Abstract
Long noncoding RNAs (lncRNAs) are dysregulated in various human cancers and control tumor development and progression. However, the upstream mechanisms underlying their dysregulation remain unclear. Here we demonstrate that the expression of hepatocyte nuclear factor 1 homeobox A antisense RNA 1 (HNF1A-AS1) is significantly upregulated in gastric cancer (GC) tissues. Overexpression of HNF1A-AS1 enhanced cell proliferation and promoted cell cycle progression, whereas knockdown of HNF1A-AS1 elicited the opposite effects. Early growth response protein 1 (EGR1) directly bound the HNF1A-AS1 promoter region and activated its transcription. Overexpression of EGR1 enhanced cell proliferation and promoted cell cycle promotion, similar to the function of HNF1A-AS1. HNF1A-AS1 functioned as competing endogenous RNA (ceRNA) by binding to miR-661, upregulating the expression of cell division cycle 34 (CDC34), which is a direct target of miR-661. EGR1 and HNF1A-AS1 enhanced the expression of cyclin-dependent kinase 2 (CDK2), CDK4, and cyclin E1 but inhibited the expression of p21 by promoting CDC34-mediated ubiquitination and degradation of p21. Taken together, these findings suggest that EGR1-activated HNF1A-AS1 regulates various pro- and anti-growth factors to promote the development of GC, implicating it as a possible target for therapeutic intervention in this disease.
Keywords: HNF1A-AS1, EGR1, CDC34, P21, ubiquitination, gastric cancer
Introduction
Gastric cancer (GC) remains the fourth most frequently diagnosed cancer and is the third leading cause of cancer-related deaths worldwide (1, 2). Cell proliferation is one characteristic of cancer development and progression, and high cellular proliferation is associated with a poor prognosis in patients with malignant tumors (3). With progress in medical care, several screening techniques including barium meal photofluorography, gastric endoscopy, and serum pepsinogen have been recommended as screening methods for the detection of early GC. Nevertheless, some patients are still diagnosed at advanced stages (4), which often have poor prognosis mainly due to the progression of the GC and tumor growth (5). Therefore, novel insights into the mechanisms underlying GC progression are warranted, which in turn may identify potential therapeutic targets that to improve patient survival.
Recent transcriptomic studies have shown that the majority of the human genome is transcribed into noncoding RNA molecules (6, 7). Long noncoding RNAs (lncRNAs) are noncoding transcripts that are larger than 200 nucleotides and lack protein-coding potential. Compared to protein-coding transcripts, lncRNA genes have fewer exons, are of lower abundance, and are highly cell- and tissue-specific (8). LncRNAs regulate gene expression at the transcriptional, post-transcriptional, and epigenetic levels by interacting with various biomolecules, including DNA, RNA, and proteins (9). There is a growing body of evidence that these noncoding transcripts participate in a wide range of cellular processes, including cell proliferation (10), apoptosis (11), differentiation (12) and the immune response (13), all of which are involved in tumorigenesis. Thus, studying the expression of lncRNAs could be of profound value with regard to understanding the mechanisms underlying GC progression.
Among all known cancer-related lncRNAs, hepatocyte nuclear factor 1 homeobox A antisense RNA 1 antisense RNA 1 (HNF1A-AS1) expression is upregulated in various malignancies (14–17) and its elevated expression is associated with an advanced stage and/or poor prognosis, indicating an oncogenic role for HNF1A-AS1 in tumor progression. Wu et al. reported that HNF1A-AS1 could regulate cyclin D1, E-cadherin, N-cadherin, and β-catenin expression, inducing the EMT process, thereby promoting lung adenocarcinoma proliferation and metastasis (18). Additionally, HNF1A-AS1 functions as an oncogene in colon cancer cells by acting as a ceRNA for miR-34a, which then leads to de-suppression of SIRT1, thereby facilitating colon cancer progression (19). However, no studies on the upstream regulatory mechanisms of HNF1A-AS1 and promoter analysis of HNF1A-AS1 have been reported to date.
In this study, we demonstrate that HNF1A-AS1 is upregulated in GC tissues, and promotes GC malignant phenotypes. Early growth response protein 1 (EGR1) transcriptionally upregulate HNF1A-AS1, which acts as a ceRNA by competitively binding to microRNA (miR)-661 to enhance the expression of cell division cycle 34 (CDC34). Additionally, we show that HNF1A-AS1 promotes GC cell proliferation and cell cycle progression, in which cyclin-dependent kinase 2 (CDK2), CDK4, and cyclin E1 are involved and induces the dysregulation of P21 protein via CDC34-mediated ubiquitination.
Methods and Materials
Tissue specimens and cell lines
A total of 99 fresh GC tissues and 8 non-tumorous gastric tissues were obtained from Qilu Hospital of Shandong University (Shandong Province, China). Our study was approved by the Institute’s Research Ethics Committee of Shandong University and conducted in accordance with ethical guidelines of the World Medical Association Declaration of Helsinki. Informed consent was written by all patients prior to this study. All the cells were purchased from the ATCC. Cell authentication was verified by short tandem repeat profiling. Human MKN-45 cells and BGC-823 cells were cultured in RPMI-1640 medium (Gibco, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS; Gibco) at 37°C, 5% CO2. HEK293 cells were cultured in Dulbecco’s Modified Eagle Medium (Gibco) supplemented with 10% FBS. Mycoplasma PCR testing of these cells was performed every month. Cells were grown for no more than 20 passages in total for any experiment.
siRNA and cell transfection
Two siRNA targeting HNF1A-AS1 (si-HNF1A-AS1–1 and si-HNF1A-AS1–2) and scrambled negative control (si-NC) were designed and synthesized by RiboBio (Guangzhou, China). The sequence of si-HNF1A-AS1–1 and si-HNF1A-AS1–2 were as follows: CCCTCCATCTAACATTCAA; CCCTCATCCTGGCATTCAA. MKN-45 and BGC-823 cells were seeded in 6- and 12-well plates and transfected 48 h later using X-tremeGENE transfection reagent (Roche Applied Science, Indianapolis, IN, USA) according to the manufacturer’s instructions.
RNA extraction and RT-qPCR
Total RNAs were extracted from the tissues and cultured cells using TRIzol® (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacturer’s instructions. Approximately 1 μg of total RNA was reverse transcribed to cDNA using a reverse transcriptase cDNA synthesis kit (Toyobo, Osaka, Japan), and qPCR was performed using a SYBR Green PCR kit (Roche, Basel, Switzerland). Comparative quantification was assessed using the 2−ΔCt method using GAPDH as the endogenous control.
Gastric cancer cell viability and proliferation assay
The MTS, colony formation, and EdU cell proliferation assays were performed to evaluate the viability and proliferative capacity of GC cells as previously described (20).
Migration and invasion assays
Transwell™ chambers either uncoated or coated with Matrigel were used to observe GC cell migration and invasion as described elsewhere (21).
Flow cytometry analysis of the cell cycle and apoptosis
MKN-45 and BGC-823 cells transfected with HNF1A-AS1-PCDNA3.1 or Si-HNF1A-AS1 were cultured in six-well plates for 48 h. To evaluate the cell cycle, the cells were stained with propidium iodide (PI, Beyotime, Shanghai, China). To evaluate apoptosis, the cells were stained with Annexin V-fluorescein isothiocyanate (FITC) and PI using an FITC Annexin V Apoptosis Detection kit. Harvested cells were analyzed by flow cytometry (FACScan; BD Biosciences, San Jose, CA, USA) according to the manufacturer’s recommendations.
Bioinformatics analysis
The HNF1A-AS1 sequence was downloaded from UCSC Genome Browser (http://genome.ucsc.edu/), from which the 2,000-bp transcription start site (TSS) upstream sequence was extracted. To identify putative transcription factors, the promoter sequence of HNF1A-AS1 was submitted to the JASPAR program (http://jaspar.genereg.net/).
Vector construction
The putative HNF1A-AS1 promoter regions (−2,000/0, −1,014/0, −503/0, −285/0, and −187/0) were PCR-amplified from the genomic DNA of MKN-45 cells, which were then inserted into the HindIII-NheI sites upstream of the firefly luciferase in the pGL3-Basic vector (Promega, Madison, WI, USA). All of the constructs were named based on the location of the promoter fragments relative to the TSS. The full-length cDNA sequence of KLF5, ELK1, and EGR1 were PCR-amplified from the cDNA of MKN-45 cells and cloned into the pcDNA3.1 vector. The HNF1A-AS1 expression vector PCDNA3.1-HNF1A-AS1 was generated in our previous study. Ubiquitin plasmid was generously provided by Dr. Hu (Shandong University). P21, CDC34 and SKP2 vector were purchased from Vigene Biosciences (Rockville, MD, USA). The 3’-UTR of CDC34 was sub-cloned into the pmirGLO vector and named pmiRGLO-CDC34. All vectors were confirmed by direct sequencing using primers shown in Supplementary Table 1.
HNF1A-AS1 short hairpin RNA stable knockdown
Short hairpin RNAs (shRNAs) targeting HNF1A-AS1 were selected and expressed using the pGPU6/GFP/Neo vector (GenePharma, Shanghai, China). The shRNA sequence was as follows: CCCTCCATCTAACATTCAA. The MKN-45 and BGC-823 cells were seeded into six-well plates and transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). To screen stable HNF1A-AS1 knockdown cells, G418 (500 μg/mL) was added to the medium 72 h after transfection. The knockdown efficiency of HNF1A-AS1 is shown in Supplementary Figures 1A–B.
Dual-luciferase reporter assays
The HEK293 cells were seeded overnight in 24-well plates and then co-transfected per well 0.5 μg of the overexpression vector, 0.5 μg of the HNF1A-AS1 promoter-luciferase reporter plasmid, and 0.01 μg of pRL-TK plasmid with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). PRL-TK was used as the internal control. Approximately 48 h after transfection, the cells were subjected to luciferase activity analysis using a Dual-Luciferase Reporter Assay System (Promega) following the manufacturer’s instructions.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays were performed using the EZ-Magna ChIP™ Chromatin Immunoprecipitation Kit (Millipore, Darmstadt, Germany) according to the manufacturer’s instructions. Briefly, the MKN-45 and BGC-823 cells in each immunoprecipitation reaction were crosslinked with 1% formaldehyde for 10 min at room temperature. After incubation with anti-EGR1 (Cell Signaling Technology [CST], Danvers, MA, USA) or IgG antibody and Protein A/G magnetic beads overnight, immunoprecipitation was performed. Co-precipitated DNA was quantified using PCR and qPCR. The HNF1A-AS1 primers used are shown in Supplementary Table 1.
Actinomycin D and cycloheximide assays
The HNF1A-AS1 and CDC34 plasmids were transiently transfected into GC cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). After 24 h, actinomycin D (ActD, 5 μg/mL) or cycloheximide (CHX, 5 μg/mL) was added to the culture medium, followed by incubation for 15 min, 30 min, 1 h, 2 h, or 3 h. P21 mRNA stability in the ActD treatment group was analyzed by qPCR. The half-life of p21 protein in the CHX treatment group was observed by western blotting.
Western blotting
The antibodies used for western blotting were rabbit anti-CDK2 (CST), rabbit anti-CDK4 (CST), rabbit anti-cyclin E1 (CST), rabbit anti-p21 (Abways, shanghai, China), rabbit anti-SKP2 (CST), and rabbit anti-P27 (CST).
In vitro ubiquitination analysis
HNF1A-AS1, ubiquitin, and p21 plasmids were transfected into cells using Lipofectamine 2000. Approximately 48 h post-transfection, the lysates were immunoprecipitated with p21 antibodies (Sigma-Aldrich, St. Louis, MO, USA) overnight at 4°C with gentle rotation. The immunoprecipitated proteins were analyzed by western blotting.
RNA binding protein immunoprecipitation assays
RNA binding protein immunoprecipitation (RIP) assays were performed using the Magna RIP RNA Binding Protein Immunoprecipitation Kit (Millipore, Burlington, MA, USA) according to the manufacturer’s instructions. Briefly, the cells were lysed in lysis buffer containing a protease inhibitor cocktail and RNase inhibitor, and the lysates were immunoprecipitated with anti-Ago2 and IgG antibodies. Finally, the retrieved RNAs were quantified by qPCR. The HNF1A-AS1 and CDC34–3’UTR primers used for amplification are listed in Supplementary Table 1.
Biotin-labeled pull-down assays
The BGC-823 cells were transfected with biotinylated miR-661 and a scrambled control miRNA (GenePharma, Shanghai, China) and collected 48 h after transfection. The cell lysates were incubated with M-280 streptavidin magnetic beads (Invitrogen) and 10 μL of yeast tRNA on a rotator at 4°C for 2 h. The bound RNAs were purified by adding 750 μL of TRIzol (Invitrogen, Carlsbad, CA, USA) per sample and 250 μL of water to the input and the pull-down beads for further RT-qPCR analysis. The 3’ biotin-labeled miR-661 sequence was as follows: 5’UGCCUGGGUCUCUGGCCUGCGCGU 3’BiO. The scrambled control miRNA sequence was as follows: 5’-UUCUCCGAACGUGUCACGUTT-3’BiO.
Immunohistochemistry staining
Fresh GC tissue sections were cut into 4-μm sections and then analyzed by immunohistochemistry (IHC) using antibodies against EGR1 (1:50; CST). Brown signals in the nuclei and cytoplasm indicated positive EGR1 immunostaining. EGR1 staining intensity was defined using a three-scale grading system: weak (score: 1), moderate (score: 2), or strong (score: 3). A staining score was calculated as follows: Score (maximum of 300) = Sum of 1 × percentage of weak, 2 × percentage of moderate and 3 × percentage of strong staining. Then, the relationship between EGR1 levels and lncRNA HNF1A-AS1 was assessed for each sample.
Tumor xenograft model
Four-week-old male Nu/nu athymic nude mice (Weitonglihua Biotechnology, Beijing, China) were raised under specific pathogen-free conditions. LV-HNF1A-AS1 and LV-NC-MKN-45-transfected cells were respectively injected subcutaneously into the flank region of the mice (7 × 105 cells/150 μL per flank, n = 8). The tumor volumes were measured every seven days after inoculation. Five weeks after injection, the mice were sacrificed, and the tumor nodules were harvested. The tumors, lungs, liver, and kidneys were isolated from the mice for further analysis. The protocol was approved by the Committee on the Ethics of Animal Experiments of the Shandong University.
Statistical analysis
All data were statistically analyzed using GraphPad Prism 5 software (San Diego, CA, USA). Analysis of the differences between the two groups was determined by Student’s t-test. The correlation between EGR1 levels and HNF1A-AS1 was assessed using Spearman’s correlation coefficient. A P value of less than 0.05 was considered statistically significant.
Results
HNF1A-AS1 promotes GC cell growth and invasion
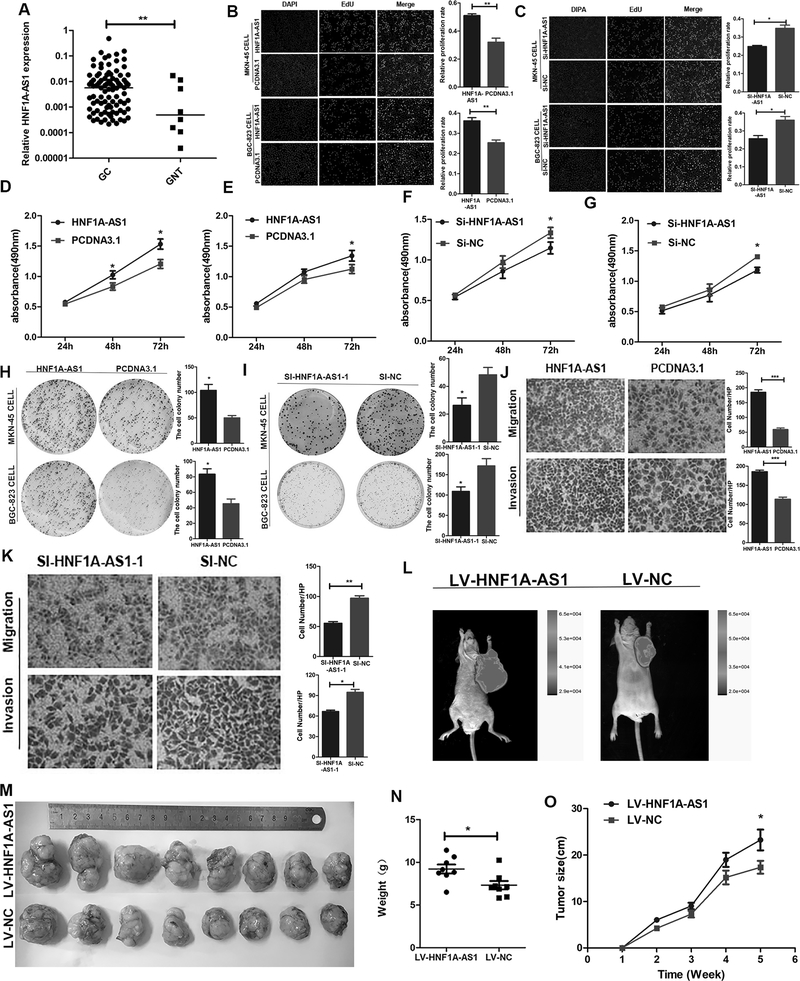
In our study, we found that HNF1A-AS1 expression was significantly upregulated in GC tumor tissues compared to non-tumorous gastric tissues (Figure 1A). To further explore the biological functions of HNF1A-AS1 in GC cells, the GC cells were transfected with the pcDNA3.1-HNF1A-AS1 plasmid or small interfering RNA (siRNA) targeting HNF1A-AS1 (si-HNF1A-AS1–1 and si-HNF1A-AS1–2). The overexpression and knockdown efficiency of HNF1A-AS1 were detected 48 h after transfection (Supplementary Figures 1C–H). EDU assays showed that HNF1A-AS1 overexpression enhanced cell proliferation (Figure 1B), whereas HNF1A-AS1 knockdown significantly suppressed cell growth (Figure 1C and Supplementary Figure 1I). Coinciding with our EDU results, the MTS assay also showed that HNF1A-AS1 enhances cell proliferation (Figures 1D–G and Supplementary Figure 1J–K). Additionally, colony formation assays revealed that the number and size of all colonies increased in the HNF1A-AS1 overexpression groups (Figure 1H), indicating that HNF1A-AS1 also promotes the anchorage-independent growth of GC cells. HNF1A-AS1 downregulation effectively repressed anchorage-independent growth (Figure 1I and Supplementary Figure 1L). In addition, the overexpression of HNF1A-AS1 significantly promoted GC cell migration and invasion (Figure 1J and Supplementary Figure 1M), whereas HNF1A-AS1 knockdown markedly suppressed migration and invasion (Figure 1K, and Supplementary Figure 1N, 2A–B). To determine whether HNF1A-AS1 regulates GC cell proliferation in our xenograft experiments, we established xenograft tumor models in nude mice using MKN-45 GC cells transfected with LV-HNF1A-AS1 or LV-NC (expressing GFP). All nude mice developed xenograft tumors at the injection site, and the xenograft tumors were harvested five weeks after injection. We observed that HNF1A-AS1 played an important role in promoting tumor growth (Figures 1L–M). The xenograft proliferation assay demonstrated that tumor nodules derived from HNF1A-AS1-overexpressing cells grew substantially faster than those in the control group (tumor weight, 9.221 ± 1.494 g vs. 7.330 ± 1.399 g; Figure 1N; p = 0.0214; tumor volume, 114.7 ± 95.13 mm3 vs. 88.17 ± 73.24 mm3; Figure 1O; p = 0.046). These findings demonstrate that HNF1A-AS1 initiates the growth and invasion of GC cells.
Figure 1. HNF1A-AS1 promotes GC cell growth and invasion.
(A). HNF1A-AS1 expression was analyzed by qRT-PCR in GC tissues (n = 99) and adjacent normal tissues (n = 8). HNF1A-AS1 expression level was normalized to that of GAPDH.
(B-C). MKN-45 and BGC-823 cells growth was enhanced as evaluated by the EdU assay (B). And HNF1A-AS1 knockdown inhibited cell proliferation activities in both two cell lines (C). Three independent experiments were performed, and data are presented as mean ± SD.
(D-G). The cell growth rates were determined with MTS proliferation assays. Three independent experiments were performed, and data are presented as mean ± SD.
(H-I). Colony formation assays were used to explore the cell colony formation ability of HNF1A-AS1 and Si-HNF1A-AS-1–transfected cells. Three independent experiments were performed, and data are presented as mean ± SD.
(J-K). Migration and invasion ability of the BGC-823 cell lines by Transwell assays. Three independent experiments were performed, and data are presented as mean ± SD. (L). Five weeks post implantation, the tumors in Nu/Nu mice was measured by an in vivo imaging system.
(M). The HNF1A-AS1 stably expressing group or the negative control were used for tumorigenesis assay. Five weeks later, the mice were killed and the tumor nodules were harvested.
(N-O). Tumor growth curves after subcutaneous injection of MKN-45 cells containing a stable overexpression of HNF1A-AS1 or the negative control are shown. The tumor weights (N) and volumes (O) were measured every 7 days after inoculation.
*P < 0.05, ** P < 0.01, *** P < 0.001
HNF1A-AS1 promotes GC cell cycle progression
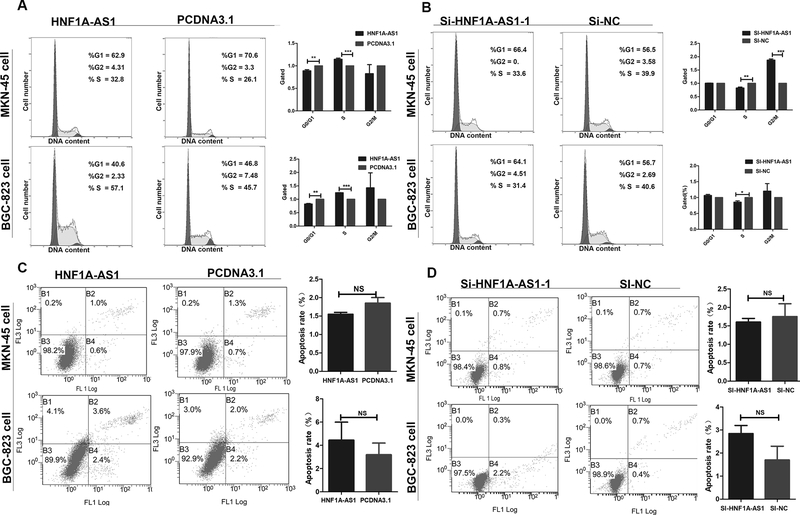
Next, we determined whether HNF1A-AS1 overexpression or knockdown affects the cycle or apoptosis in GC cells. The results indicated that HNF1A-AS1 overexpression promotes the transition from G1 to S phases of the cell cycle (Figure 2A), whereas suppression of HNF1A-AS1 induced a decrease in the number of cells in the S phase and an unstable number of cells in the G2 phase (Figure 2B and Supplementary Figure 2C). Next, we investigated the effects of HNF1A-AS1 on cell apoptosis. Annexin V-FITC assays indicated that there was no difference in the proportion of apoptotic cells between the experimental and control group following a 48-h treatment (Figures 2C–D and Supplementary Figure 2D). These results showed that HNF1A-AS1-mediated promotion of GC cell proliferation was due to cell cycle modulation, not apoptosis.
Figure 2: The effect of HNF1A-AS1 on gastric cancer cell cycle and apoptosis in vitro.
(A). Flow cytometry was performed to determine the effects of HNF1A-AS1 on cell cycle. Significant G1 phase decrease and S phase increase was observed in both cell lines.
(B). Significant G1 phase increase and S phase decrease was observed in both cell lines.
(C-D). Apoptosis was analyzed by flow cytometry, which showed that upregulation or downregulation of HNF1A-AS1 expression did not induce apoptosis in comparison with the control cells. Three independent experiments were performed, and data are presented as mean ± SD.
*P < 0.05, ** P < 0.01, *** P < 0.001
Identification of the proximal promoter regions of HNF1A-AS1
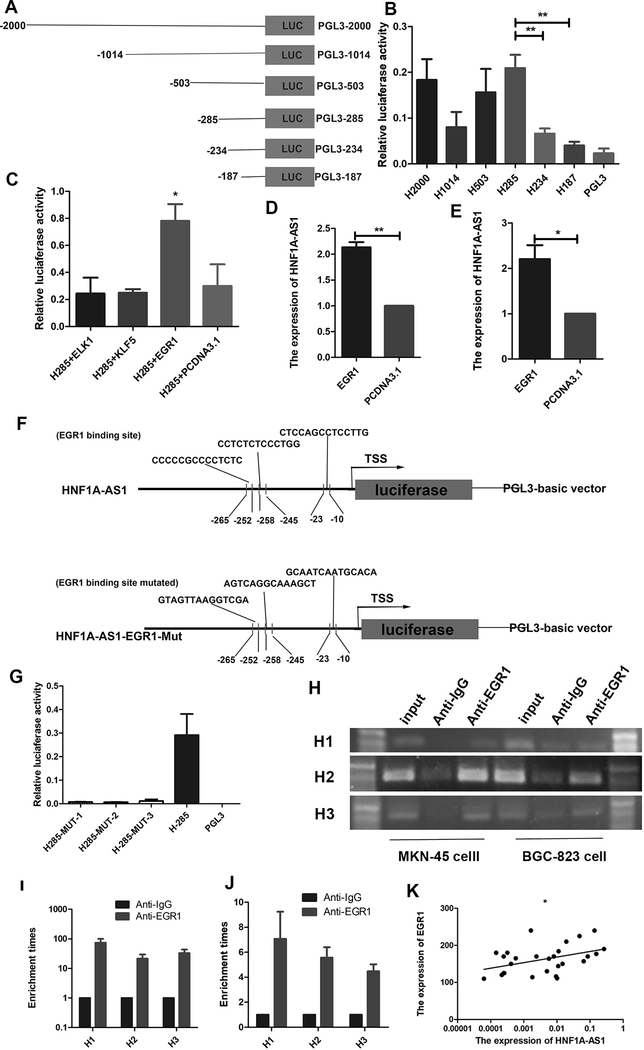
To explore the regulatory mechanisms underlying HNF1A-AS1 upregulation, detailed promoter analysis was performed. The human HNF1A-AS1 promoter sequence was obtained from the UCSC Genome Browser (http://genome.ucsc.edu/). To identify the proximal promoter region of HNF1A-AS1, the promoter activities of a~2,000-base pair (bp) region upstream of the TSS and five deletion constructs were investigated using luciferase activities assays (Figure 3A). The pGL3–2000/0 plasmid showed strong luciferase activity compared to the luciferase reporter vector pGL3-Basic in 293T cells. No statistically significant difference in luciferase activity between pGL3–2000/0 and the deletion constructs, including pGL3–1014/0, pGL3–503/0, and pGL3–285/0, were observed (Figure 3B). However, further deletion of up to −187 resulted in a 75% decrease in luciferase activity relative to that of pGL3−285/0, indicating that the region located between −285 and −187 is the basal promoter of HNF1A-AS1.
Figure 3. EGR1 activates HNF1A-AS1 transcription.
(A). Schematic diagram of the HNF1A-AS1 promoter fragments spanning from −2,000/ −1,014/ −503/ −285/ −234/ −187 to 0. This promoter fragments were cloned into the upstream of the firefly luciferase reporter gene in the pGL3-basic vector.
(B). Transcriptional activity analysis of the potential HNF1A-AS1 promoter fragments in 293T cells.
(C). Luciferase activity assay showed that EGR1 observably increased promoter activities of pGL3−285/0.
(D-E). RT-qPCR assay indicated EGR1 promoted the expression level of HNF1A-AS1 in MKN-45 cells (D) and BGC-823 cells (E).
(F). Schematic diagram of the luciferase reporter construct containing the human HNF1A-AS1 promoter and the mutant construct (H-Mut-1/2/3) containing the basal promoter in which the presumed HNF1A-AS1 binding site was mutated.
(G). Luciferase activity of the HNF1A-AS1 promoter was decreased when the three presumed HNF1A-AS1 binding site was mutated.
(H). ChIP-PCR assay showed that EGR1directly interacted with the EGR1 binding sites within HNF1A-AS1 promoter in MKN-45 cells and BGC-823 cells. A specific strong band of the expected size was detected in the input DNA. The fragment containing the EGR1 binding sites was detected. No band or very weak band was detected in the chromatin complex precipitated by antibody against IgG. H1, H2 and H3 respectively represent primer that covered the EGR1 binding sites.
(I-J). ChIP-qPCR analysis indicated higher fold enrichment of promoter amplicons of EGR1 in anti-EGR1 antibody group than that of IgG group in MKN-45 cells (I) and BGC-823 cells (J), indicating that EGR1 could directly bind to HNF1A-AS1 promoter. H1, H2 and H3 respectively represent primer that covered the EGR1 binding sites.
(K). A significant positive correlation was found between the protein levels of EGR1 and HNF1A-AS1 in GC tissues. Three independent experiments were performed, and data are presented as mean ± SD.
*P < 0.05, ** P < 0.01, *** P < 0.001
EGR1 activates HNF1A-AS1 transcription
To identify the mechanisms underlying regulation of the basal promoter of HNF1A-AS1, the transcription factor-binding site region from −285 to −187 was analyzed using the JASPER program. Three putative binding sites, including three ELK1-binding sites, six Krueppel-like factor 5 (KLF5)-binding sites, and three EGR1-binding sites were identified. To determine their roles in regulating HNF1A-AS1 transcription, ELK1, KLF5, and EGR1 were respectively overexpressed in 293T cells that were transiently transfected with the pGL3–285/0 construct. The results showed that the promoter activities of pGL3–285/0 significantly increased in the EGR1 overexpression group (Figure 3C). The RT-qPCR assay also showed that EGR1 promotes the expression of HNF1A-AS1 (Figures 3D-E). These results indicate that the EGR1-binding site is essential for HNF1A-AS1 transcription. To investigate the contribution of the three putative EGR1-binding sites to the regulation of the HNF1A-AS1 promoter, we introduced three point mutations, designated as p (−10 to −23) Luc-EGR1–1, p (−245 to −258) Luc-EGR1–2, and p (−252 to −265) Luc-EGR1–3 (Figure 3F). The three mutant plasmids were respectively transfected into the 293T cells, and luciferase assays were performed. Figure 3G shows that the three mutation vectors respectively led to an approximate 90% decrease in promoter activity compared to the wild-type control, indicating that the EGR1-binding site cluster is responsible for HNF1A-AS1 transcription. To determine whether EGR1 specifically interacts with the HNF1A-AS1 promoter, a ChIP assay was performed. Figure 3H shows that the three PCR products were amplified from the DNA fragment that was immunoprecipitated by the anti-EGR1 antibody using primers that covered the three EGR1-binding sites. Additionally, an approximate 10-fold enrichment of the promoter amplicons of HNF1A-AS1 in the three binding sites in GC cells was observed using the anti-EGR1 antibody (Figures 3I–J), suggesting that EGR1 could directly bind to the promoter of HNF1A-AS1. In the LinkedOmics database, enhanced EGR1 expression predicted poor patient prognosis (Supplementary Figure 2E), indicating that EGR1 functions as an oncogene and plays important roles in GC progression. Furthermore, our results revealed a positive correlation between EGR1 and HNF1A-AS1 expression in human GC tissue (Figure 3K; r = 0.3835, p = 0.0364), further supporting the fact that EGR1 positively regulates HNF1A-AS1 expression. Taken together, these findings demonstrate that EGR1 regulates the transcription of HNF1A-AS1 by directly binding to its promoter.
EGR1 enhances GC cell proliferation
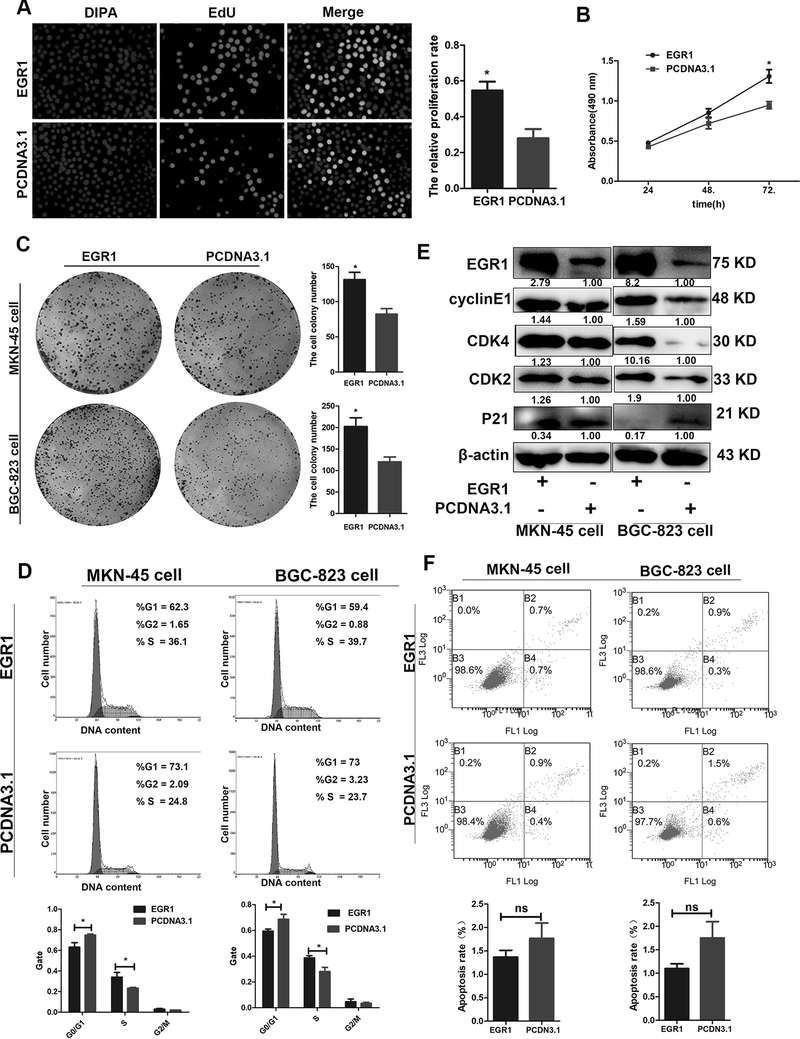
To explore the effects of EGR1 on GC cell behavior, the MKN-45 and BGC-823 cells were transiently transfected with the pcDNA3.1-EGR1 or pcDNA3.1 plasmid. The qPCR results showed that EGR1 was significantly upregulated in EGR1-transfected cell lines (Supplementary Figure 2F). The EdU assay demonstrated that EGR1 overexpression significantly promotes GC cell proliferation compared to the control group (Figure 4A). Coinciding results of the EdU assay, the MTS assay also showed that EGR1 enhances GC cell proliferation (Figure 4B). Moreover, the colony formation assays further verified these results by showing that EGR1 upregulation enhances the number and size of all cell colonies compared to the control group (Figure 4C). To elucidate the potential mechanisms underlying the growth-promoting effects of EGR1, the cell cycle and apoptosis were evaluated in the MKN-45 and BGC-823 cells by flow cytometry. The results revealed that EGR1 overexpression promotes G1 to S cell cycle transition (Figure 4D). Consistent with the results of the MTS and EdU assays, the expression of the cell cycle-related biomarkers CDK2, CDK4, and cyclin E1 increased, and p21 expression decreased when EGR1 was overexpressed in both GC cell lines (Figure 4E). However, the proportion of apoptotic cells was not affected by EGR1 overexpression compared to the control group (Figure 4F). These findings demonstrate that EGR1-induced GC cell proliferation may have been due to the promotion of cell cycle progression.
Figure 4. EGR1 enhances GC cell proliferation.
(A) EdU assay was performed to observe the effect of EGR1 overexpression on cell proliferation.
(B). Cells were seeded in 96-well plates after transfection with EGR1 overexpression or control vector, and cell number was determined every 24 h using MTS assays.
(C). Effect of EGR1 overexpression on the colony formation of MKN-45 and BGC-823 cells.
(D). Effect of EGR1 overexpression on the cell cycle progression of both cells.
(E). The expression levels of CDK2, CDK4, cylcinE1 and P21 in both MKN-45 cells and BGC-823 cells transfected with EGR1 overexpression vector were detected by western blotting assay.
(F). Apoptosis was determined by flow cytometry, which showed that EGR1 did not induce apoptosis in comparison with the control cells. Three independent experiments were performed, and data are shown as mean ± SD.
*P < 0.05, ** P < 0.01, *** P < 0.001.
HNF1A-AS1 enhances the expression of cell cycle regulators and promotes p21 degradation by CDC34-mediated ubiquitination
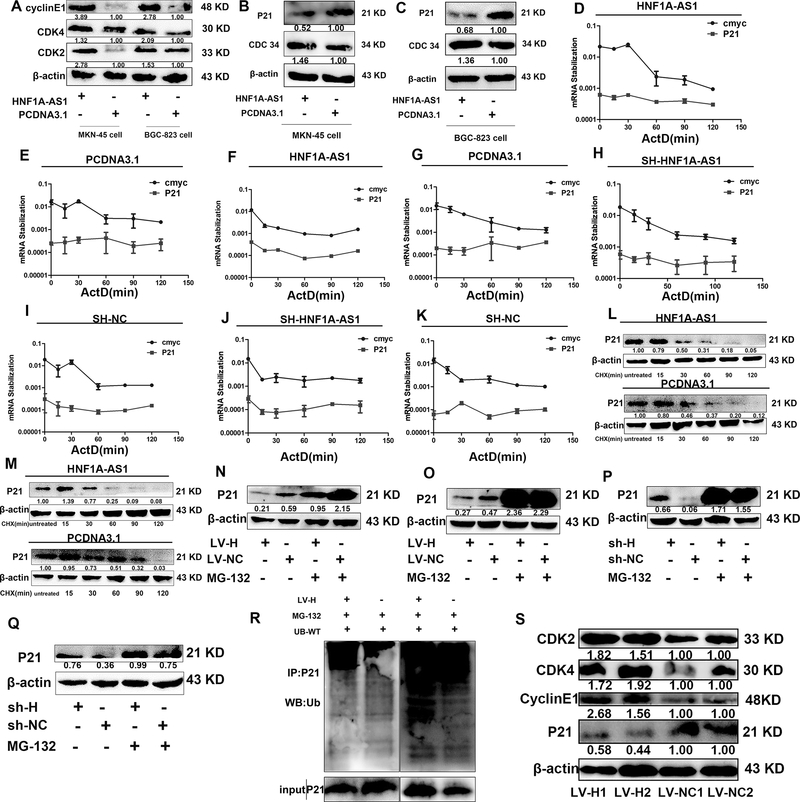
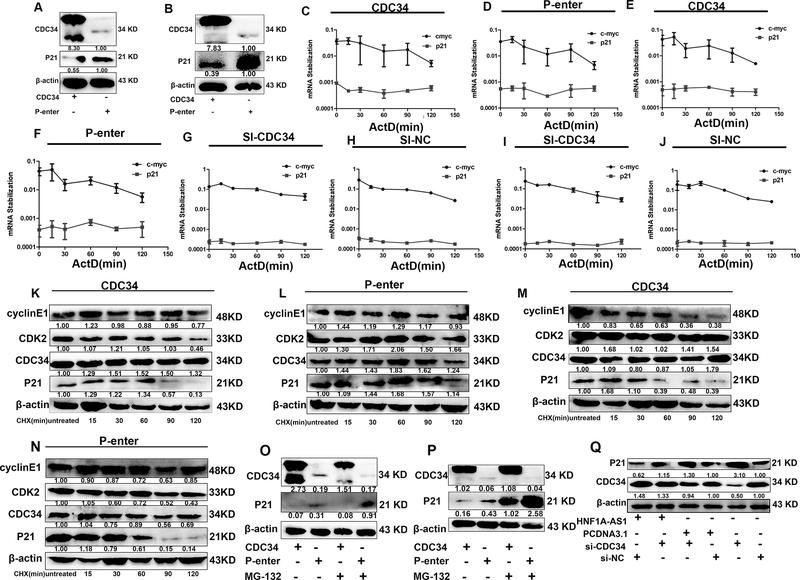
To determine how HNF1A-AS1 promotes cell cycle progression, the expression of the cell cycle regulators CDK2, CDK4, and cyclin E1 were examined by western blotting. Among these proteins, the expression of CDK2, CDK4, and cyclin E1 was significantly upregulated in the HNF1A-AS1 overexpression group compared to the control group (Figure 5A). The CDK inhibitors p21 and p27 are important regulators of the cell cycle. Thus, we hypothesized that HNF1A-AS1 promotes GC cell cycle progression via suppression of p21 or p27 expression. As expected, only p21 protein expression decreased when HNF1A-AS1 was overexpressed in the MKN-45 and BGC-823 cells (Figure 5B–C and Supplementary Figure 2G), indicating that transcriptional and post-transcriptional levels might be affected by HNF1A-AS1. Then, we performed an RNA polymerase II Inhibitor ActD chase assay to assess the effects of HNF1A-AS1 on p21 mRNA stability. The results showed that p21 mRNA levels were not significantly altered in the HNF1A-AS1 overexpression and knockdown group (Figure 5D–K). The MKN-45 cells and BGC-823 cells were treated with the protein synthesis inhibitor CHX, which showed that HNF1A-AS1 reduces the stability of the p21 protein (Figure 5L-M). To determine whether the decrease in p21 protein stability in the HNF1A-AS1 overexpression group was due to an increase in proteasomal-mediated degradation of p21, we conducted experiments with the proteasome inhibitor MG132. MG132 treatment of cells resulted in an increase in p21 protein levels compared to the control group, indicating that the ubiquitin–proteasome pathway is involved in the HNF1A-AS1-degradation of the p21 proteins (Figure 5N–Q). Consistent with our hypothesis, the ubiquitination of p21 significantly increased in the MKN-45 cells overexpressing HNF1A-AS1 compared to the control group (Figure 5R). Moreover, the expression of CDK2, CDK4, cyclin E1, and p21 was detected in mouse xenograft tumors (Figure 5S). HNF1A-AS1 overexpression increased the expression of CDK2, CDK4, and cyclin E1 and decreased the expression of p21, confirming that HNF1A-AS1 promotes cell cycle progression by inhibiting p21 expression to upregulate CDK2, CDK4, and cyclin E1 expression. Taken together, these results show that HNF1A-AS1 promotes the degradation of p21 through ubiquitination. To further investigate the mechanism by which HNF1A-AS1 regulates p21 ubiquitination, we evaluated cell cycle-related E3 ubiquitin ligases, including S-phase kinase-associated protein 2 (SKP2) and CDC34. The results showed that the expression of CDC34 but not of SKP2 was significantly upregulated in the HNF1A-AS1 overexpression group (Figures 5B–C and supplementary Figure 2G). To the best of our knowledge, no report has shown whether CDC34, a E2/E3 ubiquitin ligase that ubiquitinates substrates such as p27 (Kip1) (22), Ikappaβalpha (23), and Wee1(24), ubiquitinates p21. The present study has determined that p21 protein expression decreased in the CDC34 overexpression group (Figures 6A–B). The results of the ActD chase assay showed that p21 mRNA expression was not altered in the CDC34 overexpression or knockdown group compared to the control group (Figures 6C–J). Treatment of cells overexpressing CDC34 with CHX led to a significantly shorter p21 half-life than that in the control group (Figures 6K–N). In contrast, CDC34 increased the levels of the endogenous p21 protein in MKN-45 and BGC-823 cells treated with MG132 (Figures 6O–P). Based on these findings, we concluded that p21 is a substrate for CDC34. Additionally, western blot analysis showed that p21 expression decreased in the HNF1A-AS1 overexpression group, but CDC34 knockdown reversed the reduced p21 expression induced by HNF1A-AS1 (Figure 6Q and Supplementary Figure 2H), suggesting that CDC34 is a mediator of p21 ubiquitination of p21 protein by HNF1A-AS1. These findings suggest that HNF1A-AS1 enhances CDC34 expression, thereby promoting the ubiquitination and degradation of p21.
Figure 5. HNF1A-AS1 enhanced the expression of cell cycle regulators and promoted p21 degradation by ubiquitination.
(A). The expression levels of CDK2, CDK4 and cylcinE1 in HNF1A-AS1 over-expressed MKN-45 cells and BGC-823 cells were detected by Western Blotting assay.
(B-C). Western Blotting assay was used to detect the expression levels of CDC34 and P21 in HNF1A-AS1 over-expressed MKN-45 cells (B) and BGC-823 cells (C).
(D-G). MKN-45 cells (D-E) and BGC-823 cells (F-G) transfected with HNF1A-AS1 overexpression vector and control cells treated with Actinomycin D (ActD 5ug/ml) for the indicated periods of time. P21 mRNA levels were analyzed by RT-qPCR.
(H-K). MKN-45 cells (H-I) and BGC-823 cells (J-K) transfected with HNF1A-AS1-targeting shRNA and control cells treated with Actinomycin D (ActD 5ug/ml) for the indicated periods of time. P21 mRNA levels were analyzed by RT-qPCR.
(L-M). MKN-45 cells (L) and BGC-823 cells (M) transfected with HNF1A-AS1 overexpression vector and control cells were treated with cycloheximide (CHX 5ug/ml) or vehicle for the indicated periods. P21 protein levels were analyzed by western blotting.
(N-O). MKN-45 cells (N) and BGC-823 cells (O) transfected with HNF1A-AS1 overexpression vector and control cells were treated with MG132 (5 μM) or vehicle for 24 h. Cell lysates were analyzed by western blotting.
(P-Q). MKN-45 cells (P) and BGC-823 cells (Q) transfected with HNF1A-AS1-targeting shRNA and control cells were treated with MG132 (5 μM) or vehicle for 24 h. Cell lysates were analyzed by western blotting.
(R). HNF1A-AS1 overexpression group or control group were treated with MG-132 (10 um) for 24h. Cell lysates were immunoprecipitated with antibody against P21. The ubiquitination of p21 was notably increased in MKN-45 cells overexpressing HNF1A-AS1 compared with the control group.
(S). LV-HNF1A-AS1 enhanced the expression of CDK2, CDK4, cyclinE1, and decreased the expression of p21 in mice tumor xenografts, compared to that of the LV-NC group.
ActD and CHX is respectively short for Actinomycin D and Cycloheximide. Three independent experiments were performed, and data are shown as mean ± SD.
Figure 6. CDC34 promoted p21 degradation.
(A-B). In CDC34 over-expressed MKN-45 cells and BGC-823 cells, the expression level of P21 was detected by western blotting.
(C-F). MKN-45 cells (C-D) and BGC-823 cells (E-F) transfected with CDC34 overexpression vector and control cells treated with Actinomycin D (5ug/ml) for the indicated periods of time. P21 mRNA levels were analyzed by RT-qPCR.
(G-J). MKN-45 cells (G-H) and BGC-823 cells (I-J) transfected with si-CDC34 and control cells treated with Actinomycin D (5ug/ml) for the indicated periods of time. P21 mRNA levels were analyzed by RT-qPCR.
(K-N). MKN-45 cells (K-L) and BGC-823 cells (M-N) transfected with CDC34 overexpression vector and control cells were treated with cycloheximide (CHX 5ug/ml) or vehicle for the indicated periods. P21 protein levels were analyzed by western blotting.
(O-P). MKN-45 cells (O) and BGC-823 cells (P) transfected with CDC34 overexpression vector and control cells were treated with MG132 (5 μM) or vehicle for 24 h. Cell lysates were analyzed by western blotting.
(Q). Western blotting analysis of the p21 in BGC-823 cells overexpressing HNF1A-AS1 or control cells with or without transient transfection withCDC34 siRNA.
ActD and CHX is respectively short for Actinomycin D and Cycloheximide. Three independent experiments were performed, and data are shown as mean ± SD.
HNF1A-AS1 functions as a ceRNA with miR-661 to upregulate CDC34 expression
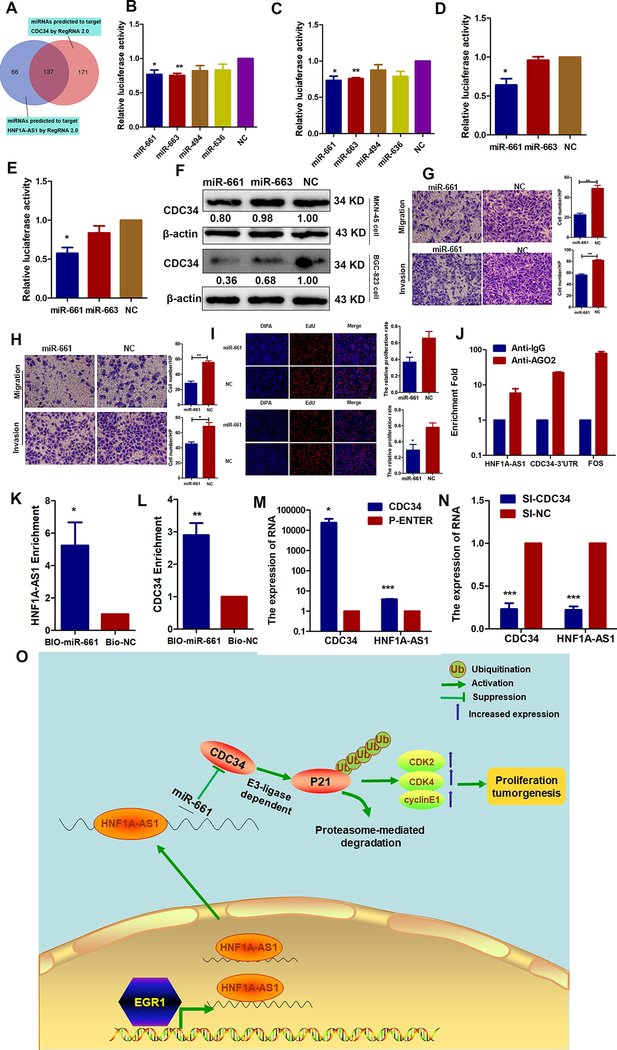
Recently, an increasing amount of evidence has demonstrated that lncRNAs can function as ceRNAs to sequester miRNAs, which decreases miRNA expression in the cytoplasm and modulates the de-repression of miRNA targets at the level of post-transcriptional regulation (25). To explore the regulatory mechanisms underlying HNF1A-AS1 enhancement of CDC34 expression and to determine whether HNF1A-AS1 works as a ceRNA, miRNAs potentially bound to both HNF1A-AS1 and CDC34 were predicted using the programs RegRNA and Targetscan in combination with miRNA functions reported in the literature (Figure 7A). MiR-661 and miR-663 exhibited lower luciferase activity in pmirGLO-HNF1A-AS1, indicating that these could directly bind to HNF1A-AS1 through their respective miRNA-binding sites (Figures 7B–C). Luciferase assays demonstrated that miR-661, not miR-663, suppressed the luciferase activity of pmirGLO-CDC34 (Figures 7D–E). Subsequently, we found that only miR-661 significantly reduces CDC34 protein levels in both GC cell lines (Figure 7F). Our study also suggested that miR-661 inhibits GC cell migration, invasion (Figures 7G–H), and proliferation (Figure 7I). MiRNAs directly downregulate their targets by repressing protein translation by forming RNA-induced silencing complexes (RISCs) that contain the argonaute-2 (Ago2) protein. To investigate whether HNF1A-AS1 and CDC34–3’ untranslated region (CDC34–3’UTR) interact with RISCs, RIP experiments were performed using antibodies against Ago2. Endogenous HNF1A-AS1 and CDC34–3’UTR were specifically enriched in Ago2 immunoprecipitates compared to the control, IgG (Figure 7J). Additionally, a biotin-labeled pulldown system was used to determine whether miR-661 could pull down HNF1A-AS1 and CDC34. Figures 7K–L show that the levels of HNF1A-AS1 and CDC34 mRNAs were markedly elevated in the pull-down product isolated from BGC-823 cells following transfection with biotin-labeled miR-661 compared to the control, suggesting that miR-661 interacts with HNF1A-AS1 and CDC34 in a sequence-specific manner. An important corollary of the ceRNA hypothesis is that ceRNA upregulation results in higher target RNA expression and vice versa (26). To assess this, RT-qPCR assays were performed to detect HNF1A-AS1 expression levels in CDC34 overexpression and knockdown GC cells. We found that HNF1A-AS1 expression was upregulated in the CDC34 overexpression group (Figure 7M and Supplementary Figure 2I) and inhibited in the CDC34 knockdown group (Figure 7N and Supplementary Figure 2J), indicating a reciprocal effect. Taken together, these results reveal that HNF1A-AS1 functions as a ceRNA for miR-661, and thereby derepresses CDC34 expression and thus provides an additional mechanism for post-transcriptional regulation.
Figure 7. HNF1A-AS1 functions as a ceRNA with miR-661 to upregulate CDC34 expression.
(A). A Venn diagram depicting 137 miRNAs that predicted to target both HNF1A-AS1 and CDC34 by RegRNA 2.0 algorithms.
(B-C). Luciferase activity was detected using the dual-luciferase assay. The data indicated a decrease in luciferase activity in MKN-45(B) and BGC-823 (C) cells transfected with PGLO-HNF1A-AS1 and miR-661 or miR-663.
(D-E). The results indicated a decrease in luciferase activity in MKN-45 (D) and BGC-823 (E) cells transfected with PGLO-CDC34–3’UTR and miR-661.
(F). Western blot analyses were performed to confirm the CDC34 gene expression in both cells transfected with miR-661 or miR-663 mimics.
(G-H). Effects of the miR-661 overexpression on migration and invasion abilities in MKN-45 cells (G) and BGC-823 cells (H).
(I). EdU assays were performed to detect the effects of miR-661 on cell proliferation in MKN-45 cells (up) and BGC-823 cells (bottom).
(J). RNA immunoprecipitation with an anti-Ago2 antibody was used to assess endogenous Ago2 binding to HNF1A-AS1 and CDC-34 3’UTR; IgG was used as the control.
(K-L). BGC-823 cells were transfected with biotinylated miR-661(BIO-miR-661) or biotinylated NC (BIO-NC). Forty-eight hours after transfection, cells were collected for a biotin-based pulldown assay. HNF1A-AS1 and CDC34 expression levels were analyzed by RT–qPCR.
(M-N). RT-qPCR assay was performed to detect HNF1A-AS1 expression in CDC34 up- or down-regulating BGC-823 cells.
(O). Proposed functional action of HNF1A-AS1 in modulating gastric cancer tumorigenesis. The transcriptional factor EGR1 activated HNF1A-AS1 expression by directly binding to its promoter. And HNF1A-AS1 functioned as ceRNA by competitively binding to miR-661 to upregulate the expression of CDC34, which is a direct target of miR-661. Then, CDC34-mediated ubiquitination decreased the stability of p21 protein via degradation of P21 protein, and subsequently increased the expressions of CDK2, CDK4, cyclinE1 to enhance the proliferation capabilities of GC cells.
Discussion
An increasing number of studies have revealed a key regulatory role of lncRNAs in the carcinogenesis and progression of human cancers (27). However, the potential biological functions and underlying mechanistic details for most lncRNAs in human GC remain elusive. In the present study, we found that HNF1A-AS1 was markedly upregulated in GC tissues compared to non-tumorous tissues. Using loss-of-function and gain-of-function assays, we demonstrated that HNF1A-AS1 promotes GC cell proliferation and invasion, suggesting that HNF1A-AS1 may function as an oncogenic lncRNA in GC. Consistent with our observations, Zhao et al. found that HNF1A-AS1 enhances cell proliferation and metastasis in osteosarcoma through activation of the Wnt/β-catenin signaling pathway (17).
Several studies have demonstrated that many lncRNAs contribute to the development and progression of various cancers. However, the regulators involved in the abnormal expression of lncRNAs in cancer cells remain unclear. Extensive evidence has demonstrated that lncRNA expression can be regulated to a certain extent, similar to protein-coding genes. For example, p53 enhances lincRNA-p21 transcription and promotes cell growth arrest and apoptosis (11). EGR1, a Cys2-His2-type zinc-finger transcription factor, belongs to the EGR family and has been implicated in the regulation of cell proliferation (28), migration (29) and apoptosis (30). An increasing amount of evidence has shown that EGR1 is involved in human cancer development and progression, particularly as a tumor promoter. In the present study, we demonstrated that HNF1A-AS1 overexpression in GC cells could be activated by the transcription factor EGR1 by binding to its promoter region. Previous studies have shown that EGR1 regulates mRNA transcription of protein-coding genes, whereas we first demonstrated that it could activate lncRNA transcription, indicating that the regulation of lncRNA and protein-coding mRNA expression share the same transcription factor. In addition, the results of the present study show that the effects of EGR1 on GC cell proliferation may be due, at least in part, to the induction of cell cycle transition from the G0/G1 to S phases, which coincides with the functions of HNF1A-AS1.
Next, flow cytometry demonstrated that HNF1A-AS1-induced GC cell proliferation may be associated with cell cycle transition from the G0/G1 to S phases. To elucidate the mechanism underlying initiation and progression of cancer cell proliferation, it is essential to determine the effects of cell cycle-related proteins such as cyclin, CDKs and CKIs on cell cycle control. The CDK2-cyclin E and CDK4/CDK6-cyclin D complexes are required for cell cycle transition from the G0/G1 to S phases (31). In the present study, HNF1A-AS1 not only promoted cyclin E1 expression, but also enhanced CDK2/CDK4 expression in vitro and in xenograft tumors. The activities of CDKs and cyclins in the G1/S phase could be inhibited by CKIs, particularly p21, which can bind to cyclin and CDK subunits through its characteristic motifs within amino-terminal moieties (32). Our study showed that HNF1A-AS1 also decreases p21 expression.
Moreover, we demonstrated that HNF1A-AS1 could promote CDC34 expression and thus enhance CDC34-mediated ubiquitination and degradation of p21. The CDC34 protein was first identified in yeast as a regulator of the G1 to S phase transition (33). CDC34 belongs to the family of ubiquitin-conjugating enzymes that can catalyze the addition of ubiquitin to other protein substrates. It usually plays an important role in the downregulation of p27, Ikappaβalpha, and Wee1 due to its E2/E3 ubiquitin ligase activity. In this study, we demonstrate that p21 is a novel substrate for human CDC34 in GC cells that overexpress HNF1A-AS1. Based on these results, we postulate that HNF1A-AS1 increases CDC34 expression, thereby enhancing CDC34-mediated ubiquitination and degradation of p21.
Recent studies have suggested that lncRNAs potentially impact the expression levels of miRNA targets by competing for miRNAs. For instance, lncRNA BC032469 sponges miR-1207–5p by mimicking its binding site, leading to the accumulation of the miR-1207–5p target hTERT in GC (34). In addition, lncRNA Unigene56159, which is highly upregulated in hepatocellular carcinoma, acts as a ‘sponge’ to downregulate miR-140–5p, leading to the reduction of translational repression of its target gene Slug (35). Thus, we hypothesized that HNF1A-AS1 acts as a ceRNA that upregulates the expression of CDC34. Potential interactions between HNF1A-AS1 and miRNAs were explored using bioinformatics analyses, luciferase assays, RIP assays and biotin-labeled pulldown assays to confirm the direct binding capabilities of the predicted miRNA responsive elements to the full-length HNF1A-AS1 transcript and CDC34–3’UTR. The results of the present study demonstrate that HNF1A-AS1 could serve as a ceRNA that sequesters miR-661, thereby protecting the target gene CDC34 from repression and in turn upregulating HNF1A-AS1. The findings of this study indicate that the crosstalk between RNAs, e.g., lncRNA and mRNA, forms a large-scale regulatory network across the transcriptome. In the context of cancer, lncRNAs have been systematically studied as potential tumor oncogenes and suppressors via their ceRNA function. Thus, this ceRNA networks could provide opportunities for novel therapeutic regimens. Previous studies have shown that miR-661 plays a tumor suppressive role in various cancers. For example, miR-661 inhibits the tumorigenicity of invasive breast cancer cells by suppressing a metastasis-related protein, MTA1 (36). Additionally, miR-661 inhibits the proliferation, migration, and invasion abilities of glioma cells by targeting hTERT (37). However, the biological roles of miR-661 in GC tumorigenesis remain unclear. Our results show that miR-661 suppresses cell proliferation and invasion, suggesting that miR-661 plays a tumor suppressive role in GC. Thus, HNF1A-AS1 enhances cell proliferation and invasion, which may be partly due to absorbed miR-661 that suppresses the effect of miR-661.
In conclusion, we have identified the core promoter region of HNF1A-AS1 and found that EGR1 plays a role in upregulating HNF1A-AS1 expression by directly binding to the HNF1A-AS1 promoter. In addition, we have demonstrated that HNF1A-AS1 acts as a ceRNA that upregulates the expression of CDC34, which in turn upregulates the level of HNF1A-AS1. Furthermore, we found that EGR1-activated HNF1A-AS1 increases GC cell proliferation, in which CDK2, CDK4, and cyclin E1 are involved, and decreases the stability of p21 protein by promoting CDC34-mediated ubiquitination and degradation (Figure 7O). These findings define the importance of HNF1A-AS1 in maintaining cellular proliferation in GC, and HNF1A-AS1 may be a promising molecular target for the treatment of GC.
Supplementary Material
Significance:
This study provides novel insights into mechanisms by which the non-coding RNA HNF1A-AS1 contributes to gastric cancer progression through modulation of the cell cycle.
Acknowledgments:
This study was supported by the National Natural Science Foundation of China (Grant No.81672842), The Taishan Scholars Program of Shandong Province (Grant No. ts201511096) and the key research and development program of Shandong Province (GG201703220053). We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Footnotes
Competing financial interests
All authors promise that there is no conflict to disclose.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: a cancer journal for clinicians. 2015;65:87–108. [DOI] [PubMed] [Google Scholar]
- 2.McLean MH, El-Omar EM. Genetics of gastric cancer. Nature reviews Gastroenterology & hepatology. 2014;11:664–74. [DOI] [PubMed] [Google Scholar]
- 3.Schipper DL, Wagenmans MJ, Peters WH, Wagener DJ. Significance of cell proliferation measurement in gastric cancer. European journal of cancer. 1998;34:781–90. [DOI] [PubMed] [Google Scholar]
- 4.Lansdorp-Vogelaar I, Kuipers EJ. Screening for gastric cancer in Western countries. Gut. 2016;65:543–4. [DOI] [PubMed] [Google Scholar]
- 5.Duraes C, Almeida GM, Seruca R, Oliveira C, Carneiro F. Biomarkers for gastric cancer: prognostic, predictive or targets of therapy? Virchows Archiv : an international journal of pathology. 2014;464:367–78. [DOI] [PubMed] [Google Scholar]
- 6.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome research. 2012;22:1775–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu P, Wang Y, Huang G, Ye B, Liu B, Wu J, et al. lnc-beta-Catm elicits EZH2-dependent beta-catenin stabilization and sustains liver CSC self-renewal. Nature structural & molecular biology. 2016;23:631–9. [DOI] [PubMed] [Google Scholar]
- 8.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nature genetics. 2011;43:621–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin Z, Guan D, Fan Q, Su J, Zheng W, Ma W, et al. lncRNA expression signatures in response to enterovirus 71 infection. Biochemical and biophysical research communications. 2013;430:629–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341:789–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma YF, Liang T, Li CR, Li YJ, Jin S, Liu Y. Long non-coding RNA HNF1A-AS1 up-regulation in non-small cell lung cancer correlates to poor survival. European review for medical and pharmacological sciences. 2016;20:4858–63. [PubMed] [Google Scholar]
- 15.Wang C, Mou L, Chai HX, Wang F, Yin YZ, Zhang XY. Long non-coding RNA HNF1A-AS1 promotes hepatocellular carcinoma cell proliferation by repressing NKD1 and P21 expression. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2017;89:926–32. [DOI] [PubMed] [Google Scholar]
- 16.Yang X, Song JH, Cheng Y, Wu W, Bhagat T, Yu Y, et al. Long non-coding RNA HNF1A-AS1 regulates proliferation and migration in oesophageal adenocarcinoma cells. Gut. 2014;63:881–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X, Xiong Y, Tang F, Bian Y, Chen Y, Zhang F. Long noncoding RNA HNF1A-AS1 indicates a poor prognosis of colorectal cancer and promotes carcinogenesis via activation of the Wnt/beta-catenin signaling pathway. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2017;96:877–83. [DOI] [PubMed] [Google Scholar]
- 18.Wu Y, Liu H, Shi X, Yao Y, Yang W, Song Y. The long non-coding RNA HNF1A-AS1 regulates proliferation and metastasis in lung adenocarcinoma. Oncotarget. 2015;6:9160–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang C, Qiu S, Sun F, Li W, Wang Z, Yue B, et al. Long non-coding RNA HNF1A-AS1 mediated repression of miR-34a/SIRT1/p53 feedback loop promotes the metastatic progression of colon cancer by functioning as a competing endogenous RNA. Cancer letters. 2017;410:50–62. [DOI] [PubMed] [Google Scholar]
- 20.Zhang H, Ma RR, Wang XJ, Su ZX, Chen X, Shi DB, et al. KIF26B, a novel oncogene, promotes proliferation and metastasis by activating the VEGF pathway in gastric cancer. Oncogene. 2017;36:5609–19. [DOI] [PubMed] [Google Scholar]
- 21.Liu HT, Xing AY, Chen X, Ma RR, Wang YW, Shi DB, et al. MicroRNA-27b, microRNA-101 and microRNA-128 inhibit angiogenesis by down-regulating vascular endothelial growth factor C expression in gastric cancers. Oncotarget. 2015;6:37458–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Butz N, Ruetz S, Natt F, Hall J, Weiler J, Mestan J, et al. The human ubiquitin-conjugating enzyme Cdc34 controls cellular proliferation through regulation of p27Kip1 protein levels. Experimental cell research. 2005;303:482–93. [DOI] [PubMed] [Google Scholar]
- 23.Vuillard L, Nicholson J, Hay RT. A complex containing betaTrCP recruits Cdc34 to catalyse ubiquitination of IkappaBalpha. FEBS letters. 1999;455:311–4. [DOI] [PubMed] [Google Scholar]
- 24.Michael WM, Newport J. Coupling of mitosis to the completion of S phase through Cdc34-mediated degradation of Wee1. Science. 1998;282:1886–9. [DOI] [PubMed] [Google Scholar]
- 25.Yang H, Liu P, Zhang J, Peng X, Lu Z, Yu S, et al. Long noncoding RNA MIR31HG exhibits oncogenic property in pancreatic ductal adenocarcinoma and is negatively regulated by miR-193b. Oncogene. 2016;35:3647–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA biology. 2012;9:703–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang D, Guan MP, Zheng ZJ, Li WQ, Lyv FP, Pang RY, et al. Transcription Factor Egr1 is Involved in High Glucose-Induced Proliferation and Fibrosis in Rat Glomerular Mesangial Cells. Cellular physiology and biochemistry : international journal of experimental cellular physiology, biochemistry, and pharmacology. 2015;36:2093–107. [DOI] [PubMed] [Google Scholar]
- 29.Tarcic G, Avraham R, Pines G, Amit I, Shay T, Lu Y, et al. EGR1 and the ERK-ERF axis drive mammary cell migration in response to EGF. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2012;26:1582–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oben KZ, Gachuki BW, Alhakeem SS, McKenna MK, Liang Y, St Clair DK, et al. Radiation Induced Apoptosis of Murine Bone Marrow Cells Is Independent of Early Growth Response 1 (EGR1). PloS one. 2017;12:e0169767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu M, Xu X, Zhao J, Tang Y. Naringenin inhibits transforming growth factor-beta1-induced cardiac fibroblast proliferation and collagen synthesis via G0/G1 arrest. Experimental and therapeutic medicine. 2017;14:4425–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes & development. 1999;13:1501–12. [DOI] [PubMed] [Google Scholar]
- 33.Goebl MG, Yochem J, Jentsch S, McGrath JP, Varshavsky A, Byers B. The yeast cell cycle gene CDC34 encodes a ubiquitin-conjugating enzyme. Science. 1988;241:1331–5. [DOI] [PubMed] [Google Scholar]
- 34.Lu MH, Tang B, Zeng S, Hu CJ, Xie R, Wu YY, et al. Long noncoding RNA BC032469, a novel competing endogenous RNA, upregulates hTERT expression by sponging miR-1207–5p and promotes proliferation in gastric cancer. Oncogene. 2016;35:3524–34. [DOI] [PubMed] [Google Scholar]
- 35.Lv J, Fan HX, Zhao XP, Lv P, Fan JY, Zhang Y, et al. Long non-coding RNA Unigene56159 promotes epithelial-mesenchymal transition by acting as a ceRNA of miR-140–5p in hepatocellular carcinoma cells. Cancer letters. 2016;382:166–75. [DOI] [PubMed] [Google Scholar]
- 36.Reddy SD, Pakala SB, Ohshiro K, Rayala SK, Kumar R. MicroRNA-661, a c/EBPalpha target, inhibits metastatic tumor antigen 1 and regulates its functions. Cancer research. 2009;69:5639–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Z, Liu YH, Diao HY, Ma J, Yao YL. MiR-661 inhibits glioma cell proliferation, migration and invasion by targeting hTERT. Biochemical and biophysical research communications. 2015;468:870–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.