Abstract
Seriola quinqueradiata (yellowtail) is the most widely farmed and economically important fish in aquaculture in Japan. In this study, we used the genome of haploid yellowtail fish larvae for de novo assembly of whole-genome sequences, and built a high-quality draft genome for the yellowtail. The total length of the assembled sequences was 627.3 Mb, consisting of 1,394 scaffold sequences (>2 kb) with an N50 length of 1.43 Mb. A total of 27,693 protein-coding genes were predicted for the draft genome, and among these, 25,832 predicted genes (93.3%) were functionally annotated. Given our lack of knowledge of the yellowtail digestive system, and using the annotated draft genome as a reference, we conducted an RNA-Seq analysis of its three digestive organs (stomach, intestine and rectum). The RNA-Seq results highlighted the importance of certain genes in encoding proteolytic enzymes necessary for digestion and absorption in the yellowtail gastrointestinal tract, and this finding will accelerate development of formulated feeds for this species. Since this study offers comprehensive annotation of predicted protein-coding genes, it has potential broad application to our understanding of yellowtail biology and aquaculture.
Keywords: Seriola quinqueradiata, genome sequencing, protein-coding genes, transcriptome analysis, digestive enzymes
1. Introduction
The genus Seriola (family Carangidae) contains nine recognized species of carnivorous, marine fish, globally distributed in tropical, subtropical and temperate ocean regions.1Seriola species have a high market value and several species are farmed around the world.2 Japan is the biggest producer of Seriola species, farming yellowtail (S. quinqueradiata), greater amberjack (S. dumerili) and yellowtail kingfish (S. lalandi). In 2016 the production of these three species was 141,000 tons, accounting for over 56% of marine finfish aquaculture in Japan.3 Most of this production is accounted for by yellowtail, which has the longest history of aquaculture, having started in 1927 and expanded rapidly in the 1960s.4 Although yellowtail aquaculture technologies have continued to improve, several challenges to the sustainable farming of this species remain to be addressed. The price of fish meal used for yellowtail feed continues to rise,5 necessitating a better understanding of yellowtail digestive pathways and nutritional requirements in order to develop more cost-effective feeds.
To improve aquaculture production efficiency, product quality, sustainability and profitability, recent efforts have focussed on the genetics and genomics of economically important aquaculture species, particularly following the development of high-throughput next-generation sequencing (NGS) technologies.6–8 Genetic maps9–11 and a whole genome radiation hybrid panel12 for yellowtail have been constructed, and the detection of quantitative trait loci (QTL) in yellowtail has recently been initiated for resistance to a monogenean parasite, Benedenia seriolae,13 and sex-determination.14,15 However, the resources for genes and protein annotations for this species are limited. Only 257 protein sequences have been deposited at NCBI Genbank, taxonomy ID: 8161 (as of 11 July 2018). A comprehensive annotation of the overall protein-coding genes for the yellowtail genome will help identify important genes and pathways involved in the regulation of economically important traits. In addition, a well-annotated genome will provide a powerful tool for better understanding yellowtail biology, and will enhance physiological studies for optimization of breeding technologies, including the development of artificial diets.
Recently, we demonstrated that use of the genome of a haploid gynogenetic yellowtail larva leads to a significant improvement in de novo assembly because allelic variation does not impede contig extension as it does for assembly of diploid genomes.16 Here, we present a draft genome sequence for yellowtail based on a haploid genome assembly and its annotation of protein-coding genes. Furthermore, to better understand molecular mechanisms of feed digestion and nutrient absorption in yellowtail, we performed RNA-Seq analysis of the three major organs of the gastrointestinal (GI) tract i.e. stomach, intestine and rectum, using the annotated draft yellowtail genome as a reference. The analysis suggested that certain proteolytic digestive enzymes play key roles in digestion and absorption, and could inform the choice of ingredients for the development of yellowtail-specific feeds for use in aquaculture.
2. Materials and methods
2.1. Genome sequencing and assembly
Preparation of genomic DNA from haploid gynogenetic yellowtail larvae induced by ultraviolet-irradiated sperm and the blood of its dam (diploid genome) has been described in.16 For contig construction, the genomic shotgun libraries were prepared from the haploid genomic DNA, and sequenced with the Ion PGM (two runs) and Proton (four runs) platforms (Life Technologies, Carlsbad, CA). These haploid reads were assembled using Newbler 3.0 (Roche Diagnostics, Basel, Switzerland). Four Illumina paired-end (PE) libraries (insert size: 240, 360, 480 and 720 bp) and three mate-pair (MP) libraries (insert size: 3–5, 10 and 20 kb) were constructed from the diploid dam genomic DNA and sequenced on a NextSeq 500 sequencer (Illumina, San Diego, CA, USA) using 2 × 150 bp chemistry. To improve sequence accuracy of the haploid contig sequences generated by Ion PGM and Proton sequencers, the Illumina PE reads were mapped against the haploid contigs using Burrows–Wheeler aligner-maximum exact matches (BWA-MEM),17 and nucleotide mismatches or short indels were overridden by the Illumina read sequences. We also carried out a de novo assembly of the four diploid Illumina PE libraries (all merged 240 bp, and 10 Gb of 360, 480 and 720 bp inserts) with Platanus 1.2.4,18 and performed a sequence comparison between the haploid and diploid contigs using BLASTN19 (E value threshold of 1E – 20 and the bit score of the top hit with a more than 2-fold difference over that of the second hit),20 and the diploid contigs absent in the haploid contigs were added to the haploid contigs. These merged contigs were subjected to scaffolding using Platanus 1.2.418 together with all the Illumina PE (unmerged 240, 360, 480 and 720 bp insert) and MP reads (3–5, 10 and 20 kb insert), and gaps in the scaffolds were filled by the Illumina PE reads (unmerged 240, 360, 480 and 720 bp inserts) with the gap-close step in Platanus. The resulting scaffolds were further assembled using RNA-Seq data sets generated in this study (described later), by L_RNA_scaffolder.21
To confirm ploidy level and estimate genome size, Jellyfish software22 was used to produce a k-mer distribution (19-mer) with the Ion Proton haploid reads and with the Illumina PE diploid reads (all merged 240 bp, and 10 Gb of 360, 480 and 720 bp inserts).
2.2. Linkage map construction and scaffold correction
To detect and correct scaffold misassemblies, we generated a double-digest restriction-site associated DNA (ddRAD)-based genetic linkage map of the yellowtail using a previously described method20 with slight modifications. Briefly, genomic DNA samples were extracted from 161 full-sib progeny and their parents’ muscles using the DNeasy Blood & Tissue Kit (Qiagen, Venlo, Netherlands). For preparation of the ddRAD libraries, genomic DNA was digested using two restriction enzymes (PstI and MspI) and ligated to Ion Plus Fragment Library Adapters using an Ion Xpress Plus gDNA Fragment Library kit (Life technologies). The ligation products from ∼25 individuals were pooled in equimolar proportions and size-selected within a range of 200–260 bp using BluePippin (Sage Science, Beverly, MA, USA). After size selection, the ddRAD libraries were amplified by eight-cycle PCR, purified using Agencourt AMPure XP beads (Beckman Coulter Inc., Brea, CA, USA), and sequenced on the Ion Proton (Life technologies). Raw sequences were filtered to discard those of low quality and demultiplexed using the process_radtags programme in Stacks.23 BWA-MEM17 was used to map the filtered ddRAD-Seq reads to the scaffold sequences, and the output SAM file was filtered using custom Perl scripts based on the following criteria: mapping quality (MQ) of ≥ 30, length fraction of 0.95 and non-specific match handling of ignore. After local realignment, single nucleotide polymorphism (SNP) calling was performed using the Unified Genotyper module in the Genome Analysis Toolkit (GATK)24 and the variants were filtered as follows: only bi-allelic sites, genotype quality (GQ) of ≥ 30, either reference (REF) or alternative (ALT) allele frequencies of ≥ 95% in homozygous sites and both REF and ALT allele frequencies of ≥ 30% in heterozygous sites. The R/qtl package25 was employed for linkage analysis and linkage map construction with the same parameters as previously described in.20 Linkage group (LG) numbering and orientation followed that of Fuji et al.10 To detect the possibility of misassembled chimeric scaffolds, the linkage marker sequences were mapped to the scaffold sequences with BLASTN using the same criteria as previously described in20; the scaffolds attributed to two or more LGs were split into consistent scaffolds based on the locations of markers. Scaffolds >2 kb in length were used for subsequent analysis.
2.3. Evaluation of completeness of the final assembly
A completeness assessment was performed with gVolante26 using the pipeline CEGMA (Core Eukaryotic Genes Mapping Approach)27 and the reference dataset Core Vertebrate Genes (CVGs).28 We also assessed the assembly quality by Benchmarking Universal Single-Copy Orthologues (BUSCO)29 version 3.0.2 with an Actinopterygii-specific set of 4,584 single copy orthologs.
2.4. cDNA sequencing
For gene predictions, normalized, full-length enriched cDNA libraries were constructed as follows: Total RNAs were isolated from eighteen yellowtail tissues (gills, skin, fins, red muscle, white muscle, heart, kidney, spleen, stomach, intestine, pyloric caeca, liver, gallbladder, retina, cerebellum, optic lobe, olfactory lobe and ovary) using RNeasy Plus Universal Mini Kit (Qiagen). The isolated total RNA samples were treated with 2 U of TURBO DNase from the TURBO DNase-free Kit (Life Technologies) at 37°C for 30 min as recommended by the manufacturer’s instructions. RNA integrity was assessed with an Agilent Bioanalyzer 2100 with the RNA 6000 Nano Kit (Agilent Technologies, Santa Clara, CA, USA). Poly(A) + RNA (mRNA) was purified using the FastTrack MAG Micro mRNA Isolation Kit (Invitrogen, Carlsbad, CA, USA). Equal amounts of mRNA were pooled from each tissue and 500 ng of the pooled mRNA was used to construct a full-length enriched cDNA library using the SMART PCR cDNA Synthesis Kit (Clontech Laboratories, Inc. Palo Alto, CA, USA) with modifications as described by Meyer et al.30 The primer used for first-strand cDNA synthesis was a modified 3’ SMART CDS Primer II A (AAGCAGTGGTATCAACGCAGAGTCGCAGTCGGTACTTTTTTCTTTTTTV) including a recognition site for the restriction enzyme BpmI,30 and the cDNA was amplified using the 5’ PCR Primer II A (AAGCAGTGGTATCAACGCAGAGT) with the KAPA library amplification kit (KAPA Biosystems, Boston, MA, USA). Amplified cDNA was purified using the QIAquick PCR Purification Kit (Qiagen) and normalized using a Trimmer-2 cDNA normalization kit (Evrogen, Moscow, Russia). The normalized cDNA was purified with the QIAquick PCR purification kit, and the purified cDNA was digested with BpmI for 1 h at 37°C to remove poly (A) tails. Finally, the digested cDNA was purified using a QIAquick PCR purification kit. In addition to the 18-tissue normalized cDNA library, additional full-length cDNA libraries for pituitary, testis, ovary haploid cells and digestive organs from larval fish (8–10 days post-hatch) were prepared individually with the SMART PCR cDNA Synthesis Kit employing the same procedure as above. Among these, the cDNA libraries for the digestive organs from the larval fish were normalized using the methods above. These cDNA libraries were sequenced with the 454 GS FLX+ (Roche Diagnostics), Ion PGM or Proton platforms (Life Technologies). Detailed information on the cDNA libraries with corresponding sequencers and sequence statistics are summarized in Supplementary Table S2.
2.5. Gene prediction and annotation
Protein-coding genes in the yellowtail genome were predicted using AUGUSTUS (version 3.1).31 First, repeat regions in the yellowtail scaffold sequences (>2 kb) were masked by RepeatMasker32 according to the yellowtail repeat database constructed by RepeatModeler.33 Next, the RNA-Seq reads of yellowtail sequenced in this study were mapped to the scaffolds using TopHat34 and assembled with Cufflinks.35 Then, the scaffold sequences were scanned with the Zebrafish model in AUGUSTUS, using the map information of RNA-Seq as hints. The predicted gene sequences (24,728 genes) were validated using BUSCO,29 and loci attributed to ‘complete’ (792 loci) were used to construct a yellowtail gene training model. Moreover, protein sequences of 11 fish species: spotted gar (Lepisosteus oculatus), Mexican tetra (Astyanax mexicanus), zebrafish (Danio rerio), Atlantic cod (Gadus morhua), Nile tilapia (Oreochromis niloticus), platyfish (Xiphophorus maculatus), Amazon molly (Poecilia formosa), medaka (Oryzias latipes), stickleback (Gasterosteus aculeatus), green puffer (Tetraodon nigroviridis) and fugu (Takifugu rubripes), were downloaded from the Ensembl database (Release 84),36 mapped to the yellowtail scaffolds by TBLASTN37 with E-value < 10−5, and the map information was used as hints in AUGUSTUS. Finally, the predicted gene sequences were analysed with InterProScan38 and those matched by any domain or supported by any AUGUSTUS hint were collected as valid protein-coding genes.
The predicted amino acid sequences were compared using BLASTP19 (E value threshold of 1E − 5) against those of well-annotated model fishes, medaka and zebrafish, in the Ensembl database (Release 84),36 and against the NCBI non-redundant (nr) protein database. Functional annotation was performed using Blast2GO39 and the Kyoto Encyclopedia of Genes and Genomes (KEGG) Automatic Annotation Server.40 Orthologous gene cluster analysis of yellowtail, Pacific bluefin tuna (Thunnus orientalis),41 croaker (Larimichthys crocea),42 Nile tilapia, medaka and zebrafish was performed using the OrthoVenn43 web server with default parameters (E-value 1e–5, inflation value 1.5). OrthoVenn was also used to assign significantly enriched GO terms of species-specific gene clusters based on a hypergeometric test (P-value < 0.05). The same analysis also applied to comparisons within the genus Seriola including yellowtail, S. dumerili (Genbank: BDQW01000000) and S. dorsalis (Genbank: PEQF01000000).
2.6. RNA-Seq analysis of yellowtail GI tract
The three major organs of the GI tract: stomach, intestine and rectum, were dissected from three healthy adult yellowtails (n = 3) obtained from brood stock maintained at the Goto laboratory of the Seikai National Fisheries Research Institute, Nagasaki, Japan and immediately immersed in RNAlater stabilized solution (Thermo Fisher Scientific, Waltham, MA, USA). Total RNA was extracted using the Maxwell RSC simply RNA Kit (Promega, Madison, WI, USA) and mRNA was purified from 3 µg total RNA using the Gene Read Pure mRNA kit (Qiagen). Sequencing libraries were constructed from 5 ng of each mRNA sample using the Ion Total RNA-Seq Kit v2 and sequenced with Ion Proton (Life technologies). RNA-Seq data analysis was carried out using CLC Genomics Workbench 9.5.2 software (CLC Bio, Aarhus, Denmark) as follows: The low-quality sequences were filtered with default parameters (quality limit = 0.05 and ambiguous limit = 2), and the filtered sequence reads were mapped to the yellowtail reference genome constructed in this study with default parameters (mismatch cost = 2, insertion cost = 3 and deletion cost = 3, length fraction = 0.8 and similarity fraction = 0.8). Subsequently, transcripts per millions (TPMs)44 values for each gene were calculated. The differentially expressed genes (DEGs) were considered significant for the following criteria: a 2-fold or greater change in expression with a false discovery rate P-value < 0.05. Visualization of the DEGs was performed by constructing a hierarchical heatmap with Euclidean distance and complete linkage in the CLC Genomics Workbench. To compare the over-represented functional categories between the highly expressed genes (500 highest TPM values) in the three tissues, a GO enrichment analysis was conducted using the functional annotation tool, PANTHER version 12.0 (released 7 Ocotber 2017)45 using the zebrafish gene list. The Bonferroni correction for multiple testing was applied with the corrected P-value < 0.05 considered significantly over-represented in the PANTHER analysis.
3. Results and discussion
3.1. Genome sequencing and assembly
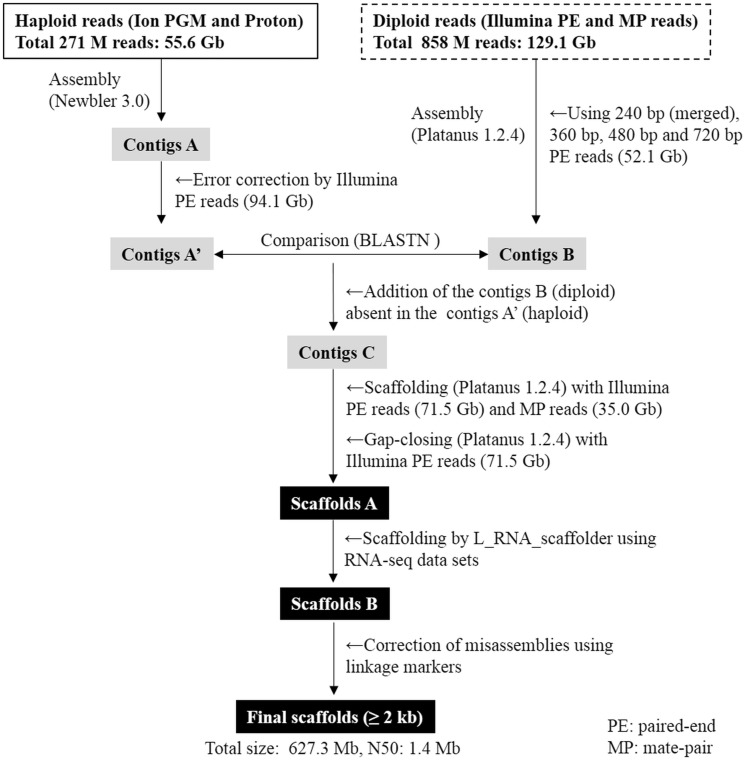
Recent advances in the speed, accuracy and cost effectiveness of NGS technologies have provided the opportunity for whole genome sequencing of even non-model organisms with no available reference genomes. However, de novo assembly of diploid species, including teleosts, remains challenging because of allelic variation. Recently, several teleost studies have used DNA samples with theoretically no allelic variations as biological starting material for genome assembly. Zhang et al.46 reported a marked improvement in genome assembly after employing doubled-haploid fugu individuals via mitotic gynogenesis. Although doubled haploid fishes are useful in genomic analysis, limited doubled haploid fish lines are available, and their establishment is labour-intensive, time-consuming and expensive. On the other hand, the use of haploid larvae is a convenient means of obtaining a non-allelic variation genome in fish if an artificial insemination technique is established, and we have already demonstrated the effectiveness of genomic DNA from a haploid gynogenetic yellowtail larva in genome assembly.16 However, there are two concerns related to the use of haploid fish larvae for the construction of reference genome sequences. To overcome these concerns, we established an assembling strategy (Fig. 1) as follows. First, defective regions may exist in the haploid genome sequence due to allelic variations. Taking this probability into account, we compared the haploid contigs with those generated from its dam (diploid), and then merged the diploid sequences absent in the haploid contigs (Fig. 1). Second, because haploid larvae die during early larval stages, the amount of DNA available from a haploid fish larva is limited, and not sufficient for the preparation of PE and MP libraries for scaffolding. To make up for this limitation, the contigs were constructed from limited amounts of DNA from a haploid fish, and scaffolding was performed using sufficiently available DNA from its dam (Fig. 1).
Figure 1.
Strategy for de novo assembly of the yellowtail (S. quinqueradiata) genome using the genome of haploid larvae. More detailed information on the sequencing libraries and the summary statistics of reads used in this study can be found in Supplementary Table S1, and contig and scaffold statistics are shown in Supplementary Tables S3 and S4, respectively.
In this study, a total of 11.2 million reads (3.7 Gb) and 259.4 million reads (51.9 Gb) from the haploid genome were generated from Ion PGM and Ion Proton, respectively, and a total of 858 million reads (129.1 Gb) were obtained from a series of Illumina PE (240 bp insert: 27.8 Gb, 360 bp insert: 31.5 Gb, 480 bp insert: 25.3 Gb and 720 bp insert: 9.5 Gb) and MP (3–5 kb insert: 10.8 Gb, 10 kb insert: 12.2 Gb and 20 kb insert: 12.0 Gb) libraries of the diploid genome (Supplementary Table S1). The genome size of the yellowtail has been calculated to be ∼685 Mb based on the c-value,10 while the genome size based on k-mer frequency was estimated to be 603.8 and 602.7 Mb from the haploid and diploid reads datasets, respectively. Thus, the generated sequence data (184.7 Gb) has 270- to 306-fold coverage of its genome size. Figure 1 provides a flow diagram of the genome assembly procedure used for this study; information on the sequencing libraries and summary statistics of reads appears in Supplementary Table S1. Because our previous study demonstrated that the overlap-layout-consensus (OLC)-based de novo assembly (i.e. Newbler) was the most efficient method for haploid reads generated from the Ion Torrent sequencing platform,16 the haploid reads (55.6 Gb) were assembled by Newbler and generated 90,827 contigs (>500 bp) with a total length with 611.9 Mb (Fig. 1, Contigs A and Supplementary Table S3). On the other hand, a 52.1 Gb of diploid reads from the four Illumina PE libraries (Supplementary Table S1) were assembled with Platanus, a de novo assembler specifically designed to reconstruct highly heterozygous genomes,18 and the resulting assembly consists of 209,882 contigs (>500 bp) with a total length with 596.7 Mb (Fig. 1, Contigs B and Supplementary Table S3). Although different types of sequencers and assemblers were used for the haploid and diploid reads, the assembly of the haploid genome significantly reduced the total number of contigs with longer average and N50 contig lengths when compared with the diploid genome assembly (Supplementary Table S3). Furthermore, the k-mer distribution of the diploid reads showed an additional small peak which corresponds to heterozygous regions, i.e. a ‘hetero-peak’18 (Supplementary Fig. S1). These heterozygous regions may disrupt the extension of contigs during the assembly process.16 Thus, these results support the argument for using the haploid genome for de novo assembly.
After sequence error correction of the haploid contigs using 94.1 Gb of Illumina PE reads (Fig. 1, Contigs A’), the diploid contigs (Fig. 1, Contigs B) absent in the haploid contigs (Fig. 1, Contigs A’) were added to the haploid contigs. This process could add 26,916 contigs (4.5 Mb) to the haploid contigs, and generated 162,987 contigs (Fig. 1, Contigs C and Supplementary Table S3). These merged contigs (Fig. 1, Contigs C) were subjected to scaffolding with 71.5 Gb of Illumina PE and 35.0 Gb of Illumina MP reads (Supplementary Table S1), and the resulting scaffolds were filled by 71.5 Gb of Illumina PE reads (Supplementary Table S1). As a result, 1,225 scaffolds (>2 kb) were obtained (Fig. 1, Scaffolds A and Supplementary Table S4). Further scaffolding using RNA-Seq datasets generated in this study (Supplementary Table S2) was conducted and 1,155 scaffolds (>2 kb) were constructed (Fig. 1, Scaffolds B and Supplementary Table S4). Finally, the linkage marker sequences from the ddRAD-based genetic linkage map of the yellowtail generated in this study were used to correct the scaffold misassemblies (Fig 1, Final scaffolds). The female and male maps (consisting of 1, 163 and 1, 147 ddRAD markers for female and male maps, respectively) for LGs (LG1–LG24) are shown in Supplementary Fig. S2, and the locations of genetic markers are listed in Supplementary Table S5. The final assembly is composed of 1, 394 scaffolds (>2 kb) and the total size of scaffolds is 627.3 Mb, with a G + C content of 40.9%, an N50 length of 1.43 Mb (number of N50 or longer: 127) and a largest scaffold size of 6.62 Mb (Table 1).
Table 1.
Assembly statistics of yellowtail genome
| Number of scaffolds (>2 kb) | 1,394 |
| Total size of scaffolds (bp) | 627,268,966 |
| Average scaffold length (bp) | 449,978 |
| N50 scaffold length (bp) | 1,433,177 |
| N50 scaffold number | 127 |
| N90 scaffold length (bp) | 404,933 |
| N90 scaffold number | 453 |
| Longest scaffold (bp) | 6,617,595 |
| GC (%) | 40.9 |
The completeness of the generated genome assembly was evaluated using CEGMA analysis, which identified 97.86% complete and 98.28% partial genes from the 233 CVGs. This percentage of complete gene hits is higher than that of model fish genome assemblies such as medaka (93.56%), zebrafish (93.13%), stickleback (92.27%) and spotted green puffer (90.56%), according to the gVolante, Completeness Score Database (https://gvolante.riken.jp/script/database.cgi (11 July 2018, date last accessed)). Recently, two yellowtail genome assemblies (Squ_1.0 and Squ_2.0) have been deposited at DDBJ/EMBL/Genbank under accession numbers BDJM01000000 (Squ_1.0) and BDMU00000000 (Squ_2.0). According to assembly information at NCBI (https://www.ncbi.nlm.nih.gov/assembly/organism/8161/all/ (11 July 2018, date last accessed)), both assemblies were generated from diploid DNA samples with an Illumina Hiseq2500 platform and a long-read sequencer PacBio RSII. Squ_2.0 is an updated version of Squ_1.0 and is representative of the genome of this species at present. Although Squ_2.0 has fewer scaffolds (384) and a larger N50 scaffold size (5.6 Mb) than our assembly, the assembly quality based on BUSCO and CEGMA analysis showed that our assembly has higher scores than those of Squ_2.0 in both analyses. Our assembly captured 97.27% (4,459 out of 4,584) of complete BUSCOs, while Squ_2.0 captured 96.81% (4,438 out of 4,584). Similarly, CEGMA evaluation showed that the completeness of our assembly and that of Squ_2.0 was 97.85% (228 out of 233) and 97.42% (227 out of 233), respectively. Despite using relatively short sequence reads for the genome assembly, we obtained a high level of completeness in BUSCO and CEGMA analyses, suggesting the advantage of utilizing the haploid genome for de novo assembly. Thus, we concluded that our assembly has a more complete gene space than Squ_2.0 and is more suitable for conducting gene-prediction and annotation of yellowtail.
3.2. Annotation of protein-coding genes
A total of 27,693 protein-coding genes were predicted for the yellowtail draft genome, based on AUGUSTUS, the yellowtail training model and the transcript + protein hints. A summary of the functional annotation of the yellowtail predicted genes is shown in Table 2 and the results of the functional annotation of the complete set of predicted protein-coding genes are available in Supplementary Table S7. Of the 27,693 predicted genes, 25,319 (91.4%) genes matched (E value threshold of 1 E − 5) at least one entry in the NCBI nr database, while 21,233 (76.7%) and 23,422 (84.6%) genes matched with model fishes, medaka and zebrafish, respectively (Table 2). A total of 18,575 (67.1%) genes mapped to at least one GO term (biological process: 12,606 genes, cellular component: 11,784 genes and molecular function: 13,094 genes). In addition, 13,880 (50.1%) genes were mapped to KEGG pathways, while 2,867 genes were identified to have at least one enzyme hit. The conserved domain search (InterPro) showed that 25,035 (90.4%) genes were identified to have at least one domain. Overall, 25,832 predicted genes (93.3%) of yellowtail were functionally annotated by at least one database, and only 1,861 predicted genes (6.7%) were unannotated (Table 2). This richness in functional annotation for yellowtail protein-coding genes will help us to better understand their biology in downstream studies, such as whole-transcriptome analysis.
Table 2.
Functional annotation of the 27,693 yelowtail predicted protein-coding genes
| Database | Number of the yellowtail genes mached | Percentage of annotated genes |
|---|---|---|
| NCBI non-redundant (nr) databasea | 25,319 | 91.4% |
| Medaka genesa | 21,233 | 76.7% |
| Zebrafish genesa | 23,422 | 84.6% |
| Gene ontrogy (GO) | 18,575 | 67.1% |
| Biological process | 12,606 | 45.5% |
| Cellular component | 11,784 | 42.6% |
| Molecular function | 13,094 | 47.3% |
| InterPro | 25,035 | 90.4% |
| Enzyme | 2,867 | 10.4% |
| KEGG pathway | 13,880 | 50.1% |
| At least one functional annotation | 25,832 | 93.3% |
| Unannotated | 1, 861 | — |
BLASTP hit of less than 1E−5.
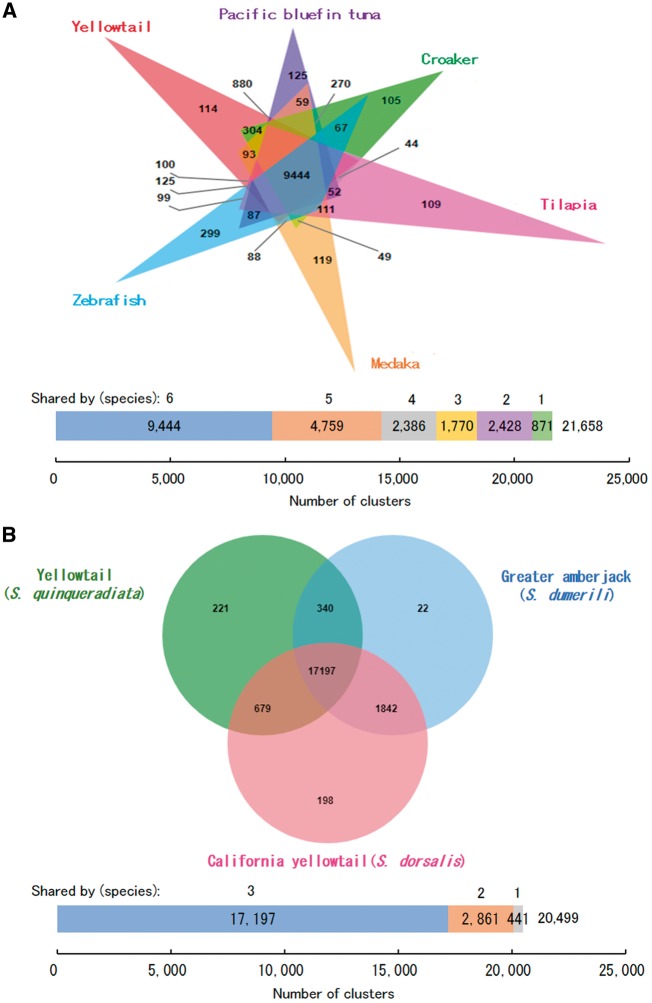
The genome-wide comparison of orthologous gene clusters was performed to identify the degree of commonality across yellowtail and five other teleost species: Pacific bluefin tuna, croaker, tilapia, medaka and zebrafish. Based on sequence similarity of proteins, all protein sequences from the six species were grouped into 21,658 clusters, from which 9,444 (43.6%) orthologous gene clusters were shared by all six species and 20,787 (96.0%) orthologous clusters contained at least two species (Fig. 2A). On the other hand, 871 (4.0%) single-species clusters: 114 clusters (336 genes) for yellowtail, 125 clusters (364 genes) for Pacific bluefin tuna, 105 clusters (236 genes) for croaker, 109 clusters (326 genes) for tilapia, 119 (490 genes) clusters for medaka and 299 (1,515 genes) clusters for zebrafish, were identified (Fig. 2A), suggesting a lineage-specific expansion of these clusters. As seen earlier, the number of zebrafish-specific clusters was 2.4- to 2.7-fold higher than those for the other five teleosts, including yellowtail, which may explain the notable gene duplication and retention of recent duplicates in zebrafish after a teleost-specific genome duplication event around 300 million years ago.47 A hypergeometric test for assessing enriched GO terms in the single-species clusters was performed. Overall, many immune-related GO terms were found in the significantly enriched GO terms of species-specific clusters, except for those in madaka, which showed that transposon-related GO terms were the most significant (P = 2.58E-4) (Supplementary Table S8).
Figure 2.
Venn diagram showing the number of unique and shared gene clusters in (A) six teleosts (yellowtail, Pacific bluefin tuna, croaker, tilapia, medaka and zebrafish) and in (B) three Seriola species: yellowtail, greater amberjack (S. dumerili) and California yellowtail (S. dorsalis). Orthologous gene clusters were identified and visualized using OrthoVenn.43 The bar charts below the Venn diagrams represent the total number of clusters that are unique to a single species or shared between 2 and 6 species.
Focussing on the yellowtail-specific clusters, one of the most significant GO terms was taurine: sodium symporter activity (GO: 0005369, P = 0.015), containing two taurine transporter (TauT/Slc6a6) genes, g27489 and g26790 (Supplementary Table S8). TauT mediates cellular uptake of taurine (2-aminoethane sulphonic acid) which is found in high concentrations in animal tissues.48 Taurine is a vital nutrient for many fish species, particularly for carnivorous fish, due to their limited taurine biosynthesis abilities.49 It has been reported that taurine synthesis is markedly low or negligible in yellowtail.50 Therefore, the absorption of taurine from feeds via TauT is arguably important for yellowtail. Further study of the two TauT genes from the yellowtail-specific cluster will provide insight into taurine absorption in this species.
Draft genomes for greater amberjack (S. dumerili)51 and California yellowtail (S. dorsalis)52 have recently been published. We compared the predicted-protein sequences of yellowtail with those of these two Seriola species. The protein sequences from yellowtail (27,693), greater amberjack (22,005) and California yellowtail (25,789) were grouped into 20,499 clusters from which 17,197 (84%) clusters were shared across all three species, and 20,058 (97.8%) clusters contained at least two species (Fig. 2B). We found 441 single-species clusters (2.2%) including 221 clusters (814 genes) from yellowtail, 22 clusters (53 genes) from greater amberjack and 198 clusters (416 genes) from California yellowtail (Fig. 2B). As in the results of the comparisons with five teleosts, the most significant enriched GO terms in yellowtail- and greater amberjack-specific clusters were related to immune processes such as antigen binding (GO: 0003823, P = 5.1E-4) in yellowtail and positive regulation of interleukin-1 beta secretion (GO: 0050718, P = 3.9E-4) in greater amberjack (Supplementary Table S9). California yellowtail-specific clusters showed regulation of osteoclast differentiation (GO: 0045670, P = 0.008) and regulation of RNA stability (GO: 0043487, P = 0.008) as the most enriched clusters, but the significance of these data is unclear (Supplementary Table S9). Together with the results of the comparison across six teleosts, most of the species-specific clusters included immune-related GO terms, suggesting that lineage-specific expansion of immune-related genes is likely to play a key role in species diversification. This may be due to the remarkable evolutionary plasticity of the teleost immune system in response to habitat, specific environmental factors and lineage-specific pathogens, and which is linked to the survival and radiation of the teleost linnage.53,54
3.3. A snapshot of yellowtail GI tract transcriptome
We used our yellowtail reference genome for RNA-Seq analysis of three major organs of the GI tract: stomach, intestine and rectum as a first step toward understanding digestion and nutrient absorption in yellowtail at the molecular level. This RNA-Seq generated 10,692,067–27,326,877 reads (mean read length: 135–167 bp) per individual tissue type (Supplementary Table S6).
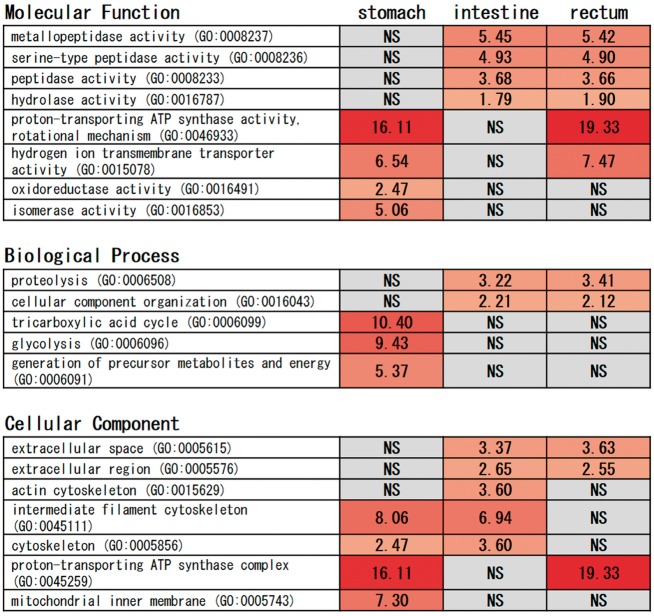
First, we characterized the highly expressed genes (top 500) in each tissue using a GO enrichment analysis, because the highly expressed genes can be considered to serve functionally important roles in the respective tissues that will help us to understand the mode of operation of each tissue. This analysis showed that most of the enriched categories were shared between the intestine and rectum, and included: ‘metallopeptidase activity’ (GO: 0008237, nine genes each in intestine and rectum), ‘serine-type peptidase activity’ (GO: 0008236, 13 genes each in intestine and rectum), ‘peptidase activity’ (GO: 0008233, 29 genes each in intestine and rectum), ‘hydrolase activity’ (GO: 0016787, 62 genes in intestine and 66 genes in rectum) under the molecular function category, and ‘proteolysis’ (GO: 0006508, 31 genes in intestine and 33 genes in rectum) under the biological process category (Fig. 3). These categories are involved in the hydrolysis of proteins into smaller polypeptides, suggesting that both the intestine and rectum function to digest protein in the yellowtail. We also found enriched GO terms common in the stomach and rectum that are related to proton transport activity, including ‘proton-transporting ATP synthase activity, rotational mechanism’ (GO: 0046933, five genes in stomach and six genes in rectum), ‘hydrogen ion transmembrane transporter activity’ (GO: 0015078, seven genes in stomach and eight genes in rectum) under the molecular function category, and ‘proton-transporting ATP synthase complex’ (GO: 0045259, five genes in stomach and six genes in rectum) under the cellular component category (Fig. 3). These genes probably contribute to the acidity of the stomach necessary for acid hydrolysis of feeds.55 In general, carnivorous fish have well developed stomachs capable of an acid digestion phase, whereas in stomach-less fish such as carp, no such phase exists.56 In the rectum, these genes may play a role in osmoregulation.57 The stomach has more non-overlapping enriched GO terms than other organs, and overexpressed ‘oxidoreductase activity’ (GO: 0016491, 24 genes), ‘isomerase activity’ (GO: 0016853, 11 genes) under the molecular function category, ‘tricarboxylic acid cycle’ (GO: 0006099, five genes), ‘glycolysis’ (GO: 0006096, six genes), ‘generation of precursor metabolites and energy’ (GO: 0006091, 19 genes) under the biological process category, and ‘mitochondrial inner membrane’ (GO: 0005743, six genes) under the cellular component category (Fig. 3). These GO terms suggest the role of the stomach in energy metabolism. Although interpretation of the roles of these genes is unclear, these results suggest that the digestive function of the stomach differs from that of the intestine and rectum in yellowtail.
Figure 3.
GO enrichment analysis of the top 500 highly expressed genes in the yellowtail stomach, intestine and rectum. This analysis was performed using PANTHER45 and the zebrafish gene list was selected. Values indicate fold enrichment (P-value < 0.05 after Bonferroni correction) and NS means not significant.
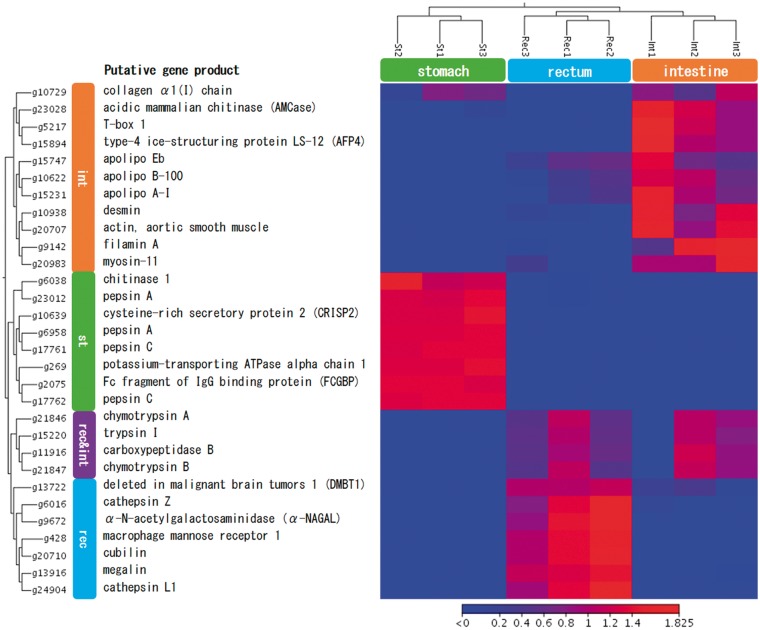
Next, we surveyed DEGs among the three organs to identify tissue-specific gene expression patterns. The number of DEGs in the stomach, when compared with those in the intestine and rectum were 6,001 and 6,040, respectively, indicating that the expression pattern of the stomach was different from those of the intestine and rectum, as expected by the GO enrichment analysis of the highly expressed genes (Fig. 3). The comparison between the intestine and the rectum also showed considerable differences in the gene expression pattern, with 2,195 DEGs, suggesting that each part has a specialized function in digestive and absorption processes. Similarly, over 1,900 DEGs (fold-change cutoff of 2) were observed between anterior-middle and posterior-intestinal segments (including the rectum) in a carnivorous fish, European sea bass (Dicentrarchus labrax) using a custom oligo microarray.58 Using a cluster analysis of the DEGs, we extracted expression patterns that are characteristic of the three tissue types (Supplementary Fig. S3). Figure 4 shows the heat map of 30 DEGs with the highest coefficient of variation among the three tissues.
Figure 4.
Heat map of 30 DEGs with the highest coefficient of variation among the yellowtail stomach (st), intestine (int) and rectum (rec).
A gene encoding potassium-transporting ATPase alpha chain 1 (g269), which has a role in acid production in the mammalian stomach55,59 was specifically expressed in the stomach (Fig. 4). As mentioned above, this gene may facilitate the acidic environment of the yellowtail stomach. In addition, four genes (g6958, g17761, g17762 and g23012) encoding pepsinogens, and activated to the active form pepsin under acidic conditions,60 were specifically expressed in the stomach (Fig. 4). In vertebrates, including fish, pepsin has been identified as the major acidic protease in the stomach, acting during the earliest stage of protein digestion in breaking down long-chain peptides.61,62 In particular, carnivorous fish have higher pepsin levels in their stomachs than omnivorous fish with stomachs and stomach-less fish,63 such as zebrafish, which lack the pepsinogen gene.64 Another digestive enzyme, encoded by a chitinase gene (g6038), was also found in the stomach-specific cluster (Fig. 4). It has been reported that the fish stomach exhibits chitinase activity, which is associated with the digestion of chitinous substances from crustaceans.65,66 We found two stomach-specific genes, the Fc fragment of IgG binding protein (FCGBP; g2075) and cysteine-rich secretory protein 2 (CRISP2; g10639). FCGBP (g2075) is abundant in the mucus of humans and mice, and may be functionally related to the gel-forming mucins,67 while CRISP2 is strongly expressed in the mammalian male reproductive tract.68 Since the function of these two genes in fish has not been elucidated, further studies are required to identify the role of these genes in the yellowtail stomach.
As expected following the results of the GO enrichment analysis (Fig. 3), proteolytic digestive enzyme genes, a trypsinogen (precursor of trypsin) gene (g15220), two chymotrypsinogen (precursor of chymotrypsin) genes (g21846 and g21847) and a carboxypeptidase B gene (g11916) were found to be specifically expressed in the intestine and the rectum (Fig. 4). In vertebrates including fish, hydrolysis and absorption of proteins occurs primarily in the intestine.69 It has been reported that trypsin activity in fish is significantly higher in the intestine than in the stomach, which showed very little or no activity,61,62 and high trypsinogen mRNA expression levels were also found in the intestine of Atlantic salmon (Salmo salar),70 Senegalese sole (Solea senegalensis)71 and pufferfish (Takifugu obscurus).72 Our RNA-Seq results suggest that both the intestine and rectum play major roles in hydrolysis and absorption of proteins in yellowtail. It should be noted that the gene expression of these proteolytic digestive enzymes was low in the intestine of one fish (Fig. 4). Calduch-Giner et al.58 suggested that fish intestine transcriptomes are not static, but change spatially, seasonally and with diet. Since the samples of GI tract examined were obtained from cultured yellowtail maintained under the same conditions and sampled at the same time, our study suggests that the expression of proteolytic enzymes also varies between individuals, and that such differences may cause different physiological conditions in individuals.
Focussing on intestine-specific gene expression, three apolipoprotein genes, apolipo Eb (g15747), apolipo B-100 (g10622) and apolipo A-1 (g15231), were included in this cluster (Fig. 4). Apolipoproteins play a crucial role in lipid transport and uptake in vertebrates and are synthesized mainly in the intestine and liver of most teleosts.73–75 In addition, another intestine-specific gene, Type-4 ice-structuring protein LS-12 (AFP4; g15894), which was first isolated as an antifreeze protein from the serum of the longhorn sculpin (Myoxocephalus octodecimspinosis),76 may also play a role in lipid metabolism because it has an apolipoprotein A-II domain (InterPro: IPR006801). It has also been reported that the expression of the AFP4 gene was higher in the anterior-middle intestinal segments than the posterior segment in European seabass.58 In contrast to mammals, which use carbohydrates as their main energy source, most fish predominantly make use of lipids.77 Therefore, these apolipoproteins and AFP4 may play important roles in lipid metabolism in the yellowtail intestine. In addition to these lipid metabolism-related genes, an acidic mammalian chitinase (AMCase) gene (g23028) was found in the intestine-specific cluster (Fig. 4), and a higher expression of this gene in the anterior-middle intestinal area as opposed to posterior segments in European seabass has been found.58 In the mouse, acidic mammalian AMCase can act in digestion of chitin polymers even in the presence of pepsin C, trypsin and chymotrypsin.78 This enzyme also plays a critical role in the human immune response to pathogens.79 It should be noted that fish intestines function as immune barriers to pathogens80,81 and that fish apolipoproteins play a role in this immune function.82–84 Thus, AMCase and apolipoproteins may have roles not only in digestion and absorption but also in defense against intestinal pathogens. Four muscle-related genes, myosin-11 (g20983), desmin (g10938), actin (g20707) and filamin A (g9142) were highly expressed in the intestine (Fig. 4). The GO enrichment analysis (Fig. 3) also showed the over-representation of muscle-related GO terms in the intestine such as ‘cytoskeleton’ (GO: 0005856, 22 genes), ‘actin cytoskeleton’ (GO: 0015629, 13 genes) and ‘intermediate filament cytoskeleton’ (GO: 0045111, 6 genes). Recently, Abrams et al.85 reported that zebrafish myosin-11 (myh11) gene mutations affected intestinal motility. In addition, a collagen α-1(I) chain gene (g10729) was found in the intestine-specific expression cluster (Fig. 4), and its role in responding to stretching of foetal human intestinal smooth muscle cells has been investigated.86 Thus, these muscle-related genes of yellowtail may have functions related to intestinal motility. Grau et al.87 suggested the importance of intestinal motility for intestinal absorption in carnivorous fish with irregular intake of large quantities of food.
The rectum of yellowtail is separated from the posterior region of the intestine by the ileo-rectal valve. As observed in the rectum of the greater amberjack,87 the mucosa are more deeply folded than in those of the intestine, and the rectum is also considered to play a role in absorption.87 The rectum-specific cluster included two cathepsin genes, cathepsin L1 (g24904) and cathepsin Z (g6016) (Fig. 4). Cathepsin L1 gene also showed a higher expression in the anterior segments than in the posterior segment of the intestinal tract in zebrafish64 and European seabass.58 Cathepsins are essential lysosomal proteolytic enzymes and the activity of cathepsin D has been shown to be involved primarily with the posterior intestine in rainbow trout (Oncorhynchus mykiss).69 However, little is known about the function of cathepsins in the fish rectum and studies are needed to elucidate their role in digestion and absorption processes. Two multiligand endocytic receptor genes, cubilin (g20710) and megalin (g13916), were also found as rectum-specific genes (Fig. 4), and each shown to mediate reabsorption of proteins and vitamins in mammals. The megalin/cubilin complex delivers its ligands (proteins and vitamins) to lysosomes, where all proteins are degraded, and the amino acids and vitamins returned to circulation.88,89 Two rectum-specific genes, deleted in malignant brain tumours 1 (DMBT1; g13722) and macrophage mannose receptor 1 (g428), are possibly involved in defense against bacterial pathogens.90,91 The gene (g9672) encoding α-N-acetylgalactosaminidase, a lysosomal glycohydrolase,92 was also specifically expressed in the rectum (Fig. 4), although the functions of this enzyme in the rectum are unknown.
The results of GO enrichment analysis of highly expressed genes and cluster analysis of the DEGs together showed that the acid secretion-related genes and acid-activated protease (pepsinogen) genes were specifically expressed in the stomach, while the proteolytic digestive enzymes, trypsin, chymotrypsin and carboxypeptidase B, were highly and commonly expressed in the intestine and the rectum. In addition, the RNA-Seq also revealed differences in gene expression patterns between the intestine and the rectum. The possible lipid metabolism-related genes (apolipoproteins and AFP4) were specifically expressed in the intestine, while the genes involved in lysosomal digestive and reabsorption processes (cathepsins, cubilin and megalin), were specifically expressed in the rectum. These transcriptional signatures exhibit the main features of the digestive tract of carnivorous fishes, which possess higher proteolytic enzyme activities than herbivorous or omnivorous species,93,94 which in general exhibit higher α-amylase (carbohydrate enzyme) activities.95 It should be noted that two α-amylase genes (g13238 and g13239) were found in the yellowtail, but their expression was low or undetected in the three GI tissues. Efficiency of food absorption and conversion can depend on the availability of digestive enzymes.96 Thus, these findings suggest that proteolytic digestive enzymes play a key role in digestion and nutrient absorption in the yellowtail GI tract.
3.4. Gene expression patterns of proteolytic digestive enzymes
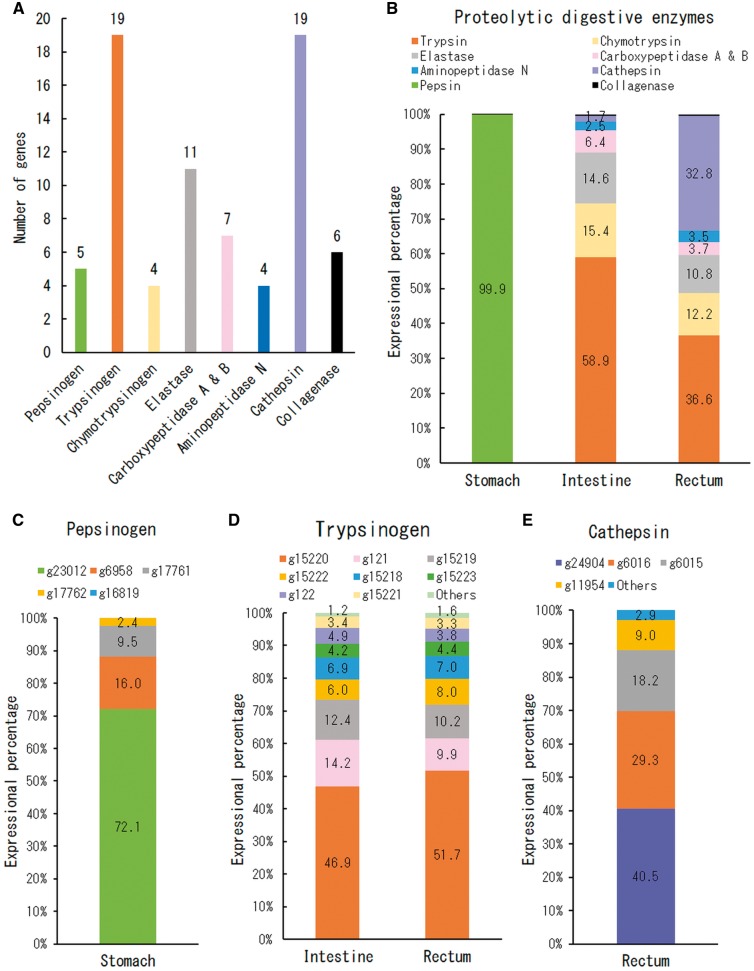
Because the RNA-Seq results suggested the importance of proteolytic processes in the yellowtail GI tract, we further analysed the expression patterns of proteolytic digestive enzymes in yellowtail. In the yellowtail genome, we found 75 candidate genes for proteolytic digestive enzymes: five pepsinogens, 19 trypsinogens, four chymotrypsinogens, 11 elastases, seven carboxypeptidases (A and B), four aminopeptidase Ns, 19 cathepsins and six collagenases (Fig. 5A). The expressional percentages of these categories were expressed as TPM values for each tissue (Fig. 5B). The distribution patterns of these enzymes varied among the three tissues. In the stomach, the expressional percentage was almost entirely occupied by pepsinogen genes, which accounted for 99.9% of the total. Surprisingly, the expression percentage of pepsinogens accounted for 45.6–65.0% of the overall stomach transcriptome in three individuals. In the intestine, the transcript levels of trypsinogens were very high (58.9%), followed by chymotrypsinogens (15.4%), elastases (14.6%) and carboxypeptidases (6.4%). These gene expression patterns are correlated with the intestinal protease activity in carnivorous fish as observed in previous studies. Eshel et al.97 estimated that trypsin contributes to 40–50% of the overall protein digestion in carnivorous fish intestines. Furthermore, it has been reported that trypsin activity in carnivorous fish is about four times (3.9 times) that of chymotrypsin activity, a relationship that is reversed in omnivorous and herbivorous species.98 Similarly, the expressional percentage of trypsinogen genes in yellowtail showed about four times (3.8 times) that of chymotrypsinogen genes. Trypsinogen genes were also expressed at the highest levels in the rectum (36.6%), but the expressional percentage was lower than that of the intestine (58.9%). Alternatively, cathepsin genes showed a high transcript level in the rectum which accounted for 32.8%, the highest among the three tissues.
Figure 5.
Transcript levels of proteolytic digestive enzyme genes in the yellowtail stomach, intestine and rectum. (A) The number of proteolytic digestive enzyme genes identified in the yellowtail genome. (B) Expressional percentages based on TPM values for different categories of proteolytic enzyme genes. The expression levels of individual genes in the highly expressed categories in each tissue are also shown in (C) pepsinogen genes in stomach, (D) trypsinogen genes in intestine and rectum and (E) cathepsin genes in the rectum. The amino acid sequence identities and similarities between these highly expressed genes can be found in Supplementary Fig. S4.
To determine whether there are differential gene usages within each category of proteolytic digestive enzymes, we further analysed the expression levels of individual genes in the highly expressed categories in each tissue (Fig. 5B): pepsinogen genes in the stomach (Fig. 5C), trypsinogen genes in the intestine and rectum (Fig. 5D) and cathepsin genes in the rectum (Fig. 5E). This analysis revealed that individual gene expression levels are different for each enzyme. Of the five pepsinogen genes in the stomach, g23012 occupied 72.1% of the total expression value, followed by g6958 which accounted for 16.0% (Fig. 5C). In teleosts, two types of pepsinogen have been identified, pepsinogen A and pepsinogen C.99 Both g23012 and g6958 genes encode pepsinogen A, and the amino acid identity and similarity between these two sequences are 64.6 and 78.1%, respectively (Supplementary Fig. S4A). Of 19 trypsinogen genes, only three, g15220, g121 and g15219, accounted for >70% of the total in the intestine and rectum (Fig. 5D). Among them, g15220 showed about half of the total expression value in the intestine (46.9%) and rectum (51.7%). Based on amino acid sequence identities, most trypsins can be classified into three basic Groups I–III.100 g15220 and g15219 encode Trypsin I, and the amino acid identity and similarity between them was 84.3 and 92.6%, respectively. On the other hand, g121 encodes Trypsin III, and it has 69.0 and 82.7% amino acid identity and similarity with g15220, and 73.9 and 86.7% amino acid identity and similarity with g15219 (Supplementary Fig. S4B). Of 19 cathepsin genes in the rectum, the transcript levels of g24904 (cathepsin L1) were highest (40.5%), followed by g6016 (cathepsin Z, 29.3%) and g6015 (cathepsin Z, 18.2%). The protein homology values among them were low or showed no significant similarity (Supplementary Fig. S4C).
Overall, these results suggest that most proteolytic digestion in the yellowtail is related to the expression of specific genes, despite the large number of proteolytic digestive enzyme genes in the genome (Fig. 5A). The highly expressed proteolytic digestive enzyme genes identified in this study may play a key role in digestion in adult yellowtail. This finding suggests that the identification of proteins digestible by the products of these genes may lead to a yellowtail feed formulation with enhanced digestibility. Lemeiux et al.96 reported that among the activities of a number of enzymes, trypsin alone showed a significant correlation with food conversion efficiency and growth rate in the carnivorous Atlantic cod. Furthermore, it has been demonstrated that trypsin isoforms showed differing substrate preferences and distinct catalytic properties.100,101 Therefore, further study of the substrate preferences of the products of the three highly expressed trypsinogen genes (g15220, g121 and g15219) may inform the makeup of yellowtail feed ingredients.
This study has focussed on adult yellowtail, and the gene expression patterns for digestive enzymes may be different for larval stages.71 Studies of gene expression patterns for digestive enzymes in larval stages of yellowtail will inform the development of formulated diets for larvae of this species. In addition, the digestive systems of fish show numerous structural and functional adaptations to their feeding habits. Thus, the transcriptomes of digestive tissues in fish are considered highly species-specific,58 suggesting that transcriptome profiling of these tissues may be instructive for the development of fish feed for species of interest in the aquaculture industry.
Availability
The draft genome sequence of S. quinqueradiata was submitted to DDBJ under accession no. BEWX01000001–BEWX01001394 (1394 entries). The complete set of predicted protein-coding genes are accessible at http://nrifs.fra.affrc.go.jp/ResearchCenter/5_BB/genomes/Yellowtail_genes/index.html. The results of the functional annotation of the predicted protein-coding genes are available in Supplementary Table S7. Raw RNA-Seq datasets for the three digestive organs of yellowtail have been deposited at DDBJ Sequence Read Archive (DRA) under accession no. DRR107933–DRR107935 (stomach), DRR107936–DRR107938 (intestine) and DRR107939– DRR107941 (rectum).
Supplementary Material
Acknowledgements
This work was partly supported by an Adaptable and Seamless Technology transfer Programme through Target-driven R&D (A-STEP: AS2815106U) from the Japan Science and Technology Agency. We thank Dr Shintaro Imamura (National Research Institute of Fisheries Science, Japan Fisheries Research and Education Agency) and Dr Susumu Uji (National Research Institute of Aquaculture, Japan Fisheries Research and Education Agency) for providing samples of larval yellowtail digestive organs and Dr Tsubasa Uchino (National Research Institute of Fisheries Science, Japan Fisheries Research and Education Agency) for his help in writing Section 2.2. Linkage map construction and scaffold correction. We also would like to thank Dr Kristian von Schalburg (Department of Molecular Biology and Biochemistry, S.F.U., Vancouver, BC, Canada) and Editage (www.editage.jp) for English language editing.
Conflict of interest
None declared.
References
- 1. Swart B.L., von der Heyden S., Bester-van der Merwe A., Roodt-Wilding R.. 2015, Molecular systematics and biogeography of the circumglobally distributed genus Seriola (Pisces: carangidae), Mol. Phylogenet. Evol., 93, 274–80. [DOI] [PubMed] [Google Scholar]
- 2. Sicuro B., Luzzana U.. 2016, The State of Seriola spp. Other Than Yellowtail (S. quinqueradiata) Farming in the World, Rev. Fish Sci. Aquac., 24, 314–25. [Google Scholar]
- 3. Ministry of Agriculture, Forestry and Fisheries of Japan. 2017, Annual Statistics of Fishery and Aquaculture Production Statistics Department. http://www.maff.go.jp/j/tokei/kouhyou/kaimen_gyosei/attach/pdf/index-7.pdf (in Japanese). [Google Scholar]
- 4. Dhirendra P.T. 2005, Cultured Aquatic Species Information Programme. Seriola quinqueradiata In: Cultured Aquatic Species Information Programme. Rome: FAO Fisheries and Aquaculture Department, pp. 1–11. [Google Scholar]
- 5. World Bank. 2013, Fish to 2030: Prospects for Fisheries and Aquaculture. Agriculture and Environmental Services Discussion Paper, No. 3. Washington DC: World Bank Group, pp. 1–102. [Google Scholar]
- 6. Abdelrahman H., ElHady M., Alcivar-Warren A., et al. 2017, Aquaculture genomics, genetics and breeding in the United States: current status, challenges, and priorities for future research, BMC Genomics, 18, 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Macqueen D.J., Primmer C.R., Houston R.D., et al. 2017, Functional Annotation of All Salmonid Genomes (FAASG): an international initiative supporting future salmonid research, conservation and aquaculture, BMC Genomics, 18, 484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yue G.H., Wang L.. 2017, Current status of genome sequencing and its applications in aquaculture, Aquaculture, 468, 337–47. [Google Scholar]
- 9. Aoki J.Y., Kai W., Kawabata Y., et al. 2015, Second generation physical and linkage maps of yellowtail (Seriola quinqueradiata) and comparison of synteny with four model fish, BMC Genomics. , 16, 406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fuji K., Koyama T., Kai W., et al. 2014, Construction of a high-coverage bacterial artificial chromosome library and comprehensive genetic linkage map of yellowtail Seriola quinqueradiata, BMC Res. Notes, 7, 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ohara E., Nishimura T., Nagakura Y., Sakamoto T., Mushiake K., Okamoto N.. 2005, Genetic linkage maps of two yellowtails (Seriola quinqueradiata and Seriola lalandi), Aquaculture, 244, 41–8. [Google Scholar]
- 12. Aoki J.Y., Kai W., Kawabata Y., et al. 2014, Construction of a radiation hybrid panel and the first yellowtail (Seriola quinqueradiata) radiation hybrid map using a nanofluidic dynamic array, BMC Genomics, 15, 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ozaki A., Yoshida K., Fuji K., et al. 2013, Quantitative trait loci (QTL) associated with resistance to a monogenean parasite (Benedenia seriolae) in yellowtail (Seriola quinqueradiata) through genome wide analysis, PloS One, 8, e64987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fuji K., Yoshida K., Hattori K., et al. 2010, Identification of the sex-linked locus in yellowtail, Seriola quinqueradiata, Aquaculture, 308, S51–5. [Google Scholar]
- 15. Koyama T., Ozaki A., Yoshida K., et al. 2015, Identification of sex-linked snps and sex-determining regions in the yellowtail genome, Mar. Biotechnol. , 17, 502–10. [DOI] [PubMed] [Google Scholar]
- 16. Iwasaki Y., Nishiki I., Nakamura Y., et al. 2016, Effective de novo assembly of fish genome using haploid larvae, Gene, 576, 644–9. [DOI] [PubMed] [Google Scholar]
- 17. Li H. 2013, Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM, arXiv Preprint, arXiv, 1303, 3997. [Google Scholar]
- 18. Kajitani R., Toshimoto K., Noguchi H., et al. 2014, Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads, Genome Res., 24, 1384–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J.. 1990, Basic local alignment search tool, J. Mol. Biol., 215, 403–10. [DOI] [PubMed] [Google Scholar]
- 20. Kai W., Nomura K., Fujiwara A., et al. 2014, A ddRAD-based genetic map and its integration with the genome assembly of Japanese eel (Anguilla japonica) provides insights into genome evolution after the teleost-specific genome duplication, BMC Genomics, 15, 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xue W., Li J.T., Zhu Y.P., et al. 2013, L_RNA_scaffolder: scaffolding genomes with transcripts, BMC Genomics., 14, 604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marcais G., Kingsford C.. 2011, A fast, lock-free approach for efficient parallel counting of occurrences of k-mers, Bioinformatics, 27, 764–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Catchen J.M., Amores A., Hohenlohe P., Cresko W., Postlethwait J.H.. 2011, Stacks: building and genotyping Loci de novo from short-read sequences, G3 (Bethesda), 1, 171–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. DePristo M.A., Banks E., Poplin R., et al. 2011, A framework for variation discovery and genotyping using next-generation DNA sequencing data, Nat. Genet., 43, 491–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Broman K.W., Wu H., Sen S., Churchill G.A.. 2003, R/qtl: qTL mapping in experimental crosses, Bioinformatics, 19, 889–90. [DOI] [PubMed] [Google Scholar]
- 26. Nishimura O., Hara Y., Kuraku S.. 2017, gVolante for standardizing completeness assessment of genome and transcriptome assemblies, Bioinformatics, 33, 3635–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Parra G., Bradnam K., Ning Z., Keane T., Korf I.. 2009, Assessing the gene space in draft genomes, Nucleic Acids Res., 37, 289–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hara Y., Tatsumi K., Yoshida M., Kajikawa E., Kiyonari H., Kuraku S.. 2015, Optimizing and benchmarking de novo transcriptome sequencing: from library preparation to assembly evaluation, BMC Genomics, 16, 977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Simao F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M.. 2015, BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs, Bioinformatics, 31, 3210–2. [DOI] [PubMed] [Google Scholar]
- 30. Meyer E., Aglyamova G.V., Wang S., et al. 2009, Sequencing and de novo analysis of a coral larval transcriptome using 454 GSFlx, BMC Genomics., 10, 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stanke M., Diekhans M., Baertsch R., Haussler D.. 2008, Using native and syntenically mapped cDNA alignments to improve de novo gene finding, Bioinformatics , 24, 637–44. [DOI] [PubMed] [Google Scholar]
- 32. Smit A., Hubley R., Green P.. 1996–2010, RepeatMasker Open-3.0. http://www.repeatmasker.org.
- 33. Smit A., Hubley R.. 2008–2010, RepeatModeler Open-1.0. http://www.repeatmasker.org.
- 34. Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L.. 2013, TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions, Genome Biol., 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Trapnell C., Williams B.A., Pertea G., et al. 2010, Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation, Nat. Biotechnol., 28, 511–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yates A., Akanni W., Amode M.R., et al. 2016, Ensembl 2016, Nucleic Acids Res., 44, D710–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Altschul S.F., Madden T.L., Schaffer A.A., et al. 1997, Gapped BLAST and PSI-BLAST: a new generation of protein database search programs, Nucleic Acids Res., 25, 3389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jones P., Binns D., Chang H.Y., et al. 2014, InterProScan 5: genome-scale protein function classification, Bioinformatics, 30, 1236–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gotz S., Garcia-Gomez J.M., Terol J., et al. 2008, High-throughput functional annotation and data mining with the Blast2GO suite, Nucleic Acids Res., 36, 3420–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Moriya Y., Itoh M., Okuda S., Yoshizawa A.C., Kanehisa M.. 2007, KAAS: an automatic genome annotation and pathway reconstruction server, Nucleic Acids Res., 35, W182–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nakamura Y., Mori K., Saitoh K., et al. 2013, Evolutionary changes of multiple visual pigment genes in the complete genome of Pacific bluefin tuna, Proc. Natl. Acad. Sci. U. S. A., 110, 11061–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xu T., Xu G., Che R., et al. 2016, The genome of the miiuy croaker reveals well-developed innate immune and sensory systems, Sci. Rep., 6, 21902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang Y., Coleman-Derr D., Chen G., Gu Y.Q.. 2015, OrthoVenn: a web server for genome wide comparison and annotation of orthologous clusters across multiple species, Nucleic Acids Res., 43, W78–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wagner G.P., Kin K., Lynch V.J.. 2012, Measurement of mRNA abundance using RNA-seq data: rPKM measure is inconsistent among samples, Theory Biosci., 131, 281–5. [DOI] [PubMed] [Google Scholar]
- 45. Mi H., Muruganujan A., Casagrande J.T., Thomas P.D.. 2013, Large-scale gene function analysis with the PANTHER classification system, Nat. Protoc., 8, 1551–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang H., Tan E., Suzuki Y., et al. 2015, Dramatic improvement in genome assembly achieved using doubled-haploid genomes, Sci. Rep., 4, 6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lu J., Peatman E., Tang H., Lewis J., Liu Z.. 2012, Profiling of gene duplication patterns of sequenced teleost genomes: evidence for rapid lineage-specific genome expansion mediated by recent tandem duplications, BMC Genomics, 13, 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pinto W., Ronnestad I., Jordal A.E., Gomes A.S., Dinis M.T., Aragao C.. 2012, Cloning, tissue and ontogenetic expression of the taurine transporter in the flatfish Senegalese sole (Solea senegalensis), Amino Acids, 42, 1317–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Takeuchi T. 2014, Progress on larval and juvenile nutrition to improve the quality and health of seawater fish: a review, Fish. Sci., 80, 389–403. [Google Scholar]
- 50. Takagi S., Murata H., Goto T., et al. 2005, The green liver syndrome is caused by taurine deficiency in yellowtail, Seriola quinqueradiata Fed diets without fishmeal, Aquacult. Sci., 53, 279–90. [Google Scholar]
- 51. Sarropoulou E., Sundaram A.Y.M., Kaitetzidou E., et al. 2017, Full genome survey and dynamics of gene expression in the greater amberjack Seriola dumerili, GigaScience, 6, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Purcell C.M., Seetharam A.S., Snodgrass O., Ortega-Garcia S., Hyde J.R., Severin A.J.. 2018, Insights into teleost sex determination from the Seriola dorsalis genome assembly, BMC Genomics, 19, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Malmstrom M., Matschiner M., Torresen O.K., et al. 2016, Evolution of the immune system influences speciation rates in teleost fishes, Nat. Genet., 48, 1204–10. [DOI] [PubMed] [Google Scholar]
- 54. Solbakken M.H., Voje K.L., Jakobsen K.S., Jentoft S.. 2017, Linking species habitat and past palaeoclimatic events to evolution of the teleost innate immune system, Proc. R Soc. B, 284, 20162810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Maeda M., Oshiman K., Tamura S., Futai M.. 1990, Human gastric (H+ + K+)-ATPase gene. Similarity to (Na+ + K+)-ATPase genes in exon/intron organization but difference in control region, J. Biol. Chem., 265, 9027–32. [PubMed] [Google Scholar]
- 56. Ogino C., Takeuchi L., Takeda H., Watanabe T.. 1979, Availability of dietary phosphorus in carp and rainbow trout, Nippon Suisan Gakkaishi, 45, 1527–32. [Google Scholar]
- 57. Guffey S., Esbaugh A., Grosell M.. 2011, Regulation of apical H(+)-ATPase activity and intestinal HCO(3)(-) secretion in marine fish osmoregulation, Am. J. Physiol. Regul. Integr. Comp. Physiol., 301, R1682–91. [DOI] [PubMed] [Google Scholar]
- 58. Calduch-Giner J.A., Sitja-Bobadilla A., Perez-Sanchez J.. 2016, Gene expression profiling reveals functional specialization along the intestinal tract of a carnivorous teleostean fish (Dicentrarchus labrax), Front. Physiol., 7, 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sachs G., Chang H.H., Rabon E., Schackman R., Lewin M., Saccomani G.. 1976, A nonelectrogenic H+ pump in plasma membranes of hog stomach, J. Biol. Chem., 251, 7690–8. [PubMed] [Google Scholar]
- 60. Foltmann B. 1981, Gastric proteinases–structure, function, evolution and mechanism of action, Essays Biochem., 17, 52–84. [PubMed] [Google Scholar]
- 61. Chong A.S.C., Hashim R., Chow-Yang L., Ali A.B.. 2002, Partial characterization and activities of proteases from the digestive tract of discus fish (Symphysodon aequifasciata), Aquaculture, 203, 321–33. [Google Scholar]
- 62. Deguara S., Jauncey K., Agius C.. 2003, Enzyme activities and pH variations in the digestive tract of gilthead sea bream, J. Fish Biol., 62, 1033–43. [Google Scholar]
- 63. Haard N.F. 1995, Digestibility and in vitro evaluation of vegetable proteins for salmonid feed In: Lim C., Sessa D.J. (eds), Nutrition and Utilization Technology in Aqualture, Champaign, IL: AOCS Press, pp. 199–219. [Google Scholar]
- 64. Wang Z., Du J., Lam S.H., Mathavan S., Matsudaira P., Gong Z.. 2010, Morphological and molecular evidence for functional organization along the rostrocaudal axis of the adult zebrafish intestine, BMC Genomics, 11, 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ikeda M., Kakizaki H., Matsumiya M.. 2017, Biochemistry of fish stomach chitinase, Int. J. Biol. Macromol., 104, 1672–81. [DOI] [PubMed] [Google Scholar]
- 66. Matsumiya M., Mochizuki A.. 1996, Distribution of chitinase and β-N-acetylhexosaminidase in the organs of several fishes, Fish. Sci., 62, 150–1. [Google Scholar]
- 67. Lang T., Klasson S., Larsson E., Johansson M.E., Hansson G.C., Samuelsson T.. 2016, Searching the evolutionary origin of epithelial mucus protein components-mucins and FCGBP, Mol. Biol. Evol., 33, 1921–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Koppers A.J., Reddy T., O’Bryan M.K.. 2011, The role of cysteine-rich secretory proteins in male fertility, Asian J. Androl., 13, 111–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sire M.F., Vernier J.-M.. 1992, Intestinal absorption of protein in teleost fish, Comp. Biochem. Physiol. A Physiol., 103, 771–81. [Google Scholar]
- 70. Lilleeng E., Froystad M.K., Ostby G.C., Valen E.C., Krogdahl A.. 2007, Effects of diets containing soybean meal on trypsin mRNA expression and activity in Atlantic salmon (Salmo salar L), Comp. Biochem. Physiol. A Mol. Integr. Physiol., 147, 25–36. [DOI] [PubMed] [Google Scholar]
- 71. Manchado M., Infante C., Asensio E., Crespo A., Zuasti E., Cañavate J.P.. 2008, Molecular characterization and gene expression of six trypsinogens in the flatfish Senegalese sole (Solea senegalensis Kaup) during larval development and in tissues, Comp. Biochem. Physiol. B, Biochem. Mol. Biol., 149, 334–44. [DOI] [PubMed] [Google Scholar]
- 72. Zhong G., Qian X., Hua X., Zhou H.. 2011, Effects of feeding with corn gluten meal on trypsin activity and mRNA expression in Fugu obscurus, Fish Physiol. Biochem., 37, 453–60. [DOI] [PubMed] [Google Scholar]
- 73. Choudhury M., Oku T., Yamada S., et al. 2011, Isolation and characterization of some novel genes of the apolipoprotein A-I family in Japanese eel, Anguilla Japonica, Cent. Eur. J. Biol., 6, 545. [Google Scholar]
- 74. Kim K.Y., Cho Y.S., Bang I.C., Nam Y.K.. 2009, Isolation and characterization of the apolipoprotein multigene family in Hemibarbus mylodon (Teleostei: cypriniformes), Comp. Biochem. Physiol. B Biochem. Mol. Biol., 152, 38–46. [DOI] [PubMed] [Google Scholar]
- 75. Qu M., Huang X., Zhang X., Liu Q., Ding S.. 2014, Cloning and expression analysis of apolipoprotein A-I (ApoA-I) in the Hong Kong grouper (Epinephelus akaara), Aquaculture, 432, 85–96. [Google Scholar]
- 76. Deng G., Laursen R.A.. 1998, Isolation and characterization of an antifreeze protein from the longhorn sculpin, Myoxocephalus octodecimspinosis, Biochim. Biophys. Acta, 1388, 305–14. [DOI] [PubMed] [Google Scholar]
- 77. Luquet P., Watanabe T.. 1986, Lipid nutrition in fish, Fish Physiol. Biochem., 2, 121., [DOI] [PubMed] [Google Scholar]
- 78. Ohno M., Kimura M., Miyazaki H., et al. 2016, Acidic mammalian chitinase is a proteases-resistant glycosidase in mouse digestive system, Sci. Rep., 6, 37756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Vannella K.M., Ramalingam T.R., Hart K.M., et al. 2016, Acidic chitinase primes the protective immune response to gastrointestinal nematodes, Nat. Immunol., 17, 538–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Martin S.A.M., Dehler C.E., Król E.. 2016, Transcriptomic responses in the fish intestine, Dev. Comp. Immunol., 64, 103–17. [DOI] [PubMed] [Google Scholar]
- 81. Rombout J.H.W.M., Yang G., Kiron V.. 2014, Adaptive immune responses at mucosal surfaces of teleost fish, Fish Shellfish Immunol., 40, 634–43. [DOI] [PubMed] [Google Scholar]
- 82. Concha M.I., Molina S., Oyarzun C., Villanueva J., Amthauer R.. 2003, Local expression of apolipoprotein A-I gene and a possible role for HDL in primary defence in the carp skin, Fish Shellfish Immunol., 14, 259–73. [DOI] [PubMed] [Google Scholar]
- 83. Concha M.I., Smith V.J., Castro K., Bastias A., Romero A., Amthauer R.J.. 2004, Apolipoproteins A-I and A-II are potentially important effectors of innate immunity in the teleost fish, Cyprinus Carpio. Eur. J. Biochem., 271, 2984–90. [DOI] [PubMed] [Google Scholar]
- 84. Villarroel F., Bastias A., Casado A., Amthauer R., Concha M.I.. 2007, Apolipoprotein A-I, an antimicrobial protein in Oncorhynchus mykiss: evaluation of its expression in primary defence barriers and plasma levels in sick and healthy fish, Fish Shellfish Immunol., 23, 197–209. [DOI] [PubMed] [Google Scholar]
- 85. Abrams J., Einhorn Z., Seiler C., Zong A.B., Sweeney H.L., Pack M.. 2016, Graded effects of unregulated smooth muscle myosin on intestinal architecture, intestinal motility and vascular function in zebrafish, Dis. Model. Mech., 9, 529–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Gutierrez J.A., Perr H.A.. 1999, Mechanical stretch modulates TGF-β1 and α1(I) collagen expression in fetal human intestinal smooth muscle cells, Am. J. Physiol. Gastrointest. Liver Physiol., 277, G1074–80. [DOI] [PubMed] [Google Scholar]
- 87. Grau A., Crespo S., Sarasquete M.C., de Canales M.L.G.. 1992, The digestive tract of the amberjack Seriola dumerili, Risso: a light and scanning electron microscope study, J. Fish Biol., 41, 287–303. [Google Scholar]
- 88. Christensen E.I., Birn H., Storm T., Weyer K., Nielsen R.. 2012, Endocytic receptors in the renal proximal tubule, Physiology (Bethesda), 27, 223–36. [DOI] [PubMed] [Google Scholar]
- 89. Haraldsson B. 2010, Tubular reabsorption of albumin: it's all about cubilin, J. Am. Soc. Nephrol., 21, 1810–2. [DOI] [PubMed] [Google Scholar]
- 90. Rodrı´guez A., Esteban M.A., Meseguer J.. 2003, A mannose-receptor is possibly involved in the phagocytosis of Saccharomyces cerevisiae by seabream (Sparus aurata L.) leucocytes, Fish Shellfish Immunol., 14, 375–88. [DOI] [PubMed] [Google Scholar]
- 91. Rosenstiel P., Sina C., End C., et al. 2007, Regulation of DMBT1 via NOD2 and TLR4 in intestinal epithelial cells modulates bacterial recognition and invasion, J. Immunol., 178, 8203–11. [DOI] [PubMed] [Google Scholar]
- 92. Wang A.M., Desnick R.J.. 1991, Structural organization and complete sequence of the human α-N-acetylgalactosaminidase gene: homology with the α-galactosidase A gene provides evidence for evolution from a common ancestral gene, Genomics, 10, 133–42. [DOI] [PubMed] [Google Scholar]
- 93. Sabapathy U., Teo L.H.. 1993, A quantitative study of some digestive enzymes in the rabbitfish, Siganus canaliculatus and the sea bass, Lates calcarifer, J. Fish Biol., 42, 595–602. [Google Scholar]
- 94. Kapoor B.G., Smit H., Verighina I.A.. 1976, The alimentary canal and digestion in teleosts In: Russell F.S., Yonge M. (eds), Advances in Marine Biology, London: Academic Press, pp. 109–239. [Google Scholar]
- 95. Hidalgo M.C., Urea E., Sanz A.. 1999, Comparative study of digestive enzymes in fish with different nutritional habits. Proteolytic and amylase activities, Aquaculture, 170, 267–83. [Google Scholar]
- 96. Lemieux H., Blier P., Dutil J.D.. 1999, Do digestive enzymes set a physiological limit on growth rate and food conversion efficiency in the Atlantic cod (Gadus morhua)?, Fish Physiol. Biochem., 20, 293–303. [Google Scholar]
- 97. Eshel A., Lindner P., Smirnoff P., Newton S., Harpaz S.. 1993, Comparative study of proteolytic enzymes in the digestive tracts of the european sea bass and hybrid striped bass reared in freshwater, Comp. Biochem. Physiol. A Physiol., 106, 627–34. [Google Scholar]
- 98. Jónás E., Rágyanszki M., Oláh J., Boross L.. 1983, Proteolytic digestive enzymes of carnivorous (Silurus glanis L.), herbivorous (Hypophthalmichthys molitrix Val.) and omnivorous (Cyprinus carpio L.) fishes, Aquaculture, 30, 145–54. [Google Scholar]
- 99. Chi L., Xu S., Xiao Z., et al. 2013, Pepsinogen A and C genes in turbot (Scophthalmus maximus): characterization and expression in early development, Comp. Biochem. Physiol. B, Biochem. Mol. Biol., 165, 58–65. [DOI] [PubMed] [Google Scholar]
- 100. Gudmundsdottir A., Palsdottir H.M.. 2005, Atlantic cod trypsins: from basic research to practical applications, Mar. Biotechnol., 7, 77–88. [DOI] [PubMed] [Google Scholar]
- 101. Fletcher T.S., Alhadeff M., Craik C.S., Largman C.. 1987, Isolation and characterization of a cDNA encoding rat cationic trypsinogen, Biochemistry, 26, 3081–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.