Abstract
Transposable elements constitute an important fraction of eukaryotic genomes. Given their mutagenic potential, host-genomes have evolved epigenetic defense mechanisms to limit their expansion. In fungi, epigenetic modifications have been widely studied in ascomycetes, although we lack a global picture of the epigenetic landscape in basidiomycetes. In this study, we analysed the genome-wide epigenetic and transcriptional patterns of the white-rot basidiomycete Pleurotus ostreatus throughout its life cycle. Our results performed by using high-throughput sequencing analyses revealed that strain-specific DNA methylation profiles are primarily involved in the repression of transposon activity and suggest that 21 nt small RNAs play a key role in transposon silencing. Furthermore, we provide evidence that transposon-associated DNA methylation, but not sRNA production, is directly involved in the silencing of genes surrounded by transposons. Remarkably, we found that nucleus-specific methylation levels varied in dikaryotic strains sharing identical genetic complement but different subculture conditions. Finally, we identified key genes activated in the fruiting process through the comparative analysis of transcriptomes. This study provides an integrated picture of epigenetic defense mechanisms leading to the transcriptional silencing of transposons and surrounding genes in basidiomycetes. Moreover, our findings suggest that transcriptional but not methylation reprogramming triggers fruitbody development in P. ostreatus.
Keywords: epigenetics, DNA methylation, transcription, life cycle, fungi
1. Introduction
The extraordinary increase in genomic data released during the last decade has made possible to begin to unravel the impact of transposable elements (TEs) on a wide range of eukaryotic genomes.1,2 TEs are ‘selfish’ genetic units that can mobilize and increase their copy number in the host genome. TEs can also interrupt genes, produce rearrangements and lead to illegitimate recombination events.3 Most TEs are usually present as defective copies that have accumulated mutations and deletions, which ultimately inactivate their transposition potential. In many organisms, TEs accumulate in centromeric and pericentromeric regions where they play a crucial role in genome plasticity and heterochromatin maintenance.4,5 Nevertheless, active transposons constitute a significant source of mutations that can lead to harmful effects in the host genome.6,7 Thus, most eukaryotes have evolved epigenetic defense mechanisms to limit TE expansion. More specifically, transcriptional and post-transcriptional gene silencing pathways (TGS and PTGS) have been described to limit transposon activity in plants and animals.8,9 TGS operates through DNA methylation, whereas PTGS is mediated by small RNAs and orchestrated by the RNA interference pathway (RNAi). DNA methylation is an epigenetic modification involved in several cellular processes and described in a variety of eukaryotic genomes.10,11 Such modification occurs by the addition of a methyl group to the C-5 position of cytosine in DNA. PTGS mediated by siRNAs (short-interfering RNAs, a class of small RNAs) starts with the production of aberrant double-stranded RNAs (dsRNAs) originated from transposons, viruses, or RNA hairpins, among other sources. The dsRNAs are processed by Dicer RNAse III producing short fragments of 21–25 nucleotides (siRNAs), which are loaded in the RNA-induced silencing complex, guided to complementary mRNA and subsequently degraded by the Argonaute slicer activity.12,13 Also, siRNAs can produce TGS by promoting DNA methylation, as described in plants and fungi.13,14 Recent studies in plants and mammals have described a link between DNA methylation and the RNAi pathways15,16 in transposon silencing, indicating that these mechanisms are functionally related.17 In fungi, epigenetic silencing mechanisms have been extensively explored in the filamentous ascomycete Neurospora crassa and are beginning to be documented in other lineages. This model organism uses TGS and PTGS mechanisms to inactivate TEs at the vegetative and sexual stages. Specifically, a repeat-induced point mutation (RIP) operates during the sexual cycle to transcriptionally silence repetitive sequences through a homology-dependent mechanism. RIP induces C: G to A: T hypermutations in these sequences during the sexual cycle, leading to their degeneration and silencing by DNA methylation.18,19 In ascomycetes, transposons can be inactivated by DNA methylation linked to RIP mutations,20 whereas in basidiomycetes this mechanism has been examined in silico in members of the Pucciniomycotina21 and in the ustilaginomycete Microbotrium violaceum.22 A related mechanism called methylation-induced premeiotically, has been detected in Ascobolus immersus and in the basidiomycete Coprinus cinereus and it displays similar hallmarks with RIP.23,24 Distinct DNA methylation patterns have been observed in some fungal lineages while in others this mechanism seems to be absent.25–27 Recent comprehensive methylome analyses carried out in five fungi belonging to Zygomycota, Ascomycota and Basidiomycota groups reported a marked preference for methylation at CG sites within transposons and other repeated sequences, in contrast to the low methylation levels found in gene-coding regions.26,27 Regarding PTGS and TGS of TEs, three RNAi mechanisms have been described in N. crassa: quelling28 (related to plant co-suppression), MSUD (meiotic silencing by unpaired DNA)29 and DNA methylation associated to disiRNA loci.14 Quelling and MSUD mechanisms are based on the production of small RNAs, and rely on the core components of the RNAi pathway. The former leads to the silencing of repetitive DNA sequences (i.e. transposons or multi-copy genes) in the vegetative phase, and the latter silences unpaired regions between two parental chromosomes during meiosis. Moreover, an RNAi-dependent mechanism has been recently discovered to silence TEs post-transcriptionally during sexual development (SIS, sex-induced silencing) in the basidiomycete Cryptococcus neoformans.30 Although epigenetic mechanisms for TE control have been extensively described in the ascomycete model N. crassa, only a few studies have analysed DNA methylation31,32 and RNAi interference mechanism33 in basidiomycetes. Beyond its biotechnological applications, the lignin-degrader Pleurotus ostreatus has gained relevance in genetic and genomics studies in recent years. Its simple life cycle, the ease of cultivation under laboratory conditions and the availability of an un-gapped telomere-to-telomere genome sequence makes P. ostreatus a good model for basidiomycete studies. Its life cycle alternates between monokaryotic (cells contain only one haploid nucleus) and dikaryotic (cells are dihaploid and contain two haploid nuclei) phases.34 A dikaryotic strain is formed when two compatible monokaryotic strains mate to form a dikaryon where the haploid nuclei remain independent throughout the vegetative growth and fruit-body development. Karyogamy takes place after both haploid nuclei are joined to form a diploid one which undergoes meiosis. Recent comprehensive analyses carried out in our group characterized the landscape of TEs in two compatible monokaryotic strains of P. ostreatus35 (PC9 and PC15). This study uncovered the presence of 80 TE families, which encompass 2.5 and 6.2% of the total genome sizes of PC9 and PC15, respectively. Most TEs were aggregated in 40 non-homologous clusters spread across the 12 chromosomes. Moreover, it was observed that genes having a TE inserted upstream or downstream the gene body have lower transcription levels than average, especially when the genes are enclosed in TE-rich clusters.35 Regarding the potential RNAi activity of P. ostreatus, preliminary data suggests the presence of the RNAi machinery core, which has been detected in silico by screening for Neurospora MSUD and quelling orthologous proteins.36 Using high-throughput sequencing, we describe the genome-wide epigenetic (DNA methylation and small RNAs) and transcriptional (mRNA) profiles of two compatible P. ostreatus monokaryons as well as dikaryons at different stages during fruit body development. Our results support the evidence of strain-specific DNA methylation and small RNA production primarily involved in the repression of transposons activity. Also, we provide evidence that the TE-associated gene silencing effect, previously described by Castanera et al.,35 is correlated to the extension of DNA methylation associated to surrounding transposons. Finally, the comparative analysis of the transcriptomes at different stages of the P. ostreatus life cycle identifies the genes and functions that might be involved in triggering fruit body primordia formation and development.
2. Materials and methods
2.1 Fungal strains and growth conditions
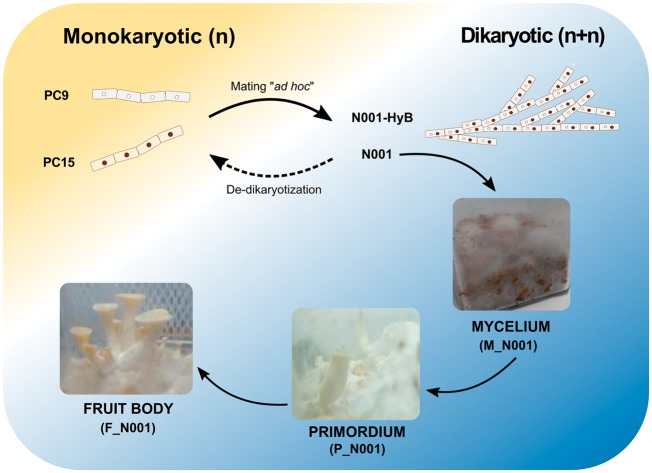
Four P. ostreatus strains were used: PC9 (Spanish Type Culture Collection accession CECT20312), PC15 (CECT20312), N001 (CECT20600) and N001-HyB. PC9 and PC15 are two compatible monokaryotic protoclones obtained by de-dikaryotization of the N001 commercial strain in 1999. PC15 and PC9 strains have been described previously by our group,37 and display slow- and fast growing phenotypes, respectively (Supplementary Fig. S1). N001-HyB is a dikaryotic strain regenerated ad hoc by mating PC9 and PC15 in 2014,36 which contains the same genetic complement as N001. All strains were cultured in Erlenmeyer flasks containing 200 ml of Malt Extract (ME, 20 g/l) in the dark, at 24°C under orbital shaking (125 rpm). After 6 days, the cultures were homogenized using an Omni mixer and used as inoculum (15 ml) for Submerged Fermentation (SmF) and Solid-state Fermentation (SSF) cultures. SmF was carried out in Erlenmeyer flasks containing 135 ml of liquid ME medium and maintained in the dark for 7 days at 24°C under orbital shaking. SSF was carried out in polycarbonate Magenta boxes containing 15 g of the total dry substrate (v/v) (88% sawdust, 10% millet and 2% CaCO3) and adjusted to an 80% water content. A total of six samples representing the main stages of the P. ostreatus life cycle were obtained for further analysis: (i) vegetative mycelium in SmF (PC9 and PC15); (ii) mycelium under fruiting induction in SSF (M_N001 and N001-HyB); (iii) primordia (P_N001); and (iv) mature fruitbodies (F_N001) (Fig. 1) in SSF. To induce fruiting conditions, completely colonized SSF cultures were maintained at 18°C under a light/dark photoperiod of 12 h until fruit-body formation (∼15 days). Three biological replicates per condition were separately sampled, ground in a sterile mortar in the presence of liquid nitrogen and stored before nucleic acids extraction.
Figure 1.
Summary of P. ostreatus samples used in this study. PC15 and PC9 represent two monokaryotic strains that fuse to generate ad hoc the dikaryon N001-HyB. Given its inability to fructify, N001-HyB is examined exclusively at the mycelium stage. The N001 dikaryotic strain, bearing both PC15 and PC9 haploid nuclei and maintained under continuing subculturing during several years, is analyzed under different developmental stages (mycelium M_N001, primordium P_N001, and fruitbodies F_N001). N001-HyB and N001 harbor the same genetic complement although they show different fruiting ability.
2.2 Construction and sequencing of whole genome bisulphite libraries
Global DNA methylation levels were estimated by performing sodium bisulphite treatment, based on the chemical conversion of unmethylated cytosines,38 followed by high-throughput sequencing (BS-seq). Whole genome bisulphite (WGBS) libraries were prepared as described in Morselli et al.39 Briefly, genomic DNA (gDNA) from the fungal samples was extracted using an E.Z.N.A Fungal DNA Mini Kit (Omega Bio-Tek, Norcross, GA). After additional RNase A treatment (10 mg/ml for 60 min at 37°C), gDNA was purified using phenol: chloroform solution (3:1), precipitated overnight with ethanol (2:1) and the pellet resuspended in nuclease-free water. For additional purification after extraction, gDNA was treated with the Genomic DNA Clean & Concentrator Kit (Zymo Research, Irvine, CA). The concentrations were quantified using a Qubit 2.0 fluorometer (Life Technology, Carlsbad, CA) and total gDNA was fragmented with a Covaris S-2 ultrasonicator to obtain fragments spanning from 150 to 300 bp size range. Library preparation was performed using the Illumina TruSeq DNA Sample Prep40 according to the manufacturer’s instruction. Bisulphite conversion was carried out with the EpiTect Kit (QIAGEN), performing two consecutive rounds of conversion for a total of 10 h of incubation. Converted DNA was amplified according to the following PCR programme: denaturation at 98°C for 2 min, 12 cycles of 98°C for 15 s, 60°C for 30 s, 72°C for 30 s and final extension at 72°C for 5 min. All libraries obtained after bisulphite treatment were sequenced by an Illumina HiSeq2000 system (Illumina, San Diego, CA, USA) using 100 bp single-end reads. Quantitative DNA methylation assays were also performed in a selected set of genes by bisulphite-free real-time PCR following the MSRE-qPCR approach (detailed protocol shown described in Supplementary Material S1).
2.3 Preparation of mRNA and small RNAs-sequencing libraries
Total RNA isolation for mRNA (mRNA-seq) and small RNA (sRNA-seq) sequencing was performed using a Fungal RNA E.Z.N.A Kit (Omega Bio-Tek, Norcross, GA, USA) according to the manufacturer’s guidelines. The integrity and quantity of RNA were validated by Bioanalyzer (version 2100) and Qubit 2.0 fluorometer. mRNA-seq libraries were prepared using the TruSeq RNA Sample Prep Kit (Illumina) following the manufacturer’s instruction. Total RNA was used for isolation of poly(A)-carrying mRNA molecules and synthesis of double-stranded cDNA before adapters ligations. For sRNA libraries, small RNAs molecules were resolved by electrophoresis on a 6% (w/v) polyacrylamide gel and the fraction corresponding to < 200 nt in length was eluted from the gel. Adapter-ligated molecules were reverse transcribed and enriched by PCR. The final libraries were quantified by real-time PCR in LightCycler 480 (Roche) and sequenced with an Illumina HiSeq 2000 system using 75 and 100 bp paired-end reads.
2.4 Analysis of sequencing data
Reads from all libraries were mapped to the P. ostreatus reference genomes (PC15 v2.0, www.genome.jgi.doe.gov/PleosPC15_2/PleosPC15_2.home.html and PC9 v1.0 https://genome.jgi.doe.gov/PleosPC9_1/PleosPC9_1.info.html). The PC15 v2.0 reference genome is completely assembled in twelve scaffolds (34.3 Mb genome size).37 In total, 12,330 genes have been annotated in this genome.41 A detailed description of the data analysis workflows used in this study is shown in Supplementary Material S1. BS-seq, sRNA-seq and mRNA-seq data-sets were obtained in triplicate, except for Primordia, where n = 2 in BS-seq analysis.
3. Results
3.1 DNA methylation and sRNA profiles during P. ostreatus’ life cycle
Whole-genome bisulphite sequencing (BS-seq) was carried out in six representative samples of the P. ostreatus life cycle (Fig. 1) to investigate the profile of 5-cytosine DNA methylation. Sequencing of BS-seq libraries yielded an average of 40 ± 6 million total reads per sample. Reads were aligned to the PC15 v2.0 reference genome, obtaining coverages ranging from 66 to 97× (Supplementary Table S2A). The global methylation levels ranged from 2.8 to 6.7% of the total cytosines, and the bisulphate non-conversion rate was 0.27%. P. ostreatus has the lowest methylation levels in the monokaryotic stage, although the two strains tested showed differences between them (2.8% in PC15 vs. 4.4% in PC9, respectively) (Supplementary Table S2B). Interestingly, we found that the reconstructed N001-HyB dikaryotic strain (obtained by mating PC15 and PC9 monokaryons, see Section 2) exhibited methylation levels substantially lower than the ‘natural’ N001 dikaryotic strain (3.96 vs. 6.48%), which displayed nearly identical levels in the three developmental stages (Fig. 2A). Cytosine methylation was clearly predominant in the CpG context in all samples (4.0 ± 2% in CpG vs. 0.5 ± 0.1% in CHG and 0.6 ± 0.2% in CHH (Fig. 2A and Supplementary Table S2B) and hereafter we focus only on this context. Reads were also aligned to the P. ostreatus PC9 reference genome, and results yielded similar methylation levels compared with the PC15 reference genome in all among samples (Supplementary Table S2C and D). Due to lower assembling quality of PC9 reference genome (572 scaffolds and a total of 476 gaps covering 9.72% of the whole assembly), the subsequent analyses were performed on the fully assembled PC15 reference genome. To validate the findings obtained by BS-seq analysis, methylation levels were also estimated by performing bisulphite-free real-time PCR method. The results of the MSRE-qPCR (methylation sensitive restriction enzyme qPCR) profiles of five genes in PC9 and PC15 monokaryotic samples confirmed the trend previously outlined by bisulphite treatment followed by NGS sequencing (Supplementary Fig. S2). The distribution of 5-methylcytosines (5mC) was analysed in two different genomic contexts: genes and TEs. The results uncovered significant differences between these two features. Genes showed patterns of hypomethylation, with average methylation levels ranging from 1.5 to 4%. In contrast, TEs were heavily methylated, with levels ranging from 20 to 60% (Fig. 2B). We also observed sharp 5mC increments in the adjacent regions of both initial and terminal TE insertion sites, reaching the maximum methylation levels along the whole transposon body. In contrast, this trend was absent in protein-coding genes. Regarding the difference between samples, the methylation levels of N001 were the highest and showed no variation during fruitbody development, neither within genes nor in TEs. The genome-wide production of small RNAs was investigated in the same six samples by sRNA sequencing (mapping statistics using PC15 and PC9 as reference genomes are shown in Supplementary Table S3A and B). Most of the small RNAs originated from non-annotated genomic features, which could match with heterochromatin regions spread in the genome of P. ostreatus (Fig. 2C). The amount of repeat-associated small interfering RNA (rasiRNA) varied between the six samples, ranging from 14% to 37% of the total mapped reads, whereas the percentage of sRNAs mapping to genes ranged from 16 to 44%. The population of small RNAs was further characterized by analysing the length distribution. A maximum peak was found at 21 nt in all strains and samples (Fig. 2D and Supplementary Fig. S3).
Figure 2.
Global DNA methylation and small RNAs profiles within the six samples. (A) Average methylation levels of the CG, CHG and CHH contexts. Error bars represent the standard deviation of three biological replicates (excepted for P_N001 where n = 2). (B) Metaplots showing DNA methylation across adjacent regions, genes and transposons body. (C) Percentage of sRNA reads mapped in transposons (TEs), genes (Genes) and other regions (Others). (D) Line chart displaying the percentage of sRNAs mapped reads ranging from 17 to 30 nt in length.
3.2 Genome-wide transcriptional profiles during P. ostreatus development
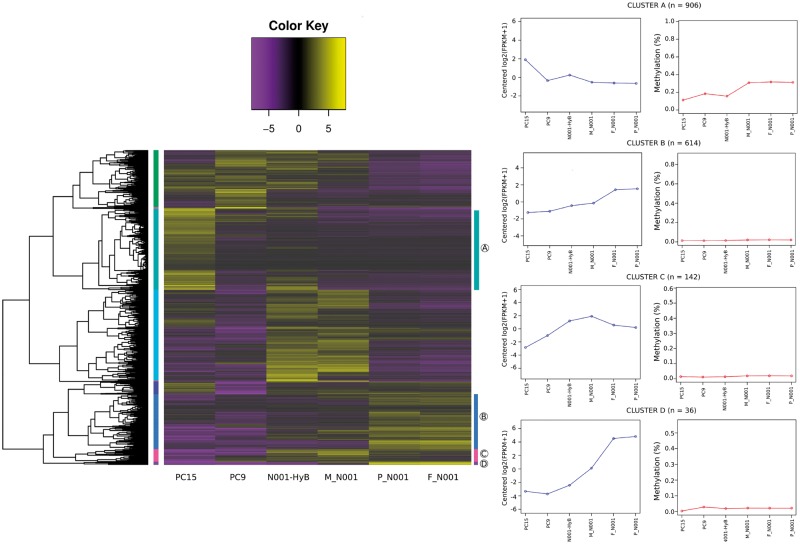
Next, we sought to perform mRNA-seq on the same six samples to analyse gene expression changes during P. ostreatus development. An average of 24.16 ± 2.6 millions of uniquely aligned reads per sample (Supplementary Table S4) were used to calculate the transcriptional levels of all non-TE genes (genes overlapping with TEs were excluded) and perform differential expression analyses. We performed all-by-all sample comparisons and found that 3,531 out of the 11,828 genes were differentially expressed in the samples analysed (3-fold Log2 cutoff, FDR-corrected P-value < 0.05). The biggest differences were found between PC15 and M_N001 (1,528 DEGs, Table 1), and the smallest between primordia (P_N001) and fruitbody (F_N001) samples of N001 (6 DEGs). To analyse the expression trends of DEGs under the six samples, we performed a hierarchical clustering of the 3,531 genes. We identified a total of nine clusters of co-expression, consisting of variable numbers of genes (heatmap vertical axis, Fig. 3). Three of the nine clusters were further analysed due to their relevance for the study of fruitbody triggering and development (Clusters B, C and D) and one for the role of dominance in the expression profiles of the dikaryotic stage (Cluster A). Specifically, the expression of genes belonging to Cluster A (906 genes) was high in PC15 and low in PC9, and the dikaryons showed either intermediate profile (N001-HyB) or PC9-like profile (M_N001, P_N001 and F_N001). Cluster C (142 genes) showed an increased transcription during fruitbody triggering and higher expression than monokaryons in primordia and fruitbodies. Finally, Clusters B and D (614 and 36 genes, respectively) were upregulated during fruitbody development. To better understand the link between transcription and DNA methylation in the different stages of P. ostreatus development, we represented the average value of these two marks in genes belonging to the selected clusters (Fig. 3). Genes belonging to Clusters B, C and D showed a nearly complete lack of methylation. Notably, genes included in Cluster A showed higher methylation levels in PC9 and N001 (about 35% average methylation level for N001 samples). Moreover, this hypermethylation pattern coincided with a low expression in the two strains. Further analysis showed that 29% of the genes belonging to Cluster A were present inside the TE-rich regions defined by Castanera et al.35 In contrast, this percentage decreased to 13% in Cluster B, 20% in Cluster C and 8% in Cluster D. (Supplementary Fig. S4). Next, we retrieved the functional annotation of all genes and performed gene ontology (GO) enrichment focussing on the selected Clusters. This approach revealed over-represented molecular functions (MFs), biological processes (BPs) and cellular components. Specifically, the most enriched ontology of cluster A was 3’-5’ exonuclease activity (MF), in Cluster B was monooxygenase activity (MF), in cluster C was structural constituent of cell wall (MF), and in Cluster D was fruiting body development (Supplementary Table S5). Interestingly, other significantly enriched BP enriched during fruiting induction was multicellular organism development (Table 2, Cluster C). These data also provided evidence of the different transcriptional profiles between the natural N001 (M_N001) and the regenerated N001-HyB. We found that a total of 395 genes were differentially transcribed between these two strains. The comparison displayed that 349 genes were upregulated in the N001-HyB and 46 in M_N001 strain (Supplementary Fig. S5). Not surprisingly, when we performed GO enrichment analysis between these two dikaryotic strains, the subset of enriched functions revealed that BP involved in fruitbody development was exclusively represented in the genes upregulated in M_N001 strain (Supplementary Table S6).
Table 1.
Differentially expressed genes (FDR-corrected P-value <0.05, Log2 Fold-change >3
| PC15 | PC9 | N001-HyB | M_N001 | P_N001 | F_N001 | |
|---|---|---|---|---|---|---|
| PC15 | 0 | 1368 | 1,156 | 1,528 | 1,437 | 1,505 |
| PC9 | 1,368 | 0 | 1,176 | 1,125 | 1,137 | 1,255 |
| N001-HyB | 1,156 | 1,176 | 0 | 395 | 1,027 | 1,230 |
| M_N001 | 1,528 | 1,125 | 395 | 0 | 595 | 819 |
| P_N001 | 1,437 | 1,137 | 1,027 | 595 | 0 | 6 |
| F_N001 | 1,505 | 1,255 | 1,230 | 819 | 6 | 0 |
Figure 3.
Hierarchical clustering of differentially expressed genes. Heatmap plot illustrating differentially expressed genes (3,531 genes). Nine main clusters are shown indicating grouping genes with similar expression profiles. Gene expression levels from lower to higher are shown. The more intensively studied Clusters A–D are highlighted at the right of the heatmap. Expression and methylation profiles of genes included in Clusters A–D are shown in the right panels. Color figure is available at online version.
Table 2.
BPs enriched in co-expression clusters under study
| GO | GO level | Name | Ratio in cluster | Ratio in genome | P-value | |
|---|---|---|---|---|---|---|
| A | ...GO: 0006139 | 4 | nucleobase-containing compound metabolic process | 5/906 | 23/11,828 | 0.0275 |
| ......GO: 0006278 | 7 | RNA-dependent DNA biosynthetic process | 2/906 | 2/11,828 | 0.00586 | |
| B | ......GO: 0006468 | 7 | protein phosphorylation | 26/614 | 284/11,828 | 0.00429 |
| ......GO: 0006032 | 7 | chitin catabolic process | 3/614 | 8/11,828 | 0.00641 | |
| ...GO: 0051341 | 4 | regulation of oxidoreductase activity | 5/614 | 31/11,828 | 0.0205 | |
| C | ...GO: 0006810 | 4 | transport | 11/142 | 299/11,820 | 0.000953 |
| .....GO: 0006865 | 6 | amino acid transport | 3/142 | 17/11,820 | 0.00102 | |
| .......GO: 0046087 | 8 | cytidine metabolic process | 1/142 | 1/11,820 | 0.012 | |
| ...GO: 0007275 | 4 | multicellular organism development | 1/142 | 2/11,820 | 0.0239 | |
| .....GO: 0009228 | 6 | thiamine biosynthetic process | 1/142 | 3/11,820 | 0.0356 | |
| .......GO: 0006817 | 8 | phosphate ion transport | 1/142 | 3/11,820 | 0.0356 | |
| D | .....GO: 0030582 | 6 | fruiting body development | 1/36 | 1/11,828 | 0.00304 |
| ......GO: 0019836 | 7 | hemolysis by symbiont of host erythrocytes | 1/36 | 1/11,828 | 0.00304 | |
| .......GO: 0009168 | 8 | purine ribonucleoside monophosphate biosynthetic process | 1/36 | 4/11,828 | 0.0121 | |
| ...GO: 0006081 | 4 | cellular aldehyde metabolic process | 1/36 | 4/11,828 | 0.0121 |
dots in the GO column reflect levels in GO hierarchy.
3.3 Different nucleus-specific methylation profiles operate in the P. ostreatus dikaryotic stage
In previous sections, we have shown how the dikaryotic mycelium of the natural N001 (represented here by M_N001 sample) and the ad hoc generated N001-HyB strains displayed different methylation profiles under identical conditions, although they share the same genetic complement. Thus, we considered the possibility of an unequal contribution of each monokaryotic nucleus to the dikaryons. To test this hypothesis, we mapped BS-seq data to a new set of pseudo-genomes consisting of the nucleus-specific regions concatenated with the common regions sharing a similarity < 90% (one unique pseudo-genome for PC15 and another for PC9). To build this reference sequences sets, we performed a whole genome alignment between PC15 and PC9 using the NUCmer software.42 The direct pairwise comparison revealed an average of 97.2% similarity in the aligned regions, which spanned 87.3% of PC15 assembly and 83.2% of PC9. Afterward, we determined the nucleus-specific methylation levels in M_N001 and N001-HyB by performing BS-Seeker2 analyses43 on the unique regions (pseudo-genomes of PC9 and PC15 used as references). We observed similar low mappability rates between the M_N001 and N001-HyB strains when aligned to each reference genome, which could reflect the presence of repetitive sequences along both pseudo-genomes. Interestingly, when we looked at the global DNA methylation values, we found that each nucleus contributed differentially in the two dikaryotic strains (Table 3). In fact, while genomic regions deriving from the PC9 nucleus exhibited comparable methylation levels in both the natural and ad hoc dikaryons (coefficient of variation of 13.4%), differences in regions associated with the PC15 nucleus were higher (coefficient of variation of 37.9%). Specifically, methylation of the PC15 nucleus in N001-HyB was considerably lower than in M_N001 (18.11 vs. 34.45%), where the methylation levels of both PC9 nuclei were similar (32.23 vs. 39.62). Next, we sought to identify genomic regions showing significant differential methylation between these two strains. Using the SMART2 software, we detected a total of 2, 199 differentially methylated regions (DMRs) of 200–5,714 bp in length (Supplementary Fig. S6A). Among the resulting DMR, 98% were significantly hypermethylated in M_N001 vs. N001-HyB (Supplementary Fig. S6 B).
Table 3.
Summary of BS-seq mapping to PC15 and PC9 unique pseudo-genomes
| Dikaryon | N001-HyB |
M_N001 |
||
|---|---|---|---|---|
| Reference genome | PC15-unique | PC9-unique | PC15-unique | PC9-unique |
| Total raw reads | 40,266,055 | 40,266,055 | 37,608,379 | 37,608,379 |
| Mapped reads | 2,821,468 | 1,589,586 | 2,434,806 | 1,553,782 |
| Mappability (%) | 7.1 | 4.2 | 6.6 | 4.3 |
| Coverage (× | 65.2 | 57.7 | 60.1 | 58.3 |
| mCG (%) | 18.11 | 32.23 | 34.45 | 39.62 |
3.4 DNA methylation, small RNA production and transcriptome landscape in P. ostreatus
The overall distribution and levels of DNA methylation, mRNA and sRNAs production were analysed along the twelve P. ostreatus chromosomes using the dikaryotic N001 strain (sample M_N001) as reference. We noticed that DNA methylation and sRNAs production were tightly associated with TE-rich clusters spread along the genome, where the transcriptional activity had dramatic depletion (Figs 4A and B). Moreover, we found that the levels of methylation and sRNA production varied regionally and between chromosomes. Considering these data, we sought to analyse the correlation of methylation levels with mRNA and sRNA expression across the entire genome. For this purpose, the genome was divided into 200 bp windows. Windows were split into three groups according to their methylation levels (Group I: 0–20%, Group II: 20–60% and Group III: > 60%), and plotted with their corresponding sRNA and mRNA expression. In addition, we analysed the correlation across genes and promoter regions according to the same methylation ranges previously mentioned. As shown in Figure 4C, DNA methylation exhibited a negative correlation with mRNA transcription. Although the strongest repression was found in windows with >60% average methylation, the sharpest decrease in expression was found to occur when methylation exceeded 20%. Regarding DNA methylation and sRNAs expression, the two marks were positively correlated (Fig. 4D). Notably, we noticed that the majority of the sRNA production (approximately 95%) derived exclusively from a very small portion of the whole genome (3.3% of entire genome). When we analysed both mRNA and sRNA abundances at genes and promoter regions, a similar negative correlation between methylation and mRNA expression was uncovered among the three methylation ranges. In particular, promoter regions displayed lower mRNA transcriptional levels when compared with genes. On the contrary, a positive correlation was detected between sRNA and methylation in both genes and promoter regions, with slightly higher sRNA expression levels in correspondence to promoter regions.
Figure 4.
Global association of CpG methylation, mRNA and sRNAs expression in the M_N001 strain. Circular genome and data visualization with Circos for BS-seq, mRNA-seq and sRNA-seq profiles (A). From inside to out: mRNA expression (yellow), small RNA production (violet) and DNA methylation (light blue). The outer track report TEs (grey bands). All tracks represent mean values of three biological replicates. The presence of TE-rich clusters is indicated by blue asterisks. (B) Integrative genomics viewer (IGV) browser visualization of a representative 260 kb region in scaffold 2 (location: 210,736–472,880 bp) of a replicate of M_N001. Plotted from top to bottom are: TEs annotation, DNA methylation, sRNAs and mRNA transcription and genes annotation (logarithmic scale). Boxplots showing the correlation of DNA methylation with mRNA (C) and sRNAs (D) transcription. Each boxplot represents the genome split in 200-bp windows, along with genes and promoters grouped based on their methylation levels (0–20, 20–60 and 60–100%). In panel D, ‘no producing’ group represents genomic regions, genes and promoters having <10 sRNA mapped reads. In panel C, genes are grouped as following according to the methylation levels: 10, 692 genes in 0–20%, 428 in 20–60% and 398 in 60–100%. In (D): 6,505 no producing, 4,969 in 0–20%, 70 in 20–60% and 104 in 60–100%.
3.5 Repeat-associated DNA methylation, expression and smallRNA profiles
A clearly opposite trend was observed in transcription and methylation levels between genes (high transcription with low methylation) and TEs (low transcription with high methylation) (Fig. 5A and B). Within the different TE orders, terminal inverted repeats (TIR), long terminal repeats (LTR) and Dictyostelium Intermediate Repeat Sequences (DIRS) were the most heavily methylated and showed the lowest expression. Helitron and long interspersed nuclear elements (LINE) elements had slightly lower methylation and higher expression values. This general trend was maintained in all samples and strains, although, PC15 had the lowest 5mC rate in all TE orders, which corresponded to the highest expression ratios. We performed a deeper analysis accounting for the 80 TE families present in P. ostreatus and found that PC15 TE families were consistently hypomethylated compared with the other strains (Fig. 5D). Hierarchical clustering of TE methylation and expression displayed that the most invasive TE families (LTR/Gypsy) were strongly methylated (26–59% on average) and transcriptionally repressed (Fig. 5D, Supplementary Fig. S7A). Nevertheless, other less abundant families, such as TIR_1, displayed higher methylation levels (35–67%) and complete transcriptional repression. Next, we analysed the production of sRNAs by genes, TE orders and families in the context previously described. The amount of sRNAs per element (i.e. gene or TE) was higher in TEs than in genes (Fig. 5C), although it varied greatly depending on the strain and TE order. sRNAs were abundantly produced by TIR transposons in all samples (reaching average levels up to ∼7,000 RPM/copy in the PC9 strain), with the only exception of PC15 which displayed much lower production of sRNA associated to TIR elements. The LTRs in the Gypsy superfamily and Helitrons also showed high sRNAs production, especially in PC15 and N001-HyB strains, and the remaining orders had low amounts of mapped sRNAs. Considering the percentage of sRNAs mapped to TEs (rasiRNAs), Gypsy, Copia retroelements and TIRs were the main sources, with great differences between families (Supplementary Fig. S7B). Next, we tested whether the production of rasiRNAs by TE families was related to (i) size (family copy number) or (ii) age (divergence between TE copies and family consensus). Despite detecting differences between strains, we found that the TE families containing more than 10 copies tend to have the highest rasiRNAs expression (Supplementary Fig. S8A). This effect was especially relevant in abundant sRNAs-producing samples, such as PC9, where rasiRNA abundance (RPM/copy) was positively correlated with family copy number (Pearson correlation coefficient = 0.42, P-value = 1.542−5) (Fig. 6A). To explore the relationship between rasiRNAs production and family age, we correlated rasiRNAs data to the average divergence rate, which increases linearly with family age (Supplementary Fig. S8B). Using PC9 as a reference, we found that the average family divergence and rasiRNAs expression were negatively correlated (Pearson correlation coefficient = −0.25, P-value = 0.01617) (Fig. 6B).
Figure 5.
DNA methylation, transcriptome and sRNAs expression over annotated genomic features. Histograms showing average methylation levels (A), mRNA transcription (B) and sRNA production (C) overlapping with genes and TE orders belonging to Classes I and II. All tracks represent the mean values of three biological replicates. In (C), the y-axis is reported at different scale in the lower and upper part. Percentage values below each order term indicate the relative occupancy in the total P. ostreatus TEs landscape. (D) Hierarchical clustering reporting DNA methylation levels within 80 TE families. Blue and yellow colours indicated high and low values respectively, expressed as mCG (%).
Figure 6.

Association of transposons size and age with rasiRNA production. Line charts showing the correlation of sRNAs production with copy number (A) and divergence rate (B) in 80 TE families in the PC9 strain. The coefficient of determinations is reported on the top right of each graph.
3.6 Role of epigenetic modifications on TE-mediated gene silencing
As discussed earlier, TEs represent the primary target of cytosine methylation in P. ostreatus. Our previous investigations found that TE insertions lead to a significant reduction of the expression levels of surrounding genes within 1 kb.35 Also, we found that this TE-mediated gene silencing effect was stronger when genes were located inside any of the 40 TE clusters described in P. ostreatus. We explored the possibility that this phenomenon had an epigenetic explanation. We tested the hypothesis that methylation could spread outside TE boundaries reaching the surrounding genes and blocking their transcription. Therefore, we compared the methylation and expression levels of genes surrounded by transposons (Fig. 7) (within a window of 1 kb, either upstream or downstream the gene body, Group I, labelled as +TE) and genes not surrounded by TEs (Group II, labelled as Ctl). Additionally, to uncover the impact of TE clusters on this phenomenon, we split the first group of genes in two contexts: (i) genes located inside a TE cluster (cluster) and (ii) genes located outside TE clusters (isolated). As shown in Figure 7A and B using N001_HyB and M_N001 strains as a model, genes carrying a TE insertion displayed higher methylation levels than the control groups (P < 0.05). In the case of genes in TE cluster (Fig. 7B), we found a bimodal distribution of methylation, with approximately half of the genes having high 5mC levels (up to 40% in N001-HyB and 70% in M_N001). Regarding transcriptional profiles (Fig. 7C and D), genes surrounded by TEs had lower expression than controls (P < 0.05), especially genes present inside a TE cluster (Fig. 7D). This phenomenon was present in the six strains, although with different intensities (Supplementary Fig. S9A–D and Table S7). Furthermore, we analysed the distribution of sRNA in genes grouped into the contexts described earlier. Notably, no significant differences in sRNAs production were found among genes surrounded by a TE and the group control (Supplementary Fig. S9E and F). Next, we studied the distribution of methylation across the gene body and regions adjacent to the TSS and TTS. The aim was to understand if the methylation extension was spreading from the TE to the whole gene body or only to the promoter sequence in both contexts (TE cluster or isolated TE). We found that the methylation levels mildly decreased from the adjacent regions to the gene body. Nevertheless, gene body methylation of genes in both contexts was much higher than the control, reaching average values around 13% for genes isolated and with TE insertions and up to 25% for genes with TE insertions present in a TE cluster (Fig. 8A). Further analyses on the relationship between transcription and gene body methylation found that genes carrying TE insertions (at 1 kb upstream or downstream) had only two activity state. As represented in Figure 8B and C, the clear majority of genes surrounded by a TE held methylated and silenced activity state, whereas only a few copies displayed unmethylated and transcriptionally active state, similarly to almost all genes included in the control group not enclosed by TE insertions (Fig. 8D).
Figure 7.
Influence of TE-associated methylation on nearby gene transcription. Violin plots showing methylation (A and B) and transcription (C and D) levels of genes in N001-HyB and M_N001 dikaryotic strains. Genes are classified as surrounded (+TE) or not surrounded (Ctl) by a transposon within a 1 kb-window (either upstream or downstream). Isolated (A and C) and Cluster (B and D) contexts indicate genes located outside and inside TE-rich clusters, respectively. White dot inside each plot represents the median. Below each violin plot, n represents the number of genes, and P represents the P-value of the Mann-Whitney-Wilcoxon test.
Figure 8.
Association between methylation and transcription in the M_N001 strain. (A) Metagene plot showing the average methylation levels across the genes body and adjacent region: (i) surrounded by TEs at 1 kb upstream and downstream localized inside a TE-rich cluster (Cluster, upper line); (ii) surrounded by TEs at 1 kb up- stream and downstream localized outside a TE-rich cluster (Isolated, intermediate line); and (iii) isolated and not surrounded by TEs (Control, bottom line). Each of these groups is represented by a dashed (upper), dotted (intermediate) and solid (bottom) line, respectively. Scatterplot distribution at different scale reporting the relationship between methylation levels and expression of genes classified in the cluster (B), isolated (C) and control (D) groups.
4. Discussion
4.1 Epigenetic factors contribute to P. ostreatus genome regulation
Previous findings have illustrated the presence and importance of epigenetic modifications in fungi (for review, see44). Most of the data on fungal epigenetics comes from DNA methylation and RNAi studies performed in ascomycete fungi. However, few studies have reported evidence of the presence of such mechanisms in basidiomycetes.26,32 In N. crassa, it was observed that Dim-2 DMTase is responsible for methylation at both symmetrical and asymmetrical sites and is required for de novo and maintenance of DNA methylation.45,46 In plants, asymmetric methylation is maintained by the activity of de novo methyltransferases drm1, drm2 and chromomethylase 3 (cmt3). In fact, Cao et al.47 reported the absence of asymmetric methylation only in drm1 drm2 and cmt3 triple mutant plants, suggesting that the regulation of non-CG methylation is complex. Castanera et al.35 reported that Dim-2 DMTase was transcriptionally active in the genome of P. ostreatus, and it also carries a transcriptionally active cmt3. Nevertheless, it lacks dmr1 and dmr2 homologs, as it happens in N. crassa. According to these results, we hypothesize that the differences in the non-CG methylation levels between basidiomycetes and ascomycetes might be associated with the different evolutionary trajectories of their respective Dim-2 DMTases. Our results confirm the presence of 5-cytosine methylation in this basidiomycete, predominantly localized within repetitive regions, in the symmetric CpG context. This predominance has also been detected by methylome analyses of five species belonging to the three major fungal groups (ascomycetes, basidiomycetes and zygomycetes).26 Despite an estimated divergence time of >1 billion years, similar methylation patterns were found between basidiomycetes and zygomycetes, displaying a marked trend toward CpG methylation of regions located within TEs or other repetitive loci, and global weak methylation associated to genes. The methylation pattern of P. osteratus was similar to what has been described for other basidiomycete species, showing substantial amount of CG methylation and a small amount of non-CG methylation both concentrated in repetitive regions. These divergent patterns might underline evolutionary differences in mechanisms related to DNA methylation within fungal species characterized by different genome structure and lifestyle. Selker et al48, stated that in N. crassa, asymmetric methylation might be associated to methylation events occurring during premeiosis in the parental nuclei. This condition is maintained during the vegetative state in those genomic regions exhibiting RIP mutations and where the methylation is present. Based on these observations, the low asymmetric methylation levels found in P. ostreatus could be due to its negligible RIP levels (data not shown), which could make up a strategy to avoid silencing of genes involved in growth and development processes in monokaryon as well as dikaryons mycelia, the most common developmental stage in nature. In this regard, Chan et al.49 suggested that non-CG DNA methylation can be inherited via a network of different and persistent signals that have been co-opted to regulate developmentally important genes, though we have not carried out in Pleurotus experiments to test such hypothesis. Thus, this statement is only a speculation. Our findings showed that cytosine methylation ranged from 2 to 6% in the six samples analysed at different growing stages. These methylation levels are comparable to those described for other fungi so far,26,27,50 except the highly methylated genome of Tuber melanosporum, displaying up to 44% cytosine methylation.51 Previous studies in plants52 and fungi27 have demonstrated that the activity of transposons can be efficiently shut down by chromatin modifications linked to 5mC, as a defense mechanism aimed at controlling their expansion. In higher fungi, this trend has been reported in T. melanosporum, suggesting that methylation principally targets TEs.51 Consistent with this observation, we found that P. ostreatus transposons are highly methylated whereas gene-coding regions are hypomethylated, independently of the strain and developmental stage (Fig. 2B). It is worth noting that the highest methylation levels are maintained across the whole transposon body, while they sharply decrease beyond the transposon borders. A plausible explanation for this locally-restricted methylation might be the ‘mosaic methylation’ described in invertebrates and plants. As reviewed by Suzuki et al.,10 this strategy attempts to delimit potential detrimental changes induced by methylation and prevent spurious transcriptional silencing across the genome. In addition to 5-cytosine methylation, we describe the genomic hallmark associated with the production of small RNAs. We show that endogenous sRNAs produced by P. ostreatus are enriched in the 21 nt fraction (Fig. 2D) which could reflect the presence of RNAi-like silencing mechanisms such as those described in other eukaryotes.5,53 In the basidiomycete C. neoformans, experimental analyses showed the presence of 21–23 nt endogenous sRNAs involved in the control of mobile elements.30 Also, the plant-pathogen Puccinia striiformis produces 20–22 nt sRNA likely involved in PTGS.33 In this sense, P. ostreatus contains a highly conserved, transcriptionally active RNAi machinery composed of Argonaute, Dicer and RNA-dependent RNA polymerase proteins36 (18 proteins sharing 93–100% similarity between PC9 and PC15 strains).
4.2 TEs are targeted by DNA methylation and RNAi machinery
TEs play a major role in genome stability and evolution54 but are also a source of mutation that can result in deleterious or lethal effects to the host. Thus, epigenetic mechanism encoded by the host genome has been developed to silence their expression at the transcriptional and post-transcriptional levels.5 Here, we found that DNA methylation and sRNA production were correlated with the presence of silent TEs, suggesting a role in their activity leading to transcriptional suppression. This association was even more striking in TE-rich regions. Specifically, TE clusters were shown to be transcriptionally silenced and highly methylated, similarly to that described in the basidiomycete Laccaria bicolor.26 In this sense, the accumulation of silent transposon knobs could correspond to heterochromatic regions matching to centromeric and pericentromeric zones, as described in plants.55 Regarding DNA methylation, both Classes I and II transposons had higher methylation compared with genes. This observation supports the hypothesis that transcriptional silencing mediated by DNA methylation could be primarily responsible for TE inactivation in P. ostreatus. Similarly to the ascomycete T. melanosporum,51 we found that both LTR-retrotransposons and Class II transposons were abundantly methylated. It is interesting to note that the most methylated orders, TIR and LTR/Gyspy, also accounted for the larger portion of 21 nt rasiRNAs (Fig. 5C), especially TIR transposons belonging to the Mariner superfamily, the only cut-and-paste elements described in P. ostreatus. An example of this association has been reported in wheat, where most of the 21–22 nt sRNAs perfectly targeted within TIR regions of MITE elements, indicating that they are subjected to post-transcriptional control.56 An explanation for such an impressive amount of rasiRNA might reside in the hairpin RNA structure that elements from the TIR1 family can adopt due to the presence of Terminal Inverted Repeats (Supplementary Fig. S10), a conformation that promotes the presence of dsRNA and triggers RNAi machinery. In Caenorhabditis elegans, it has been proposed that such conformations lead to rasiRNA production from Mariner elements.5 Previous studies have described that Class I transposons predominate in basidiomycetes genomes whereas Class II transposons show limited expansion. This is especially relevant in the agaricomycotina subphylum,57 to which P. ostreatus belongs, and might be a consequence of the efficient replicative mechanism of Class I elements. According to our results, the under-representation of Class II transposons in the P. ostreatus genome might also be related to a stronger post-transcriptional inactivation in comparison to Class I elements. Based on our results, the production of rasiRNAs positively correlates with family size and negatively correlates with mean family divergence (Fig. 6A). Thus, the production of sRNA molecules might reflect an attempt to limit the expansion of the youngest, most invasive TE families of the genome. This observation is reminiscent of co-suppression studies carried out in the basidiomycete C. neoformans. In this species, mitotic-induced silencing pathway, a quelling-like asexual mechanism operating in trans, lead to RNAi-mediated silencing of homologous sequences and repeated elements during vegetative growth.58 Also, a similar mechanism was shown to induce post-transcriptional inactivation of repetitive transgenes and transposons mediated by 21–23-nt sRNAs during the sexual reproduction (SIS).30 Interestingly, the production of sRNAs in this fungal model increased according to the transgenes copy number with both mechanisms. Our finding suggests that P. ostreatus can inactivate transposons at the transcriptional and post-transcriptional level, by epigenetic modifications associated with DNA methylation and 21 nt sRNAs production. In this scenario, we speculate that sRNAs might be involved in the methylation of repetitive regions spread in the genome, guiding their silencing at the transcriptional level similarly to what has been reported in plants,59 where non-canonical RdDM has been described to be mediated by 21–22 nt long sRNAs.16
4.3 Methylated-TEs induce transcriptional silencing of nearby genes
According to our results, we propose that TE-associated transcriptional silencing of nearby genes occurs due to the extension of DNA methylation from TEs to the surrounding genes, which contain significantly higher methylation levels compared with controls. This is consistent with the results described in Magnaporthe oryzae, where genes with upstream, downstream or gene body methylation show lower expression than controls.50 Similarly, in T. melanosporum TEs close to highly expressed genes (1 kb upstream/downstream the TSS or TTS) tend to be less methylated than transposons located in the proximity of lowly expressed genes.51 In our model, methylation of genes surrounded by TE insertions decreases from regions adjacent to the TSS and TTS, reaching the lowest levels in the gene body. This observation fits with the hypothesis of methylation being extended from the closest TE, repressing the transcriptional activity of neighbour genes as documented in plants60 and animals.61 Another intriguing point is to understand how much methylation is needed to impact the transcriptional activity. In light of our findings, we propose that low to intermediate methylation levels (<20%) can prompt transcriptional silencing, although the strongest repression is found when methylation exceeds 60% (Figs 4C and 8B). Another striking observation is that genes displaying TEs insertion upstream or downstream of gene bodies have equal or even lower sRNA levels than control genes (Supplementary Fig. S9E and F). This suggests that TE insertions are presumably not involved in post-transcriptional silencing of nearby genes, and indicates that TE-mediated gene silencing is promoted by DNA methylation. Nevertheless, other mechanisms associated to silent heterochromatin structures such as methylation of histone H3 at Lys9 (H3K9me3)62 should be further investigated to understand their putative role in this phenomenon.
4.4 P. ostreatus fruiting stage is associated with methylation-independent transcriptional reprogramming
The exploratory analyses carried out during P. ostreatus development yielded some insights into the transcriptional changes underlying fruitbody induction and development. The transition from vegetative mycelium to primordium stage is a complex process that requires the aggregation of cells into compact hyphal knots which later experience tissue differentiation. In P. ostreatus, fruiting is triggered (among other environmental conditions) by lowering temperature and introducing light-dark cycles.63 In our study, these conditions lead to fruiting induction in N001, accompanied by the activation of a set of genes (especially Cluster D) expressed at low levels in monokaryons and also in N001-HyB (a strain unable to fruit), suggesting their putative role in the fruiting process. According to the number of DEGs between M_N001 and N001-HyB, at most 3% of the P. ostreatus genes are necessary for the early induction, while very few genes are presumably involved in changes from primordial to mature fruit bodies, similarly to what has been found in the basidiomycete Coprinopsis cinerea.64 Within this relatively small gene pool, the impressive overexpression of the pleurotolysin B (GO: 0030582, > 15-fold increase in M_N001, P_N001 and F_N001 vs. any of the monokaryotic stages and N001_Hyb), suggests its important role in the fruiting process. In this sense, previous experimental work has shown that the expression of these hemolytic proteins is activated during the formation of primordia and young fruit bodies in P. ostreatus and Agrocybe aegerita.65 Within the clusters of genes expressed during fruiting induction, we also found enriched functions related to multicellular development (Cluster C), although the most enriched biological function was related to transport, similarly to what was reported for C. cinerea, were such activity is upregulated prior to enlargement of fruiting bodies.64 The analysis of MFs activated during fruitbody triggering and development in P. ostreatus suggest that oxidorreductase and binding activities are the most enriched. Glycoside hydrolases were also found to be enriched in Cluster B, along with other proteins such as oxidative enzymes, hydrophobins involved in the aggregation of aerial hyphae and lectins, previously described to be involved in this process in basidiomycetes.64,66 This indicates that the core genes involved in the fruiting process are conserved across the Basidiomycota phylum. Interestingly, despite the differences in transcription of fruiting-associated genes, methylation levels were invariably low along the six samples. These results suggest that fruiting body formation in P. ostreatus is not triggered by epigenetic modifications linked to DNA methylation.
4.5 Nucleus-specific methylation is compensated in the long-term dikaryotic stage
The sample-specific profiles presented in this study can provide a framework for understanding the epigenetic and transcriptomic differentiation observed during the P. ostreatus life cycle (Fig. 1). Also, our experimental design allowed us to compare the epigenetic profiles of short-term (N001-HyB) vs. long-term (N001) cultured dikaryons. In fact, striking differences were found between these two strains, which share the same genetic complement but have clearly different methylation profiles. In particular, N001 has heterotic TE methylation levels higher than the parental strains PC15 and PC9, whereas N001-HyB shows mid-parent level values (Fig. 5A). Our results suggest that this difference can be explained by the different contribution of each nucleus to the overall methylation levels in the dikaryon, as in N001-HyB the PC9 nucleus is more methylated than PC15, whereas in N001 both nuclei show similar levels (Table 3). The N001 strain has been sub-cultured for >20 years as a dikaryon, whereas N001-HyB is the result of a very recent mating (< 10 sub-cultures) between the compatible protoclones of PC15 and PC9, which were stored as isolated strains in a Culture Collection longer than 15 years. In fungi, the dikaryotic stage is accomplished through the migration of nuclei from one cell to another cell. Thus, the impaired contribution in the dikaryotic formation, could suggest that the dikaryotization was not completely established in the N001-HyB mycelium, besides the presence of clamp connections in the culture. The two nuclei present in dikaryons experience co-evolution in the long-term.67 In this context, the dikaryotic stage is thought to be favored over the monokaryotic, as deleterious mutations in one nucleus can be compensated with the healthy allele of the other, or even reverted by compensatory mutations such as described for Schizophyllum commune.68 Our results suggest that in addition to permanent modifications (such as mutations), epigenetic profiles are also compensated in long-lasting dikaryons. This compensation led to higher methylation in the N001 strain, which can account for their better defense against TEs than their single monokaryotic counterparts. A similar phenomenon was observed in Arabidopsis, where the distribution of epialleles inherited from the parental lines may reflect the selection against demethylated traits possibly influencing plant fitness.69 Nevertheless, this phenomenon is not observed in recently formed dikaryons such as N001-HyB, where methylation profiles of independent nuclei resemble those of their original monokaryotic parentals. To our knowledge, no similar study has reported this event in fungi. Nevertheless, in plant hybrids, an siRNA-mediated mechanism called trans-chromosomal methylation is responsible for equilibrating the methylation levels of alleles, leading to an increase of methylation in the ‘low parent allele’ and resulting in overall higher methylation levels in the F1 hybrids.70 The N001 profile mimics this phenomenon, where the increase in methylation of PC15 alleles would lead to the balanced nucleus-specific methylation that we have described. The intermediate values found in N001-HyB suggest that this phenomenon could be the result of a slow process requiring progressive co-adaptation of the two nuclei in an unique cytoplasm, something that seems reasonable as nuclei in dikaryons remain un-fused during most of their lives. The case of Cluster A (Fig. 3) is an interesting example of how long-term compensation can impact the methylation levels of many genes. This group of genes is present in TE-rich clusters showing opposite methylation profiles in PC15 and PC9, which leads to the presence of epialleles in the early formed dikaryon N001-HyB. In this case, the hypomethylated profile of PC15 is dominant in many Cluster A genes of the latter strain but shifts to a hypermethylated and transcriptionally silent PC9-like profile in the long-term dikaryon N001. This observation suggests that the two nuclei that coexist in the recently formed dikaryon retain some degree of independence prior to establishing crosstalk interactions. Based on our findings, we propose that the TE dynamics could rewire the epigenetic landscape of the fungal genome, promoting gene silencing at their surroundings and generating epialleles in the dikaryotic stage. The methylation profile of these epialleles is subjected to compensation after long-term culture, leading to balanced methylation levels in each nucleus.
Funding
This work was supported by Spanish National Research Plan (Project AGL2014-55971 R) and FEDER funds; Public University of Navarre. AB and LLV hold a PhD scholarship from the Public University of Navarre, RC holds a FPI-PhD scholarship from the Ministry of Economy and Competitiveness and M.M. holds a Philip Whitcome and Dissertation Year Fellowship. M.P. and M.M. were supported by a cooperative agreement with the US Department of Energy Office of Science, Office of Biological and Environmental Research programme under Award DE-FC02-02ER63421.
Conflict of interest: None declared. Accession number Accession GSE112588
Supplementary Material
References
- 1. Feschotte C., Pritham E. J.. 2007, DNA Transposons and the Evolution of Eukaryotic Genomes, Annu. Rev. Genet., 41, 331–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chénais B., Caruso A., Hiard S., Casse N.. 2012, The impact of transposable elements on eukaryotic genomes: from genome size increase to genetic adaptation to stressful environments, Gene, 509, 7–15. [DOI] [PubMed] [Google Scholar]
- 3. Doolittle W. F., Sapienza C.. 1980, Selfish genes, the phenotype paradigm and genome evolution, Nature, 284, 601–3. [DOI] [PubMed] [Google Scholar]
- 4. Fukui K. N., Suzuki G., Lagudah E. S.. 2001, Physical arrangement of retrotransposon-related repeats in centromeric regions of wheat, Plant Cell Physiol., 42, 189–96. [DOI] [PubMed] [Google Scholar]
- 5. Slotkin R. K., Martienssen R.. 2007, Transposable elements and the epigenetic regulation of the genome, Nat. Rev. Genet., 8, 272–85. [DOI] [PubMed] [Google Scholar]
- 6. McGinnis W., Shermoen A. W., Beckendorf S. K.. 1983, A transposable element inserted just 5’ to a Drosophila glue protein gene alters gene expression and chromatin structure, Cell, 34, 75–84. [DOI] [PubMed] [Google Scholar]
- 7. Lonnig W.-E., Saedler H.. 2002, Chromosome rearrangements and transposable elements, Annu. Rev. Genet., 36, 389–410. [DOI] [PubMed] [Google Scholar]
- 8. Klahre U., Crété P., Leuenberger S. A., Iglesias V. A., Meins F. Jr. and 2002, High molecular weight RNAs and small interfering RNAs induce systemic posttranscriptional gene silencing in plants, Proc. Natl. Acad. Sci. U. S. A., 99, 11981–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Malone C. D., Hannon G. J.. 2009, Small RNAs as guardians of the genome, Cell, 136, 656–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Suzuki M. M., Bird A.. 2008, DNA methylation landscapes: provocative insights from epigenomics, Nat. Rev. Genet., 9, 465. [DOI] [PubMed] [Google Scholar]
- 11. Law J. A., Jacobsen S. E.. 2010, Establishing, maintaining and modifying DNA methylation patterns in plants and animals, Nat. Rev. Genet., 11, 204–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Valencia-Sanchez M. A., Liu J., Hannon G. J., Parker R.. 2006, Control of translation and mRNA degradation by miRNAs and siRNAs, Genes Dev, 20, 515–24. [DOI] [PubMed] [Google Scholar]
- 13. Carthew R. W., Sontheimer E. J.. 2009, Origins and Mechanisms of miRNAs and siRNAs, Cell, 136, 642–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dang Y., Li L., Guo W., Xue Z., Liu Y.. 2013, Convergent Transcription Induces Dynamic DNA Methylation at disiRNA Loci, PLoS Genet., 9, e1003761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huang Y.-Z., Sun J.-J., Zhang L.-Z.. 2015, Genome-wide DNA Methylation Profiles and Their Relationships with mRNA and the microRNA Transcriptome in Bovine Muscle Tissue (Bos taurine), Sci. Rep., 4, 6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhao J.-H., Fang Y.-Y., Duan C.-G., Fang R.-X., Ding S.-W., Guo H.-S.. 2016, Genome-wide identification of endogenous RNA-directed DNA methylation loci associated with abundant 21-nucleotide siRNAs in Arabidopsis, Sci. Rep., 6, 36247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zilberman D., Cao X., Jacobsen S. E.. 2003, ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation, Science, 299, 716–9. [DOI] [PubMed] [Google Scholar]
- 18. Selker E. U., Cambareri E. B., Jensen B. C., Haack K. R.. 1987, Rearrangement of duplicated DNA in specialized cells of Neurospora, Cell, 51, 741–52. [DOI] [PubMed] [Google Scholar]
- 19. Selker E. U., Tountas N. A., Cross S. H., et al. 2003, The methylated component of the Neurospora crassa genome, Nature, 422, 893–7. [DOI] [PubMed] [Google Scholar]
- 20. Galagan J. E., Selker E. U.. 2004, RIP: the evolutionary cost of genome defense, Trends Genet., 20, 417–23. [DOI] [PubMed] [Google Scholar]
- 21. Horns F., Petit E., Yockteng R., Hood M. E.. 2012, Patterns of repeat-induced point mutation in transposable elements of basidiomycete fungi, Genome Biol. Evol., 4, 240–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Johnson L. J., Giraud T., Anderson R., Hood M. E.. 2010, The impact of genome defense on mobile elements in Microbotryum, Genetica, 138, 313–9. [DOI] [PubMed] [Google Scholar]
- 23. Barry C., Faugeron G., Rossignol J. L.. 1993, Methylation induced premeiotically in Ascobolus: coextension with DNA repeat lengths and effect on transcript elongation, Proc. Natl. Acad. Sci. U. S. A., 90, 4557–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Freedman T., Pukkila P. J.. 1993, De novo methylation of repeated sequences in Coprinus cinereus, Genetics, 135, 357–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Malagnac F., Silar P.. 2010, Epigenetics of eukaryotic Microbes, Handbook of Epigenetics, 185–201.
- 26. Zemach A., McDaniel I. E., Silva P., Zilberman D.. 2010, Genome-wide evolutionary analysis of eukaryotic DNA methylation, Science, 328, 916–9. [DOI] [PubMed] [Google Scholar]
- 27. Zemach A., Zilberman D.. 2010, Evolution of eukaryotic DNA methylation and the pursuit of safer sex, Curr. Biol., 20, R780. [DOI] [PubMed] [Google Scholar]
- 28. Romano N., Macino G.. 1992, Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences, Mol. Microbiol., 6, 3343–53. [DOI] [PubMed] [Google Scholar]
- 29. Shiu P. K., Raju N. B., Zickler D., Metzenberg R. L.. 2001, Meiotic silencing by unpaired DNA, Cell, 107, 905–16. [DOI] [PubMed] [Google Scholar]
- 30. Wang X., Hsueh Y. P., Li W., Floyd A., Skalsky R., Heitman J.. 2010, Sex-induced silencing defends the genome of Cryptococcus neoformans via RNAi, Genes Dev., 24, 2566–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Binz T., D’Mello N., Horgen P. A.. 1998, A comparison of DNA methylation levels in selected isolates of higher fungi, Mycologia, 90, 785. [Google Scholar]
- 32. Foulongne-Oriol M., Murat C., Castanera R., Ramírez L., Sonnenberg A. S. M.. 2013, Genome-wide survey of repetitive DNA elements in the button mushroom Agaricus bisporus, Fungal Genet. Biol., 55, 6–21. [DOI] [PubMed] [Google Scholar]
- 33. Mueth N. A., Ramachandran S. R., Hulbert S. H.. 2015, Small RNAs from the wheat stripe rust fungus (Puccinia striiformis f.sp. tritici), BMC Genomics, 16, 718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Eugenio C. P., Anderson N. A.. 1968, The Genetics and Cultivation of Pleurotus ostreatus, Mycologia, 60, 627. [Google Scholar]
- 35. Castanera R., López-Varas L., Borgognone A., et al. 2016, Transposable elements versus the fungal genome: impact on whole-genome architecture and transcriptional profiles, PLoS Genet., 12, e1006108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Borgognone A., Castanera R., Muguerza E., Pisabarro A. G., Ramírez L.. 2017, Somatic transposition and meiotically driven elimination of an active helitron family in Pleurotus ostreatus, DNA Res., 24, 103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Larraya L. M., Pérez G., Peñas M. M., et al. 1999, Molecular karyotype of the white rot fungus Pleurotus ostreatus, Appl. Environ. Microbiol., 65, 3413–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Frommer M., McDonald L. E., Millar D. S., et al. 1992, A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands, Proc. Natl. Acad. Sci. U. S. A., 89, 1827–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Morselli M., Pastor W. A., Montanini B., et al. 2017, In vivo targeting of de novo DNA methylation by histone modifications in yeast and mouse, Elife, 6, e06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Feng S., Rubbi L., Jacobsen S. E., Pellegrini M.. 2011, Determining DNA methylation profiles using sequencing, Methods Mol. Biol., p 223–38. [DOI] [PubMed] [Google Scholar]
- 41. Riley R., Salamov A. A., Brown D. W., et al. 2014, Extensive sampling of basidiomycete genomes demonstrates inadequacy of the white-rot/brown-rot paradigm for wood decay fungi, Proc. Natl. Acad. Sci. U. S. A., 111, 14959–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kurtz S., Phillippy A., Delcher A. L., et al. 2004, Versatile and open software for comparing large genomes, Genome Biol., 5, R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Guo W., Fiziev P., Yan W., et al. 2013, BS-Seeker2: a versatile aligning pipeline for bisulfite sequencing data, BMC Genomics. , 14, 774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Smith K. M., Phatale P. A., Bredeweg E. L., et al. 2012, Epigenetics of Filamentous Fungi Encyclopedia of Molecular Cell Biology and Molecular Medicine. Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany. [Google Scholar]
- 45. Kouzminova E., Selker E. U.. 2001, Dim-2 encodes a DNA methyltransferase responsible for all known cytosine methylation in Neurospora, Embo J., 20, 4309–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Freitag M., Williams R. L., Kothe G. O., Selker E. U.. 2002, A cytosine methyltransferase homologue is essential for repeat-induced point mutation in Neurospora crassa, Proc. Natl. Acad. Sci. U. S. A., 99, 8802–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cao X., Jacobsen S. E.. 2002, Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes, Proc. Natl. Acad. Sci. U. S. A., 99, 16491–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Selker E. U. 1997, Epigenetic phenomena in filamentous fungi: useful paradigms or repeat-induced confusion?, Trends Genet., 13, 296–301. [DOI] [PubMed] [Google Scholar]
- 49. Chan S. W. L., Henderson I. R., Zhang X., Shah G., Chien J. S. C., Jacobsen S. E.. 2006, RNAi, DRD1, and histone methylation actively target developmentally important Non-CG DNA methylation in Arabidopsis, PLoS Genet., 2, e83–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jeon J., Choi J., Lee G. W., et al. 2015, Genome-wide profiling of DNA methylation provides insights into epigenetic regulation of fungal development in a plant pathogenic fungus, Magnaporthe oryzae, Sci. Rep., 5, 8567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Montanini B., Chen P.-Y., Morselli M., et al. 2014, Non-exhaustive DNA methylation-mediated transposon silencing in the black truffle genome, a complex fungal genome with massive repeat element content, Genome Biol., 15, 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Saze H., Tsugane K., Kanno T., Nishimura T.. 2012, DNA Methylation in plants: relationship to small RNAs and histone modifications, and functions in transposon inactivation, Plant Cell Physiol., 53, 766–84. [DOI] [PubMed] [Google Scholar]
- 53. Schwach F., Moxon S., Moulton V., Dalmay T.. 2009, Deciphering the diversity of small RNAs in plants: the long and short of it, Brief. Funct. Genomic Proteomic, 8, 472–81. [DOI] [PubMed] [Google Scholar]
- 54. Kumekawa N., Ohmido N., Fukui K., Ohtsubo E., Ohtsubo H.. 2001, A new gypsy-type retrotransposon, RIRE7: preferential insertion into the tandem repeat sequence TrsD in pericentromeric heterochromatin regions of rice chromosomes, Mol. Genet. Genomics, 265, 480–8. [DOI] [PubMed] [Google Scholar]
- 55. Gao D., Jiang N., Wing R. A., Jiang J., Jackson S. A.. 2015, Transposons play an important role in the evolution and diversification of centromeres among closely related species, Front. Plant Sci., 6, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cantu D., Vanzetti L. S., Sumner A., et al. 2010, Small RNAs, DNA methylation and transposable elements in wheat, BMC Genomics., 11, 408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Castanera R., Borgognone A., Pisabarro A. G., Ramírez L.. 2017, Biology, dynamics, and applications of transposable elements in basidiomycete fungi, Appl. Microbiol. Biotechnol., 101, 1337–50. [DOI] [PubMed] [Google Scholar]
- 58. Wang X., Wang P., Sun S., Darwiche S., Idnurm A., Heitman J.. 2012, Transgene induced co-suppression during vegetative growth in Cryptococcus neoformans Copenhaver, G. P., (ed.), PLoS Genet., 8, e1002885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Qi Y., He X., Wang X.-J., Kohany O., Jurka J., Hannon G. J.. 2006, Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation, Nature, 443, 1008–12. [DOI] [PubMed] [Google Scholar]
- 60. Arnaud P., Goubely C., Pélissier T., Deragon J. M.. 2000, SINE retroposons can be used in vivo as nucleation centers for de novo methylation, Mol. Cell. Biol., 20, 3434–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jähner D., Haase K., Mulligan R., Jaenisch R.. 1985, Insertion of the bacterial gpt gene into the germ line of mice by retroviral infection, Proc. Natl. Acad. Sci. U. S. A., 82, 6927–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Honda S., Lewis Z. A., Shimada K., Fischle W., Sack R., Selker E. U.. 2012, Heterochromatin protein 1 forms distinct complexes to direct histone deacetylation and DNA methylation, Nat. Struct. Mol. Biol., 19, 471–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Arjona D., Aragón C., Aguilera J. A., Ramírez L., Pisabarro A. G.. 2009, Reproducible and controllable light induction of in vitro fruiting of the white-rot basidiomycete Pleurotus ostreatus, Mycol. Res., 113, 552–8. [DOI] [PubMed] [Google Scholar]
- 64. Muraguchi H., Umezawa K., Niikura M., et al. 2015, Strand-specific RNA-seq analyses of fruiting body development in Coprinopsis cinerea, PLoS One, 10, e0141586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Vidic I., Berne S., Drobne D., et al. 2005, Temporal and spatial expression of ostreolysin during development of the oyster mushroom (Pleurotus ostreatus), Mycol. Res., 109, 377–82. [PubMed] [Google Scholar]
- 66. Wösten H. A. B., Wessels J. G. H.. 2006, The Emergence of Fruiting Bodies in Basidiomycetes Growth, Differentiation and Sexuality. Springer-Verlag, Berlin/Heidelberg, pp. 393–414. [Google Scholar]
- 67. Anderson J. B., Kohn L. M.. 2007, Dikaryons, diploids, and evolution Sex in Fungi: Molecular Determination and Evolutionary Implications. ASM Press, Washington, DC, pp. 333–48. [Google Scholar]
- 68. Clark T. A., Anderson J. B.. 2004, Dikaryons of the basidiomycete fungus Schizophyllum commune: evolution in long-term culture, Genetics, 167, 1663–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Reinders J., Wulff B. B. H., Mirouze M., et al. 2009, Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes, Genes Dev., 23, 939–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Greaves I. K., Groszmann M., Dennis E. S., James Peacock W.. 2012, Trans-chromosomal methylation, Epigenetics, 7, 800–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.