Abstract
Lactate, a product of aerobic glycolysis in astrocytes, is required for memory formation and consolidation, and has recently emerged as a signaling molecule for neurons and various cell types in peripheral tissues. In particular lactate stimulates mRNA expression of a few plasticity-related genes. Here, we describe a RNA-seq study that unravels genome-wide transcriptomic responses to this energy metabolite in cortical neurons. Our results show that mRNA expression of 20 immediate-early genes involved in the MAPK signaling pathway and in synaptic plasticity were increased by more than twofold following 1 h of lactate stimulation. This effect was dependent on NMDA receptor (NMDAR) activity since it was prevented by pre-treatment with MK-801. Comparison with published datasets showed that a significant proportion of genes modulated by lactate were similarly regulated by a stimulation protocol activating specifically synaptic NMDARs known to result in upregulation of pro-survival and downregulation of pro-death genes. Remarkably, transcriptional responses to lactate were reproduced by NADH (for 74 of the 113 genes, FDR < 0.05), suggesting a redox-dependent mechanism of action. Longer-term gene expression changes observed after 6 h of lactate treatment affected genes involved in regulating neuronal excitability and genes coding for proteins localized at synapses. Gene set enrichment analyses performed with ranked lists of expressed genes revealed effects on molecular functions involved in epigenetic modulation, and on processes relevant to sleep physiology and behavioral phenotypes such as anxiety and hyperactivity. Overall, these results strengthen the notion that lactate effectively regulates activity-dependent and synaptic genes, and highlight new signaling effects of lactate in plasticity and neuroprotection.
Keywords: lactate, synaptic plasticity, neuroprotection, NMDA receptor, transcriptome, NADH, gene expression
Introduction
Lactate is a metabolic end-product of glycolysis with established roles as an energy substrate in multiple tissues, including skeletal muscle, heart, liver, and the brain (Adeva-Andany et al., 2014). In the brain, lactate is mainly produced by astrocytes, a type of glial cells, and shuttled to neurons in response to synaptic activity – a mechanism known as the astrocyte-to-neuron L-lactate shuttle, ANLS (Pellerin and Magistretti, 1994, 2012).
In addition to its energetic role, L-Lactate has emerged as a valuable signaling molecule in different tissues and cell types, including circulating stem/progenitor cells (SPCs) (Milovanova et al., 2008), muscle (Hashimoto et al., 2007), T cells, monocytes and dendritic cells (Gottfried et al., 2006; Dietl et al., 2010; Haas et al., 2016). Lactate is also an active metabolite for cancer cells promoting angiogenesis, immune escape and cell migration (Kim and Dang, 2006; San-Millan and Brooks, 2017).
Recent work has demonstrated that lactate also acts as a signaling molecule in the brain (Magistretti and Allaman, 2018), modifying the excitability of neurons (Tang et al., 2014; Sada et al., 2015), playing an important role in learning and memory (Suzuki et al., 2011) and favoring neuroprotection (Jourdain et al., 2016).
The essential role of lactate in long-term memory formation in mice (Suzuki et al., 2011) is likely to rely on modulation of synaptic plasticity and neuronal activity-dependent genes. Yang et al. (2014) have shown in primary cultured neurons and in the mouse sensory motor cortex that L-lactate stimulates the expression of genes such as Arc, c-Fos, Bdnf and Zif268 (Egr1). These genes are important for synaptic plasticity and are induced by neuronal activity (Guzowski et al., 2001; Flavell and Greenberg, 2008). The effect of lactate on these genes was dependent in particular on NMDA receptor (NMDAR) activity, being abolished in the presence of NMDAR antagonist MK801.
NMDA receptors are glutamate-gated ion channels with important roles in excitatory neurotransmission, experience-dependent synaptic remodeling, and long-lasting changes in synaptic efficacy such as LTP, a key process in learning and memory (Cull-Candy and Leszkiewicz, 2004; Lynch, 2004; Lau and Zukin, 2007).
Lactate was also shown to regulate gene expression in other cell systems and tissues, such as L6 muscle cells (Hashimoto et al., 2007), HCT 116 colon cancer cells (Latham et al., 2012), glioma cells (Baumann et al., 2009), tumor-associated macrophages (Colegio et al., 2014), T cells (Haas et al., 2015), and mesenchymal stem cells (Zieker et al., 2008).
In the context of the observed effects and in order to gain further insights into the signaling mechanisms regulated by lactate in the brain, we have evaluated genome-wide transcriptional responses to this metabolite in primary neuron cultures.
Results show that L-lactate modulates key genes important for different forms of synaptic plasticity in an NMDAR-dependent manner and genes under the genomic pro-survival program activated specifically by synaptic NMDARs. Furthermore, NADH, but not pyruvate, primarily reproduces these effects on gene expression. In addition, functional bioinformatics analysis of gene expression data points to the involvement of the MAPK signaling pathway, the serum response factor (SRF) and transcription factor Zif268 (Egr1).
The transcriptome study also unravels groups of genes modulated by L-lactate in an NMDAR-independent manner, for which other signaling pathways and gene regulatory mechanisms could be responsible. These results open avenues for further investigations that can contribute to a better understanding of the physiological coupling of energy metabolism to synaptic activity in the brain.
Materials and Methods
Reagents
Sodium L-lactate and sodium L-pyruvate were purchased from Sigma-Aldrich. (5S,10R)-(+)-5-methyl-10,11-dihydro-5H-dibenzo[a,d]cyclohepten-5,10-imine maleate (MK801 maleate) and N-methyl-D-aspartic acid (NMDA) were purchased from Tocris. NADH was obtained from Roche.
Cell Cultures
Experiments were conducted in accordance with the Swiss Federal Guidelines for Animal Experimentation and were approved by the Cantonal Veterinary Office for Animal Experimentation (Vaud, Switzerland). Primary cultures of cortical neurons were prepared from embryonic day 17 (E17) OF1 mice embryos (Charles River Laboratories) as previously described (Allaman et al., 2010). Briefly, minced pieces (1–2 mm3) of isolated cerebral cortices were enzymatically digested with papain and gently triturated to a single-cell suspension. Dissociated cells were plated onto poly-L-ornithine-coated cell culture dishes at a density of 4 × 104 cells/cm2 and maintained in Neurobasal medium supplemented with B27, GlutaMax, penicillin (50 U/mL), and streptomycin (50 μg/mL) (Invitrogen) at 37°C in a humidified atmosphere of 5% CO2 and 95% air. These culture conditions typically produced 93–95% pure neuronal cultures (Belanger et al., 2011). Neurons were used for experiments at day in vitro 11.
Cell Treatments
Neuronal cultures were treated by directly adding lactate (20 mM), pyruvate (20 mM), NADH (4 mM) or NMDA (50 μM) into the culture medium from 50 to 100x stock solutions prepared in milli-Q® water. When indicated, MK-801 (40 μM) was added 30 min prior to lactate. For each independent culture and treatment condition, three biological replicates were included.
RNA Extraction and Sequencing
Total RNA was isolated from cultured cells using Nucleospin RNA II kit (Macherey-Nagel) according to the manufacturer’s instructions. As much as 5 μg of RNA, quantified using NanoDrop spectrophotometer (Nanodrop 2000, Thermo Scientific), were preserved in RNAstable (Biomatrica) and subsequently shipped to KAUST’s next-generation sequencing core lab where it was reconstituted in RNAse-free water. The integrity and level of degradation of the purified RNA was assessed with the Agilent BioAnalyzer 2100 (Agilent Technologies, Inc.). RIN value was above 8 for all the samples, on a scale from 1 (degraded) to 10 (intact).
Illumina RNA-Seq was performed according to manufacturer’s instructions. Library was prepared using the TruSeq stranded mRNA library protocol. Briefly, mRNA was isolated from total RNA with poly-T oligo-attached magnetic beads and then fragmented. First-strand cDNA was synthesized using SuperScript III reverse transcriptase and random primers. Second strand cDNA was subsequently synthesized and double-stranded cDNA generated. The cDNA was adenylated and ligated to adapters. cDNA fragments were purified and size-selected with AMPure XP beads (Beckman Coulter).
Sequencing was performed on the HiSeqTM 2000 (Illumina) with a paired-end (PE) sequencing strategy. The read length was set at 100 bp (PE) and the sequencing depth was 40–50 million reads.
qRT-PCR
The first strand of cDNA was synthesized from 1,000 ng of total RNA (10 min at 25°C followed by 120 min at 37°C and 5 min at 85°C) using the High-capacity cDNA reverse transcription kit (Applied Biosystems). The resulting cDNA was amplified by quantitative PCR (qPCR) with an ABI Prism 7900 system (Applied Biosystems). The PCR mix was composed of 16 ng of cDNA, 250 nM of forward and reverse primers in 10 μL of 1× SYBR-Green PCR MasterMix (Applied Biosystems). Primer sequences were designed using Primer Express 3.0 software (Applied Biosystems) and salt-free purified oligonucleotides were synthesized by Integrated DNA Technologies (IDT). A full list of the primer sequences used is available in Supplementary Table S6.
The specificity of PCR amplification for each set of primers was checked by the presence of a single sharp peak in the melting curve analysis. Data were computed using the sequence detector software SDS 2.3 (Applied Biosystems) and analyzed using the delta-Ct relative quantification (ΔΔ Ct) method (Livak and Schmittgen, 2001) with cyclophilin and β-actin as reference genes.
RNA-Seq Data Analysis
Raw sequencing data quality was evaluated with the application FastQC1.
The raw RNA-seq reads were preprocessed to remove adapter sequences using trimmomatic2 with the following parameters: ILLUMINACLIP: ../TruSeq3-PE-2.fa:2:151:10, LEADING:3, TRAILING:3, SLIDINGWINDOW:4:15, MINLEN:36.
For each sample, reads that passed filtering were mapped to the UCSC mouse reference genome [build mm10] using TopHat3 with the following changes to the default parameters described in the protocol of Trapnell et al. (2012): -G genes.gtf –no-novel-juncs –max-multihits 1. These options specified mapping to the reference genome without novel splice discovery and selection of uniquely mapped reads.
The resulting aligned reads were then analyzed with the Cufflinks suite4 that assembles aligned reads into transcripts and measures their relative abundance. The expression of each protein-coding gene was quantified by adding up the expression levels of all transcripts for that particular gene. Abundance is reported as the number of reads mapping to that gene divided by the gene length in kilobases and the total number of mapped reads in millions, which is called fragments per kilobase of exon per million fragments mapped (fpkm).
For further analysis, only protein-coding genes differentially expressed with a fpkm value >1 in at least one of the two conditions compared were included. The aim was to remove genes with very low expression levels, in order to minimize “background noise” and focus on more robust gene expression.
The number of independent cultures (n) per treatment condition used for the differential gene expression analysis differed as follows for the 1 h time point: L-lactate (n = 3), MK-801 (n = 2), MK-801 and L-lactate (n = 2), pyruvate (n = 2), NADH (n = 1), NMDA (n = 1), NMDA and L-lactate (n = 1); and was n = 2 for the 6 h time point and treatment with L-lactate.
Where more than one independent culture was used for the analysis in response to a given treatment, log2FC values for genes in common among the independent experiments were averaged.
GO and Pathway Enrichment Analysis
Gene ontology (GO) and pathway enrichment analysis were performed using the clusterProfiler package (Yu et al., 2012) in the R programming environment. The functions enrichGO and enrichKEGG were utilized with Benjamini-Hochberg (BH) p-value adjustment (cutoff < 0.1) and the background set of genes comprised all genes with a fpkm value ≥1 in one of the two treatment conditions compared. The resulting GO terms were analyzed for semantic similarity (cut-off value of 0.7) with GOSemSim R package (Yu et al., 2010) to reduce redundancy. Selections of non-redundant enriched terms from the GO analysis were plotted. All p-values for plotted terms are corrected for multiple testing.
Visualization of results was performed using the dotplot function of the clusterProfiler package, and the Bioconductor packages pathview and GOplot.
Gene Set Enrichment Analysis (GSEA)
Gene set enrichment analysis (GSEA) was conducted using WebGestalt 2017 online platform (Wang et al., 2017) run on pre-ranked list of all the genes expressed in cortical neurons with fpkm expression values >1, using the default parameters and identification of enriched categories based on false discovery rate (FDR) threshold of 0.05. If no terms were enriched below this threshold, additional terms were identified below a less stringent threshold of 0.1. Ranking was based on fold-change induction in L-lactate-treated cells compared to control cells. GSEA results were assessed as being statistically significant by permutation of 1,000 samples.
The enrichment was indicated by the net enrichment score (NES), which accounts for differences in gene set size and in correlations between gene sets and the expression dataset (Wang et al., 2017).
The leading core genes identified are genes that show high correlation between their expression level and the phenotype to which they are associated and tend to be at the extremes of the distribution, rather than randomly distributed. This subset is essentially the genes responsible for the enrichment score for a given biological process or cellular component.
Transcription Factor Binding Site Enrichment Analysis
Detection of over-represented conserved transcription factor binding sites (TFBS) was performed with the Single Site Analysis tool on the oPOSSUM-3 website (Kwon et al., 2012) using a conservation cutoff of 0.60 and a matrix score threshold of 85%. The amount of upstream/downstream sequence analyzed was 3,000/1,000 bp. A list of all expressed genes with fpkm values >1 in cortical neurons was used for background. oPOSSUM analyzes TFBS enrichment through both Z-score and F-score, which is the negative log of the Fisher one-tailed exact probability. The Z-score used compares the rate of occurrence of a TFBS in the target set of genes to the expected rate estimated from the pre-computed background. The F-score is based on the one-tailed Fisher exact probability, which compares the proportion of co-expressed genes containing a particular TFBS to the proportion contained in the background set. Thus the F-score/p-value does not consider the number of times a TFBS appears near a gene beyond once. For enrichment of TFBS, we employed a combined threshold of 10 for Z-scores and 7 for F-scores, based on available recommendations and literature (Ahmed et al., 2017; Taylor et al., 2017).
Cell Type-Enrichment Analysis
To estimate cell-type sources, including cell culture contaminants such as astrocytes, endothelial cells, oligodendrocytes and microglia, from which genes may originate, we used a recently published cell-type deconvolution analysis (Saul et al., 2017) using, as reference sets, RNA-seq data of sorted cells from the mouse frontal cortex (Zhang et al., 2014). Genes enriched for each cell type were selected on the basis of expression values greater than 1 fpkm and on expression in a given cell type compared to all others greater than fivefold. Overlap with the lists of differentially expressed genes in our study was evaluated.
Results
L-Lactate Modulates Expression of Immediate-Early Genes Involved in Transcription Regulation and Early Signaling Events
Lactate induces plasticity-related genes in cortical neurons (Yang et al., 2014). In order to gain an exhaustive insight into the transcriptional response to lactate in neurons, we have performed a genome-wide RNA-seq study.
RNA sequencing was performed on RNA isolated from DIV 11 cultured cortical neurons 1 h after addition of sodium L-lactate to cell culture media.
All treated samples used in this study had RIN values >8 (range 8.2–10) confirming that all RNA samples were of excellent quality (Ibberson et al., 2009). In total, we obtained an average of 44 million reads per sample treated with lactate for 1 h (n = 3 independent cultures, three biological replicates per culture). Low quality bases and adapters were filtered out from the datasets prior to read mapping. An average of 42 million of reads per sample were mapped to the mouse genome (UCSC, mm10).
Overall, around 12,000 genes were detected in neurons with expression levels above the threshold fpkm value of 1, used to separate genes with very low expression levels; this total number of expressed genes is representative for brain tissue which is characterized by the expression of more genes with less dominance of few highly expressed genes (Ramskold et al., 2009).
Differential gene expression analysis revealed that 113 genes were differentially regulated in cortical neurons treated with lactate for 1 h when compared with the control (no treatment), of which 80 genes were up-regulated and 33 were down-regulated (Supplementary Table S1). These genes were identified in each independent culture using a q-value < 0.05. The q-value represents the Benjamini-Hochberg multiple testing corrected p-value and is a measure of the FDR. Fold change values for the 113 genes are the average of the three independent cultures.
To account for possible contribution of non-neuronal cell types to gene expression changes mediated by L-lactate in our cultures, we have used a cell-type specific classification method of genes (Saul et al., 2017) based on RNA-seq data sets obtained from sorted cells of the mouse frontal cortex (Zhang et al., 2014) and examined the overlap with our list of differentially expressed genes.
Out of the 113 differentially expressed genes in response to 1 h L-lactate treatment, no astrocyte- or oligodendrocyte-enriched genes were identified in the overlaps. Two (Apold1, Kcnj2) and 6 (Calhm2, Egr2, Egr3, Gadd45b, Gm13889, and Txnip) genes were found to be, respectively, enriched in endothelial and microglial cells. Interestingly, six of these genes were also robustly expressed in neurons, while only two genes, Calhm2 and Kcnj2, had expression values below 1 fpkm in neurons (Zhang et al., 2014).
Some of the genes previously reported as modulated by lactate in neurons after a treatment of 1 h (Arc, Egr1/Zif268, c-Fos) were among the 113 genes identified.
Out of the 113 genes differentially regulated by lactate, 22 genes showed an average fold change above 2, indicating a strong modulation.
We next performed a GO enrichment analysis to identify over-represented biological processes, molecular functions and cellular components corresponding to the list of differentially expressed genes.
For this analysis, all differentially expressed genes with a q-value < 0.1 were used (135 genes – Supplementary Table S1). This q-value threshold is more lenient and can be used when aiming to maximize the discovery of relevant biological processes.
Around 35 of the genes modulated by L-lactate at 1 h are immediate-early genes, with transcription factor activity, or genes coding for proteins involved in the regulation of phosphorylation and early signaling events, as emphasized upon conducting a GO over-representation test (Figure 1A). Interestingly, five out of ten of the most enriched molecular function terms point to regulation of RNA polymerase II activity. Among the most enriched GO terms representing biological processes, muscle tissue development and differentiation are included. A full list of enriched GO terms is described in Supplementary Table S2.
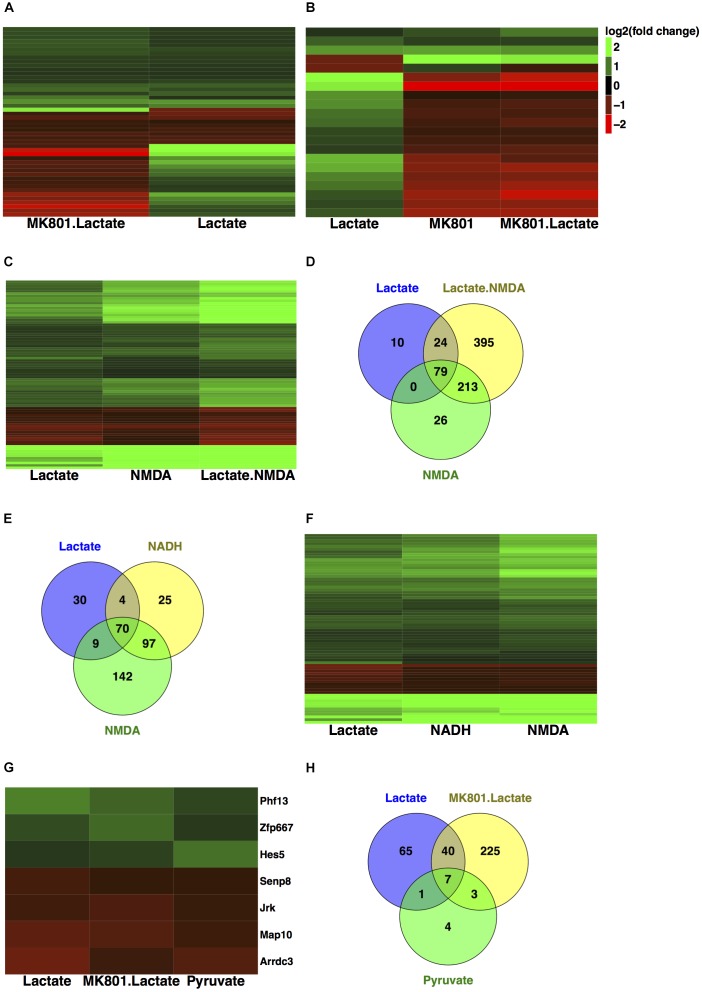
FIGURE 1.
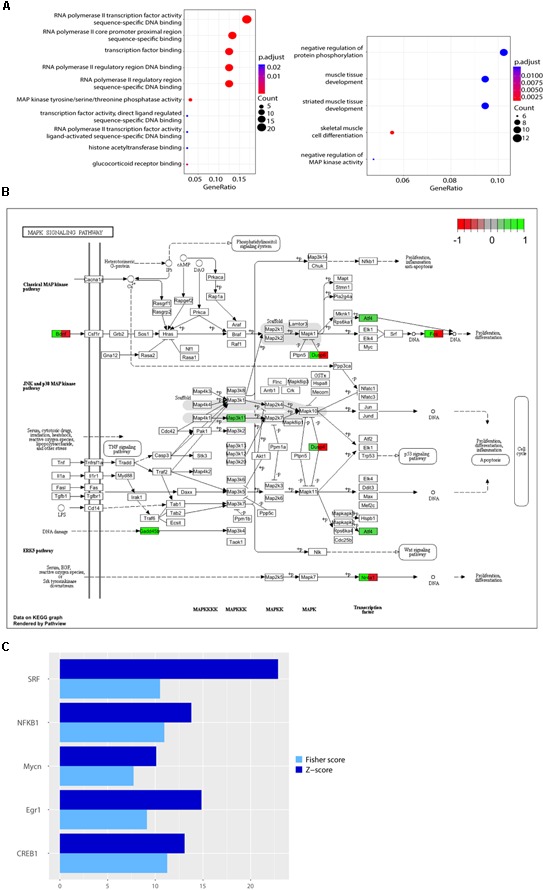
Functional analysis of genes differentially expressed in response to L-lactate after 1 h. (A) GO plots showing enrichment of biological processes and molecular functions for the genes differentially expressed after 1 h stimulation with L-lactate (135 genes, q-value < 0.1, three biological replicates, n = 3 independent cultures). Statistical significance for the enriched terms is based on the multiple testing-corrected p-value, which is color-coded. The number of genes associated with each respective term is indicated by the dot size. (B) A detailed KEGG graph of the MAPK signaling pathway with genes modulated by L-lactate at 1 h highlighted in green indicating that they are up-regulated. Dusp10 and Gadd45g are not indicated on the pathway map. Genes highlighted in red are genes down-regulated in the co-treatment condition with MK801 and L-lactate. (C) Bar plot showing fold enrichment of specific transcription factor binding sites in the promoters of genes modulated by L-lactate at 1 h (FDR < 0.001). Z-score is the main enrichment indicator and is obtained by dividing the rate of occurrence of a TFBS in the target set of genes to the expected rate estimated from the pre-computed background. The Fisher score is calculated as the negative natural logarithm of the one-tailed Fisher exact probability.
Ten out of the 135 genes differentially regulated by lactate, namely c-Fos, Bdnf, Atf4, Nr4a1, Gadd45b, Gadd45g, Map3k11, Dusp4, Dusp6, Dusp10 are associated with the MAPK signaling pathway, a key-signaling cascade mediating, in neurons, cytoplasmic and nuclear responses to synaptic activity, and NMDAR-dependent neuronal plasticity (Sweatt, 2001; Yang et al., 2014; Bluthgen et al., 2017). They are all up-regulated by L-lactate (highlighted in green; Figure 1B).
Genes Modulated by L-Lactate Are Under the Regulatory Influence of Transcription Factors That Are Induced by Synaptic Activity and NMDAR Signaling
In the context of gene expression changes by 1 h lactate treatment, we have searched for transcription factors with over-represented DNA binding site motifs in promoters of differentially expressed genes with a q-value < 0.1, using the oPOSSUM web application and the JASPAR database of transcription factor matrices.
The transcription factors with the highest over-representation (Z-score > 10 and Fisher score > 7), including SRF, Egr1, CREB1, NF-κB, and Mycn, are known to be modulated by neuronal activity and to be involved in plasticity. The SRF shows the highest enrichment in promoters of genes modulated by L-lactate at 1 h (Figure 1C). Interestingly, this transcription factor was previously reported in literature to play important roles in synaptic plasticity, memory and survival (Knoll and Nordheim, 2009; Stern et al., 2012). Both SRF and the cAMP response element-binding protein (CREB1), another transcription factor that is enriched in the set of genes regulated by lactate, respond to synaptic activity triggered by activation of NMDARs and the subsequent intracellular calcium increase (Sala et al., 2000; Knoll and Nordheim, 2009). Egr1, one of the most strongly up-regulated genes by lactate at 1 h, is induced in association with long-term potentiation (LTP) and after specific learning experiences, and is implicated in regulating plasticity and survival (Cole et al., 1989; Hall et al., 2001; Jones et al., 2001). NF-κB activation was also shown to be important for induction of synaptic plasticity and memory formation (Gutierrez and Davies, 2011; Engelmann and Haenold, 2016). The p50 subunit (encoded by Nfkb1, the transcription factor motif enriched in our analysis) is a subunit of the classical NF-κB pathway and is implicated in regulating axonogenesis (Haenold et al., 2014). In addition, another enriched transcription factor, Mycn, was previously reported as being induced by neuronal activity (Jopling and Willis, 2001).
A full list of enriched transcription factor motif binding sites is provided in Supplementary Table S3.
The Effect of L-Lactate on Immediate-Early Gene Expression Regulation Is NMDAR Dependent
It was previously reported that L-lactate can potentiate the responses of activated NMDARs (Yang et al., 2014). To determine whether the effects of L-lactate on gene expression were mediated by NMDARs, we stimulated neurons for 1 h with lactate in the presence of the NMDARs non-competitive antagonist MK-801 (Wong et al., 1986; Huettner and Bean, 1988) or with the blocker alone, and compared the gene expression profiles. This selective NMDAR blocker was used at a concentration of 40 μM and was applied 30 min before the addition of lactate. Under these conditions NMDARs are fully blocked for the duration of the subsequent treatment (McKay et al., 2013).
MK-801 treatment alone modulated 165 genes (q-value < 0.05), of which 90 were up-regulated and 75 down-regulated. Among the down-regulated genes, we identified Arc, Egr1, and c-Fos, as well as Rgs4, Rgs2, and Dusp6, previously reported to be induced by co-treatment with bicuculline (BiC) and 4-aminopyridine (4-AP) and regulated in the opposite direction by MK-801 (Papadia et al., 2008). 4-AP is a K+ channel antagonist that enhances burst frequency (Papadia et al., 2008), and BiC is a GABAA receptor antagonist that indirectly activates synaptic NMDAR by blocking synaptic inhibition (Avoli et al., 1997). These findings demonstrate that MK-801 treatment in the cortical neurons cultures down-regulates several activity-dependent genes.
A total of 275 genes were differentially regulated in cortical neurons treated with lactate and MK-801 (q-value < 0.05), of which 160 genes were up-regulated and 115 were down-regulated. Five genes were up-regulated with fold changes above 2 – Txnip, Gadd45b, Bbc3, Ier5l, and Lpin1. Txnip, Bbc3, and Ier5 were previously identified as up-regulated in cortical neurons treated for 2 h with APV, a selective NMDAR antagonist (Chen and Hall, 2017).
Arc, Egr1, and c-Fos, genes up-regulated by lactate treatment alone, were down-regulated by the MK-801/lactate co-treatment, with fold changes below -1.5.
When comparing expression profiles modulated by lactate and lactate together with MK-801, we found that 47 out of 113 (more than 41%) of all the genes modulated by L-lactate at 1 h were also differentially expressed when NMDARs were blocked (Figure 2A). Interestingly, 21 out of the 47 genes, including those with the most pronounced fold changes, showed an opposite directionality of response in the co-treatment condition with MK-801 and the condition with the blocker alone (Figure 2B).
FIGURE 2.
Comparison of transcriptional responses to L-lactate, MK-801, NADH, NMDA, and pyruvate. (A) Heatmap with genes regulated by L-lactate in absence and presence of MK-801 channel blocker in cortical neurons. Colors red and green indicate down-regulated and, respectively, up-regulated genes with the corresponding log2(fold change) values. (B) Heatmap with genes regulated by L-lactate, MK-801/L-lactate co-treatment and MK-801 alone. (C) Heatmap with genes modulated by L-lactate, NMDA, and L-lactate/NMDA co-treatment, after 1 h. All genes represented in the heatmap are differentially expressed in each of the treatment conditions (q < 0.05). (D) Total numbers of genes differentially expressed in response to L-lactate, NMDA, and L-lactate/NMDA co-treatment. (E) Total numbers of genes differentially expressed in response to L-lactate, NADH, and NMDA. (F) Comparison of transcriptional responses to L-lactate, NADH, and NMDA after 1 h. (G) Heatmap depicting genes modulated by L-lactate alone, L-lactate/MK-801 co-treatment, and pyruvate, after 1 h. (H) Venn diagram depicting total number of genes modulated by L-lactate, MK-801/L-lactate co-treatment and pyruvate.
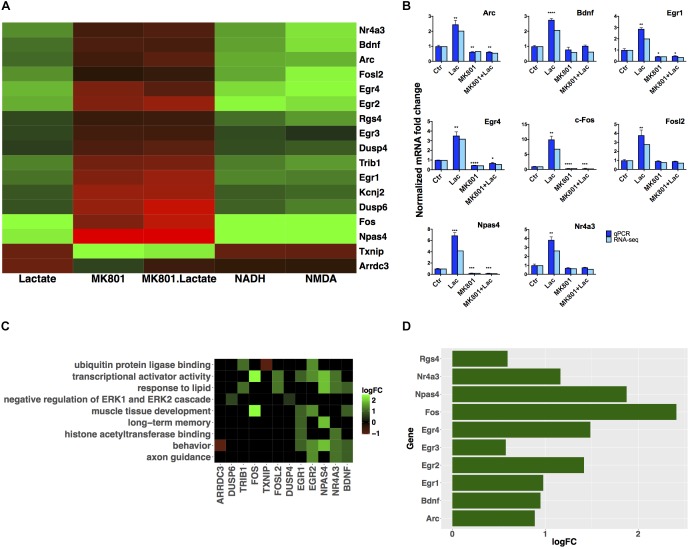
Gene expression validation by quantitative real time-polymerase chain reaction (qRT-PCR) was performed for eight of these genes, and it showed good correlation with RNA-seq data (Figure 3B).
FIGURE 3.
Subset of genes modulated by L-lactate. (A) Fold change modulation of genes regulated by L-lactate in presence and absence of MK-801, and by NADH, NMDA, and MK-801 alone. (B) Validation of RNA-seq data with qRT-PCR, comparison of fold expression changes between RNA-seq and qRT-PCR data for genes is shown. Error bars correspond to standard error of the mean (SEM) values. T-test conducted for qRT-PCR analysis, between the different treatments and Ctr condition; significance (p < 0.05∗, p < 0.01∗∗, p < 0.001∗∗∗, p < 0.0001∗∗∗∗). (C) Genes-terms relationship heatmap emphasizing genes associated with multiple gene ontology terms of interest and showing their fold changes expression in response to L-lactate treatment at 1 h. Up-regulated genes are indicated in green and down-regulated genes in red. All terms displayed are over-represented with a false discovery rate (FDR) < 0.05. (D) Bar plot showing fold induction of a few selected known synaptic plasticity-related genes. The x-axis shows gain of expression log2(fold change) 1 h after L-lactate stimulation.
Five of the previously identified genes involved in the MAPK signaling pathway (Bdnf, c-Fos, Dusp4, Dusp6, Nr4a1) that were up-regulated by lactate treatment alone were down-regulated following co-treatment with MK-801 and L-lactate (Figure 1B). These observations suggest that lactate modulates selective MAPK pathway genes in a NMDAR-dependent manner.
Transcriptional Responses to L-Lactate Are Reproduced by NADH and NMDA
Next, we aimed to validate the effects of L-lactate on gene expression mediated through the NMDAR by testing the effects of the NMDAR agonist NMDA and comparing it to that of L-lactate. Transcriptional responses 1 h after treatment with selective receptor agonist NMDA alone (50 μM) or L-lactate and NMDA co-treatment were assessed. NMDA is an established agonist of the NMDAR, which was previously shown to induce neuronal activity and modulate gene expression in cortical neurons (Thakurela et al., 2015).
We found that 79 out of the 113 genes (70%) modulated by L-lactate are similarly differentially expressed in response to NMDA alone (q < 0.05; Figure 2D). Remarkably, the genes in common between the two conditions have the same directionality of response (Figure 2C).
More than 91% of the genes modulated by L-lactate alone are also modulated by L-lactate and NMDA co-treatment (103 out of 113 genes; Figure 2D). All the fold changes for the genes in common between the two conditions are more pronounced in the case of L-lactate and NMDA co-treatment indicating additive effects (Figure 2C).
These observations further confirm that lactate modulates gene expression in a NMDAR-dependent manner.
Yang et al. (2014) have shown that NADH, a by-product of the metabolic processing of L-lactate to pyruvate, mimics the effect of L-lactate on the expression of Arc, Zif268, Bdnf and c-Fos genes, and that its effect is antagonized by MK-801 and follows NMDAR activation (Yang et al., 2014).
In line with these observations, we hypothesized that NADH could regulate a large proportion of the genes we see modulated by L-lactate at 1 h. Indeed, we found that approximately 65% of the total number of genes regulated by L-lactate is also modulated by NADH (74 out of 113 genes) (Figure 2E).
As many as 95% of the genes modulated by both L-lactate and NADH at 1 h (70 out of 74 genes), were also differentially expressed in response to NMDA treatment, an established agonist of the NMDAR (Figure 2E).
A very close resemblance of gene expression changes, by directionality and fold change values, was observed between the NMDA and NADH treatments (Figure 2F).
This suggests that for a significant proportion of genes (70 out of 113 genes), a mechanism through which L-lactate could affect gene expression is by generating an increase in NADH, which could in turn have a positive modulatory affect on NMDAR signaling.
A subgroup of genes that showed same directionality of response to L-lactate, NADH and NMDA, and an opposite directionality upon co-treatment with L-lactate and MK-801 or the blocker alone, was identified (Figure 3A). These 17 genes have been differentially expressed in each of the treatment conditions described above with a q-value < 0.05.
For these genes, we suggest that they are modulated by L-lactate through downstream NADH increase, which in turn potentiates NMDAR activity via a possible redox-dependent mechanism (Jourdain et al., 2018). The effect of L-lactate on these genes, with one exception only, is MK-801-sensitive and NMDAR-dependent.
The exception is Arrdc3, a gene coding for the arrestin-domain containing protein 3, which is modulated more strongly by L-lactate compared to both NADH and NMDA, and for which the effect of L-lactate on its expression is diminished but still persists upon NMDAR blockade (Figure 3A).
We further conducted a GO over-representation analysis for this subgroup of 17 genes. We found that several genes are transcriptional activators, of which Egr1, Egr2, Npas4, and Nr4a3 are implicated in processes such as memory and behavior, and that also, some of these genes are associated with muscle tissue development: Egr1, Egr2, c-Fos (Figure 3C).
Next, in order to gain further insights into the biological functions of the NMDAR- and NADH-dependent genes, and based on the previous observations that L-lactate stimulates synaptic plasticity genes (Yang et al., 2014), we have performed a literature search to identify other genes from this gene set which could be involved in promoting synaptic plasticity. We were able to identify six additional genes (Egr2, Egr3, Egr4, Npas4, Nr4a3, Rgs4), which participate in synaptic plasticity-related processes (Atluri et al., 2013; Gerber et al., 2016). A list with corresponding fold changes is included in Figure 3D.
Pyruvate Reproduces Only a Few of the Modulatory Effects of L-Lactate on Immediate-Early Genes
When comparing transcriptional responses to L-lactate alone and to L-lactate together with MK-801, we observed that about half of the genes differentially expressed in both conditions showed the same directionality of response (Figure 2A). This provides an indication that there could be non-NMDAR-dependent mechanisms responsible for the effects of L-lactate on gene expression.
As L-lactate is converted into pyruvate by L-lactate dehydrogenase, some of these effects could be mediated by pyruvate level increases. It is well known that astrocyte-derived lactate is taken up by neurons to fuel their energy needs, a process in which lactate is converted to pyruvate, which is then oxidized to CO2 and water in the TCA cycle to produce ATP (Laughton et al., 2007; Wyss et al., 2011; Brooks and Martin, 2015).
To determine whether some of the effects of lactate on gene expression could be due to changes in energy metabolism, we tested whether pyruvate could mimic effects of L-lactate on gene expression.
In cortical neurons, 1 h of pyruvate treatment modulated the expression of 15 genes, of which 11 were down-regulated and 4 genes up-regulated (q-value < 0.05).
Among the genes regulated by pyruvate, we found that eight genes were also significantly modulated by L-lactate (Figure 2H). Interestingly, however, as shown in Figure 2G, the expression of seven of these genes (Phf13, Zfp667, Hes5, Senp8, Jrk, Map10, Arrdc3) was not affected by the NMDAR blocker when co-applied with lactate. This comparison suggests that the effect of L-lactate on the expression of these genes could be due to an increase in the oxidative energy metabolism via pyruvate and its subsequent use by the TCA cycle, but not through activation of NMDARs.
Gene Set Enrichment Analysis Reveals Epigenetic Mediators That Are Regulated by L-Lactate and Gene Targets Linked to Various Phenotypes
In order to achieve additional insights into biological mechanisms modulated by L-lactate, gene set enrichment analysis (GSEA) was performed on the full list of genes robustly expressed (fpkm > 1) in cortical neurons treated with L-lactate after 1 h (12,259 genes). This analysis takes into account genes with low or non-differentially expressed values that can participate in final enrichment through cumulative effects (Subramanian et al., 2005; Fruzangohar et al., 2017).
The GSEA conducted on the 12,259 genes ranked by fold change and using the GO functional database, revealed a positive enrichment of histone acetyltransferase binding genes and a negative enrichment of DNA packaging complex and protein-DNA complex genes. The leading core genes, genes which contributed the most to the enrichment signal calculation for a given GO term (described in “Materials and Methods” section), are listed in Table 1. A full list of ontology terms enriched is included in Supplementary Table S4.
Table 1.
Enriched gene ontology terms as assessed by gene set enrichment analysis of genes expressed after 1 h of L-lactate treatment.
| Gene ontology term | Net enrichment score (NES) | False discovery Rate (FDR) | Leading core genes |
|---|---|---|---|
| Histone acetyltransferase binding | 1.92 | 0.04 | Cebpb; Egrl; Zbtb7a; Cited2; Nr4a3 |
| DNA packaging complex | -2.38 | 0.00 | Histlh4m; Hlf0; Hist2h3cl; Hist2h2aal; Histlh2bk; Histlh2bm; Histlh2bp; Hist2h2bb; Hist2h2be; Histlhlc |
| Protein–DNA complex | -1.99 | 0.01 | Histlh4m; Gins4; Hlf0; Hist2h3cl; Hist2h2aal; Nfe212; Nfya; Nfyb; Poldl; Pole2; Gtf2h3; Terfl; Xpa; Gins2; Hist3h2a; Histlh2bk; Histlh2bm; Histlh2bp; Hist2h2bb; Hist2h2be; Histlhlc; Rpa3 |
A positive net enrichment score indicates that genes over-represented associated with the respective term are up-regulated in the expression dataset. Negative NES indicates the opposite.
It was previously reported that L-lactate is a weak histone deacetylase inhibitor (Latham et al., 2012) and that it can induce histone H3 and H4 hyperacetylation and decrease chromatin compactness in HeLa cells (Wagner et al., 2015).
The same type of analysis conducted on the 12,259 genes using the mammalian phenotype ontology database, revealed enrichment of a series of phenotypes which have been previously linked to the Astrocyte Neuron Lactate Shuttle (ANLS) (Magistretti and Allaman, 2018) and lactate, such as learning (Tadi et al., 2015; Shannon et al., 2016; Jin et al., 2017) and sleep (Naylor et al., 2012; Petit et al., 2013; Haydon, 2017), and others for which there are indications of lactate involvement such as hyperactivity and anxiety disorders (Dillon et al., 1987; Russell et al., 2006; Johnson et al., 2008; Killeen et al., 2013).
The leading core genes accounting for the phenotypes enrichment signal are listed in Table 2. A full list of terms enriched is included in Supplementary Table S4.
Table 2.
Gene targets of L-lactate linked to selected phenotypes by gene set enrichment analysis.
| Phenotype | NES | FDR | Leading core genes |
|---|---|---|---|
| Abnormal NMDA-mediated synaptic currents | 2.10 | 0.03 | Fosb |
| Impaired spatial learning | 2.13 | 0.03 | Fosb |
| Abnormal paradoxical sleep pattern | 2.08 | 0.03 | Fos; Fosb |
| Abnormal sleep pattern | 2.28 | 0.00 | Fos; Fosb; Perl; Per2; Ptchdl; Slcl8a2 |
| Impaired learning | 2.29 | 0.00 | Arx; Fmrl; Fosb; Nr4a2; Syt4; Ptchdl; Tnc |
| Abnormal sleep behavior | 2.14 | 0.03 | Fos; Fosb; Perl; Per2; Ptchdl; Slcl8a2; Foxo3 |
| Abnormal interleukin-4 secretion | 2.09 | 0.03 | Atf3; Cish; Gadd45b; Nfatcl; Nfil3; Dusp4; Rnfl28 |
| Decreased dopamine level | 2.11 | 0.03 | Gpr37; Hipk2; Nr4a2; Perl; Ptgerl; Sstr2; Slcl8a2; Tnc |
| Abnormal tumor necrosis factor level | 2.21 | 0.01 | Ticam; Cebpb; Crebbp; Egrl; Fos; Ier3; Ldlr; Mapkapk2; Mertk; Ncoa3; Nfil3; Npylr; Pcskl; Duspl; Relb; Arid5a; Tnfrsfla; Pde4d; Sh3bp2; Rc3hl; Tbkl; Apba3; Pelil |
| Abnormal fear/anxiety-related behavior | 2.15 | 0.03 | Spred; Adcyap1; Arc; Arx; Bdnf; Crem; Arid3a; Fmr1; Fos; Fosb; Gpr19; Grin2a; Nr4a2; Penk; Dusp1; Rgs2; Slc2a3; Sstr2; Cpeb3; Slc18a2; Git1; Tnc; Npas4; Gpr26; Dcaf10; B3galt2; Park2; Lrrtm1 |
| Hyperactivity | 2.13 | 0.03 | Braf; Adcyapl; Arx; Bdnf; Chrm4; Crem; Fmrl; Fos; Fosb; Ldlr; Nr4a3; Npylr; Nr4a2; Perl; Lrrc4; Rgs4; Slcl2a2; Ptchdl; Gitl; Tnc; Npas4; Sik2; B3galt2; Dnajb9; Vgf; Usp2; Fezf2; Cystml; Adipor2; Ssfa2; Gpr22 |
On the Long-Term L-Lactate Modulates Genes Involved in Receptor Signaling and Neuronal Excitability
In order to investigate how the transcriptome of cortical neurons is regulated by L-lactate on the long-term, differentially expressed genes in cortical neurons after 6 h treatment with L-lactate were identified.
In total, 313 genes were differentially expressed, of which 112 were down-regulated and 201 were up-regulated (Benjamini-Hochberg adjusted p-value < 0.05). Fold change values for the majority of these genes were below 1.5, which indicates weak modulation.
Among these 313 genes, 8 are enriched in astrocytes (Dio2, Fras1, Gfap, Mtmr11, Ngef, Notch3, Ppp1r3c, Thbs1), 9 in endothelial cells (Apln, Cd55, Lrrc55, Net1, Odc1, Slc39a10, Tes, Tfrc, Usp43), 3 in microglia (Rtn4rl1, St6galnac4, Txnip), and 3 in oligodendrocytes (Bcas1, Dnph1, Plp1).
Some of the up-regulated genes with the strongest modulation were Vegfa (vascular endothelial growth factor-coding gene), Synj2 (synaptojanin 2) and Baiap2l2 (brain specific angiogenesis inhibitor 1-associated protein 2-like protein 2).
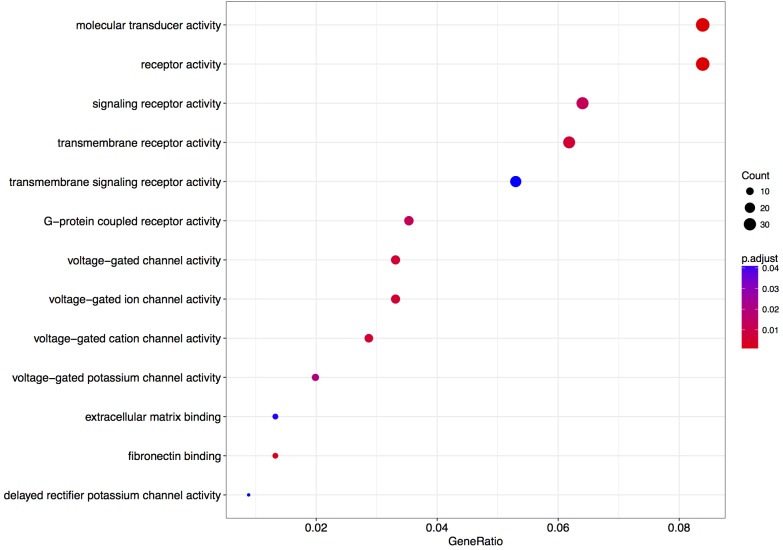
We next performed a GO over-representation test on all differentially expressed genes with a q-value < 0.1 (in total 484 genes – Supplementary Table S5).
The enrichment of genes coding for voltage-gated cation channel activity proteins and potassium channels was observed (Figure 4). Few of the terms are associated with receptor-mediated signaling events, which could be associated with synaptic plasticity phenomena.
FIGURE 4.
Enriched gene ontology terms 6 h after L-lactate stimulation. GO plot shows enrichment of biological processes and molecular functions for genes modulated in cortical neurons by L-lactate after 6 h.
In addition to the GO analysis, we have performed a gene set enrichment analysis on all genes robustly expressed (fpkm > 1) in cortical neurons in the treatment condition with L-lactate after 6 h (12,462 genes).
A positive enrichment of genes located at the synaptic membrane and dendritic processes was observed, along with a negative enrichment of genes coding for members of the cytochrome complex, the mitochondrial matrix, and the DNA packaging complex (Table 3).
Table 3.
Enriched gene ontology terms as assessed by gene set enrichment analysis of genes expressed after 6 h of L-lactate treatment.
| Gene ontology ID | Description | NES | FDR | Core genes |
|---|---|---|---|---|
| GO:0030425 | Dendrite | 1.62 | 0.07 | Supplementary Table S4 |
| GO:0097060 | Synaptic membrane | 1.62 | 0.07 | Supplementary Table S4 |
| GO:0070069 | Cytochrome complex | -1.73 | 0.08 | Uqcc3; Cox6a2; Cox7c; Cox8b; Ndufa4; Coxl5; Uqcrl0; Coxl0 |
| GO:0005759 | Mitochondrial matrix | -1.79 | 0.08 | Supplementary Table S4 |
| GO:0044815 | DNA packaging complex | -1.79 | 0.08 | Cenpa; Smc2; Hist2h2aal; H2afj; Hlfx; Hist3h2a; Histlh2bk; Histlh2bm; Histlh2bp; Hist2h2bb; Hist2h2be; Cenpt; H2afy2; Histlhlc; Ncapd2; Hist3h2ba; Ncapd3 |
Discussion
In this study, we report a transcriptional analysis of gene expression modulation in neurons evoked by exposure to L-lactate. Taken together, the results obtained expand in an unbiased manner and refine the findings of the previous investigation made on candidate genes which demonstrated that L-lactate potentiates NMDA signaling in neurons, promotes expression of selected plasticity genes, and that NADH reproduces the effects of L-lactate on NMDA signaling (Yang et al., 2014). In addition, the current study shows that L-lactate and NADH regulate a majority of immediate early genes (70 out of 113 genes) in a similar way as the NMDAR agonist, NMDA.
This study has identified six synaptic plasticity-promoting genes – in addition to Arc, Egr1, c-Fos and Bdnf (Yang et al., 2014) – namely: Egr2, Egr3, Egr4, Npas4, Nr4a3, Rgs4 which are modulated by L-lactate in cortical neurons, and 10 genes associated with the MAPK signaling pathway, a key-signaling cascade mediating cytoplasmic and nuclear responses to synaptic activity, and NMDAR-dependent neuronal plasticity, namely: c-Fos, Bdnf, Atf4, Nr4a1, Gadd45b, Gadd45g, Map3k11, Dusp4, Dusp6, Dusp10 (Sweatt, 2001; Yang et al., 2014; Bluthgen et al., 2017).
In a previous study, Zhang et al. (2007) showed data indicating that the sub-cellular location of the activated NMDARs (synaptic vs. extra-synaptic) specifies the phenotype of the transcriptional response. Stimulation of synaptic NMDARs up-regulates pro-survival genes and down-regulates pro-death genes, while activation of extra-synaptic NMDARs leads to an opposite effect (Zhang et al., 2007).
Interestingly, among the 113 genes significantly modulated by L-lactate after 1 h (q-value < 0.1), 13 genes (1190002N15Rik, Atf4, Dusp6, Dusp10, Fosl2, Frmd6, Junb, Maff, Nfil3, Npas4, Nr4a1, Pim3, Trib1) were previously reported as regulated by a stimulation protocol known to specifically activate synaptic NMDARs (Zhang et al., 2007), whereas none are common with genes regulated by stimulating extra-synaptic NMDARs (Zhang et al., 2007).
In addition, five out of the eight genes previously reported (Papadia et al., 2008) as modulated by BiC and 4-AP in a MK-801 sensitive manner, both in vitro and in vivo, are significantly modulated by 1 h L-lactate treatment with the same pattern of regulation: Dusp6, Egr1, Rgs2, Rgs4, and Txnip. It was previously shown that BiC/4-AP treatment triggers activation of synaptic but not extra-synaptic NMDARs (Hardingham et al., 2002). Together, these observations suggest that lactate may act preferentially on synaptically localized NMDARs regulating transcriptome profile favoring neuroprotection and plasticity.
A transcription factor motifs binding sites analysis revealed that a majority of the genes modulated by L-lactate at 1 h are under the regulatory influence of transcription factors, which are induced by synaptic activity and mediate plasticity events (SRF, CREB1, Egr1, NF-κB, Mycn). Remarkably, the gene coding for Egr1, one of the transcription factors with over-represented binding sites, is itself up-regulated by L-lactate after 1 h (fold change of 1.97), and could well represent a master regulator of gene expression in response to L-lactate.
The set of 16 genes identified (excluding Arrdc3) for which there is strong evidence of a NADH-mediated and MK-801-sensitive mechanism (Figure 3A), appear as representing a “canonical” set of early genes modulated by L-lactate in cortical neurons in a NMDAR-dependent manner.
Arrdc3 modulation showed an intriguing feature, presenting an opposite directionality of response with the blocker alone compared to L-lactate treatment, but the same directionality of response in the L-lactate treatment condition and the co-treatment condition with MK-801. This partial insensitivity to MK-801 in presence of L-lactate could be explained by the fact that pyruvate was also found to modulate this gene and thus the effect of L-lactate on its regulation could be a metabolic and not an NMDAR/NADH-dependent one. Arrdc3 regulates the endosomal residence time and intracellular signaling of the β2 adrenergic receptor in HEK293 and BT549 cancer cell lines (Patwari et al., 2011; Tian et al., 2016), however, to our knowledge, there is no evidence in the literature for a role in the brain, where its transcript levels were found to be very low (Patwari et al., 2011).
In this study, we have identified in total seven genes whose expression is modulated by pyruvate: Phf13, Zfp667, Hes5, Senp8, Jrk, Map10, and Arrdc3.
One mechanism through which NADH reproduces L-lactate-mediated gene expression could rely on its ability to positively modulate NMDAR activity through its redox-sensitive regulatory sites (Choi et al., 2001; Jourdain et al., 2018). Consistent with this, NADH has been previously shown to trigger an MK-801-sensitive (hence NMDAR-mediated) rise in intracellular calcium and to amplify NMDAR-mediated currents (Yang et al., 2014).
A previous study reported that rapid activity-induced transcription of IEGs in neurons relies on elevated RNA polymerase II presence at promoters through stalling (Saha et al., 2011). It is notable, that some of the most enriched GO terms for genes modulated after 1 h by L-lactate are associated with RNA polymerase II activity, binding, and regulation. This evidence could point to a mechanism responsible for IEGs induction by L-lactate, upon NMDAR potentiation.
It is also of note that among the most enriched GO terms for biological processes, muscle tissue development and differentiation are included. This may reflect the presence of neurotrophin-coding genes which have also been reported to play an important role in muscle differentiation and regeneration (Clow and Jasmin, 2010), and could indicate a conserved gene regulatory role of L-lactate in muscle and the brain.
The enrichment of genes coding for voltage-gated cation channel activity proteins and potassium channels after 6 h, point to long-term signaling effects of L-lactate on regulation of neuronal excitability (Sjostrom et al., 2008; Shah and Aizenman, 2014). The positive enrichment of genes located at the synaptic membrane and dendritic processes, points to effects on synaptic plasticity processes. Among the genes with the highest fold change response to L-lactate after 6 h, Vegfa is known to stimulate neuronal survival and axonal outgrowth (Carmeliet and Storkebaum, 2002; Theis and Theiss, 2018), while Synj2 is implicated in recycling neurotransmitter vesicles (Planchart, 2013), and Baiap2l2’s homolog Baiap2 has been implicated in dendritic spine development and NMDAR regulation (Kang et al., 2016).
Apart from Vegfa, other genes implicated in neuronal survival and neuroprotection were modulated by L-lactate after 1 h, namely Nr4a2, Npas4/Nxf, Bdnf, and Bcl2l11/Bim, and after 6 h: Adcyap1, Bdnf, Apaf1, Bim, Gfra2, Hrk (Table 4). Interestingly, Hrk, Apaf1, and Bim that are key factors in promoting neuronal cell death (Cheung et al., 2005; Hetz et al., 2007; Steckley et al., 2007; Ghosh et al., 2011) were down-regulated by L-lactate, while Bdnf, Nr4a2, Npas4, Adcyap1, and Gfra2, that facilitate neuronal survival (Lahteenmaki et al., 2007) and protection (Vaudry et al., 2002; Husson et al., 2005; Shintani et al., 2005; Ooe et al., 2009; Volakakis et al., 2010) were up-regulated by L-lactate. Furthermore, L-lactate treatment for 1 h strongly down- regulated (absolute fold change of 2) Txnip, a gene coding for an endogenous inhibitor of the antioxidative function of thioredoxin. Txnip upregulation was associated with stroke and other neurological conditions and its inhibition was shown to be neuroprotective (Al-Gayyar et al., 2011; Nasoohi et al., 2018). These observations collectively suggest a role for L-lactate in modulating a neuroprotective transcriptional program.
Table 4.
Neuroprotection and pro-apoptotic genes modulated by L-lactate after 1 h and 6 h treatment.
| Function | Gene symbol | Log2FC | Time |
|---|---|---|---|
| Neuroprotection | Nr4a2 | 1.91 | 1 h |
| Npas4 | 1.88 | 1 h | |
| Bdnf | 0.95 0.40 | 1 h 6 h | |
| Vegfa | 0.83 | 6 h | |
| Adcyapl | 0.53 | 6 h | |
| Gfra2 | 0.30 | 6 h | |
| Cell death | Txnip | -0.99 | 1 h |
| Apafl | -0.23 | 6 h | |
| Bcl2111 | -0.35 | 6 h | |
| Hrk | -0.34 | 6 h |
A number of genes significantly modulated by L-lactate after 1 h are not mediated through NMDAR signaling, or through NADH or pyruvate increases. They could be regulated through epigenetic mechanisms linked to L-lactate’s weak histone deacetylase activity inhibitory role, previously reported in literature (Latham et al., 2012). Gene set enrichment analysis results have provided indications that L-lactate modulates genes coding for histone acetyltransferase binding proteins, and histone protein complexes, which could in turn impact chromatin compactness and organization, and gene expression.
The phenotypes identified as enriched for the list of genes modulated by L-lactate at 1 h point to a series of genes relevant to sleep physiology and psychiatric conditions such as anxiety and hyperactivity that open avenues for further investigation.
Conclusion
The present study provides a molecular dissection of the transcriptome-level responses to L-lactate in cortical neurons leading to identification of several groups of genes that are responsive to this energy metabolite and signaling molecule (Magistretti and Allaman, 2018) and that are important for the physiology of neurons and processes like synaptic plasticity and neuroprotection (Figure 5).
FIGURE 5.
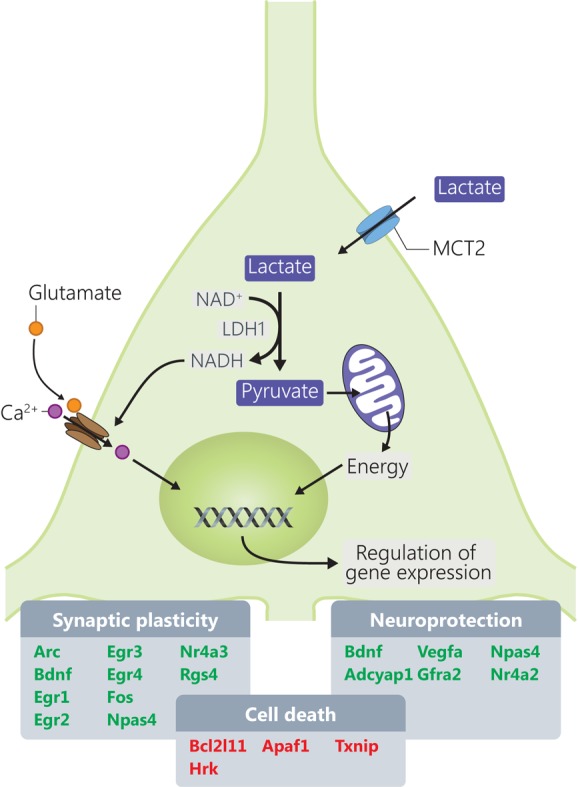
Mechanisms through which L-lactate modulates mRNA expression of genes involved in synaptic plasticity and neuroprotection. Genes highlighted in red and green are downregulated, respectively, upregulated by L-lactate.
Accession Code
Fastq files were uploaded to NCBI SRA database with accession number SRP150704.
Author Contributions
HF and PM designed the research. HF and MM performed the research. MM, HF, and HM analyzed the data. MM, HF, and PM wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Jonathan Besuchet for generating initial data analysis scripts and Evelyne Ruchti for expert technical assistance.
Funding. This research was supported by King Abdullah University of Science and Technology (KAUST), the NCCR Synapsy and by Prefargier Foundation.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2018.00375/full#supplementary-material
References
- Adeva-Andany M., López-Ojén M., Funcasta-Calderón R., Ameneiros-Rodríguez E., Donapetry-García C., Vila-Altesor M., et al. (2014). Comprehensive review on lactate metabolism in human health. Mitochondrion 17 76–100. 10.1016/j.mito.2014.05.007 [DOI] [PubMed] [Google Scholar]
- Ahmed A., Wan X., Mitxitorena I., Lindsay A. J., Paolo Pandolfi P., McCaffrey M. W., et al. (2017). Regulation of NF-κB by PML and PML-RARα. Sci. Rep. 7:44539. 10.1038/srep44539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Gayyar M. M., Abdelsaid M. A., Matragoon S., Pillai B. A., El-Remessy A. B. (2011). Thioredoxin interacting protein is a novel mediator of retinal inflammation and neurotoxicity. Br. J. Pharmacol. 164 170–180. 10.1111/j.1476-5381.2011.01336.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allaman I., Gavillet M., Belanger M., Laroche T., Viertl D., Lashuel H. A., et al. (2010). Amyloid-beta aggregates cause alterations of astrocytic metabolic phenotype: impact on neuronal viability. J. Neurosci. 30 3326–3338. 10.1523/JNEUROSCI.5098-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atluri V. S. R., Kanthikeel S. P., Reddy P. V. B., Yndart A., Nair M. P. N. (2013). Human synaptic plasticity gene expression profile and dendritic spine density changes in HIV-infected human CNS cells: role in HIV-associated neurocognitive disorders (HAND). PLoS One 8:e61399. 10.1371/journal.pone.0061399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avoli M., Hwa G., Louvel J., Kurcewicz I., Pumain R., Lacaille J. C. (1997). Functional and pharmacological properties of GABA-mediated inhibition in the human neocortex. Can. J. Physiol. Pharmacol. 75 526–534. 10.1139/y97-037 [DOI] [PubMed] [Google Scholar]
- Baumann F., Leukel P., Doerfelt A., Beier C. P., Dettmer K., Oefner P. J., et al. (2009). Lactate promotes glioma migration by TGF-β2–dependent regulation of matrix metalloproteinase-2. Neuro Oncol. 11 368–380. 10.1215/15228517-2008-106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belanger M., Yang J., Petit J. M., Laroche T., Magistretti P. J., Allaman I. (2011). Role of the glyoxalase system in astrocyte-mediated neuroprotection. J. Neurosci. 31 18338–18352. 10.1523/JNEUROSCI.1249-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluthgen N., van Bentum M., Merz B., Kuhl D., Hermey G. (2017). Profiling the MAPK/ERK dependent and independent activity regulated transcriptional programs in the murine hippocampus in vivo. Sci. Rep. 7:14. 10.1038/srep45101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks G. A., Martin N. A. (2015). Cerebral metabolism following traumatic brain injury: new discoveries with implications for treatment. Front. Neurosci. 8:408. 10.3389/fnins.2014.00408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P., Storkebaum E. (2002). Vascular and neuronal effects of VEGF in the nervous system: implications for neurological disorders. Semin. Cell Dev. Biol. 13 39–53. 10.1006/scdb.2001.0290 [DOI] [PubMed] [Google Scholar]
- Chen F. D., Hall B. J. (2017). Synaptic activity suppresses expression of neurogenic differentiation factor 2 in an NMDA receptor-dependent manner. Synapse 71:e21986. 10.1002/syn.21986 [DOI] [PubMed] [Google Scholar]
- Cheung E. C. C., Melanson-Drapeau L., Cregan S. P., Vanderluit J. L., Ferguson K. L., McIntosh W. C., et al. (2005). Apoptosis-inducing factor is a key factor in neuronal cell death propagated by BAX-dependent and BAX-independent mechanisms. J. Neurosci. 25 1324–1334. 10.1523/JNEUROSCI.4261-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y., Chen H. V., Lipton S. A. (2001). Three pairs of cysteine residues mediate both redox and zn2+ modulation of the NMDA receptor. J. Neurosci. 21 392–400. 10.1523/JNEUROSCI.21-02-00392.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clow C., Jasmin B. J. (2010). Brain-derived neurotrophic factor regulates satellite cell differentiation and skeltal muscle regeneration. Mol. Biol. Cell 21 2182–2190. 10.1091/mbc.E10-02-0154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole A. J., Saffen D. W., Baraban J. M., Worley P. F. (1989). Rapid increase of an immediate early gene messenger RNA in hippocampal neurons by synaptic NMDA receptor activation. Nature 340 474–476. 10.1038/340474a0 [DOI] [PubMed] [Google Scholar]
- Colegio O. R., Chu N.-Q., Szabo A. L., Chu T., Rhebergen A. M., Jairam V., et al. (2014). Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature 513 559–563. 10.1038/nature13490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cull-Candy S. G., Leszkiewicz D. N. (2004). Role of distinct NMDA receptor subtypes at central synapses. Sci. STKE 2004:re16. [DOI] [PubMed] [Google Scholar]
- Dietl K., Renner K., Dettmer K., Timischl B., Eberhart K., Dorn C., et al. (2010). Lactic acid and acidification inhibit TNF secretion and glycolysis of human monocytes. J. Immunol. 184 1200–1209. 10.4049/jimmunol.0902584 [DOI] [PubMed] [Google Scholar]
- Dillon D. J., Gorman J. M., Liebowitz M. R., Fyer A. J., Klein D. F. (1987). Measurement of lactate-induced panic and anxiety. Psychiatry Res. 20 97–105. 10.1016/0165-1781(87)90002-3 [DOI] [PubMed] [Google Scholar]
- Engelmann C., Haenold R. (2016). Transcriptional control of synaptic plasticity by transcription factor NF-κB. Neural Plast. 2016:7027949. 10.1155/2016/7027949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell S. W., Greenberg M. E. (2008). Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu. Rev. Neurosci. 31 563–590. 10.1146/annurev.neuro.31.060407.125631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruzangohar M., Ebrahimie E., Adelson D. L. (2017). A novel hypothesis-unbiased method for Gene Ontology enrichment based on transcriptome data. PLoS One 12:e0170486. 10.1371/journal.pone.0170486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber K. J., Squires K. E., Hepler J. R. (2016). Roles for regulator of G protein signaling proteins in synaptic signaling and plasticity. Mol. Pharmacol. 89 273–286. 10.1124/mol.115.102210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A. P., Cape J. D., Klocke B. J., Roth K. A. (2011). Deficiency of pro-apoptotic Hrk attenuates programmed cell death in the developing murine nervous system but does not affect Bcl-x deficiency-induced neuron apoptosis. J. Histochem. Cytochem. 59 976–983. 10.1369/0022155411424311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottfried E., Kunz-Schughart L. A., Ebner S., Mueller-Klieser W., Hoves S., Andreesen R., et al. (2006). Tumor-derived lactic acid modulates dendritic cell activation and antigen expression. Blood 107 2013–2021. 10.1182/blood-2005-05-1795 [DOI] [PubMed] [Google Scholar]
- Gutierrez H., Davies A. M. (2011). Regulation of neural process growth, elaboration and structural plasticity by NF-kappaB. Trends Neurosci. 34 316–325. 10.1016/j.tins.2011.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzowski J. F., Setlow B., Wagner E. K., McGaugh J. L. (2001). Experience-dependent gene expression in the rat hippocampus after spatial learning: a comparison of the immediate-early genes Arc, c-fos, and zif268. J. Neurosci. 21 5089–5098. 10.1523/JNEUROSCI.21-14-05089.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas R., Cucchi D., Smith J., Pucino V., Macdougall C. E., Mauro C. (2016). Intermediates of metabolism: from bystanders to signalling molecules. Trends Biochem. Sci. 41 460–471. 10.1016/j.tibs.2016.02.003 [DOI] [PubMed] [Google Scholar]
- Haas R., Smith J., Rocher-Ros V., Nadkarni S., Montero-Melendez T., D’Acquisto F., et al. (2015). Lactate regulates metabolic and pro-inflammatory circuits in control of T cell migration and effector functions. PLoS Biol. 13:e1002202. 10.1371/journal.pbio.1002202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenold R., Weih F., Herrmann K.-H., Schmidt K.-F., Krempler K., Engelmann C., et al. (2014). NF-κB controls axonal regeneration and degeneration through cell-specific balance of RelA and p50 in the adult CNS. J. Cell Sci. 127 3052–3065. 10.1242/jcs.140731 [DOI] [PubMed] [Google Scholar]
- Hall J., Thomas K. L., Everitt B. J. (2001). Cellular imaging of zif268 expression in the hippocampus and amygdala during contextual and cued fear memory retrieval: selective activation of hippocampal CA1 neurons during the recall of contextual memories. J. Neurosci. 21 2186–2193. 10.1523/JNEUROSCI.21-06-02186.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardingham G. E., Fukunaga Y., Bading H. (2002). Extrasynaptic NMDARs oppose synaptic NMDARs by triggering CREB shut-off and cell death pathways. Nat. Neurosci. 5 405–414. 10.1038/nn835 [DOI] [PubMed] [Google Scholar]
- Hashimoto T., Hussien R., Oommen S., Gohil K., Brooks G. A. (2007). Lactate sensitive transcription factor network in L6 cells: activation of MCT1 and mitochondrial biogenesis. FASEB J. 21 2602–2612. 10.1096/fj.07-8174com [DOI] [PubMed] [Google Scholar]
- Haydon P. G. (2017). Astrocytes and the modulation of sleep. Curr. Opin. Neurobiol. 44 28–33. 10.1016/j.conb.2017.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C., Thielen P., Fisher J., Pasinelli P., Brown R. H., Korsmeyer S., et al. (2007). The proapoptotic BCL-2 family member BIM mediates motoneuron loss in a model of amyotrophic lateral sclerosis. Cell Death Differ. 14 1386–1389. 10.1038/sj.cdd.4402166 [DOI] [PubMed] [Google Scholar]
- Huettner J. E., Bean B. P. (1988). Block of N-methyl-D-aspartate-activated current by the anticonvulsant MK-801: selective binding to open channels. Proc. Natl. Acad. Sci. U.S.A. 85 1307–1311. 10.1073/pnas.85.4.1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husson I., Rangon C. M., Lelievre V., Bemelmans A. P., Sachs P., Mallet J., et al. (2005). BDNF-induced white matter neuroprotection and stage-dependent neuronal survival following a neonatal excitotoxic challenge. Cereb. Cortex 15 250–261. 10.1093/cercor/bhh127 [DOI] [PubMed] [Google Scholar]
- Ibberson D., Benes V., Muckenthaler M. U., Castoldi M. (2009). RNA degradation compromises the reliability of microRNA expression profiling. BMC Biotechnol. 9:102. 10.1186/1472-6750-9-102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C., Gao L., Li Y., Wu S., Lu X., Yang J., et al. (2017). Lanthanum damages learning and memory and suppresses astrocyte-neuron lactate shuttle in rat hippocampus. Exp. Brain Res. 235 3817–3832. 10.1007/s00221-017-5102-5 [DOI] [PubMed] [Google Scholar]
- Johnson P. L., Truitt W. A., Fitz S. D., Lowry C. A., Shekhar A. (2008). Neural pathways underlying lactate-induced panic. Neuropsychopharmacology 33 2093–2107. 10.1038/sj.npp.1301621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. W., Errington M. L., French P. J., Fine A., Bliss T. V., Garel S., et al. (2001). A requirement for the immediate early gene Zif268 in the expression of late LTP and long-term memories. Nat. Neurosci. 4 289–296. 10.1038/85138 [DOI] [PubMed] [Google Scholar]
- Jopling C. L., Willis A. E. (2001). N-myc translation is initiated via an internal ribosome entry segment that displays enhanced activity in neuronal cells. Oncogene 20 2664–2670. 10.1038/sj.onc.1204404 [DOI] [PubMed] [Google Scholar]
- Jourdain P., Allaman I., Rothenfusser K., Fiumelli H., Marquet P., Magistretti P. J. (2016). L-Lactate protects neurons against excitotoxicity: implication of an ATP-mediated signaling cascade. Sci. Rep. 6:21250. 10.1038/srep21250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourdain P., Rothenfusser K., Ben-Adiba C., Allaman I., Marquet P., Magistretti P. J. (2018). Dual action of L-Lactate on the activity of NR2B-containing NMDA receptors: from potentiation to neuroprotection. Sci. Rep. 8:13472. 10.1038/s41598-018-31534-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J., Park H., Kim E. (2016). IRSp53/BAIAP2 in dendritic spine development, NMDA receptor regulation, and psychiatric disorders. Neuropharmacology 100 27–39. 10.1016/j.neuropharm.2015.06.019 [DOI] [PubMed] [Google Scholar]
- Killeen P. R., Russell V. A., Sergeant J. A. (2013). A behavioral neuroenergetics theory of ADHD. Neurosci. Biobehav. Rev. 37 625–657. 10.1016/j.neubiorev.2013.02.011 [DOI] [PubMed] [Google Scholar]
- Kim J. W., Dang C. V. (2006). Cancer’s molecular sweet tooth and the Warburg effect. Cancer Res. 66 8927–8930. 10.1158/0008-5472.CAN-06-1501 [DOI] [PubMed] [Google Scholar]
- Knoll B., Nordheim A. (2009). Functional versatility of transcription factors in the nervous system: the SRF paradigm. Trends Neurosci. 32 432–442. 10.1016/j.tins.2009.05.004 [DOI] [PubMed] [Google Scholar]
- Kwon A. T., Arenillas D. J., Worsley Hunt R., Wasserman W. W. (2012). oPOSSUM-3: advanced analysis of regulatory motif over-representation across genes or ChIP-Seq datasets. G3 2 987–1002. 10.1534/g3.112.003202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahteenmaki M., Kupari J., Airaksinen M. S. (2007). Increased apoptosis of parasympathetic but not enteric neurons in mice lacking GFRalpha2. Dev. Biol. 305 325–332. 10.1016/j.ydbio.2007.02.016 [DOI] [PubMed] [Google Scholar]
- Latham T., Mackay L., Sproul D., Karim M., Culley J., Harrison D. J., et al. (2012). Lactate, a product of glycolytic metabolism, inhibits histone deacetylase activity and promotes changes in gene expression. Nucleic Acids Res. 40 4794–4803. 10.1093/nar/gks066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau C. G., Zukin R. S. (2007). NMDA receptor trafficking in synaptic plasticity and neuropsychiatric disorders. Nat. Rev. Neurosci. 8 413–426. 10.1038/nrn2153 [DOI] [PubMed] [Google Scholar]
- Laughton J. D., Bittar P., Charnay Y., Pellerin L., Kovari E., Magistretti P. J., et al. (2007). Metabolic compartmentalization in the human cortex and hippocampus: evidence for a cell- and region-specific localization of lactate dehydrogenase 5 and pyruvate dehydrogenase. BMC Neurosci. 8:35. 10.1186/1471-2202-8-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lynch M. A. (2004). Long-term potentiation and memory. Physiol. Rev. 84 87–136. 10.1152/physrev.00014.2003 [DOI] [PubMed] [Google Scholar]
- Magistretti P. J., Allaman I. (2018). Lactate in the brain: from metabolic end-product to signalling molecule. Nat. Rev. Neurosci. 19 235–249. 10.1038/nrn.2018.19 [DOI] [PubMed] [Google Scholar]
- McKay S., Bengtson C. P., Bading H., Wyllie D. J. A., Hardingham G. E. (2013). Recovery of NMDA receptor currents from MK-801 blockade is accelerated by Mg2+ and memantine under conditions of agonist exposure. Neuropharmacology 74 119–125. 10.1016/j.neuropharm.2013.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milovanova T. N., Bhopale V. M., Sorokina E. M., Moore J. S., Hunt T. K., Hauer-Jensen M., et al. (2008). Lactate stimulates vasculogenic stem cells via the thioredoxin system and engages an autocrine activation loop involving hypoxia-inducible factor 1. Mol. Cell. Biol. 28 6248–6261. 10.1128/MCB.00795-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasoohi S., Ismael S., Ishrat T. (2018). Thioredoxin-interacting protein (TXNIP) in cerebrovascular and neurodegenerative diseases: regulation and implication. Mol. Neurobiol. 55 7900–7920. 10.1007/s12035-018-0917-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor E., Aillon D. V., Barrett B. S., Wilson G. S., Johnson D. A., Johnson D. A., et al. (2012). Lactate as a biomarker for sleep. Sleep 35 1209–1222. 10.5665/sleep.2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooe N., Motonaga K., Kobayashi K., Saito K., Kaneko H. (2009). Functional characterization of basic helix-loop-helix-PAS type transcription factor NXF in vivo: putative involvement in an “on demand” neuroprotection system. J. Biol. Chem. 284 1057–1063. 10.1074/jbc.M805196200 [DOI] [PubMed] [Google Scholar]
- Papadia S., Soriano F. X., Leveille F., Martel M. A., Dakin K. A., Hansen H. H., et al. (2008). Synaptic NMDA receptor activity boosts intrinsic antioxidant defenses. Nat. Neurosci. 11 476–487. 10.1038/nn2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwari P., Emilsson V., Schadt E. E., Chutkow W. A., Lee S., Marsili A., et al. (2011). The arrestin domain containing 3 (ARRDC3) protein regulates body mass and energy expenditure. Cell Metab. 14 671–683. 10.1016/j.cmet.2011.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellerin L., Magistretti P. J. (1994). Glutamate uptake into astrocytes stimulates aerobic glycolysis: a mechanism coupling neuronal activity to glucose utilization. Proc. Natl. Acad. Sci. U.S.A. 91 10625–10629. 10.1073/pnas.91.22.10625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellerin L., Magistretti P. J. (2012). Sweet sixteen for ANLS. J. Cereb. Blood Flow Metab. 32 1152–1166. 10.1038/jcbfm.2011.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit J. M., Gyger J., Burlet-Godinot S., Fiumelli H., Martin J. L., Magistretti P. J. (2013). Genes involved in the astrocyte-neuron lactate shuttle (ANLS) are specifically regulated in cortical astrocytes following sleep deprivation in mice. Sleep 36 1445–1458. 10.5665/sleep.3034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planchart A. (2013). Analysis of an intronic promoter within Synj2. Biochem. Biophys. Res. Commun. 440 640–645. 10.1016/j.bbrc.2013.09.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramskold D., Wang E. T., Burge C. B., Sandberg R. (2009). An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput. Biol. 5:e1000598. 10.1371/journal.pcbi.1000598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell V. A., Oades R. D., Tannock R., Killeen P. R., Auerbach J. G., Johansen E. B., et al. (2006). Response variability in attention-deficit/hyperactivity disorder: a neuronal and glial energetics hypothesis. Behav. Brain Funct. 2:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sada N., Lee S., Katsu T., Otsuki T., Inoue T. (2015). Epilepsy treatment. Targeting LDH enzymes with a stiripentol analog to treat epilepsy. Science 347 1362–1367. 10.1126/science.aaa1299 [DOI] [PubMed] [Google Scholar]
- Saha R. N., Wissink E. M., Bailey E. R., Zhao M., Fargo D. C., Hwang J. Y., et al. (2011). Rapid activity-induced transcription of Arc and other IEGs relies on poised RNA polymerase II. Nat. Neurosci. 14 848–856. 10.1038/nn.2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala C., Rudolph-Correia S., Sheng M. (2000). Developmentally regulated NMDA receptor-dependent dephosphorylation of cAMP response element-binding protein (CREB) in hippocampal neurons. J. Neurosci. 20 3529–3536. 10.1523/JNEUROSCI.20-10-03529.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- San-Millan I., Brooks G. A. (2017). Reexamining cancer metabolism: lactate production for carcinogenesis could be the purpose and explanation of the Warburg Effect. Carcinogenesis 38 119–133. 10.1093/carcin/bgw127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saul M. C., Seward C. H., Troy J. M., Zhang H., Sloofman L. G., Lu X., et al. (2017). Transcriptional regulatory dynamics drive coordinated metabolic and neural response to social challenge in mice. Genome Res. 27 959–972. 10.1101/gr.214221.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah N. H., Aizenman E. (2014). Voltage-gated potassium channels at the crossroads of neuronal function, ischemic tolerance, and neurodegeneration. Transl. Stroke Res. 5 38–58. 10.1007/s12975-013-0297-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon B. J., Vaishnavi S. N., Vlassenko A. G., Shimony J. S., Rutlin J., Raichle M. E. (2016). Brain aerobic glycolysis and motor adaptation learning. Proc. Natl. Acad. Sci. U.S.A. 113 E3782–E3791. 10.1073/pnas.1604977113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani N., Suetake S., Hashimoto H., Koga K., Kasai A., Kawaguchi C., et al. (2005). Neuroprotective action of endogenous PACAP in cultured rat cortical neurons. Regul. Pept. 126 123–128. 10.1016/j.regpep.2004.08.014 [DOI] [PubMed] [Google Scholar]
- Sjostrom P. J., Rancz E. A., Roth A., Hausser M. (2008). Dendritic excitability and synaptic plasticity. Physiol. Rev. 88 769–840. 10.1152/physrev.00016.2007 [DOI] [PubMed] [Google Scholar]
- Steckley D., Karajgikar M., Dale L. B., Fuerth B., Swan P., Drummond-Main C., et al. (2007). Puma is a dominant regulator of oxidative stress induced bax activation and neuronal apoptosis. J. Neurosci. 27 12989–12999. 10.1523/JNEUROSCI.3400-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern S., Sinske D., Knoll B. (2012). Serum response factor modulates neuron survival during peripheral axon injury. J. Neuroinflammation 9:78. 10.1186/1742-2094-9-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102 15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A., Stern S. A., Bozdagi O., Huntley G. W., Walker R. H., Magistretti P. J., et al. (2011). Astrocyte-neuron lactate transport is required for long-term memory formation. Cell 144 810–823. 10.1016/j.cell.2011.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweatt J. D. (2001). The neuronal MAP kinase cascade: a biochemical signal integration system subserving synaptic plasticity and memory. J. Neurochem. 76 1–10. 10.1046/j.1471-4159.2001.00054.x [DOI] [PubMed] [Google Scholar]
- Tadi M., Allaman I., Lengacher S., Grenningloh G., Magistretti P. J. (2015). Learning-induced gene expression in the hippocampus reveals a role of neuron -astrocyte metabolic coupling in long term memory. PLoS One 10:e0141568. 10.1371/journal.pone.0141568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F., Lane S., Korsak A., Paton J. F. R., Gourine A. V., Kasparov S., et al. (2014). Lactate-mediated glia-neuronal signalling in the mammalian brain. Nat. Commun. 5:13. 10.1038/ncomms4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J., Reynolds L., Hou L., Lohman K., Cui W., Kritchevsky S., et al. (2017). Transcriptomic profiles of aging in naïve and memory CD4+ cells from mice. Immun. Ageing 14:15. 10.1186/s12979-017-0092-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakurela S., Sahu S. K., Garding A., Tiwari V. K. (2015). Dynamics and function of distal regulatory elements during neurogenesis and neuroplasticity. Genome Res. 25 1309–1324. 10.1101/gr.190926.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theis V., Theiss C. (2018). VEGF - a stimulus for neuronal development and regeneration in the CNS and PNS. Curr. Protein Pept. Sci. 19 589–597. 10.2174/1389203719666180104113937 [DOI] [PubMed] [Google Scholar]
- Tian X., Irannejad R., Bowman S. L., Du Y., Puthenveedu M. A., von Zastrow M., et al. (2016). The alpha-arrestin ARRDC3 regulates the endosomal residence time and intracellular signaling of the beta2-adrenergic receptor. J. Biol. Chem. 291 14510–14525. 10.1074/jbc.M116.716589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7 562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaudry D., Pamantung T. F., Basille M., Rousselle C., Fournier A., Vaudry H., et al. (2002). PACAP protects cerebellar granule neurons against oxidative stress-induced apoptosis. Eur. J. Neurosci. 15 1451–1460. 10.1046/j.1460-9568.2002.01981.x [DOI] [PubMed] [Google Scholar]
- Volakakis N., Kadkhodaei B., Joodmardi E., Wallis K., Panman L., Silvaggi J., et al. (2010). NR4A orphan nuclear receptors as mediators of CREB-dependent neuroprotection. Proc. Natl. Acad. Sci. U.S.A. 107 12317–12322. 10.1073/pnas.1007088107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner W., Ciszewski W. M., Kania K. D. (2015). L- and D-lactate enhance DNA repair and modulate the resistance of cervical carcinoma cells to anticancer drugs via histone deacetylase inhibition and hydroxycarboxylic acid receptor 1 activation. Cell Commun. Signal. 13:36. 10.1186/s12964-015-0114-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Vasaikar S., Shi Z., Greer M., Zhang B. (2017). WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 45 W130–W137. 10.1093/nar/gkx356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong E. H., Kemp J. A., Priestley T., Knight A. R., Woodruff G. N., Iversen L. L. (1986). The anticonvulsant MK-801 is a potent N-methyl-D-aspartate antagonist. Proc. Natl. Acad. Sci. U.S.A. 83 7104–7108. 10.1073/pnas.83.18.7104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss M. T., Jolivet R., Buck A., Magistretti P. J., Weber B. (2011). In vivo evidence for lactate as a neuronal energy source. J. Neurosci. 31 7477–7485. 10.1523/JNEUROSCI.0415-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. Y., Ruchti E., Petit J. M., Jourdain P., Grenningloh G., Allaman I., et al. (2014). Lactate promotes plasticity gene expression by potentiating NMDA signaling in neurons. Proc. Natl. Acad. Sci. U.S.A. 111 12228–12233. 10.1073/pnas.1322912111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Li F., Qin Y., Bo X., Wu Y., Wang S. (2010). GOSemSim: an R package for measuring semantic similarity among GO terms and gene products. Bioinformatics 26 976–978. 10.1093/bioinformatics/btq064 [DOI] [PubMed] [Google Scholar]
- Yu G., Wang L.-G., Han Y., He Q.-Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16 284–287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. J., Steijaert M. N., Lau D., Schutz G., Delucinge-Vivier C., Descombes P., et al. (2007). Decoding NMDA receptor signaling: identification of genomic programs specifying neuronal survival and death. Neuron 53 549–562. 10.1016/j.neuron.2007.01.025 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Chen K., Sloan S. A., Bennett M. L., Scholze A. R., O’Keeffe S., et al. (2014). An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 34 11929–11947. 10.1523/JNEUROSCI.1860-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieker D., Schäfer R., Glatzle J., Nieselt K., Coerper S., Northoff H., et al. (2008). Lactate modulates gene expression in human mesenchymal stem cells. Langenbecks Arch. Surg. 393 297–301. 10.1007/s00423-008-0286-6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.