Abstract
Executive functions are a diverse and critical suite of cognitive abilities that are often disrupted in individuals with psychiatric disorders. Despite their moderate to high heritability, little is known about the molecular genetic factors that contribute to variability in executive functions and how these factors may be related to those that predispose to psychiatric disorders. We examined the relationship between polygenic risk scores built from large genome-wide association studies of psychiatric disorders and executive functioning in typically developing children. In our discovery sample (N = 417), consistent with previous reports on general cognitive abilities, polygenic risk for autism spectrum disorder was associated with better performance on the Dimensional Change Card Sort test from the NIH Cognition Toolbox, with the largest effect in the youngest children. Polygenic risk for major depressive disorder was associated with poorer performance on the Flanker test in the same sample. This second association replicated for performance on the Penn Conditional Exclusion Test in an independent cohort (N = 3681). Our results suggest that the molecular genetic factors contributing to variability in executive function during typical development are at least partially overlapping with those associated with psychiatric disorders, although larger studies and further replication are needed.
Keywords: autism, depression, development, executive function, neuropsychology, polygenic risk scores
1 | INTRODUCTION
Executive functions encompass diverse cognitive abilities, including flexibility, inhibitory control, abstraction, fluency, selective attention and working memory. The quintessentially human instantiation of these skills not only sets us apart from the rest of the animal kingdom, but also plays an integral role in cognitive development. Perhaps because they exhibit protracted maturation, executive functions are particularly variable and vulnerable during childhood and adolescence. Furthermore, deficits in executive functioning are widely reported in psychiatric populations including those affected by attention deficit/hyperactivity disorder1,2 (ADHD), autism spectrum disorder1 (ASD), bipolar disorder3 (BIP), major depressive disorder4 (MDD), schizophrenia5 (SCZ) and others.6 Whether factors associated with risk for psychiatric disorders and resulting executive function disruptions also correlate with executive function performance during typical development remains unknown.
Heritability estimates are moderate to high, ranging from 0.29 to 0.76, for performance on many individual executive function tests.7–9 Correlations in performance across tests are often summarized as having “unity and diversity”10 to note both a task-domain general performance factor that cannot be explained by general cognitive abilities (unity) and also task-domain specific factors (diversity).11,12 The heritability of executive function exhibits this same pattern. Twin and family studies suggest domain-general and domain-specific genetic contributions that appear separable from those affecting general cognitive abilities. Estimates of heritability for some of these latent factors has been as high as 1.7–9 Contrary to quantitative genetics reports, molecular genetic studies are less revealing. A number of candidate genes have been proposed13 but single gene studies provide mixed results and their reliability in small to moderate samples is questionable.14 Genome-wide association studies (GWASs) aiming to scan all common genetic variants are also yet to add significantly to our understanding of individual differences in executive functioning.15–18 It appears that the genetic architecture of executive functions, like other complex cognitive phenotypes,19,20 is diffuse across very many variants (polygenic). Extremely large sample sizes for reliable single variant studies or alternative approaches will be needed to advance understanding of the molecular genetic contributions to executive functioning.
The use of polygenic risk scores (PRSs) is a powerful approach for gaining insights into the genetic architecture of cognitive phenotypes.21,22 PRSs are quantitative scores that index, for each individual subject in a study sample, their aggregate genetic risk for a trait of interest. Specifically, a PRS is computed as the weighted sum counting all risk alleles for a selected set of single nucleotide polymorphisms (SNPs) carried by an individual. The weight used for each risk allele is the SNP log odds ratio estimated out of sample in a large GWAS of the given trait. PRSs are demonstrated to be powerful and reliable indicators not only for genetic contributions to single traits but also for genetic correlations between traits.22 Associating psychiatric PRS with cognitive performance in healthy populations may advance our understanding of the overlap among genetic factors contributing to cognitive deficits emerging through psychiatric illness and those affecting variability in unaffected individuals.23 Although PRSs do not provide the molecular specificity of single locus studies, they can provide important insights into broader aspects of genetic architectures. These broader relationships are important for informing newer analytic approaches exploiting functional hypotheses for improved power at finer scales.24,25
A growing body of work has begun investigating the association between psychiatric PRS and measures of cognitive performance in the general population. Higher ADHD PRSs have been associated with lower performance in IQ, educational achievement, working memory, and language skills in children26–28 and lower IQ, educational attainment and verbal-numerical reasoning in adults.28–30 A number of reports link increased SCZ PRS with lower IQ across the age range,31 but also include negative correlations with attention, reaction time, memory and verbal numerical reasoning in adults30,32 and language skills, verbal reasoning and social cognition in children.33,34 In adults,30 MDD and BIP PRS were negatively associated with reaction time and memory, while MDD was additionally associated with worse verbal-numerical reasoning. While reported PRS for ADHD, BIP, MDD and SCZ have typically produced negative correlations with cognitive performance, ASD is an exception. ASD PRS has been associated with higher IQ in children and adults35,36 and higher educational attainment, memory and verbal fluency in adults.30,35 Although interesting relationships are emerging the results are not yet definitive as inconsistencies exists31,37 and only a limited number of domains have been considered.
Only 2 studies, to our knowledge, have examined the association between PRS for psychiatric outcomes and executive function, per se. Germine et al33 considered the relationship between SCZ PRS and performance on a suite of tests, including 1 executive task. However, in a population of children ascertained from hospitals (Philadelphia Neurodevelopmental Cohort [PNC], described below) only associations with other domains were reported as significant (see above), but a nominally significant trend for SCZ PRS and speed performance on an executive task was observed. Benca et al38 used a population sample of young adults to consider the relationship between PRS for 5 psychiatric disorders and 3 latent executive factors derived from performance on 9 executive tasks. Although the authors did not declare any findings significant study-wide, they did report nominally significant trends for MDD, ADHD and SCZ PRS. While the frequency of studies using PRS to probe the genetic architecture of cognition is growing, their associations with executive functions during typical development, in particular, remain speculative.
The aim of this study was to investigate the association between PRS for psychiatric disorders and executive function performance in typically developing children and adolescents. We used the results from GWAS of 5 psychiatric conditions (ADHD, ASD, BIP, MDD and SCZ) provided by the Psychiatric Genomics Consortium (PGC) to compute disorder-specific PRS. In our primary hypothesis test, we examined the aggregate effects of all psychiatric PRS and their interactions with age on executive function in 417 typically developing individuals from the Pediatric Imaging, Neurocognition and Genetics (PING) study. Because of “unity and diversity” described by quantitative genetic studies, we followed our primary analysis with descriptive, post hoc analyses. The goal of these analyses was to generate novel hypotheses about the potential specificity of the strongest disorder-specific PRS to each of 2 executive function tasks and their independence from effects on more general cognitive abilities. Finally, we selected our strongest findings for replication in a second, complimentary cohort, the PNC. We hypothesized that multiple PRSs would show associations with variability in executive functions, revealing plausible evidence for domain specificity. Importantly, we also explored the understudied, moderating effect of age.
2 | MATERIAL AND METHODS
2.1 | Psychiatric GWAS
The PGC published per SNP summary statistics for GWAS of 5 psychiatric conditions (https://www.med.unc.edu/pgc/results-and-downloads). We obtained the statistics for ADHD39ADHD, ASD40, BIP41, MDD42 and SCZ43 GWAS. The numbers of cases/controls in each study was 2960/4519 (ADHD), 3303/3428 (ASD), 7481/9250 (BIP), 9240/9519 (MDD) and 34 241/45 604 (SCZ). The statistics provide per SNP odds ratios for 1 206 462, 9 499 590, 2 427 221, 1 235 110 and 9 444 231 SNPs, respectively. Odds ratios were natural log transformed (reconstituting the beta estimate from a logistic regression) for downstream analysis. Table 1 describes these data.
TABLE 1.
GWAS summaries
| Trait | GWAS | SNPs for PRS | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||
| GWAS | h2 | h2chip | PRS r2 | Cases | Controls | SNPs | PING | PNC |
| ADHDa | 0.76f | 0.28k | 0.001l | 2960 | 4519 | 1 206 462 | 5363 | — |
|
| ||||||||
| ASDb | 0.90g | 0.17k | *0.008m | 3303 | 3428 | 9 499 590 | 10 179 | 3787 |
|
| ||||||||
| BIPc | 0.90h | 0.25k | 0.028 | 7481 | 9250 | 2 427 221 | 13 965 | — |
|
| ||||||||
| MDDd | 0.31–0.42i | 0.21k | *0.006 | 9240 | 9519 | 1 235 110 | 5622 | 3752 |
|
| ||||||||
| SCZe | 0.81j | 0.23k | 0.184 | 34 241 | 45 604 | 9 444 231 | 17 119 | — |
The heritability (h2) and chip heritability (h2chip) are representative values for each disorder and suggest a large contribution of common genetic factors to disease liability. The within-trait PRS predictive power (PRS r2) of the GWAS results varies approximately according to the sample sizes.
Note that the VE by PRS reported here are likely underestimates for the PRS used here. The ASD and MDD GWAS used in these studies to create PRS were subsets (40% and 80%, respectively) of the sample used for this study.
Neale et al39.
Cross-Disorder Group of the Psychiatric Genomics40.
The Psychiatric GWAS Consortium Bipolar Disorder Working Group41.
Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium et al42.
Schizophrenia Working Group of the Psychiatric Genomics Consortium43.
Faraone and Mick44.
Freitag45.
Craddock and Sklar46.
Sullivan et al47.
Sullivan et al48.
Cross-Disorder Group of the Psychiatric Genomics Consortium et al49.
Hamshere et al50.
Anney et al51.
2.2 | The PING cohort
The PING study (http://pingstudy.ucsd.edu/Data.php)52 created a comprehensive, publicly shared, data resource for studying standardized assessments of neurocognition, neuroimaging and genetics in typically developing children. Cross-sectional measurements on 1493 individuals ranging in age from 3 to 21 years were aggregated from sites across the United States. The cohort is described fully elsewhere.52,53 Relevant to this study, subjects were excluded only for known history of neurological disorders, head trauma, preterm birth, severe psychiatric diagnosis (autism spectrum, SCZ or BIPs), intellectual disability, pregnancy, maternal daily drug or alcohol use during pregnancy or incompatibility with MRI (i.e. braces, pregnancy, claustrophobia, etc.). ADHD, general or specific learning disabilities, and/or depression, confirmed or suspected, were not exclusionary as these are fairly common in developing populations. However, no testing was conducted to screen for these conditions and therefore verification of a diagnosis or identification of additional participants who may have met criteria was not possible. Subjects were enrolled from the greater metropolitan areas of Baltimore, Boston, Honolulu, Los Angeles, New Haven, New York, Sacramento and San Diego. Each subject's medical, developmental, behavioral history, as well as family medical history and environment were obtained from parental questionnaires. Socioeconomic status (SES) was recorded as a 7-point scale rating parental education from “less than seven years” to “professional degree,” and a 12-point scale rating annual familial income from “less than $5,000” to “over $300 000.”
Neurocognitive performance was assessed using the NIH Toolbox Cognition Battery (NTCB, http://www.nihtoolbox.org/,53,54), a computerized battery designed for administration across the life span. The NTCB includes 8 subtests spanning 6 domains. In this study, we included the 2 measures of executive function, the Flanker Inhibitory Control and Attention test (Flanker) and the Dimensional Change Card Sort (DCCS) test, and the 2 language measures, the Picture Vocabulary and Oral Reading Recognition tests. Intraclass correlation coefficients of 0.92 for both the DCCS and Flanker tests,55 0.97 for Oral Reading Recognition, and 0.81 for Picture Vocabulary56 indicate that all 4 NTCB tests show excellent test-retest reliability. Test scores were adjusted by Blom rank order normalization.57 The executive function composite (EFC) score is the average of DCCS and Flanker scores and the verbal composite (VC) score is the average of the Picture Vocabulary and Oral Reading Recognition scores. Details on the Flanker and DCCS are in the Appendix S1, Supporting Information.
A total of 550 000 SNPs were genotyped from saliva samples using the Illumina Human660W-Quad BeadChip. Genotyped SNPs were imputed58 to 6 492 742 expected allelic dosages. Imputations were performed with MaCH,59 minimac60 and phased haplotypes from European subjects in 1000 Genomes Project Phase 1.61 Included dosages had r2 quality >0.3, minor allele frequency >1%, per subject missingness <1%, were autosomal, and had unambiguous strand alignment (A/T, C/G SNPs removed). Of the 1493 subjects, 1019 had acceptable NTCB, genotype and covariate data.
Analyses were restricted to PING subjects of European genetic ancestry (the same as the psychiatric GWAS) with no familial relationships. Genetic ancestry was determined using smartPCA routines in EIGENSTRAT62 on the 1019 PING subjects combined with 1224 individuals with known genetic ancestry. Reference individuals sampled HapMap,63 1000 Genomes61 and IntraGen64 databases. Subjects with European genetic ancestry had scores on the first 10 principal components (PCs) of genetic similarity within 5 SDs of the mean of reference individuals with known European ancestry, leaving 463 subjects (Figure S1). Familial relatedness was determined from estimates of genome-wide identity by descent (IBD) among the remaining subjects using GCTA.65 The 417 final subjects (191 female) were selected such that no pair had estimated IBD above 0.08 (Figure S2). The first 10 PCs recomputed with smartPCA on the final subjects were kept as covariates for residual genetic ancestry.62
PRSs were computed for each psychiatric GWAS following a standard approach22 with parameters chosen to mimic a recent exemplar.43 We intersected the 6 492 742 imputed SNPs in PING with each GWAS, randomly pruning the 5 sets so no pair of SNPs within 500 kb had r2 linkage disequilibrium above 0.1. Only those with a P value <.05 in the GWAS were retained leaving 5363 (ADHD), 10 179 (ASD), 13 965 (BP), 5622 (MDD) and 17 119 (SCZ) SNPs. For each subject, we computed PRS as the log odds weighted sum of imputed SNP minor allele counts, for each psychiatric GWAS as the PRS. Computations were carried out with the “score” function in plink1.9.66,67 Correlations among scores are in Figure S5.
Associations were performed using R version 3.1.68 A “baseline” regression model predicted EFC from 23 covariates: age at neuropsychological testing, age squared, gender, 8 dummy variables for 9 acquisition sites, the 2 SES measures and 10 ancestry PCs. The “full” model included the 23 covariates plus all PRSs and their interactions with age and age squared (15 additional terms). Coefficient estimates are reported from the full model. Age, age squared and PRS were mean-centered prior to fitting. The primary hypothesis test was a likelihood ratio test (LRT) comparing the full model with the baseline model. For descriptive purposes, the covariates and PRS terms are divided into categories and presented hierarchically in Table 2 with P values from LRTs on nested hierarchical models.
TABLE 2.
Primary hypothesis test
| Model | Res. DF | RSS | DF | SS | R2 | P (LRT) | |
|---|---|---|---|---|---|---|---|
| Intercept only | 416 | 384.5 | |||||
| +Age | 414 | 119.8 | 2 | 264.61 | 0.6883 | 3.16 × 10−199 | |
| +Gender | 413 | 119.8 | 1 | 7.19 × 10−5 | 0.6883 | 0.9874 | |
| +Environment | 403 | 111.1 | 10 | 8.71 | 0.711 | 4.7 × 10−4 | |
| +Genetic background | 393 | 105.1 | 10 | 6.01 | 0.7266 | 0.0128 | 4.92 × 10−206 |
| +PRS main effects | 388 | 102.1 | 5 | 3.07 | 0.7346 | 0.0397 | |
| +PRS age interactions | 378 | 97.6 | 10 | 4.5 | 0.7463 | 0.0654 | 0.0147 |
| −Omitted PRS terms | 389 | 101.3 | −11 | −3.72 | 0.7366 | 0.2117 |
Adding the joint effects of the 5 PRS (+PRS main effects, +PRS age interactions) is a significant improvement to the explanatory power of the model when compared to the baseline model (intercept, +age covariates, +gender covariates, +environmental covariates, +genetic background covariates). Breaking down the terms into themed groups suggests all covariates except gender are important aspects of the baseline model and the main effects of the PRS are more predictive than their age interactions. Removing the terms not considered for post hoc analysis (−omitted PRS terms) did not significantly reduce the fit of the model.
Post hoc tests were conditional on significance in the primary test and presented to describe the effects of specific PRS along with age and age squared interactions. Variables with P < .1 in the full model were selected for follow-up. These analyses (Figure 1) depict the differences in variance explained (VE) (r2) when adding the PRS or PRS plus interaction terms to the baseline model, with P values reported from nested LRTs. Analyses were repeated including VC in the baseline model according to the same procedure. Post hoc test P values are presented uncorrected for multiple testing. The tests are dependent on significance in the primary test, highly intercorrelated (i.e. the ASD PRS effect on DCCS with and without VC are essentially redundant tests) and meant to be interpreted as descriptive, generating novel hypotheses to be confirmed in independent studies.
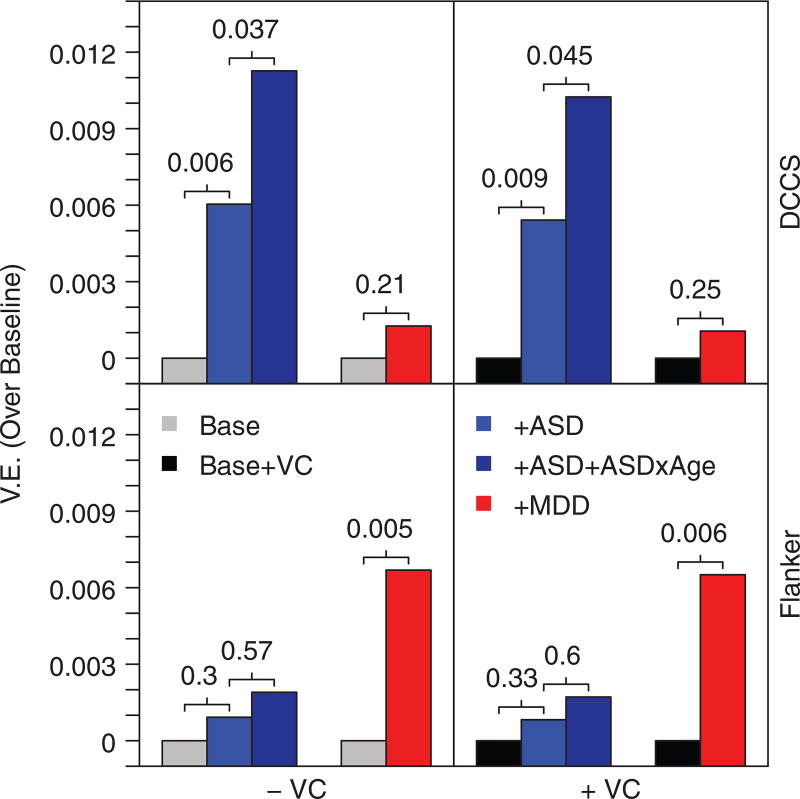
FIGURE 1.
ASD and MDD PRS associate with different executive components in the PING cohort. Bar heights reflect increments in VE over the baseline (covariates only) model. Numbers indicate P values obtained from LRTs that compare the models sequentially, left to right. VE, variance explained; ASD, autism spectrum disorder PRS; MDD, major depressive disorder PRS; VC, verbal composite; DCCS, dimensional change card sort
2.3 | The PNC cohort
The PNC was accessed through dbGAP (accession number phs000607.v1.p1). Full descriptions of the cohort, subject acquisition and protocols are available elsewhere.69,70 Briefly, 8741 subjects ages 8 to 21 were selected from approximately 50 000 recruited from the Children's Hospital of Philadelphia or affiliated clinics in the greater Philadelphia area. Inclusion required only an ability to consent and complete interviews and testing. A computerized structured screen modeled after the Kiddie-Schedule for Affective Disorders and Schizophrenia71 was administered to each subject to assess the symptoms of potential psychopathology including mood, anxiety, behavioral, eating, psychosis or substance use issues. Subjects were given a medical rating derived from this interview to summarize the relative severity of symptom reports from none (0) or minor without CNS impact (1) to major (4). Clinical administered medical diagnoses for psychiatric conditions, however, were not available.
Neurocognitive abilities were assessed using the Penn Computerized Neurocognitive Battery.69,72,73 Executive function was assessed by the Penn Conditional Exclusion Test (PCET; details in Supporting Information).74 Direct measures of reliability are not currently available for the PNC version of the PCET. Scores from the Reading subtest from the Wide Range Achievement Test (WRAT Reading score) were also available for each subject.
Six arrays were used for genotyping: Affymetrix Human SNP Array 6.0 (N = 65, SNPs = 826 525), Affymetrix Axiom Genotyping Array (N = 711, SNPs = 517 744), Illumina Omni Array (N = 1653, SNPs = 699 239), Illumina Human 610 (N = 3702, SNPs = 480 247), Illumina Human Hap 550v1 (N = 548, SNPs = 522 609) and Illumina Human Hap 550v3 (N = 1861, SNPs = 488 715). We excluded subjects genotyped on the Affymetrix arrays for low numbers and low SNP overlap and subjects with the Illumina Human Hap 550v1 array for unresolvable artifacts. The same smartPCA and GCTA routines selected unrelated, European genetic ancestry subjects (Figure S3) and created ancestry covariate PCs. A total of 3681 subjects (1884 female) with 224 444 overlapping genotypes were used for replication. ASD and MDD PRS were computed according to the same protocol as above from 3787 and 3752 SNPs, respectively.
The PNC “baseline” model included 17 covariates: age at neuropsychological testing, age squared, gender, 4 dummy variables for 5 medical ratings and 10 genetic ancestry PCs. Age, age squared and PRS were centered prior to fitting. Replication tests followed the same procedure as in the discovery phase, comparing the fit of the PRS with baseline and interaction with PRS + baseline, sequentially. Coefficients are reported from the most saturated model. Analyses were repeated with the WRAT Reading score in the baseline model.
3 | RESULTS
Tested in aggregate, the PRS for ASD, ADHD, BIP, MDD and SCZ and their interactions with linear and quadratic age explained a small but significant proportion of variance in composite executive function among PING subjects (VE = 1.97%; LRT with 15 degrees of freedom (DF) P = .01; Table 2). Variables in the full model (Table S1) with P < .10 were chosen for follow-up: the main effect of MDD PRS (β = −0.0429, SE = 0.0167, P = .011), the main effect of ASD PRS (β = 0.0269, SE = 0.0107, P = .012) and the linear (β = 0.0210, SE = 0.0119, P = .078) and quadratic (β = −0.0009, SE = 0.0004, P = .064) age interactions with ASD PRS. Removing all PRSs but these from the full model did not result in a significant loss of fit, suggesting that results of the primary analysis are driven by the ASD, ASD × Age and MDD variables (VE = −1%, LRT with −11 DF P = .21; Table 2).
Comparative post hoc analyses examined the specificity of PRS effects across tasks and independence from more general cognitive abilities (Figure 1). ASD PRS and PRS-age interactions were significantly associated with performance on the DCCS task (PRS: VE = 0.6%, LRT with 1 DF P = .006; Interactions: VE = 0.5%, LRT with 2 DF P = .038), but not the Flanker (PRS: VE = 0.1%, LRT with 1 DF P = .31; Interactions: VE = 0.1%, LRT with 2 DF P = .57). An increased ASD PRS was associated with better performance on both the DCCS (β = 0.0325, SE = 0.0119), and although not significant, the Flanker (β = 0.0126, SE = 0.0125) in the ASD PRS and interactions models. The ASD age interaction suggests a larger effect in the younger subjects (Figure S6). The effect size and direction of the ASD PRS were essentially unchanged when adding VC to the baseline model for both the main effects of ASD PRS (DCCS: VE = 0.5%, LRT 1 DF P = .009; β = 0.0308, SE = 0.0118; Flanker: VE = 0.1%, LRT 1 DF P = .33; β = 0.0120, SE = 0.0125) and the age interactions (DCCS: VE = 0.5%, LRT 2 DF P = .046; Flanker: VE = 0.1%, LRT 2 DF P = .60).
The MDD PRS was significantly associated with Flanker performance (VE = 0.7%, LRT with 1 DF P = .005), but not DCCS (VE = 0.1%, LRT with 1 DF P = .21) where increased PRS coincided with decreased performance on the tasks (Flanker: β = −0.0542, SE = 0.0194; DCCS: β = −0.0236, SE = 0.0190). These associations were also essentially unchanged by including VC in the baseline model (DCCS: VE = 0.1%, LRT 1 DF P = .25; β = −0.0217, SE = 0.0188; Flanker: VE = 0.7%, LRT 1 DF P = .006; β = −0.0535, SE = 0.0194).
The power of our discovery tests is described in the Supporting Information Note and in Figures S10 and S11, and sensitivity analyses were conducted to examine the effect of reported ADHD and learning disability diagnoses on our results (Supporting Information Note, Tables S14–S19).
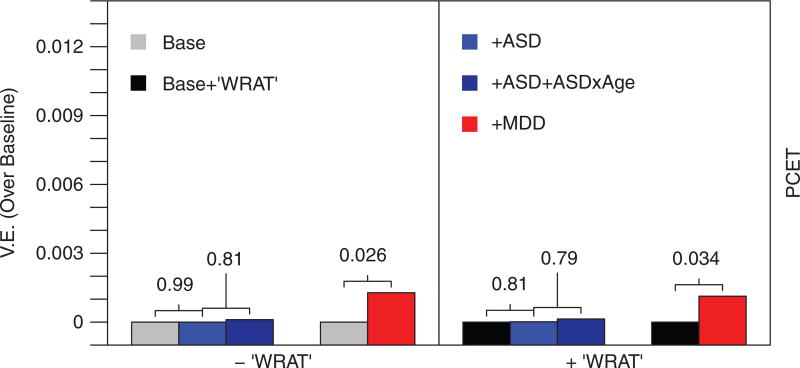
Replication (Figure 2) in the PNC showed a significant association between MDD PRS and performance on the PCET (VE = 0.1%, LRT with 1 DF P = .026) with increased MDD PRS corresponding to decreased executive function performance (β = −74.15, SE = 33.31). Neither the main ASD PRS effect (VE = 0%, LRT with 1 DF P = .99), nor the age interactions (VE = 0.01%, LRT with 2 DF P = .82) significantly associated with PCET performance. When including WRAT Reading scores into the baseline model, the main effect of MDD PRS remained significant (VE = 0.1%, LRT with 1 DF P = .034, β = −69.63, SE = 32.85), and the ASD main effect (VE = 0%, LRT with 1 DF P = .81) and interactions (VE = 0.01%, LRT with 2 DF P = .79) remained nonsignificant. Statistics from all models are provided in Tables S2–S13.
FIGURE 2.
MDD PRS is associated with executive functioning in the PNC. Bar heights reflect increments in VE over the baseline (covariates only) model. Numbers indicate P values are obtained from LRTs that compare the models sequentially, left to right. VE, variance explained; ASD, autism spectrum disorder PRS; MDD, major depressive disorder PRS; WRAT, Wide Range Achievement Test, reading subtest; PCET, Penn Conditional Exclusion Test
4 | DISCUSSION
In this study, we report an initial association between PRS for MDD and executive functioning during typical development using performance on a test of inhibition (the Flanker task). We replicated this association in an independent developmental cohort via performance on a test of cognitive flexibility/shifting (the PCET). This across-domain replication may be supported by the “unity and diversity” model of executive functions where latent factor heritability analyses have suggested genetic effects on inhibition are redundant with a common executive factor.11 Our result finds published support in a negative relationship between MDD PRS and inhibition using reaction time performance on a “Go/No-Go” task as reported by Hagenaars et al30 in a sample of 111 484 adults. In a similar study, Benca et al38 tested PRS for the same 5 disorders for associations with latent executive factors derived directly from the unity and diversity framework in a population sample of young adults that was similarly modest (n = 398). Although not declared significant at the experiment-wide threshold, they did report a nominal trend between the common executive factor and MDD PRS; however, it was in the opposite direction of our report. These results highlight a trend in the current literature describing the PRS effects on cognition where large samples often employ limited cognitive batteries and deeply phenotyped studies are limited by moderate sample sizes and power. While our result suggests a common, across-domain effect of genetic risk for major depression on executive functions during typical development, our sample size and the sparse published support necessitate further investigations.
We also report a positive association between ASD PRS and performance on the DCCS task in the PING cohort, however this effect did not replicate for performance on the PCET in the PNC, despite both targeting flexibility/shifting. In the only other study to directly consider this relationship, Benca et al38 also reported null associations between ASD PRS and all 3 of common executive, updating specific and shifting specific latent factors. These mixed results are contrasted by more consistent reports in larger samples using PRS30,35 and genetic correlations30,36,75,76 suggesting a positive correlation between genetic risk for ASD and higher cognitive functioning in unaffected individuals; although a few null reports exist as well.36,37
Given these mixed results, our initial associations should be replicated and a few important features of the executive function study cohorts, in particular, could add context and motivate further research. First, PING, the PNC and Benca et al each employed different instruments to measure executive function and direct comparisons of the overlap in genetic contributions to each have not been performed. It is possible that the test-retest reliability of the PCET could be lower than for the battery used in PING,55 reducing the power in our replication sample, although the most informative data to this point are currently lacking. “Task impurity” has been noted for measures of executive function12 and quantitative genetics studies show cognitive tests targeting the same domain can have different sensitivities to underlying genetic effects.11,77 For example, the positive correlation between general cognitive abilities and ASD PRS is present in our discovery data (Appendix S1; Figure S9) but the effect appears to be driven by a shifting/flexibility executive function component as measured by the DCCS. We cannot, however, rule out differing contributions of nontargeted domains among the studies. In this regard, the latent factor design of Benca et al38 is the strongest. Second, each study targets a different age range, with PING extending to the youngest population where, for the ASD result, a trending PRS × age interaction suggests the effects are largest. The sensitivity of neuropsychological tests to particular subdomains likely changes with age78 which could compound concerns of task impurity. As such, the differing ages of participants could obscure the expected homology across cohorts. Third, the sample sizes of each cohort are modest by current standards. In this context, the absence of an association should not be taken as a definitive null finding, although it is unlikely large effects exist. It remains possible that psychiatric risk for multiple disorders is associated with cognitive performance broadly or with varying specificity and future studies with wide-reaching cognitive batteries and larger samples will be needed to definitively characterize these effects. Finally, larger and more informative GWAS of psychiatric conditions are also needed. The sensitivity and specificity of PRS vary due to differences in sample size, power and reliability of odds ratios taken from training GWAS (Table 1) which also limits the power of current studies for defining the connections between genetic liability to psychiatric disorders and cognitive endophenotypes in the general population.
Broadly, our findings are consistent with well-established trends in research on learning (dis)abilities that suggest extensive overlap among the genetic factors contributing to normal variability in neurocognitive performance and those associated with learning disability diagnoses.23 The presence of so-called Generalist Genes23 among psychiatric disorders, both in terms of quantitative co-heritability49 and shared molecular genetic factors40 has been widely reported, as has extensive pleiotropy among cognitive abilities, most succinctly captured by the extremely high heritability of the “g” construct.79 Relationships among genetic factors affecting variability in cognitive abilities in healthy individuals and those associated with psychiatric disease are only more recently emerging.22,30,35,80 Because our results were not dependent on the presence of any disorder, we feel they can speak to 2 important themes in this discussion. First, it appears there is some overlap among molecular genetic factors contributing to differences in executive functioning and risk for disorders with executive functioning atypicalities. Second, perhaps executive brain systems dysfunctional in psychiatric disorders are components of a primary neurodevelopmental basis in which susceptibility arises, as opposed to targets of upstream dysfunctions defining the affected states.
Given the opposite directions for the PRS relationships observed in PING and described in previous reports, one could speculate that genetic and/or neurodevelopmental architectures creating ASD and MDD susceptibility may be qualitatively dissimilar. A negative association of MDD PRS and executive function performance is consistent with observations in affected individuals4 and their unaffected family members.81 ASD PRS, however, associates paradoxically in the opposite direction as reported for affected individuals,1 unaffected family members82 and healthy carriers of rare, large effect, often de novo copy number variants.83 PRSs capture only a small portion of genetic liability for ASD (Table 1) and the directional inconsistency could resolve with more informative genetic instruments. An alternative hypothesis is that components of ASD risk captured by PRS (common polygenic risk) interact with rare genetic risk factors, altered neurodevelopment or environmental exposures to induce dysfunction in the ASD affected state. That ASD results from an imbalance or interaction among individually performance enhancing neurodevelopmental features has been proposed by others.84
Finally, new association methods prioritize single gene and single variant associations based on explicit pleiotropic hypotheses. For example, a proxy-phenotype approach used reports of genetic overlap between general cognitive abilities and educational attainment24 to identify novel candidate associations for cognitive ability. Likewise, conditional approaches have suggested novel candidates by exploring genetic overlap of intuitively and unintuitively related phenotypes.85–87 Our results suggest these approaches may aid studies of executive function, especially with smaller samples and extensive neurocognitive testing.
Supplementary Material
Acknowledgments
The authors thank Armin Schwartzman for his helpful statistical advice. The authors gratefully thank the children, adolescents, adults and parents who participated in the research. This work was primarily funded by a grant from the National Institute on Drug Abuse awarded to NA and TTB (R01DA038958). AJS was also supported by a KAVLI Institute for Brain and Mind innovative research grant (#2012-032) and the Annette Merle-Smith CARTA Graduate Fellowship in Anthropogeny. Data used in this study were obtained from the Pediatric Imaging, Neurocognition, and Genetics (PING) Study database. As such, the investigators within PING contributed to the design and implementation of PING and/or provided data but did not necessarily participate in the analysis or writing of this report. A complete listing of PING investigators can be found at http://ping.chd.ucsd.edu. Data collection and sharing for this project was funded by PING (RC2DA029475; National Institute on Drug Abuse, Eunice Kennedy Shriver National Institute of Child Health and Human Development). PING data are disseminated by the PING Coordinating Center at the Center for Human Development, University of California, San Diego. Data from the Philadelphia Neurodevelopmental Cohort (PNC) was accessed through dbGAP, accession number phs000607.v1.p1, referencing the project: Neurodevelopmental Genomics: Trajectories of Complex Phenotypes. Drs Gur, Hakonarson, and collaborators request that publications resulting from these data cite their original publications (PMIDs: 22251308, 23921101). Support for the collection of the data sets was provided by grant RC2MH089983 awarded to Raquel Gur and RC2MH089924 awarded to Hakon Hakonarson. All subjects were recruited through the Center for Applied Genomics at The Children's Hospital in Philadelphia.
Data used in preparation of this article were obtained from the Pediatric Imaging, Neurocognition and Genetics (PING) study database (http://ping.chd.ucsd.edu). As such, the investigators within PING contributed to the design and implementation of PING and/or provided data but did not participate in analysis or writing of this report. A complete listing of PING investigators can be found at https://ping-dataportal.ucsd.edu/sharing/Authors10222012.pdf.
Footnotes
Conflict of interest
Anders Dale is a founder of and holds equity interest in CorTechs Labs, La Jolla, CA and serves on its scientific advisory board. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies. All other authors declare no competing financial interests in relation to the work described.
Additional supporting information may be found online in the Supporting Information section at the end of the article.
References
- 1.Craig F, Margari F, Legrottaglie AR, Palumbi R, de Giambattista C, Margari L. A review of executive function deficits in autism spectrum disorder and attention-deficit/hyperactivity disorder. Neuropsychiatr Dis Treat. 2016;12:1191–1202. doi: 10.2147/NDT.S104620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Willcutt EG, Doyle AE, Nigg JT, Faraone SV, Pennington BF. Validity of the executive function theory of attention-deficit/hyperactivity disorder: a meta-analytic review. Biol Psychiatry. 2005;57:1336–1346. doi: 10.1016/j.biopsych.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Kurtz MM, Gerraty RT. A meta-analytic investigation of neurocognitive deficits in bipolar illness: profile and effects of clinical state. Neuropsychology. 2009;23:551–562. doi: 10.1037/a0016277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Snyder HR. Major depressive disorder is associated with broad impairments on neuropsychological measures of executive function: a meta-analysis and review. Psychol Bull. 2013;139:81–132. doi: 10.1037/a0028727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fioravanti M, Carlone O, Vitale B, Cinti ME, Clare L. A meta-analysis of cognitive deficits in adults with a diagnosis of schizophrenia. Neuropsychol Rev. 2005;15:73–95. doi: 10.1007/s11065-005-6254-9. [DOI] [PubMed] [Google Scholar]
- 6.Millan MJ, Agid Y, Brune M, et al. Cognitive dysfunction in psychiatric disorders: characteristics, causes and the quest for improved therapy. Nat Rev Drug Discov. 2012;11:141–168. doi: 10.1038/nrd3628. [DOI] [PubMed] [Google Scholar]
- 7.Friedman NP, Miyake A, Young SE, Defries JC, Corley RP, Hewitt JK. Individual differences in executive functions are almost entirely genetic in origin. J Exp Psychol Gen. 2008;137:201–225. doi: 10.1037/0096-3445.137.2.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee T, Mosing MA, Henry JD, et al. Genetic influences on four measures of executive functions and their covariation with general cognitive ability: the Older Australian Twins Study. Behav Genet. 2012;42:528–538. doi: 10.1007/s10519-012-9526-1. [DOI] [PubMed] [Google Scholar]
- 9.Vasilopoulos T, Franz CE, Panizzon MS, et al. Genetic architecture of the delis-Kaplan executive function System Trail making test: evidence for distinct genetic influences on executive function. Neuropsychology. 2012;26:238–250. doi: 10.1037/a0026768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teuber HL. Unity and diversity of frontal lobe functions. Acta Neurobiol Exp (Wars) 1972;32:615–656. [PubMed] [Google Scholar]
- 11.Miyake A, Friedman NP. The nature and organization of individual differences in executive functions: four general conclusions. Curr Dir Psychol Sci. 2012;21:8–14. doi: 10.1177/0963721411429458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyake A, Friedman NP, Emerson MJ, Witzki AH, Howerter A, Wager TD. The unity and diversity of executive functions and their contributions to complex "Frontal Lobe" tasks: a latent variable analysis. Cogn Psychol. 2000;41:49–100. doi: 10.1006/cogp.1999.0734. [DOI] [PubMed] [Google Scholar]
- 13.Barnes JJ, Dean AJ, Nandam LS, O'Connell RG, Bellgrove MA. The molecular genetics of executive function: role of monoamine system genes. Biol Psychiatry. 2011;69:e127–e143. doi: 10.1016/j.biopsych.2010.12.040. [DOI] [PubMed] [Google Scholar]
- 14.Chabris CF, Hebert BM, Benjamin DJ, et al. Most reported genetic associations with general intelligence are probably false positives. Psychol Sci. 2012;23:1314–1323. doi: 10.1177/0956797611435528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ibrahim-Verbaas CA, Bressler J, Debette S, et al. GWAS for executive function and processing speed suggests involvement of the CADM2 gene. Mol Psychiatry. 2016;21:189–197. doi: 10.1038/mp.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.LeBlanc M, Kulle B, Sundet K, et al. Genome-wide study identifies PTPRO and WDR72 and FOXQ1-SUMO1P1 interaction associated with neurocognitive function. J Psychiatr Res. 2012;46:271–278. doi: 10.1016/j.jpsychires.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 17.Need AC, Attix DK, McEvoy JM, et al. A genome-wide study of common SNPs and CNVs in cognitive performance in the CANTAB. Hum Mol Genet. 2009;18:4650–4661. doi: 10.1093/hmg/ddp413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seshadri S, DeStefano AL, Au R, et al. Genetic correlates of brain aging on MRI and cognitive test measures: a genome-wide association and linkage analysis in the Framingham study. BMC Med Genet. 2007;8(Suppl 1):S15. doi: 10.1186/1471-2350-8-S1-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benyamin B, Pourcain B, Davis OS, Davies G, Hansell NK, Brion MJ, Kirkpatrick RM, Cents RA, Franic S, Miller MB, Haworth CM, Meaburn E, Price TS, Evans DM, Timpson N, Kemp J, Ring S, McArdle W, Medland SE, Yang J, Harris SE, Liewald DC, Scheet P, Xiao X, Hudziak JJ, de Geus EJ, Wellcome Trust Case Control, C. Jaddoe VW, Starr JM, Verhulst FC, Pennell C, Tiemeier H, Iacono WG, Palmer LJ, Montgomery GW, Martin NG, Boomsma DI, Posthuma D, McGue M, Wright MJ, Davey Smith G, Deary IJ, Plomin R, Visscher PM. Childhood intelligence is heritable, highly polygenic and associated with FNBP1L. Mol Psychiatry. 2014;19:253–258. doi: 10.1038/mp.2012.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le Hellard S, Steen VM. Genetic architecture of cognitive traits. Scand J Psychol. 2014;55:255–262. doi: 10.1111/sjop.12112. [DOI] [PubMed] [Google Scholar]
- 21.Plomin R. Commentary: missing heritability, polygenic scores, and gene-environment correlation. J Child Psychol Psychiatry. 2013;54:1147–1149. doi: 10.1111/jcpp.12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wray NR, Lee SH, Mehta D, Vinkhuyzen AA, Dudbridge F, Middeldorp CM. Research review: polygenic methods and their application to psychiatric traits. J Child Psychol Psychiatry. 2014;55:1068–1087. doi: 10.1111/jcpp.12295. [DOI] [PubMed] [Google Scholar]
- 23.Plomin R, Kovas Y. Generalist genes and learning disabilities. Psychol Bull. 2005;131:592–617. doi: 10.1037/0033-2909.131.4.592. [DOI] [PubMed] [Google Scholar]
- 24.Rietveld CA, Esko T, Davies G, et al. Common genetic variants associated with cognitive performance identified using the proxy-phenotype method. Proc Natl Acad Sci USA. 2014;111:13790–13794. doi: 10.1073/pnas.1404623111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schork AJ, Wang Y, Thompson WK, Dale AM, Andreassen OA. New statistical approaches exploit the polygenic architecture of schizophrenia—implications for the underlying neurobiology. Curr Opin Neurobiol. 2016;36:89–98. doi: 10.1016/j.conb.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin J, Hamshere ML, Stergiakouli E, O'Donovan MC, Thapar A. Genetic risk for attention-deficit/hyperactivity disorder contributes to neurodevelopmental traits in the general population. Biol Psychiatry. 2014;76:664–671. doi: 10.1016/j.biopsych.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin J, Hamshere ML, Stergiakouli E, O'Donovan MC, Thapar A. Neurocognitive abilities in the general population and composite genetic risk scores for attention-deficit hyperactivity disorder. J Child Psychol Psychiatry. 2015;56:648–656. doi: 10.1111/jcpp.12336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stergiakouli E, Martin J, Hamshere ML, et al. Association between polygenic risk scores for attention-deficit hyperactivity disorder and educational and cognitive outcomes in the general population. Int J Epidemiol. 2017;46:421–428. doi: 10.1093/ije/dyw216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Du Rietz E, Coleman J, Glanville K, Choi SW, O'Reilly PF, Kuntsi J. Association of polygenic risk for attention-deficit/hyperactivity disorder with co-occurring traits and disorders. Biol Psychiatry. 2018 doi: 10.1016/j.bpsc.2017.11.013. In Press. [DOI] [PMC free article] [PubMed]
- 30.Hagenaars SP, Harris SE, Davies G, et al. Shared genetic aetiology between cognitive functions and physical and mental health in UK biobank (N=112 151) and 24 GWAS consortia. Mol Psychiatry. 2016;21:1624–1632. doi: 10.1038/mp.2015.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mistry S, Harrison JR, Smith DJ, Escott-Price V, Zammit S. The use of polygenic risk scores to identify phenotypes associated with genetic risk of schizophrenia: systematic review. Schizophr Res. 2017 doi: 10.1016/j.schres.2017.10.037. [DOI] [PubMed]
- 32.Liebers DT, Pirooznia M, Seiffudin F, Musliner KL, Zandi PP, Goes FS. Polygenic risk of schizophrenia and cognition in a population-based survey of older adults. Schizophr Bull. 2016;42:984–991. doi: 10.1093/schbul/sbw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Germine L, Robinson EB, Smoller JW, et al. Association between polygenic risk for schizophrenia, neurocognition and social cognition across development. Transl Psychiatry. 2016;6:e924. doi: 10.1038/tp.2016.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riglin L, Collishaw S, Richards A, et al. Schizophrenia risk alleles and neurodevelopmental outcomes in childhood: a population-based cohort study. Lancet Psychiatry. 2017;4:57–62. doi: 10.1016/S2215-0366(16)30406-0. [DOI] [PubMed] [Google Scholar]
- 35.Clarke TK, Lupton MK, Fernandez-Pujals AM, et al. Common polygenic risk for autism spectrum disorder (ASD) is associated with cognitive ability in the general population. Mol Psychiatry. 2016;21:419–425. doi: 10.1038/mp.2015.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hill WD, Davies G, Group CCW, Liewald DC, McIntosh AM, Deary IJ. Age-dependent pleiotropy between general cognitive function and major psychiatric disorders. Biol Psychiatry. 2016;80:266–273. doi: 10.1016/j.biopsych.2015.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krapohl E, Euesden J, Zabaneh D, et al. Phenome-wide analysis of genome-wide polygenic scores. Mol Psychiatry. 2016;21:1188–1193. doi: 10.1038/mp.2015.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benca CE, Derringer JL, Corley RP, et al. Predicting cognitive executive functioning with polygenic risk scores for psychiatric disorders. Behav Genet. 2017;47:11–24. doi: 10.1007/s10519-016-9814-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neale BM, Medland SE, Ripke S, et al. Meta-analysis of genome-wide association studies of attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry. 2010;49:884–897. doi: 10.1016/j.jaac.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cross-Disorder Group of the Psychiatric Genomics Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.The Psychiatric GWAS Consortium Bipolar Disorder Working Group. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43:977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium. Ripke S, Wray NR, et al. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry. 2013;18:497–511. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Faraone SV, Mick E. Molecular genetics of attention deficit hyperactivity disorder. Psychiatr Clin North Am. 2010;33:159–180. doi: 10.1016/j.psc.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Freitag CM. The genetics of autistic disorders and its clinical relevance: a review of the literature. Mol Psychiatry. 2007;12:2–22. doi: 10.1038/sj.mp.4001896. [DOI] [PubMed] [Google Scholar]
- 46.Craddock N, Sklar P. Genetics of bipolar disorder: successful start to a long journey. Trends Genet. 2009;25:99–105. doi: 10.1016/j.tig.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 47.Sullivan PF, Neale MC, Kendler KS. Genetic epidemiology of major depression: review and meta-analysis. Am J Psychiatry. 2000;157:1552–1562. doi: 10.1176/appi.ajp.157.10.1552. [DOI] [PubMed] [Google Scholar]
- 48.Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60:1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- 49.Cross-Disorder Group of the Psychiatric Genomics Consortium. Lee SH, Ripke S, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45:984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hamshere ML, Langley K, Martin J, et al. High loading of polygenic risk for ADHD in children with comorbid aggression. Am J Psychiatry. 2013;170:909–916. doi: 10.1176/appi.ajp.2013.12081129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anney R, Klei L, Pinto D, et al. Individual common variants exert weak effects on the risk for autism spectrum disorders. Hum Mol Genet. 2012;21:4781–4792. doi: 10.1093/hmg/dds301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jernigan TL, Brown TT, Hagler DJ, Jr, Akshoomoff N, Bartsch H, Newman E, Thompson WK, Bloss CS, Murray SS, Schork N, Kennedy DN, Kuperman JM, McCabe C, Chung Y, Libiger O, Maddox M, Casey BJ, Chang L, Ernst TM, Frazier JA, Gruen JR, Sowell ER, Kenet T, Kaufmann WE, Mostofsky S, Amaral DG, Dale AM, Pediatric Imaging, N. & Genetics, S The pediatric imaging, neurocognition, and genetics (PING) data repository. Neuroimage. 2016;124:1149–1154. doi: 10.1016/j.neuroimage.2015.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Akshoomoff N, Newman E, Thompson WK, et al. The NIH toolbox cognition battery: results from a large normative developmental sample (PING) Neuropsychology. 2014;28:1–10. doi: 10.1037/neu0000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weintraub S, Bauer PJ, Zelazo PD, et al. I. NIH toolbox cognition battery (CB): introduction and pediatric data. Monogr Soc Res Child Dev. 2013;78:1–15. doi: 10.1111/mono.12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zelazo PD, Anderson JE, Richler J, Wallner-Allen K, Beaumont JL, Weintraub S. II. NIH toolbox cognition battery (CB): measuring executive function and attention. Monogr Soc Res Child Dev. 2013;78:16–33. doi: 10.1111/mono.12032. [DOI] [PubMed] [Google Scholar]
- 56.Gershon RC, Slotkin J, Manly JJ, et al. IV. NIH toolbox cognition battery (CB): measuring language (vocabulary comprehension and reading decoding) Monogr Soc Res Child Dev. 2013;78:49–69. doi: 10.1111/mono.12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blom G. Statistical Estimates and Transformed Beta-Variables. New York: Wiley; 1958. [Google Scholar]
- 58.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.The 1000 Genomes Project Consortium. Auton A, Brooks LD, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 63.The International HapMap Consortium. The international HapMap project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 64.Mitchell MK, Gregersen PK, Johnson S, Parsons R, Vlahov D New York Cancer P. The New York Cancer Project: rationale, organization, design, and baseline characteristics. J Urban Health. 2004;81:301–310. doi: 10.1093/jurban/jth116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Team RC. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2016. [Google Scholar]
- 69.Gur RC, Richard J, Calkins ME, et al. Age group and sex differences in performance on a computerized neurocognitive battery in children age 8–21. Neuropsychology. 2012;26:251–265. doi: 10.1037/a0026712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Satterthwaite TD, Elliott MA, Ruparel K, et al. Neuroimaging of the Philadelphia neurodevelopmental cohort. Neuroimage. 2014;86:544–553. doi: 10.1016/j.neuroimage.2013.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kaufman J, Birmaher B, Brent D, et al. Schedule for affective disorders and schizophrenia for school-age children-present and lifetime version (K-SADS-PL): initial reliability and validity data. J Am Acad Child Adolesc Psychiatry. 1997;36:980–988. doi: 10.1097/00004583-199707000-00021. [DOI] [PubMed] [Google Scholar]
- 72.Gur RC, Richard J, Hughett P, et al. A cognitive neuroscience-based computerized battery for efficient measurement of individual differences: standardization and initial construct validation. J Neurosci Methods. 2010;187:254–262. doi: 10.1016/j.jneumeth.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moore TM, Reise SP, Gur RE, Hakonarson H, Gur RC. Psychometric properties of the Penn computerized neurocognitive battery. Neuropsychology. 2015;29:235–246. doi: 10.1037/neu0000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kurtz MM, Ragland JD, Moberg PJ, Gur RC. The Penn conditional exclusion test: a new measure of executive-function with alternate forms of repeat administration. Arch Clin Neuropsychol. 2004;19:191–201. doi: 10.1016/S0887-6177(03)00003-9. [DOI] [PubMed] [Google Scholar]
- 75.Grove J, Ripke S, Als TD, et al. Common risk variants identified in autism spectrum disorder. bioRxiv. 2017 doi: 10.1101/224774. [DOI]
- 76.Sniekers S, Stringer S, Watanabe K, et al. Genome-wide association meta-analysis of 78,308 individuals identifies new loci and genes influencing human intelligence. Nat Genet. 2017;49:1107–1112. doi: 10.1038/ng.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kremen WS, Panizzon MS, Franz CE, et al. Genetic complexity of episodic memory: a twin approach to studies of aging. Psychol Aging. 2014;29:404–417. doi: 10.1037/a0035962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Best JR, Miller PH. A developmental perspective on executive function. Child Dev. 2010;81:1641–1660. doi: 10.1111/j.1467-8624.2010.01499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Plomin R, Deary IJ. Genetics and intelligence differences: five special findings. Mol Psychiatry. 2015;20:98–108. doi: 10.1038/mp.2014.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Power RA, Steinberg S, Bjornsdottir G, et al. Polygenic risk scores for schizophrenia and bipolar disorder predict creativity. Nat Neurosci. 2015;18:953–955. doi: 10.1038/nn.4040. [DOI] [PubMed] [Google Scholar]
- 81.Hsu KJ, Young-Wolff KC, Kendler KS, Halberstadt LJ, Prescott CA. Neuropsychological deficits in major depression reflect genetic/familial risk more than clinical history: a monozygotic discordant twin-pair study. Psychiatry Res. 2014;215:87–94. doi: 10.1016/j.psychres.2013.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Delorme R, Gousse V, Roy I, et al. Shared executive dysfunctions in unaffected relatives of patients with autism and obsessive-compulsive disorder. Eur Psychiatry. 2007;22:32–38. doi: 10.1016/j.eurpsy.2006.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stefansson H, Meyer-Lindenberg A, Steinberg S, et al. CNVs conferring risk of autism or schizophrenia affect cognition in controls. Nature. 2014;505:361–366. doi: 10.1038/nature12818. [DOI] [PubMed] [Google Scholar]
- 84.Crespi BJ. Autism as a disorder of high intelligence. Front Neurosci. 2016;10:300. doi: 10.3389/fnins.2016.00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Andreassen OA, Djurovic S, Thompson WK, et al. Improved detection of common variants associated with schizophrenia by leveraging pleiotropy with cardiovascular-disease risk factors. Am J Hum Genet. 2013a;92:197–209. doi: 10.1016/j.ajhg.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Andreassen OA, Thompson WK, Schork AJ, et al. Improved detection of common variants associated with schizophrenia and bipolar disorder using pleiotropy-informed conditional false discovery rate. PLoS Genet. 2013b;9:e1003455. doi: 10.1371/journal.pgen.1003455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Le Hellard S, Wang Y, Witoelar A, et al. Identification of gene loci that overlap between schizophrenia and educational attainment. Schizophr Bull. 2016;43:654–664. doi: 10.1093/schbul/sbw085. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.