Abstract
To address the challenges associated with glycan analyses, we have implemented a structures for lossless ion manipulations (SLIM) serpentine ultra-long path with extended routing (SUPER) ion mobility-mass spectrometry (i.e. SLIM SUPER IM-MS) platform to achieve much higher resolution of isomeric glycoforms. We have demonstrated the potential of this platform as a future component of the glycomics toolbox.
TOC Graphic:
Glycans are one of the most ubiquitous biomolecules, where their specific structures dictate myriad biological and pathological functions in plants, fungi, mammals, and bacteria1. As compared to other biomolecules (e.g., nucleic acids, peptides), glycan synthesis is performed in a non-template driven fashion, where their composition can be varied according to immunological responses, cellular signaling, and disease state1. This variation in glycan composition, termed isomeric heterogeneity, leads to numerous potential structural permutations (e.g., regioisomers, stereoisomers, and diastereomers), further complicating analyses2. This isomeric heterogeneity is further magnified by added structural permutations with key examples including glycosidic linkage position (e.g., 1–4 versus 1–6 linkage), α/β anomericity, and monosaccharide subunit composition (e.g., galactose versus glucose), as well as variation conformers (e.g., five-membered furanose versus six-membered pyranose rings), as shown in Figure 1. In order to better understand how glycan sequence and composition affect a wide array of biological factors, there is a pressing need for more effective, faster, and higher-resolution analytical methods to address this isomerism puzzle, as underscored in the 2012 National Academy of Sciences Report3.
Figure 1.
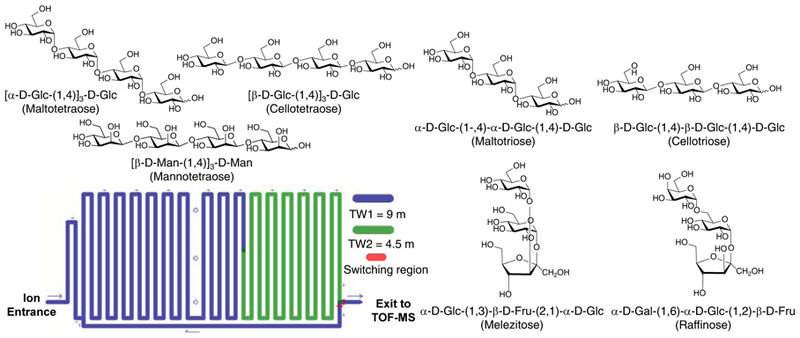
Illustration of some of the isomeric heterogeneity of glycans in various tri/tetra-saccharides (top), and a simplified schematic of the SLIM device ion path used in these experiments (bottom).
Currently, the most commonly used analytical technique for glycan separations is liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS)1,4. Chemical derivatization is often a prerequisite for reversed-phase separations so as to increase the hydrophobicity of the inherently polar glycans1,2. While reversed-phase separations remain the preferred choice in glycan separations, HILIC or normal stationary phases have seen some recent use2. Even with the growing suite of MSn methods, the deconvolution and identification of co-eluting species remains a challenge since isomers exhibit very similar fragmentation pathways4,5. Additionally, while spectroscopic approaches have shown promise for glycan fingerprinting, these methods remain of limited utility for accurately characterizing isomeric mixtures6.
Ion mobility-mass spectrometry (IM-MS)-based approaches are quickly becoming an attractive, and rapid, alternative to 1-D chromatographic separations2, and even have shown utility as a front-end separation prior to spectroscopic methods6. In IM-MS ions are separated in the gas-phase based on their mobilities (size/shape) as well as mass-to-charge (m/z). While IM-MS results have been demonstrated for rapid glycan separations, conventional approaches still suffer from limited sensitivity and resolution for analyses of isomeric glycoforms2,7, limiting their potential for broader utility. Metal adduction8, and use of other ligands9, have seen some effectiveness in increasing IM glycan resolution, but these approaches are still unable to resolve all isomeric glycoforms of interest. This resolution bottleneck, highlights the tremendous need for further development of higher resolution IM-based approaches for teasing apart the subtle differences amongst isomeric glycoconjugates.
Recently, IM-based structures for lossless ion manipulations (SLIM) has demonstrated promise for providing greatly higher resolution and increased sensitivity compared to conventional IM platforms10. Specifically, SLIM serpentine ultra-long path with extended routing (SUPER) separations are enabled by a switching region that sends ions on additional passes through the ion path, resulting in much higher resolution measurements (Figure 1)10. Increased sensitivity is simultaneously accomplished by the in-SLIM trapping of large populations of ions in the extended volume provided by the first of two or more SLIM regions (Figure 1), permitting over a billion ions to be introduced and trapped (a 2–3 orders of magnitude increase compared to conventional approaches)10. More information on the capabilities of SLIM are provided in the Supporting Information and these references10. In this work the most intense precursor for each glycan ion was selected for further SLIM SUPER IM separations in helium. See the Supporting Information for detailed experimental conditions and instrument parameters.
With our goal of developing IM-based methods for the separation of isomeric glycoforms, we began by evaluating separations for various classes of glycans that differ in their monosaccharide composition, glycosidic linkage positioning, anomericity, number of monosaccharide subunits present, and linear/branching points. Initially, we assessed the separation of four common isomeric disaccharides (Figure 2). After a 112.5 m SLIM SUPER IM separation the four disaccharide isomers were well resolved. Interestingly, non-reducing (sucrose) was higher in mobility, i.e. smaller in collision cross section or more compact in nature than the reducing species, lactose, maltose, and cellobiose. We hypothesize that the observation of only one mobility peak from sucrose may be attributed to its inability to mutarotate, and present two distinct α/β anomers at the C-1 OH group. The maltose and cellobiose reducing species both displayed two mobility features, presumably due to its α/β mutarotation, while lactose only displayed one such feature, perhaps due to the influence of the galactose moiety on mutarotation, and leading to a preference for only a single α or β anomer. (We note that the multiple mobility peaks observed for these disaccharides may alternatively be rationalized by different sodium cation attachment locations.) Additionally, we observed that cellobiose (β−1–4 linkage) was lower in mobility (more elongated) than maltose (α−1–4 linkage), while the axial OH group at C-4 of galactose moiety in lactose caused it to be higher in mobility than both maltose and cellobiose, even having the same 1–4 linkage.
Figure 2.
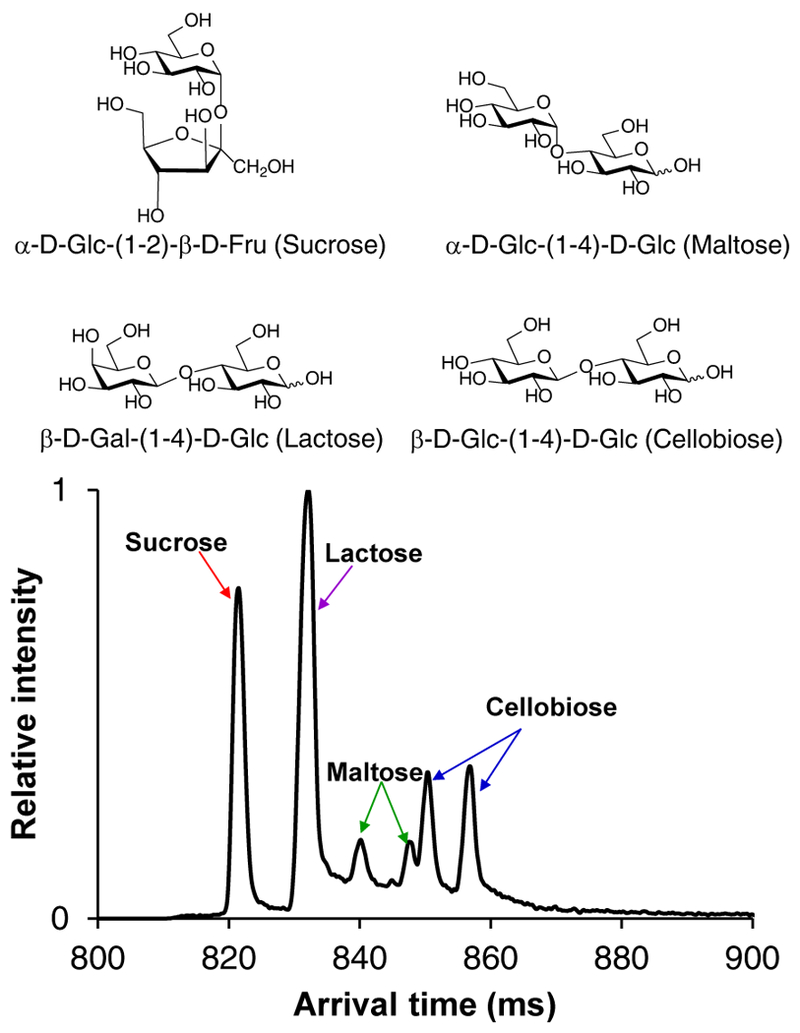
112.5 m SLIM SUPER IM separation of isomeric disaccharides, 365.1 m/z [M+Na]+.
Based on the IM trends observed for these disaccharides, we set out to see if similar patterns would hold for isomeric trisaccharides (Figure 3). Similar to the disaccharides, it was observed that the non-reducing glycans (melezitose and raffinose) were higher in mobility than their reducing counterparts, and also exhibited only one mobility peak. As speculated above, we attribute this observation to the ‘locked’ configuration at the anomeric carbon, while the reducing trisaccharides each exhibit two mobility peaks, potentially from α/β anomeric configurations. Again, it is also possible that the multiple mobility peaks are caused by multiple cation attachment sites, leading to multiple, distinct, ion conformations, that can only be teased apart with the high-resolution SLIM SUPER capabilities. In comparing the separation between α versus β linked trisaccharides (maltotriose versus cellotriose), it was observed that the β linkage was more elongated (lower mobility) than the α-linked, as also found for the isomeric disaccharides (maltose versus cellobiose). Surprisingly, this α/β anomericity trend did not apply for isomeric tetrasaccharides (Figure 3), where it was observed that cellotetraose (β-linkages) was more compact than maltotetraose (α-linkages). This suggests that a conformational change may occur when the number of monosaccharide constituents exceeds three, and where the β-linked glycans become more compact than their α-linked ones. Previous work10 showed cellopentaose was more compact than maltopentaose, indicating this potential anomericity trend also holds for pentasaccharides. Mannotetraose was seen to have the most elongated structure from the SLIM SUPER IM separations, potentially due to its axially oriented C-2 OH group on each monosaccharide, as compared to the equatorially oriented ones for the glucose tetrasaccharides. As also evident in Figure 3, these informative high-resolution separations of both isomeric tri- and tetrasaccharides could be performed on a rapid timescale (< 1 second).
Figure 3.
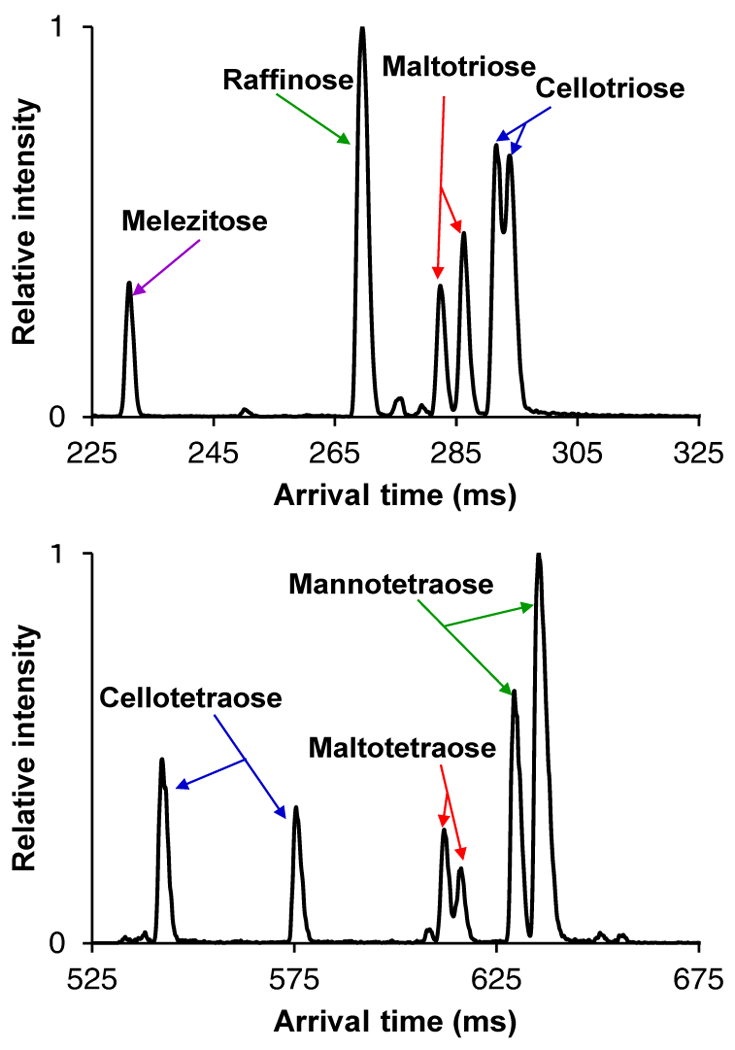
45 m SLIM SUPER IM separation of isomeric trisaccharides, 527.2 m/z (top) and 72 m separation of isomeric tetrasaccharides 689.2 m/z (bottom), both as [M+Na]+. For IM peak assignments, see Supporting Information.
With such encouraging separations for various isomeric di/tri/tetra-saccharides, it was unclear whether the approach would be effective for even more challenging, isomeric, human milk oligosaccharides (HMOs). HMOs have received widespread attention for their roles in potentially protecting the gut microbiome, promoting growth and immunological development in infants, and their anti-adhesive and probiotic properties6,11. Lacto-N-tetraose and lacto-N-neotetraose (LNT and LNnT, respectively), which only differ in their linkage position (1–3 versus 1–4) between the D-galactose and N-acetyl D-glucosamine residues, were able to be baseline resolved after 31.5 meters of separation as their protonated adducts (Figure 4). While we hypothesized that two mobility features could exist for each HMO isomer (due to their α/β anomers at the C-1 OH group undergoing mutarotation), we observed four distinct mobility features for the LNT species. This suggests that monosaccharide sites in addition to the N-acetyl D-glucosamine residue may be protonated, or potentially that the LNT ion species exists in multiple, unique, conformations. The striking difference in peak widths may potentially arise from LNnT conformers interconverting on a faster timescale than LNT, which may interconvert more slowly, allowing their resolution. In the separation of the lacto-N-fucopentaose isomers (Figure 4), it was seen that the linear isomer (LNFP i) was the most elongated and lowest mobility species, while the branching isomers (LNFP ii and iii) had more compact structures. This result supported our hypothesis that the structural difference (linear versus branched) of these isomers would have a major effect on their mobilities. LNFP i and ii shared a common mobility peak, perhaps due to similar cation attachment site, but their other observed conformers could be baseline resolved.
Figure 4.
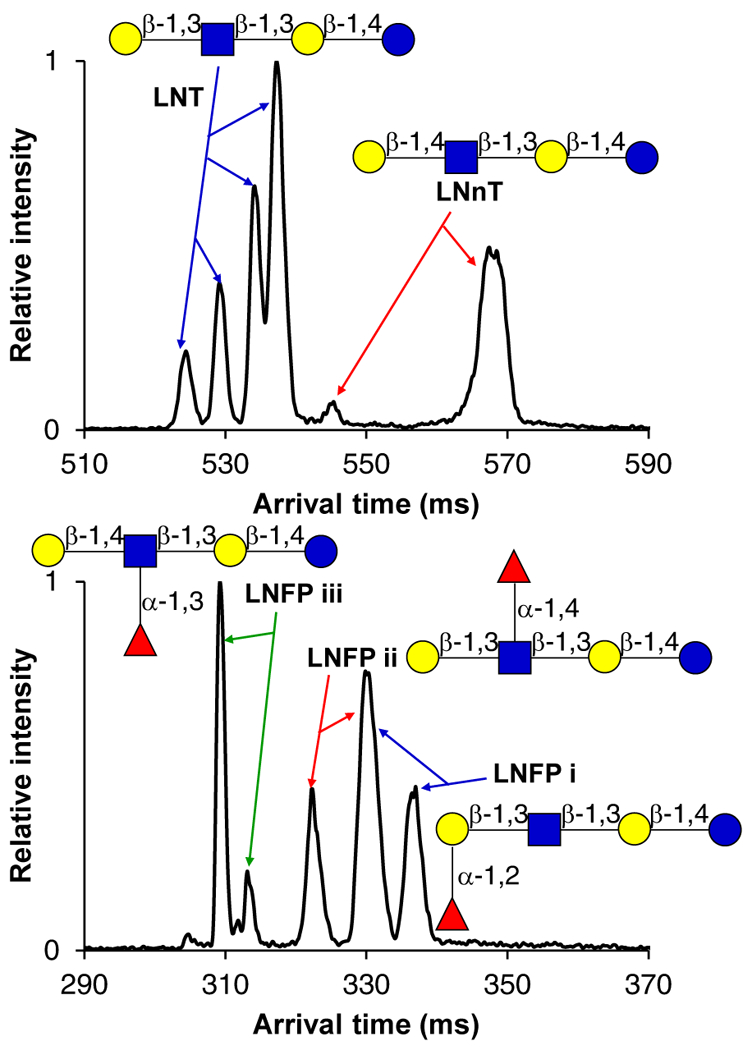
31.5 m SLIM SUPER IM separation of human milk oligosaccharide isomers, LNT and LNnT (top) as 708.3 m/z [M+H]+. 45 m separation of lacto-N-fucopentaose isomers (bottom) as 446.6 m/z [M+H+K]2+. Structures are drawn according to conventional glycan symbol nomenclature13. For IM peak assignments, see the Supporting Information.
These new results demonstrated that the combination of in-SLIM accumulation of large ion populations in conjunction with SLIM SUPER IM separations, provided previously unachieved resolution, speed, and sensitivity in the separation of isomeric HMOs and other glycan species. As compared to the separation of isomeric glycans in a 89 cm drift tube ion mobility platform8, we were not only able to much better resolve isomeric mixtures, but also tease apart other conformers and/or substructures for the individual glycan species. Future work can now e.g. probe whether the observed HMO conformers are correlated to their microbial bioactivities12. We also observed that isomers were easier to resolve as they increased in size (i.e. the number of monosaccharide constituents). The positive ion mode cation attachments enable IM separations and MS analyses that potentially provide diagnostic glycan mobility fingerprints; future work will assess whether the negative ion mode provides complementary information. Additionally, we will explore methods for annealling these glycan species into a single mobility peak, potentially through the use of fixed charge modification reagents, such as quaternary ammonium ones, in lieu of metal adduction, potentially further increasing their IM resolution. This new approach provides large gains in resolution over existing IM-MS-based glycomics platforms, suggesting it will have broad utility for improved characterization of isomeric, biologically-relevant, glycans.
We would like to acknowledge the National Institute of General Medical Sciences (P41 GM103493) and National Institutes of Health (R21 1R21CA199744-01). Experiments were performed in the W. R. Wiley Environmental Molecular Sciences Laboratory (EMSL), a DOE national scientific user facility at the Pacific Northwest National Laboratory (PNNL). PNNL is operated by Battelle under contract DE-AC05–76RL0 1830 for the DOE.
Supplementary Material
Footnotes
Electronic Supplementary Information (ESI) available: [experimental conditions and individual IM peak assignments].
Conflicts of interest
There are no conflicts to declare.
Notes and references
- 1.Gaunitz S, Nagy G, Pohl NLB and Novotny MV, Anal. Chem 2017, 89, 389; [DOI] [PMC free article] [PubMed] [Google Scholar]; Marth JD, Nat. Cell. Biol, 2008, 10, 1015; [DOI] [PMC free article] [PubMed] [Google Scholar]; Moremen KW, Tiemeyer M and Nairn AV, Nat. Rev. Mol. Cell. Biol, 2012, 13, 448; [DOI] [PMC free article] [PubMed] [Google Scholar]; Panza M, Pistorio SG, Stine KJ and Demchenko AV, Chem. Rev, 2018, DOI: 10.1021/acs.chemrev.8b00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hofmann J and Pagel K, Angew. Chemie. Int. Ed, 2017, 56, 8342–8349; [DOI] [PubMed] [Google Scholar]; Hofmann J, Hahm HS, Seeberger PH and Pagel K, Nature, 2015, 526, 241; [DOI] [PubMed] [Google Scholar]; Manz C and Pagel K, Curr. Opin. Chem. Biol, 2018, 42, 16; [DOI] [PubMed] [Google Scholar]; Chen Z, Glover MS and Li L, Curr. Opin. Chem. Biol, 2018, 42, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.in Transforming Glycoscience: A Roadmap for the Future, Washington (DC), 2012, DOI: 10.17226/13446. [DOI] [PubMed] [Google Scholar]
- 4.Lareau NM, May JC and McLean JA, Analyst, 2015, 140, 3335; [DOI] [PMC free article] [PubMed] [Google Scholar]; Tang Y, Wei J, Costello CE and Lin C, J. Am. Soc. Mass Spectrom, 2018, 29, 1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kailemia MJ, Ruhaak LR, Lebrilla CB and Amster IJ, Anal. Chem, 2014, 86, 196–212; [DOI] [PMC free article] [PubMed] [Google Scholar]; Hoffmann W, Hofmann J and Pagel K, J. Am. Soc. Mass Spectrom, 2014, 25, 471; [DOI] [PubMed] [Google Scholar]; Huang Y and Dodds ED, Anal. Chem, 2015, 87, 5664; [DOI] [PubMed] [Google Scholar]; Li H, Bendiak B, Siems William F, Gang David R and Hill Herbert H, Rapid Commun. Mass Spectrom, 2013, 27, 2699; [DOI] [PMC free article] [PubMed] [Google Scholar]; Liu X, Cool LR, Lin K, Kasko AM and Wesdemiotis C, Analyst, 2015, 140, 1182; [DOI] [PMC free article] [PubMed] [Google Scholar]; Schindler B, Barnes L. c., Renois G, Gray C, Chambert S. p., Fort S. b., Flitsch S, Loison C, Allouche A-R and Compagnon I, Nat. Comm, 2017, 8, 973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khanal N, Masellis C, Kamrath MZ, Clemmer DE and Rizzo TR, Analyst, 2018, 143, 1846; [DOI] [PMC free article] [PubMed] [Google Scholar]; Masellis C, Khanal N, Kamrath MZ, Clemmer DE and Rizzo TR, J. Am. Soc. Mass Spectrom, 2017, 28, 2217; [DOI] [PMC free article] [PubMed] [Google Scholar]; Mucha E, Gonzalez-Florez Ana I, Marianski M, Thomas Daniel A, Hoffmann W, Struwe Weston B, Hahm Heung S, Gewinner S, Schollkopf W, Seeberger Peter H, von Helden G and Pagel K, Angew. Chemie. Int. Ed, 2017, 56, 11248. [DOI] [PubMed] [Google Scholar]
- 7.Clowers BH, Dwivedi P, Steiner WE, Hill HH and Bendiak B, J. Am. Soc. Mass Spectrom, 2005, 16, 660; [DOI] [PubMed] [Google Scholar]; Fasciotti M, Sanvido Gustavo B, Santos Vanessa G, Lalli Priscila M, McCullagh M, Daroda Romeu S. í. G. F, J., Peter Martin G and Eberlin Marcos N, J. Mass Spectrom, 2012, 47, 1643-; [DOI] [PubMed] [Google Scholar]; Fenn LS and McLean JA, Phys. Chem. Chem. Phys, 2011, 13, 2196–2205; [DOI] [PubMed] [Google Scholar]; Hinneburg H, Hofmann J, Struwe WB, Thader A, Altmann F, Varon Silva D, Seeberger PH, Pagel K and Kolarich D, Chem. Commun, 2016, 52, 4381; [DOI] [PubMed] [Google Scholar]; Li H, Bendiak B, Siems WF, Gang DR and Hill HH, Anal. Chem, 2015, 87, 2228; [DOI] [PubMed] [Google Scholar]; Pagel K and Harvey DJ, Anal. Chem, 2013, 85, 5138. [DOI] [PubMed] [Google Scholar]
- 8.Huang Y and Dodds ED, Anal. Chem, 2013, 85, 9728; [DOI] [PubMed] [Google Scholar]; Huang Y and Dodds ED, Analyst, 2015, 140, 6912; [DOI] [PubMed] [Google Scholar]; Zheng X, Zhang X, Schocker NS, Renslow RS, Orton DJ, Khamsi J, Ashmus RA, Almeida IC, Tang K, Costello CE, Smith RD, Michael K and Baker ES, Anal. Bioanal. Chem, 2017, 409, 467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaye MM, Nagy G, Clemmer DE and Pohl NLB, Anal. Chem, 2016, 88, 2335. [DOI] [PubMed] [Google Scholar]
- 10.Deng L, Garimella SVB, Hamid AM, Webb IK, Attah IK, Norheim RV, Prost SA, Zheng X, Sandoval JA, Baker ES, Ibrahim YM and Smith RD, Anal. Chem, 2017, 89, 6432; [DOI] [PMC free article] [PubMed] [Google Scholar]; Deng L, Ibrahim YM, Baker ES, Aly NA, Hamid AM, Xhang X, Zheng X, Garimella SVB, Webb IK, Prost SA, Sandoval JA, Norheim RV, Anderson GA, Tolmachev AV and Smith RD, ChemistrySelect, 2016, 1, 2396; [DOI] [PMC free article] [PubMed] [Google Scholar]; Deng L, Webb IK, Garimella SVB, Hamid AM, Zheng X, Norheim RV, Prost SA, Anderson GA, Sandoval JA, Baker ES, Ibrahim YM and Smith RD, Anal. Chem, 2017, 89, 4628; [DOI] [PMC free article] [PubMed] [Google Scholar]; Garimella SV, Hamid AM, Deng L, Ibrahim YM, Webb IK, Baker ES, Prost SA, Norheim RV, Anderson GA and Smith RD, Anal. Chem, 2016, 88, 11877; [DOI] [PMC free article] [PubMed] [Google Scholar]; Deng L, Ibrahim YM, Hamid AM, Garimella SV, Webb IK, Zheng X, Prost SA, Sandoval JA, Norheim RV, Anderson GA, Tolmachev AV, Baker ES and Smith RD, Anal. Chem, 2016, 88, 8957; [DOI] [PMC free article] [PubMed] [Google Scholar]; Chouinard CD, Nagy G, Webb IK, Garimella SVB, Baker ES, Ibrahim YM and Smith RD, Anal. Chem, 2018, DOI: 10.1021/acs.analchem.8b02990; [DOI] [PMC free article] [PubMed] [Google Scholar]; Chouinard CD, Nagy G, Webb IK, Shi T, Baker ES, Prost SA, Liu T, Ibrahim YM and Smith RD, Anal. Chem, 2018, DOI: 10.1021/acs.analchem.8b02397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bode L, Glycobiology, 2012, 22, 1147; [DOI] [PMC free article] [PubMed] [Google Scholar]; Mantovani V, Galeotti F, Maccari F and Volpi N, ELECTROPHORESIS, 2016, 37, 1514. [DOI] [PubMed] [Google Scholar]
- 12.Hunt KM, Preuss J, Nissan C, Davlin CA, Williams JE, Shafii B, Richardson AD, McGuire MK, Bode L and McGuire MA, Appl. Environ. Microbiol, 2012, 78, 4763; [DOI] [PMC free article] [PubMed] [Google Scholar]; Smilowitz JT, Lebrilla CB, Mills DA, German JB and Freeman SL, Annu. Rev. Nutr, 2014, 34, 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Varki A, Cummings RD, Aebi M, Packer NH, Seeberger PH, Esko JD, Stanley P, Hart G, Darvill A, Kinoshita T, Prestegard JJ, Schnaar RL, Freeze HH, Marth JD, Bertozzi CR, Etzler ME, Frank M, Vliegenthart JFG, Lutteke T, Perez S, Bolton E, Rudd P, Paulson J, Kanehisa M, Toukach P, Aoki-Kinoshita KF, Dell A, Narimatsu H, York W, Taniguchi N and Kornfeld S, Glycobiology, 2015, 25, 1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


