Abstract
Background
Promoter DNA methylation of Cysteine dioxygenase type1 (CDO1) gene has been clarified as a molecular diagnostic and prognostic indicator in various human cancers. The aim of this study is to investigate the clinical relevance of CDO1 methylation in primary biliary tract cancer (BTC).
Methods
CDO1 DNA methylation was assessed by quantitative methylation-specific PCR in 108 BTC tumor tissues and 101 corresponding normal tissues. BTC was composed of extrahepatic cholangiocarcinoma (EHCC) (n = 81) and ampullary carcinoma (AC) (n = 27).
Results
The CDO1 methylation value in the tumor tissues was significantly higher than that in the corresponding normal tissues (p<0.0001). The overall survival (OS) in EHCC patients with hypermethylation was poorer than those with hypomethylation (p = 0.0018), whereas there was no significant difference in AC patients. Multivariate analysis identified that CDO1 hypermethylation, preoperative serum CA19-9 and perineural invasion were independent prognostic factors in EHCC. The EHCC patients with CDO1 hypermethylation exhibited more dismal prognosis than those with hypomethylation even in low group of CA19-9 level (p = 0.0006).
Conclusions
Our study provided evidence that promoter DNA methylation of CDO1 gene could be an excellent molecular diagnostic and prognostic biomarker in primary EHCC. The combination of CDO1 methylation and preoperative serum CA19-9 effectively enriched EHCC patients who showed the most dismal prognosis. These markers would be beneficial for clinical clarification of the optimal strategies in EHCC.
Introduction
Biliary tract cancer (BTC) is uncommon malignancy with an unfavorable prognosis. According to cancer statistics, 2017, BTC patients including gallbladder cancer were estimated as new cases of 11,740 and deaths of 3,830 in the United States.[1] Recent studies have reported that the global incidence has shown increasing tendency worldwide.[2,3] BTC is epithelial cancer that arise from biliary tree and can be classified by anatomic location. This classification differs not only from location but epidemiology, origin, treatment and prognosis. Extrahepatic cholangiocarcinoma (EHCC) is the most common type of BTC.[3] Among them, EHCC and ampullary carcinoma (AC) produce similar clinical presentation, and they are often diagnosed by the onset of obstructive jaundice. However, the prognosis of all subtypes of BTC has not improved due to the difficulty of early diagnosis and the limitation of effective treatment. Surgical resection remains the only potentially curative treatment, while many patients have experienced the recurrence. Such poor outcome has prompted interest in the use of adjuvant chemotherapy and radiation therapy. However, there is a paucity of high-quality evidence to support clinical effects of adjuvant therapy following resection in BTC, and patients should be encouraged to participate in clinical trials evaluating new strategies, and molecular biomarkers for the evaluation of BTC are highly demanded in clinical practice, with establishment of optimal guidelines for adjuvant or neoadjuvant therapy.
Epigenetic gene silencing of tumor suppresser genes through promoter DNA hypermethylation is a common feature in human cancers, whereas cancer-specific methylation is a relatively rare event.[4,5] We have developed pharmacologic reversal of epigenetic silencing and thereby uncovered a myriad of transcriptionally repressed genes in human cancers.[6] Using this technique, we have identified novel tumor suppressor gene candidates including the cysteine dioxygenase type1 (CDO1) gene. We and others have previously described that aberrant DNA methylation of the CDO1 promotor region is diagnostic and/or prognostic biomarker in various cancers, such as breast, esophageal, gastric, colorectal, renal, prostate, and gallbladder cancer. [7–16] Additional study has further shown the clinical utility of tumor diagnosis using CDO1 promoter DNA methylation in an endoscopic retrograde cholangiography (ERCP) solution of BTC as alternate cytology test.[17] Nevertheless, there have no reports with regard to clinico-pathological relevance of CDO1 methylation in primary BTC.
In the present study, we for the first time investigated the clinico-pathological and prognostic relevance of promotor DNA methylation of the CDO1 gene assessed by the quantitative PCR in primary BTC.
Materials and methods
Patients and tissue samples
This study investigated 108 patients who underwent surgical resection for primary BTC at the Kitasato university hospital, Japan between October 1988 and November 2012. We extracted DNA from 108 tumor tissues and 101 corresponding normal tissues. These tissue samples were collected from all patients who obtained written informed consent to use their pathological specimens. The present study was approved by the Ethics Committee of Kitasato University. Patient characteristics are shown in Table 1. Patients with neoadjuvant chemotherapy was not included.
Table 1. Univariate and multivariate analysis of clinico-pathological factors affecting 5-year overall survival in primary BTC.
| (A) Extrahepatic cholangiocarcinoma | ||||||||
| Clinico-pathological parameter | Number | Univariate analysis | Multivariate analysis | |||||
| 5yOS (%) | p value | HR* | 95%CI* | p value | ||||
| Age | ≥65 / <65 | 46 / 35 | 41 / 55 | 0.31 | ||||
| Gender | Male / Female | 59 / 22 | 42 / 64 | 0.16 | ||||
| Preoperative jaundice | absence / presence | 40 / 41 | 47 / 47 | 0.81 | ||||
| Biliary drainage | absence / presence | 13 / 68 | 64 / 44 | 0.19 | ||||
| Preoperative serum CA19-9 | ≥37 / <37 | 52 / 29 | 35 / 74 | 0.0084 | 2.3 | 1.0–5.9 | 0.047 | |
| Preoperative serum CEA | ≥5 / <5 | 4 / 77 | 0 / 49 | 0.12 | ||||
| Tumor location | Bp / Bd | 18 / 63 | 43 / 48 | 0.79 | ||||
| Lymphatic permeation | absence / presence | 18 / 63 | 76 / 40 | 0.0047 | 1.6 | 0.52–7.4 | 0.42 | |
| Vascular permeation | absence / presence | 25 / 56 | 59 / 43 | 0.22 | ||||
| Portal venous invasion | absence / presence | 77 / 4 | 49 / 25 | 0.0003 | ||||
| Arterial system invasion | absence / presence | 78 / 3 | 48 / 33 | 0.49 | ||||
| Perineural invasion | absence / presence | 22 / 59 | 80 / 36 | 0.0021 | 3.1 | 1.2–11 | 0.018 | |
| Macroscopic growth pattern | invasive / others | 71 / 10 | 45 / 68 | 0.25 | ||||
| Histology | tub1, pap / others | 49 / 32 | 53 / 39 | 0.23 | ||||
| Resection status | R0 / R1, 2 | 43 / 38 | 57 / 36 | 0.04 | 1.4 | 0.71–2.7 | 0.34 | |
| CDO1 TaqMeth value | ≥28.9 / <28.9 | 22 / 59 | 22 / 56 | 0.0018 | 2.4 | 1.2–4.7 | 0.016 | |
| the sixth TNM classification | ||||||||
| pT | Tis,T1 / T2 / T3 / T4 | 10 / 12 / 31 / 28 | 100 / 56 / 40 / 34 | 0.01 | ||||
| pN | absence / presence | 43 / 38 | 66 / 27 | 0.0002 | ||||
| pStage | 0.016 | 0.079 | ||||||
| 0, IA, IB | 15 | 76 | Reference | |||||
| IIA | 17 | 61 | 1.0 | 0.25–5.1 | ||||
| IIB | 21 | 33 | 2.0 | 0.58–9.7 | ||||
| III | 28 | 34 | 3.0 | 0.90–14 | ||||
| Operative procedure | PD / Liver resection / others | 64 / 14 / 3 | 49 / 42 / 33 | 0.64 | ||||
| Postoperative jaundice | absence / presence | 70 /11 | 47 / 55 | 0.75 | ||||
| Postoperative serum CA19-9 | ≥37 / <37 | 19 / 62 | 31 / 52 | 0.077 | ||||
| Postoperative serum CEA | ≥5 / <5 | 2 / 79 | 100 / 46 | 0.21 | ||||
| Postoperative chemotherapy | absence / presence | 20 / 61 | 62 / 43 | 0.34 | ||||
| (B) Ampullary carcinoma | ||||||||
| Clinico-pathological parameter | Number | Univariate analysis | Multivariate analysis | |||||
| 5yOS (%) | p value | HR* | 95%CI* | p value | ||||
| Age | ≥65 / <65 | 16 / 11 | 65 / 82 | 0.57 | ||||
| Gender | Male / Female | 16 / 11 | 70 / 81 | 0.66 | ||||
| Preoperative jaundice | absence / presence | 16 / 11 | 88 / 61 | 0.24 | ||||
| Biliary drainage | absence / presence | 4 / 23 | 100 / 70 | 0.31 | ||||
| Preoperative serum CA19-9 | ≥37 / <37 | 11 / 16 | 57 / 88 | 0.19 | ||||
| Preoperative serum CEA | ≥5 / <5 | 1 / 26 | 0 / 76 | 0.0068 | 1.5 | 0.06–20 | 0.76 | |
| Lymphatic permeation | absence / presence | 11 / 16 | 100 / 59 | 0.036 | 1.6E+08 | 0.04- | 0.52 | |
| Vascular permeation | absence / presence | 10 / 17 | 100 / 60 | 0.047 | 1.2E+08 | 0.03- | 0.58 | |
| Portal venous invasion | absence / presence | 27 / 0 | 74 / - | |||||
| Arterial system invasion | absence / presence | 27 / 0 | 74 / - | |||||
| Perineural invasion | absence / presence | 21 / 6 | 86 / 25 | 0.0008 | 1.0 | 0.04–15 | 0.99 | |
| Macroscopic growth pattern | invasive / others | 11 / 16 | 32 / 94 | 0.014 | 4.0 | 0.28–97 | 0.29 | |
| Histology | tub1, pap / others | 21 / 6 | 75 / 67 | 0.49 | ||||
| Resection status | R0 / R1, 2 | 24 / 3 | 83 / 0 | <0.0001 | 4.3 | 0.37–101 | 0.24 | |
| CDO1 TaqMeth value | ≥28.9 / <28.9 | 9 / 18 | 66 / 78 | 0.89 | ||||
| the sixth TNM classification | ||||||||
| pT | Tis,T1 / T2 / T3 / T4 | 9 / 4 / 14 / 0 | 100 / 100 / 54 / - | 0.052 | ||||
| pN | absence / presence | 11 / 16 | 91 / 65 | 0.25 | ||||
| pStage | 0, IA, IB / IIA / IIB / III | 8 / 3 / 16 / 0 | 100 / 67 / 65 / - | 0.26 | ||||
| Postoperative jaundice | absence / presence | 27 / 0 | 74 / - | |||||
| Postoperative serum CA19-9 | ≥37 / <37 | 5 / 22 | 50 / 82 | 0.45 | ||||
| Postoperative serum CEA | ≥5 / <5 | 1 / 26 | 100 / 73 | 0.68 | ||||
| Postoperative chemotherapy | absence / presence | 10 / 17 | 90 / 66 | 0.29 | ||||
*HR: hazard ratio, CI: confidence interval
DNA extraction and sodium bisulfite conversion
Tissue sections from primary tumors and corresponding normal tissues were stained with hematoxylin and eosin, and dissected under microscope. Genomic DNA was extracted from formalin-fixed paraffin embedded (FFPE) tissues using a QIAamp DNA FFPE Tissue Kit (QIAGEN Sciences, Hilden, Germany). Bisulfite treatment was done according to the manufacturer’s instructions of an EZ DNA Methylation-GoldTM Kit (Zymo Research, Orange, CA).
Quantitative-methylation-specific PCR (Q-MSP)
Quantitative-TaqMan methylation specific PCR (Q-MSP) was carried out using iQ Supermix (Bio-Rad Laboratories, Hercules, CA) in triplicate on the C1000 TouchTM Thermal Cycler CFX96 Real Time System (Bio-Rad). Bisulfite-treated DNA was amplified by the following PCR conditions: 95°C for 3 min, followed by 40 cycles at 95°C for 20 sec, annealing temperature (60°C) for 30 sec, and 72°C for 30 sec. The sequences of primers and probes are provided in S1 Table.[8–10,12,16] Serial dilutions of bisulfite modified DNA from human colon carcinoma cell line DLD1 was used to construct the calibration curve on each plate as a methylation positive control, and human hepatoblastoma cell line HepG2 was used as a negative control as previous described.[8–10,12,16] The methylation value was defined as a TaqMeth value by the ratio of the amplified signal value of methylated CDO1 to the value for β-actin, which was then multiplied by 100.[7]
Conventional MSP
Conventional MSP was performed using Platinum Taq DNA Polymerase (Invitrogen) according to the manufacturer’s protocol. Methylated and unmethylated primers for PCR amplification were designed to partially cover the CpG island of CDO1 promoter region. Primers sequences are also included in S1 Table. The PCR conditions were the same as Q-MSP. The PCR products were separated on 1.5–2.0% agarose gel, then visualized by ethidium bromide staining. Distilled water was used as negative control.
Immunohistochemistry
Immunostaining was performed on formalin-fixed, paraffin-embedded sections (4 μm thick). Sections were incubated using the anti-CDO1 rabbit polyclonal antibody (dilution of 1:100) (ATLAS ANTIBODIES, Bromma, Sweden). Immune complexes were detected with a Histofine Simple Stain MAX PO (MULTI) (Nichirei, Tokyo, Japan), following the manufacturer’s protocol, and visualized using the 3,3’-diaminobenzidine (DAB) substrate. Sections were counter-stained with Hematoxylin solution.
Statistical analysis
Continuous variables were analyzed using Mann-Whitney’s U test and Kruskal-Wallis test, as appropriate. Categorical variables were analyzed using X2 test. Clinico-pathological characteristics and follow up data were evaluated in terms of 5-year overall survival (5yOS). The follow up time was calculated from the date of surgery to death or end-point. 5yOS was estimated by Kaplan-Meier method, and compared using a log-rank test. Variables suggesting potential prognostic factors on univariate analyses were subjected to a multivariate analysis using a Cox proportional-hazards model. P-value <0.05 was considered to indicate statistical significance. All statistical analyses were conducted using the SAS software package (JMP Pro11, SAS Institute, Cary, NC).
Results
Promotor DNA methylation level of CDO1 gene and its correlation with clinico-pathological factors in primary BTC
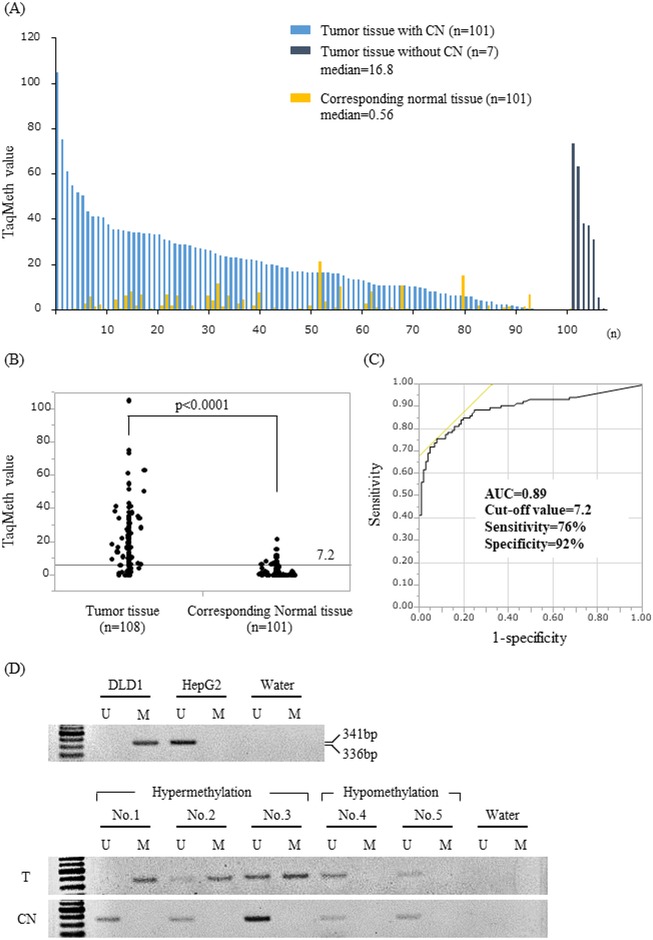
A total of 108 primary tumor specimens of BTC patients who underwent surgical resection were assessed by Q-MSP to evaluate the clinical relevance of the CDO1 methylation level. The median TaqMeth value was 16.8, ranging from 0 to 105 in the 108 tumor tissues (T) and 0.56, ranging from 0 to 21.3 in the 101 corresponding normal tissues (CN) (Fig 1A). There was significant difference in CDO1 methylation value between T and CN (p<0.0001) (Fig 1B). The most optimal cut-off value of 7.2 was calculated from receiver operating characteristic (ROC) analysis for maximizing both sensitivity and specificity of BTC detection as compared to CN, where sensitivity was 76%, and specificity was 92% (Fig 1C). We also confirmed differential methylation with conventional MSP in 10 tissue samples. T and CN are TaqMeth values of 1.2–105 and 0.0–1.4, respectively. T with hypermethylation amplified by methylated primers, but hypomethylation and CN samples were not methylated bands (Fig 1D).
Fig 1. Quantitative assessment of CDO1 methylation and representative conventional MSP in primary BTC.
(A) CDO1 TaqMeth value of the 108 primary BTC tumor tissues (T) and 101 corresponding normal tissues (CN). The median of T and CN were 16.8 and 0.56, respectively. (B) There was a significant difference in CDO1 methylation values between T and CN (p< 0.0001). (C) ROC curve of CDO1 methylation for detection of BTC. Area under the curve (AUC) represents the accuracy in discriminating normal from tumor in term of sensitivity and specificity (p< 0.0001). (D) Representative conventional MSP of CDO1 gene in T and CN (U: unmethylation, M: methylation).
Correlation of each clinico-pathological factor to TaqMeth value of CDO1 gene in primary BTC was evaluated by Mann-Whitney’s U test or Kruskal-Wallis test. There was significant correlation of CDO1 TaqMeth value to vascular permeation (p = 0.035). The other clinico-pathological factors showed no significant association with CDO1 TaqMeth value (S2 Table).
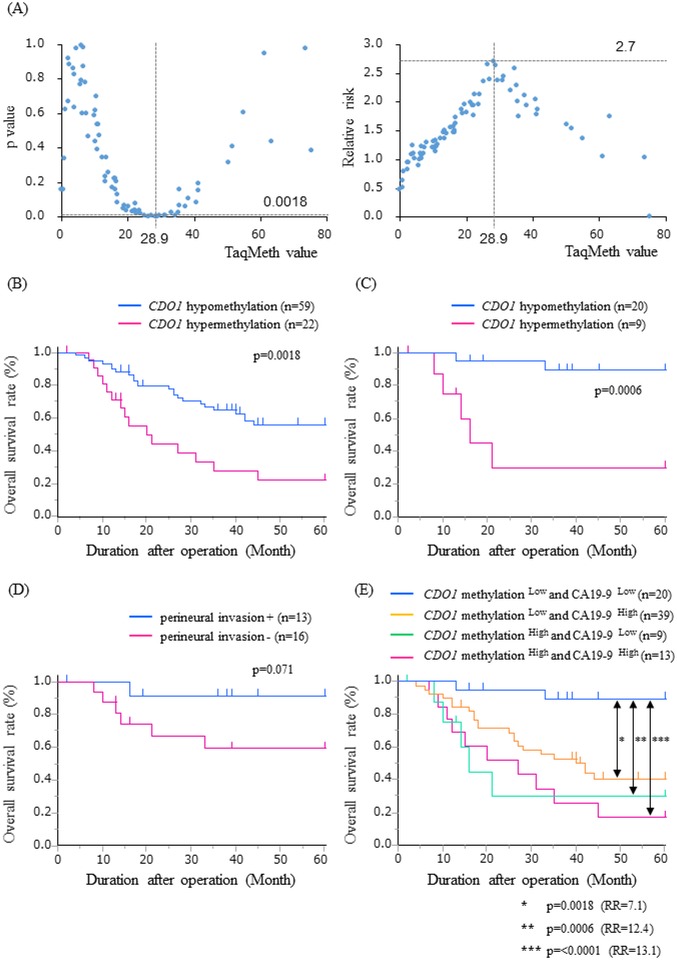
We further investigated the correlation of CDO1 TaqMeth value to OS in primary BTC. To determine the optimal cut-off values for predicting prognosis, we assessed each p value and relative risk by the log rank plot analysis. The most optimal cut-off value was defined as 28.9, which showed the highest relative risk with statistical significance in EHCC (Fig 2A). The EHCC patients with CDO1 hypermethylation (≥28.9) showed poor prognosis than those with hypomethylation (<28.9) (5yOS: 22% vs 56%, p = 0.0018) (Fig 2B). However, any cut-off values could not represent prognostic stratification in AC (S1 Fig).
Fig 2. Optimal cut-off value and survival analysis in primary extrahepatic cholangiocarcinoma.
(A) Identification of an optimal cut-off value for the prognosis using the log-rank plot analysis. Kaplan-Meier curves for 5-year overall survival stratified by CDO1 methylation status (B), and CDO1 methylation status and perineural invasion in low levels of CA19-9 (C), (D). Kaplan-Meier curves for the combination of CDO1 methylation and CA19-9 (E). RR: relative risk.
Univariate and multivariate prognostic analysis including CDO1 promoter DNA methylation status in primary BTC
The clinico-pathological factors related to prognosis were examined separately in 81 EHCC and 27 AC patients, because there was large difference of their prognosis (5yOS: 48% vs 74%, p = 0.029).
Univariate analysis showed that preoperative serum values of CA19-9 (p = 0.0084), lymphatic permeation (p = 0.0047), portal venous invasion (p = 0.0003), perineural invasion (p = 0.0021), resection status (p = 0.04), CDO1 TaqMeth value (p = 0.0018), pT (p = 0.01), pN (p = 0.0002), and pStage (p = 0.016) were significantly associated with poor prognosis in EHCC. The identified univariate prognostic factors were subjected to multivariate analysis, in which staging factors were excluded, i.e., portal venous invasion, pT and pN, because they were confounding factors for pStage. Multivariate analysis indicated that preoperative serum value of CA19-9 (p = 0.047), perineural invasion (p = 0.018), and CDO1 TaqMeth value (p = 0.016) were finally remnant independent prognostic factors related to OS in EHCC (Table 1A).
On the other hand, univariate analysis in AC showed preoperative serum value of CEA (p = 0.0068), lymphatic permeation (p = 0.036), vascular permeation (p = 0.047), perineural invasion (p = 0.0008), macroscopic growth pattern (p = 0.014), and resection status (p<0.0001) were significantly poor prognosis. Multivariate analysis revealed that there was no independent factor in AC (Table 1B).
Correlation of clinico-pathological factors to promoter DNA methylation status of the CDO1 gene divided by the prognostically optimized cut-off value in primary EHCC
Correlation between clinico-pathological factors and CDO1 methylation status divided by cut-off value of 28.9 in primary EHCC was determined by a X2 test. Preoperative serum CEA was the only significant parameter associated with CDO1 methylation value (p = 0.027) (Table 2).
Table 2. Correlation of clinico-pathological chracterestics and CDO1 methylation in primary EHCC.
| Factor category | Clinico-pathological parameter | CDO1 TaqMeth value | |||||
|---|---|---|---|---|---|---|---|
| Low (<28.9) | High (≥28.9) | p value | |||||
| No. | % | No. | % | ||||
| Preoperative factor | Age | ≥65 | 30 | 65 | 16 | 35 | 0.08 |
| <65 | 29 | 83 | 6 | 17 | |||
| Gender | Male | 41 | 69 | 18 | 31 | 0.27 | |
| Female | 18 | 82 | 4 | 18 | |||
| Preoperative jaundice | absence | 27 | 68 | 13 | 33 | 0.29 | |
| presence | 32 | 78 | 9 | 22 | |||
| Biliary drainage | absence | 9 | 69 | 4 | 31 | 0.75 | |
| presence | 50 | 74 | 18 | 26 | |||
| Preoperative serum CA19-9 | ≥37 | 39 | 75 | 13 | 25 | 0.56 | |
| <37 | 20 | 69 | 9 | 31 | |||
| Preoperative serum CEA | ≥5 | 1 | 25 | 3 | 75 | 0.027 | |
| <5 | 58 | 75 | 19 | 25 | |||
| Pathological factor | Tumor location | Bp | 11 | 61 | 7 | 39 | 0.20 |
| Bd | 48 | 76 | 15 | 24 | |||
| Lymphatic permeation | absence | 16 | 89 | 2 | 11 | 0.083 | |
| presence | 43 | 68 | 20 | 32 | |||
| Vascular permeation | absence | 20 | 80 | 5 | 20 | 0.33 | |
| presence | 39 | 70 | 17 | 30 | |||
| Portal venous invasion | absence | 57 | 74 | 20 | 26 | 0.29 | |
| presence | 2 | 50 | 2 | 50 | |||
| Arterial system invasion | absence | 56 | 72 | 22 | 28 | 0.28 | |
| presence | 3 | 100 | 0 | 0 | |||
| Perineural invasion | absence | 17 | 77 | 5 | 23 | 0.58 | |
| presence | 42 | 71 | 17 | 29 | |||
| Macroscopic growth pattern | invasive | 52 | 73 | 19 | 27 | 0.83 | |
| others | 7 | 70 | 3 | 30 | |||
| Histology | tub1,pap | 38 | 78 | 11 | 22 | 0.24 | |
| others | 21 | 66 | 11 | 34 | |||
| Resection status | R0 | 31 | 72 | 12 | 28 | 0.87 | |
| R1,2 | 28 | 74 | 10 | 26 | |||
| pT | Tis,T1 | 9 | 90 | 1 | 10 | 0.40 | |
| T2 | 10 | 83 | 2 | 17 | |||
| T3 | 21 | 68 | 10 | 32 | |||
| T4 | 19 | 68 | 9 | 32 | |||
| pN | absence | 35 | 81 | 8 | 19 | 0.07 | |
| presence | 24 | 63 | 14 | 37 | |||
| pStage | 0,IA,IB | 14 | 93 | 1 | 7 | 0.26 | |
| IIA | 12 | 71 | 5 | 29 | |||
| IIB | 14 | 67 | 7 | 33 | |||
| III | 19 | 68 | 9 | 32 | |||
| Treatment factor | Operative procedure | PD | 49 | 77 | 15 | 23 | 0.19 |
| (postoperative factor) | Liver resection | 9 | 64 | 5 | 36 | ||
| others | 1 | 33 | 2 | 67 | |||
| Postoperative jaundice | absence | 50 | 71 | 20 | 29 | 0.47 | |
| presence | 9 | 82 | 2 | 18 | |||
| Postoperative serum CA19-9 | ≥37 | 11 | 58 | 8 | 42 | 0.09 | |
| <37 | 48 | 77 | 14 | 23 | |||
| Postoperative serum CEA | ≥5 | 2 | 100 | 0 | 0 | 0.38 | |
| <5 | 57 | 72 | 22 | 28 | |||
| Postoperative chemotherapy | absence | 15 | 75 | 5 | 25 | 0.80 | |
| presence | 44 | 72 | 17 | 28 | |||
Prognostic relevance of CDO1 TaqMeth value in EHCC
We then examined prognostic relevance of the combination of the independent prognostic factors, CDO1 methylation and preoperative serum CA19-9 level, and perineural invasion in EHCC. Kaplan-Meier curves showed that patients with CDO1 hypermethylation showed significantly more dismal prognosis than those with hypomethylation even among the patients with low preoperative serum value of CA19-9 (5yOS: 30% vs 89%, p = 0.0006) (Fig 2C). There was no significant difference between patients with and without perineural invasion in low value of CA19-9 (p = 0.071) (Fig 2D). Finally, EHCC patients with CDO1 hypomethylation and low preoperative serum value of CA19-9 exhibited significantly much better prognostic outcome rather than those with either CDO1 hypermethylation or high preoperative value of CA19-9 (Fig 2E).
Promoter methylation of CDO1 gene critically affects CDO1 expression in tumor tissues
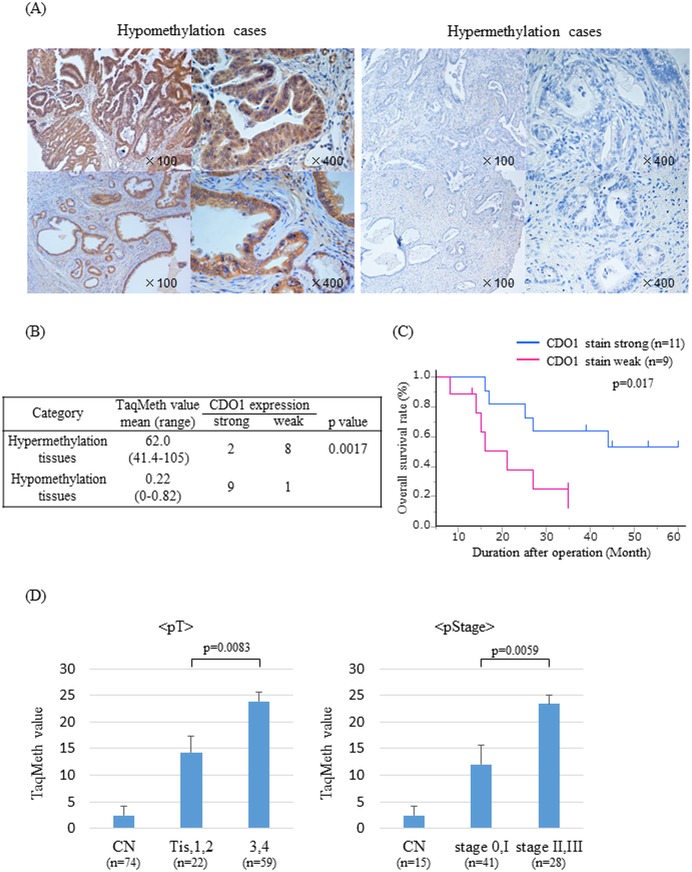
CDO1 protein expression was examined in the 10 tissues with the highest and lowest of the methylation value using immunostaining with anti-CDO1 polyclonal antibody (Fig 3A). Strong expression of CDO1 protein was observed in 90% of hypomethylation tissues, whereas weak expression was dominant in 80% of hypermethylation tissues. The difference of CDO1 protein expression between these two groups was identified as statistical significance (p = 0.0017) (Fig 3B), suggesting that CDO1 protein expression is significantly associated with promoter DNA methylation of the CDO1 gene. Additionally, there was also significant difference between CDO1 protein expression and poor survival (p = 0.017) (Fig 3C).
Fig 3. Immunohistochemical staining for CDO1 and correlation of CDO1 TaqMeth value to clinico-pathological factors in primary tumor.
(A) Representative images of immunostaining with an anti-CDO1 antibody in methylation-high or low BTC tumor tissues (original magnification, X100, X400). (B) Correlation between expression of CDO1 (via IHC) and methylation status, (C) and survival. (D) Correlation of CDO1 TaqMeth value to pT and pStage according to 6th UICC in EHCC. Data are expressed as the mean ±SE.
Discussion
We have discovered that promoter DNA methylation of CDO1 gene is extra-ordinarily specific to various human cancers by pharmacological unmasking microarray.[6,7] Recently, Andresen and Vedeld et al.[18,19] also reported cancer-specific aberrations of CDO1 in BTC. However there has been no report on CDO1 methylation with regard to clinico-pathological relevance in primary BTC, particularly by accurate quantitative assessment. In our current study, we for the first time assessed the clinical and prognostic relevance of CDO1 methylation status in primary BTC.
Although CDO1 TaqMeth value was significantly associated with vascular invasion among clinico-pothological factors of total BTC, it tended to increase in a stepwise manner toward higher depth of invasion and higher pathological stage, if restricted to EHCC (Fig 3D). When T factor divided Tis/T1/T2 from T3/T4, and UICCstage divided stage0/I from stageII/III in primary EHCC, there were statistically significant differences between groups, which again recapitulate the results of adenoma-carcinoma sequence of colorectal cancer and those of gallbladder cancer.[12,16]
We have demonstrated that other types of cancer have also exhibited clinical and prognostic relevance of aberrant cancer-specific promoter DNA methylation of CDO1 gene, such as breast, esophageal, colorectal and gallbladder cancer.[8–10,12,16] Among such well-documented literatures, promoter DNA hypermethylation of CDO1 reproducibly represents more aggressive phenotypes independently of tumor stage in a multivariate prognostic analysis, and our current study also supported the hypothesis that CDO1 gene may have predictive value of prognosis in primary BTC.
There is great difference between the cut-off values that differentiated the tumor tissues from the corresponding normal tissues and the most optimal prognostic values. The former one is always lower than the latter one in various cancer such as colon (15.6, 20.5) and gallbladder (5.4, 17.7)[12,16], as in primary BTC (7.2, 28.9). It is hard to believe that a cut-off value discriminating normal and tissue in Taqman assay, which is from Q-PCR analysis, can represent a functional involvement of a protein expression.
Recent studies describing prognostic factors in primary BTC were R0 resection, lymph node metastasis, perineural invasion, portal vein and hepatic artery invasion, and preoperative value of CA19-9 [20–26], and our latest data identified independent prognostic factors of preoperative value of CA19-9 and perineural invasion.[25] However, there has been no established molecular biomarkers to improve management of primary BTC.[27] In this study, CDO1 methylation was able to clearly stratify the prognostic outcome of EHCC patients after excluding AC using the optimal cut off value of 28.9 according to log-rank plot analysis. Moreover, promoter DNA methylation status of the CDO1 gene is well associated with CDO1 protein expression in EHCC tumor tissues. These findings indicated that the CDO1 TaqMeth value of 28.9 has great clinical value to affect prognosis, reflecting its functional contribution in EHCC.
To further clarify the clinical utility in EHCC clinics, we focused on the EHCC patients with low preoperative serum value of CA19-9. Concurrent low group of CDO1 methylation and CA19-9 exhibited very excellent prognosis, and 5yOS rate reached as much as 89%. On the other hand, both high group and either high group of CDO1 methylation or CA19-9 level exhibited significantly dismal survival, and the prognosis after surgery remains unsatisfactory with the current treatment of surgery alone. Therefore, these poor groups should consider more effective adjuvant treatment after surgical resection than ever.
Recent study by Andresen et al. has described that cancer detection from ERCP biliary brush samples was robust by using CDO1 hypermethylation.[17] Furthermore, they rigorously explored epigenetic biomarkers of cholangiocarcinoma, and have identified 13 candidate genes which displayed high methylation frequencies using gene expression profiles of primary cholangiocarcinoma and representative genes methylated across multiple gastrointestinal cancer types.[17–19] The top 5 genes of AUC value were CNR1P1, TMEFF2, CDO1, MAL and SFRP1, and individual AUC of 0.93, 0.93, 0.91, 0.91 and 0.80, respectively, in the primary tumor tissues, while the best performance was seen in CDO1 gene in the liquid biopsy (ERCP solution) (AUC = 0.93 as compared to those of other genes, ranged from 0.8 to 0.9). More importantly, COD1 gene alone is comparable even with the combination analysis (AUC = 0.94) as a single methylation marker. In this study, we didn’t evaluate any other tumor suppressor genes, however we believe that CDO1 gene would be one of the most excellent cancer-specific biomarkers for diagnostic exploration of BTC at present.
We herein assessed CDO1 methylation in the tumor tissues and corresponding normal tissues using TaqMeth value as quantitative method.[7] Our quantitative analysis clearly showed high accuracy of diagnosis of cancer tissues from the corresponding normal tissues in AUC of 0.89. In this study, we have to concern that 8 out of 101 corresponding normal tissues were higher methylation than cut off value of 7.2 (this methylation contamination in the normal tissues reduced AUC in our study as compared to Anderson’s data, 0.91). Frequent CDO1 methylation of the corresponding normal samples from BTC individuals may suggest the existence of a potentially precancerous lesion.
The limitations of this study are a retrospective study design and low statistical power in AC patients. So, this study may suffer from bias, i.e., change in treatment strategy such as surgical procedure, range of lymph node dissection and application of chemoradiotherapy. Prospective validation is thus further needed to clarify the relationship of CDO1 promoter DNA methylation to prognosis in primary BTC.
In conclusion, we for the first time demonstrated that promotor DNA methylation of CDO1 gene could be a potential candidate of molecular diagnostic and prognostic biomarker in primary EHCC. CDO1 methylation status accurately indicates poor prognosis by combination with preoperative serum CA19-9 in the context of the modern treatment strategy. This information would be beneficial to identify patients with recurrence or long-term survival in the outpatient center, and to select the optimal strategies and postoperative surveillance for this type of cancer.
Supporting information
(XLSX)
(XLSX)
(TIF)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- 2.Tyson GL, Ilyas JA, Duan Z, Green LK, Younes M, El-Serag HB, et al. Secular trends in the incidence of cholangiocarcinoma in the USA and the impact of misclassification. Dig Dis Sci. 2014;59(12):3103–10. 10.1007/s10620-014-3276-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flemming JA, Zhang-Salomons J, Nanji S, Booth CM. Increased incidence but improved median overall survival for biliary tract cancers diagnosed in Ontario from 1994 through 2012: A population-based study. Cancer. 2016;122(16):2534–43. 10.1002/cncr.30074 [DOI] [PubMed] [Google Scholar]
- 4.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349(21):2042–54. 10.1056/NEJMra023075 [DOI] [PubMed] [Google Scholar]
- 5.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128(4):683–92. 10.1016/j.cell.2007.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamashita K, Upadhyay S, Osada M, Hoque MO, Xiao Y, Mori M, et al. Pharmacologic unmasking of epigenetically silenced tumor suppressor genes in esophageal squamous cell carcinoma. Cancer Cell. 2002;2(6):485–95. [DOI] [PubMed] [Google Scholar]
- 7.Brait M, Ling S, Nagpal JK, Chang X, Park HL, Lee J, et al. Cysteine dioxygenase 1 is a tumor suppressor gene silenced by promoter methylation in multiple human cancers. PLoS One. 2012;7(9):e44951 10.1371/journal.pone.0044951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Minatani N, Waraya M, Yamashita K, Kikuchi M, Ushiku H, Kojo K, et al. Prognostic Significance of Promoter DNA Hypermethylation of cysteine dioxygenase 1 (CDO1) Gene in Primary Breast Cancer. PLoS One. 2016;11(1):e0144862 10.1371/journal.pone.0144862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ushiku H, Yamashita K, Katoh H, Ema A, Minatani N, Kikuchi M, et al. Promoter DNA methylation of CDO1 gene and its clinical significance in esophageal squamous cell carcinoma. Dis Esophagus. 2017;30(2):1–9. [DOI] [PubMed] [Google Scholar]
- 10.Kojima K, Yamashita K, Ushiku H, Katoh H, Ishii S, Tanaka T, et al. The clinical significance of cysteine dioxygenase type 1 methylation in Barrett esophagus adenocarcinoma. Dis Esophagus. 2017;30(3):1–9. [DOI] [PubMed] [Google Scholar]
- 11.Ushiku H, Yamashita K, Ema A, Minatani N, Kikuchi M, Kojo K, et al. DNA diagnosis of peritoneal fluid cytology test by CDO1 promoter DNA hypermethylation in gastric cancer. Gastric Cancer. 2017;20(5):784–92. 10.1007/s10120-017-0697-6 [DOI] [PubMed] [Google Scholar]
- 12.Kojima K, Nakamura T, Ohbu M, Katoh H, Ooizumi Y, Igarashi K, et al. Cysteine dioxygenase type 1 (CDO1) gene promoter methylation during the adenoma-carcinoma sequence in colorectal cancer. PLoS One. 2018;13(5):e0194785 10.1371/journal.pone.0194785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamashita K, Waraya M, Kim MS, Sidransky D, Katada N, Sato T, et al. Detection of methylated CDO1 in plasma of colorectal cancer; a PCR study. PLoS One. 2014;9(12):e113546 10.1371/journal.pone.0113546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deckers IA, Schouten LJ, Van Neste L, van Vlodrop IJ, Soetekouw PM, Baldewijns MM, et al. Promoter Methylation of CDO1 Identifies Clear-Cell Renal Cell Cancer Patients with Poor Survival Outcome. Clin Cancer Res. 2015;21(15):3492–500. 10.1158/1078-0432.CCR-14-2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meller S, Zipfel L, Gevensleben H, Dietrich J, Ellinger J, Majores M, et al. CDO1 promoter methylation is associated with gene silencing and is a prognostic biomarker for biochemical recurrence-free survival in prostate cancer patients. Epigenetics. 2016;11(12):871–80. 10.1080/15592294.2016.1241931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Igarashi K, Yamashita K, Katoh H, Kojima K, Ooizumi Y, Nishizawa N, et al. Prognostic significance of promoter DNA hypermethylation of the cysteine dioxygenase 1 (CDO1) gene in primary gallbladder cancer and gallbladder disease. PLoS One. 2017;12(11):e0188178 10.1371/journal.pone.0188178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andresen K, Boberg KM, Vedeld HM, Honne H, Jebsen P, Hektoen M, et al. Four DNA methylation biomarkers in biliary brush samples accurately identify the presence of cholangiocarcinoma. Hepatology. 2015;61(5):1651–9. 10.1002/hep.27707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andresen K, Boberg KM, Vedeld HM, Honne H, Hektoen M, Wadsworth CA, et al. Novel target genes and a valid biomarker panel identified for cholangiocarcinoma. Epigenetics. 2012;7(11):1249–57. 10.4161/epi.22191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vedeld HM, Andresen K, Eilertsen IA, Nesbakken A, Seruca R, Gladhaug IP, et al. The novel colorectal cancer biomarkers CDO1, ZSCAN18 and ZNF331 are frequently methylated across gastrointestinal cancers. Int J Cancer. 2015;136(4):844–53. 10.1002/ijc.29039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kondo S, Hirano S, Ambo Y, Tanaka E, Okushiba S, Morikawa T, et al. Forty consecutive resections of hilar cholangiocarcinoma with no postoperative mortality and no positive ductal margins: results of a prospective study. Ann Surg. 2004;240(1):95–101. 10.1097/01.sla.0000129491.43855.6b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kayahara M, Nagakawa T, Ohta T, Kitagawa H, Tajima H, Miwa K. Role of nodal involvement and the periductal soft-tissue margin in middle and distal bile duct cancer. Ann Surg. 1999;229(1):76–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakamoto Y, Kosuge T, Shimada K, Sano T, Ojima H, Yamamoto J, et al. Prognostic factors of surgical resection in middle and distal bile duct cancer: an analysis of 55 patients concerning the significance of ductal and radial margins. Surgery. 2005;137(4):396–402. 10.1016/j.surg.2004.10.008 [DOI] [PubMed] [Google Scholar]
- 23.Nimura Y, Kamiya J, Kondo S, Nagino M, Uesaka K, Oda K, et al. Aggressive preoperative management and extended surgery for hilar cholangiocarcinoma: Nagoya experience. J Hepatobiliary Pancreat Surg. 2000;7(2):155–62. 10.1007/s005340000070155.534 [DOI] [PubMed] [Google Scholar]
- 24.Nagakawa T, Kayahara M, Ikeda S, Futakawa S, Kakita A, Kawarada H, et al. Biliary tract cancer treatment: results from the Biliary Tract Cancer Statistics Registry in Japan. J Hepatobiliary Pancreat Surg. 2002;9(5):569–75. 10.1007/s005340200076 [DOI] [PubMed] [Google Scholar]
- 25.Kawamata H, Yamashita K, Nakamura K, Katagiri H, Ishii K, Takahashi Y, et al. Perineural invasion and preoperative serum CA19-9 as predictors of survival in biliary tract cancer. Anticancer Res. 2013;33(2):583–94. [PubMed] [Google Scholar]
- 26.Cai WK, Lin JJ, He GH, Wang H, Lu JH, Yang GS. Preoperative serum CA19-9 levels is an independent prognostic factor in patients with resected hilar cholangiocarcinoma. Int J Clin Exp Pathol. 2014;7(11):7890–8. [PMC free article] [PubMed] [Google Scholar]
- 27.Valle JW, Borbath I, Khan SA, Huguet F, Gruenberger T, Arnold D. Biliary cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2016;27(suppl 5):v28–v37. 10.1093/annonc/mdw324 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.