Abstract
The mitotic spindle is the microtubule-based apparatus that reliably segregates chromosomes during cell division. Recently, it was discovered that microtubules originate within the mitotic spindle by nucleating off of existing spindle microtubules. This mechanism, termed branching microtubule nucleation, allows the efficient amplification of microtubules while preserving their original polarity as required in the spindle. Three molecular players are known to be involved in this process, namely, the protein TPX2, the protein complex augmin, and the gamma-tubulin ring complex; however, little is known about the assembly of the protein complexes. Here, we use the eight-subunit augmin complex as an example of how to dissect the function and assembly of a protein complex using meiotic Xenopus egg extracts. Specifically, immunodepletion combined with total internal reflection fluorescence (TIRF) microscopy is used to identify the role of the protein complex. In parallel, immunoprecipitation (IP) and tandem mass spectrometry (MS/MS) are used to infer how it is assembled. This approach can be applied to investigate the assembly of other multisubunit protein complexes that function in branching microtubule nucleation and mitotic spindle assembly.
MATERIALS
It is essential that you consult the appropriate Material Safety Data Sheets and your institution’s Environmental Health and Safety Office for proper handling of equipment and hazardous materials used in this protocol.
RECIPES: Please see the end of this protocol for recipes indicated by <R>. Additional recipes can be found online at http://cshprotocols.cshlp.org/site/recipes.
Reagents
Antibodies targeting augmin subunits
Generate antibodies targeting augmin subunits, or order from a commercial vendor. Use immunoglobulin G (IgG) from rabbit serum (Sigma-Aldrich) as a control antibody.
CSF-arrested Xenopus laevis egg extract
Prepare CSF-arrested Xenopus laevis egg extract as described in Protocol: Preparation of Cellular Extracts from Xenopus Eggs and Embryos (Good and Heald 2018) with the following modification: The packing spin, during which the interstitial buffer is removed without lysing the eggs, should be performed in a table centrifuge at 150g for 60 sec, followed by 598g for 25 sec.
CSF-XB <R>
Cy5-labeled porcine tubulin (20 μm in CSF-XB) (Peloquin et al. 2005)
Dynabeads Protein A (Thermo Scientific)
Glycine (0.1 m [pH 2.5]) (optional; see Step 24)
mCherry-EB1 (2 μm in CSF-XB, purified as described by Petry et al. [2011])
RanQ69L (200 μm in CSF-XB, purified as described by Weis et al. [1996] and Petry et al. [2013])
Sodium orthovanadate (Vanadate) (10 mm in CSF-XB)
TBS-T (Tris-buffered saline [pH 7.4] with 0.1% Tween 20)
Equipment
Forceps
Glass coverslips (22 mm × 22 mm)
Immersion oil
Magnetic separation rack
Microcentrifuge tubes (1.5-mL)
Microscope slides
Nail polish
Tape (double-sided)
TIRF (total internal reflection fluorescence) microscope, equipped with a 100 × 1.49 NA objective, an electron-multiplying charge-coupled device (EM-CCD) or complementary metal-oxide semiconductor (CMOS) camera, and laser power of at least 20 mW out-of-fiber
METHOD
Coupling Antibodies to Magnetic Protein A Beads
Transfer 150 μL of Dynabeads Protein A suspension to a microcentrifuge tube.
Retrieve the beads using a magnetic separation rack. Exchange the buffer with 150 μL of TBS-T and resuspend the beads.
Repeat Step 2 twice.
-
Prepare a suspension of 35 μg of antibody targeting augmin subunits in TBS-T. Add the suspension to the beads and gently mix using a rotator overnight at 4°C or for 2 h at room temperature.
This antibody amount is above the maximum binding capacity of the beads.
After mixing, keep beads at 4°C until ready to proceed with Step 19.
Preparing the Flow Cell
-
5.
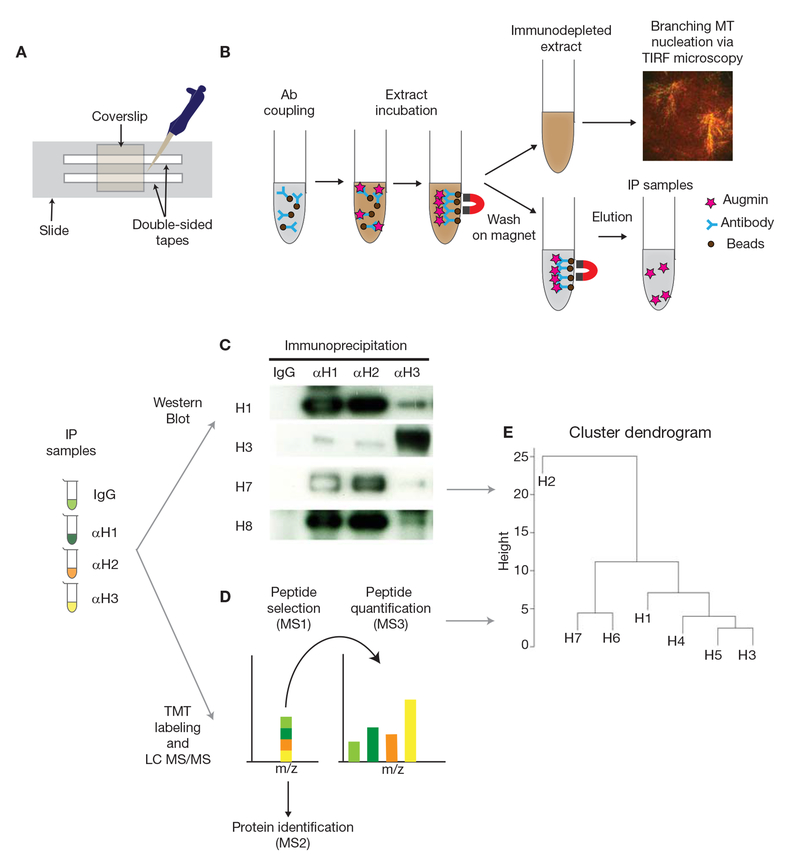
Place two strips of double-sided tape onto a microscope slide to make a flow channel with a 2- to 3-mm gap, which will accommodate ~5 μL of extract (Fig. 1A).
-
6.
Place a glass coverslip over the flow channel.
-
7.
Seal the flow cell by gently pressing onto the glass coverslip with the blunt end of a forceps.
FIGURE 1.
Schematic diagram of the protocol. (A) Preparation of the flow cell. (B) Immunoprecipitation (IP) and immunodepletion (ID) using Protein A Dynabeads and Xenopus egg extracts, followed by the branching microtubule nucleation assay via TIRF microscopy. (C) IP samples obtained using IgG, anti-HAUS1 (αH1), -anti-HAUS2 (αH2), and anti-HAUS3 (αH3) antibodies are analyzed by western blot to quantify the intensities of augmin subunits (H1-8). (D) Quantitative LC MS/MS using IP samples. Sequential mass spectrometry analyses allow identifying augmin subunits and relative amounts in each IP sample. (E) Hierarchical clustering dendrogram is generated by hclust package of R using normalized intensities of augmin subunits in each sample. Seven augmin subunits are depicted as H1-7 (H8 is not identified), showing a putative assembly model for augmin.
Performing the Branching Microtubule Nucleation Assay
-
8.
Aliquot 8.0 μL of CSF-arrested egg extract into a 1.5-mL tube on ice, pipetting as gently as possible.
-
9.
Gently add 0.5 μL of mCherry EB1, 0.5 μL of Vanadate, 0.5 μL of RanQ69L, and 0.5 μL of Cy5-tubulin to the extract to prepare a 10.0-μL reaction mixture.
Reagents should be individually and freshly added to the extract. Do not make a premixture of reagents.
-
10.
Gently pipette the extract up and down once or twice on ice to mix the reagents.
-
11.
Add the extract mixture to the flow cell.
-
12.
(Optional) Time the start of the reaction.
The reaction begins as soon as the extract mixture is introduced to the flow channel and warmed to room temperature.
-
13.
(Optional) Seal the flow cell with nail polish to prevent drying.
-
14.
Place the coverslip face-down on the 100× 1.49N objective of a TIRF microscope with immersion oil. Image the sample.
-
15.
Optimize the TIRF angle, laser power, and exposure time to visualize mCherry-EB1 and Cy5-microtubules at the best contrast and without photobleaching.
-
16.
Begin imaging the branching microtubule nucleation (Petry et al. 2013).
Microtubule nucleation activity can vary based on the surrounding temperature. A constant temperature of 18°C is ideal for imaging this reaction.
-
17.
(Optional) Collect images every 2 sec for 15–30 min.
-
18.
If the extract quality is sufficient to activate the branching reaction while maintaining low background nucleation, proceed to immunodepletion.
Performing Immunodepletion and Immunoprecipitation
A schematic diagram is provided in Figure 1B.
-
19.
Using the magnetic separation rack, wash the antibody-coupled beads from Step 4 three times with 150 μL of TBS-T followed by twice with 150 μL of CSF-XB.
-
20.
Remove the CSF-XB from the beads and place the tubes on ice.
-
21.
Add 65 μL of CSF-arrested egg extract to the antibody-conjugated beads. Mix the extract with the beads by gentle pipetting.
Wide-bore tips are preferred for mixing the extract.
-
22.
Incubate the extract with the beads on ice for a total of 45 min, gently mixing every 15 min.
Do not use a rotator to mix the extract, as mechanical stress to the extract should be minimized.
-
23.
Using the magnetic separation rack, retrieve the immunodepleted extract from the beads and transfer it to a fresh tube on ice until ready to proceed with Step 25.
Because of the viscosity of the extract, it takes 10 min to completely separate the extract from the beads on the magnetic separation rack.
-
24.
(Optional) For further analysis of the immunoprecipitation samples, wash the beads three times with 150 μL of TBS-T and then elute with 100 μL of 0.1 m glycine (pH 2.5).
Samples can be further analyzed by SDS-PAGE and western blot or mass spectrometry (see Discussion).
Performing Branching Microtubule Nucleation Using Immunodepleted Extracts
-
25.
Prepare 8.0-μL aliquots of immunodepleted extract samples in tubes on ice.
-
26.
Perform the branching reaction and initiate imaging using TIRF microscopy as described in Steps 9–17.
-
27.
Compare the activities of the branching reactions among immunodepleted extract samples.
DISCUSSION
This method provides an initial approach to analyzing the endogenous assembly of augmin (or other protein complexes), which is essential for branching microtubule nucleation, in Xenopus laevis egg extracts. Additional approaches include the analysis of immunoprecipitated samples by western blot, which can be used to calculate the relative intensities of subunits in a protein complex (Fig. 1C). For better quantification, liquid chromatography (LC) MS/MS analysis can be performed. The best technique for this purpose is multiplexed tandem mass tag labeling, in which each IP sample is individually labeled with the TMTsixplex isobaric label reagent set (Thermo Scientific). Orbitrap LC MS/MS is performed to identify peptides and quantify proteins (Fig. 1D; Eng et al. 1994; Ting et al. 2011; Wuhr et al. 2012; McAlister et al. 2014). After normalizing the protein signals to the total protein intensity across all samples, hierarchical clustering analysis can be performed using the normalized intensities of augmin subunits by each IP sample to understand the assembly of the protein complex (Fig. 1E; here hclust package of Ris used to generate the hierarchical clustering dendrogram). Last, the LC MS/MS results can be compared with the quantified Western blot data to obtain a more accurate model for the architecture of the protein complex studied.
With a single experimental setup, this protocol includes immunodepletion for functional investigation and immunoprecipitation for assembly analysis of the protein complex. Furthermore, these results guide the design of subunit constructs for in vitro reconstitution and structural studies. Although this protocol is described for augmin, it is applicable for any multisubunit protein complex in Xenopus extract that is involved in mitotic spindle assembly.
RECIPES
CSF-XB
| Reagent | Amount to add |
|---|---|
| 1 m HEPES (pH 7.7, adjusted with KOH) | 5 mL |
| XB salts (20×) <R> | 25 mL |
| 0.5 m EGTA (pH 8.0, adjusted with KOH) | 5 mL |
| Sucrose | 50 g |
Combine the listed reagents in abeaker. Add β-mercaptoethanol to a final concentation of 6 mM and bring the volume to 500 mL with deionized H2O (18.2 MΩ-cm). Stir with a magnetic bar to dissolve sucrose and mix reagents. Adjust to pH 7.8 if necessary. Store at 4° C.
XB Salts (20×)
2 mm CaCl2
2 m KCl
20 mm MgCl2
Filter-sterilize. Store at 4°C
ACKNOWLEDGMENTS
We thank Matthew R. King for critical reading of the manuscript and helpful comments. This work was supported by the National Institutes of Health New Innovator Award (DP2), the Pew Scholars Program in the Biomedical Sciences, and the David and Lucile Packard Foundation (all to S.P.).
REFERENCES
- Eng JK, McCormack AL, Yates JR. 1994. An approach to correlate tandem mass-spectral data of peptides with amino-acid-sequences in a protein database. J Am Soc Mass Spectrom 5: 976–989. [DOI] [PubMed] [Google Scholar]
- Good MC, Heald R. 2018. Preparation of cellular extracts from Xenopus eggs and embryos. Cold Spring Harb Protoc doi: 10.1101/pdb.prot097055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlister GC, Nusinow DP, Jedrychowski MP, Wuhr M, Huttlin EL, Erickson BK, Rad R, Haas W, Gygi SP. 2014. MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal Chem 86: 7150–7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peloquin J, Komarova Y, Borisy G. 2005. Conjugation of fluorophores to tubulin. Nat Methods 2: 299–303. [DOI] [PubMed] [Google Scholar]
- Petry S, Pugieux C, Nedelec FJ, Vale RD. 2011. Augmin promotes meiotic spindle formation and bipolarityin Xenopus egg extracts. Proc Natl Acad Sci 108: 14473–14478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petry S, Groen AC, Ishihara K, Mitchison TJ, Vale RD. 2013. Branching microtubule nucleation in Xenopus egg extracts mediated by augmin and TPX2. Cell 152: 768–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting L, Rad R, Gygi SP, Haas W. 2011. MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat Methods 8: 937–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis K, Dingwall C, Lamond AI. 1996. Characterization of the nuclear protein import mechanism using Ran mutants with altered nucleotide binding specificities. EMBO J 15: 7120–7128. [PMC free article] [PubMed] [Google Scholar]
- Wuhr M, Haas W, McAlister GC, Peshkin L, Rad R, Kirschner MW, Gygi SP. 2012. Accurate multiplexed proteomics at the MS2 level using the complement reporter ion cluster. Anal Chem 84: 9214–9221. [DOI] [PMC free article] [PubMed] [Google Scholar]