SEM)
Hydrogel samples were lyophilized for characterization by SEM. After mounted on a copper plate and coated with platinumto form a 10–20 nm thick layer, the samples were observed using an SEM (S-4800, Hitachi, Tokyo, Japan) at 3 kV.32
FTIR
The secondary structures of the various samples in solid state were analyzed with FTIR on a Nicolet FTIR 5700 spectrometer (Thermo Scientific, FL, USA). For each measurement, 64 scans were coded with a resolution of 4 cm−1, with the wavenumber ranging from 400 to 4000 cm−1. Fourier self deconvolution (FSD) of the infrared spectra covering the amide I region (1595–1705 cm−1) was performed using Peakfit software to identify silk secondary structures.33
Circular Dichroism (CD)
Jasco-815 CD spectrophotometer (Jasco Co., Japan) was used to measure the secondary structures of the silk hydrogels.30 CD spectra from 250 to 190 nm wave lengths were recorded with an accumulation of five scans at the scanning rate of 100 nm min−1. The experiments repeated three times.
Atomic Force Microscopy (AFM)
The composite RSF-SNF solutions containing HRP were spin-coated onto freshly cleaved 4 × 4 mm mica surfaces and cultured for above 1 h for gel formation. The morphology of hydrogels was then observed through AFM (Nanoscope V, Veeco, NY, USA) in air. One 225 µm long silicon cantilever with a spring constant of 3 N m−1 was used in tapping mode.34
Dynamic Oscillatory Rheology
A Rheometer (AR2000, TA Instruments, New Castle, USA) fitted with a 20 mm cone plate (Ti, 20/1°) was used to evaluate the rheological properties of the hydrogels.29 All the crosslinked hydrogels were cut into same cylinder structure (diameter 1.5 cm, height 1 cm). Frequency sweeps were collected continuously with wide frequency range from 100 to 0.1 rads−1 at 37°C. Before measurement, all samples were stabilized for 20 minutes.
In Vitro Biocompatibility of the RSF-SNF hydrogels
Rat bone marrow mesenchymal stem cells (rBMSCs) were isolated from Sprague Dawley (SD) rats at 8 weeks as described.35 The use of rats was carried out following approval and granted by the animal ethics committee of Soochow University. Cells were maintained in DMEM medium (Dulbecco’s modified Eagle’s medium, Invitrogen) with 10% FBS and 1% antibiotic/antimycotic (Life Technologies, Grand Island, NY) in a humidified incubator at 37°C in 5% CO2. Cells at passage three were used for experiments. The hydrogels with thickness of 400 µm were cultured in DMEM medium solution for 72 h at 37°C to remove residual H2O2 and HRP. Cells were then seeded on the samples and incubated at 37°C in 5% CO2.
Three groups (R3-SN1, R12-SN1, and R100) were used to culture the cells in vitro and to assess cell proliferation. The passaged cells were re-suspended and cultured in DMEM medium containing 10% FBS after digestion with trypsin. The cell concentration was adjusted to 1 × 105 cells/ml and cultured on samples with 2 ml medium at 37°C in 5% CO2. After culturing for 1, 3, 7 and 14 d, the cells were counted with the CCK-8 method.36 Aliquots (200µl) of CCK-8 (CK04-500, Dojindo, Japan) were added to each well and then incubated for 4 hours. Supernatant (100 µl) was removed from each well and transferred to a new 96-well plate. The absorbance at a wavelength of 450 nm was measured with microplate reader (Infinite M200, Tecan, Switzerland).
Cell staining (Calcein-AM) count
The cells (1 × 105 cells/ml) were cultured in 6 well-plates with 2 ml medium at 37°C in 5% CO2. At day 14, cells were stained with Calcein. The storage concentration of calcein-AM was 1mM in DMSO, and the storage solution was diluted to 50 µM with PBS. Aliquots (200 ul) were added into each well and incubated at 37°C for 20 min. Then the cells were washed with PBS twice. Fluorescence intensity of the cells was tested under a fluorescent microscope with an excitation wavelength of 490 nm and an emission wavelength of 515 nm.
Quantitative Real-Time PCR
The in vitro differentiation behaviors of the cells cultured on the silk hydrogels were evaluated with quantitative real-time PCR (qRT-PCR), western blot and immunofluorescence staining.37,38
Total RNA was extracted from SD rat bone mesenchymal stem cells (rBMSCs) using TRIzol (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. First-strand cDNA was synthesized with reverse transcriptase kit. Real-Time PCR was performed with SYBR Green PCR reagent system (SYBR-green, Invitrogen, Carlsbad, CA, USA). Primers of the endothelial marker CD31 and Flt1 (Fms related tyrosine kinase 1), early myogenic markers MyoD1 (Myogenic differentiation 1) and osteogenic marker BMP2 (Bone morphogenetic protein 2), ALP (Alkaline phosphates) and Runx2 (Runt-related transcription factor 2) are shown in Table 1:
Table 1.
Primers used in quantitative real-time PCR
| Genes | Primer sequence(F,R,5’-3’) |
|---|---|
|
| |
| β-actin | AACCCTAAGGCCAACCGTGAAAAG |
| (NM_031144.3) | CGACCAGAGGCATACAGGGACAAC |
|
| |
| CD31 | GGACTGCGCCCATCACTTACC |
| (NM_031591) | TCATCCACCGGGGCTATTACCTT |
|
| |
| Flt1 | GGGCAAAGAATAGCGTGGGACAG |
| (NM_019306.2) | ACGGGGCTCGGTGGGCTTATTT |
|
| |
| MyoD1 | GCGCCGCCTGAGCAAAGTGAAC |
| (NM_176079.1) | CAGCGGGCCAGGTGCGTAGAAG |
|
| |
| BMP2 | AAAGCGTCAAGCCAAACACAAACA |
| (NM_017178.1) | RGAGGGGCCACGATCCAGTCAT |
|
| |
| ALP | AGAAGCCGCCAACCTGTG |
| (NM_053650.1) | TGCGCTTTCCCATCTTCC |
|
| |
| Runx-2 | GAACCCACGGCCCTCCCTGAACTC |
| (NM_001278483.1) | AGCGGCGTGGTGGAATGGATGGAT |
Western Blot
Total cell protein was obtained with the cell lysis buffer. The protein concentration was obtained with BCA protein assay (Pierce, IL, USA) based on the manufacturer's instructions. Protein samples (10µl/lane) were separated by 11% SDS-PAGE. The primary antibodies included anti-CD31, anti-Flt1, anti-MyoD1, anti-ALP, anti-BMP2 and anti-Runx2 (Santacruz, CA, USA). The secondary antibody was rabbit anti-mouse IgG (1:3000, Santacruz, CA, USA). Relative protein levels were quantified by scanning densitometry and the gray value of protein level were measured with NIH Image J Software.
Immunofluorescence Staining
Immunofluorescence staining of cells with CD31, MyoD1 and Runx2 were used to characterize endothelial, myoblastic and osteogenic differentiation of BMSCs. Briefly, the samples were fixed in 4% formaldehyde (Sigma-Aldrich, St. Louis, MO, USA) for 30 min, permeabilized with 1% Triton X-100 for 10 min, and blocked in PBS containing 3% BSA for 1 h. Cells were incubated with anti-CD31, anti-MyoD1 and anti-Runx2 primary antibodies (Santacruz, CA, USA) for 1 h. Samples were then rinsed and incubated with secondary antibodies. DNA and silk hydrogels were stained with DAPI (Sigma-Aldrich, St. Louis, MO, USA). FITC-phalloidin (Invitrogen, Grand Island, NY, USA) was used to stain F-actin. The images were collected by confocal laser scanning microscopy (CLSM, Olympus FV10 inverted microscope, Nagano, Japan).
Statistical Analysis
All in vitro experiments were repeated at least three times. All numerical data were expressed as Mean ± SD as depicted. The statistical software SPSS 17.0 was used for analysis. All data was analyzed using one-way ANOVA.
Results and discussion
Formation of the RSF hydrogels containing NSF
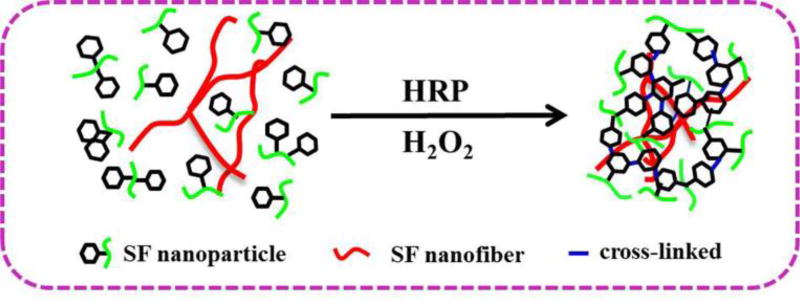
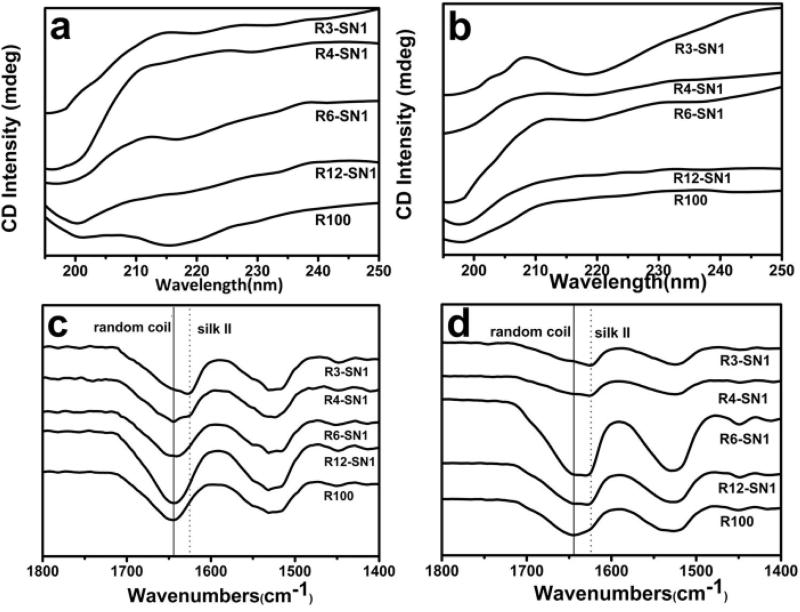
Unlike traditional RSF solutions, the SNF solution cannot easily translate into hydrogels in reactions with HRP and H2O2 (Figure S-1). The composition and ECM-biomimic nanofibrous structures make SNF a desirable additive to improve the mechanical and morphological properties of SF hydrogels without sacrificing biocompatibility and crosslinking. Traditional RSF molecules in aqueous solutions were crosslinked in the presence of HRP and H2O2 to form dityrosine bonds, resulting in hydrogel formation, while the SNF without the crosslinking reaction dispersed homogeneously in the hydrogels (Figure 1). Following the increase of SNF in the system, the hydrogels transformed from transparent to opaque (Figure S-2). The gelation time was similar for the systems containing same amount of RSF but different contents of SNF, suggesting that the HRP catalyzed reaction only occurred among the RSF molecules. Circular dichroism (CD) was used to evaluate the protein secondary structures before (Figure 2a) and after the reaction (Figure 2b). A more distinct minimum at 218 nm appeared following the increase of SNF amount, due to the beta sheet rich structure of the SNF. Similar to previous studies,39,40 a minimum near 218 nm remained unchanged after the reaction for the systems containing various contents of SNF, confirming that the SNF did not appear to significantly influence the HRP reaction.
Figure 1.
The mechanisms and structure of silk fibroin nanofiber-reinforced hydrogels prepared via the enzymatically crosslinked process. HRP is horseradish peroxidase for the crosslinking reaction.
Figure 2.
Secondary conformational changes of silk fibroin after the crosslinking process: (a) CD curves of different silk fibroin solutions before crosslinking; (b) CD curves of different silk fibroin hydrogels after crosslinking; (c) FTIR spectra of freeze-dried silk fibroin solutions before crosslinking; and (d) FTIR spectra of freeze-dried silk fibroin hydrogels after crosslinking. The samples were as follows: R3-SN1, the ratio of RSF and SNF was 3:1; R4-SN1, the ratio of RSF and SNF was 4:1; R6-SN1, the ratio of RSF and SNF was 6:1; R12-SN1, the ratio of RSF and SNF was 12:1; R100, pure RSF hydrogel.
Structure and morphology of RSF-SNF hydrogels
Secondary structure of the various hydrogels was assessed with FTIR. Because of absorption in the amide I region of the peptide chains, the infrared spectral region is within 1700–1600 cm−1, which has been usually used for analyzing different secondary structures of silk. Both 1648–1652 cm−1 and 1635–1645 cm−1 peaks are indicative of the random coil, while the 1610–1630 cm−1 peaks is characteristic of silk II (beta sheet) conformation. For the systems with different contents of SNF, no significant conformational changes appeared by FTIR for RSF and SNF in the hydrogel state and in aqueouos solutions (Figure 2c, 2d), indicating that the crosslinking reaction had no significant influence on the conformation of SF. Due to the beta sheet rich structure of the SNF, the peak near 1625 cm−1 gradually appeared for the hydrogels with higher contents of SNF, suggesting that the SNF remained stable in the hydrogels.
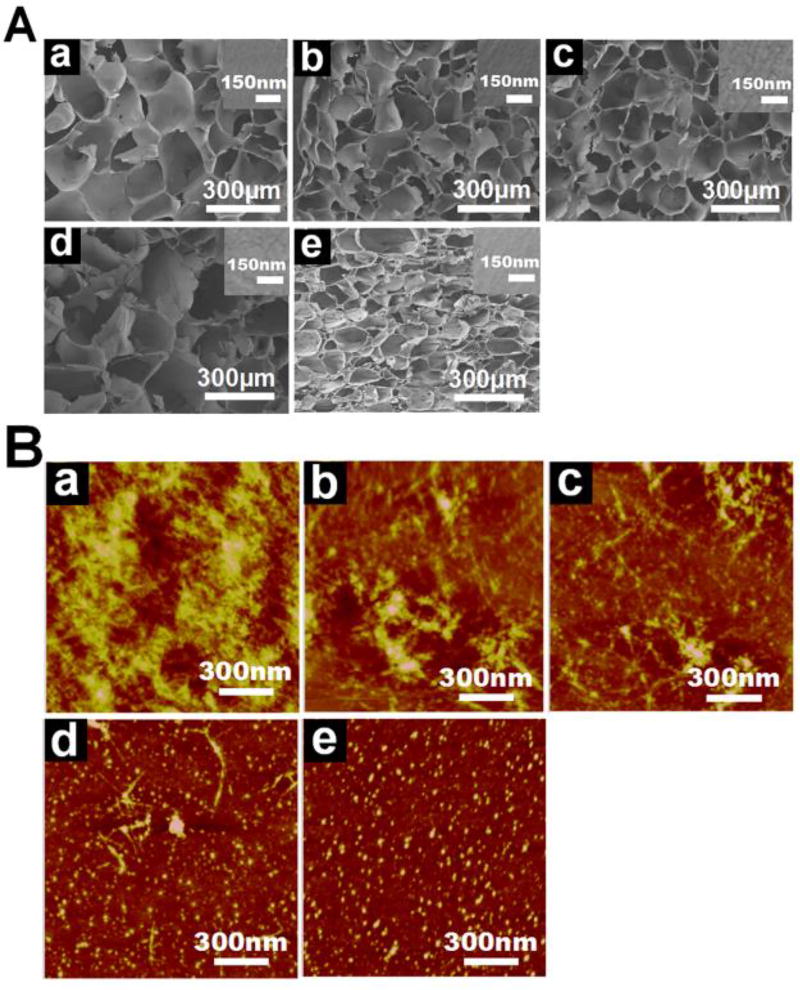
The micromorphology of the freeze-dried hydrogels was investigated with SEM (Figure 3A). The SEM images showed homogeneous porous structures for all the hydrogels, suggesting that the SNF was dispersed homogeneously in the hydrogels without phase separation. The different pore sizes of the hydrogels containing different amounts of SNF implied altered interactions between RSFs and SNFs. The pore walls revealed nano-topographic structures in the hydrogels with no phase separation observed on the walls, confirming the homogeneous distribution of the SNF inside the hydrogels. Considering that SNF and RSF have different nanomorphologies, AFM was used to reveal the distribution of SNFs inside the hydrogels (Figure 3B). More nanofibrous features appeared in the RSF-SNF hydrogels with enriched SNF, but this feature disappeared in the pure RSF hydrogels. This finding suggeted that the SNF retained the nanofirous structures in the hydrogels. Three-dimensional height images showed homogeneous distributions of the SNF nanofibers in the composite hydrogels, which may provide improved physical cues for cells.
Figure 3.
Microstructures of the crosslinked silk fibroin hydrogels: (A) SEM images of the freeze-dried hydrogels, the insets were the magnified surface morphologies of the porous walls; (B) AFM images of the different hydrogels. The samples were as follows: (a) R3-SN1, the ratio of RSF and SNF was 3:1; (b) R4-SN1, the ratio of RSF and SNF was 4:1; (c) R6-SN1, the ratio of RSF and SNF was 6:1; (d) R12-SN1, the ratio of RSF and SNF was 12:1; (e) R100, pure RSF hydrogel.
Mechanical properties of the RSF-SNF hydrogels
A persisten need remains for versatile, tunable and biocompatible hydrogels to match the mechanical properties of native ECMs of different tissues. Several strategies towards this goal include optimzing crosslinking reactions, changing SF concentration and controlling the secondary composition of SF to tune the mechanical properties of SF hydrogels.26, 41–45 The mechanical properties of SF hydrogels have been broadened to the MPa range using these approaches.26, 28, 46 However, the regulation of the differentiation of MSCs into different cell lineages using SF hydrogels and control of material stiffness has not been explored.
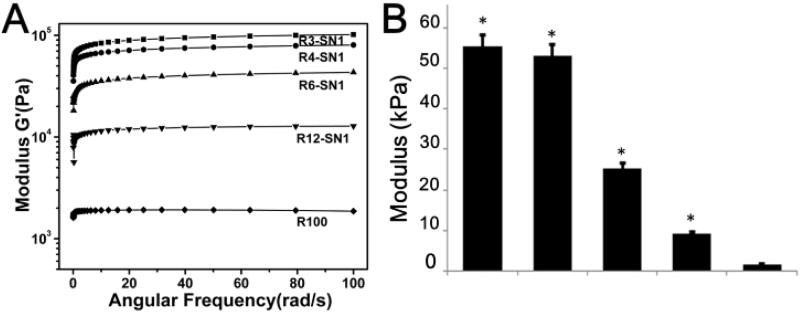
The rheometer was used to measure the storage modulus of hydrogels based on previous studies.6, 29 Unlike previous strategies, SNFs, stable nanofibers composed of pure silk fibroin, could be added directly to the RSF solutions, providing a simple way to tune the fine stiffness of the materials. As shown in Figure 4, the SNF-RSF hydrogels exhibited different moduli in the range from 9.2 KPa to 55 KPa, while the modulus of pure RSF hydrogels at same concentration was about 1 KPa. These results indicated that the introduction of SNF could significantly enhance the mechanical properties of the crosslinked hydrogels. Previous modification methods such as optimizing crosslinking process and altering SNF concentrations could be used in the present system, further improving the stiffness to above 100 KPa (data not shown). The delicate adjustment of the mechanical properties was then achieved through tuning the content of SNF, suggesting the feasibility of actively controlling the mechanical properties of the hydrogels. These results indicated that programmable modulation of stiffness was easily obtained by the introduction of SNFs, making it possible to develop bioactive SF hydrogels with tunable features for the differentiation of stem cells.
Figure 4.
Rheological properties of the different hydrogels: (a) Frequency sweeps; (b) The storage modulus. The samples were as follows: R100, pure RSF hydrogel; R12-SN1, the ratio of RSF and SNF was 12:1; R6-SN1, the ratio of RSF and SNF was 6:1; R4-SN1, the ratio of RSF and SNF was 4:1; R3-SN1, the ratio of RSF and SNF was 3:1. Significantly higher and increased stiffness was achieved for the hydrogels following the introduction and content increase of SNF. *P < 0.05
Cytocompatibility of the RSF-SNF hydrogels in Vitro
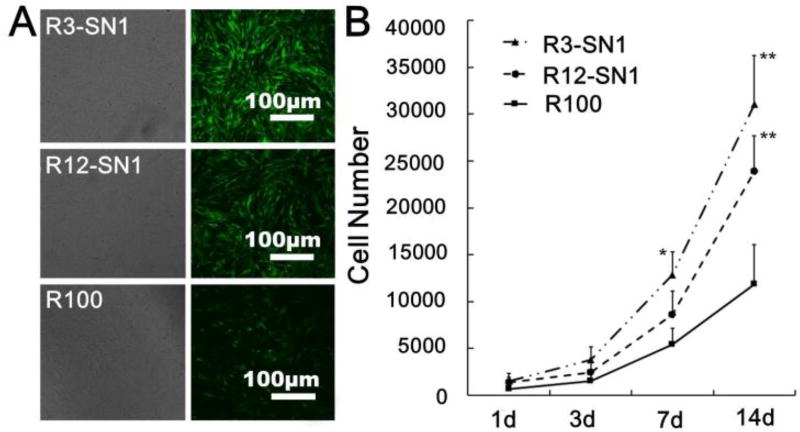
To verify that the introduction of SNF had no negative influence on cell compatibility, BMSCs were cultured on the surface of pure RSF hydrogels and RSF-SNF hydrogels with different contents of SNF. The cells seeded on the different hydrogels showed excellent adhesion and proliferation over 14 days of culture (Figure 5). At day 14, significantly more cells proliferated on the RSF-SNF hydrogels than on the pure RSF hydrogels (Figure 5A). Cell numbers increased steadily up to 14 days without reaching a plateau (Figure 5B). Further improvement of cell proliferation occurred for the cells on the RSF-SNF hydrogels with higher content of SNF, confirming the promotion of cell compatibility after the introduction of SNF. Although multiple cues such as stiffness, compositions and nanostructures have significant influence on cell proliferation, the cell culture results on RSF-SNF hydrogels indicated that the introduction of SNF resulted in a better niche for cell response in vitro, which is similar to several studies that revealed an enhanced influence of nanofibrous SF materials on cell compatibility.28,47–49
Figure 5.
The in vitro cell compatibility of the different hydrogels: (a) Cell proliferation and morphologies after cultured on the hydrogels for 14 days; (b) The cell numbers measured with the CCK8 method. The samples were as follows: R3-SN1, the ratio of RSF and SNF was 3:1; R12-SN1, the ratio of RSF and SNF was 12:1; R100, pure RSF hydrogel. *P<0.05, **P<0.01.
Differentiation Control of BMSCs on the RSF-SNF hydrogels
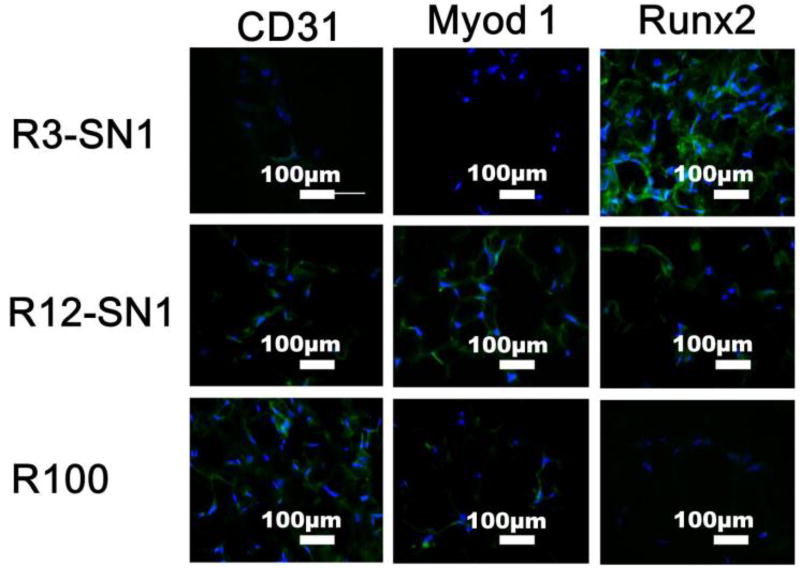
In cell-matrix interactions, cells are sensitive to the stiffness of the matrix and show different differentiation behavior according to the mechanical cues of the microenvironment.50,51 Considering the broad range of the stiffness from KPa to MPa for different tissues, it is preferable to finely tune the stiffness of the SF hydrogels. Unlike previous crosslinked SF hydrogels,52 the RSF-SNF hydrogels could be finely tuned to achieve suitable stiffness for various tissues. Three RSF-SNF hydrogels (R100, R12-SN1, R3-SN1) with stiffness of 1.7 KPa, 9.2 KPa and 55 KPa, suitable for blood vessel, muscle and bone tissues, respectively, were chosen to evaluate the differentiation behavior of MSCs into endothelial cells, muscle cells and osteoblasts.
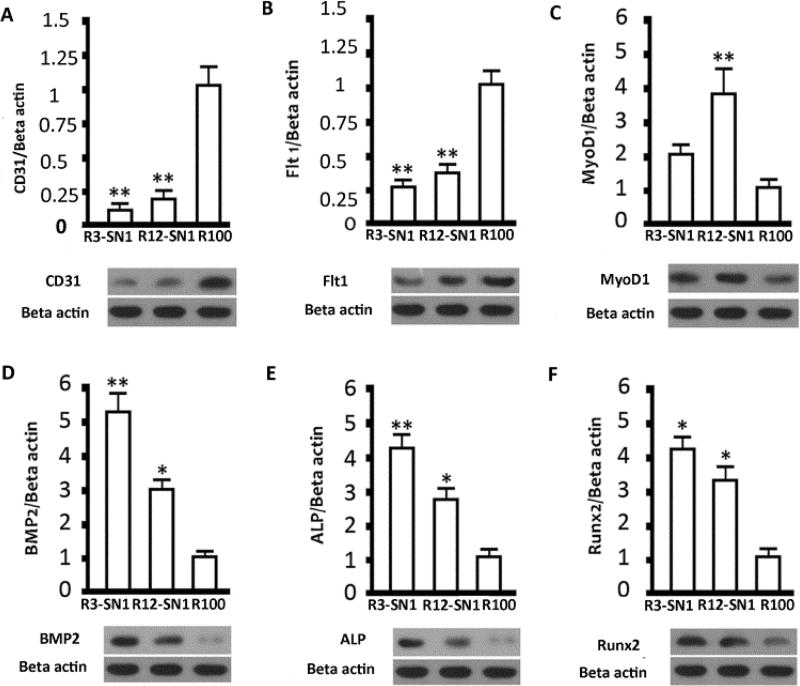
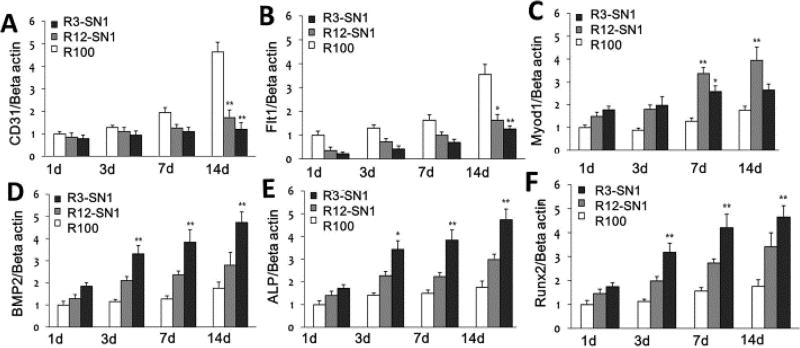
Flt1/CD31, MyoD1, and ALP/BMP2/Runx2 are important markers of endothelial, myoblast and osteoblasts, respectively.53–55 The differentiation behaviour of BMSCs were evaluated with immunofluorescent staining and mRNA expression (Figures 6 and 7). Significantly higher expression of the markers for endothelial, myoblast and osteogenic cells appeared for the MSCs cultured on the three hydrogels with stiffness of 1.7 KPa, 9.2 KPa and 55 KPa, respectively, suggesting that differentiation capacity was successfully achieved through the introduction of SNF into the SF hydrogels. Western blots were used to quantify the production of the different markers (Figure 8). Significantly higher levels of CD31/Flt1, MyoD1, and ALP/BMP2/Runx2 appeared with the cells cultured on R100, R12-SN1, and R3-SN1 hydrogels, respectively, confirming that SF hydrogels with tunable differentiation capacity into various tissue cells could be achieved by tuning the stiffness to match various tissues. Tuning the stiffness of the biomaterials to control differentiation behaviors of the stem cells has been considered as a powerful way of achieving bioactive biomaterials. Although the mechanical properties of silk hydrogels could be changed through various processes or different crosslinking methods, it remains a great challenge to finely tune their stiffness to match different tissues. Therefore, only SF scaffolds rather than hydrogels with various differentiation capacities for MSCs have been reported in several studies.56–59 As far as we know, this is the first time tunable differentiation for the growth factor free SF hydrogels was found. More interestingly, the differentiation behaviors of the stem cells on our SF hydrogels are consistent with that cultured on other biomaterial hydrogels.3–5 However, significant deviation appeared for the cells cultured on SF scaffolds with tunable stiffness. For example, although the study based on hydrogel system suggested that the stiffness in the range of 1–7 KPa could induced the differentiation of stem cells into endothelial cells,4 the stem cells cultured on the SF scaffolds with modulus of 1–4 KPa failed to exhibit endothelial differentiation capacity.60 It is reasonable since the modulus of the scaffolds not only depends on the stiffness of the scaffolds but also is influenced by the porous structures. Other factors such as nanostructures and orientation could also tune the differentiation behaviours of the stem cells, suggesting the possibility of optimizing the control of cell behaviors through designing suitable microenvironments. Considering that various factors could be introduced in our present hydrogel systems, the present study provides a feasible way of developing bioactive SF hydrogels with tunable differentiation behavior without the introduction of growth factors or other polymers. Further optimization of the specific differentiation capacity can be anticipated through subtle further tuning of the stiffness.
Figure 6.
Immunofluorescence staining of the cells cultured on SF hydrogels with different stiffness over time. The specific marker proteins (CD31, MyoD1 and Runx2) were used to evaluate endothelial, myoblast and osteogenic differentiation of rBMSCs, respectively. The cell culture times for CD 31, MyoD1 and Runx2 measurements were 7, 7 and 14 days, respectively. The samples were as follows: R3-SN1, the ratio of RSF and SNF was 3:1; R12-SN1, the ratio of RSF and SNF was 12:1; R100, pure RSF hydrogel. *P<0.05, **P<0.01.
Figure 7.
Q-PCR assessment of the differentiation behavior of rBMSCs cultured on silk hydrogels with different stiffness for different time points: (A, B) vascular endothelial cells markers CD31 and Flt1 after 7 days; (C) muscle marker MyoD1 after 7 days, (D, E, F) Osteogenesis markers BMP 2, ALP and Runx2 after 7 days. The samples were as follows: R3-SN1, the ratio of RSF and SNF was 3:1; R12-SN1, the ratio of RSF and SNF was 12:1; R100, pure RSF hydrogel. *indicates significant difference between groups (p < 0.05),** indicates significant difference between groups (p < 0.01).
Figure 8.
Western Blot analysis of the differentiation of rBMSCs cultured on the hydrogels with different stiffness for the screened time points. (A, B) The expression of vascular endothelial cells markers CD31 and Flt after the cells were cultured for 7 days; (C) the expression of muscle marker MyoD1 after the cells were cultured for 7 days; (D, E, F) Osteogenesis markers BMP2, ALP and Runx2 after the cells were cultured for 14 days. The samples were as follows: R3-SN1, the ratio of RSF and SNF was 3:1; R12-SN1, the ratio of RSF and SNF was 12:1; R100, pure RSF hydrogel. Results were shown as mean ± SD of gray value. * means P<0.05 and ** means P<0.01
Conclusions
Bioactive SF hydrogels with tunable mechanical properties were prepared through introducing SF nanofibers into HRP catalyzed crosslinking processes. The stiffness of the hydrogels was significantly improved and finely adjusted by changing the amount of SF nanofibers, providing a feasible fabrication method. The differentiation behavior of MSCs into endothelial, myoblast and osteoblast cells, as well as better cytocompatibility of the hydrogels, suggests useful features for these hydrogels in different tissue regeneration applications.
Supplementary Material
Acknowledgments
The authors thank the National Key Research and Development Program of China (2016YFE0204400), Shanghai Jiao Tong university Medical professionals crossing project (YG2015QN09) and the NIH (R01NS094218, R01EB021264).
References
- 1.Watt FM, Huck WT. Nat. Rev. Mol. Cell. Biol. 2013;14:467–473. doi: 10.1038/nrm3620. [DOI] [PubMed] [Google Scholar]
- 2.Watt FM, Fujiwara H. Cold Spring Harb Perspect. Biol. 2011;3:1–14. doi: 10.1101/cshperspect.a005124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abagnale G, Steger M, Nguyen VH, Hersch N, Sechi A, Joussen S, Denecke B, Merkel R, Hoffmann B, Dreser A, Schnakenberg U, Gillner A, Wagner W. Biomaterials. 2015;61:316–326. doi: 10.1016/j.biomaterials.2015.05.030. [DOI] [PubMed] [Google Scholar]
- 4.Bai SM, Han H, Huang X, Xu W, Kaplan DL, Zhu H, Lu Q. Acta Biomater. 2015;20:22–31. doi: 10.1016/j.actbio.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai SM, Zhang W, Lu Q, Ma Q, Kaplan DL, Zhu H. J. Mater. Chem. B. 2014;2:6590–6600. doi: 10.1039/C4TB00878B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 7.Park JS, Chu JS, Tsou AD, Diop R, Tang Z, Wang A, Li S. Biomaterials. 2011;32:3921–3930. doi: 10.1016/j.biomaterials.2011.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das S, Sharma M, Saharia D, Sarma KK, Muir EM, Bora U. Biomed. Mater. 2017;12:045025. doi: 10.1088/1748-605X/aa7802. [DOI] [PubMed] [Google Scholar]
- 9.Orlova AA, Kotlyarova MS, Lavrenov VS, Volkova SV, Arkhipova AY. Bull. Exp. Biol. Med. 2014;158:88–91. doi: 10.1007/s10517-014-2699-2. [DOI] [PubMed] [Google Scholar]
- 10.Naskar D, Ghosh AK, Mandal M, Das P, Nandi SK, Kundu SC. Biomaterials. 2017;136:67–85. doi: 10.1016/j.biomaterials.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 11.Ko E, Lee JS, Kim H, Yang SY, Yang D, Yang K, Lee J, Shin J, Yang HS, Ryu W, Cho SW. ACS Appl. Mater. Interfaces. 2017;10:7614–7625. doi: 10.1021/acsami.7b03328. [DOI] [PubMed] [Google Scholar]
- 12.Shi W, Sun M, Hu X, Ren B, Cheng J, Li C, Duan X, Fu X, Zhang J, Chen H, Ao YS. Adv. Mater. 2017;29:1701089. doi: 10.1002/adma.201701089. [DOI] [PubMed] [Google Scholar]
- 13.Xie SY, Peng LH, Shan YH, Niu J, Xiong J, Gao JQ. J. Nanosci. Nanotechnol. 2016;16:5498–5505. doi: 10.1166/jnn.2016.11730. [DOI] [PubMed] [Google Scholar]
- 14.Xue C, Zhu H, Tan D, Ren H, Gu X, Zhao Y, Zhang P, Sun Z, Yang Y, Gu J, Gu Y, Gu X. J. Tissue Eng. Regen. Med. 2017;12:e1143–e1153. doi: 10.1002/term.2449. [DOI] [PubMed] [Google Scholar]
- 15.Zhan K, Bai L, Wu Q, Lei D, Wang G. J. Biomed. Mater. Res. A. 2017;105:2276–2290. doi: 10.1002/jbm.a.36094. [DOI] [PubMed] [Google Scholar]
- 16.Stoppato M, Stevens HY, Carletti E, Migliaresi C, Motta A, Guldberg RE. Biomaterials. 2013;34:4573–4581. doi: 10.1016/j.biomaterials.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun JY, Zhao X, Illeperuma WR, Chaudhuri O, Oh KH, Mooney DJ, Vlassak JJ, Suo Z. Nature. 2012;489:133–136. doi: 10.1038/nature11409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang HN. Korea-Aust. Rheol. J. 2013;25:185–196. [Google Scholar]
- 19.Zhao Y, Nakajima T, Yang JJ, Kurokawa T, Liu J, Lu J, Mizumoto S, Sugahara K, Kitamura N, Yasuda K, Daniels AU, Gong JP. Adv. Mater. 2014;26:436–442. doi: 10.1002/adma.201303387. [DOI] [PubMed] [Google Scholar]
- 20.Partlow BP, Hanna CW, Rnjak-Kovacina J, Moreau JE, Applegate MB, Burke KA, Marelli B, Mitropoulos AN, Omenetto FG, Kaplan DL. Adv. Funct. Mater. 2014;24:4615–4624. doi: 10.1002/adfm.201400526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou B, Wang P, Cui L, Yu Y, Deng C, Wang Q, Fan X. Appl. Biochem. Biotechnol. 2017;182:1548–1563. doi: 10.1007/s12010-017-2417-4. [DOI] [PubMed] [Google Scholar]
- 22.Chirila TV, Suzuki S, Papolla C. Biotechnol. Appl. Biochem. 2017;64:771–781. doi: 10.1002/bab.1552. [DOI] [PubMed] [Google Scholar]
- 23.Fini M, Motta A, Torricelli P, Giavaresi G, Nicoli Aldini N, Tschon M, Giardino R, Migliaresi C. Biomaterials. 2005;26:3527–3536. doi: 10.1016/j.biomaterials.2004.09.040. [DOI] [PubMed] [Google Scholar]
- 24.Silva SS, Motta A, Rodrigues MT, Pinheiro AF, Gomes ME, Mano JF, Reis RL, Migliaresi C. Biomacromolecules. 2008;9:2764–2774. doi: 10.1021/bm800874q. [DOI] [PubMed] [Google Scholar]
- 25.Morita Y, Tomita N, Aoki H, Wakitani S, Tamada Y, Suguro T, Ikeuchi K. Biomed. Mater. Eng. 2002;12:291–298. [PubMed] [Google Scholar]
- 26.Luo K, Yang Y, Shao Z. Adv. Funct. Mater. 2016;26:872–880. [Google Scholar]
- 27.Dong X, Zhao Q, Xiao L, Lu Q, Kaplan DL. Biomacromolecules. 2016;17:3000–3006. doi: 10.1021/acs.biomac.6b00863. [DOI] [PubMed] [Google Scholar]
- 28.Gandhimathi C, Venugopal JR, Tham AY, Ramakrishna S, Kumar SD. Mater. Sci. Eng. C. Mater. Biol. Appl. 2015;49:776–785. doi: 10.1016/j.msec.2015.01.075. [DOI] [PubMed] [Google Scholar]
- 29.Leisk GG, Lo TJ, Yucel T, Lu Q, Kaplan DL. Adv. Mater. 2010;22:711–715. doi: 10.1002/adma.200902643. [DOI] [PubMed] [Google Scholar]
- 30.Bai SM, Liu SS, Zhang C, Xu W, Lu Q, Han H, Kaplan DL, Zhu H. Acta. Biomater. 2013;9:7806–7813. doi: 10.1016/j.actbio.2013.04.033. [DOI] [PubMed] [Google Scholar]
- 31.Bai SM, Zhang X, Lu Q, Sheng W, Liu L, Dong B, Kaplan DL, Zhu H. Biomacromolecules. 2014;15:3044–3051. doi: 10.1021/bm500662z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han F, Liu SS, Liu X, Pei YZ, Bai SM, Zhao H, Lu Q, Ma F, Kaplan DL, Zhu H. Acta Biomater. 2014;10:921–930. doi: 10.1016/j.actbio.2013.09.026. [DOI] [PubMed] [Google Scholar]
- 33.Yao D, Dong S, Lu Q, Hu X, Kaplan DL, Zhang B, Zhu H. Biomacromolecules. 2012;13:3723–3729. doi: 10.1021/bm301197h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu Q, Zhu H, Zhang C, Zhang F, Zhang B, Kaplan DL. Biomacromolecules. 2012;13:826–832. doi: 10.1021/bm201731e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sukul M, Nguyen TB, Min YK, Lee SY, Lee BT. Tissue Eng. Part A. 2015;21:1822–1836. doi: 10.1089/ten.tea.2014.0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y, Liu R, Yang F, Cheng R, Chen X, Cui S, Gu Y, Sun W, You C, Liu Z, Sun F, Wang Y, Fu Z, Ye C, Zhang C, Li J, Chen X. Mol. Cancer. 2017;16:53. doi: 10.1186/s12943-017-0625-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu X, Park SH, Gil ES, Xia XX, Weiss AS, Kaplan DL. Biomaterials. 2011;32:8979–8989. doi: 10.1016/j.biomaterials.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mathur AB, Collinsworth AM, Reichert WM, Kraus WE, Truskey GA. J. Biomech. 2001;34:1545–1553. doi: 10.1016/s0021-9290(01)00149-x. [DOI] [PubMed] [Google Scholar]
- 39.Yang M, Shuai Y, Zhou G, Mandal N, Zhu L, Mao C. ACS Appl. Mater. Interfaces. 2014;6:13782–13789. doi: 10.1021/am503214g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang M, He W, Shuai Y, Min S, Zhu L. J. Polym. Sci. Part B Polym. Phys. 2013;51:742–748. [Google Scholar]
- 41.Brown JE, Partlow BP, Berman AM, House MD, Kaplan DL. Am. J. Obstet. Gynecol. 2016;214:118, e1–e9. doi: 10.1016/j.ajog.2015.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Numata K, Yamazaki S, Katashima T, Chuah JA, Naga N, Sakai T. Macromol. Biosci. 2014;14:799–806. doi: 10.1002/mabi.201300482. [DOI] [PubMed] [Google Scholar]
- 43.McGill M, Coburn JM, Partlow BP, Mu X, Kaplan DL. Acta Biomater. 2017;63:76–84. doi: 10.1016/j.actbio.2017.09.020. [DOI] [PubMed] [Google Scholar]
- 44.Bradner SA, Partlow BP, Cebe P, Omenetto FG, Kaplan DL. Biopolymers. 2017;107:e23030. doi: 10.1002/bip.23030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vu T, Xue Y, Vuong T, Erbe M, Bennet C, Palazzo B, Popielski L, Rodriguez N, Hu X. Int. J. Mol. Sci. 2016;17:1497. doi: 10.3390/ijms17091497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li J, Illeperuma WRK, Suo Z, Vlassak JJ. ACS Macro Lett. 2014;3:520–523. doi: 10.1021/mz5002355. [DOI] [PubMed] [Google Scholar]
- 47.Chen D, Yin Z, Wu F, Fu H, Kundu SC, Lu S. J. Appl. Polym. Sci. 2017;134:45050. [Google Scholar]
- 48.Wray LS, Hu X, Gallego J, Georgakoudi I, Omenetto FG, Schmidt D, Kaplan DL. J. Biomed. Mater. Res. B Appl. Biomater. 2011;99:89–101. doi: 10.1002/jbm.b.31875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suganya S, Venugopal J, Ramakrishna S, Lakshmi BS, Dev VR. Int. J. Biol. Macromo. 2014;68:135–143. doi: 10.1016/j.ijbiomac.2014.04.031. [DOI] [PubMed] [Google Scholar]
- 50.Lin X, Shi Y, Cao Y, Liu W. Biomed. Mater. 2016;11:014109. doi: 10.1088/1748-6041/11/1/014109. [DOI] [PubMed] [Google Scholar]
- 51.Even-Ram S, Artym V, Yamada KM. Cell. 2006;126:645–647. doi: 10.1016/j.cell.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 52.Su D, Yao M, Liu J, Zhong Y, Chen X, Shao Z. ACS Appl. Mater. Interfaces. 2017;9:17489–17498. doi: 10.1021/acsami.7b04623. [DOI] [PubMed] [Google Scholar]
- 53.Nicklin SA, Reynolds PN, Brosnan MJ, White SJ, Curiel DT, Dominiczak AF, Baker AH. Hypertension. 2001;38:65–70. doi: 10.1161/01.hyp.38.1.65. [DOI] [PubMed] [Google Scholar]
- 54.Yang S, Wei D, Wang D, Phimphilai M, Krebsbach PH, Franceschi RT. J. Bone Miner. Res. 2003;18:705–715. doi: 10.1359/jbmr.2003.18.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gang EJ, Jeong JA, Hong SH, Hwang SH, Kim SW, Yang IH, Ahn C, Han H, Kim H. Stem Cells. 2004;22:617–624. doi: 10.1634/stemcells.22-4-617. [DOI] [PubMed] [Google Scholar]
- 56.Li DW, Lei X, He FL, He J, Liu YL, Ye YJ, Deng X, Duan E, Yin DC. Int. J. Biol. Macromol. 2017;105:584–597. doi: 10.1016/j.ijbiomac.2017.07.080. [DOI] [PubMed] [Google Scholar]
- 57.Navone SE, Pascucci L, Dossena M, Ferri A, Invernici G, Acerbi F, Cristini S, Bedini G, Tosetti V, Ceserani V, Bonomi A, Pessina A, Freddi G, Alessandrino A, Ceccarelli P, Campanella R, Marfia G, Alessandri G, Parati EA. Stem Cell Res. Ther. 2014;5:7. doi: 10.1186/scrt396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luo Z, Jiang L, Xu Y, Li H, Xu W, Wu S, Wang Y, Tang Z, Lv Y, Yang L. Biomaterials. 2015;52:463–475. doi: 10.1016/j.biomaterials.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 59.Yang Y, Yuan X, Ding F, Yao D, Gu Y, Liu J, Gu X. Tissue Eng. Part A. 2011;17:2231–2244. doi: 10.1089/ten.TEA.2010.0633. [DOI] [PubMed] [Google Scholar]
- 60.Sang Y, Li M, Liu J, Yao Y, Ding Z, Wang L, Xiao L, Lu Q, Fu X, Kaplan DL. ACS Appl. Mater. Interfaces. 2018 doi: 10.1021/acsami.7b19204. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.