Abstract
Mobile elements (MEs) collectively contribute to at least 50% of the human genome. Due to their past incremental accumulation and ongoing DNA transposition, MEs serve as a significant source for both inter- and intra-species genetic and phenotypic diversity during primate and human evolution. By making use of the most recent genome sequences for human and many other closely related primates and robust multi-way comparative genomic approach, we identified a total of 14,870 human-specific MEs (HS-MEs) with more than 8,000 being newly identified. Collectively, these HS-MEs contribute to a total of 14.2 Mbp net genome sequence increase. Several new observations were made based on these HS-MEs, including the finding of Y chromosome as a strikingly hot target for HS-MEs and a strong mutual preference for SINE-R/VNTR/Alu (SVAs). Furthermore, ∼8,000 of these HS-MEs were found to locate in the vicinity of ∼4,900 genes, and collectively they contribute to ∼84 kb sequences in the human reference transcriptome in association with over 300 genes, including protein-coding sequences for 40 genes. In conclusion, our results demonstrate that MEs made a significant contribution to the evolution of human genome by participating in gene function in a human-specific fashion.
Keywords: mobile elements, human genome, evolution
1. Introduction
Like in most genomes of higher organisms, transposable elements or mobile elements (‘MEs’ hereafter) constitute a major component in the human genome. The percentage of MEs in the human genome was reported to be from 45 to 48.5%,1–3 and in this study we revised it to 52.1% based on the most recent reference genome sequences and ME annotation. This percentage is likely to increase slightly with further improvements of the human genome sequences, especially for the constitutive heterochromatin regions, and with tools capable of detecting more diverged and novel MEs.
In the human genome, MEs are mainly represented by retrotransposons, which propagate in a copy-and-paste fashion via transcribed RNAs as the intermediates, and they are divided into long-terminal repeat (LTR) and non-LTR retrotransposons.2,3 The LTR retrotransposon group is characterized by the presence of LTRs at the two ends of the internal viral sequences, and it is mainly represented by the endogenous retrovirus (ERVs) or human ERVs (HERVs). These ERVs came from exogenous virus affecting germline cells and integrating into the genomes during different stages of primate and human evolution, and they constitute ∼8% of the human genome.4 The non-LTR retrotransposon group consists of several very distinct types, including Short-INterspersed Elements (SINEs), Long INterspersed Elements (LINEs), and a chimeric type of repeat elements (SINE-R/VNTR/Alu or SVA). Non-LTR retrotransposons share the common properties of having a 3′ poly(A) tail and L1-based target-primed reverse transcription (TPRT) mechanism for retrotransposition.4,5 Among non-LTR MEs, Alu elements, representing the relative young and most successful SINEs by number in primates, have more than 1 million copies and contribute to ∼13% of the human genome.4,6 L1s, representing the dominant group of the LINEs, have more than half million copies and make the largest contribution to the human genome by sequence length (∼17%). L1s with full coding capacity are also responsible for transposing all non-LTR MEs.7,8 SVAs, emerged as the newest and most active group of retrotransposons that are mainly found within the hominid group of primates, have ∼5,000 copies and contribute to ∼0.1% of the human genome.9,10
Despite initially being considered to junk DNA, implying that they had no function,11 research work, mostly from the last two decades, has convincingly demonstrated that MEs make very significant contributions in shaping the evolution of genomes and impacting gene function via a variety of mechanisms. These mechanisms range from generation of insertional mutations and causing genomic instability, creation of new genes and splicing isoforms, and exon shuffling to alteration of gene expression and epigenetic regulation.12–22 Functioning also as endogenous insertional mutagens, MEs are known to be responsible for causing certain genetic diseases in humans.23,24
More recently, MEs are also shown to contribute to the generation of tandem repeats and providing definitive regulatory function or potentials. It was shown that at least 23% of all tandem repeats, another type of repetitive elements, were derived from transposable elements.25 In a recent study, Ward et al. demonstrated that under a heterologous regulatory environment, regulatory sites in MEs, including those specific to humans, can be activated to alter histone modifications and DNA methylation as well as expression of nearby genes in both germline and somatic cells.26 A profound implication of this observation is that lineage- and species-specific MEs can provide novel regulatory sites to the host genome, which can potentially regulate nearby genes’ expression in a lineage- and species-specific manner and lead to phenotypic differences. A very recent study added such an example by showing that an ERV element is responsible for regulating innate immunity in humans by controlling the expression of adjacent IFN-induced genes.27
Past and ongoing retrotransposition generates genetic diversity among species and among individuals within the same species.3,28–30 Therefore, analysis of species-specific MEs can help understand the roles of MEs in speciation and species-specific phenotypes. In the human genome, certain members from L1, Alu, SVA, and HERV families are still active in retrotransposition, and they are responsible for generating HS-MEs and MEs that are polymorphic among humans by the presence or absence of the insertions.8,31,32 So far, a few studies have also examined HS-MEs as being present in the human genome, but not in the orthologous regions of any other primate genomes.30,33,34 Among these, the study by Mills et al., representing the most comprehensive analysis of species-specific at the genome-scale, identified a total of 7,786 and 2,933 MEs that are specific to human and chimpanzee, respectively.30 This study was done with earlier versions of the human and chimpanzee genome sequences (GRHc35/hg17 and CGSC1.1/panTrol1.1), which contained more unsequenced regions and assembly errors, in particular for the chimpanzee genome, and with no other primate genome sequences available. Since then, the genome sequences of human and chimpanzee both have been subjected to several major improvements, and the genome sequences of many additional closely related primates have also become available.35–40 These additional primate genomes can be useful in providing complementary information to chimpanzee genome sequences for the analysis of HS-MEs. Motivated by these factors, we developed a more robust multi-way comparative genomic approach to compare the human genome with that of nine non-human primate genomes. We identified a total of ∼15,000 HS-MEs, among which more than 8,000 were reported as HS-MEs for the first time. This dataset represents a major improvement over prior studies and permitted us to provide a more complete and accurate picture about the recent DNA transposition in the human genome with several novel observations.
2. Materials and methods
2.1. Sources of primate genomic sequences
The human genome sequences used in this study was the latest version released in December 2013 (GRCh38/UCSC hg38). The most recent versions of genome sequences for nine other primate species were also included. These include chimpanzee (May 2016, CSAC Pan_tro 3.0/panTro5), gorilla (December 2014, NCBI project 31265/gorGor4.1), orangutan (July 2007, WUSTL version Pongo_albelii-2.0.2/ponAbe2), gibbon (October 2012, GGSC Nleu3.0/nomLeu3), green monkey (March 2014, Chlorocebus_sabeus 1.1/chlSab2), crab-eating macaque (June 2013, WashU Macaca_fascicularis_5.0/macFas5), rhesus monkey (November 2015, BCM Mmul_8.0.1/rheMac8), baboon (anubis) (March 2012, Baylor Panu_2.0/papAnu2), and marmoset (March 2009, WUGSC 3.2/calJac3). All genome sequences in fasta format and the corresponding RepeatMasker annotation files were downloaded from the UCSC genomic website (http://genome.ucsc.edu) onto our local servers for in-house analyses.
2.2. Identification of HS-MEs
2.2.1. Pre-processing of human MEs
Our starting list of MEs is those annotated in the human genome using RepeatMasker (http://hgdownload.soe.ucsc.edu/goldenPath/hg38/database/rmsk.txt.gz). Since RepeatMasker reports fragments of MEs interrupted by other sequences and internal inversions/deletions as individual ME entries, we performed a pre-process to integrate these fragments back to ME sequences representing the original retrotransposition events. Briefly, we examined partial MEs that are next to each other with a distance up to 50 kb, and checked their mapping positions in the repeat consensus sequence and orientation. If two neighbouring partial MEs map to the same repeat consensus sequence in the neighbouring non-overlapping regions and in the same orientation and show the presence of target-site duplications (TSDs) at the revised ME ends, we then treat these ME segments as one ME entry with the start and end positions adjusted accordingly. For LTR retrotransposons, RepeatMasker reports the two LTRs and the internal viral sequences each as a separate entry, and we grouped all components of a full-length LTR as one entry.
2.2.2. Identification of HS-MEs
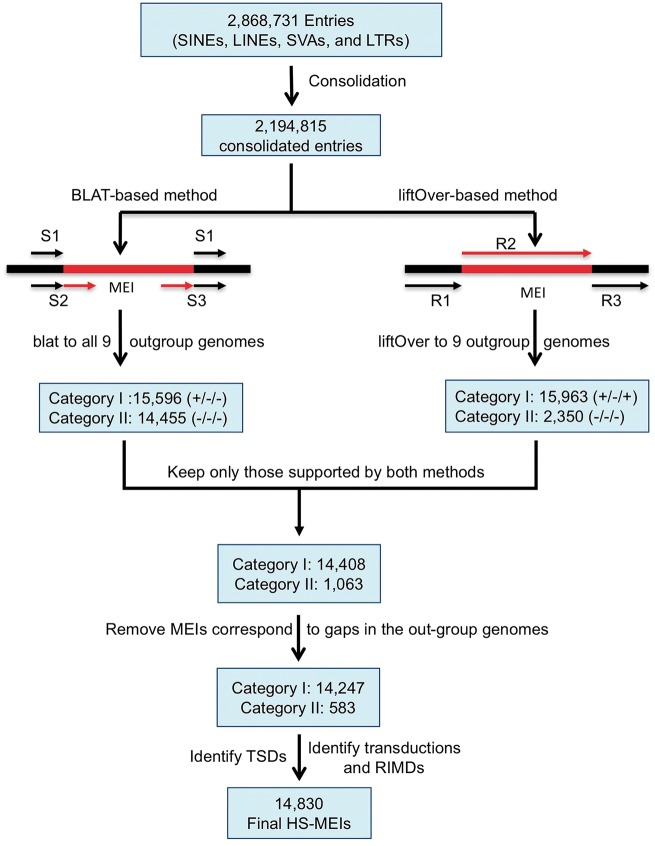
As shown in Fig. 1, our strategy for identifying HS-MEs is to examine the sequences at the insertion site and its two flanking regions for each of the MEs (after integration) annotated in the human reference genome and compare with the sequences of the orthologous regions in each of the nine non-human primate genomes. If an ME is determined with confidence to be absent from the orthologous regions of all other primate genomes, then it is considered human-specific. Briefly, we used two tools, BLAT41 and liftOver (http://genomes.ucsc.edu), for determining the orthologous sequences and the human-specific status of MEs using the aforementioned integrated RepeatMasker ME list as input. Only MEs supported to be unique to human by both tools were included in the final list of HS-MEs. Based on whether the MEs’ flanking sequences were detected in one or more of the out-group genomes, we divided our HS-MEs into two categories. Those with one or both of the flanking sequences detected in the orthologous region of the outgroup genomes are placed in Category I, representing HS-MEs with high confidence. Those with neither of the flanking sequences detected in the orthologous region of the out-group genomes are placed in Category II, likely representing HS-MEs that are located inside another human-specific sequences. A flow chart of the method is shown in Fig. 1, and further detailed description is provided in the Supplementary materials and methods.
Figure 1.
Flow chart of identifying HS-MEs.
2.3. Validation of HS-MEs
To assess the accuracy of our HS-MEs both in terms of false-positive and false-negative errors, we performed manual inspection using the UCSC genome browser with a set of 100 randomly sampled HS-ME candidates, and performed further validations using the following 3 methods.
2.3.1. Method 1
We compared our list against the previous HS-MEs list reported by Mills et al.30 and the human-specific HERV-Ks reported by Shin et al.34
2.3.2. Method 2
A total of 3,110 polymorphic ME entries present in the reference genome, representing 2,221 or 50% of entries documented in dbRIP,42 plus additional 779 entries identified by the 1000 Genome Project team43,44 were collected and cross-matched with the list of HS-ME candidates.
2.3.3. Method 3
A total of 15 randomly selected HS-ME entries were subjected to PCR validation using primate DNA samples (Details are provided in Supplementary materials and methods).
2.4. Identification of TSDs and analysis of sequence motifs at the ME integration sites
The TSDs as well as transduction and retrotransposon insertion mediated-deletions (RIMDs) for all HS-MEs were identified using in-house Perl scripts incorporating the utility of the NCBI bl2seq and UCSC liftOver tools. The method takes the human genomic sequence covering an HS-ME, plus the flanking sequence on each side and aligns them to the corresponding orthologous sequence from the chimpanzee genome (or the next closest genome with available orthologous sequences), which represents the pre-integration allele. In order to retrieve the orthologous pre-integration allele sequences, the orthologous positions of both flanking sequences of an HS-ME in the out-group genomes were identified using liftOver. For a typical Category I HS-ME, liftOver can find the orthologous region of the immediate flanking regions. However, the immediate flanking sequences of an HS-ME in human genome may not represent the pre-integration sequence due to transduction or RIMD events. Therefore, our scripts liftOver multiple subsequent blocks of flanking sequences retrieved in human genome onto the out-group genomes (100 bp per block for up to 5 kb). The liftOver results were then grouped together to identify the shortest orthologous region containing the pre-integration site, which was used to align against the two human flanking sequences using the NCBI bl2seq tool. The overlapped region in the pre-integration allele between the two aligned regions with the flanking sequences of the ME represents the TSD. For those with TSDs, a 30-bp sequence centred at each insertion site at the pre-integration alleles was extracted after removing the ME sequence and one copy of the TSDs from the ME alleles. The sequence motif for the regions covering 15 bp before and after the insertion sites was obtained using the weblogo tool (http://weblogo.berkeley.edu/logo.cgi).
Entries with identified TSDs and extra sequences between the ME and either copy of the TSDs were considered candidates for ME insertion-mediated transductions and were subject to further validation (see details in the Supplementary material). For entries without TSDs, if there are extra sequences at the pre-integration site in the out-group genomes, they were considered to be candidates for RIMD in the human genome, which were subject to further validation as described in the Supplementary materials and methods.
2.5. Analysis of HS-MEs in Y chromosome
The analysis of HS-MEs for Y chromosome required some special considerations for two reasons. First, authentic Y chromosome sequences are currently available only for human, chimpanzee, green monkey, rhesus and marmoset, and they are not as complete as for autosomal chromosomes.45 Also, since the sequences for the pseudoautosomal regions were basically copied from X chromosome of the same genome, we excluded these regions from the analysis for Y chromosome.
2.6. Analysis of HS-MEs’ association with genes and regulatory elements
The genomic coordinates of genes down to individual exons and coding regions were based on GENCODE46 and NCBI RefSeq annotation47 provided in the UCSC Genome Browser. The entire genome was divided into a non-redundant list of categorized regions in gene context as coding sequence (CDS), non-coding RNA, 5′-UTR, 3′-UTR, promoter (1 kb), intron, and intergenic regions using an in-house Perl script. This order of genic region categories as listed above was used to set the priority from high to low in handling overlaps between splice forms of the same gene or different genes. For example, if a region is a CDS for one transcript/gene and is a UTR or intron for another, then this region would be categorized as CDS.
For identifying HS-MEs overlapping with known transcriptional factor binding sites (TFBS), we used the ENCODE46 Transcription Factor ChIP-seq contained in the wgEncodeRegTfbsClusteredV3.bed.gz available from the UCSC Genome Browser site.
3. Results
3.1. Revised ME composition in the human genome and a summary of HS-MEs
To improve the accuracy in identifying HS-MEs and their TSDs and calculating the rate of retrotransposition, we pre-processed MEs annotated by RepeatMasker to integrate fragmented MEs to represent original ME events. This integration process led to a reduction of almost one million (1,180,428) in ME counts from the 5,520,017 RepeatMasker ME entries in NCBI38/hg38 to 4,339,589 (Supplementary Table S1). In the meantime, it resulted an increase of full-length MEs by 21% (data not shown). As seen in Supplementary Table S1, the number of annotated MEs showed a consistent increase in newer versions of the human reference genomes, especially between earlier updates, mainly due to improved coverage of sequenced regions. Notably, the proportion of MEs in the human reference genome increased from 48.8% for the earlier version3 to 52.1% in the latest version (GRCh38, December 2013) (Supplementary Table S1). Based on GRCh38, the copy numbers (after integration) and percentages of the genome for DNA transposons, L1s, Alus, LTRs, SVAs, and the ‘Others’ (all remaining MEs consisting of mostly non-L1 LINEs) are 295,956 (3.5%), 564,195 (17.8%), 1,132,541 (10.5%), 488,208 (9.1%), 4,933 (0.1%), and 1,733,490 (11.2%), respectively. To our knowledge, these data represent the most updated ME composition in the human reference genome.
Using a multi-way comparative genomic approach involving comparison of human genome sequences to that of nine other closely related primates as outlined in Fig. 1, we identified a total of 14,870 HS-MEs, consisting of 8,817 Alus, 3,912 L1s, 1,571 SVAs, and 530 HERVs. By the presence/absence of flanking sequence in the non-human primate genomes, there are 14,247 in category I and 583 category II (Table 1). Among these HS-MEs, a total of 8,049 MEs were reported as HS-MEs for the first time, while 6,738 entries were shared with the previously reported HS-MEs48 (Supplementary Table S2). The complete list of HS-MEs is provided in Supplementary file S1 and in the dbRIP database (http://dbrip.org).42
Table 1.
Summary of human-specific mobile elements (HS-MEs)
| ME type | RM count | Integrated count | HS-MEs |
|||
|---|---|---|---|---|---|---|
| Category I | Category II | Total | HS% | |||
| L1 | 962,085 | 563,594 | 3,654 | 258 | 3,912 | 0.7 |
| Alu | 1,181,072 | 1,129,987 | 8,626 | 191 | 8,817 | 0.8 |
| SVA | 5,397 | 4,928 | 1,512 | 59 | 1,571 | 31.9 |
| LTR | 720,177 | 496,306 | 455 | 75 | 530 | 0.1 |
| Total | 2,868,731 | 2,194,815 | 14,247 | 583 | 14,830 | 0.7 |
3.2. Validation of HS-MEs
Due to the complexity of the task in identifying species-specific MEs as further discussed later, data generated by computational methods are very likely to have both false positives and false negatives. While it is cost-prohibitive to experimentally validate all 14,830 HS-MEs due to the extremely large number, we managed to validate the accuracy of our HS-ME list in three ways in addition to manual checking of 100 randomly selected entries on the UCSC Genome Browser, which showed an accuracy of 98% (data not shown). First, we used polymorphic MEs as a test dataset to assess the sensitivity of method [true positive/(true positive +false negative)]. Here, we reason that polymorphic MEs represent most recent insertions into the human genomes, and thus they should be mostly, if not all, HS-MEs. Thus, any polymorphic MEs failed to be identified as human specific ME are considered as false-negatives. By cross-checking the HS-ME list with the 3,110 polymorphic MEs present in the reference genome (2,331 from dbRIP not used in training set and 779 from the 1000 genome project), we obtained a sensitivity of 95.5% (2,972/3,110) (data not shown).
Second, we cross-checked the HS-MEs with the 7,786 previously reported HS-MEs by Mills et al.,30 and we obtained 6,738 entries as shared (Supplementary Table S2). Among the 1,048 entries not on our list, we found that 904 actually represented false positives from the previous study and 26 were absent in the new version of the genome sequences, 47 entries for ME types not included in our input list (mainly due to the differences between different versions of Repeatmasker), and 71 entries represent false negatives in our list (these were added to the final list of HS-MEs; Supplementary Table S2). This converts to a sensitivity of 99.0% (6,738/6,738 + 71)) for our method. Since both the polymorphic MEs and those reported by Mills et al.30 are mostly from non-repetitive regions, which are less susceptible to errors in sequencing and computational analysis, the high ratio of overlapping is expected, and these were mostly identified as category I HS-MEs in our analysis. We further compared our list with the human-specific HERV-K list published by Shin et al.34 Among the 28 ME_IN entries, 26 of them overlap with our list while the other entries were not included in the final list due to low confidence.
Third, we performed validation using PCR, which is the gold standard for ascertaining MEs. We were able to validate 12 of the 13 randomly selected HS-ME loci, for which PCR was successful. This converts to a true positive rate (precision) of 92.3% or a 7% of false positive rate. Representative PCR results can be seen in Supplementary Fig. S1 and full details of all examined loci are provided in Supplementary Table S3.
Overall, combining the results from the above three methods of validation, we showed that our method has a minimal sensitivity of 95.5% and a precision rate of 92.3%.
3.3. Main sources for newly identified HS-MEs
We examined the contributions of different factors to the 8,049 novel HS-MEs, among ‘HS-MEs in MEs’, ME integration, non-canonical MEs (transduction, RMID, no-TSD), extra MEs in hg38 (over hg17) and use of multiple genomes, with each of these factors considered independently (redundancy exist among categories, Supplementary Table S4a) and combined in a step-wise order (Supplementary Table S4b). When each source considered independently, the top contributors of novel HS-MEs include ‘MEs in MEs’ (3,744) and non-canonical cases (3,256), followed by ME integration (972), use of multiple genomes (822), and new human genome (161) (Supplementary Table S4a). ME integration is the top contributor for novel HS-Alus, while the non-canonical MEs is the top contributor for L1, SVA, and LTR (Supplementary Table S4a). These sources collectively contributed to more than 6,000 non-redundant entries or 75% of the novel HS-MEs (Supplementary Table S4b). The remaining 25% of the novel HS-MEs are likely due to other factors such as improved version of chimpanzee genome. By percentage of novel HS-MEs among all HS-MEs across four ME types, LTR and L1 showed much higher ratios (78 and 73%, respectively) than Alu and SVA (46 and 44%, respectively) (Supplementary Table S4b).
3.4. HS-MEs contributed 14 Mbp net size increase to the human genome
ME insertions can lead to genome size increase via insertion of MEs, generation of TSDs, and transductions, and they can also reduce genome size via RIMD. As shown in Table 2, the occurrences of all four mechanisms and a net genome sequence increase were observed for each ME type. Collectively, all HS-MEs contributed to a total of 14.2 Mbp net genome size increase since the last common ancestor (LCA) with chimpanzee. Among ME types, L1s made the largest net increase (∼8.3 Mbp), followed by Alus (∼2.6 Mbp), SVAs (∼2.4 Mbp), and HERVs (0.8 Mbp). As expected, HS-L1s contributed to the largest size changes from insertion, IMD, and transductions due to their large insert size and high copy number, while HS-Alus contributed to more size increase via TSDs than any other ME types due to the largest insertions in number of HS-Alus. Despite having a low retrotransposition activity, LTRs did contribute ∼750 kb size increase to the genome with 530 human-specific copies. It is also interesting to note that by ratio of size increase in relation to all MEs in a type, SVA showed the highest ratio among all (Table 2), seemingly attributed by the combination of a large average size of insertions, the highest ratio of transduction, and the highest ratio of HS-MEs.
Table 2.
Impact of human-specific mobile elements (HS-MEs) on genome size (kb)
| ME type | HS-MEs | TSDs | Trans# | RIMD* | Net size | Increase ratio |
Transd# | |
|---|---|---|---|---|---|---|---|---|
| /1K ME | /Mbp ME | /1K ME | ||||||
| L1 | 8,422.3 | 38.5 | 380.7 | −521.5 | 8,320.0 | 14.7 | 15.9 | 0.7 |
| Alu | 2,567.5 | 111.4 | 187.9 | −265.7 | 2,601.0 | 2.3 | 8.4 | 0.2 |
| SVA | 2,417.9 | 20.2 | 194.5 | −75.0 | 2,557.5 | 518.5 | 604.8 | 39.4 |
| LTR | 834.3 | 2.0 | 33.4 | −117.5 | 752.2 | 1.5 | 2.8 | 0.1 |
| Total | 14,242.0 | 172.0 | 796.5 | −979.7 | 14,230.8 | 6.5 | 12.9 | 0.4 |
Transd#, transduction; RIMD*, retrotransposition insertion-mediated deletion.
3.5. Retrotransposition activity level of MEs during early human evolution
The comprehensive list of HS-MEs provided us an opportunity to re-examine more accurately the activities of MEs in the human genome, especially during the earlier part of human evolution after separation from chimpanzees, which cannot be assessed using the polymorphic MEs. As shown in Supplementary Table S5, while many observations were similar to what we learned from prior studies based on younger polymorphic MEs.3,8,30,31 These include that (i) Alu, L1, SVA, and HERVs are the only four types of MEs with retrotransposition during human evolution and (ii) AluYa5 and AluYb8/9, L1HS, SVA_E/D, and HERV_K are the most active subfamilies for Alu, L1, SVA, and ERV, respectively. In addition, several notable differences were also observed. First, the ratio of HS-MEs in relation to all MEs in the most active subfamilies were much higher (10 times or more) than the corresponding ratios of polymorphic MEs8,31 (Supplementary Table S5). Second, a few active subfamilies not seen among polymorphic MEs were seen for HS-MEs, and these include AluYf, AluYk subfamilies, L1P4, L1M, SVA_A, and ERV1 subfamilies (Supplementary Table S5).
3.6. Patterns of MEs and HS-MEs in the repetitive regions: SVAs are strongly biased for inserting into SVAs
With HS-MEs in repetitive regions available for the first time, we were interested in knowing whether there are any biases among MEs both as the sources and targets of retrotransposition insertions. We first examined the percentage of MEs inserting into other MEs for all MEs and for HS-MEs separately. For all MEs as the sources of insertions in MEs, there seems to be an increasing trend from LINEs (17.8%), DNA transposons (22.2%), LTRs (28.2%), SINEs (31.2%), and SVAs (36.5%) with the ratio of SVAs being more than doubled of that for LINEs (Supplementary Table S6a). When only HS-MEs were considered (Supplementary Table S6b), a similar trend (with DNA transposons absent as they were not included in this study) was seen, but the degree of difference among ME types is much smaller than for all MEs. Nevertheless, significant differences among ME types were still observed. For example, HS-Alus show the highest percentage inserted into MEs (46.5%), followed by HS-SVAs (40.7%), while HS-L1s and HS-LTRs have lower rates (35.5 and 35.3%) (Supplementary Table S6b).
For each ME type as the insertion targets of HS-MEs, we calculated the frequency of HS-MEs as the number of HS-MEs per Mbp of host ME sequences (Supplementary Table S6c). Among the ME types as targets, the HS-ME density follows a decreasing trend from high to low among LINEs (7.3), DNA transposons (5.0), LTRs (4.0), and SINEs (1.8), seemly correlating with their overall age in the genome from old to new. However, very interestingly, SVA seems to be the striking outlier to this trend by having the highest density of HS-MEs (13.0) despite being the youngest ME type (Supplementary Table S6c). HS-Alus seem to be more frequently inserting into MEs than all other three ME types when all MEs are considered as targets seemly due to the highest number of HS-Alus. HS-Alus showed the highest ratio in LINEs, LTRs, and SINEs, but not for SVAs. Remarkably, HS-SVAs seem to show an unusually high preference for SVAs as targets at a frequency that is at least 18 times higher than the ratio of HS-SVAs into any other ME types (12.7 vs. 0.7 for LINE) (Supplementary Table S6c). In the meantime, HS-SVAs are also the most frequent seen HS-MEs in SVAs at a frequency that is more than 60 times higher than for other MEs (12.7 vs. 0.2 for HS-Alus and 0 for HS-L1s and HS-LTRs) (Supplementary Table S6c).
3.7. Y chromosome is a hot target for HS-LTRs
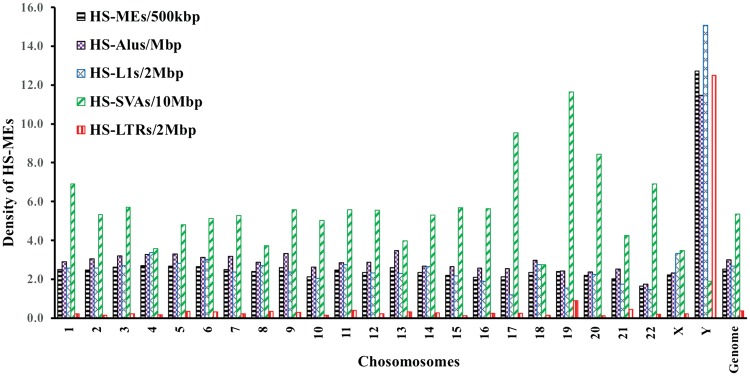
To examine the distribution pattern of HS-MEs in the genome, we measured the HS-ME density (the number of HS-MEs per million base pairs of non-gaped chromosome sequences) and the ratio of HS-MEs among the same type of MEs in the chromosomes for each ME type or all types combined. As shown in Fig. 2 and Supplementary Table S7, with all types combined, the HS-ME density varies across chromosomes with Y chromosome showing the highest density (25.4 copies/Mbp), more than 5 times higher than the genome average (5 copies/Mbp). The pattern is different among the 4 ME types (Fig. 2). Both Alus and L1s showed a more or less homogenous density among autosomes. However, Y chromosome showed the highest density being four to five times higher than the genome average or more than 10 times higher than that of the chromosomes with the lowest density (Supplementary Table S7). SVAs showed relatively similar densities among autosomes, except for chromosome 19, which has a density of HS-SVA that is 2 times higher than the genome average. But for Y chromosome, opposite to what seen for HS-L1s and HS-Alus, the HS-SVA density is more than two times lower than the genome average. LTRs showed the most variable distribution among chromosomes, with very low densities for most chromosomes, but with high and extremely high density in chromosome 19, and Y, respectively. The HS-LTR density in Y chromosome (12.5 copies/2 Mbp) is more than 30 times higher than the genome average (0.4 copies /2 Mbp) (Supplementary Table S7 and Fig. S2). Therefore, Y chromosome seems to be a hot target for Alus, L1s, and LTRs with LTRs showing an extreme high preference over the rest of the genome, while it is the least preferred target for SVAs (Supplementary Fig. S2). Among other chromosomes, chromosome 19 stands out as having the highest density of MEs, genes, HS-SVAs, and HS-LTRs, while chromosome 22 seems to have the least HS-Alus, and HS-L1s. Correlation analysis showed a strong positive correlation of HS-SVA density with that of genes and with that of all MEs (R = 0.9 in both cases), while HS-L1s showed a moderate negative correlation with gene density (R = −0.4) (data not shown).
Figure 2.
Genome distribution of HS-MEs. Bar plots showing the density of HS-MEs for each type of MEs among 24 chromosomes and the entire genome.
3.8. The pattern of TSD length and integration site sequence motifs for HS-MEs
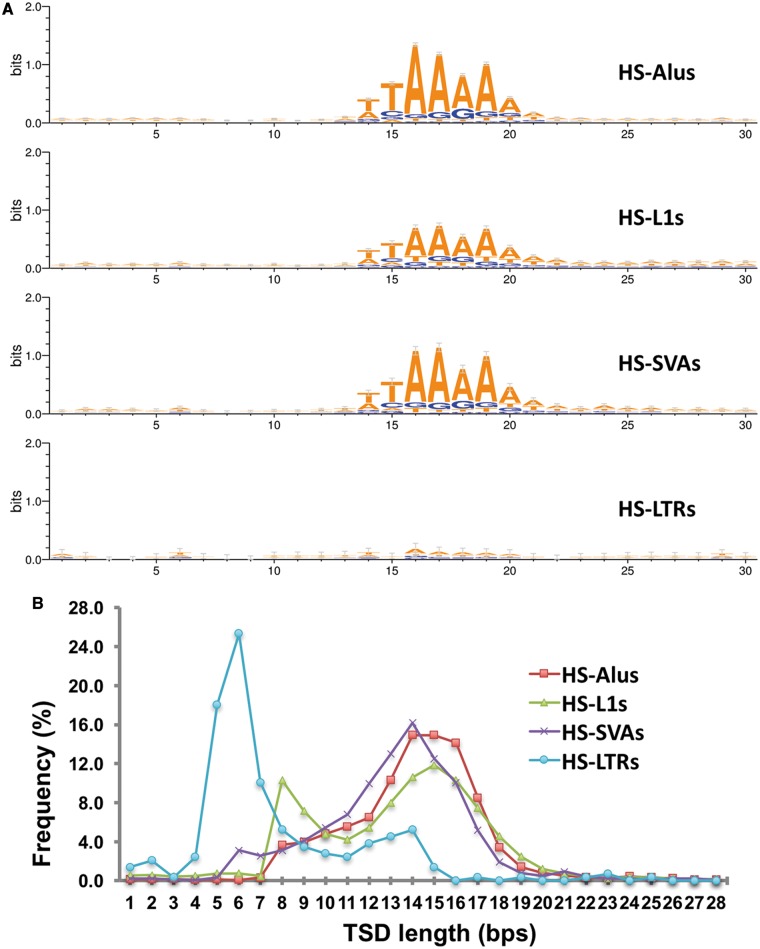
We also surveyed TSD length and insertion site sequence motifs for HS-MEs, and compared among the four ME types. As in Fig. 3A, Alus, L1s, and SVAs showed an identical core sequence motif of ‘TT/AAAA’, confirming that all non-LTR retrotransposition use the same TPRT mechanisms.9,42,49,50 LTRs showed basically no recognizable motif signal, an observation not reported before despite reported having certain site preferences.51 For the pattern of TSD length, as shown in Fig. 3B, all three non-LTR ME types showed a more or less similar distribution pattern with the TSD length peaking at the 15 bp. However, minor differences in the detailed pattern of the length distribution are also noticeable. For example, SVAs had a narrower peak, while Alus showed a flatter peak covering 14–16 bp. Interestingly, L1s showed a secondary peak at 8 bp in addition to the main peak at 15 bp, while LTRs also showed a minor peak at 14 bp in addition to the dominant peak at 6 bp. These data clearly indicate the differences of the retrotransposition mechanisms used by LTR and non-LTR insertions, as well as some minor differences among different types of non-LTR insertions.
Figure 3.
Characteristics of pre-integration sites and TSD lengths for HS-MEs. (A) Sequence logos for each HS-ME type at the integration sites. (B) Line plots showing the frequencies of TSDs at each length for each type of HS-MEs.
3.9. HS-MEs contribute to genes and regulatory elements
To predict the functional impact of HS-MEs, we analysed the gene context of their insertion sites based the most updated version (release 23 July 2015) of gene annotation data from the GENCODE project46 combined with those in NCBI RefGene.47 As shown in Table 3, a total of 7,547 HS-MEs, representing more than half of all HS-MEs, are located in protein coding genes and non-coding RNAs as well as transcribed pseudogenes, representing a total of 4,607 unique genes/transcripts (data not shown). While the majority of these HS-MEs are located inside introns (Table 3), a significant number (304) also directly participated in the transcriptomes as part of exon regions of protein coding genes, non-coding RNAs, and transcribed pseudogenes. Collectively, these 304 HS-MEs contributed to a total of 84 kb sequence in the reference transcriptome, which totals in ∼134 Mbp in length (Supplementary Table S8). Among those involved in the exons of protein-coding genes, 40 contribute to protein coding. In all these 40 cases, the HS-ME transcripts represent rare splice forms that have not been documented in the NCBI’s RefSeq genes, but reported in the GENCODE dataset, and interestingly SVAs contributed to 32 or 80% of these CDS HS-MEs (Supplementary Table S9).
Table 3.
Distribution of human-specific mobile elements (HS-MEs) in genic regions
| Gene region | Protein coding | Non-coding RNA | Transcribed pseudogenes | Total counts | % all HS-MEs |
|---|---|---|---|---|---|
| 1kb Promoter | 60 | 178 | 4 | 242 | 3.2 |
| CDS exon | 40 | NA | NA | 40 | 0.5 |
| Non-coding exons | 64 | 195 | 5 | 264 | 3.5 |
| Intron | 5,266 | 1,622 | 113 | 7,001 | 92.8 |
| Total | 5,430 | 1,995 | 122 | 7,547 | 100.0 |
We also examined the contribution of HS-MEs in regulatory elements, specifically the ENCODE ChIP-seq TFBS for 161 factors.52 Among the HS-MEs, 1,167 have sequence overlap with a total of 3,032 TFBS for 142 transcriptional factors (Supplementary Table S10). Interestingly, proportionally HS-LTRs showed the highest ratio (14.7%), followed by HS-SVAs (12.5%), which are much higher than that for LINEs (7.9%) and SINEs (6.6%), albert SINEs contribute to the largest number of TFBS (1,621), followed by LINEs (690), SVAs (504), and LTRs (217). Our data suggest that these young HS-MEs have started making their ways in participating in transcriptome, protein coding, and regulation of splicing and transcription.
3.10. Deposition and access of the HS-ME data
We deposited the HS-ME data into the dbRIP database (htpp://dbrip.org) under a study ID of 2018-01 (available for hg19). In dbRIP, the HS-MEs can be visualized in the same as the polymorphic MEs, i.e. in the UCSC genome browser along with other available data tracks or in detailed data page. HS-MEs were distinguished from polymorphic MEs in the dbRIP with a letter ‘h’ at the end of the ID. The HS-ME data (for hg38) in fasta format with the sequences organized into left flanking (400 bp), TSD1, ME insertion, TSD2, and right flanking (400 bp) and all other related information provided in the definition lines are available for downloading from the dbRIP data download page. The HS-ME data are also made available along with the polymorphic MEs data via Track Hub (dbRIP) at the UCSC genome browser site at http://genome.ucsc.edu.
4. Discussions
In this study, we aimed to provide a comprehensive compilation of MEs that are uniquely present in the human genomes. This is necessary resource for a comprehensive assessment of the MEs’ impacts on human biology. By taking the advantages of the much-improved reference genome sequences for humans and the closely related non-human primates and by tackling the MEs inside repetitive sequences and utilizing a more robust multi-way comparative genomics strategy, we were able to identify a total of 14,870 HS-MEs. Among HS-MEs, more than half (8,049) were reported as HS-MEs for the first time, thus representing a significant improvement. Such a comprehensive list of HS-MEs provides unique opportunities in examining the pattern and trend of retrotransposition during human genome evolution since the divergence from chimpanzee and gaining new insights regarding the roles of MEs in human evolution.
4.1. Challenges in identifying HS-MEs
As shown in Supplementary Table S1, the number of MEs annotated in the human reference genome increased significantly in the most recent version (GRC38, December 2013) compared with an earlier version released in 2004 (GRC35, UCSC hg17),53 which covered the first major updates since the initial publication of the human draft genome in 2001.1 MEs increased ∼289,000 in number and ∼140 Mb in sequence, leading to the increase of ME percentage in the human genome from 48.8% in GRC35 to 52.1% in GRC38 (based on non-gap sequences). The larger increase among ME type is for LTRs (∼34,000 in number and ∼16.5 Mbp in size. L1s had an increase of 42,000 in number and 21.8 Mbp in size. The increase for Alus is ∼12,300 in number and ∼675 kb in size (Supplementary Table S1).
Despite constant improvements in the quality of the reference genome sequences for human and other primates and of the related bioinformatics tools, obtaining a precise list of MEs uniquely present in the human genomes remains difficult with many challenges. Several factors contribute to the complications in this task, and these include but are not limited to: (i) incomplete coverage of the reference genome sequences for human and more so for other primates as exemplified in a few recent publications related to human and other primates36,54; (ii) assembly errors, particularly in regions rich of MEs; (iii) genome rearrangements occurring in a lineage- and species-specific fashion, often involving or mediated by MEs17–20; (iv) mis-annotation of MEs.
Prior similar studies on HS-MEs, best represented by Mills et al.,30 were limited by all these factors as well as by the limitation of methodologies and study scope that either focused on a specific type of MEs or part of the genome.8,33,55,56 Most of the MEs in the repetitive regions, including these in the ME-rich regions, were excluded from all previous studies.
4.2. Improvements for identification of HS-MEs
In our study, we tried to address most of the issues mentioned above. In addition to the use of the best available reference genome sequences for human and other primates, we took an unbiased approach to cover all annotated MEs in the human reference genome, thus representing the first study to include MEs in the repetitive regions, particularly those within the ME-rich regions. This has contributed to 3,744 HS-MEs or 47% of the 8,049 novel HS-MEs (Supplementary Table S4b). Furthermore, the process of integrating MEs in the original RepeatMasker provided a more accurate counting of the transposition events represented by the MEs in the human genome and locations of their flanking sequences. The former leads to more accurate DNA transposition rate, while the latter is critical for identifying HS-MEs and characterizing their sequences including the TSDs. As shown in Supplementary Table S1, the fragmentation affects a significant proportion of MEs (as high as ∼60% in L1s) with the rates seemly to be positively correlated with the element length and relative ages (data not shown). The integration led to the identification of 998 HS-MEs that would not have been identified otherwise, and this has in part contributed to the higher ratio of novel HS-MEs for LTRs and L1s than SVAs and Alus (Supplementary Table S4b). LTRs and L1s are much longer in average than SVAs and Alus, so they have a high chance of being interrupted by genomic rearrangement events, while full-length LTRs were also reported arbitrarily as three entries by RepeatMasker. The use of multiple non-human primate genomes not only helped reduce the false positive by providing orthologous sequences for gaps or regions of rearrangements in the chimpanzee genome, but also helped increase the sensitivity via contributing to 822 novel HS-MEs (Supplementary Table S4a).
The use of two sequence alignment tools, blat and liftOver, in identifying the orthologous sequences also helped reduce the false positives that can be caused by many of the aforementioned complicating factors. LiftOver uses alignment data linking closely related genomes through blastz,57 which focuses on large-scale synteny conservation, and is basically the strategy used in the previous analysis of human- and chimp-specific MEs by Mills et al.30 The method may work well for well conserved regions, but usually performs poorly for regions involving local rearrangements or with high density of repetitive sequences or misplacement of contigs during assembly. Therefore, it may miss to detect the presence of orthologous MEs and generate high false positives, most likely as category II HS-MEs (missing orthologous flanking regions). In contrast, the blat method focuses on identifying local alignments and performs better in handling regions with species-specific sequence changes near MEs. But it may miss some true HS-MEs by generating some false positive detection of orthologous MEs due to non-orthologous sequence similarity. Therefore, by requiring support of both methods for calling an HS-ME status, we were able to achieve a minimal level of false positive rate in HS-ME detection with an estimated sensitivity and specificity of 95.4 and 97%, respectively. We would like to believe that our current list of HS-MEs may still represent an underestimate of all HS-MEs that may exist in the human genomes due to many complications associated with ME analysis and our emphasis on reducing the false positives. Notably, our list does not cover the processed pseudogenes, which represent copies of mRNAs that were copied back into the genome as a side-product of L1-based transposition,58,59 and the human-specific processed pseudogenes are likely to be in the order of hundreds based on the level of detected polymorphic levels.60,61
4.3. The pattern of HS-MEs in MEs
MEs in repetitive regions have been mostly ignored by prior studies since it is much more difficult to analyse than those inserted into unique genomic regions. Not only the repetitive regions are more prone to sequence assembly errors, it is also more challenging for computational analysis aiming at specifies-specific and polymorphic ME entries. In a sense, the ignorance of these MEs may be justified for locating in regions assumed to be ‘less functionally important’. However, this may not be true, and by ignoring them, we might be missing useful information. In this study, by taking an unbiased approached, we identified more than 3,700 HS-MEs in the ME regions, which count for approximately a quarter of all HS-MEs. These MEs allowed us to observe some interesting patterns regarding the trend of DNA transposition in the human genome and the different behaviours of different ME types.
When all MEs were considered, the rates of MEs that inserted inside MEs by ME type showed an increasing trend in the order of L1, DNA, LTR, SINE, and SVA (Supplementary Table S6a). We think these differential ratios reflect the relative overall ages of these ME types with the older groups (e.g. LINEs) having less MEs than the younger groups (e.g. SVAs) in the genome as the targets for insertions.
In comparison, the ratios of HS-MEs in MEs are more or less similar and lack a clear trend among the ME types ranging from ∼35% for both L1 and LTR to ∼46% for Alu and ∼41% for SVA (Supplementary Table S6b). This is expected since HS-MEs from all types share approximately the same time span in the human genome and thus they had the same amount of MEs as insertion targets. The differences we see here with HS-MEs may represent a more accurate reflection of the preferences for insertions into MEs among ME types. In other words, Alus are more likely to insert to MEs than L1s, LTRs, and SVAs.
An unusual observation was made when we examined each ME type as the targets of HS-ME insertion individually (Supplementary Table S6c). Here we examined the preference of each HS-ME type for each type of MEs as the targets. By the density of HS-MEs, L1, LTR, and Alu all showed a more or less consistent decreasing trend for inserting into LINEs, DNA transposons, LTRs, SINEs, and SVAs, seemly correlating with their overall age in the genome from old to new. This is expected as older MEs have more chance to be inserted by later MEs. For HS-SVAs, while the trend seems to be the same among LINEs, DNAs, LTRs, and SINEs as the targets, they showed a density in SVAs that is more than 120, 40, 30, 16 times higher than that in SINEs, LTRs, DNAs and LINEs, respectively (Supplementary Table S6c). Furthermore, SVAs have the highest density of HS-MEs among the ME types and with over 98% of HE-MEs in SVAs being SVAs and the rest 2% being Alus. This extreme bias of HS-SVAs for SVAs and vice versa is remarkable since SVAs are the youngest ME type in the human genome, and by random chance we would expect to see the lowest ratio of HS-MEs in SVAs. In fact, very few or no insertion of HS-MEs from L1s, LTRs, and Alus were seen in SVAs. The reason for this extremely strong preference for SVAs inserting into SVAs and its functional implication are to be investigated in future studies.
4.4. The retrotransposition activity level during early human evolution
In the last decade, we and others have extensively studied the profiles of active MEs and their relative retrotransposition levels in the human genome based on analysis of polymorphic MEs.8,31,32,34,42–44,56,62–66 These studies show that L1s, Alus, SVAs, and HERV-K are ME types remaining ongoing retrotransposition activity. The most active subfamilies for each ME type have also been established. It is worth to point out that these data reflect the retrotransposition profiles in the most recent phase of modern human evolution, during which humans migrates and established as distinct populations, and the rate of retrotransposition in this period might not necessarily be the same as for the earlier part of human evolution. The availability of a comprehensive list of HS-MEs in relation to chimpanzee and other non-human primates can be used to fill this gap.
As expected, our data confirmed the active retrotransposon subfamilies identified by prior studies, including, Ya5 and Yb8/9 for Alus, L1HS for L1s, the F-subfamily for SVAs, and HERV-K for LTR retrotransposons (Supplementary Table S5). In the meantime, notable differences were also observed. First, the ratios of HS-MEs in relation to all MEs in the same subfamilies are much higher than that for polymorphic MEs. Second, many extra active subfamilies were observed, and these include the AluYf and AluYk subfamilies among Alus, the L1P4 and L1M subfamilies for L1s, SVA_A subfamily, and ERV1 subfamily for LTR retrotransposons (Supplementary Table S5). While the higher ratios of HS-MEs compared with that of polymorphic MEs in each active subfamily may be explained merely by the longer time span covered by HS-MEs, the presence of the extra active subfamilies not seen based on polymorphic MEs might be a result of the longer time-span for HS-MEs and the dropping retrotransposition activity in the modern human genomes.
4.5. Y chromosome as a hot target for HS-LTRs
As shown in Supplementary Table S7, Fig. S2, and Fig. 2, the HS-ME density on Y chromosome is significantly higher than all other chromosomes. By ME type, HS-MEs from Alus, L1s, and LTRs all showed a higher density for Y chromosome. For Alus and L1s, the HS-ME density in Y chromosome is at least 4 times higher than the genome average, while for HS-LTRs, the density in Y chromosome is more than 30 times higher than the genome average. In contrary, HS-SVAs showed a strong bias against Y chromosome with the density being only ∼35% of the genome average. This differential bias for Y chromosome among different types of HS-MEs cannot be a result of artefacts, such as poorer sequence quality or lack of Y-chromosome-specific sequences for other primates, as such factors would lead to the same trend of bias for all ME types. It does not seem to be explainable by the gene density either, since chromosome 13 has a lower gene density than Y chromosome, but its HS-ME densities for all 4 types of HS-MEs do not show a strong deviation from the genome average (Supplementary Table S7). The extreme contrast between the densities of SVAs and LTRs for Y chromosome as well as the differences between Y chromosome and all other chromosomes for the density of all HS-MEs are well visualized in the genome plots of HS-MEs shown in Supplementary Fig. S2. The high preference for Y chromosome by HS-MEs for Alus and L1s may be partially explained by the lack of homologous recombination-based deletion and lack of selection pressure and relative longer time in male germline.67,68 However, we cannot explain the extreme strong positive bias for HS-LTRs and negative bias for HS-SVAs. Therefore, the exact reason behind the significant differential bias for different types of HS-MEs in Y chromosome is unknown, and so is the impact of such biases. Nevertheless, our observation does seem to support a recent notion that remodelling and regeneration have dominated chimpanzee and human male specific chromosome evolution, while genetic decay seems to be a general trend in the evolution of Y chromosomes.45 In addressing the recent heated debate among the science community about whether chromosome Y is disappearing,69 our result seems to hint that Y chromosome is certainly not disappearing for human and HS-MEs may have contributed to the observed fast evolving pattern on Y chromosome after the human and chimpanzee divergence.45
4.6. The impact of MEs on human genome size and gene function
The 14,870 HS-MEs collectively contributed a net increase of the genome size by 14.2 Mbp since LCA with chimpanzee (Table 2). This size of genome increase is close to one-quarter of the Y chromosome and is larger than the genomes of free-living eukaryotic organisms, such as yeast.70 This size increase is significant for the relative short period of time and by a single molecular mechanism, and it may represent only well-defined significant change in the human genome during human evolution. The only other molecular mechanism that could contribute a significant size change to the human genome would be genomic segmental duplication.71 However, no data are available about the exact amount of human-specific segmental duplications occurred for the same period of human evolution.
We attempted to assess the overall functional impact of these HS-MEs on genes by examining their location in the genome in relation to known genes and functional regulatory elements. A total of 7,547 or 50.7% of these HS-MEs are located inside or in the 1 kb promoter regions of genes for protein coding, non-coding RNAs, as well as transcribed pseudogenes (Table 3), which represent 4,607 unique genes/transcripts (data not shown). Among these, 240 HS-MEs are part of transcripts, representing mostly alternative splice forms. Interestingly, in 40 of these cases, an HS-ME contributes to part of the coding region in the transcript, albeit all of these transcripts represent rare splice forms documented the GENCODE, but are not yet included in NCBI Ref Gene list (Supplementary Table S9). A profound observation here is that 80% of these CDS HS-MEs are contributed by SVAs, which represent the youngest and most active group of MEs in the human genome.10,31 This pattern seems to hold true when all MEs were included in the analysis of MEs’ contribution to human transcriptome (Joshi et al., manuscript in preparation). These data suggest to us that SVAs as the youngest and most active group of MEs in the human genome might have played a significant role in past human evolution and have the highest potential among all ME types in impacting future human genome evolution.
Outside of the promote and exon regions, 1,167 of the HS-MEs contribute to 3,032 binding sites for 142 of the 161 examined transcriptional factors (Supplementary Table S10). While their specific functional impact would be hard to predict computationally and can only be more accurately assessed/validated experimentally, many examples of such functional impact have been demonstrated.14,21
In summary, our data suggest that, despite being very young in the genome, many of these HS-MEs have already participated in gene function via regulation of transcription, splicing, and protein coding, and there may be more potentials for their future participation as demonstrated by Ward et al.26
4.7. Future directions
Due to the technical challenges associated with the analysis of MEs and deficiencies of the reference genome sequences for human and other primates, our list of HS-MEs still suffers a certain level of false negatives and false positives. We can expect that the number of HS-MEs continue to increase from regions with sequencing gaps, especially regions highly rich of repetitive sequences, such as the centromere and telomere regions, which may be hot spots for certain types of ME insertion, such as LTRs.72 A good proportion of the HS-MEs would be shared by other archaic human species/subspecies, notably the Neanderthals and Denisovans.73–75 Once high quality genome sequences become available for these genomes, we can further break down the human evolution into more phases and examine and compare the DNA transposition profiles among these periods.76 It would be interesting to find out see how many of these HS-MEs are truly unique to Homo sapiens. Furthermore, a certain portion of the HS-MEs are polymorphic, and it should be useful to generate a list of HS-MEs common to all humans (minimal set of HS-MEs) and another list of HS-MEs that are polymorphic (including also those outside of the reference genome). The former would be useful for analysing MEs’ impact on evolution of all modern humans, while the latter would be useful for studying MEs’ contribution to genetic and phenotypic diversity among human populations and individuals. Additionally, we are extending similar analysis to all other primate genomes, for which genome sequences are available, to access the impact of retrotransposition on primate evolution (work in progress).
Supplementary Material
Acknowledgements
This work is in part supported by grants from the Canadian Research Chair program, Canadian Foundation of Innovation, Ontario Ministry of Research and Innovation, Canadian Natural Science and Engineering Research Council (NSERC), and Brock University to PL, and was made possible by Compute Canada/SHARCNET high performance computing facilities.
Conflict of interest
None declared.
References
- 1. Lander E.S., Linton L.M., Birren B., et al. 2001, Initial sequencing and analysis of the human genome, Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 2. Deininger P.L., Moran J.V., Batzer M.A., Kazazian H.H. Jr. 2003, Mobile elements and mammalian genome evolution, Curr. Opin. Genet. Dev., 13, 651–8. [DOI] [PubMed] [Google Scholar]
- 3. Cordaux R., Batzer M.A.. 2009, The impact of retrotransposons on human genome evolution, Nat. Rev. Genet., 10, 691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kazazian H.H., Jr 2004, Mobile elements: drivers of genome evolution, Science, 303, 1626–32. [DOI] [PubMed] [Google Scholar]
- 5. Ostertag E.M., Kazazian H.H. Jr. 2001, Biology of mammalian L1 retrotransposons, Annu. Rev. Genet., 35, 501–38. [DOI] [PubMed] [Google Scholar]
- 6. Batzer M.A., Deininger P.L.. 2002, Alu repeats and human genomic diversity, Nat. Rev. Genet., 3, 370–9. [DOI] [PubMed] [Google Scholar]
- 7. Kazazian H.H. Jr, Goodier J.L.. 2002, LINE drive. retrotransposition and genome instability, Cell, 110, 277–80. [DOI] [PubMed] [Google Scholar]
- 8. Mills R.E., Bennett E.A., Iskow R.C., Devine S.E.. 2007, Which transposable elements are active in the human genome? Trends Genet., 23, 183–91. [DOI] [PubMed] [Google Scholar]
- 9. Ostertag E.M., Goodier J.L., Zhang Y., Kazazian H.H. Jr. 2003, SVA elements are nonautonomous retrotransposons that cause disease in humans, Am. J. Hum. Genet., 73, 1444–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang H., Xing J., Grover D., et al. 2005, SVA elements: a hominid-specific retroposon family, J. Mol. Biol., 354, 994–1007. [DOI] [PubMed] [Google Scholar]
- 11. Doolittle W.F., Sapienza C.. 1980, Selfish genes, the phenotype paradigm and genome evolution, Nature, 284, 601–3. [DOI] [PubMed] [Google Scholar]
- 12. Symer D.E., Connelly C., Szak S.T., et al. 2002, Human l1 retrotransposition is associated with genetic instability in vivo, Cell, 110, 327–38. [DOI] [PubMed] [Google Scholar]
- 13. Szak S.T., Pickeral O.K., Landsman D., Boeke J.D.. 2003, Identifying related L1 retrotransposons by analyzing 3' transduced sequences, Genome Biol., 4, R30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Han J.S., Szak S.T., Boeke J.D.. 2004, Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes, Nature, 429, 268–74. [DOI] [PubMed] [Google Scholar]
- 15. Wheelan S.J., Aizawa Y., Han J.S., Boeke J.D.. 2005, Gene-breaking: a new paradigm for human retrotransposon-mediated gene evolution, Genome Res., 15, 1073–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mita P., Boeke J.D.. 2016, How retrotransposons shape genome regulation, Curr. Opin. Genet. Dev., 37, 90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Callinan P.A., Wang J., Herke S.W., Garber R.K., Liang P., Batzer M.A.. 2005, Alu retrotransposition-mediated deletion, J. Mol. Biol., 348, 791–800. [DOI] [PubMed] [Google Scholar]
- 18. Han K., Sen S.K., Wang J., et al. 2005, Genomic rearrangements by LINE-1 insertion-mediated deletion in the human and chimpanzee lineages, Nucleic Acids Res., 33, 4040–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sen S.K., Han K., Wang J., et al. 2006, Human genomic deletions mediated by recombination between Alu elements, Am. J. Hum. Genet., 79, 41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Han K., Lee J., Meyer T.J., et al. 2007, Alu recombination-mediated structural deletions in the chimpanzee genome, PLoS Genet., 3, 1939–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Quinn J.P., Bubb V.J.. 2014, SVA retrotransposons as modulators of gene expression, Mob. Genet. Elements., 4, e32102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Konkel M.K., Batzer M.A.. 2010, A mobile threat to genome stability: the impact of non-LTR retrotransposons upon the human genome, Semin. Cancer Biol., 20, 211–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hancks D.C., Kazazian H.H. Jr. 2012, Active human retrotransposons: variation and disease, Curr. Opin. Genet. Dev., 22, 191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Callinan P.A., Batzer M.A.. 2006, Retrotransposable elements and human disease, Genome Dyn., 1, 104–15. [DOI] [PubMed] [Google Scholar]
- 25. Ahmed M., Liang P.. 2012, Transposable elements are a significant contributor to tandem repeats in the human genome, Comp. Funct. Genomics, 2012, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ward M.C., Wilson M.D., Barbosa-Morais N.L., et al. 2013, Latent regulatory potential of human-specific repetitive elements, Mol. Cell, 49, 262–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chuong E.B., Elde N.C., Feschotte C.. 2016, Regulatory evolution of innate immunity through co-option of endogenous retroviruses, Science, 351, 1083–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang C.R., Schneider A.M., Lu Y., et al. 2010, Mobile interspersed repeats are major structural variants in the human genome, Cell, 141, 1171–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kazazian H.H. Jr, Moran J.V.. 1998, The impact of L1 retrotransposons on the human genome, Nat. Genet., 19, 19–24. [DOI] [PubMed] [Google Scholar]
- 30. Mills R.E., Bennett E.A., Iskow R.C., et al. 2006, Recently mobilized transposons in the human and chimpanzee genomes, Am. J. Hum. Genet., 78, 671–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang J., Song L., Gonder M.K., et al. 2006, Whole genome computational comparative genomics: a fruitful approach for ascertaining Alu insertion polymorphisms, Gene, 365, 11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ahmed M., Li W., Liang P.. 2013, Identification of three new Alu Yb subfamilies by source tracking of recently integrated Alu Yb elements, Mob. DNA, 4, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Buzdin A., Ustyugova S., Khodosevich K., et al. 2003, Human-specific subfamilies of HERV-K (HML-2) long terminal repeats: three master genes were active simultaneously during branching of hominoid lineages, Genomics, 81, 149–56. [DOI] [PubMed] [Google Scholar]
- 34. Shin W., Lee J., Son S.Y., Ahn K., Kim H.S., Han K.. 2013, Human-specific HERV-K insertion causes genomic variations in the human genome, PLoS One, 8, e60605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chimpanzee S., Analysis C.. 2005, Initial sequence of the chimpanzee genome and comparison with the human genome, Nature, 437, 69–87. [DOI] [PubMed] [Google Scholar]
- 36. Gordon D., Huddleston J., Chaisson M.J., et al. 2016, Long-read sequence assembly of the gorilla genome, Science, 352, aae0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Locke D.P., Hillier L.W., Warren W.C., et al. 2011, Comparative and demographic analysis of orang-utan genomes, Nature, 469, 529–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Carbone L., Harris R.A., Gnerre S., et al. 2014, Gibbon genome and the fast karyotype evolution of small apes, Nature, 513, 195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rhesus Macaque Genome, S., Analysis C., Gibbs R.A., et al. 2007, Evolutionary and biomedical insights from the rhesus macaque genome, Science, 316, 222–34. [DOI] [PubMed] [Google Scholar]
- 40. Zimin A.V., Cornish A.S., Maudhoo M.D., et al. 2014, A new rhesus macaque assembly and annotation for next-generation sequencing analyses, Biol. Direct., 9, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kent W.J. 2002, BLAT—the BLAST-like alignment tool, Genome Res., 12, 656–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang J., Song L., Grover D., Azrak S., Batzer M.A., Liang P.. 2006, dbRIP: a highly integrated database of retrotransposon insertion polymorphisms in humans, Hum. Mutat., 27, 323–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stewart C., Kural D., Stromberg M.P., et al. 2011, A comprehensive map of mobile element insertion polymorphisms in humans, PLoS Genet., 7, e1002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sudmant P.H., Rausch T., Gardner E.J., et al. 2015, An integrated map of structural variation in 2, 504 human genomes, Nature, 526, 75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hughes J.F., Skaletsky H., Pyntikova T., et al. 2010, Chimpanzee and human Y chromosomes are remarkably divergent in structure and gene content, Nature, 463, 536–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Harrow J., Frankish A., Gonzalez J.M., et al. 2012, GENCODE: the reference human genome annotation for The ENCODE Project, Genome Res., 22, 1760–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pruitt K.D., Tatusova T., Maglott D.R.. 2007, NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins, Nucleic Acids Res., 35, D61–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mills R.E., Luttig C.T., Larkins C.E., et al. 2006, An initial map of insertion and deletion (INDEL) variation in the human genome, Genome Res., 16, 1182–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cost G.J., Boeke J.D.. 1998, Targeting of human retrotransposon integration is directed by the specificity of the L1 endonuclease for regions of unusual DNA structure, Biochemistry, 37, 18081–93. [DOI] [PubMed] [Google Scholar]
- 50. Jurka J. 1997, Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons, Proc. Natl. Acad. Sci. U.S.A., 94, 1872–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bushman F.D. 2003, Targeting survival: integration site selection by retroviruses and LTR-retrotransposons, Cell, 115, 135–8. [DOI] [PubMed] [Google Scholar]
- 52. Abecasis G.R., Altshuler D., Auton A., et al. 2010, A map of human genome variation from population-scale sequencing, Nature, 467, 1061–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. International Human Genome Sequencing, C. 2004, Finishing the euchromatic sequence of the human genome, Nature, 431, 931–45. [DOI] [PubMed] [Google Scholar]
- 54. Kim J.I., Ju Y.S., Park H., et al. 2009, A highly annotated whole-genome sequence of a Korean individual, Nature, 460, 1011–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Barbulescu M., Turner G., Seaman M.I., Deinard A.S., Kidd K.K., Lenz J.. 1999, Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans, Curr. Biol., 9, 861–8. [DOI] [PubMed] [Google Scholar]
- 56. Bennett E.A., Keller H., Mills R.E., et al. 2008, Active Alu retrotransposons in the human genome, Genome Res., 18, 1875–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Schwartz S., Kent W.J., Smit A., et al. 2003, Human-mouse alignments with BLASTZ, Genome Res., 13, 103–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kazazian H.H., Jr 2014, Processed pseudogene insertions in somatic cells, Mob. DNA, 5, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Casola C., Betran E.. 2017, The genomic impact of gene retrocopies: what have we learned from comparative genomics, population genomics, and transcriptomic analyses? Genome Biol. Evol., 9, 1351–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ewing A.D., Ballinger T.J., Earl D., et al. 2013, Retrotransposition of gene transcripts leads to structural variation in mammalian genomes, Genome Biol., 14, R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kabza M., Kubiak M.R., Danek A., et al. 2015, Inter-population differences in retrogene loss and expression in humans, PLoS Genet., 11, e1005579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Konkel M.K., Wang J., Liang P., Batzer M.A.. 2007, Identification and characterization of novel polymorphic LINE-1 insertions through comparison of two human genome sequence assemblies, Gene, 390, 28–38. [DOI] [PubMed] [Google Scholar]
- 63. Lee J., Cordaux R., Han K., et al. 2007, Different evolutionary fates of recently integrated human and chimpanzee LINE-1 retrotransposons, Gene, 390, 18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wildschutte J.H., Williams Z.H., Montesion M., Subramanian R.P., Kidd J.M., Coffin J.M.. 2016, Discovery of unfixed endogenous retrovirus insertions in diverse human populations, Proc. Natl. Acad. Sci. U.S.A., 113, E2326–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Iskow R.C., McCabe M.T., Mills R.E., et al. 2010, Natural mutagenesis of human genomes by endogenous retrotransposons, Cell, 141, 1253–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Witherspoon D.J., Zhang Y., Xing J., et al. 2013, Mobile element scanning (ME-Scan) identifies thousands of novel Alu insertions in diverse human populations, Genome Res., 23, 1170–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Abrusan G., Krambeck H.J., Junier T., Giordano J., Warburton P.E.. 2008, Biased distributions and decay of long interspersed nuclear elements in the chicken genome, Genetics, 178, 573–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Boissinot S., Entezam A., Furano A.V.. 2001, Selection against deleterious LINE-1-containing loci in the human lineage, Mol. Biol. Evol., 18, 926–35. [DOI] [PubMed] [Google Scholar]
- 69. Griffin D.K. 2012, Is the Y chromosome disappearing? Both sides of the argument, Chromosome Res., 20, 35–45. [DOI] [PubMed] [Google Scholar]
- 70. Cherry J.M., Ball C., Weng S., et al. 1997, Genetic and physical maps of Saccharomyces cerevisiae, Nature, 387, 67–73. [PMC free article] [PubMed] [Google Scholar]
- 71. Bailey J.A., Eichler E.E.. 2006, Primate segmental duplications: crucibles of evolution, diversity and disease, Nat. Rev. Genet., 7, 552–64. [DOI] [PubMed] [Google Scholar]
- 72. Zahn J., Kaplan M.H., Fischer S., et al. 2015, Expansion of a novel endogenous retrovirus throughout the pericentromeres of modern humans, Genome Biol., 16, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Prufer K., Racimo F., Patterson N., et al. 2014, The complete genome sequence of a Neanderthal from the Altai Mountains, Nature, 505, 43–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Fu Q., Hajdinjak M., Moldovan O.T., et al. 2015, An early modern human from Romania with a recent Neanderthal ancestor, Nature, 524, 216–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Disotell T.R. 2012, Archaic human genomics, Am. J. Phys. Anthropol., 149(Suppl 55), 24–39. [DOI] [PubMed] [Google Scholar]
- 76. Ahmed M., Liang P.. 2013, Study of modern human evolution via comparative analysis with the Neanderthal Genome, Genomics Inform., 11, 230–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.