Abstract
Organogenesis is regulated by mesenchymal-epithelial signaling events that induce expression of cell-type specific transcription factors critical for cellular proliferation, differentiation and appropriate tissue patterning. While mesenchymal transcription factors play a key role in mesenchymal-epithelial interactions, transcriptional networks in septum transversum and splanchnic mesenchyme remain poorly characterized. Forkhead Box F1 (FOXF1) transcription factor is expressed in mesenchymal cell lineages; however, its role in organogenesis remains uncharacterized due to early embryonic lethality of Foxf1−/− mice. In the present study, we generated mesenchyme-specific Foxf1 knockout mice (Dermol-Cre Foxf1−/−) and demonstrated that FOXF1 is required for development of respiratory, cardiovascular and gastrointestinal organ systems. Deletion of Foxf1 from mesenchyme caused embryonic lethality in the middle of gestation due to multiple developmental defects in the heart, lung, liver and esophagus. Deletion of Foxf1 inhibited mesenchyme proliferation and delayed branching lung morphogenesis. Gene expression profiling of micro-dissected distal lung mesenchyme and ChIP sequencing of fetal lung tissue identified multiple target genes activated by FOXF1, including Wnt2, Wnt11, Wnt5A and Hoxb7. FOXF1 decreased expression of the Wnt inhibitor Wif1 through direct transcriptional repression. Furthermore, using a global Foxf1 knockout mouse line (Foxf1−/−) we demonstrated that FOXF1-deficiency disrupts the formation of the lung bud in foregut tissue explants. Finally, deletion of Foxf1 from smooth muscle cell lineage (smMHC-Cre Foxf1−/−) caused hyper-extension of esophagus and trachea, loss of tracheal and esophageal muscle, mispatterning of esophageal epithelium and decreased proliferation of smooth muscle cells. Altogether, FOXF1 promotes lung morphogenesis by regulating mesenchymal-epithelial signaling and stimulating cellular proliferation in fetal lung mesenchyme.
Keywords: FOXF1, lung development, transgenic mice, mesenchyme, smooth muscle cells
INTRODUCTION
Lung development in the mouse begins at 9.5 days post coitum (E9.5) when the foregut endoderm invades the splanchnic mesenchyme and undergoes dichotomous branching (Cardoso, 2001; Morrisey and Hogan, 2010; Warburton et al., 1999; Whitsett et al., 2004). Lung morphogenesis depends on mesenchymal-epithelial crosstalk between splanchnic mesenchyme and endoderm-derived epithelial cells, which is mediated by SHH, WNTs, FGFs, TGF-β, BMP4 and NOTCH (Bolte et al., 2018; Hogan and Yingling, 1998; Morrisey and Hogan, 2010). These signaling molecules regulate proximal-distal patterning, cellular proliferation and differentiation of various respiratory cell types by inducing expression of cell-type specific transcription factors. The developing respiratory epithelium produces SHH, which acts via PTCH receptor and the GLI family of zinc finger transcription factors to promote mesenchyme survival and stimulate its differentiation into multiple stromal cell lineages, including fibroblasts, pericytes and smooth muscle cells (Weaver et al., 2003). Conditional inactivation of Shh in respiratory epithelium or deletion of either Gli2 or Gli3 results in lung hypoplasia, diminished epithelial branching and loss of mesenchyme and smooth muscle cells (Grindley et al., 1997; Miller et al., 2004; Motoyama et al., 1998). Compound mutations in Glil/2 cause severe lung hypoplasia, whereas Gli2/3 null mutants completely fail to specify the respiratory lineage (Grindley et al., 1997; Motoyama et al., 1998; Rankin et al., 2016). While these studies implicate SHH/PTCH/GLI signaling in regulation of lung development, downstream targets of the SHH signaling pathway in lung mesenchyme remain poorly characterized.
FOXF1 (previously known as HFH-8 or Freac-1) is a transcription factor from the Forkhead Box (FOX) family, which is expressed in septum transversum and splanchnic mesenchyme prior to the initiation of lung development (Mahlapuu et al., 2001b; Peterson et al., 1997). Later in development, FOXF1 is expressed in lung mesenchyme, endothelial cells and airway smooth muscle cells (Cai et al., 2016; Kalin et al., 2008; Kalinichenko et al., 2001b; Mahlapuu et al., 1998; Ren et al., 2014). FOXF1 induces migration of lung mesenchymal cells in vitro by activating transcription of Integrin β3 and Notch-2 (Kalinichenko et al., 2004; Malin et al., 2007). Fox/ΓA mice exhibit an embryonic lethal phenotype due to lack of vasculature in the yolk sac and allantois (Mahlapuu et al., 2001b). Foxf1 haploinsufficiency (Foxf1+/-) causes lung hypoplasia, decreased formation of pulmonary capillaries and increased mortality after birth (Kalinichenko et al., 2001a; Kalinichenko et al., 2002a; Kalinichenko et al., 2002b; Lim et al., 2002; Mahlapuu et al., 2001a; Ormestad et al., 2006). Inactivating mutations in the FOXF1 gene are often found in patients with Alveolar Capillary Dysplasia with Misalignment of Pulmonary Veins (ACD/ MPV) (Sen et al., 2013; Szafranski et al., 2013), a fatal congenital disorder of newborns and infants associated with lung hypoplasia and the loss of peripheral pulmonary capillaries. Mouse genetic studies demonstrated that FOXF1 acts downstream of GLI to promote SHH signaling during cardiac, intestinal and craniofacial development (Hoffmann et al., 2014; Mahlapuu et al., 2001b; Xu et al., 2016). Shhmouse embryos exhibit decreased Foxf1 expression in mesenchyme (Mahlapuu et al., 2001a). SHH/GLI signaling induces Foxf1 expression through GLI-mediated transcriptional activation of the Foxf1 promoter (Kim et al., 2005; Madison et al., 2009; Szafranski et al., 2013). Mutations in GLI-binding sites in the human FOXF1 promoter cause ACD/MPV in spite of the preservation of normal FOXF1 coding sequences (Szafranski et al., 2013), emphasizing the importance of the SHH/GLI signaling pathway in regulation of FOXF1 gene expression. While previous studies mostly focused on the role of FOXF1 in endothelial cells during lung development, FOXF1 functions in lung mesenchyme remain uncharacterized.
In the present study, we demonstrated that Foxf1 expression in mesenchymal cells is required for development of respiratory, cardiovascular and gastrointestinal organ systems as deletion of Foxf1 from Dermol-expressing mesenchyme (Dermol-Cre Foxf1−/−) causes embryonic lethality and a variety of developmental defects. Furthermore, deletion of Foxf1 inhibits mesenchyme proliferation and delays branching lung morphogenesis. FOXF1 promotes mesenchymal-epithelial signaling and stimulates cellular proliferation by inducing expression of Wnt2 and Hoxb7 and suppressing the Wnt-inhibitor Wifi in developing lung mesenchyme. Deletion of Foxf1 from smooth muscle cells (smMHC-Cre Foxf1−/−) causes perinatal lethality due to enlargement of the esophagus and trachea, thinning of muscle layers and decreased proliferation of smooth muscle cells. Altogether, our data indicate that FOXF1 promotes lung morphogenesis by regulating mesenchymal-epithelial signaling and stimulating cellular proliferation in mesenchyme-derived cells.
METHODS
Mouse strains
The generation of Foxf1flox/flox(Foxf1fl/fl) mice was described previously (Ren et al., 2014). Foxf1-floxed allele contains LoxP sequences flanking the DNA binding domain (exon 1) (Cai et al., 2016; Ren et al., 2014). Foxf1fl/fl mice were bred with Dermol-Cretg/- mice (Jackson Laboratory, (Yu et al., 2003)) to generate Dermol-Cretg/- Foxf1fl/fl double transgenic mice. Foxf1fl/fl and Dermol-Cretg/- Foxf1fl/+ littermates were used as controls. Generation of Foxf1+/− and smMHC-Cretg/- Foxf1fl/+ mouse lines was described previously (Hoggatt et al., 2013; Kalinichenko et al., 2001a; Sen et al., 2014). Foxf1fl/fl and Foxf1+/− mice were generated as C57Bl/6 x 129/SVEV hybrids (Ren et al., 2014) and were bred into C57Bl/6 background for 10 generations. mTmG mice (Ren et al., 2014) were purchased from the Jackson Laboratory. Animal studies were reviewed and approved by the Animal Care and Use Committee of Cincinnati Children’s Research Foundation.
Immunohistochemical staining
Embryos were harvested, fixed overnight with 4% buffered paraformaldehyde and then embedded into paraffin blocks. Five μm paraffin sections were used for staining with hematoxylin and eosin (H&E), immunohistochemistry and immunofluorescent co-staining as described previously (Ren et al., 2013; Wang et al., 2010; Wang et al., 2003). The following antibodies were used for immunostaining: FOXF1 (Ren et al., 2014); αSMA (A5228, Sigma); γSMA (LMAB-B4, Seven Hills Bioreagents); FLK1 (2479, Cell signaling); FOXM1 (Cheng et al., 2014; Weiler et al., 2017); BrdU (G3G4, DSHB); Ki-67 (clone Tec-3, Dako); PH3 (sc8656r, Santa Cruz Biotechnology); Cleaved Caspase 3 (MAB835, R&D Systems); SOX2 (WRAB-1236, Seven Hills Bioreagents); SOX9 (AB5535, Millipore); NKX2.1 (WRAB-1231, Seven Hills Bioreagents); E-Cadherin (MAB7481, R&D Systems; and 4065, Cell Signaling); CCSP (sc9772, Santa Cruz Biotechnology); β-tubulin (MU 178-UC, BioGenex); p63 (sc71827, Santa Cruz Biotechnology); Cytokeratin 13 (ab92551, Abcam); Cytokeratin 14 (ab7800, Abcam); and activated β-catenin (phospho-Ser552) (5651, Cell Signaling). Antibody-antigen complexes were detected using biotinylated secondary antibody followed by avidin-horseradish peroxidase (HRP) complex and DAB substrate (all from Vector Lab) as described (Balli et al., 2012; Ren et al., 2010; Wang et al., 2012; Wang et al., 2014). Sections were counterstained with nuclear fast red (Vector Labs, Burlingame, CA). BrdU incorporation was performed according to the manufacturer’s protocol as previously described (Ren et al., 2014; Sengupta et al., 2013). BrdU was injected intraperitoneally into pregnant females 2 hours before embryo harvest. For colocalization experiments, secondary antibodies conjugated with Alexa Fluor 488, Alexa Fluor 594 or Alexa Fluor 647 (Invitrogen) were used as previously described (Bolte et al., 2011; Bolte et al., 2012; Ustiyan et al., 2012; Ustiyan et al., 2016). Slides were counterstained with DAPI (Vector Laboratory). Images were obtained using a Zeiss Axioplan 2 microscope with AxioCam MRc5 and AxioCam MRm digital cameras and Axio Vision 4.8 Software (Carl Zeiss). The number of αSMA+Foxf1+ cells and the percentage of the airway epithelium circumference covered by αSMA+cells were calculated as described (Hines et al., 2013).
Ex vivo culture of foregut and lung explants
Mouse foregut regions (E8.5) or whole lungs (E11.5) were dissected from embryos in Hank’s balanced salt solution, explanted onto 8 μm pore Whatman Nucleopore Track-Etch Membranes (Millipore) and cultured in DMEM (Gibco) with 5% fetal bovine serum (FBS, Sigma) for lung explant culture or 20% FBS for foregut explant culture (Havrilak et al., 2017; Havrilak and Shannon, 2015). Explants were cultured at 37°C in a 5% CO2 incubator for 1-3 days. Whole-mount immunostaining was performed as previously described (Havrilak and Shannon, 2015). Images were captured on a Nikon A1Rsi inverted laser confocal microscope and composed using Bitplane Imaris software.
Quantitative real-time RT-PCR (qRT-PCR)
Lungs were dissected from embryos or newborn mice and homogenized in STAT60. Total lung RNA was isolated as described previously (Milewski et al., 2017a; Milewski et al., 2017b; Sun et al., 2017) and analyzed by qRT-PCR using a StepOnePlus Real-Time PCR system (Applied Biosystems, Foster City, CA) (Bolte et al., 2015; Pradhan et al., 2016; Xia et al., 2015). TaqMan Gene Expression Master Mix (Applied Biosystems) was used to amplify samples in combination with inventoried TaqMan gene expression assays for the gene of interest (Suppl. Table S1). Reactions were analyzed in triplicate and expression levels were normalized to β-actin mRNA.
ChIPseq and RNAseq
Analysis of FOXF1 binding sites for genes of interest was performed using a published ChIPseq data set generated from E18.5 mouse lungs (Accession number GSE77951) (Dharmadhikari et al., 2016). Data analysis was performed using the Biowardrobe platform as previously described (Bolte et al., 2017). RNAseq analysis was performed using distal lung mesenchyme micro-dissected from control and Dermol-Cre Foxf1−/− mouse embryos as previously described (Bolte et al., 2017; Cai et al., 2016; Sen et al., 2014). DESeq package was used to determine and analyze differences in gene expression. Differentially expressed genes were used to build heat map with JMP Genomics 6.0 software. RNAseq data is available as GEO accession GSE78184.
Statistical analysis
Student’s T-test was used to determine statistical significance. P values ≤ 0.05 were considered significant. Values for all measurements were expressed as the mean ± standard deviation (SD).
RESULTS
Deletion of Foxf1 from mesenchymal cells causes embryonic lethality and defects in multiple organ systems
Immunohistochemistry was performed to examine FOXF1 expression in the developing mouse lung. At E11.5, FOXF1 staining was detected in endothelial cells of pulmonary blood vessels and pulmonary mesenchyme adjacent to respiratory epithelial tubules (Fig. 1A), a finding consistent with published studies (Kalinichenko et al., 2003; Ren et al., 2014). FOXF1 was also expressed in visceral smooth muscle and mesenchyme surrounding esophagus, trachea, bronchi, stomach and intestine (Fig. 1A, Suppl. Fig. S1A-C). FOXF1 was not detected in vascular smooth, cardiac or skeletal muscles (Suppl. Fig. S1D-F). To examine requirements for FOXF1 in mesenchyme and its derivatives, we crossed Foxf1fl/fl mice with mice containing Dermol-Cre, a mesenchyme-specific Cre driver (Yu et al., 2003). In Dermol-Cre Foxf1−/− embryos (abbreviated as Dermol-Foxf1−/−), the exon 1, encoding the FOXF1 DNA-binding domain, was deleted (Fig. 1B). The presence of Dermol-Cre and Foxf1-floxed alleles were confirmed by PCR (Fig. 1C). Dermol-Foxf1−/− embryos exhibited severe growth retardation and died in utero after E16.5 (Fig. 1D). Genotype analysis of embryos from crosses between Dermol-Cre Foxf1fl/+ males and Foxf1fl/fl females revealed a gradual reduction in percentages of Dermol-Foxf1−/− embryos between E13.5 and E16.5 (Table 1). No Dermol-Foxf1−/− embryos were recovered after E16.5.
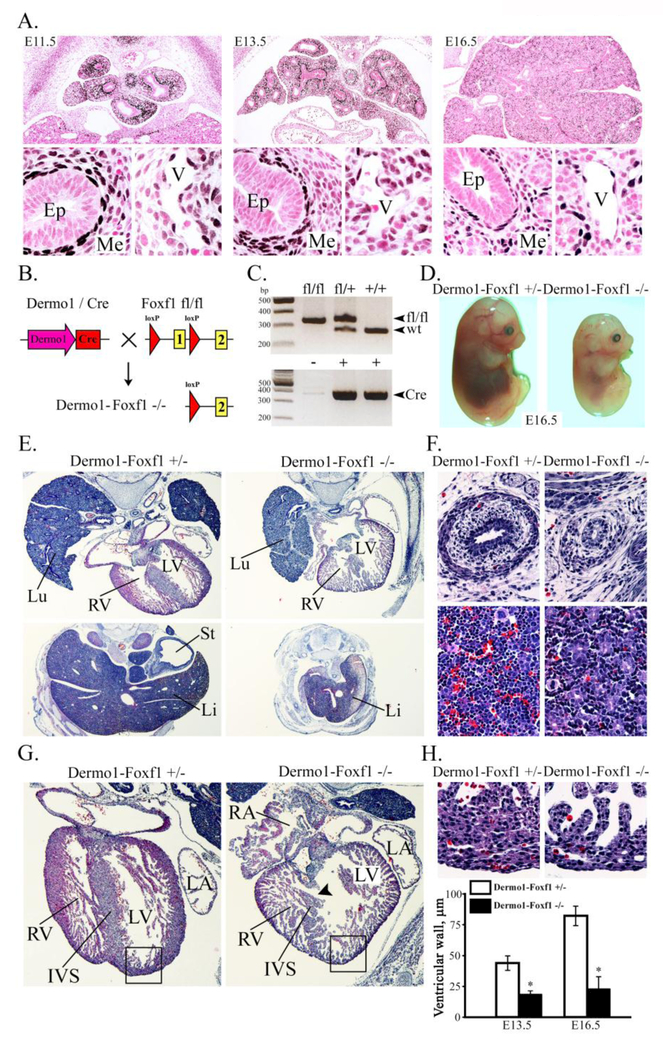
Figure 1. Deletion of Foxf1 from mesenchyme causes growth retardation and heart ventricular hypoplasia.
A, Foxf1 is expressed in lung mesenchyme and endothelial cells during embryonic development. Paraffin lung sections from wild type mouse embryos (E11.5, E13.5 and E16.5) were stained with FOXF1 antibodies (dark brown) and counterstained with nuclear fast red (red). B-C, Schematic drawing illustrates deletion of Foxf1-floxed allele from mesenchymal cells. Foxf1fl/fl mice were bred with Dermo1-Cretg/- mice to generate Dermol-Cretg/- /Foxm1fl/fl double transgenic mice (Dermo1-Foxf1−/−). PCR shows deletion of Foxf1 and presence of Cre transgene. D, Foxf1 deletion from mesenchyme causes growth retardation. Representative photographs of E16.5 Dermo1-Foxf1−/− and Dermo1-Foxf1+/− embryos are shown. E-H, Hematoxylin and eosin (H&E) staining shows hypoplasia of the lung (E), liver (E), esophagus (F, top panels), lack of blood islands in liver tissue (F, bottom panels), interventricular septum defect (G) and myocardial hypoplasia (G-H) in E16.5 Dermo1-Foxf1−/− embryos. Thickness of the compact layer of ventricular myocardium (H) was quantitated using 10 random x400 microscope fields (n = 3 embryos in each group; * indicates p < 0.05). Abbreviations: Ep, epithelium; Me, mesenchyme; V, vein; Lu, lung; Li, liver; St, stomach; RV, right ventricle; LV, left ventricle; RA, right atrium; LA, left atrium; IVS, interventricular septum. Magnification: A (top panels) and G, x50; A (bottom panels), F and H, x400; E (top panels), x32; E (bottom panels), x16.
Table 1.
Genotypes of embryos from crosses between Dermo1-Cre Foxf1fl+ males and Foxf1fl/fl females were identified using PCR analysis of genomic DNA.
| Embryos | Foxf1fl/+ | Foxf1fl/fl | Dermo1-Foxf1fl+ | Dermo1-Foxf1−/− | |
|---|---|---|---|---|---|
| E9.5-10.5 | 34 | 8 (23.5%) | 9 (26.5%) | 9 (26.5%) | 8 (23.5%) |
| E11.5-12.5 | 70 | 14 (20.0%) | 16 (23.0%) | 21 (30.0%) | 19 (27.0%) |
| E13.5-14.5 | 70 | 13 (18.6%) | 21 (30.0%) | 22 (31.4%) | 14 (20.0%) |
| E15.5-16.5 | 23 | 9 (39.1%) | 4 (17.4%) | 7 (30.4%) | 3 (13.0%) |
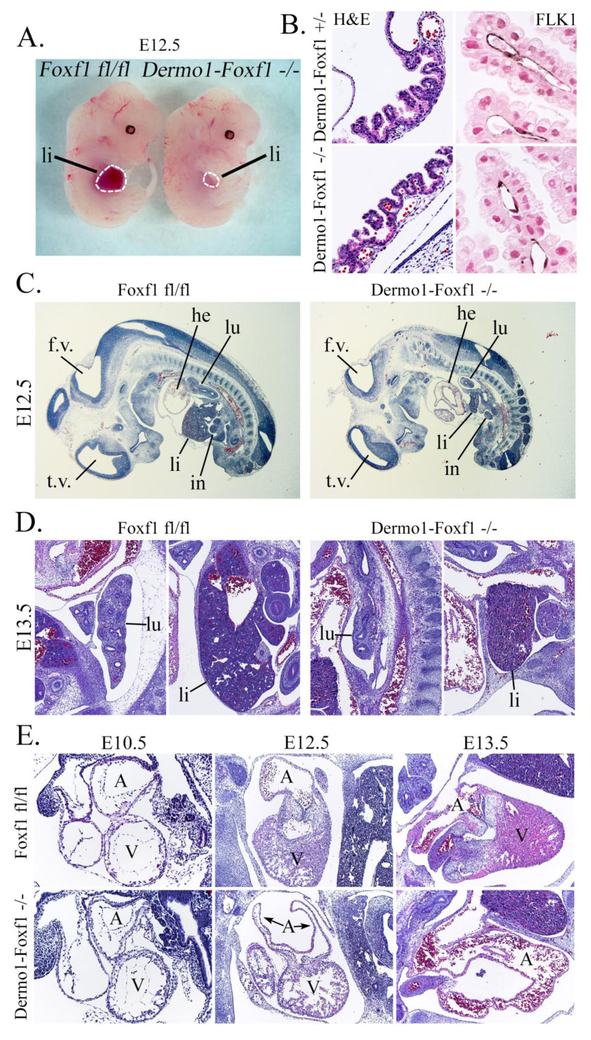
Histological studies were carried out to identify the cause of growth retardation and lethality in Dermol-Foxf1−/− embryos. At E16.5, the liver, lung and esophagus in Dermol-Foxf1−/− embryos were smaller compared to control littermates (Fig. 1E-F). The lack of blood islands in the liver and the pale appearance of Dermol-Foxf1−/− embryos were consistent with hematopoietic deficiency (Fig. 1D and Fig. 1F). Dermol-Foxf1−/− E16.5 embryos exhibited heart abnormalities, including ventricular hypoplasia, inter-ventricular septal defect, thinning of the compact layer of the myocardium and atrial chamber malformations (Fig. 1G-H and Suppl. Fig. S2A). The trabecular layer of the myocardium was unaltered in Dermol-Foxf1−/− mutants (Fig. 1G-H). Earlier time-points were examined to determine timing of the organ abnormalities. At E12.5-E13.5, there were no differences in body size between Dermol-Foxf1−/− and control embryos (Fig. 2A). However, Dermol-Foxf1−/− livers and lungs were smaller (Fig. 2C-D), and heart defects were already present (Fig. 2E). Distal parts of Dermol-Foxf1−/− atrial chambers were collapsed (Suppl. Fig. S3). FOXF1 staining was reduced in septum transversum and venous pole mesenchyme, and the dorsal mesenchymal protrusion was absent in Dermol-Foxf1−/− embryos (Suppl. Fig. S4). Heart defects in Dermol-Foxf1−/− embryos were consistent with venous pole mesenchymal defects during heart morphogenesis (Snarr et al., 2007). There were no histological abnormalities in the yolk sac of Dermol-Foxf1−/− embryos as shown by H&E staining and immunostaining for the endothelial marker FLK1 (Fig. 2B). At E13.5, developing limbs in Dermol-Foxf1−/− embryos had all 5 digits (Suppl. Fig. S2B). Altogether, deletion of Foxf1 from mesenchymal cells causes growth retardation and embryonic lethality due to defects in multiple organ systems.
Figure 2. Developmental abnormalities in Dermo1-Foxf1−/− embryos at E12.5-E13.5.
A, Representative photographs of Dermo1-Foxf1−/− and control Foxf1fl/fl embryos show no differences in body sizes at E12.5. B, H&E and immunostaining for FLK1 show no vascular abnormalities in yolk sacs of E13.5 Dermo1-Foxf1−/− embryos. FLK1-stained slides (dark brown) were counterstained with nuclear fast red (red nuclei). C-E, H&E staining of E10.5-E13.5 embryos show lung and liver hypoplasia (C-D) and heart defects (E) after deletion of Foxf1. Abbreviations: lu, lung; li, liver; in, intestine; he, heart; A, heart atrium; V, heart ventricle; t.v., third brain ventricle; f.v., fourth brain ventricle. Magnification: B (left panels), x200; B (right panels), x400; C, x16; D and E (middle and right panels), x50; E (left panels), x100.
Deletion of Foxf1 causes lung hypoplasia
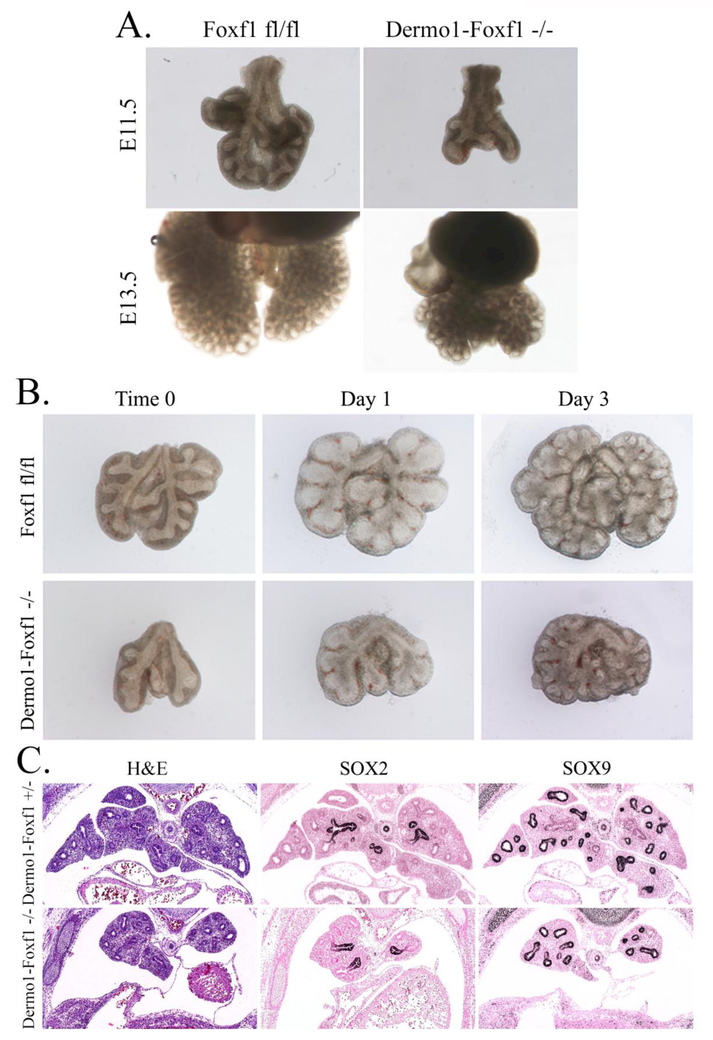
Next, we focused on lung development in Dermol-Foxf1−/− embryos. At E11.5, Dermol-Foxf1−/− lungs were hypoplastic and contained less branching points at the time of harvest (Fig. 3 A and Suppl. Fig. S5). Isolated Dermol-Foxf1−/− lungs were capable of branching ex vivo but remained hypoplastic during the tissue culture period (Fig. 3B). Immunohistochemistry was carried out to examine cell-specific markers in Dermol-Foxf1−/− lungs. SOX2 and SOX9 were present in proximal and distal epithelial progenitors (Fig. 3C and Suppl. Fig. S6) and there were no changes in Sox2 and Sox9 mRNAs (Fig. 4A). At E16.5, the terminal tips of airways were not dilated in Dermol-Foxf1−/− mutants and lung histology was consistent with the pseudoglandular stage of lung development (Suppl. Fig. S6).
Figure 3. Lung hypoplasia in Dermo1-Foxf1−/− embryos.
A, Representative photographs of E11.5 and E13.5 mouse lungs show lung hypoplasia in Dermo1-Foxf1−/− embryos. B, E11.5 lungs from Dermo1-Foxf1−/− and control Foxf1fl/fl embryos were cultured ex vivo for three days. Dermo1-Foxf1−/− lungs were capable of branching but remained hypoplastic. C, Paraffin lung sections from E13.5 embryos were used for H&E staining or immunostained with antibodies against SOX2 and SOX9 (dark brown). Slides were counterstained with nuclear fast red (red). Magnification in panels C is x50.
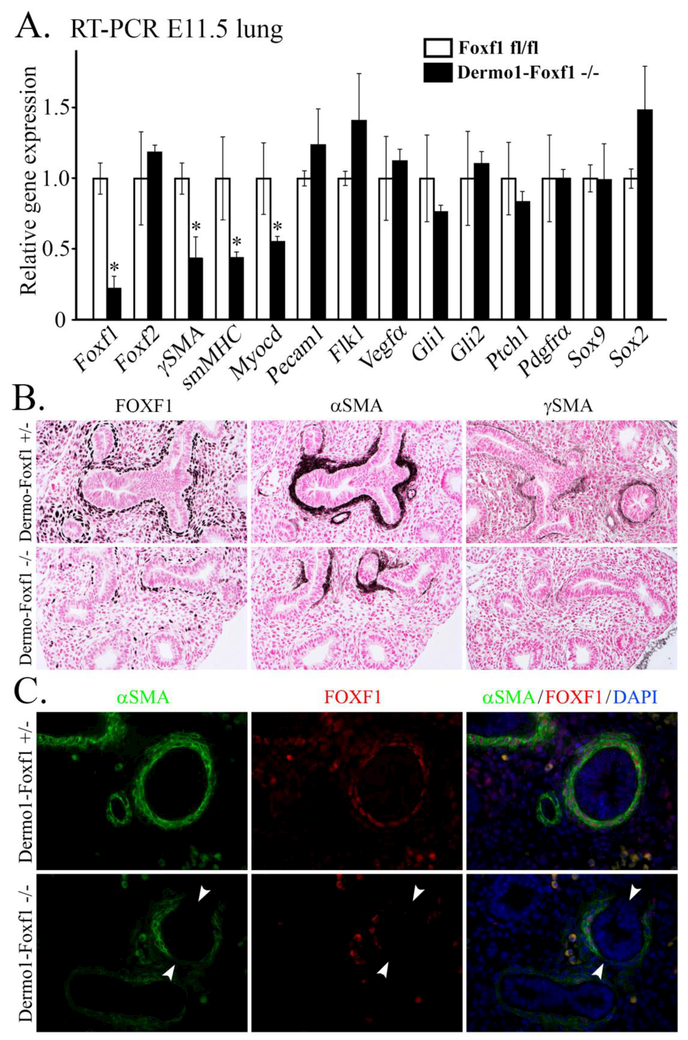
Figure 4. Foxf1 deletion from mesenchyme inhibits smooth muscle cell lineage during lung development.
A, E11.5 lungs from Dermo1-Foxf1−/− and control embryos were used for qRT-PCR. mRNAs were analyzed in triplicate and expression levels were normalized to β-actin. Data are presented as means ± SD (* indicates p < 0.05). qRT-PCR shows decreased expression of Foxf1 and several smooth muscle specific genes (γSMA, smMHC and Myocardin) in E11.5 Dermo1-Foxf1−/− lungs. B, Immunostaining shows decreased expression of FOXF1, αSMA and γSMA in E13.5 Dermo1-Foxf1−/− lungs (dark brown). Slides were counterstained with nuclear fast red (red). C, Co-localization studies were performed using E13.5 lung sections and antibodies against FOXF1 and αSMA. Loss of peribronchial αSMA-positive cells was observed in Dermo1-Foxf1−/− lungs. Decreased αSMA staining was observed in areas with the highest deletion of Foxf1 (white arrowheads). Magnification: B panels, x200; C panels, x400.
FOXF1 staining and Foxf1 mRNA were decreased in Dermol-Foxf1−/− lungs (Fig. 4A-B), a finding consistent with efficient deletion of Foxf1 by the Dermol-Cre transgene. Expression of smooth muscle markers αSMA, γSMA and smMHC was decreased in Dermol-Foxf1−/− lungs as shown by immunostaining and qRT-PCR (Fig. 4A-B). Loss of αSMA occurred in mesenchymal regions with the highest Foxf1 deletion efficiency (Fig. 4C). Loss of αSMA was also evident in the trachea and bronchi of Dermol-Foxf1−/− embryos (Suppl. Fig. S7A and S8). Deletion of Foxf1 from mesenchyme significantly decreased the number of αSMA+FOXF1+ cells (Suppl. Fig. S7B) and reduced the percentage of the airway epithelium circumference covered by αSMA+ cells (Suppl. Fig. S7C). Vascular smooth muscle was not affected in Dermol-Foxf1−/− mutants (Suppl. Fig. S7D).
Foxf1 deletion from mesenchyme led to mispatterning of tracheal epithelium and cartilage. While the tracheal epithelium of Dermol-Foxf1−/− embryos expressed E-cadherin, NKX2.1 and SOX2, the number of p63-positive basal cells was lower (Suppl. Fig. S8). SOX9 staining was reduced in the developing cartilage of Dermol-Foxf1−/− embryos (Suppl. Fig. S8). There were no differences in expression of endothelial markers FLK1 and PECAM-1 in Dermol-Foxf1−/− lungs (Fig. 4A and Suppl. Fig. S9). mRNAs of Foxf2, Gli1, Gli2, Ptchl, Vegfa and Pdgfra were unchanged after deletion of Foxf1 (Fig. 4A). Altogether, deletion of Foxf1 from mesenchyme decreases smooth muscle cell markers and causes mispatterning in tracheal epithelium and mesenchyme.
Deletion of Foxf1 decreases mesenchyme proliferation
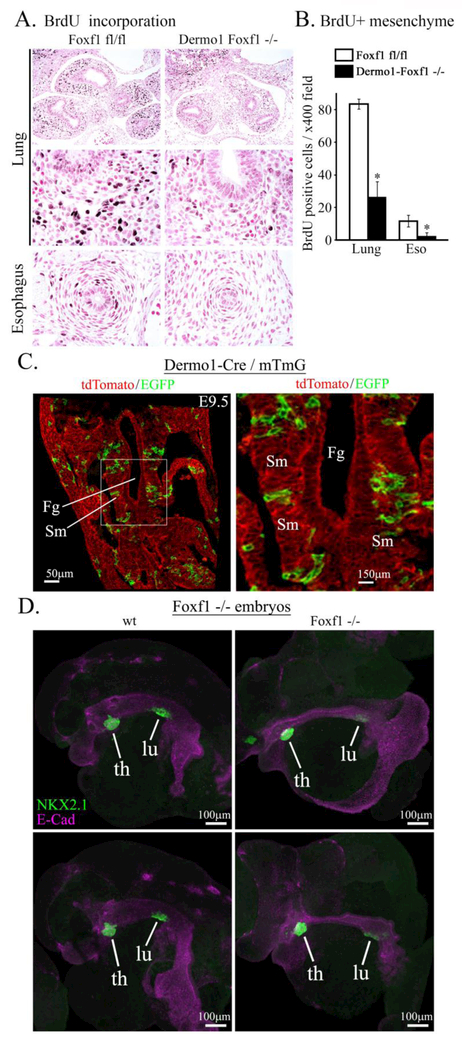
To measure cellular proliferation in Dermol-Foxf1−/− lungs, we used BrdU incorporation to label cells undergoing DNA replication. The number of BrdU-positive mesenchymal cells was significantly decreased in Dermol-Foxf1−/−embryos (Fig. 5A-B), a finding consistent with reduced proliferation rates in FOXF1-deficient endothelial cells during embryogenesis (Ren et al., 2014) and lung regeneration (Bolte et al., 2017). There was no difference in cell apoptosis in Dermol-Foxf1−/− lungs as shown by immunostaining for activated (cleaved) Caspase 3 (Suppl. Fig. S9). Thus, deletion of Foxf1 reduces mesenchyme proliferation but does not influence cell survival.
Figure 5. Foxf1 deletion decreases mesenchyme proliferation.
A, Immunostaining of paraffin sections from E11.5 embryos show decreased incorporation of BrdU into mesenchyme of Dermo1-Foxf1−/− lungs and esophagus (dark brown). Slides were counterstained with nuclear fast red (red). B, The number of BrdU-positive cells was decreased in Dermo1-Foxf1−/− lung and esophagus. Ten random x400 microscope fields were counted (n = 5 embryos in each group; * indicates p < 0.05). Magnification: top panels, x100; remaining panels, x400. C, mT/mG transgene was used to show a mosaic pattern of Dermo1-Cre-mediated recombination in mesenchyme surrounding foregut of E9.5 embryos. Images show GFP and tdTomato fluorescence in Dermo1-Cre mTmG embryos. Scale bars are 50μm (left panel) and 150 μm (right panel). D, Global deletion of Foxf1 inhibits lung budding ex vivo. Foreguts with surrounding mesenchyme were micro-dissected from E8.5 Foxf1−/− and control WT embryos and cultured ex vivo for two days. Lung and thyroid buds were visualized by immunostaining for NKX2.1 (TTF1, green) and E-Cadherin (purple). Scale bars are 100 μm. Abbreviations: Fg, foregut; Sm, splanchnic mesenchyme; th, thyroid; lu, lung.
Deletion of Foxf1 disrupts lung budding
Next, we examined whether FOXF1 regulates the formation of lung buds, which normally occurs at E9.5 in the mouse (Morrisey and Hogan, 2010; Rankin et al., 2016). Dermol-Cre driven recombination was ineffective to delete Foxf1 from splanchnic mesenchyme at E9.5 as shown by GFP fluorescence in Dermol-Cre mTmG embryos (Fig. 5C). Therefore, we used a global Foxf1 knockout mouse line (Foxf1−/−) to examine the formation of lung bud. Since Foxf1−/− mice die in utero after E8.5, which is approximately one day earlier than the initiation of the lung bud (Mahlapuu et al., 2001b), we isolated foregut regions from Foxf1−/− and WT E8.5 embryos and cultured them ex vivo. NKX2.1 staining was used to visualize lung and thyroid buds formed in the foregut. While lung buds were present in Foxf1−/− foregut explants, they were smaller compared to WT (Fig. 5D). There were no differences in sizes of thyroid buds (Fig. 5D). Thus, Foxf1 expression in mesenchyme is critical for lung budding ex vivo.
FOXF1 regulates mesenchymal genes critical for mesenchymal-epithelial signaling and cell proliferation
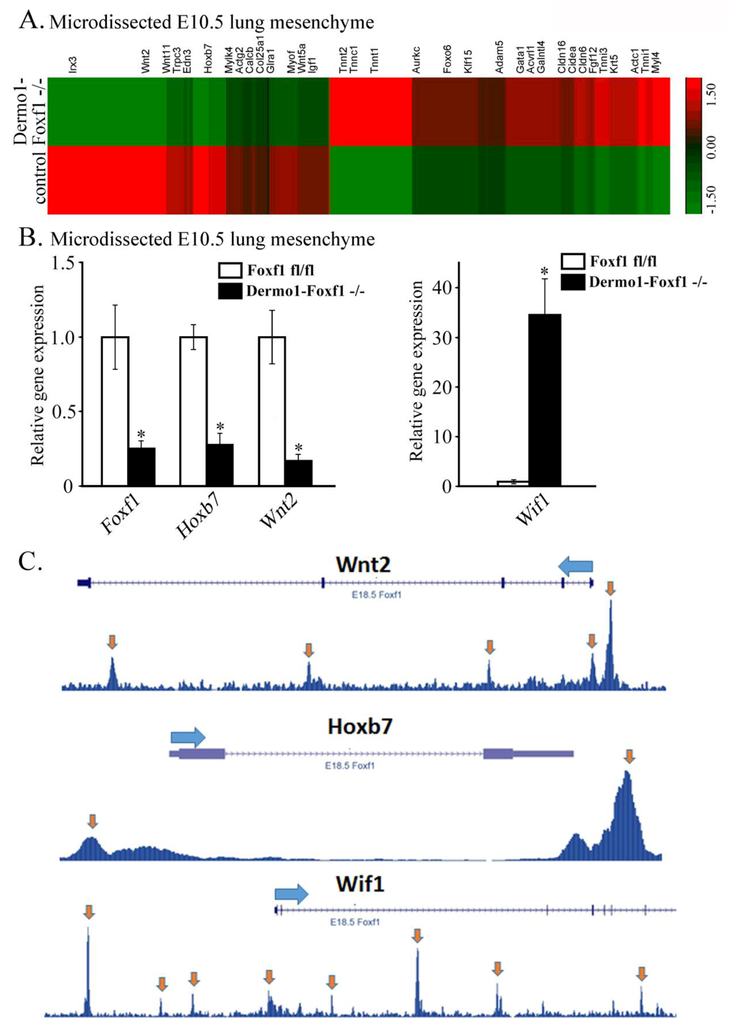
To identify FOXF1 target genes in lung mesenchyme, we used Dermol-Foxf1−/− and control Foxf1fl/fl embryos to micro-dissect mesenchyme from E10.5 distal lung tips and perform RNAseq analysis. Significant changes in expression of 1714 genes were found in lung mesenchyme from Dermol-Foxf1−/− embryos compared to Foxf1fl/fl embryos (Fig. 6A). These include 721 genes in which expression was significantly decreased (> 1.5-fold, p < 0.05) and 993 genes with increased expression (> 1.5-fold, p < 0.05) (Table 2 and GEO accession GSE78184). Differentially expressed genes in Dermol-Foxf1−/− mesenchyme were functionally classified according to Gene Ontology. Functional categories of “mesenchymal cell development“, “smooth muscle contraction”, “fibroblast proliferation” and “canonical Wnt signaling pathway” were significantly enriched in the subset of down-regulated genes (Suppl. Fig. S10). Expression levels of selected genes were confirmed by RT-PCR (Fig. 6B). Foxf1 mRNA was decreased by 75% in Dermol-Foxfl−/− lung mesenchyme compared to Foxf1fl/fl controls (Fig. 6B). Consistent with diminished cellular proliferation (Fig. 5A-B), mRNAs of Igf1, Egfr and Hoxb7 were reduced in Dermol-Foxf1−/− mesenchyme (Table 2 and Fig. 6A-B). Interestingly, expression of genes regulating canonical Wnt signaling pathway was significantly decreased in Dermol- Foxf1−/− lung mesenchyme (Table 2 and Fig. 6A-B). These include Wnt ligands Wnt2, Wnt5a and Wnt11, as well as known regulators of the canonical Wnt signaling pathway, including Snai2, Siah2, Zfp 703, Biccl, Nog, Rspol, Is11, Trpm4 and Nkd2 (DiMeo et al., 2009; Morrisey and Hogan, 2010; Peng et al., 2013). In contrast, expression of the Wnt inhibitor Wif1 was significantly increased in Dermol-Foxf1−/− lungs (Table 2), a finding confirmed by qRT-PCR (Fig. 6B). Cross-reference of the RNAseq data with previously published FOXF1 ChIPseq of mouse fetal lung tissue (Dharmadhikari et al., 2016) demonstrated that FOXF1 directly bound to gene promoters and introns of Wnt2, Wnt11, Wnt5a, Wif1, Hoxb7, Igf1 and Egfr (Fig. 6C, Suppl. Fig. S11 and Suppl. Table S2), implicating FOXF1 in regulation of these genes. Using antibodies against the activated form of β-catenin, we found that activated β-catenin was reduced in respiratory epithelium and mesenchyme of Dermol-Foxf1−/− lungs (Suppl. Fig. S12), a finding consistent with reduced Wnt/β-catenin signaling. Thus, FOXF1 regulates multiple mesenchymal genes critical for lung morphogenesis, canonical Wnt signaling and mesenchyme proliferation.
Figure 6. FOXF1 regulates expression of Hoxb7, Wnt2 and Wif1 in lung mesenchyme.
A, RNAseq heat-map shows gene expression in distal lung mesenchyme of Dermo1-Foxf1−/− and control Foxf1fl/fl embryos. RNA was extracted from micro-dissected mesenchyme of distal lung tips of E10.5 Dermo1-Foxf1−/− and control embryos (n = 3 embryos were used in each group). B, qRT-PCR of E10.5 lung mesenchyme shows decreased expression of Foxf1, Hoxb7 and Wnt2 and increased Wif1 mRNA in Dermo1-Foxf1−/− embryos. mRNA was analyzed in triplicate and expression levels were normalized to β-actin mRNA (n = 3 embryos in each group; * indicates p < 0.05). C, ChIPseq of mouse embryonic lungs shows FOXF1 binding to DNA regions in Hoxb7, Wnt2 and Wif1 genes. Statistically significant binding is indicated by arrows.
Table 2.
RNAseq of distal tip lung mesenchyme from E10.5 embryos.
| Gene | Folds | p-value | Gene name |
|---|---|---|---|
| Wnt signaling pathway | |||
| Wnt2 | −2.17 | 1.07E-14 | Wnt family member 2 |
| Wnt 11 | −2.14 | 2.67E-07 | Wnt family member 11 |
| Nog | −1.94 | 0.0062 | Noggin |
| Trpm4 | −1.92 | 7.64E-06 | Transient receptor potential cation channel, subfamily M, member 4 |
| Nkd2 | −1.90 | 0.00026 | Naked cuticle homolog 2 |
| Biccl | −1.83 | 1.55E-13 | BicC family RNA binding protein 1 |
| Isl1 | −1.82 | 9.96E-06 | ISL LIM homeobox 1 |
| Snai2 | −1.79 | 0 | Snail family zinc finger 2 |
| Wnt5a | −1.77 | 0.0006 | Wnt family member 5A |
| Siah2 | −1.71 | 0.0043 | Siah E3 ubiquitin protein ligase 2 |
| Zfp703 | −1.65 | 0.0006 | Zinc finger protein 703 |
| Rspo1 | −1.59 | 4.44E-11 | R-spondin 1 |
| Wif1 | 4.87 | 0 | Wnt inhibitory factor 1 |
| Regulation of smooth muscle proliferation and differentiation | |||
| Cdh13 | −10.7 | 0 | Cadherin13 |
| Hoxb7 | −10.47 | 2.72E-10 | Homeobox B7 |
| Trpc3 | −4.62 | 5.77E-15 | Transient receptor potential cation channel subfamily C member 3 |
| Calcb | −3.98 | 0.00028 | Calcitonin related polypeptide beta |
| Cacna1c | −3.81 | 0 | Calcium voltage-gated channel subunit alpha 1C |
| Kcnb2 | −2.95 | 1.75E-11 | Potassium voltage-gated channel subfamily B member 2 |
| Twist2 | −2.52 | 1.18E-08 | Twist family bHLH transcription factor 2 |
| Actg2 | −2.27 | 0.0024 | Gamma2 actin |
| Hand2 | −2.24 | 0.0067 | Heart and neural crest derivatives expressed 2 |
| Edn3 | −2.07 | 2.8E-6 | Endothelin 3 |
| Egfr | −2.05 | 0 | Epidermal growth factor receptor |
| Gucy1a3 | −1.84 | 0 | Guanylate cyclase 1 soluble subunit alpha |
| Chrm3 | −1.77 | 0.011 | Cholinergic receptor muscarinic 3 |
| Igf1 | −1.7 | 0 | Insulin like growth factor 1 |
| Klf4 | −1.68 | 1.77E-05 | Kruppel like factor 4 |
| Cald1 | −1.63 | 0 | Caldesmon 1 |
| Cardiovascular system development | |||
| Bmp4 | 1.86 | 0 | Bone morphogenic protein 4 |
| Tnnt2 | 2.12 | 1.11E-16 | Troponin T2, cardiac type |
| Tnni3 | 2.9 | 2.31E-05 | Troponin I3, cardiac type |
| Tnni1 | 3.46 | 0 | Troponin I1, slow skeletal; cardiac during embryonic development |
| Tnnc1 | 5.09 | 7.24E-13 | Troponin C1, slow skeletal and cardiac type |
| Csrp3 | 13.26 | 0 | Cysteine and glycine rich protein 3 |
Deletion of Foxf1 from smooth muscle cells causes tracheal and esophageal abnormalities
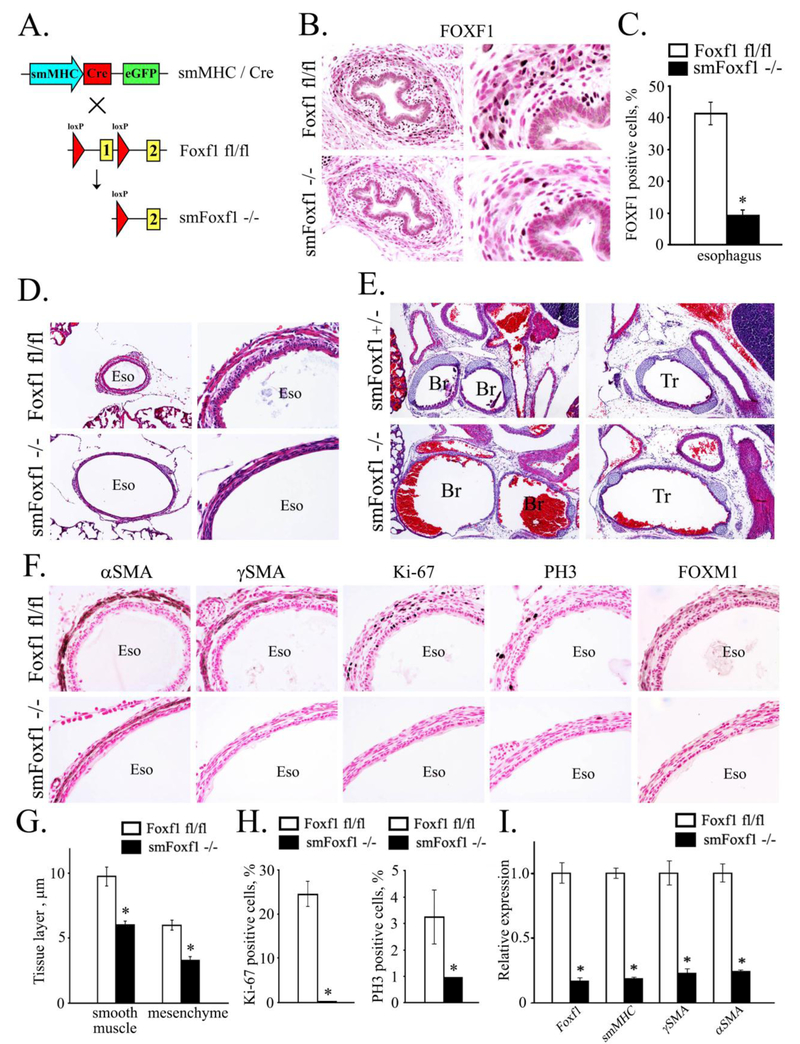
To examine the requirements for FOXF1 in smooth muscle cell lineage, we used the smMHC-Cre transgene (Fig. 7A), which is active after E13.5 (Xin et al., 2002). FOXF1 staining was decreased in esophageal smooth muscle of E17.5 smMHC-Cre Foxf1−/− embryos, consistent with efficient Foxf1 deletion from smooth muscle cells (Fig. 7B-C). smMHC-Cre Foxf1−/− mice died immediately after birth due to postnatal hemorrhage, causing accumulation of blood inside airways (Fig. 7E). smMHC-Cre Foxf1−/− newborn mice exhibited hyperextension of esophagus, trachea and bronchi (Fig. 7D-E), which was associated with thinning of smooth muscle layers and loss of smooth muscle markers αSMA and γSMA (Fig. 7F-G). Decreased mRNAs of αSMA, γSMA and smMHC were found in micro-dissected smMHC-Cre Foxf1−/− esophagi, consistent with the loss of smooth muscle cells (Fig. 7I). Proliferation of smooth muscle cells was reduced in smMHC-Cre Foxf1−/− mice as shown by immunostaining for proliferation markers Ki-67, phospho-histone H3 (PH3) and FOXM1 (Fig. 7F and Fig. 7H).
Figure 7. Deletion of Foxf1 from smooth muscle cells causes tracheal and esophageal abnormalities.
A, Schematic drawing shows deletion of Foxf1-floxed allele from smooth muscle cells. Foxf1fl/fl mice were bred with smMHC-Cretg/- mice to generate smMHC-Cretg/- Foxf1fl/fl double transgenic mice (smMHC-Foxf1−/−; abbreviated as smFoxf1−/−). B, Immunostaining for FOXF1 was performed using paraffin sections from E17.5 smMHC-Foxf1−/− and control Foxf1fl/fl embryos. FOXF1 staining (dark brown) was decreased in esophageal smooth muscle of smMHC-Foxf1−/− embryos. Slides were counterstained with nuclear fast red (red). C, The number of FOXF1 -positive cells was decreased in esophageal smooth muscle of smMHC-Foxf1−/− embryos. Ten random x400 microscope fields were used for quantification (n = 3 mice per group; * indicates p < 0.05). D-E, H&E staining shows thinning and hyper-extension of esophagus and trachea in newborn smMHC-Foxf1−/− mice. F, Immunostaining shows reduced smooth muscle cell proliferation in smMHC-Foxf1−/− newborn mice. Immunostaining was performed using antibodies against αSMA, γSMA, Ki-67, PH3 and FOXM1. G, Thickness of esophageal smooth muscle and mesenchymal layers was decreased smMHC-Foxf1−/− newborn mice compared to control Foxf1fl/fl mice (n=3 mice per group). H, Percentage of proliferating cells was reduced in esophageal smooth muscle of smMHC-Foxf1−/− newborn mice. Cells were counted using 10 random x400 microscope fields (n = 3 mice per group). I, qRT-PCR of RNA isolated from esophagus of smMHC-Foxf1−/− newborn mice shows decreased expression of Foxf1, αSMA, γSMA and smMHC. Abbreviations: Eso, esophagus; Br, bronchus; Tr, trachea. Magnification: B (left panels), D (right panels) and F, x400; E and D (left panels), x100; B (right panels), x800.
Esophageal epithelium was hypoplastic in smMHC-Cre Foxf1−/− mice (Supp. Fig. S13). While CK13-positive suprabasal cells were present in smMHC-Cre Foxf1−/− tracheas, the basal cell layer had diminished expression of SOX2, p63 and CK14 (Supp. Fig. S13). CCSP-positive Clara and β-tubulin-positive ciliated cells were present in proximal and distal airways of both smMHC-Cre Foxf1−/− and control mice (Suppl. Fig. S14). αSMA staining was reduced in proximal bronchioles of smMHC-Cre Foxf1−/− lungs (Suppl. Fig. S14), a finding consistent with the loss of peribronchial smooth muscle cells. The number of proliferative cells was significantly reduced in peribronchial smooth muscle of smMHC-Cre Foxf1−/− mice compared to controls (Suppl. Fig. S15). There were no differences in the thickness of smooth muscle layers surrounding intestine, and vascular smooth muscle was normal in smMHC-Cre Foxf1−/− mice (Suppl. Fig. S16). Taken together, deletion of Foxf1 decreases cell proliferation and disrupts proper development of smooth muscle in esophagus, trachea and bronchi.
DISCUSSION
Published studies demonstrate that FOXF1 is expressed in extraembryonic mesoderm, the second heart field and lateral mesoderm (Mahlapuu et al., 2001b; Peterson et al., 1997). Later in development, FOXF1 is detected in septum transversum, venous pole mesenchyme and splanchnic mesoderm, which are critical for mesenchymal-epithelial signaling during formation of the heart, lung, liver, esophagus, stomach and intestine (Kalinichenko et al., 2002a). Consistent with this expression pattern, we found that FOXF1 is critical for proper development of respiratory, cardiovascular and gastrointestinal organ systems as Dermol-Cre-mediated deletion of Foxf1 from mesenchyme caused multiple developmental defects in the heart, lung, liver and esophagus. The presence of atrioventricular septal defects in Dermol-Foxf1−/− hearts is consistent with published studies that reported similar findings in hearts of Foxf1+/− Foxf2+/− embryos (Hoffmann et al., 2014), implicating FOXF1 in mesenchymal signaling during cardiac septation. Compared to Foxf1+/− Foxf2+/− embryos, heart defects in Dermol-Foxf1−/− embryos were more severe and included ventricular hypoplasia, thinning of myocardium and atrial chamber malformations, the latter of which could be a result of abnormal venous pole mesenchymal development. In addition to heart defects, Dermol-Foxf1−/− embryos had small livers that lacked blood islands. This can result in hematopoietic deficiency and contribute to embryonic lethality in Dermol-Foxf1−/− embryos. Since FOXF1 is expressed in septum transversum mesenchyme prior to liver development (Kalinichenko et al., 2002a), it is possible that mesenchymal Foxf1 deletion inhibits proper liver morphogenesis by disrupting signaling between the mesenchyme and endoderm-derived hepatic progenitors. In contrast to Foxf1−/− and Tie2-Cre Foxf1−/− embryos that exhibited vascular defects in the yolk sac (Mahlapuu et al., 2001b; Ren et al., 2014), there were no abnormalities in the yolk sac of Dermol-Foxf1−/− embryos, suggesting that the Dermol-Cre transgene is inefficient in targeting the yolk sac vasculature. Our results also suggest that combined heart/liver insufficiency causes growth retardation and lethality in Dermol-Foxf1−/− embryos.
Published studies demonstrate that haploinsufficiency of Foxf1 in mice (Foxf1+/−) causes alveolar capillary dysplasia, fusion of the lung lobes and various developmental defects in mesenchyme of the gallbladder, esophagus, intestine and trachea (Kalinichenko et al., 2001a; Kalinichenko et al., 2002a; Lim et al., 2002; Mahlapuu et al., 2001a; Ormestad et al., 2006). Interestingly, inactivating mutations in FOXF1 gene were found in patients with Alveolar Capillary Dysplasia with Misalignment of Pulmonary Veins (ACD/MPV), a fatal congenital disorder which is often associated with lung hypoplasia, liver abnormalities and congenital heart defects, including AVS (Bishop et al., 2011). The lung, heart and liver defects in ACD/MPV patients are consistent with mouse genetic studies described in this manuscript, suggesting that FOXF1 deficiency in mesenchymal cells contributes to pathogenesis of ACD/MPV. While our study did not focus on FOXF1 requirements in the heart and liver, our results implicate FOXF1-expressing mesenchymal cells in development of these organs.
Lung morphogenesis requires crosstalk between respiratory epithelium and pulmonary mesenchyme mediated by SHH, WNTs, FGFs and BMPs (Bolte et al., 2018; Hogan and Yingling, 1998; Morrisey and Hogan, 2010). SHH is secreted by respiratory epithelial cells and acts via mesenchymal PTCH receptor and GLI transcription factors to induce FOXF1 through direct binding of GLI to the Foxf1 promoter (Madison et al., 2009; Szafranski et al., 2013). In the present study, we found that Dermol-Foxf1−/− embryos exhibited severe lung hypoplasia, a phenotype similar to Gli2+/−;Gli3+/− mutants and embryos with epithelial-specific inactivation of Shh (Grindley et al., 1997; Miller et al., 2004; Motoyama et al., 1998). Therefore, FOXF1 could be an important downstream mediator of SHH signaling in lung mesenchyme. Furthermore, deletion of Foxf1 from mesenchyme disrupted epithelial development in the trachea and esophagus, which is consistent with previous studies demonstrating a critical role of mesenchymal-epithelial signaling in epithelial differentiation (Hines et al., 2013; Lee et al., 2017). Our data also suggest that FOXF1 deficiency causes lung hypoplasia, at least in part by inhibiting cell proliferation in fetal lung mesenchyme, leading to diminished expansion of mesenchyme-derived tissues. These data are consistent with recent reports showing decreased cellular proliferation in FOXF1-deficient endothelial cells (Bolte et al., 2017; Ren et al., 2014) and rhabdomyosarcomas (Milewski et al., 2017b). Altogether, our findings in Foxf1−/− foregut explants and Dermol-Foxf1−/− embryos indicate that FOXF1 expression in mesenchymal cells is critical for mesenchyme proliferation, as well as lung budding and branching.
In addition to proliferation defects, FOXF1-deficient mesenchyme had diminished expression of several WNT ligands and downstream targets of canonical WNT signaling pathway. Activated β-catenin was reduced in lung epithelium and mesenchyme of Dermol-Foxf1−/− embryos, consistent with diminished WNT/β-catenin signaling. The lack of mesenchymal WNT ligands could lead to proliferation defects in respiratory epithelium and smooth muscle cells, contributing to lung hypoplasia in Dermol-Foxf1−/− embryos. It is possible that FOXF1 directly induces transcription of Wnt2, Wnt11 and Wnt5A in lung mesenchyme since Foxf1-binding sites were detected by ChIPseq in promoters and enhancers of these genes. Canonical WNT signaling is required for specification of the lung bud (Herriges and Morrisey, 2014). Mice lacking Wnt2/2b or β-catenin fail to generate a lung field, whereas over-activation of the canonical WNT pathway expands the specified lung field into the anterior foregut (Goss et al., 2009). Therefore, reduced expression of Wnt2 and other mesenchymal Wnt ligands can disrupt lung budding and contribute to lung hypoplasia in Dermol-Foxf1−/−embryos.
In summary, FOXF1 directly activates expression of genes critical for WNT signaling, mesenchyme proliferation and formation of the lung bud. FOXF1 acts downstream of SHH signaling and is required for development of multiple organ systems. Targeting FOXF1 or its effectors could be beneficial for treatment of human congenital disorders, including ACD/MPV.
Supplementary Material
FOXF1 staining was observed in mesenchyme and smooth muscle cells surrounding esophagus and trachea (A), stomach (B) and intestine (C) of E13.5 wild type embryos. FOXF1 was not expressed in smooth muscle cells surrounding artery (D). There was no FOXF1 staining in myocardium (E) or skeletal muscle (F). Abbreviations: Eso, esophagus; Tr, trachea; St, stomach; In, intestine; Ar, artery; LV, left ventricle; RV, right ventricle; Sk, skeletal muscle. Magnification: A, C, D and F, x200; B, x100; E, x50.
A, H&E staining shows intraventricular septum defect (arrow) and lack of blood in atriums of E13.5 Dermol-Foxf1−/− hearts. B, Limbs in E13.5 Dermol-Foxf1−/− embryos had all 5 digits (indicated by numbers). Abbreviations: eso, esophagus; tr, trachea; a, artery; LV, left ventricle; RV, right ventricle; RA, right atrium; LA, left atrium. Magnification: A, x20; B, x50.
H&E was used to stain consecutive paraffin sections of E12.5 Dermol-Foxf1−/− and control Foxf1fl/fl hearts. Sections are shown from the top to bottom and from the left to right. Distal parts of Dermol-Foxf1−/− atrial chambers are collapsed (arrows). Magnification is x50.
FOXF1 staining (dark brown) is observed in venous pole mesenchyme (VPM), dorsal mesenchymal protrusion (DMP) and septum transversum mesenchyme (STM) of E12.5 embryos. Slides were counterstained with nuclear fast red (red). High magnification images (boxes) are shown in middle and bottom panels. DMP is absent in Dermol-Foxf1−/− embryos. FOXF1 staining is decreased in VMP and STM of Dermol-Foxf1−/− embryos. Abbreviations: A, atrium; V, ventricle; Li, liver. Magnification: top panels, x50; middle and bottom panels, x400.
Whole lungs were dissected from E11.5 Dermol-Foxf1−/− and control Foxf1fl/fl embryos. Photographs show lung hypoplasia in Dermol-Foxf1−/− embryos (n = 3 embryos in each group).
Representative H&E images of Dermol-Foxf1−/− and control Dermol-Foxf1+/− lungs show no differences in histology at E16.5 (top and middle panels). Immunostaining for SOX2 (red) and SOX9 (green) was used to visualize proximal and distal airways (bottom panels). Magnification: top panels, x200; middle and bottom panels, x400.
A, Immunostaining shows decreased expression of αSMA in trachea and bronchi of Dermol-Foxf1−/− embryos. Co-localization studies were performed using E13.5 lung sections and antibodies against FOXF1, SOX9 and αSMA. Slides were counterstained with DAPI. B-C, The number of αSMA+FOXF1+ cells and the percentage of the airway epithelium circumference covered by αSMA+ cells were calculated using ten x200 microscope fields (n = 3 embryos in each group; * indicates p < 0.05). D, Immunostaining shows normal expression of αSMA in pulmonary arteries (arrows). Slides were counterstained with nuclear fast red. Abbreviations: Tr, trachea; Br, bronchi; Ar, artery. Magnification: top panels in A, x400; bottom panels in A, x300; panels in D, x400.
Co-localization studies were performed using E16.5 paraffin sections and antibodies against FOXF1, αSMA, SOX9, SOX2, E-Cadherin, p63, Ki-67 and NKX2.1. Loss of peritracheal αSMA-positive cells was observed in Dermol-Foxf1−/− embryos. p63 and SOX9 were reduced in trachea (Tr) of Dermol-Foxf1−/− E16.5 embryos compared to Dermol-Foxf1+/− controls. Magnification: top panels in each staining, x300; remaining panels, x1000.
Immunostaining was performed using paraffin sections from El3.5 Dermol-Foxf1−/− and control embryos. FLK1-positive blood vessels were observed in Dermol-Foxfl−/− and control lungs. Apoptosis was not observed in lungs or esophagi of Dermol-Foxf1−/− or control embryos. Inserts show rare apoptotic cells (positive for Cleaved caspase-3) in ganglions adjacent to the spinal cord. Magnification is x400 for panels and x800 for inserts.
RNA-seq was performed using distal tip lung mesenchyme micro-dissected from E10.5 Dermol-Foxf1−/− and control Foxf1fl/fl embryos (n=3 embryos in each group). Biological processes influenced by Dermol-Cre-mediated Foxf1 deletion were identified using ToppGene Suite (http://toppgene.cchmc.org). Statistical significance of each bioprocess is shown using negative log2 transformation of P value.
ChIPseq shows Foxf1-binding sites (arrows) in Egfr, Igf1, Wnt5a and Wnt11 genes. Whole E18.5 lungs were used for ChIPseq. Data were aligned using the Biowardrobe platform.
Immunostaining was performed using antibody to the activated form of β-catenin (phospho-Ser552) and paraffin sections from E13.5 (A) and E16.5 (B) Dermol-Foxf1−/− embryos. Dermol-Foxf1+/− littermates were used as controls. Arrowheads show epithelial cells containing nuclear β-catenin. Magnification: top panels in A, x200; bottom panels in A, x800; panels in B, x400; inserts in B, x1000.
Immunostaining shows decreased expression of p63, SOX2 and CK14 in esophagus (Eso) of smMHC-Cre Foxf1−/− newborn mice (abbreviated as smFoxf1−/−). smMHC-Cre Foxf1+/− littermates (abbreviated as smFoxf1+/−) were used for comparison. Slides were counterstained with nuclear fast red. Magnification is x1000.
Immunostaining was performed using lung paraffin sections from smMHC-Cre Foxf1−/− newborn mice (abbreviated as smFoxf1−/−). smMHC-Cre Foxf1+/− littermates (abbreviated as smFoxf1+/−) were used as controls. Proximal (top panels) and distal airways (bottom panels) are shown for comparison. Slides were counterstained with nuclear fast red. Abbreviations: Br, bronchi; Ar, artery. Magnification is x400.
A, Immunostaining shows decreased Ki-67 staining in peribronchial smooth muscle of smMHC-Cre Foxf1−/− newborn mice (abbreviated as smFoxf1−/−). smMFIC-Cre Foxf1+/− littermates (abbreviated as smFoxf1+/−) were used as controls. Slides were counterstained with nuclear fast red. Magnification is x400. B, The number of Ki-67+ smooth muscle cells was counted using ten x400 microscope fields (n = 3 embryos in each group; * indicates p < 0.05). Abbreviations: Br, bronchi.
Paraffin sections from smMFIC-Cre Foxf1−/− newborn mice (abbreviated as smFoxf1−/−) were used for H&E staining and immunostaining for αSMA and γSMA, Abbreviations: A, artery; In, intestine. Magnification: left panels, x200; middle and right panels, x400.
Highlights:
-
-
FOXF1 is required for embryonic development of respiratory, cardiovascular and gastrointestinal organ systems.
-
-
FOXF1 expression in mesenchymal cells is critical for lung budding and branching.
-
-
FOXF1 stimulates cellular proliferation in fetal lung mesenchyme.
-
-
Deletion of Foxf1 from mesenchyme causes the loss of visceral smooth muscle cells and disrupts epithelial development in the trachea and esophagus.
-
-
FOXF1 induces canonical WNT signaling in respiratory epithelium and lung mesenchyme.
ACKNOWLEDGMENTS
This work was supported by NIH Grants HL84151, HL123490 and HL141174 (to V. V. K.) and HL132849 (to T. V. K.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Balli D, Ren X, Chou FS, Cross E, Zhang Y, Kalinichenko VV, Kalin TV, 2012. Foxml transcription factor is required for macrophage migration during lung inflammation and tumor formation. Oncogene 31, 3875–3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop NB, Stankiewicz P, Steinhom RH, 2011. Alveolar capillary dysplasia. American journal of respiratory and critical care medicine 184, 172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte C, Flood HM, Ren X, Jagannathan S, Barski A, Kalin TV, Kalinichenko VV, 2017. FOXF1 transcription factor promotes lung regeneration after partial pneumonectomy. Sci Rep 7, 10690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte C, Ren X, Tomley T, Ustiyan V, Pradhan A, Hoggatt A, Kalin TV, Herring BP, Kalinichenko VV, 2015. Forkhead box F2 regulation of platelet-derived growth factor and myocardin/serum response factor signaling is essential for intestinal development. The Journal of biological chemistry 290, 7563–7575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte C, Whitsett JA, Kalin TV, Kalinichenko VV, 2018. Transcription Factors Regulating Embryonic Development of Pulmonary Vasculature. Adv Anat Embryol Cell Biol 228, 1–20. [DOI] [PubMed] [Google Scholar]
- Bolte C, Zhang Y, Wang IC, Kalin TV, Molkentin JD, Kalinichenko VV, 2011. Expression of Foxml transcription factor in cardiomyocytes is required for myocardial development. PloS one 6, e22217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte C, Zhang Y, York A, Kalin TV, Schultz Jel J, Molkentin JD, Kalinichenko VV, 2012. Postnatal ablation of Foxml from cardiomyocytes causes late onset cardiac hypertrophy and fibrosis without exacerbating pressure overload-induced cardiac remodeling. PloS one 7, e48713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Bolte C, Le T, Goda C, Xu Y, Kalin TV, Kalinichenko VV, 2016. FOXF1 maintains endothelial barrier function and prevents edema after lung injury. Sci Signal 9, ra40. [DOI] [PubMed] [Google Scholar]
- Cardoso WV, 2001. Molecular regulation of lung development. Annual review of physiology 63, 471–494. [DOI] [PubMed] [Google Scholar]
- Cheng XH, Black M, Ustiyan V, Le T, Fulford L, Sridharan A, Medvedovic M, Kalinichenko VV, Whitsett JA, Kalin TV, 2014. SPDEF inhibits prostate carcinogenesis by disrupting a positive feedback loop in regulation of the Foxm1 oncogene. PLoS genetics 10, e1004656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmadhikari AV, Sun JJ, Gogolewski K, Carofino BL, Ustiyan V, Hill M, Majewski T, Szafranski P, Justice MJ, Ray RS, Dickinson ME, Kalinichenko VV, Gambin A, Stankiewicz P, 2016. Lethal lung hypoplasia and vascular defects in mice with conditional Foxf1 overexpression. Biol Open 5, 1595–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMeo TA, Anderson K, Phadke P, Fan C, Perou CM, Naber S, Kuperwasser C, 2009. A novel lung metastasis signature links Wnt signaling with cancer cell self-renewal and epithelial-mesenchymal transition in basal-like breast cancer. Cancer research 69, 5364–5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss AM, Tian Y, Tsukiyama T, Cohen ED, Zhou D, Lu MM, Yamaguchi TP, Morrisey EE, 2009. Wnt2/2b and beta-catenin signaling are necessary and sufficient to specify lung progenitors in the foregut. Developmental cell 17, 290–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley JC, Bellusci S, Perkins D, Hogan BL, 1997. Evidence for the involvement of the Gli gene family in embryonic mouse lung development. Developmental biology 188, 337–348. [DOI] [PubMed] [Google Scholar]
- Havrilak JA, Melton KR, Shannon JM, 2017. Endothelial cells are not required for specification of respiratory progenitors. Developmental biology 427, 93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havrilak JA, Shannon JM, 2015. Branching of lung epithelium in vitro occurs in the absence of endothelial cells. Dev Dyn 244, 553–563. [DOI] [PubMed] [Google Scholar]
- Herriges M, Morrisey EE, 2014. Lung development: orchestrating the generation and regeneration of a complex organ. Development (Cambridge, England) 141, 502–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines EA, Jones MK, Verheyden JM, Harvey JF, Sun X, 2013. Establishment of smooth muscle and cartilage juxtaposition in the developing mouse upper airways. Proceedings of the National Academy of Sciences of the United States of America 110, 19444–19449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AD, Yang XH, Burnicka-Turek O, Bosman JD, Ren X, Steimle JD, Vokes SA, McMahon AP, Kalinichenko VV, Moskowitz IP, 2014. Foxf genes integrate tbx5 and hedgehog pathways in the second heart field for cardiac septation. PLoS genetics 10, e1004604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan BL, Yingling JM, 1998. Epithelial/mesenchymal interactions and branching morphogenesis of the lung. Curr Opin Genet Dev 8, 481–486. [DOI] [PubMed] [Google Scholar]
- Hoggatt AM, Kim JR, Ustiyan V, Ren X, Kalin TV, Kalinichenko VV, Herring BP, 2013. The transcription factor Foxf1 binds to serum response factor and myocardin to regulate gene transcription in visceral smooth muscle cells. The Journal of biological chemistry 288, 28477–28487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalin TV, Meliton L, Meliton AY, Zhu X, Whitsett JA, Kalinichenko VV, 2008. Pulmonary mastocytosis and enhanced lung inflammation in mice heterozygous null for the Foxf1 gene. American journal of respiratory cell and molecular biology 39, 390–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinichenko VV, Gusarova GA, Kim I-M, Shin B, Yoder HM, Clark J, Sapozhnikov AM, Whitsett JA, Costa RH, 2004. Foxf1 Haploinsufficiency Reduces Notch-2 Signaling during Mouse Lung Development. Am J Physiol Lung Cell Mol Physiol. 286, L521–L530. [DOI] [PubMed] [Google Scholar]
- Kalinichenko VV, Gusarova GA, Shin B, Costa R, 2003. The Forkhead Box F1 Transcription Factor is Expressed in Brain and Head Mesenchyme during Mouse Embryonic Development. Gene Expression Patterns 3, 153–158. [DOI] [PubMed] [Google Scholar]
- Kalinichenko VV, Lim L, Beer-Stoltz D, Shin B, Rausa FM, Clark J, Whitsett JA, Watkins SC, Costa RH, 2001a. Defects in Pulmonary Vasculature and Perinatal Lung Hemorrhage in Mice Heterozygous Null for the Forkhead Box f1 transcription factor. Developmental biology 235, 489–506. [DOI] [PubMed] [Google Scholar]
- Kalinichenko VV, Lim L, Shin B, Costa RH, 2001b. Differential Expression of Forkhead Box Transcription Factors Following Butylated Hydroxytoluene Lung Injury. American journal of physiology 280, L695–L704. [DOI] [PubMed] [Google Scholar]
- Kalinichenko VV, Zhou Y, Bhattacharyya D, Kim W, Shin B, Bambal K, Costa RH, 2002a. Haploinsufficiency of the Mouse Forkhead Box f1 Gene Causes Defects in Gall Bladder Development. The Journal of biological chemistry 277, 12369–12374. [DOI] [PubMed] [Google Scholar]
- Kalinichenko VV, Zhou Y, Shin B, Beer-Stoltz D, Watkins SC, A. WJ, Costa RH, 2002b. Wild Type Levels of the Mouse Forkhead Box f1 Gene are Essential for Lung Repair. Am J Physiol Lung Cell Mol Physiol. 282, L1253–L1265. [DOI] [PubMed] [Google Scholar]
- Kim IM, Zhou Y, Ramakrishna S, Hughes DE, Solway J, Costa RH, Kalinichenko VV, 2005. Functional characterization of evolutionary conserved DNA regions in forkhead box f1 gene locus. The Journal of biological chemistry 280, 37908–37916. [DOI] [PubMed] [Google Scholar]
- Lee JH, Tammela T, Hofree M, Choi J, Marjanovic ND, Han S, Canner D, Wu K, Paschini M, Bhang DH, Jacks T, Regev A, Kim CF, 2017. Anatomically and Functionally Distinct Lung Mesenchymal Populations Marked by Lgr5 and Lgr6. Cell 170, 1149–1163 e1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L, Kalinichenko VV, Whitsett JA, Costa RH, 2002. Fusion of right lung lobes and pulmonary vessels in mice heterozygous for the Forkhead Box f1 targeted allele. American journal of physiology 282, L1012–L1022. [DOI] [PubMed] [Google Scholar]
- Madison BB, McKenna LB, Dolson D, Epstein DJ, Kaestner KH, 2009. FoxF1 and FoxL1 link hedgehog signaling and the control of epithelial proliferation in the developing stomach and intestine. The Journal of biological chemistry 284, 5936–5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlapuu M, Enerback S, Carlsson P, 2001a. Haploinsufficiency of the forkhead gene Foxf1, a target for sonic hedgehog signaling, causes lung and foregut malformations. Development (Cambridge, England) 128, 2397–2406. [DOI] [PubMed] [Google Scholar]
- Mahlapuu M, Ormestad M, Enerback S, Carlsson P, 2001b. The forkhead transcription factor Foxf1 is required for differentiation of extra-embryonic and lateral plate mesoderm. Development (Cambridge, England) 128, 155–166. [DOI] [PubMed] [Google Scholar]
- Mahlapuu M, Pelto-Huikko M, Aitola M, Enerback S, Carlsson P, 1998. FREAC-1 contains a cell-type-specific transcriptional activation domain and is expressed in epithelial-mesenchymal interfaces [Errata published Dev. Biol. (1999) 207:476]. Developmental biology 202, 183–195. [DOI] [PubMed] [Google Scholar]
- Malin D, Kim IM, Boetticher E, Kalin TV, Ramakrishna S, Meliton L, Ustiyan V, Zhu X, Kalinichenko VV, 2007. Forkhead box F1 is essential for migration of mesenchymal cells and directly induces integrin-beta3 expression. Molecular and cellular biology 27, 2486–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milewski D, Balli D, Ustiyan V, Le T, Dienemann H, Warth A, Breuhahn K, Whitsett JA, Kalinichenko VV, Kalin TV, 2017a. FOXM1 activates AGR2 and causes progression of lung adenomas into invasive mucinous adenocarcinomas. PLoS genetics 13, e1007097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milewski D, Pradhan A, Wang X, Cai Y, Le T, Turpin B, Kalinichenko VV, Kalin TV, 2017b. FoxF1 and FoxF2 transcription factors synergistically promote rhabdomyosarcoma carcinogenesis by repressing transcription of p21Cip1 CDK inhibitor. Oncogene 36, 850–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller LA, Wert SE, Clark JC, Xu Y, Perl AK, Whitsett JA, 2004. Role of Sonic hedgehog in patterning of tracheal-bronchial cartilage and the peripheral lung. Dev Dyn 231, 57–71. [DOI] [PubMed] [Google Scholar]
- Morrisey EE, Hogan BL, 2010. Preparing for the first breath: genetic and cellular mechanisms in lung development. Developmental cell 18, 8–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motoyama J, Liu J, Mo R, Ding Q, Post M, Hui CC, 1998. Essential function of Gli2 and Gli3 in the formation of lung, trachea and oesophagus. Nature genetics 20, 54–57. [DOI] [PubMed] [Google Scholar]
- Ormestad M, Astorga J, Landgren H, Wang T, Johansson BR, Miura N, Carlsson P, 2006. Foxf1 and Foxf2 control murine gut development by limiting mesenchymal Wnt signaling and promoting extracellular matrix production. Development (Cambridge, England) 133, 833–843. [DOI] [PubMed] [Google Scholar]
- Peng T, Tian Y, Boogerd CJ, Lu MM, Kadzik RS, Stewart KM, Evans SM, Morrisey EE, 2013. Coordination of heart and lung co-development by a multipotent cardiopulmonary progenitor. Nature 500, 589–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RS, Lim L, Ye H, Zhou H, Overdier DG, Costa RH, 1997. The winged helix transcriptional activator HFH-8 is expressed in the mesoderm of the primitive streak stage of mouse embryos and its cellular derivatives. Mech. Dev 69, 53–69. [DOI] [PubMed] [Google Scholar]
- Pradhan A, Ustiyan V, Zhang Y, Kalin TV, Kalinichenko VV, 2016. Forkhead transcription factor FoxF1 interacts with Fanconi anemia protein complexes to promote DNA damage response. Oncotarget 7, 1912–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin SA, Han L, McCracken KW, Kenny AP, Anglin CT, Grigg EA, Crawford CM, Wells JM, Shannon JM, Zorn AM, 2016. A Retinoic Acid-Hedgehog Cascade Coordinates Mesoderm-Inducing Signals and Endoderm Competence during Lung Specification. Cell Rep 16, 66–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Shah TA, Ustiyan V, Zhang Y, Shinn J, Chen G, Whitsett JA, Kalin TV, Kalinichenko VV, 2013. FOXM1 promotes allergen-induced goblet cell metaplasia and pulmonary inflammation. Molecular and cellular biology 33, 371–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Ustiyan V, Pradhan A, Cai Y, Havrilak JA, Bolte CS, Shannon JM, Kalin TV, Kalinichenko VV, 2014. FOXF1 transcription factor is required for formation of embryonic vasculature by regulating VEGF signaling in endothelial cells. Circulation research 115, 709–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Zhang Y, Snyder J, Cross ER, Shah TA, Kalin TV, Kalinichenko VV, 2010. Forkhead box M1 transcription factor is required for macrophage recruitment during liver repair. Molecular and cellular biology 30, 5381–5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen P, Dharmadhikari AV, Majewski T, Mohammad MA, Kalin TV, Zabielska J, Ren X, Bray M, Brown HM, Welty S, Thevananther S, Langston C, Szafranski P, Justice MJ, Kalinichenko VV, Gambin A, Belmont J, Stankiewicz P, 2014. Comparative analyses of lung transcriptomes in patients with alveolar capillary dysplasia with misalignment of pulmonary veins and in foxf1 heterozygous knockout mice. PloS one 9, e94390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen P, Yang Y, Navarro C, Silva I, Szafranski P, Kolodziejska KE, Dharmadhikari AV, Mostafa H, Kozakewich H, Kearney D, Cahill JB, Whitt M, Bilic M, Margraf L, Charles A, Goldblatt J, Gibson K, Lantz PE, Garvin AJ, Petty J, Kiblawi Z, Zuppan C, McConkie-Rosell A, McDonald MT, Peterson-Carmichael SL, Gaede JT, Shivanna B, Schady D, Friedlich PS, Hays SR, Palafoll IV, Siebers-Renelt U, Bohring A, Finn LS, Siebert JR, Galambos C, Nguyen L, Riley M, Chassaing N, Vigouroux A, Rocha G, Fernandes S, Brumbaugh J, Roberts K, Ho-Ming L, Lo IF, Lam S, Gerychova R, Jezova M, Valaskova I, Fellmann F, Afshar K, Giannoni E, Muhlethaler V, Liang J, Beckmann JS, Lioy J, Deshmukh H, Srinivasan L, Swarr DT, Sloman M, Shaw-Smith C, van Loon RL, Hagman C, Sznajer Y, Barrea C, Galant C, Detaille T, Wambach JA, Cole FS, Hamvas A, Prince LS, Diderich KE, Brooks AS, Verdijk RM, Ravindranathan H, Sugo E, Mowat D, Baker ML, Langston C, Welty S, Stankiewicz P, 2013. Novel FOXF1 mutations in sporadic and familial cases of alveolar capillary dysplasia with misaligned pulmonary veins imply a role for its DNA binding domain. Human mutation 34, 801–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta A, Kalinichenko VV, Yutzey KE, 2013. FoxO1 and FoxM1 Transcription Factors Have Antagonistic Functions in Neonatal Cardiomyocyte Cell-Cycle Withdrawal and IGF1 Gene Regulation. Circulation research 112, 267–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snarr BS, Wirrig EE, Phelps AL, Trusk TC, Wessels A, 2007. A spatiotemporal evaluation of the contribution of the dorsal mesenchymal protrusion to cardiac development. Dev Dyn 236, 1287–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Ren X, Wang IC, Pradhan A, Zhang Y, Flood HM, Han B, Whitsett JA, Kalin TV, Kalinichenko VV, 2017. The FOXM1 inhibitor RCM-1 suppresses goblet cell metaplasia and prevents IL-13 and STAT6 signaling in allergen-exposed mice. Sci Signal 10, aai8583. [DOI] [PubMed] [Google Scholar]
- Szafranski P, Dharmadhikari AV, Brosens E, Gurha P, Kolodziejska KE, Zhishuo O, Dittwald P, Majewski T, Mohan KN, Chen B, Person RE, Tibboel D, de Klein A, Pinner J, Chopra M, Malcolm G, Peters G, Arbuckle S, Guiang SF 3rd, Hustead VA, Jessurun J, Hirsch R, Witte DP, Maystadt I, Sebire N, Fisher R, Langston C, Sen P, Stankiewicz P, 2013. Small noncoding differentially methylated copy-number variants, including lncRNA genes, cause a lethal lung developmental disorder. Genome research 23, 23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustiyan V, Wert SE, Ikegami M, Wang IC, Kalin TV, Whitsett JA, Kalinichenko VV, 2012. Foxm1 transcription factor is critical for proliferation and differentiation of Clara cells during development of conducting airways. Developmental biology 370, 198–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustiyan V, Zhang Y, Perl AK, Whitsett JA, Kalin TV, Kalinichenko VV, 2016. beta-catenin and Kras/Foxm1 signaling pathway are critical to restrict Sox9 in basal cells during pulmonary branching morphogenesis. Dev Dyn 245, 590–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IC, Snyder J, Zhang Y, Lander J, Nakafuku Y, Lin J, Chen G, Kalin TV, Whitsett JA, Kalinichenko VV, 2012. Foxm1 Mediates Cross Talk between Kras/Mitogen-Activated Protein Kinase and Canonical Wnt Pathways during Development of Respiratory Epithelium. Molecular and cellular biology 32, 3838–3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IC, Ustiyan V, Zhang Y, Cai Y, Kalin TV, Kalinichenko VV, 2014. Foxml transcription factor is required for the initiation of lung tumorigenesis by oncogenic Kras(G12D.). Oncogene 33, 5391–5396. [DOI] [PubMed] [Google Scholar]
- Wang IC, Zhang Y, Snyder J, Sutherland MJ, Burhans MS, Shannon JM, Park HJ, Whitsett JA, Kalinichenko VV, 2010. Increased expression of FoxM1 transcription factor in respiratory epithelium inhibits lung sacculation and causes Clara cell hyperplasia. Developmental biology 347, 301–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Bhattacharyya D, Dennewitz MB, Zhou Y, Kalinichenko VV, Lepe R, Costa RH, 2003. Rapid Hepatocyte Nuclear Translocation of the Forkhead Box M1B (FoxM1B) Transcription factor Causes a Transient Increase in Size of Regenerating Transgenic Hepatocytes. Gene Expression 11, 149–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton D, Zhao J, Berberich MA, Bernfield M, 1999. Molecular embryology of the lung: then, now, and in the future. The American journal of physiology 276, L697–704. [DOI] [PubMed] [Google Scholar]
- Weaver M, Batts L, Hogan BL, 2003. Tissue interactions pattern the mesenchyme of the embryonic mouse lung. Developmental biology 258, 169–184. [DOI] [PubMed] [Google Scholar]
- Weiler SME, Pinna F, Wolf T, Lutz T, Geldiyev A, Sticht C, Knaub M, Thomann S, Bissinger M, Wan S, Rossler S, Becker D, Gretz N, Lang H, Bergmann F, Ustiyan V, Kalin TV, Singer S, Lee JS, Marquardt JU, Schirmacher P, Kalinichenko VV, Breuhahn K, 2017. Induction of Chromosome Instability by Activation of Yes-Associated Protein and Forkhead Box M1 in Liver Cancer. Gastroenterology 152, 2037–2051 e2022. [DOI] [PubMed] [Google Scholar]
- Whitsett JA, Wert SE, Trapnell BC, 2004. Genetic disorders influencing lung formation and function at birth. Hum Mol Genet 13 Spec No 2, R207–R215. [DOI] [PubMed] [Google Scholar]
- Xia H, Ren X, Bolte CS, Ustiyan V, Zhang Y, Shah TA, Kalin TV, Whitsett JA, Kalinichenko VV, 2015. Foxm1 regulates resolution of hyperoxic lung injury in newborns. American journal of respiratory cell and molecular biology 52, 611–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin HB, Deng KY, Rishniw M, Ji G, Kotlikoff MI, 2002. Smooth muscle expression of Cre recombinase and eGFP in transgenic mice. Physiol Genomics 10, 211–215. [DOI] [PubMed] [Google Scholar]
- Xu J, Liu H, Lan Y, Aronow BJ, Kalinichenko VV, Jiang R, 2016. A Shh-Foxf-Fgf18-Shh Molecular Circuit Regulating Palate Development. PLoS genetics 12, e1005769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Xu J, Liu Z, Sosic D, Shao J, Olson EN, Towler DA, Ornitz DM, 2003. Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development (Cambridge, England) 130, 3063–3074. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FOXF1 staining was observed in mesenchyme and smooth muscle cells surrounding esophagus and trachea (A), stomach (B) and intestine (C) of E13.5 wild type embryos. FOXF1 was not expressed in smooth muscle cells surrounding artery (D). There was no FOXF1 staining in myocardium (E) or skeletal muscle (F). Abbreviations: Eso, esophagus; Tr, trachea; St, stomach; In, intestine; Ar, artery; LV, left ventricle; RV, right ventricle; Sk, skeletal muscle. Magnification: A, C, D and F, x200; B, x100; E, x50.
A, H&E staining shows intraventricular septum defect (arrow) and lack of blood in atriums of E13.5 Dermol-Foxf1−/− hearts. B, Limbs in E13.5 Dermol-Foxf1−/− embryos had all 5 digits (indicated by numbers). Abbreviations: eso, esophagus; tr, trachea; a, artery; LV, left ventricle; RV, right ventricle; RA, right atrium; LA, left atrium. Magnification: A, x20; B, x50.
H&E was used to stain consecutive paraffin sections of E12.5 Dermol-Foxf1−/− and control Foxf1fl/fl hearts. Sections are shown from the top to bottom and from the left to right. Distal parts of Dermol-Foxf1−/− atrial chambers are collapsed (arrows). Magnification is x50.
FOXF1 staining (dark brown) is observed in venous pole mesenchyme (VPM), dorsal mesenchymal protrusion (DMP) and septum transversum mesenchyme (STM) of E12.5 embryos. Slides were counterstained with nuclear fast red (red). High magnification images (boxes) are shown in middle and bottom panels. DMP is absent in Dermol-Foxf1−/− embryos. FOXF1 staining is decreased in VMP and STM of Dermol-Foxf1−/− embryos. Abbreviations: A, atrium; V, ventricle; Li, liver. Magnification: top panels, x50; middle and bottom panels, x400.
Whole lungs were dissected from E11.5 Dermol-Foxf1−/− and control Foxf1fl/fl embryos. Photographs show lung hypoplasia in Dermol-Foxf1−/− embryos (n = 3 embryos in each group).
Representative H&E images of Dermol-Foxf1−/− and control Dermol-Foxf1+/− lungs show no differences in histology at E16.5 (top and middle panels). Immunostaining for SOX2 (red) and SOX9 (green) was used to visualize proximal and distal airways (bottom panels). Magnification: top panels, x200; middle and bottom panels, x400.
A, Immunostaining shows decreased expression of αSMA in trachea and bronchi of Dermol-Foxf1−/− embryos. Co-localization studies were performed using E13.5 lung sections and antibodies against FOXF1, SOX9 and αSMA. Slides were counterstained with DAPI. B-C, The number of αSMA+FOXF1+ cells and the percentage of the airway epithelium circumference covered by αSMA+ cells were calculated using ten x200 microscope fields (n = 3 embryos in each group; * indicates p < 0.05). D, Immunostaining shows normal expression of αSMA in pulmonary arteries (arrows). Slides were counterstained with nuclear fast red. Abbreviations: Tr, trachea; Br, bronchi; Ar, artery. Magnification: top panels in A, x400; bottom panels in A, x300; panels in D, x400.
Co-localization studies were performed using E16.5 paraffin sections and antibodies against FOXF1, αSMA, SOX9, SOX2, E-Cadherin, p63, Ki-67 and NKX2.1. Loss of peritracheal αSMA-positive cells was observed in Dermol-Foxf1−/− embryos. p63 and SOX9 were reduced in trachea (Tr) of Dermol-Foxf1−/− E16.5 embryos compared to Dermol-Foxf1+/− controls. Magnification: top panels in each staining, x300; remaining panels, x1000.
Immunostaining was performed using paraffin sections from El3.5 Dermol-Foxf1−/− and control embryos. FLK1-positive blood vessels were observed in Dermol-Foxfl−/− and control lungs. Apoptosis was not observed in lungs or esophagi of Dermol-Foxf1−/− or control embryos. Inserts show rare apoptotic cells (positive for Cleaved caspase-3) in ganglions adjacent to the spinal cord. Magnification is x400 for panels and x800 for inserts.
RNA-seq was performed using distal tip lung mesenchyme micro-dissected from E10.5 Dermol-Foxf1−/− and control Foxf1fl/fl embryos (n=3 embryos in each group). Biological processes influenced by Dermol-Cre-mediated Foxf1 deletion were identified using ToppGene Suite (http://toppgene.cchmc.org). Statistical significance of each bioprocess is shown using negative log2 transformation of P value.
ChIPseq shows Foxf1-binding sites (arrows) in Egfr, Igf1, Wnt5a and Wnt11 genes. Whole E18.5 lungs were used for ChIPseq. Data were aligned using the Biowardrobe platform.
Immunostaining was performed using antibody to the activated form of β-catenin (phospho-Ser552) and paraffin sections from E13.5 (A) and E16.5 (B) Dermol-Foxf1−/− embryos. Dermol-Foxf1+/− littermates were used as controls. Arrowheads show epithelial cells containing nuclear β-catenin. Magnification: top panels in A, x200; bottom panels in A, x800; panels in B, x400; inserts in B, x1000.
Immunostaining shows decreased expression of p63, SOX2 and CK14 in esophagus (Eso) of smMHC-Cre Foxf1−/− newborn mice (abbreviated as smFoxf1−/−). smMHC-Cre Foxf1+/− littermates (abbreviated as smFoxf1+/−) were used for comparison. Slides were counterstained with nuclear fast red. Magnification is x1000.
Immunostaining was performed using lung paraffin sections from smMHC-Cre Foxf1−/− newborn mice (abbreviated as smFoxf1−/−). smMHC-Cre Foxf1+/− littermates (abbreviated as smFoxf1+/−) were used as controls. Proximal (top panels) and distal airways (bottom panels) are shown for comparison. Slides were counterstained with nuclear fast red. Abbreviations: Br, bronchi; Ar, artery. Magnification is x400.
A, Immunostaining shows decreased Ki-67 staining in peribronchial smooth muscle of smMHC-Cre Foxf1−/− newborn mice (abbreviated as smFoxf1−/−). smMFIC-Cre Foxf1+/− littermates (abbreviated as smFoxf1+/−) were used as controls. Slides were counterstained with nuclear fast red. Magnification is x400. B, The number of Ki-67+ smooth muscle cells was counted using ten x400 microscope fields (n = 3 embryos in each group; * indicates p < 0.05). Abbreviations: Br, bronchi.
Paraffin sections from smMFIC-Cre Foxf1−/− newborn mice (abbreviated as smFoxf1−/−) were used for H&E staining and immunostaining for αSMA and γSMA, Abbreviations: A, artery; In, intestine. Magnification: left panels, x200; middle and right panels, x400.