Abstract
Fruit ripening is a developmental process regulated by a complex network of endogenous and exogenous cues. Sea buckthorn is an excellent material for fruit ripening studies due to its dramatic ripening process and high contents of nutritional and anti-oxidant compounds in berries. Here, the whole transcriptome of sea buckthorn fruit at three development stages were analysed using multiple high-throughput sequencings. We assembled and annotated 9,008 long non-coding RNAs (lncRNAs) in sea buckthorn fruits, and identified 118 differentially expressed lncRNAs (DE-lncRNAs) and 32 differentially expressed microRNAs in fruit developmental process. In addition, we predicted 1,061 cis-regulated and 782 trans-regulated targets of DE-lncRNAs, and these DE-lncRNAs are specifically enriched in the biosynthesis of ascorbic acid, carotenoids and flavonoids. Moreover, the silencing of two lncRNAs (LNC1 and LNC2) in vivo and expression analysis revealed that LNC1 and LNC2 can act as endogenous target mimics of miR156a and miR828a to reduce SPL9 and induce MYB114 expression, respectively, which lead to increased and decreased anthocyanin content as revealed by high-performance liquid chromatography analysis. Our results present the first global functional analysis of lncRNA in sea buckthorn and provide two essential regulators of anthocyanin biosynthesis, which provides new insights into the regulation of fruit quality.
Keywords: anthocyanin biosynthesis, endogenous target mimics, fruit ripening, long non-coding RNAs, sea buckthorn
1. Introduction
Fruits are unique plant developmental systems representing an important constituent of human and animal diets due to their high mineral, vitamin and sugar contents.1 In addition, fruits are rich sources of anti-oxidant compounds, such as carotenoids, anthocyanins and flavonoids, and can therefore be used as dietary nutraceutics for human health.2,3 The textures, flavours and nutritional qualities of fruits are determined at the ripening stage.4 In fruit trees and crops, various genes and regulators have been identified as encoding or regulating enzymes in the biosynthetic pathways of fruit components during ripening.5 For example, in tomato, MADS-box genes, ripening inhibitor (RIN) and agamous-like 1 (TAGL1) dramatically affect fruit ripening.6,7 However, fruit ripening is a developmental process characterized by a series of transitions that are coordinated by a network of interacting genes and signalling pathways,8 and thus, the genetic and molecular factors that regulate ripening must be deciphered to improve fruit quality at the genome-wide level.
With the rapid development of transcriptome sequencing, a large proportion of non-coding RNAs (ncRNAs) have been found in the genome.9 Regulatory ncRNAs, including small ncRNAs and long ncRNAs (lncRNAs), play vital roles in plant growth and development.10 In the past decade, the functions of small ncRNAs including microRNAs and small interfering RNAs in plant development have been intensively studied.11 For example, in transcript regulation, sly-miR1534 plays a major role in the synthesis of plant hormones in tomato.12 Although many studies on fruit ripening have been performed,13–15 only a few studies have reported on the role of lncRNAs in fruit qualities and fruit pigmentation. Generally, lncRNAs are >200 nucleotides in length and mainly transcribed by RNA polymerase II, and they are involved in the regulation of various biological processes, including plant growth and development and the response to stress.16–18 According to their positions with respect to protein-coding genes, lncRNAs can be classified into intergenic lncRNAs (lincRNAs), anti-sense lncRNAs (lncNATs) and intronic lncRNAs.19 Currently, lncRNAs are regarded as star RNAs in many respects, including the regulation of gene expression, protein binding and the maintenance of chromosome stability.20 LncRNAs also function as precursors and endogenous target mimics (eTMs) for certain miRNAs, providing a new mechanism for the regulation of miRNA activity.21 With the development of next-generation sequencing, thousands of lncRNAs have been identified in a small number of plants, such as Arabidopsis,17 tomato,22Populus,23 wheat24 and maize.25 Recently, the regulatory mechanism of vernalization in Arabidopsis by COOLAIR (cool-assisted intronic ncRNA) and COLDAIR (cold-assisted intronic ncRNA), two lncRNAs transcribed from Flowering Locus C, has been illustrated.26 However, the functions of the majority of lncRNAs have not been fully studied to date.
Sea buckthorn (Hippophae rhamnoides L.) is a pioneer plant for land reclamation, and its fruits have been used for nutritional purposes for many centuries in Russia, Europe and Asia due to their high contents of fat-soluble vitamins (A, K and E), fatty acids, amino acids, vitamins C, B1 and B2, folic acid and phenols. The berries of sea buckthorn contain high amounts of natural anti-oxidants including ascorbic acid (ASA), tocopherols, carotenoids and flavonoids, which are known to have beneficial effects on human health.27 Because of its various beneficial contents and significant ecological value, sea buckthorn has become an ideal material for both basic research and application. However, the number, expression pattern, and characteristics of lncRNAs in sea buckthorn remain largely unknown. Therefore, it is necessary and urgent to identify novel lncRNAs as well as their functions in sea buckthorn fruit development.
Here, the whole transcriptome of three development stages fruit, including mature green (MG), breaker (BR) and red-ripe (RR), were analysed using multiple high-throughput sequencings and diverse bioinformatic platforms. Function and expression analysis revealed that the lncRNAs LNC1 and LNC2 can reduce and induce the expression of SPL9 and MYB114, respectively, to regulate anthocyanin biosynthesis and accumulation in sea buckthorn fruit.
2. Materials and methods
2.1. Plant materials
The sea buckthorn plant ‘hongguo’ (H. rhamnoides L. subsp. Mongolica Rousi × chinensis Rousi) was used in this study. All sea buckthorn plants were planted in the desert forest experimental centre in Inner Mongolia, China. Healthy fresh sea buckthorn fruits were harvested at 46, 63 and 76 days post-anthesis, which respectively considered as MG, BR and RR stages. The whole fruit was quick-frozen in liquid nitrogen and stored at -80°C. The sea buckthorn ‘hongguo’ was also planted for virus-induced gene silencing (VIGS) in fruits.
2.2. RNA extraction, library construction and sequencing
Total RNA was isolated from the MG, BR and RR fruits of ‘hongguo’ (three biological replicates per stages from two fruits each) using TRIzol reagent (Invitrogen, USA) according to the manufacturer’s protocol. Agarose gel electrophoresis was used for checking the RNA integrity. Then, total RNA was treated to remove rRNA using a kit. An Agilent 2100 Bioanalyzer was used to measure the quantity and quality of retaining RNA without rRNA. Then, nine transcriptome libraries were constructed using the Ribo-Zero Kit (Illumina, USA) for mRNA and lncRNAs sequencing. In addition, total RNA was also isolated from the MG, BR and RR fruits of ‘hongguo’ using TRIzol reagent (Invitrogen, USA). And, small RNA library construction of each stage and Illumina high-throughput sequencing were performed according to past research.28 These libraries were run on an Illumina HiSeq 2500 sequencer (Illumina, USA).
2.3. Transcriptome assembly and lncRNA identification
The raw reads were trimmed and quality-filtered using FastQC tools (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Each RNA-seq sequence dataset was aligned to the whole sea buckthorn genome (unpublished) by using TopHat2.29 The transcripts from each dataset were assembled using the Cufflinks 2.0 program.30 Cuffmerge was used to pool and merge the final transcripts. Then, the abundance of all transcripts from the BAM output files, estimated using fragments per kilobase of transcript per million mapped reads (FPKM) values, were calculated using Cuffdiff.
On the basis of the merged results, the remaining 654, 750 transcripts were used to identify the intergenic lncRNAs (lincRNAs), anti-sense lncRNAs (lncNATs) and intronic ncRNAs. The process was as follows: (i) Transcripts with an FPKM score <2 in a single exon or 0.5 in multiple exons in at least one sample were discarded; (ii) Transcripts with a length longer than 200 bp and an open reading frame (ORF) length shorter than 100 aa were retained; (iii) The CPC program was used to calculate the coding potential of the retained transcripts,31 and the transcripts with CPC scores >0 were discarded; (iv) HMMER32 was employed to scan the protein coding potential of each transcript against the Pfam protein family database with the transcript length >200 nucleotides, and no ORF encoding >100 amino acids; (v) The transcripts were compared against the NCBI non-redundant (NR) protein database, the Kyoto Encyclopedia of Genes and Genomes (KEGG) database and the Swiss-Protein database (Swiss-Prot) by BLASTX (E-value = 1e-10, coverage > 85%, and identity > 95%) to retain transcripts without significant homology to known proteins. The raw reads generated have been deposited in the NCBI SRA database under accession numbers SRP124171 for the miRNA-Seq reads and the ssRNA-Seq reads.
2.4. Differential expression of lncRNAs and mRNAs
Differentially expressed lncRNAs (DE-lncRNAs) between the samples at different developmental stages were identified using Cuffdiff. Any lncRNA exhibiting a log2-transformed |fold change| ≥ 1 and adjusted P-value < 0.05 was selected as a DE-lncRNA. On the basis of the features of the lncRNAs, their localization and abundance were shown using Circos.33
2.5. Prediction and functional analysis of DE-lncRNA target
To explore the functions of lncRNAs, we first predicted their cis and trans targets. In the cis role, lncRNAs act on neighbouring target genes. In this study, we searched for coding genes 100 kb upstream and downstream of a lncRNA. The trans role refers to the influence of lncRNAs on other genes at the expression level. Here, we calculated Pearson’s correlation coefficients between the expression levels of lncRNAs and mRNAs with custom scripts (|r| > 0.95). Then, we performed functional enrichment analysis of the target genes of lncRNAs by using goseq34 and KOBAS.35
2.6. Bioinformatics analysis of small RNAs
After small RNA sequencing, low-quality reads, adapter-contaminating tags and reads with lengths smaller than 18 nt were discarded. All unique clean reads were considered for ncRNA (rRNA, scRNA, snoRNA, snRNA and tRNA) identification in a BLAST all search against the Rfam (version 10.1) database.36 Next, the remaining reads were compared with known miRNAs of plants deposited in miRbase.37 Then, the secondary structures of these miRNAs were predicted by RNAfold. The frequency of the miRNAs was normalized to transcripts per million (TPM) in each sample. Differentially expressed miRNAs were identified by log2-transformed |fold change| > 1 and P-value < 0.05. To explore whether mRNAs functioned as targets of miRNAs, the transcripts and identified miRNAs were submitted to psRobot with an expectation ≤ 2.38 Transcripts containing a total of no more than four mismatches and G/U pairs within the complementary regions were considered as miRNA targets. The target mimics were predicted using psRNATarget combined with local scripts and the rules established by Wu et al.21
2.7. Construction of DE-lncRNA-miRNA-mRNA network
To infer the functions of lncRNAs, networks were constructed based on the complementary pairs of miRNAs-lncRNAs and miRNAs-mRNAs. The DE-lncRNA–miRNA–mRNA network was reconstructed based on ceRNA theory. First, expression correlation between lncRNAs and mRNAs was evaluated using the Pearson correlation coefficient (PCC). The lncRNA-mRNA pairs were considered as co-expression pairs with threshold PCC > 0.9 and P < 0.05. Then, for a lncRNA-mRNA pair, both mRNA and lncRNA, which as common miRNA targets and co-expressed negatively with this miRNA, were selected as co-expression competing triplet. Finally, all these co-expression competing triplets were used for constructing lncRNA–miRNA–mRNA network and visualized using Cytoscape software.39
2.8. qRT-PCR analysis
The total RNA from sea buckthorn fruit (MG, BR, RR) was used for quantitative PCR analysis. The RNA was then reverse transcribed to cDNA using ReverTra Ace reverse transcriptase (Toyobo). qRT-PCR of the randomly selected DE-lncRNAs was performed using a Bio-Rad CFX96 Touch™ RealTime PCR Detection System (Bio-Rad) with SYBR Green RealTime PCR Master Mix (ABI) according to the standard protocol. Specific primers for DE-lncRNAs and DE-miRNAs are listed in Supplementary Table S1. The 18S rRNA and U6 were used as the internal control genes for DE-lncRNAs and DE-miRNAs in these experiments. All reactions were conducted in triplicate for both technical and biological repetitions. The 2–ΔΔCT method was used to calculate the relative gene expression levels.
2.9. Subcellular localization of LNC1/2
The subcellular localization of two sea buckthorn DE-lncRNAs was examined using fluorescence in situ hybridization (FISH). Sea buckthorn (‘hongguo’) fruits at RR stage were collected for FISH. We selected three healthy fresh fruits for subcellular localization of LNC1/2. The pipeline of subcellular localization was followed as previously described,40 including probe synthesis, sample fixation and probe hybridization. Firstly, the whole fruit are fixed with fixative: n-Heptan (1:1) solution. Then, to preserve tissue morphology, samples were rinsed in 75% (v/v) ethanol, 50% (v/v) ethanol/PBS and 25% (v/v) ethanol/PBS each for 10 min. Tissues are incubated in Hybridization Mix at 52°C. Afterwards, samples were rinsed in Washing Solution to remove non-specific and/or repetitive RNA hybridization, and analysed by microscopy. Two probe sequences are listed in Supplementary Table S2.
2.10. VIGS of sea buckthorn fruits
According to previous study,16 VIGS of sea buckthorn fruit was performed using tobacco rattle virus (TRV). To avoid off-target silencing, we analysed LNC1 and LNC2 using the VIGS tool.16 A pTRV2-LNC1/2 construct was generated by inserting the EcoRI-digested PCR fragment of lncRNA into the pTRV2 vector. Agrobacterium strain GV3101 constructs containing pTRV1, pTRV2 and pTRV2-LNC1/LNC2 vectors were grown at 28°C in Luria Broth medium containing 10 mM MES, 20 μM acetosyringone, 50 μg ml−1 kanamycin and 50 μg ml−1 rifampicin. After shaking for 12 h, agrobacterium cells were harvested through centrifugation, resuspended in infiltration buffer (10 mM MgCl2, 200 μM acetosyringone and 5% sucrose) to a final OD600 of 1.0. Resuspensions of pTRV1 and pTRV2 or pTRV2-lncRNA were mixed at a ratio of 1:1. After 3 h of incubation, the agrobacterium was infiltrated into the fruits with a micro syringe. Sea buckthorn fruits infiltrated with pTRV1 and pTRV2 were used as controls (CK). Three different plants from each sample were used for infiltration. When the VIGS phenotype was visible, the sea buckthorn fruits were collected and stored at –80°C.
2.11. qRT-PCR of lncRNAs, miRNAs and genes involved in anthocyanin biosynthesis
Total RNA was extracted from the CK, TRV-LNC1 and TRV-LNC2 fruits using the Total RNA Kit (Aidlab, Beijing, China). Briefly, the first cDNA strand was obtained using the Prime Script™ RT Master Mix (Takara, Dalian, China). All primers used in this study are listed in Supplementary Table S1. qRT-PCR was performed on the Bio-Rad CFX96 Touch™ RealTime PCR Detection System using a standard SYBR Green PCR Kit (Bio-Rad). The conditions for quantitative PCR and the primers for the lncRNAs and genes are listed in Supplementary Table S1. Sample cycle threshold (Ct) values were determined and standardized relative to the endogenous control gene 18S rRNA, and the 2–ΔΔCT method was used to calculate the relative changes in gene expression based on the qRT-PCR data. All reactions were carried out in three biological repetitions. qRT-PCR was used to validate the sequencing results of two miRNAs and the genes involved in anthocyanin biosynthesis. The quantitative PCR conditions and the primers for the miRNAs and genes are listed in Supplementary Table S1.
2.12. Determination of anthocyanins by high-performance liquid chromatography analysis
Sea buckthorn fruit from CK, TRV-LNC1 and TRV-LNC2 plants was collected to quantify the content of anthocyanins. Each group included three biological repetitions. The method of anthocyanin determination was described in previous research.41
3. Results
3.1. Identification and characterization of sea buckthorn lncRNAs
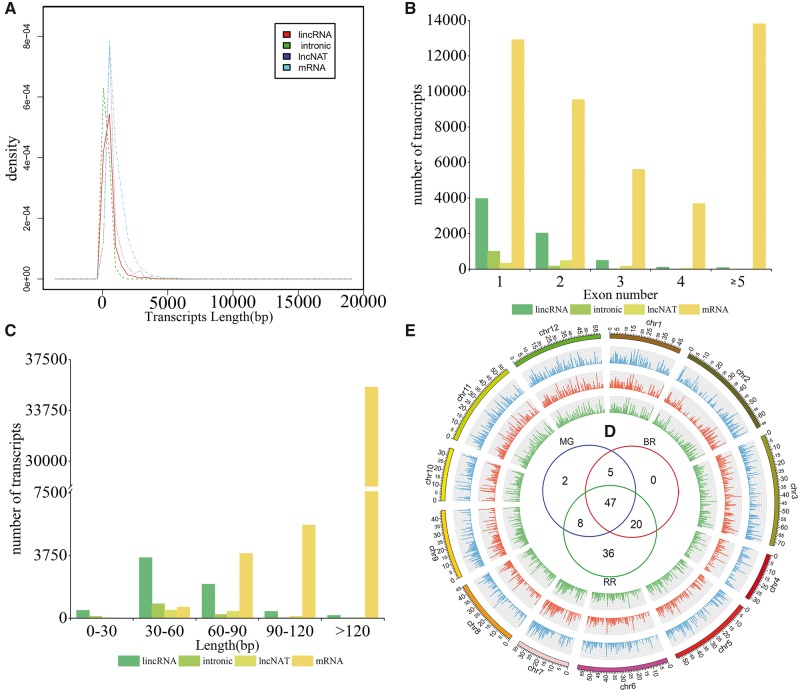
By integrating lncRNA computational identification methods, we developed a de novo lncRNA prediction pipeline using the resulting strand-specific RNA-seq (ssRNA-seq) data sets from three developmental stages of sea buckthorn (Supplementary Table S3; Supplementary Fig. S1). After mapping the RNA-seq data to the whole genome of sea buckthorn, we identified 9,008 lncRNAs in the fruits. The obtained lncRNAs were further classified into three types: lincRNAs, lncNATs and intronic lncRNAs, based on their locations to the protein coding genes. Whereas the majority of all lncRNAs (6,750) was located in intergenic regions, only 13.4% and 11.7% of all lncRNAs were intronic lncRNAs and lncNATs of protein-coding genes, respectively. A circos plot clearly showed that sea buckthorn lncRNAs were not evenly distributed across chromosomes (Fig. 1E), similar to the distribution of lncRNAs observed in other species.18,23,42 In addition, the expression level of lncRNAs varied between chromosomes. For instance, the average expression levels of lncRNAs generated from chromosomes 4 and 7 were 67.69 and 2.1415, respectively.
Figure 1.
Characteristics of sea buckthorn lncRNAs. (A) Length distributions of lncRNAs and transcripts. (B) Number of exons per lncRNA and transcript. (C) Distribution of the number of open reading frame (ORFs) in lncRNAs and transcripts. (D) A Venn diagram showing DE-lncRNAs that are commonly expressed in the mature green (MG), breaker (BR) and red-ripe (RR) stages as well as those specifically expressed in one but not another. (E) The expression levels of lncRNAs (log10FPKM) along the twelve sea buckthorn chromosomes (generated using Circos). Each of the three concentric rings corresponds to different samples. In the second, third and fourth track (outer to inner), each vertical line reports the expression of lncRNAs in sea buckthorn fruit in the MG, BR and RR stages, respectively.
We compared the lengths of transcript and ORF and exon counts of the 9,008 putative lncRNAs with identified mRNAs. The length distribution of the identified lncRNAs (median 271 bp; average 463.28 bp) was shorter than that of the mRNAs (median 714 bp; average 976.08 bp), while the length distribution showed no significant differences among intergenic lncRNAs, intronic lncRNAs and lncNATs (Fig. 1A). The exon counts of the putative lncRNAs (average 1.56 exons per transcript) were also lower than those of the mRNAs (average 4.19 exons per transcript), while the exon counts of the three kinds of lncRNA showed no differences (Fig. 1B). We found the ORF lengths of the majority of lncRNAs were shorter than mRNAs (Fig. 1C).
A total of 118 known lncRNAs and 108 novel transcripts were differentially expressed in the three samples (Supplementary Table S4). We also found fewer DE-lncRNAs (2.46%) than DE-mRNAs (5.64%, total mRNAs 45,546) in fruits at different development stages (Supplementary Table S5). This distinction indicated that lncRNAs might have a different expression pattern from protein-coding genes. Only two and 36 DE-lncRNAs were expressed specifically in the MG and RR stage, respectively (Fig. 1D). In contrast to MG, 38 and 72 lncRNAs were upregulated in BR or RR, and the remaining 44 and 46 lncRNAs were downregulated.
3.2. Functional analysis of DE-lncRNAs
To investigate the functions of DE-lncRNAs, we predicted the potential cis and trans targets of the lncRNAs. For the cis action of lncRNAs, computational prediction identified 1,061 potential target genes for 84 DE-lncRNAs (Supplementary Table S6). Gene Ontology (GO) analysis revealed that cis lncRNA targets significantly represented in the developmental process (Supplementary Fig. S2). Moreover, pathway analysis showed that these cis target genes of lncRNAs were enriched in plant hormone signal transduction, fatty acid and unsaturated fatty acid metabolism, carotenoid biosynthesis and flavonoid biosynthesis (Supplementary Table S7). On the other hand, the trans functions of 67 DE-lncRNAs were examined based on their expression correlation coefficients (|Pearson correlation| ≥ 0.95) with protein-coding genes. A total of 21,772 interactions were detected in trans between 67 DE-lncRNAs and 782 protein-coding genes in the sea buckthorn genome (Supplementary Table S8). Functional analysis showed that the trans target genes were also enriched in a variety of biological processes. Importantly, we observed a few plant growth terms, including the regulation of growth, cell wall organization or biogenesis, and regulation of cellular component organization. Of the identified 110 KEGG pathways, five were associated with fruit development, namely, ascorbate and aldarate metabolism, flavonoid biosynthesis and brassinosteroid biosynthesis (Supplementary Table S9).
3.3. DE-lncRNAs participate in miRNA-lncRNA-mRNA networks
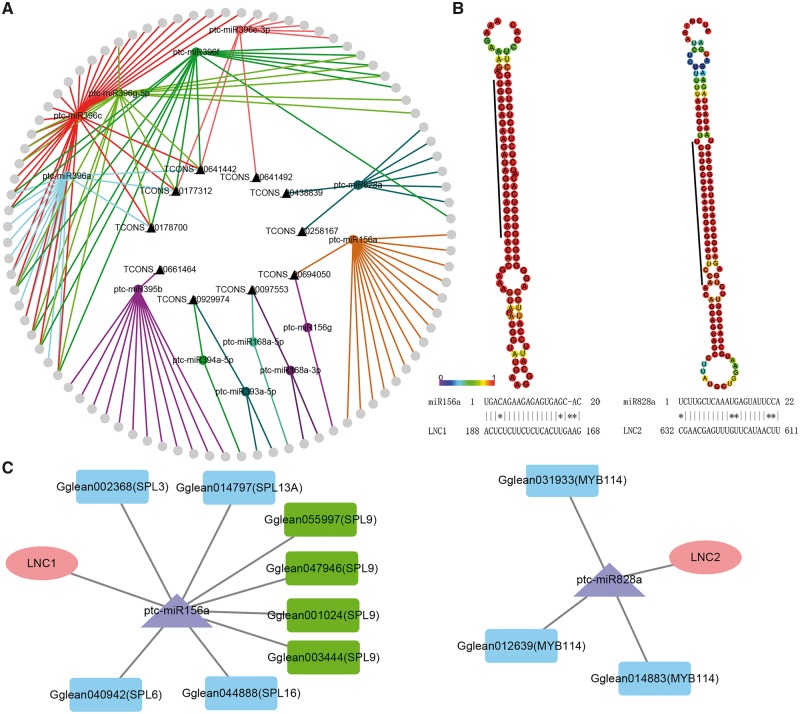
Recent evidence suggested the roles of miRNAs and lncRNAs in regulating the expression of mRNAs in many species,43–45 but the comprehensive patterns of miRNA and lncRNAs in sea buckthorn remain unknown. We predicted 68 conserved miRNAs and 79 pre-miRNAs, which were identified as belonging to 31 known miRNA families deposited in miRBase. A total of 32 DE-miRNAs were predicted to target 67 genes in this study (Supplementary Table S10). The target prediction analysis revealed that 22 lncRNAs may act as eTMs of 25 DE-miRNAs (Supplementary Table S11). Recently, some researches have shown that lncRNAs can affect the regulation of miRNAs as eTMs in plants.21,46 To investigate the function of lncRNAs as miRNA targets, a comprehensive genome-wide network mediated by miRNAs was constructed. The network consisted of 109 nodes and 201 edges, including 13 DE-miRNAs, 10 DE-lncRNAs and 87 DE-mRNAs (Fig. 2A). The majority of the nodes in the network belonged to the miR396 family, which regulates growth by targeting growth regulation factors. Then, the function of each lncRNA was inferred based on the functions of the connected mRNAs. We found that lncRNAs mainly participate in cellular, metabolic and some biological processes, especially highly enriched in cellular component, binding including ‘ion binding’, ‘nucleotide binding’ and ‘small molecule binding’ (Supplementary Table S12).
Figure 2.
Analysis of lncRNA-miRNA-mRNA networks. (A) The networks of lncRNA-miRNA-mRNA. The triangle represents lncRNAs, the circle with label represents miRNAs, and the circle without label represents mRNAs. (B) LNC1/2 as endogenous target mimics (eTMs) of miRNAs. Secondary structure of miRNAs precursors and the basepairing relationship between lncRNA and miRNAs. The predicted secondary structure was generated using RNAfold. (C) Sub interaction networks of LNC1/2 from lncRNA-miRNA-mRNA networks. The ellipse represents lncRNAs, the triangle represents miRNAs, and the rectangle represents mRNAs.
3.4. Analysis of lncRNAs related to the anthocyanin biosynthesis of fruits
According to previous studies, the module miR156-SPL affects anthocyanin biosynthesis in plants.47 Additionally, R2R3-MYBs (MYB114) are known to combine with bHLHs and WDR to form a ternary complex that activates late biosynthesis genes and transfers protein genes.48 Here, based on the miRNA-lncRNA-mRNA network, we identified two anthocyanin biosynthesis-related lncRNAs (TCONS_00694050 and TCONS_00438839, termed LNC1 and LNC2) for further analysis (Fig. 2). On the basis of the network, we found that LNC1 and LNC2 may regulate the expression of SPL9 and MYB114 by acting as eTMs of miR156a and miR828a.
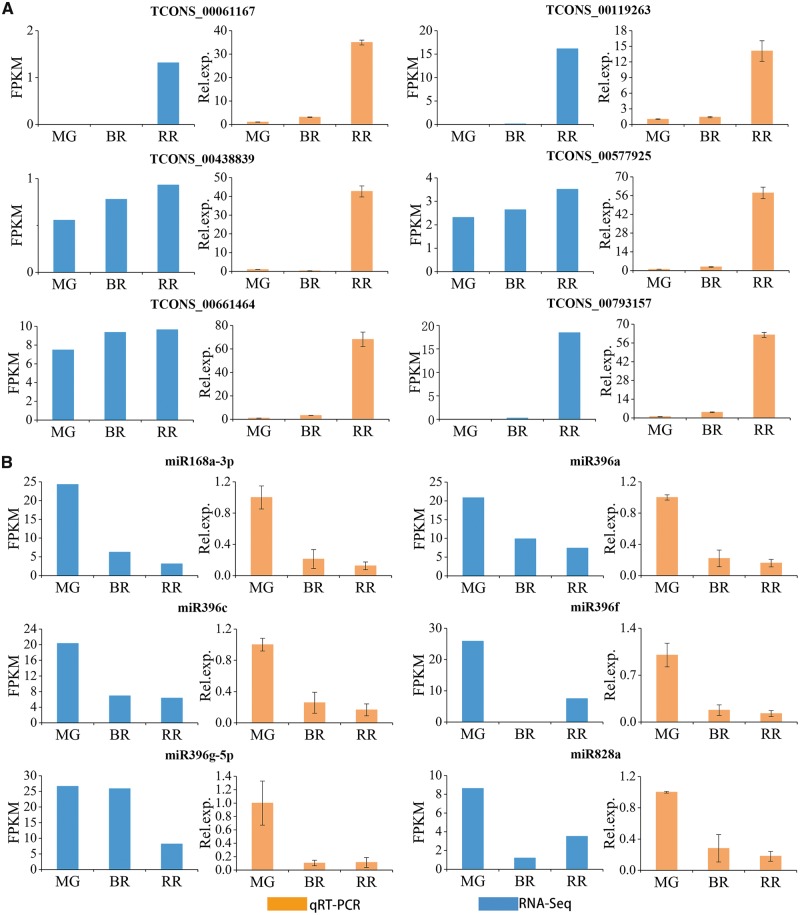
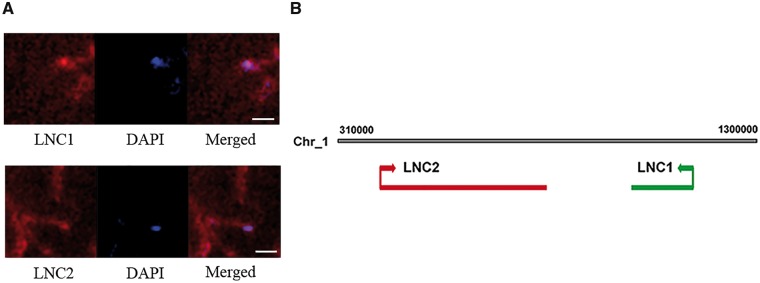
To validate the putative relationships between the miRNAs and lncRNAs, their expression levels were examined by qRT-PCR (Fig. 3). The qRT-PCR results were consistent with our RNA-seq data regarding lncRNA and miRNA expression, suggesting that these two lncRNAs identified by next-generation sequencing might have negative regulation relation with miR156a and miR828a. Distribution analysis is an important step towards functional research on lncRNAs as eTMs. To demonstrate the subcellular localization of LNC1 and LNC2, FISH was performed in fruits using a fluorescence probe specific to LNC1 and LNC2. In combination with the DAPI signal, the probe showed that these two lncRNAs were mainly enriched in cytoplasm (Fig. 4A). Genome chromosomal location analyses revealed that both LNC1 and LNC2 reside in chromosome 1 and lack homologs compared with other plants (Fig. 4B).
Figure 3.
qRT-PCR validation of putative lncRNAs and miRNAs in sea buckthorn. The expression levels of lncRNAs (A) and miRNAs (B) in three developmental stages: mature green (MG), breaker (BR) and red-ripe (RR).
Figure 4.
Subcellular localization and chromosome location of LNC1 and LNC2 in sea buckthorn fruit. (A) Visualization of LNC1 and LCN2 in sea buckthorn fruit by RNA fluorescence in situ hybridization using an antisense probe. Scale bar, 20 μm. (B) Location regions of LNC1 and LNC2 on chromosome.
3.5. LNC1 and LNC2 regulate anthocyanin biosynthesis in fruits
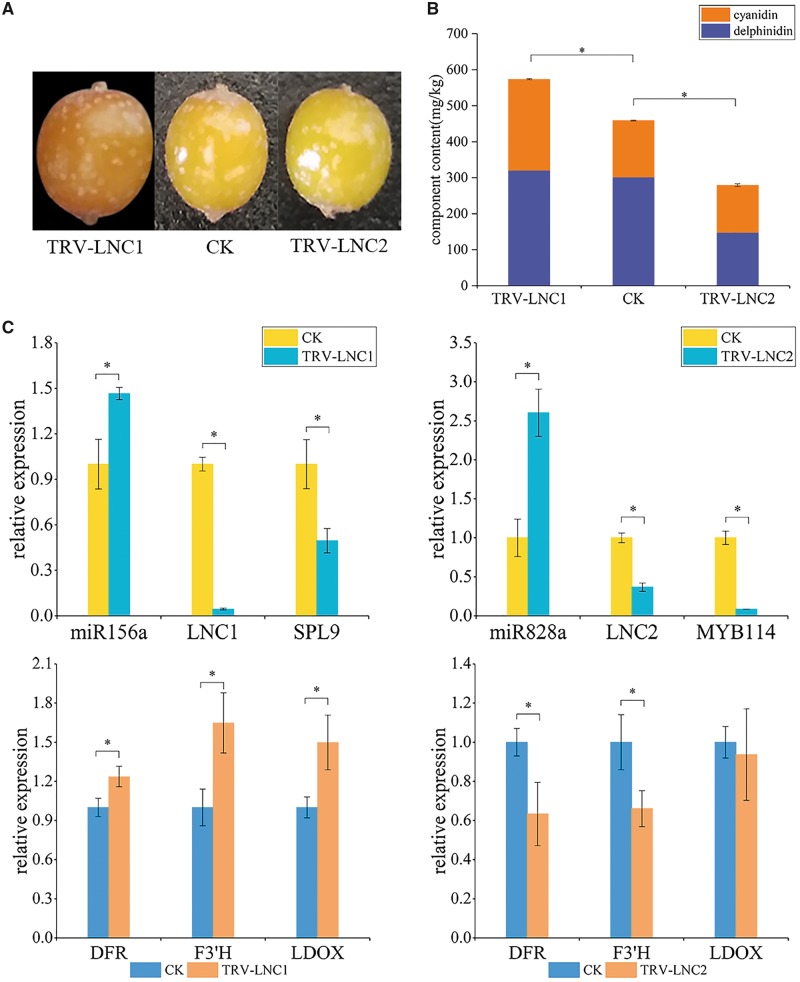
To test the regulation role of LNC1 and LNC2 in anthocyanin biosynthesis, VIGS was performed to silence LNC1 and LNC2 in sea buckthorn fruits (Fig. 5A). Intriguingly, two or three weeks after infiltration, anthocyanin contents of sea buckthorn fruits injected with TRV-LNC1 or TRV-LNC2 showed significant difference compared with that of control fruits injected with TRV (Fig. 5B). Semi-quantitative PCR analysis suggested that a recombinant virus could spread from carpogonia to fruits, resulting in the VIGS of lncRNAs in sea buckthorn fruits. Compared with that in TRV control fruits, the transcript level of LNC1 in TRV-LNC1 fruits was dramatically decreased by 96% (Fig. 5C). Similarly, the level of LNC2 in TRV-LNC2 fruits decreased by 63.3% compared with that in TRV control fruits (Fig. 5C). Consistently, the anthocyanin content of TRV-LNC1 fruit increased by 25.23% with respect to TRV control fruits, and the anthocyanin content of TRV-LNC2 fruit decreased by 39.12% with respect to TRV control fruits (Fig. 5C). Previous studies have reported that SPL and MYB transcription factors can affect anthocyanin content by regulating the expression of genes involved into anthocyanin biosynthesis pathway, such as ANS, DFR, F3′H, LDOX, UGT78D2 and UGT75C1.48 Similarly, three genes involved in anthocyanin biosynthesis were increased or decreased in TRV-LNC1 compared with their levels in TRV control sea buckthorn fruits (Fig. 5C). In contrast, DFR, F3′H and LDOX were decreased in TRV-LNC2 compared with their levels in TRV control sea buckthorn fruits (Fig. 5C). These results strongly suggested the essential role of the two novel lncRNAs in the regulation of the anthocyanin accumulation of sea buckthorn fruits.
Figure 5.
Silencing of LNC1/2 in sea buckthorn fruit. (A) Sea buckthorn fruits (3 weeks after infiltration). CK: sea buckthorn fruits infiltrated with pTRV1 and pTRV2; TRV-LNC1: sea buckthorn infiltrated with pTRV1 and pTRV2-LNC1; TRV-LNC2: sea buckthorn infiltrated with pTRV1 and pTRV2-LNC2. (B) High-performance liquid chromatography analysis of total anthocyanin content in sea buckthorn fruits. (C) qRT -PCR analysis of the expression of related genes in sea buckthorn fruits. The expression of 18S rRNA was used as an internal reference. The relative level was normalized to that in TRV control plants. Error bars indicate SD of three biological replicates, each sample measured in three replicates. Asterisks indicate a significant difference as determined by Student's t-test (*P < 0.01).
4. Discussion
4.1. A reliable list of lncRNAs from sea buckthorn fruits
Recently, plenty of studies have revealed the crucial roles of lncRNAs in various biological processes in plants.17,45,49 Although many lncRNAs have been identified in model plants, such as Arabidopsis,50 rice,51 and maize,25 little research has been performed on sea buckthorn. In this study, a total of 9008 lncRNA loci were identified in the fruit of sea buckthorn, an ideal material for the study of fruit development. Furthermore, structural analysis showed that the mean length of lncRNAs was 1.1-fold shorter than that of coding transcripts, and the lncRNA expression level was lower than that of coding transcripts, in agreement with previous studies.16,42,52 LncRNAs also have specific expression patterns in tissue types and subcellular compartments. In our study, among the significant DE-lncRNAs, we found five expression patterns in MG, BR and RR.
4.2. DE-lncRNAs contribute to fruit development of sea buckthorn fruit
Transcriptomic sequencing of different varieties of sea buckthorn revealed a lot of ncRNAs in many biological processes.28,53 In this study, we predicted potential cis and trans target genes based on physical location and expression relationships with mRNA, respectively. Although the functions of the majority of lncRNAs are unknown, previous studies have suggested that lncRNAs may play different roles in a variety of biological processes. According to previous study,21 we analysed the functions of DE-lncRNAs based on their target mRNAs, which involved in many processes, including carotenoid, ASA, flavonoid, sugar and hormone pathways.
We identified 10 DE-lncRNAs involved in carotenoid biosynthesis pathways, including β-carotenoid and lycopene biosynthesis. Among these DE-lncRNAs, one lncRNA (TCONS_00082246) targeted the phytoene synthase (PSY) gene, which is involved in the first committed step in carotenoid biosynthesis (Supplementary Table S12). This reaction has been reported to be the rate-limiting step controlling the metabolic flux to carotenoid biosynthesis in many plants.54 For example, the PSY gene plays an important role in carotenoid biosynthesis in tomato.12 Therefore, this study opened the door to understanding the role of lncRNAs in the regulation of genes involved in carotenoid biosynthesis in sea buckthorn fruit. Recently, many studies have shown that carotenoids are considered as the main fruit pigments in tomato, bilberry, citrus, papaya, watermelon and sea buckthorn.55–57 In addition, lycopene accumulation is the most important reason for the pigmentation of red fruit in sea buckthorn. Therefore, these results revealed that lncRNAs may play roles in the development of sea buckthorn fruit pigmentation.
Sea buckthorn fruit contains abundant ASA and is known as the king of vitamin C, and ASA is a prominent product of the ascorbate and aldarate metabolism pathway. In this pathway, we found seven lncRNAs targeting GDP-mannose-epimerase (GME) and SKS12, which encodes l-ascorbate oxidase. Several studies have revealed that GME and SKS12 are associated with the accumulation of ASA in the fruits of many plants, including blueberry, Arabidopsis, tomato and peach.58–61 For instance, the importance of GME in the major ascorbate biosynthesis pathway in tomato was confirmed using gene-silencing technology.62 Thus, our results provide a resource for further analysis of the regulation of ASA biosynthesis in sea buckthorn fruit.
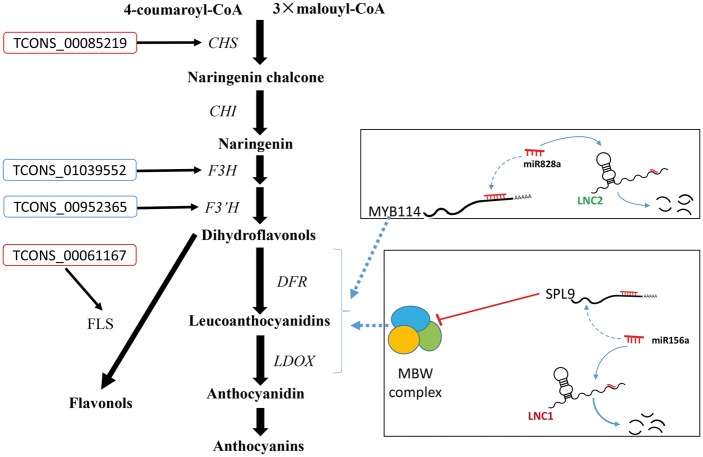
Flavonoids are a large group of natural products that are widely present in the leaves, flowers, fruits and seeds of various plants. According to their basic chemical structures, flavonoids can be further divided into different subgroups, including flavonols, anthocyanins and proanthocyanidins. Our results consistent with those of previous studies, reveal that sea buckthorn fruit contains various flavonoids. In the flavonoid biosynthesis pathway, chalcone synthase (CHS) is the first committed enzyme that facilitates the stepwise synthesis of chalcone, and it is also pivotal for the biosynthesis of flavonoid and anthocyanin pigments in plants.63 TCONS_00085219 was predicted to target CHS, indicating a possible regulatory relationship between lncRNA and CHS (Fig. 6). Another important enzyme involved in this pathway is flavanone 3-hydroxylase (F3H), which was reported in rice.58 TCONS_01039552 was predicted to target F3H, implying a potential role in flavonol and anthocyanin biosynthesis through the regulation of F3H. In addition to CHS and F3H, other important genes, such as the gene encoding flavonol synthase 1, were predicted as target genes of two lncRNAs (TCONS_00061167 and TCONS_00061354) identified in our study. All these results implied the widespread involvement of lncRNAs in the regulation of flavonoid biosynthesis and fruit pigment in the development of sea buckthorn fruit, which will provide novel insight into the regulation of flavonoid biosynthesis in other plants.
Figure 6.
Model of lncRNA regulation in sea buckthorn fruit. This pathway begins with the general phenylpropanoid metabolism, and subsequent steps are catalyzed by a series of structural enzymes, leading to the biosynthesis of flavonols and anthocyanins. The expression of the late biosynthetic genes (LBGs) requires the transcriptional activation activity of the R2R3-MYB/bHLH/WDR (MBW) complex. LncRNAs target FLS and CHS: functions in trans regulation. LncRNAs target F3H and F3′H: functions in cis regulation. CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3-hydroxylase; F3′H, flavanone 3′-hydroxylase; DFR, dihydroflavonol 4-reductase; LDOX, leucoanthocyanidin dioxygenase.
4.3. LNC1 and LNC2 regulate anthocyanin biosynthesis in the fruit development of sea buckthorn by eTMs
Previous studies suggest that lncRNAs function via interacting with miRNAs.21 The reverse miRNA-lncRNA regulation, first discovered in Arabidopsis, was termed target mimicry.64 Previous research systematically identified 36 eTMs for 11 Arabidopsis miRNAs and 189 target mimics for 19 rice miRNAs.21,51 In Populus, seven miRNAs target TCONS_00013609, including pto-miR6462a/b/c.60 Therefore, the prediction and analysis of miRNAs that interact with lncRNAs provide a useful way to explore the functions of the corresponding lncRNAs. In this study, we identified 19 DE-lncRNA eTMs for 23 DE-miRNAs (seven families). These results first showed that miRNAs target lncRNAs in sea buckthorn. Furthermore, our investigation constructed a comprehensive RNA-mediated network, including DE-miRNA–DE-lncRNA and DE-miRNA–DE-mRNA interactions.
However, a mechanistic understanding of the roles of lncRNAs in plants, especially trees, has remained extremely lacking. lncRNA1459 and lncRNA1840 were found to regulate the ripening of tomato.16 For mammal, lnc-mg was identified to promote myogenesis by functioning as a ceRNA/eTM for microRNA-125b to control the protein abundance of insulin-like growth factor 2.65 In Arabidopsis, a representation of the classical TM model, miR399: IPS1, was discovered by Franco-Zorrilla.64 Few studies have reported on the mechanism of lncRNA involvement in anthocyanin biosynthesis during fruit development. In this study, we identified two lncRNAs (LNC1 and LNC2) involved in the anthocyanin biosynthesis pathway (Fig. 6). Additionally, we first used FISH and VIGS for function analysis of lncRNAs in sea buckthorn. The VIGS results provide strong evidence that these two lncRNAs function in fruit anthocyanin biosynthesis. When LNC1 and LNC2 expression is reduced, anthocyanin accumulation is increased or decreased, respectively. This mechanism shows that anthocyanin accumulation is regulated through the LNC1-induced downregulation of SPL9, which affects the stability of the MYB, bHLH and WDR (MBW) complex, thus promoting anthocyanin biosynthesis in sea buckhorn fruit. In contrast, the LNC2-induced downregulation of MYB114 reduced anthocyanin biosynthesis in sea buckhorn fruit. Thus, this study illuminated the complex regulation of fruit anthocyanin biosynthesis, which might instigate more comprehensive studies on sea buckthorn lncRNAs. Furthermore, the functional motifs and target genes of lncRNAs in trees need to be investigated further to fully elucidate the regulatory mechanisms of lncRNAs in trees.
Conflict of interest
None declared.
Funding
This study was supported by grants from Special Fund for Forest Scientific Research in the Public Welfare (201504103) and the National Natural Science Foundation of China (31470616).
Supplementary Material
References
- 1. Giovannoni J.J. 2003, Genetic regulation of fruit development and ripening, Plant Cell, 16(Suppl), S170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Winkel-Shirley B. 2001, It takes a garden. How work on diverse plant species has contributed to an understanding of flavonoid metabolism, Plant Physiol., 127, 1399–404. [PMC free article] [PubMed] [Google Scholar]
- 3. Liu C.J., Deavours B.E., Richard S.B.. 2006, Structural basis for dual functionality of isoflavonoid O-methyltransferases in the evolution of plant defense responses, Plant Cell, 18, 3656–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carrari F., Fernie A.R.. 2006, Metabolic regulation underlying tomato fruit development, J. Exp. Bot., 57, 1883–97. [DOI] [PubMed] [Google Scholar]
- 5. Boss P.K., Davies C., Robinson S.P.. 1996, Analysis of the expression of anthocyanin pathway genes in developing Vitis vinifera L. cv shiraz grape berries and the implications for pathway regulation, Plant Physiol., 111, 1059–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nguyen C.V., Vrebalov J.T., Gapper N.E., et al. 2014, Tomato GOLDEN2-LIKE transcription factors reveal molecular gradients that function during fruit development and ripening, Plant Cell, 26, 585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ito Y., Nishizawa-Yokoi A., Endo M., Mikami M., Toki S.. 2015, CRISPR/Cas9-mediated mutagenesis of the RIN locus that regulates tomato fruit ripening, Biochem. Biophys. Res. Commun., 467, 76–82. [DOI] [PubMed] [Google Scholar]
- 8. Liu M., Pirrello J., Chervin C., Roustan J.P., Bouzayen M.. 2015, Ethylene control of fruit ripening: revisiting the complex network of transcriptional regulation, Plant Physiol., 169, 2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wilhelm B.T., Marguerat S., Watt S., et al. 2008, Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution, Nature, 453, 1239. [DOI] [PubMed] [Google Scholar]
- 10. Kim E.D., Sung S.. 2012, Long noncoding RNA: unveiling hidden layer of gene regulatory networks, Trends Plant Sci., 17, 16. [DOI] [PubMed] [Google Scholar]
- 11. Liu J., Jung C., Xu J., et al. 2012, Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis, Plant Cell, 24, 4333–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Din M., Barozai M.Y.K.. 2014, Profiling microRNAs and their targets in an important fleshy fruit: tomato (Solanum lycopersicum), Gene, 535, 198–203. [DOI] [PubMed] [Google Scholar]
- 13. Kallio H., Yang B., Peippo P.. 2002, Effects of different origins and harvesting time on vitamin C, tocopherols, and tocotrienols in sea buckthorn (Hippophaë rhamnoides) berries, J. Agric. Food Chem., 50, 6136–42. [DOI] [PubMed] [Google Scholar]
- 14. Karlova R., Chapman N., David K., Angenent G.C., Seymour G.B., De Maagd R.A.. 2014, Transcriptional control of fleshy fruit development and ripening, J. Exp. Bot., 65, 4527–41. [DOI] [PubMed] [Google Scholar]
- 15. Giovannoni J., Nguyen C., Ampofo B., Zhong S., Fei Z.. 2017, The epigenome and transcriptional dynamics of fruit ripening, Annu. Rev. Plant Biol., 68, 61. [DOI] [PubMed] [Google Scholar]
- 16. Zhu B., Yang Y., Li R., et al. 2015, RNA sequencing and functional analysis implicate the regulatory role of long non-coding RNAs in tomato fruit ripening, J. Exp. Bot., 66, 4483–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Seo J.S., Sun H.X., Park B.S., et al. 2017, ELF18-INDUCED LONG NONCODING RNA associates with Mediator to enhance expression of innate immune response genes in Arabidopsis, Plant Cell, 29, 1024., tpc.00886.02016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li S., Yu X., Lei N., et al. 2017, Genome-wide identification and functional prediction of cold and/or drought-responsive lncRNAs in cassava, Sci. Rep., 7, 45981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chekanova J.A. 2015, Long non-coding RNAs and their functions in plants, Curr. Opin. Plant Biol., 27, 207–16. [DOI] [PubMed] [Google Scholar]
- 20. Shafiq S., Li J., Sun Q.. 2016, Functions of plants long non-coding RNAs, Biochim. Biophys. Acta, 1859, 155–62. [DOI] [PubMed] [Google Scholar]
- 21. Wu H.-J., Wang Z.-M., Wang M., Wang X.-J.. 2013, Widespread long noncoding RNAs as endogenous target mimics for microRNAs in plants, Plant Physiol., 161, 1875–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mou W., Li D., Bu J., et al. 2016, Comprehensive analysis of ABA effects on ethylene biosynthesis and signaling during tomato fruit ripening, PLoS One, 11, e0154072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen M., Wang C., Bao H., Chen H., Wang Y.. 2016, Genome-wide identification and characterization of novel lncRNAs in Populus under nitrogen deficiency, Mol. Genet. Genomics, 291, 1663–80. [DOI] [PubMed] [Google Scholar]
- 24. Huang D., Feurtado J.A., Smith M.A., Flatman L.K., Koh C., Cutler A.J.. 2017, Long noncoding miRNA gene represses wheat beta diketone waxes, Proc. Natl. Acad. Sci. U.S.A., 114, E3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lv Y., Liang Z., Min G., et al. 2016, Genome-wide identification and functional prediction of nitrogen-responsive intergenic and intronic long non-coding RNAs in maize (Zea mays L.), BMC Genomics, 17, 350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Csorba T., Questa J.I., Sun Q., Dean C.. 2014, Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization, Proc. Natl. Acad. Sci. U.S.A., 111, 16160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zakynthinos G., Varzakas T., Petsios D.. 2016, Sea Buckthorn (Hippophae rhamnoides) lipids and their functionality on health aspects, Curr. Res. Nutr. Food Sci., 4, 182–94. [Google Scholar]
- 28. Ding J., Ruan C., Guan Y., Krishna P.. 2018, Identification of microRNAs involved in lipid biosynthesis and seed size in developing sea buckthorn seeds using high-throughput sequencing, Sci. Rep., 8, 4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L.. 2013, TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions, Genome Biol., 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Trapnell C., Roberts A., Goff L., et al. 2012, Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks, Nat. Protoc., 7, 562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kong L., Zhang Y., Ye Z.Q., et al. 2007, CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine, Nucleic Acids Res., 35, W345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Finn R.D., Clements J., Eddy S.R.. 2011, HMMER web server: interactive sequence similarity searching, Nucleic Acids Res., 39, W29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Krzywinski M., Schein J., Birol İ., et al. 2009, Circos: an information aesthetic for comparative genomics, Genome Res., 19, 1639–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Young M.D., Wakefield M.J., Smyth G.K., Oshlack A.. 2010, Gene ontology analysis for RNA-seq: accounting for selection bias, Genome Biol., 11, R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu J., Mao X., Cai T., Luo J., Wei L.. 2006, KOBAS server: a web-based platform for automated annotation and pathway identification, Nucleic Acids Res., 34, W720–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Griffithsjones S., Moxon S., Marshall M., Khanna A., Eddy S.R., Bateman A.. 2004, Rfam: annotating non-coding RNAs in complete genomes, Nucleic Acids Res., 33, D121–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kozomara A., Griffiths-Jones S.. 2014, miRBase: annotating high confidence microRNAs using deep sequencing data, Nucleic Acids Res., 42, D68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu H.J., Ma Y.K., Chen T., Wang M., Wang X.J.. 2012, PsRobot: a web-based plant small RNA meta-analysis toolbox, Nucleic Acids Res., 40, W22–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shannon P., Markiel A., Ozier O., et al. 2003, Cytoscape: a software environment for integrated models of biomolecular interaction networks, Genome Res., 13, 2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bleckmann A., Dresselhaus T.. 2016, Fluorescent whole-mount RNA in situ hybridization (F-WISH) in plant germ cells and the fertilized ovule, Methods, 98, 66–73. [DOI] [PubMed] [Google Scholar]
- 41. Fei L., Giusti M.M.. 2016, Quantification of purple corn (Zea mays L.) anthocyanins using spectrophotometric and HPLC approaches: method comparison and correlation, Food Anal. Methods, 9, 1367–80. [Google Scholar]
- 42. Kang C., Liu Z.. 2015, Global identification and analysis of long non-coding RNAs in diploid strawberry Fragaria vesca during flower and fruit development, BMC Genomics, 16, 815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. José R.J., Bailey L.J., Mai Q.A., et al. 2015, microRNA regulation of fruit growth, Nat. Plants, 1, 15036. [DOI] [PubMed] [Google Scholar]
- 44. Wang C.Y., Liu S.R., Zhang X.Y., Ma Y.J., Hu C.G., Zhang J.Z.. 2017, Genome-wide screening and characterization of long non-coding RNAs involved in flowering development of trifoliate orange (Poncirus trifoliata L. Raf.), Sci. Rep., 7, 43226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cui J., Luan Y., Jiang N., Bao H., Meng J.. 2017, Comparative transcriptome analysis between resistant and susceptible tomato allows the identification of lncRNA16397 conferring resistance to Phytophthora infestans by co-expressing glutaredoxin, Plant J., 89, 577. [DOI] [PubMed] [Google Scholar]
- 46. Fan C., Hao Z., Yan J., Li G.. 2015, Genome-wide identification and functional analysis of lincRNAs acting as miRNA targets or decoys in maize, BMC Genomics, 16, 793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gou J.Y., Felippes F.F., Liu C.J., Weigel D., Wang J.W.. 2011, Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor, Plant Cell, 23, 1512–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gonzalez A., Zhao M., Leavitt J.M., Lloyd A.M.. 2008, Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings, Plant J., 53, 814–27. [DOI] [PubMed] [Google Scholar]
- 49. Qin T., Zhao H., Cui P., Albesher N., Xiong L.. 2017, A nucleus-localized long non-coding RNA Enhances Drought and Salt Stress Tolerance, Plant Physiol., 175, 1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhu Q.H., Stephen S., Taylor J., Helliwell C.A., Wang M.B.. 2014, Long noncoding RNAs responsive to Fusarium oxysporum infection in Arabidopsis thaliana, New Phytol., 201, 574–84. [DOI] [PubMed] [Google Scholar]
- 51. Wang H., Niu Q.W., Wu H.W., et al. 2015, Analysis of non-coding transcriptome in rice and maize uncovers roles of conserved lncRNAs associated with agriculture traits, Plant J., 84, 404–16. [DOI] [PubMed] [Google Scholar]
- 52. Tang W., Zheng Y., Dong J., et al. 2016, Comprehensive transcriptome profiling reveals long noncoding RNA expression and alternative splicing regulation during fruit development and ripening in Kiwifruit (Actinidia chinensis), Front. Plant Sci., 7, 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang G., Duan A., Zhang J., He C.. 2017, Genome-wide analysis of long non-coding RNAs at the mature stage of sea buckthorn (Hippophae rhamnoides Linn) fruit, Gene, 596, 130–6. [DOI] [PubMed] [Google Scholar]
- 54. Cazzonelli C.I., Pogson B.J.. 2010, Source to sink: regulation of carotenoid biosynthesis in plants, Trends Plant Sci., 15, 266–74. [DOI] [PubMed] [Google Scholar]
- 55. Kato M., Ikoma Y., Matsumoto H., Sugiura M., Hyodo H., Yano M.. 2004, Accumulation of carotenoids and expression of carotenoid biosynthetic genes during maturation in citrus fruit, Plant Physiol., 134, 824–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lv P., Li N., Liu H., Gu H., Zhao W.E.. 2015, Changes in carotenoid profiles and in the expression pattern of the genes in carotenoid metabolisms during fruit development and ripening in four watermelon cultivars, Food Chem., 174, 52–9. [DOI] [PubMed] [Google Scholar]
- 57. He C., Zhang G., Zhang J., Zeng Y., Liu J.. 2017, Integrated analysis of multiomic data reveals the role of the antioxidant network in the quality of sea buckthorn berry, FASEB J., 31, 1929–38. [DOI] [PubMed] [Google Scholar]
- 58. Kim J.H., Lee Y.J., Kim B.G., Lim Y., Ahn J.H.. 2008, Flavanone 3beta-hydroxylases from rice: key enzymes for favonol and anthocyanin biosynthesis, Mol. Cells, 25, 312. [PubMed] [Google Scholar]
- 59. Wang J., Yu W., Yang Y., et al. 2015, Genome-wide analysis of tomato long non-coding RNAs and identification as endogenous target mimic for microRNA in response to TYLCV infection, Sci. Rep., 5, 16946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shuai P., Liang D., Tang S., et al. 2014, Genome-wide identification and functional prediction of novel and drought-responsive lincRNAs in Populus trichocarpa, J. Exp. Bot., 65, 4975–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kwenda S., Birch P.R.J., Moleleki L.N.. 2016, Genome-wide identification of potato long intergenic noncoding RNAs responsive to Pectobacterium carotovorum subspecies brasiliense infection, BMC Genomics, 17, 614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gilbert L., Alhagdow M., Nunes-Nesi A., et al. 2009, GDP-D-mannose 3, 5-epimerase (GME) plays a key role at the intersection of ascorbate and non-cellulosic cell-wall biosynthesis in tomato, Plant J., 60, 499–508. [DOI] [PubMed] [Google Scholar]
- 63. Yu H.N., Wang L., Sun B., Gao S., Cheng A.X., Lou H.X.. 2015, Functional characterization of a chalcone synthase from the liverwort Plagiochasma appendiculatum, Plant Cell Rep., 34, 233. [DOI] [PubMed] [Google Scholar]
- 64. Franco-Zorrilla J.M., Valli A., Todesco M., et al. 2007, Target mimicry provides a new mechanism for regulation of microRNA activity, Nat. Genet., 39, 1033–7. [DOI] [PubMed] [Google Scholar]
- 65. Zhu M., Liu J., Xiao J., et al. 2017, Lnc-mg is a long non-coding RNA that promotes myogenesis, Nat. Commun., 8, 14718. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.