Abstract
Specification of dorsoventral regional identity in progenitors of the developing telencephalon is a first pivotal step in the development of the cerebral cortex and basal ganglia. Previously, we demonstrated that the two zinc finger doublesex and mab-3 related (Dmrt) genes, Dmrt5 (Dmrta2) and Dmrt3, which are coexpressed in high caudomedial to low rostrolateral gradients in the cerebral cortical primordium, are separately needed for normal formation of the cortical hem, hippocampus, and caudomedial neocortex. We have now addressed the role of Dmrt3 and Dmrt5 in controlling dorsoventral division of the telencephalon in mice of either sex by comparing the phenotypes of single knock-out (KO) with double KO embryos and by misexpressing Dmrt5 in the ventral telencephalon. We find that DMRT3 and DMRT5 act as critical regulators of progenitor cell dorsoventral identity by repressing ventralizing regulators. Early ventral fate transcriptional regulators expressed in the dorsal lateral ganglionic eminence, such as Gsx2, are upregulated in the dorsal telencephalon of Dmrt3;Dmrt5 double KO embryos and downregulated when ventral telencephalic progenitors express ectopic Dmrt5. Conditional overexpression of Dmrt5 throughout the telencephalon produces gene expression and structural defects that are highly consistent with reduced GSX2 activity. Further, Emx2;Dmrt5 double KO embryos show a phenotype similar to Dmrt3;Dmrt5 double KO embryos, and both DMRT3, DMRT5 and the homeobox transcription factor EMX2 bind to a ventral telencephalon-specific enhancer in the Gsx2 locus. Together, our findings uncover cooperative functions of DMRT3, DMRT5, and EMX2 in dividing dorsal from ventral in the telencephalon.
SIGNIFICANCE STATEMENT We identified the DMRT3 and DMRT5 zinc finger transcription factors as novel regulators of dorsoventral patterning in the telencephalon. Our data indicate that they have overlapping functions and compensate for one another. The double, but not the single, knock-out produces a dorsal telencephalon that is ventralized, and olfactory bulb tissue takes over most remaining cortex. Conversely, overexpressing Dmrt5 throughout the telencephalon causes expanded expression of dorsal gene determinants and smaller olfactory bulbs. Furthermore, we show that the homeobox transcription factor EMX2 that is coexpressed with DMRT3 and DMRT5 in cortical progenitors cooperates with them to maintain dorsoventral patterning in the telencephalon. Our study suggests that DMRT3/5 function with EMX2 in positioning the pallial-subpallial boundary by antagonizing the ventral homeobox transcription factor GSX2.
Keywords: Dmrt, dorsoventral patterning, Emx2, Gsx2, pallium–subpallium boundary, telencephalon
Introduction
The mammalian telencephalon is the largest and most complex region of the mammalian brain, controlling cognitive processes and purposeful actions. It comprises the cerebral cortex dorsally and the amygdala and basal ganglia structures ventrally. Defects in telencephalon development are associated with many human neuropsychiatric and neurological disorders (Gaitanis and Walsh, 2004; Hu et al., 2014). Related to this study, a loss-of-function mutation in the human DMRT5/DMRTA2 gene has been found to be associated with microcephaly (Urquhart et al., 2016).
Specification of dorsoventral (DV) regional identity in progenitors of the developing telencephalon is a pivotal step in the development of the cerebral cortex and basal ganglia. In the developing telencephalon, as in the spinal cord, bone morphogenetic proteins (BMPs) and Wingless-Int proteins (WNTs) produced dorsally and Sonic Hedgehog (SHH) secreted from ventral sources are implicated in DV specification of the telencephalon (Ericson et al., 1995; Chiang et al., 1996; Backman et al., 2005; Fernandes et al., 2007). Opposition between these morphogens alone does not establish DV telencephalic identity. Rather, interactions among the transcription factor GLI3, FGF signaling from the rostral telencephalic patterning center, and ventral sources of SHH regulate DV patterning (Ohkubo et al., 2002; Shimogori et al., 2004; Hasenpusch-Theil et al., 2017; Grove et al., 1998; Theil et al., 1999; Tole et al., 2000a; Aoto et al., 2002; Kuschel et al., 2003). GLI3 is a transcriptional activator in the presence of high levels of SHH, and a repressor, GLI3R, when levels of SHH are low. SHH promotes ventral identity by maintaining FGF signaling at the rostral telencephalic patterning center in part by suppressing formation of Gli3R, which represses Fgf8 expression (Ohkubo et al., 2002; Rallu et al., 2002; Rash and Grove, 2007). FGFs at the rostral telencephalic patterning center are in turn required to establish or maintain Shh expression in the ventral telencephalon (Storm et al., 2006). FGF signaling further promotes ventral telencephalon development independently of SHH, through regulating expression of the Foxg1 transcription factor gene (Gutin et al., 2006; Tole and Hébert, 2013). Pax6 and other homeobox genes, such as Emx2 (Muzio et al., 2002a,b) expressed throughout the pallium, and the homeobox gene Gsx2 expressed in the subpallium are also involved in positioning the pallium–subpallium boundary (PSB) (Stoykova et al., 2000; Toresson et al., 2000; Yun et al., 2001; Kroll and O'Leary, 2005; Carney et al., 2009). How these different transcription factors function together to control telencephalon DV patterning and whether there are other players involved remain unknown.
Dmrt3 and Dmrt5 (Dmrta2) encode related transcription factors expressed in a similar high caudomedial to low rostrolateral gradient in the cortical primordium. Their loss leads to a similar phenotype, more severe in Dmrt5 than in Dmrt3 mutants. In either single null mutant, Wnt and BMP expression at the cortical hem is decreased, and adjacent hippocampus and caudal neocortical areas are reduced in size. In conditional Dmrt5 mouse models, Dmrt5 loss or gain of function after hem formation also leads to a reduction of hippocampal size and alters neocortical area map formation, indicating that DMRT5 is not only required for hem formation but also directly controls cortical progenitor proliferation and specification. DMRT3 and DMRT5 are thus crucial regulators of cortical development, acting at different steps of its formation (Konno et al., 2012; Saulnier et al., 2013; Young et al., 2017; De Clercq et al., 2018). DMRT3 and DMRT5 have similar DNA-binding properties (Murphy et al., 2007), suggesting they act redundantly in telencephalic development, implying that analysis of single knock-out (KO) embryos did not reveal their full function. We therefore generated double KO and compared their telencephalic development with that of single KO embryos. Further, we conditionally overexpressed Dmrt5 in the telencephalon. As Emx2 is coexpressed with Dmrt3 and Dmrt5 in cortical progenitors, we also generated Dmrt5;Emx2 double KO embryos. Our new findings reveal that DMRT3, DMRT5, and EMX2 cooperate to repress Gsx2 and maintain DV patterning in the telencephalon.
Materials and Methods
Animals.
All mice were maintained on a C57BL/6 or CD1/C57BL6 mixed background and mice of either sex were used. Midday of the day of the vaginal plug discovery was defined as embryonic day (E) 0.5. Animal care was in accordance with Institutional guidelines, and the policies of the National Institutes of Health.
Dmrt3 (Saulnier et al., 2013), Dmrt5 (De Clercq et al., 2018), Emx2 (Pellegrini et al., 1996), and Gsx2EGFP mice (Wang et al., 2009) were genotyped by PCR as described, respectively, in these articles. Dmrt3+/− or Dmrt3−/− animals, which are viable, were crossed with Dmrt5+/− mice to obtain Dmrt3+/−;Dmrt5+/− mice. These double heterozygotes were then crossed to obtain Dmrt3−/−;Dmrt5−/− homozygous double KO embryos. Dmrt5−/−;Emx2−/− homozygous double mutants were obtained by intercrossing Dmrt5+/− heterozygous mutants with Emx2+/− heterozygous mutants. Dmrt5 conditional transgenic (Dmrt5Tg) mice were maintained and genotyped as described previously (De Clercq et al., 2018) and crossed to Foxg1-IRES-Cre (Kawaguchi et al., 2016) mice to overexpress Dmrt5 throughout the telencephalon or to Gsx2-Cre-IRES-EGFP (Gsx2-CIE) (Qin et al., 2016) to overexpress it in ventral telencephalon progenitors. tetO-Gsx2-IRES-EGFP mice were crossed to Foxg1TA/+ mice to obtain Foxg1TA/+;tetO-Gsx2-IRES-EGFP embryos overexpressing Gsx2 throughout the telencephalon (Waclaw et al., 2009).
Histology, immunofluorescence (IF), and ISH.
Standard H&E staining was performed on 6–8 μm sections of embryos or brains fixed overnight in 4% PFA/PBS, dehydrated, and paraffin-embedded. For IF, embryos were fixed overnight at 4°C in 4% PFA/PBS, infused in 30% sucrose/PBS overnight, frozen in gelatin (7.5% gelatin, 15% sucrose/PBS) or NEG-50, and cryosectioned (12–20 μm). Antigen retrieval was performed by boiling the sections in Target Retrieval Solution Citrate, pH 6.0 (Dako). Slides were the blocked with 10% normal goat serum, 0.3% Triton X-100 in PBS, and incubated with primary antibodies overnight at 4°C. The incubation with secondary antibodies was performed for 2 h at room temperature. Samples were then mounted in Dako mounting medium. The following primary antibodies were used: rabbit anti-TH (1:500, Immunostar), rabbit anti-DMRT5 (1:2000) (De Clercq et al., 2018), rabbit anti-GSX2 (1:500) (Toresson et al., 2000), mouse anti-ASCL1 (1:100, gift from C. Parras laboratory), goat anti-PAX6 (1:200, Santa Cruz Biotechnology), rabbit anti-TBR2 (1/500, Abcam), and chicken anti-GFP (1:1000, Aves Labs). The following secondary antibodies were used: anti-mouse AlexaFluor-488 (1:400, Invitrogen), anti-mouse AlexaFluor-594 (1:400,Invitrogen), anti-rabbit AlexaFluor-488 (1:400, Invitrogen), anti-rabbit AlexaFluor-488 (1:400, Invitrogen), and anti-rabbit AlexaFluor-594 (1:400, Invitrogen). Sections were counterstained with DAPI. Images were acquired with a Carl Zeiss LSM 70 confocal microscope using ZenBlack software or Nikon A1R gallium arsenide phosphide inverted Confocal Microscope and processed using ImageJ and Photoshop software.
ISH on sections and whole-mount ISH of embryos were performed using antisense digoxigenin-labeled riboprobes as described previously (Saulnier et al., 2013; De Clercq et al., 2018). The other antisense probes were generated from the previously described cDNA clones: Emx1 (Theil et al., 1999), Ascl1 and Gad67 (Fode et al., 2000), Dlx2 (Porteus et al., 1991), Gsx2 (Toresson et al., 2000), Sp8 (Sahara et al., 2007), Isl1 (Huber et al., 2013), ER81 (Chotteau-Lelièvre et al., 1997), and sFrp2 and TGF-α (Assimacopoulos et al., 2003). Images were acquired with an Olympus SZX16 stereomicroscope and a XC50 camera, using the Imaging software CellSens.
Confocal imaging.
Confocal imaging used an LSM-710 confocal microscope (Carl Zeiss) using both ×10/0.3 EC Plan Neofluor and ×20/0.8 PlanApochromat objectives (Carl Zeiss) and with specific excitation using a 405 nm laser diode, a 488 nm Argon ion laser, and a 594 nm Helium/Neon laser. A specific gallium arsenide phosphide Airyscan detector was used when necessary to increase signal detection and signal/noise ratio. Images were acquired using ZenBlack software (Carl Zeiss). For Tile scan imaging, acquisitions were performed with a 10% overlap of fields, and images were reconstruct using ZenBlack software. For nuclei counting in Figure 5, a homemade automated macro was developed on Fiji software. Briefly, background of images was reduced using a “rolling ball radius” function, and nuclei were segmented through fluorescence intensity using an automated threshold. Nuclei segmented from both “green” and “red” channels were counted automatically through a size selection, and nuclei present in both channels were considering as colocalizing. Brightness and contrast adjustments and image processing were done using Fiji and Photoshop software.
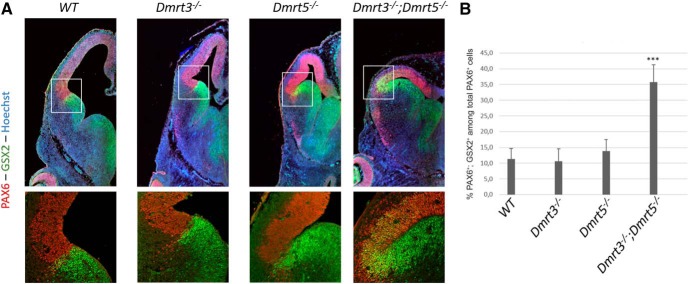
Figure 5.
Dorsal telencephalon cells express ventral markers in Dmrt3;Dmrt5 double KO embryos. A, Coronal section though E12.5 brains of the indicated genotypes processed by IF for Pax6 and Gsx2. Bottom, high-magnification views of the PSB. B, Histograms showing the number of double-positive cells among Pax6-positive cells in the boxed area. ***p < 0.001.
RNA sequencing (RNAseq).
RNA was extracted using the RNeasy mini kit from QIAGEN and prepared for sequencing using the TruSeq RNA Sample Prep Kit version 2. Four WT samples and five samples of each genotype were analyzed. Sequence reads have been obtained using an Illumina Hiseq 2000 in single-end mode (51 bp). Sequencing quality was checked and approved using the FastQC software (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and one Dmrt5 KO sample that had too low number of reads was removed. Reads were aligned to the mouse genome reference (genome assembly mm10 downloaded from www.ensembl.org) using the STAR alignment software (https://www.ncbi.nlm.nih.gov/pubmed/23104886, version 2.3) allowing for 2 mismatches within 51 bp. Gene expression quantification was performed using the featureCounts tool (https://www.ncbi.nlm.nih.gov/pubmed/24227677). Count normalization and differential gene expression analyses were conducting using R/Bioconductor (version 3.2) and the DESeq2 package (https://www.ncbi.nlm.nih.gov/pubmed/25516281/, version 1.8.2). PCA and hierarchical clustering were applied to normalized count data. Two outlier samples were removed (one from control group after inspection of sample cluster plots and the one from the Dmrt5 KO group with low number of reads). Genes were annotated using the biomaRt package (https://www.ncbi.nlm.nih.gov/pubmed/19617889, version 2.24). Differentially expressed genes were selected based on a log2-fold-change and false discovery rate cutoff of 1 and 0.05, respectively. Gene Ontology analysis was performed using DAVID 6.7 (http://david.abcc.ncifcrf.gov) in the annotation category BP-FAT. Strongly enriched categories had a score of ≤0.05 after Benjamini multiple test correction. RNAseq data have been deposited at NCBI GEO (GSE108611).
Plasmid construct and transgenesis.
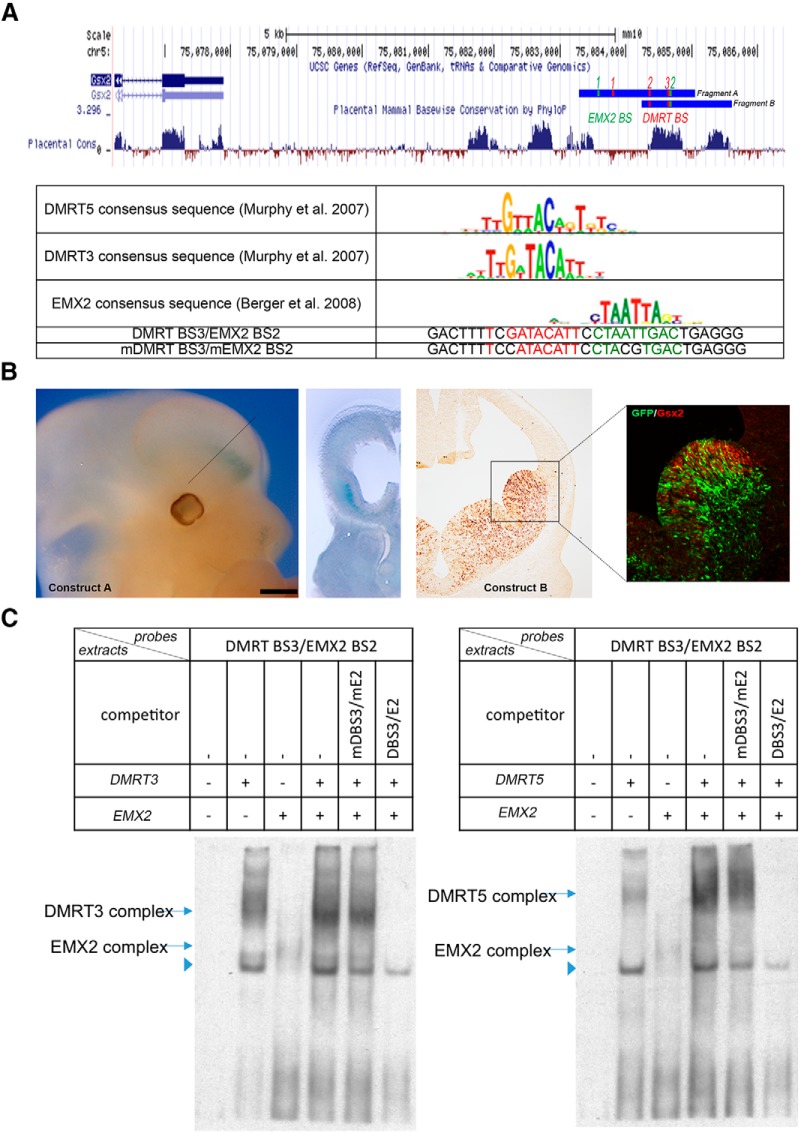
The 1.8 kb of the Gsx2 locus (chr5: 75481187–75479361, mm9 assembly) was amplified by PCR with the oligonucleotides forward 5′-CGCACCGTTGGGGATTCTAA-3′ and reverse 5′ TCTCTCAATTCCCAGGGGTCA-3′ and using DNA from BAC clone RP23–382i10 as template. The resulting fragment was subcloned 5′ of the globin basal promoter in the SpeI and NotI sites of a reporter vector (BGZA) containing the chick β-globin promoter, the lacZ reporter gene, and an SV40 polyadenylation cassette (Yee and Rigby, 1993). The 1.3 kb fragment of the Gsx2 locus (chr5: 75480316–75481596, mm9 assembly) was amplified by PCR with the oligonucleotides forward 5′-GGCGCGCCCACCCTTTTGTTTTGTTGTTAAGACTTAG-3′ and reverse 5′-ACAAAAAAGCAGGCTGTTGTCGTTCAGGTGGCAAGG-3′ using DNA from BAC clone RP24–223O21 as template, and was inserted into a vector containing Cre-IRES-EGFP. Transgenic embryos were generated by pronuclear injection at the Transgenesis platform of the De Duve Institute (UCL, Leuven) using fertilized eggs from B6CBAF1/Crl crosses (for fragment A) or at the Transgenic Animal and Genome Editing Core at Cincinnati Children's Hospital Medical Center by microinjection of fertilized eggs from FVN mice (for fragment B). Transgenic embryos were identified by PCR using tail DNA. Expression of the transgene was analyzed in E12.5 embryos. LacZ activity in transgenic embryos was detected by whole-mount X-gal staining. Expression of the GFP transgene was examined by DAB staining of a E12.5 brain sections with chicken anti-GFP antibodies (1:1000, Aves Labs), followed by biotinylated donkey anti-chicken IgY (1:200, Jackson ImmunoResearch Laboratories) and ABC solution (Vector Laboratories).
Electrophoretic mobility shift assay (EMSA).
EMSAs were performed using extracts from HEK293 cells transfected with Dmrt3, Dmrt5, and/or Emx2 pEFX expression vectors. The double-stranded oligonucleotide used the Dmrt and/or Emx2 binding sites (BSs) (italicized) as follows: Dmrt BS1, 5′-GCTGGGTTACACTGGAAGGA-3′; Dmrt BS2, 5′-TAGTCACTGTTTCATTAGGC-3′; Dmrt BS3, 5′-GACTTTTCGATACATTCCTA-3′; Emx2 BS1, ACCTCCCCTCTTCCTTCCTAATTAATGACCAT-3′; and DMRT BS3/EMX2 BS2, 5′-GACTTTTCGATACATTCCTAATTGACTGAGGG-3′. Briefly, double-stranded biotin-labeled probes and nonlabeled competitors were prepared by mixing equal molar ratio of two complimentary oligos in annealing buffer (10 mm Tris-HCl, pH 7.9, 10 mm MgCl2, and 50 mm NaCl). The paired oligos were heated to 95°C for 5 min; then the temperature was reduced slowly to room temperature on hot blot. DNA binding reaction was performed in binding buffer (10 mm Tris-acetate, pH 7.9, 5 mm magnesium acetate, 25 mm potassium acetate, 0.1 μm ZnSO4, and 50 μg/ml BSA), including 0.5 μg sonicated herring sperm DNA and 1 pmol double-stranded DNA probe. A total of 23 pmol of competitor (23-fold molar excess to probe) was used in each reaction as indicated. Protein-DNA complexes were separated by electrophoresis at 50 V for 3 h on 5% polyacrylamide mini-gels in 0.5 × TBE at 4°C. DNA was transferred to Hybond+ membrane using Bio-Rad Trans-blot SD SemiDry Electrophoretic Transfer Cell and crosslinked by UV light for 5 min. Biotin-labeled DNA was detected using the Chemiluminescent Nucleic Acid Detection Module (Thermo Scientific Pierce).
Experimental design and statistical analysis.
Quantification of the dorsal surface area of the cortical hemisphere of E12.5 and E18.5 animals was obtained by taking measurements from images of whole brains. Photographs were taken with an Olympus SZX16 stereomicroscope and a XC50 camera. Measurements were done using ImageJ software. All quantified data are expressed as mean ± SD values with the value obtained for WT set to 1. Significance tests were performed using a two-tailed Student's t test; p values < 0.05 were regarded as statistically significant. For each genotype, at least 5 embryos were examined.
In Dmrt single and double KO and transgenic Dmrt5-overexpressing embryos, for each marker and age, 2–4 embryos were analyzed. In Dmrt5;Emx2 double mutants, markers were only examined in 2 embryos due to the difficulties of obtaining them. For each of the markers used and in embryos of the different genotypes analyzed, photographs of sections from the rostral to caudal or from the lateral to medial part of the telencephalic vesicles were taken and assembled into series. Comparable coronal sections, taken at rostral to mid telencephalon levels (based on landmarks provided by the ganglionic eminences), and sagittal sections, taken at the level of the olfactory bulb (OB), are shown.
Mice with conditional misexpression of Dmrt5 throughout the telencephalon (using the Foxg1-IRES-Cre driver line) show a ventral expansion of the normally dorsally expressed gene, Emx1. The amount of expansion was measured in images of coronal brain sections through E12.5 conditional transgenic and control embryos. Images were obtained with an Axioscope microscope fitted with Axiovision software (Carl Zeiss). Five 12.5 transgenic embryos and 5 controls, from 3 different litters were used. To quantify the ectopic spread of Emx1 expression, a consistent landmark between control and mutant mice was essential. The demarcation between the LGE and MGE was such a landmark, given that the MGE appeared essentially unaltered in Dmrt5Tg/+;Foxg1IRES-Cre/+ embryos. We therefore measured how close the expression of Emx1 came to this landmark, focusing on expression in the medial and central parts of the LGE where Emx1 is not expressed in WT mice. Measurements were made using ImageJ, and the two groups were statistically compared with the paired-sample Student's t test.
Results
Dmrt3;Dmrt5 double KO shows greater defects in the cerebral cortex than either single KO embryos
We generated Dmrt3;Dmrt5 double mutants by intercrossing the Dmrt3 and Dmrt5 single mutants (Konno et al., 2012; Saulnier et al., 2013; De Clercq et al., 2018). Dmrt3;Dmrt5 double KO die at birth, as do Dmrt5 single KO mice (Saulnier et al., 2013); therefore, embryos were analyzed just before birth (E18.5). We found that the dorsal surface area approximately correlated with the number of null Dmrt3 and Dmrt5 alleles in an individual genotype, such that cortical hemispheres in Dmrt3;Dmrt5 double KO were strikingly smaller than either single KO embryos (−63 ± 3.4% compared with −43 ± 7.6% in Dmrt5 KO and −13 ± 10.4% in Dmrt3 KO embryos) (Fig. 1A,B). This more severe reduction of the cerebral hemispheres in double KO embryos was already apparent at E12.5 (−52.5 ± 0.04% compared with −43.7 ± 0.06% in Dmrt5 KO and with −19.9 ± 0.32% in Dmrt3 KO embryos).
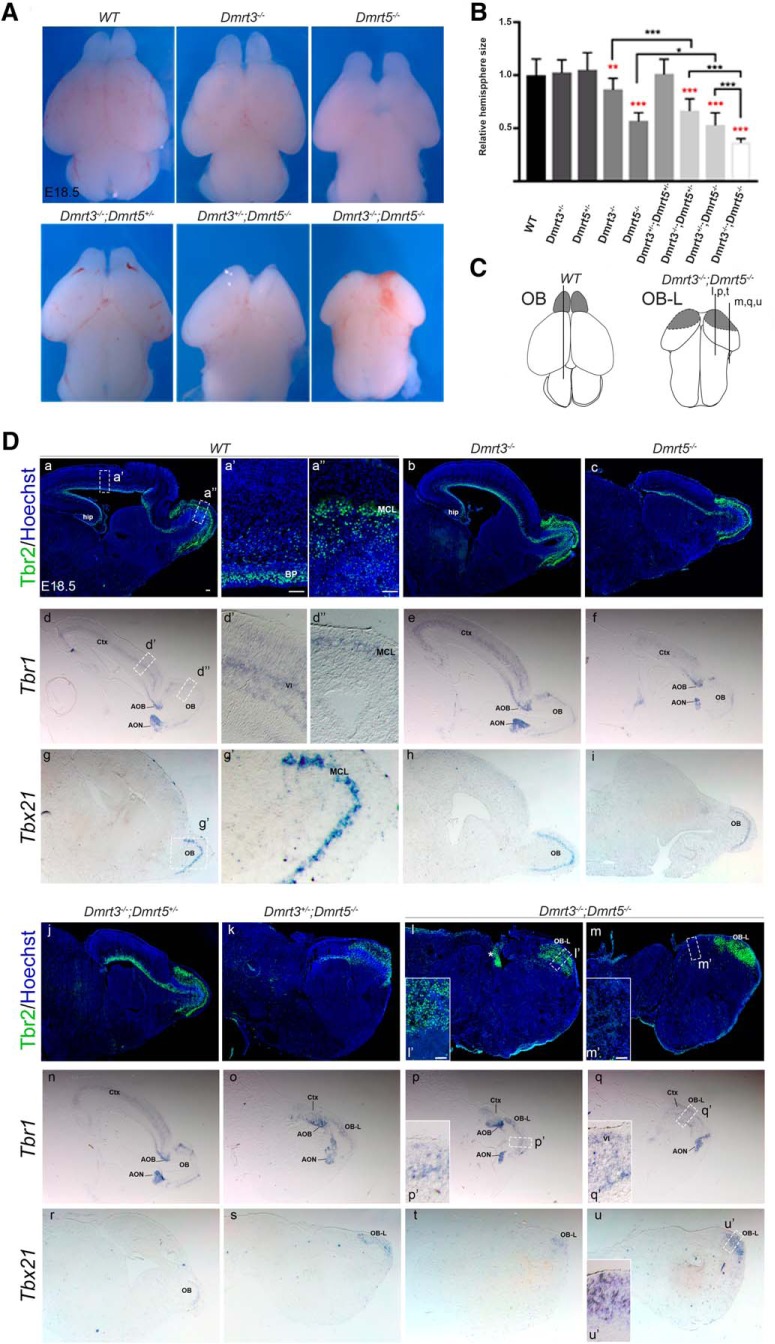
Figure 1.
Cortices of Dmrt3;Dmrt5 double KO embryos are more severely reduced in size than either single KO embryos and contain a prominent OBLS. A, Dorsal views of E18.5 brains of the indicated genotypes. B, Quantification of dorsal cortical surface area compared with WT controls (red) or as indicated. **p < 0.01, ***p < 0.001. n ≥ 5. C, Diagram showing the telencephalon of E18.5 WT embryos and the size reduction and absence of the OBs observed in Dmrt3−/−;Dmrt5−/−. D, Sagittal sections through E18.5 brains processed by IF or ISH for Tbr1, Tbr2, and Tbx21 OB mitral projection neuron markers. In the cortex, Tbr2 is also detected in SVZ progenitors and Tbr1 in layer VI neurons. For WT and double KO embryos, high-magnification views of the cortex (a′, d′, m′, q′) and OBs (a′, d′, l′, p′, u′) are shown. For the double KO brains, two sections are shown (i, j, m, n), taken at the levels indicated in the diagram. In the Dmrt3+/−;Dmrt5−/− and double KO embryos, the presence of a prominent OBLS occupying most of the reduced cortex (shown schematically in the diagram compared with controls) in which Tbr1, Tbr2, and Tbx21 are expressed and form a cluster or appear as a disorganized layer (l′, p′, u′). Note also in the cortex the disappearance of cortical Tbr2+ basal progenitors (compare a′ and m′) and more diffuse Tbr1 expression (compare d′ and q′). *Region of strong ectopic expression of Tbr2 outside the cortex in the double KO embryos. AOB, Accessory OB; AON, anterior olfactory nucleus; BP, basal progenitor; Ctx, cortex, OB-L, OB-like structure; Hip, hippocampus; MCL, mitral cell layer; OB, olfactory bulb. Scale bars, 50 μm.
In Dmrt3;Dmrt5 double KO, the reduction of caudal neocortical areas, hippocampus, and other dorsomedial telencephalic structures, such as the cortical hem, is more dramatic than in single KO embryos (data not shown). An obvious external difference between single KO and Dmrt3;Dmrt5 double KO embryos was that the telencephalic vesicles of the double KO embryos show no discernible OB (Fig. 1A,C). However, when analyzed with gene and protein expression markers of the OB, including Tbx21 whose expression is limited to OB mitral cells (Faedo et al., 2002; Kahoud et al., 2014), the residual pallium of the double KO embryos appears dominated by an OB-like structure (OBLS) (Fig. 1D). Thus, the Dmrt3;Dmrt5 double KO have more substantial defects than either single KO embryos, suggesting that Dmrt3 and Dmrt5 compensate for the loss of one another.
Loss of both Dmrt3 and Dmrt5 causes ventralization of the pallial neuroepithelium
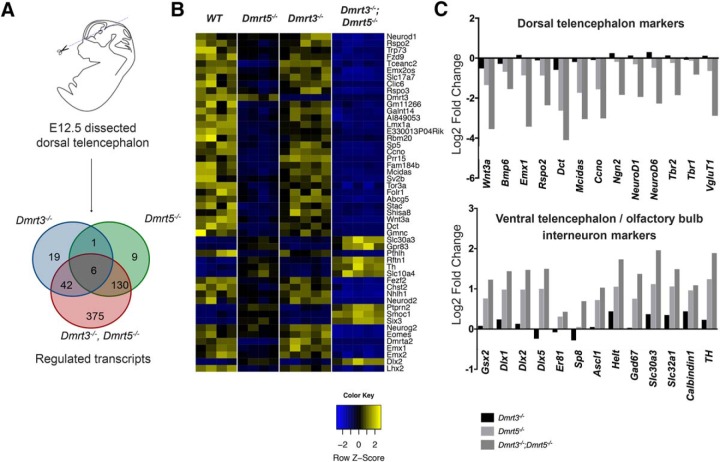
To determine how DMRT3 and DMRT5 interact to regulate gene expression in the developing telencephalon, we dissected the dorsal telencephalon from E12.5 Dmrt3 KO, Dmrt5 KO, Dmrt3;Dmrt5 double KO and WT embryos and performed a transcriptome analysis of the dissected tissue by RNAseq (Fig. 2A). Applying a minimal cutoff of twofold change and a significance level of p < 0.05, 68 differentially expressed genes were identified in the Dmrt3 single KO, 146 in the Dmrt5 single KO embryos, and 553 in the double KO embryos. The majority of differentially expressed genes were downregulated (67 in Dmrt3 KO, 126 in Dmrt5 KO, and 422 in Dmrt3;Dmrt5 double KO), and the overlap between the single and double KO embryos was very high (Fig. 2A). Gene Ontology analysis of the deregulated genes identified in the double KO embryos revealed an enrichment in genes involved in neuron differentiation, forebrain development, neuron fate, and cell fate commitment (data not shown). Among the downregulated genes in the double KO embryos were, as expected (Saulnier et al., 2013; De Clercq et al., 2018), genes of the WNT and BMP signaling pathways associated with the cortical hem and downstream targets genes linked to hippocampus and choroid plexus development (Wnt3a, Bmp6, Lmx1, Emx1, Lhx2, Nfix, Rspo1–3, Foxj1, Ttr, Mcidas, Gmnc, Ccno, Dnah6). Partially overlapping this list were transcription regulators of dorsal telencephalic fates, including Emx1, Emx2, Lhx2, Tbr2/Eomes, the proneural factors Ngn2/Neurog2, NeuroD1, Math2/NeuroD6, Nhlh1, and the glutamatergic neuronal gene Vglut1. Among upregulated genes were unexpectedly, many transcriptional determinants of ventral telencephalic cell fates, including Gsh2/Gsx2, Dlx2, Dlx5, Sp8, Sp9, Ascl1, ventral neuronal telencephalic genes (Gad1/Gad67, Gad2, Scl30a3, Slc32a1), and OB interneuron genes (Foxp2, TH, calbindin) (Fig. 2B,C). The dorsal determinant Pax6 was also slightly increased, but its change of expression level was below the applied twofold change cutoff. Genes encoding general regulators of neurogenesis, such as MyT1, Myt1l, and general neuronal markers (Dcx, Mapt, Nefh) were, in contrast, not differentially expressed. The most highly upregulated and downregulated genes together with the change of their expression in the different KO genotypes are shown in Figure 2B, C. For the vast majority of the deregulated genes identified, the changes observed in the Dmrt3;Dmrt5 double KO embryos relative to WT controls were in the same direction as those observed in the two single KO embryos, suggesting that Dmrt3 and Dmrt5 are redundant in several aspects of cortical development. The changes were generally greater than additive and the deregulation observed in Dmrt5 KO was always much stronger than in Dmrt3 KO embryos. For a given target, the difference between the WT and Dmrt3 KO expression levels (EWT − EDmrt3 KO) and between the Dmrt5 KO and Dmrt3;Dmrt5 double KO (EDmrt5KO − EDmrt3/5 DKO) represent the contribution of Dmrt3 to gene expression in the presence or absence of Dmrt5. For many of the transcripts, this EWT − EDmrt3 KO value is smaller than the EDmrt5KO − EDmrt3/5 DKO value. The greater effect of the loss of Dmrt3 in the double KO embryos than in the WT context suggests that Dmrt5 indeed partially compensates for the loss of Dmrt3.
Figure 2.
Genome-wide transcriptome analysis reveals that DMRT3 and DMRT5 cooperate to regulate cortical gene expression and play a role in early telencephalon DV patterning. A, Dissected dorsal telencephalic tissues analyzed by RNA-seq and Venn diagram showing the overlap of differentially expressed genes (both upregulated and downregulated) identified in Dmrt3 KO, Dmrt5 KO, and double KO E12.5 embryos. B, Heatmap for the 50 most significantly regulated genes (according to p value) in a comparison between WT and double KOs. Yellow represents upregulated genes. Blue represents downregulated genes. C, Examples of identified differentially expressed dorsal and ventral genes with log2-fold changes observed in each genotype.
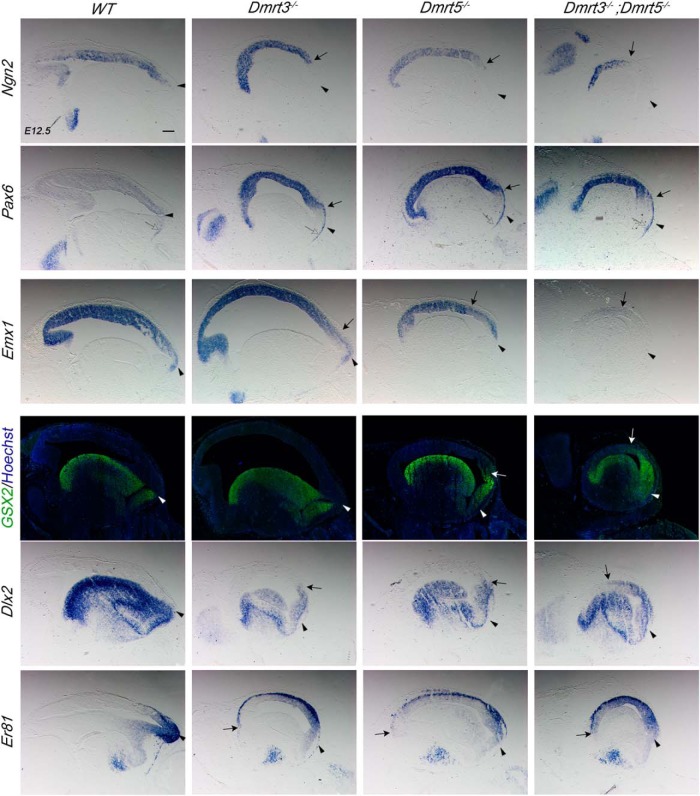
Distinct domains are distinguished in the early telencephalic neuroepithelium (Fig. 3). The dorsal telencephalon, or pallium, is subdivided into the medial, dorsal, lateral, and ventral pallium (MP, DP, LP, and VP). MP generates the hippocampus, and the DP gives rise to the neocortex. The LP and the VP contribute neurons to olfactory cortex and the amygdala. The ventral telencephalon, or subpallium, has three proliferative subdomains, the lateral, medial, and caudal ganglionic eminences (LGE, MGE, and CGE), which develop into the striatum, globus pallidus, and parts of the amygdala. The MGE and CGE are also the source of GABAergic interneurons, which migrate tangentially into the cortex (Anderson et al., 1997; Puelles et al., 2000; Flames et al., 2007). To confirm the differential gene expression revealed by the RNA-seq analysis, we performed ISH of selected deregulated genes on brain sections of E12.5 embryos (Fig. 3). We first examined patterning genes, such as Emx1, Emx2, Pax6, and Ngn2, expressed in the cortex of WT mice, and Dbx1 expressed selectively in the VP, which lies adjacent to the PSB (Fig. 3A). For genes expressed in the subpallium, we analyzed Gsx2, Dlx2, and Ascl1 (Toresson et al., 2000; Yun et al., 2001; Tole et al., 2005).
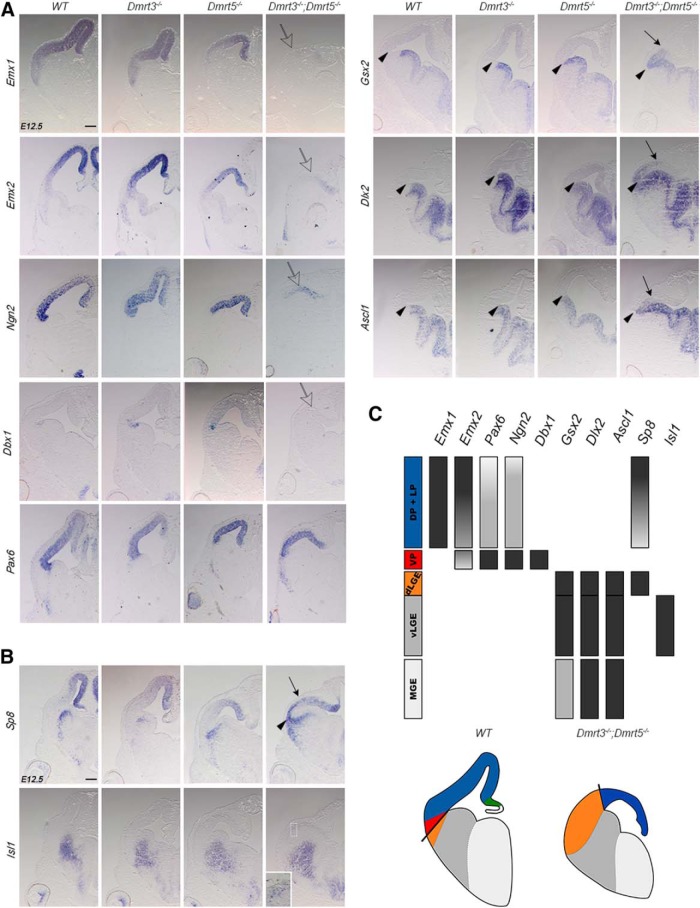
Figure 3.
Downregulation of dorsal determinants and expansion of dLGE markers in the lateral telencephalic neuroepithelium of Dmrt3;Dmrt5 double KO embryos. A, B, Coronal brain sections of E12.5 embryos hybridized with the indicated markers. Arrowheads indicate the region of the PSB. Arrows point to the shifted dorsal limit of ventral markers expressed ectopically in the pallium of the double KO embryos. Open arrows indicate the strongly downregulated expression of Emx1, Emx2, and Ngn2 that remains detectable only in the dorsomedial telencephalon and the absence of Dbx1. Inset in the Isl1 panel of the Dmrt double KO represents a high-magnification view of the boxed region where some Isl1 ectopic staining in the pallium is observed. C, Diagram showing the expression domain of the different markers used in the telencephalon of E12.5 WT embryos and the reduction of the ventral pallium and an expansion of the dLGE domain as observed in Dmrt3−/−;Dmrt5−/−. Scale bar, 200 μm.
The dorsal regulators Emx1, Emx2, and Ngn2 were strongly decreased in cortical progenitors of the double KO, more severely than in either single KO embryos. Expression of these genes remained detectable only in the dorsomedial telencephalon (Fig. 3A, open arrows). Dbx1 expression, which remains detectable in the VP of both single KO embryos, was lost in the double KO embryos (Fig. 3A, open arrow). In contrast, Pax6, which is upregulated in the cortex of Dmrt single KO (Saulnier et al., 2013; De Clercq et al., 2018), remained expressed in the cortex of the double KO embryos but was reduced at the PSB. Gsx2, Dlx2, and Ascl1 whose expression is restricted to the subpallium in WT embryos, extended dorsally into the lateral part of the telencephalon in the double KO (Fig. 3A, arrows), a phenotype that was not observed in either single Dmrt KO embryos.
The LGE is subdivided into a dorsal (dLGE) and a ventral domain (vLGE) (Yun et al., 2001). dLGE progenitors express Sp8 and generate OB interneurons, whereas vLGE progenitors express Isl1 and give rise to striatal GABAergic projection neurons (Stenman et al., 2003; Waclaw et al., 2006, 2009; Ehrman et al., 2013). We examined their expression in single and double KO embryos. Sp8, normally restricted to the dLGE SVZ, showed a similar expression in single KO but expanded dorsally the double KO embryos (Fig. 3B, arrow). Isl1 expression in single KO embryos was similar to WT controls. In double KO, Isl1 appears only moderately affected, with only very weak staining detectable in the cortex (Fig. 3B).
We also examined expression of dorsal and ventral regulator genes in sagittal sections (Fig. 4). Expression of Ngn2 and Emx1 retracted caudodorsally. Pax6 expression, upregulated in the single KO, also appeared to retract in double KO embryos. This was accompanied by a dorsal expansion of Gsx2 and Dlx2 expression in the anterior telencephalon. In the brain of E12.5 Dmrt3;Dmrt5 double KO embryos, subpallial gene expression expand dorsally both in the lateral and anterior telencephalon (Fig. 4, arrows). We found that Er81 expression, restricted at E12.5 in WT embryos to the OB primordium (OBP) (Stenman et al., 2003; Allen et al., 2007; Besse et al., 2011), was also extensively expanded caudally (Fig. 4, arrow). Together, these results indicated that in E12.5 Dmrt3;Dmrt5 double KO, there is an expansion of dLGE and OBP domains, as characterized by gene expression, into the dorsal telencephalon, and a concomitant shrinkage of the dorsal pallium (Figs. 1C, 3C).
Figure 4.
Subpallial gene expression expands in the rostral telencephalon of Dmrt3;Dmrt5 double KO embryos. Sagittal sections of E12.5 brains processed by ISH or IF with the indicated markers, with the rostral part to the right. Arrowheads indicate PSB. Arrows indicate the rostral or caudal boundaries of the gene expression domains. These boundaries are shifted dorsally in the Dmrt3;Dmrt5 double KO embryos. Open arrows indicate stronger Pax6 staining in a stream of postmitotic cells that derive from dLGE progenitors in the mantle zone of the rostral part of cortical neuroepithelium of the single and double KO embryos. Scale bar, 200 μm.
The expansion of dLGE and OBP domains in E12.5 Dmrt3;Dmrt5 double KO could be the consequence of the collapse of the dorsal telencephalon and invasion of ventral progenitors or of a respecification, namely, ventralization of dorsal telencephalic progenitors. To test the second hypothesis, we performed double immunostaining with antibodies for both the dorsal PAX6 and ventral GSX2 markers on coronal brain sections of E12.5 single and double KO embryos and WT controls. In the lateral cortex of the double KO embryos, but not in the single KO and WT embryos, many cells were found to be positive for both markers (Fig. 5), suggesting that indeed dorsal telencephalic progenitors were acquiring a ventral identity, perhaps corresponding to that in WTs of the few cells present at the PSB that coexpress Pax6 and Gsx2, or a hybrid fate.
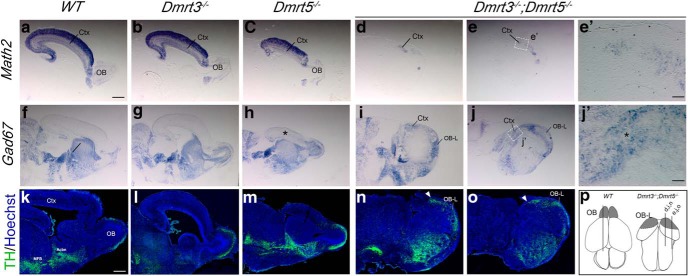
To assess the consequences of the ventralization of dorsal progenitors in Dmrt3;Dmrt5 double KO embryos, we examined TH, found in a subpopulation of OB interneurons, Gad67, a GABAergic interneuron marker, and Math2, a marker of glutamatergic cortical neurons in sagittal sections of E18.5 brains. In accordance with our observation of an expansion of the dLGE and OBP at E12.5, compared with single KO and controls, TH-positive cells were detected more caudally and Gad67 was increased in the dorsal telencephalon of the double KO embryos. Compared with single KO and controls, Math2 expression was in contrast dramatically reduced overall (Fig. 6), suggesting some respecification of cortical neuroblasts.
Figure 6.
Expression of GABAergic, glutamatergic, and OB interneuron markers in the cortex of E18.5 Dmrt3 and Dmrt5 single KO and in double KO embryos. a, o, Sagittal sections through brains of the indicated genotypes processed by ISH or IF for the indicated markers. p, The line in the schematic of the brain of WT and double KO embryos indicates the positions of the sections. For the double KO brains, two sections are shown for each marker. e′, j′, A high magnification of the Math2 and Gad67 staining in the residual cortex of the double KO. There is slight upregulation of Gad67 in both single and double mutants (*) and a dramatic loss of Math2 in the cortex of the double KO embryos. TH-positive cells that are not correctly laminated and form a large cluster in the OB-L structures of the double KO that extend more caudally in the dorsal telencephalon than in WT embryos (arrowheads). Ctx, Cortex; MFB, medial forebrain bundle; Acbn, accumbens nucleus. Scale bars: 500 μm; h, p, 50 μm. OB, olfactory bulb; OB-L, olfactory bulb like.
Misexpression of Dmrt5 in the subpallium represses ventral telencephalic markers
To perform Dmrt5 gain-of-function experiments, we used a Dmrt5 Cre/loxP conditional transgenic mouse model we recently generated. In our previous work, the Dmrt5 conditional transgenics (designated here Dmrt5Tg mice) were crossed with Emx1-Cre mice to study the role of DMRT5 in neocortical area map formation (De Clercq et al., 2018). Emx1-Cre drives excess Dmrt5 only in the dorsal, not ventral, telencephalon, and Cre lox recombination begins in the most medial telencephalon at approximately E10, spreading laterally so that full recombination in lateral cortex is as late as E11.5. In the present study, Dmrt5Tg mice were crossed with different Cre driver lines, Foxg1-IRES-Cre or Gsx2-Cre-IRES-EGFP (Gsx2-CIE). In Dmrt5Tg/+;Foxg1IRES-Cre/+ embryos, Dmrt5 is overexpressed in both the dorsal and ventral telencephalon from E8.5 (Kawaguchi et al., 2016), thus much earlier and more broadly than with Emx1-Cre. In Dmrt5Tg/Tg;Gsx2-CIE embryos, Dmrt5 is overexpressed selectively in the ventral telencephalon from E10.5 (Qin et al., 2016).
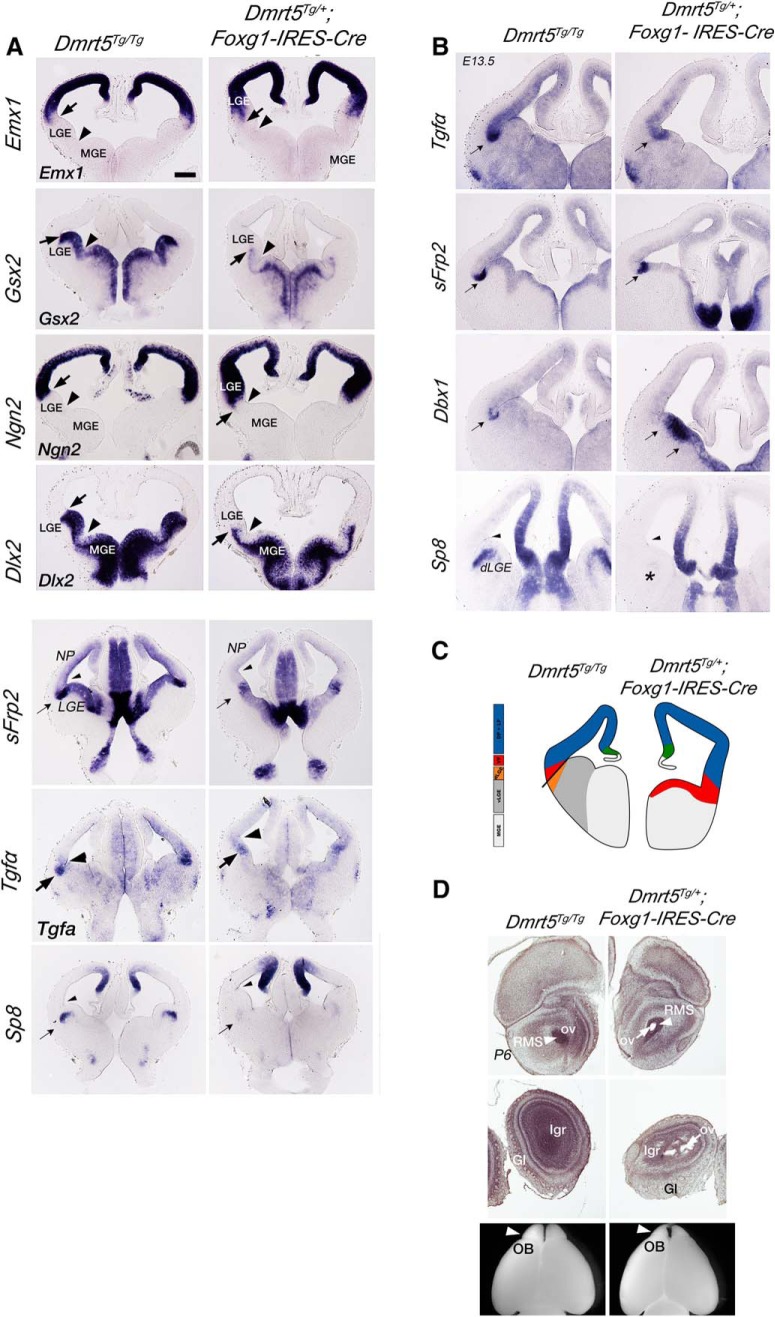
As expected, in Dmrt5Tg/+;Foxg1IRES-Cre/+ embryos, Dmrt5 transgene expression detected by GFP ISH filled the entire telencephalon (data not shown). In E12.5 Dmrt5Tg/+;Foxg1IRES-Cre/+ embryos, we examined the expression of ventral (Gsx2, Dlx2, and Ascl1) and dorsal (Emx1 and Ngn2) telencephalic markers. We also analyzed sFrp2, Tgf-α, Dbx1 expressed in the VP (Assimacopoulos et al., 2003) and Sp8 marking the dLGE (Fig. 7).
Figure 7.
Repression of ventral and expansion of dorsal telencephalic and ventral pallium markers in the subpallium of Dmrt5Tg/+;Foxg1-IRES-Cre embryos. A, B, Coronal sections through the telencephalon of transgenic and control embryos at E12.5 (A) and E13.5 (B) processed by ISH for the indicated markers. A, Top 8 panels, Arrowheads indicate the boundary of the MGE and LGE, a constant landmark between control and mutant mice, used for quantification of Emx1 expansion. Arrows point to the ventral limit of Emx1 and Ngn2 expression, or the dorsal limit of Gsx2 and Dlx2 expression. Notably, in Foxg1-IRES-Cre embryos, the arrows and arrowheads are closer together than in controls, indicating the dorsalization of the telencephalon when Dmrt5 is overexpressed. A, Bottom 6 panels, arrowheads indicate the PSB, arrows indicate the mis-located expression of sFrp2 and Tgfa when Dmrt5 is overexpressed, and the near loss of Sp8 expression. B, Arrowheads indicate the PSB. Arrows indicate restored expression of sFrp2 and Tgf-α to the PSB, and upregulated expression of Dbx1. Asterisk marks continued absence of Sp8. C, Diagram showing the ventral expansion of DP and VP markers as observed in the telencephalon of Dmrt5Tg/Tg;Foxg1-IRES-Cre embryos. D, Dorsal view of P6 brains and eosin staining of OBs. Arrowheads point to the RMS. Arrow indicates the OV. RMS, Rostral migrating stream; OV, olfactory ventricle; GL, glomerular layer; Igr, internal granule layer; LGE, lateral ganglionic eminence; dLGE, dorsal lateral ganglionic eminence; MGE, medial ganglionic eminence; OB, olfactory bulb; NP, neocortical primordium.
In E12.5 control embryo sections, Emx1 expression did not enter the LGE and only extended along the far lateral edge of the LGE in a territory that may be presumptive olfactory cortex. By contrast, in Dmrt5Tg/+;Foxg1IRES-Cre/+ E12.5 embryos, the Emx1 expression domain expanded ventrally into the central and medial LGE (Fig. 7A, arrow). This expansion of Emx1 expression in Dmrt5Tg/+;Foxg1IRES-Cre/+ E12.5 embryos, compared with controls, was quantified by measuring how close the central sector of Emx1 expression came to the boundary between the LGE and the MGE. The latter boundary served as a consistent landmark because the MGE appeared morphologically unaffected by overexpression of Dmrt5. For Dmrt5Tg/+;Foxg1IRES-Cre/+ E12.5 embryos the mean distance from the Emx1 expression front and the LGE/MGE boundary was 133.3 μm (SEM, 25.3 μm; SD 75.9 μm), and for control embryos, 273.8 μm (SEM, 18.2 μm; SD, 54.6 μm). The two groups were significantly different (p = 0.0004). Notable by eye, but not quantified, the extension of Emx1 expression increased from rostral to caudal, reaching the MGE/LGE boundary in more caudal sections. Similarly, Ngn2 expression, normally confined to the cerebral cortical primordium, expanded into almost all the LGE territory (Fig. 7A, arrow).
In contrast, Gsx2 and Dlx2 expression was downregulated overall and did not reach along the entire DV length of the LGE (Fig. 7A, arrow). The strongest expression of both sFrp2 and Tgf-α was shifted ventrally, and Sp8 in the dLGE was virtually absent (Fig. 7A, arrow). At E13.5, Tgf-α and sFrp2 appeared to have recovered a normal expression level at the PSB, but expression of Dbx1, a selective marker of the VP, was greatly expanded and shifted ventrally. Sp8 remained almost absent in the dLGE (Fig. 7B). Thus, at E13.5, the PSB boundary expressed some appropriate genes, yet the VP area as marked by Dbx1 had expanded (Fig. 7C). An enlarged VP was previously observed in the Gsx2 null mutant (Yun et al., 2001; Carney et al., 2009; Waclaw et al., 2010).
Sp8 in the dLGE is required for OB interneurons to begin migration through the embryonic rostral migratory stream (Waclaw et al., 2006). In Dmrt5Tg/+;Foxg1IRES-Cre/+ brains at P6, consistent with the virtual loss of Sp8 expression in the dLGE, the ventricle of the OB was slightly open rather than filled with migrating interneurons as in WT mice, suggesting that interneurons were not reaching the OB correctly (Fig. 7D).
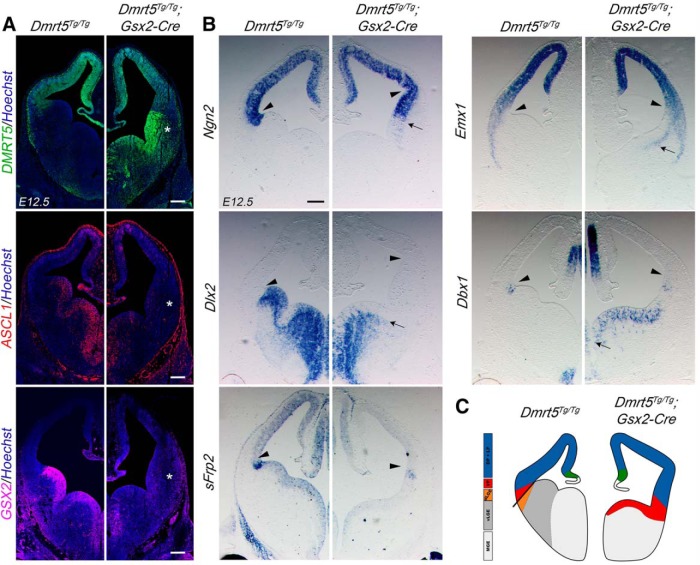
In Dmrt5Tg/Tg;Gsx2-CIE embryos, Dmrt5 overexpression was only detected in the ventral telencephalon, as expected. As observed in Dmrt5Tg/+;Foxg1IRES-Cre/+ embryos, Dmrt5 ectopic expression induces loss of the ventral (Gsx2, Dlx2, Ascl1) and gain of the dorsal (Emx2, Ngn2, Dbx1) markers tested (Fig. 8), suggesting that the ventral expansion of dorsal telencephalic and ventral pallial markers is largely caused by the downregulation of ventral determinants.
Figure 8.
Repression of ventral and expansion of dorsal and ventral pallium markers in the subpallium of Dmrt5Tg/Tg;Gsx2-Cre embryos. A, B, Coronal sections through the telencephalon of E12.5 embryos processed by IF (A) or ISH (B) with the indicated markers. A, *The ectopic expression of Dmrt5 and downregulation of Ascl1 and Gsx2 in the ventral telencephalon. B, Arrowheads indicate the region of the PSB. Arrows point to the expansion and shifted ventral limit of Ngn2, Emx1, and Dbx1 and downregulation and shifted dorsal limit of Dlx2 expression in the subpallium of the transgenics. C, Diagram showing the expansion of the ventral pallium as observed in Dmrt5Tg/Tg;Gsx2-CIE embryos. Scale bar, 200 μm.
EMX2 interacts with DMRT5 to control telencephalic DV patterning, and both transcription factors can bind to a ventral telencephalic-specific enhancer of the Gsx2 locus
Emx2 is coexpressed with Dmrt3 and Dmrt5 in cortical progenitors in a similar graded manner. As in Dmrt5 or in Dmrt3 single mutants, the hippocampus and caudomedial neocortex are reduced in Emx2-deficient mice (Yoshida et al., 1997; Tole et al., 2000b). This reduction too is more dramatic in Dmrt5/Emx2 double KO than in single KO embryos (Fig. 9A), suggesting that Dmrt5 and Emx2 genes interact in controlling telencephalic growth and patterning. Supporting such an interaction, ISH showed that, in E12.5 Dmrt5/Emx2 double KO brains, the pallial transcription factor gene Ngn2 was switched off, whereas gene expression domains of Gsx2, Dlx2, and Ascl1, normally confined to the subpallium, extended dorsally, a phenotype not observed in either Dmrt5 or Emx2 single KO embryos (Fig. 9B).
Figure 9.
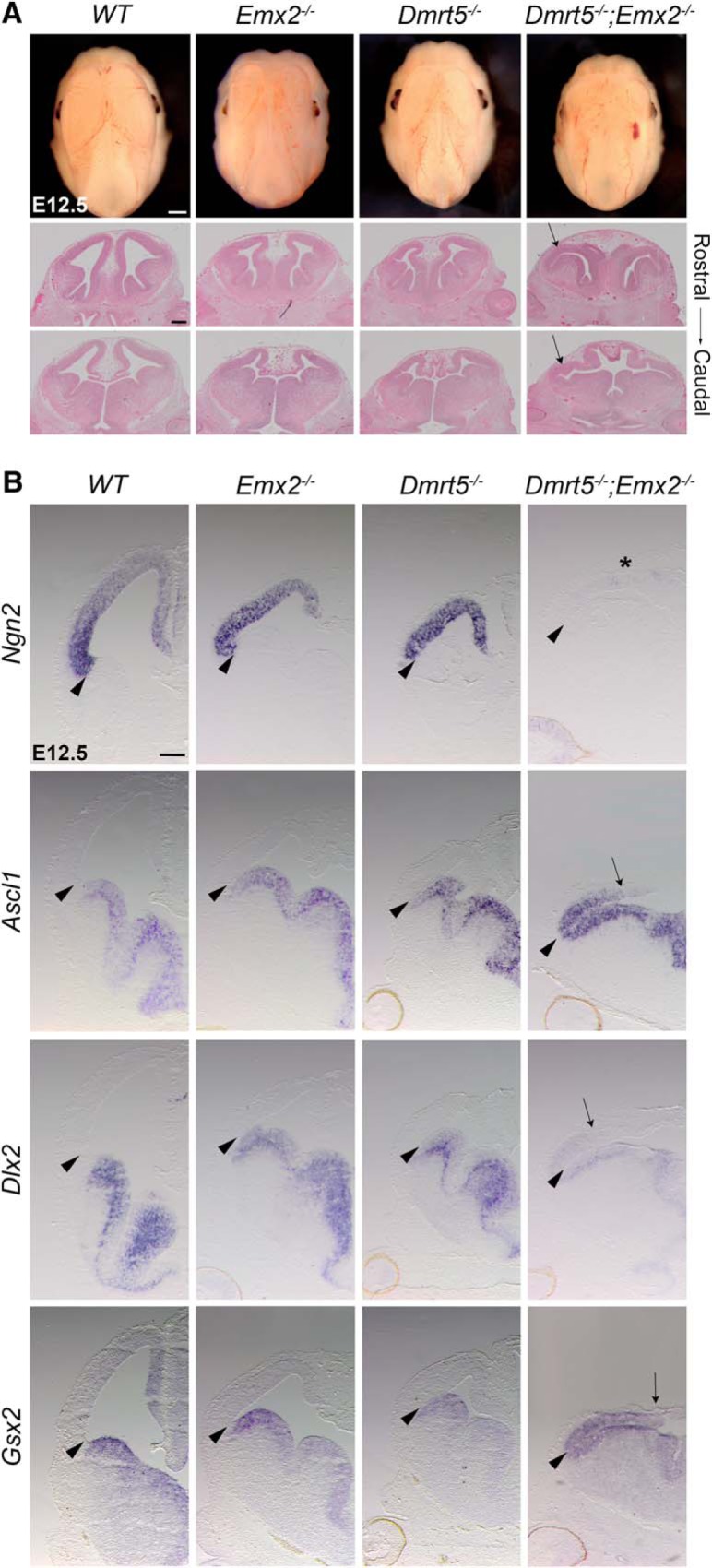
DMRT5 and EMX2 cooperate in telencephalon DV patterning. A, Dorsal views of the head of E12.5 embryos. H&E staining of E12.5 brain coronal sections of the telencephalon, at rostral and caudal levels. Arrows point to the caudomedial cortex of the Dmrt5;Emx2 double mutants, more severely reduced than in single mutants. B, Coronal brain sections of E12.5 embryos processed by ISH with the indicated markers. Arrowheads indicate the region of the PSB. Arrows point to the dorsal limit of Gsx2, Dlx2, and Ascl1 expression, shifted dorsally in the Dmrt5−/−;Emx2−/− double KO embryos. *Dramatic reduction of Ngn2 in the cortex of the double KO embryos. Scale bar, 200 μm.
GSX2 is crucial for ventral identity in the telencephalon. In Gsx2 mutants, the VP expands into the dLGE (Toresson et al., 2000; Yun et al., 2001; Carney et al., 2009; Waclaw et al., 2010). Conversely, Gsx2 misexpression in the pallium results in increased expression of ventral telencephalic markers and repression of dorsal ones (Toresson et al., 2000; Yun et al., 2001; Carney et al., 2009; Waclaw et al., 2009), including Dmrt5 (Fig. 10).
Figure 10.
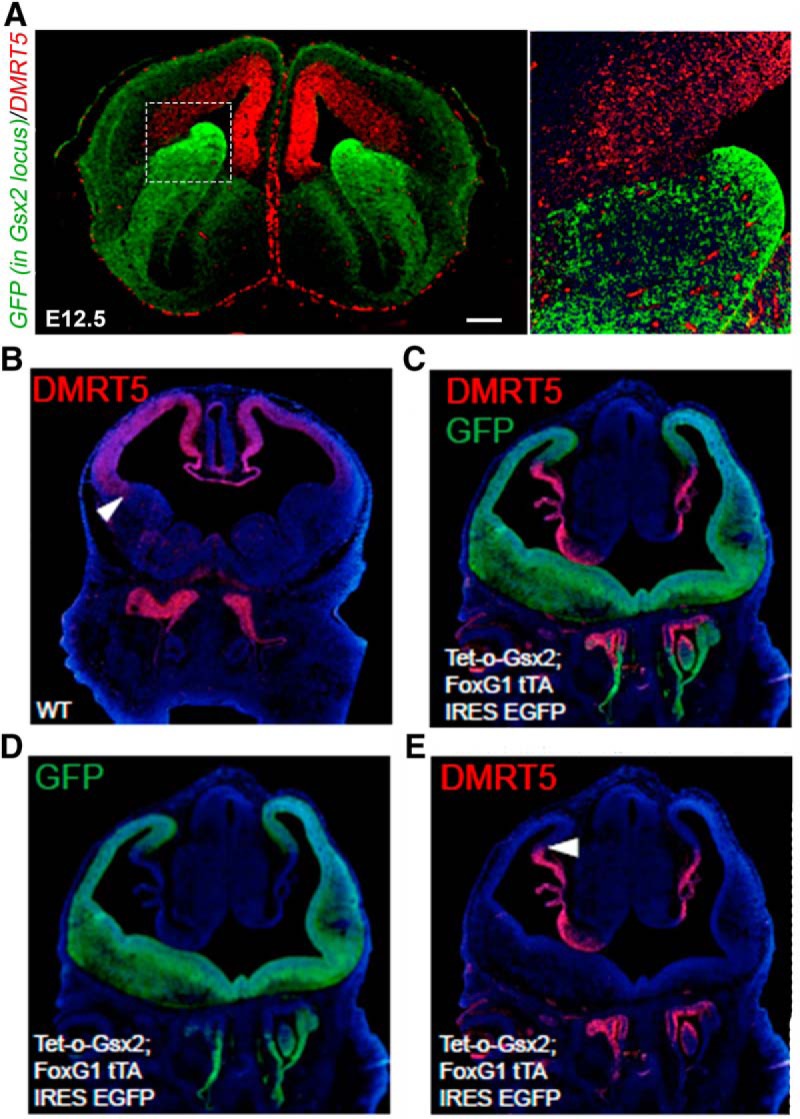
DMRT5 forms a boundary with GSX2 at the PSB, and the overexpression of Gsx2 represses Dmrt5 expression. A, Coronal sections of the head of E14.5 Gsx2GFP/+ knock-in embryos processed by IF with DMRT5 and GFP antibodies showing that, at the PSB, cells expressing DMRT5 do not express GFP, hence, GSX2. Boxed area is shown at a high magnification on the left. Oe, Olfactory epithelium. Scale bar, 200 μm. B–E, Coronal sections through the telencephalon of E12.5 WT or Foxg1tTA/+; tet-O-Gsx2-IRES-EGFP double-transgenic embryos immunostained with the indicated antibodies. In these transgenic embryos, Gsx2 is misexpressed throughout the embryonic telencephalon and Dmrt5 is reduced. B, E, Arrows point to the ventral limit of high Dmrt5 expression.
To investigate how GSX2 and DMRT5 interact, we first compared by IF their expression in Gsx2GFP/+ knock-in embryos and found that the two proteins abut at the PSB (Fig. 10). Their exclusive expression at the PSB, the upregulation of Gsx2 observed in Dmrt3;Dmrt5 and Dmrt5;Emx2 double KO embryos, along with its downregulation in response to overexpression of Dmrt5, suggest that DMRT5 may regulate cortical identity in telencephalic progenitors via repression of Gsx2 transcription. We therefore searched the Gsx2 locus for noncoding, evolutionarily conserved potential enhancer sequences (Bejerano et al., 2004; Pennacchio et al., 2006) with potential DMRT3/5 BSs (Murphy et al., 2007). We focused on one such conserved site of ∼0.5 kb that has been described as an enhancer bound by PAX6 that is active in the forebrain but not the lens (Sun et al., 2015), and is located 8.8 kb downstream of Gsx2 transcribed sequences. Two overlapping fragments, encompassing this conserved region, one of 1.8 kb including upstream sequences and the other of 1.3 kb containing downstream sequences and including another conserved region (fragments A and B, respectively, Fig. 11A) were cloned and fused to a reporter gene (Constructs A and B). We found that both fragments tested exhibit specific enhancer activity in the ventral forebrain in E12.5 transgenic embryos, suggesting that it is the conserved region common to the two fragments that is responsible for the activity (Fig. 11B). This conserved region contains two potential DMRT3/5 BSs as well as one 5′-ATTA-3′, which represent the core of potential homeobox transcription factor-binding motifs (Berger et al., 2008). This 5′-ATTA-3′motif is adjacent to the second Dmrt3/5 BS. Upstream of this conserved region, within the cloned genomic fragments, we found one additional Dmrt3/5 BS and one additional 5′-ATTA-3′ motif (Fig. 11A). To test for direct binding of DMRT3/5 and EMX2 to these putative BSs, we performed EMSAs using extracts prepared from HEK293 cells transfected with Dmrt3, Dmrt5, or Emx2 expression plasmids. In first EMSA assays, using oligonucleotides containing the different Dmrt and Emx2 potential BSs, we found that DMRT5 and DMRT3 both bind strongly to BS3 but not to the other sites. EMX2 can bind to both identified BSs (data not shown). We then examined the ability of DMRT3, DMRT5, and EMX2 to bind to an extended oligonucleotide encompassing both Dmrt BS3 and Emx2 BS2, in the presence or absence of an identical competitor oligonucleotide containing mutated Dmrt and Emx2 BSs. Figure 11C shows that, as expected, the binding of DMRT3, DMRT5, and EMX2 to the Dmrt BS3/Emx2 BS2 oligonucleotide is abolished in the presence of WT but not mutated competitors. Interestingly, in the presence of EMX2, the complex of DMRT3 appears more intense and to run faster than in its absence. Although no change in mobility shift was observed, DMRT5 binding in the presence of EMX2 appears also more intense than in its absence, suggesting cooperative interactions between DMRT3/5 and EMX2.
Figure 11.
DMRT5 and EMX2 bind a Gsx2 ventral-specific telencephalon enhancer. A, UCSC genome browser view of the Gsx2 locus with the location of the two cloned fragments tested in transgenic embryos. The identified putative DMRT3/5 and EMX2 BSs are shown. B, Left, A lateral view and a coronal section of the head of a E12.5 Gsx2 1.8 kb enhancer-LacZ reporter transgenic embryo (Construct A). Scale bars: lateral view, 500 μm; coronal section, 200 μm. Dashed line indicates the level of the section. Right, A coronal section of the brain of a Gsx2 1.3 kb enhancer-GFP reporter transgenic embryo (Construct B) processed by DAB immunostaining for GFP and a high-magnification view of the LGE region processed by IF for both GSX2 (red) and GFP (green). C, EMSA showing in vitro binding of cellular extracts containing DMRT3, DMRT5, and EMX2 or control extracts to BS3 of the Gsx2 enhancer. DMRT3/5 and EMX2 complex formation is competed by WT enhancer oligonucleotides but not by oligonucleotides containing mutations in the DMRT and EMX2 BSs. Arrowhead indicates a nonspecific band.
Discussion
We have previously identified DMRT5 and DMRT3 as important regulators of the development of caudomedial cortical structures acting downstream of dorsal WNT midline signals and controlling Wnt expression in a feedback loop (Hasenpusch-Theil et al., 2012; Saulnier et al., 2013; De Clercq et al., 2018). In the present study, we show that the size of the cortical hemispheres is drastically smaller in the Dmrt3;Dmrt5 double KO than in the single KO embryos, further highlighting their importance in the control of the proliferative state of progenitor cells. This dramatic reduction in size of the cortex of the double KO embryos may due to premature differentiation and exhaustion of the pool of progenitors as observed in Dmrt5−/− embryos (Young et al., 2017), which remains to be explored. We also show that the development of caudomedial telencephalic structures, such as the cortical hem and hippocampus, is more severely reduced in the Dmrt3;Dmrt5 double KO than in the single KO embryos. Together, these observations indicate that DMRT3 and DMRT5 have similar overlapping function in several aspects of cortical development and partially compensate for one another. Dmrt5 is upregulated in the absence of Dmrt3, and Dmrt3 is downregulated in Dmrt5 KO embryos. The two factors appear thus to function in the same cascade, but it remains unclear which one is upstream of the other.
More unexpectedly, we observed that double KOs have a dorsal telencephalon that is ventralized. In addition, the OBs, the most rostral cortical structure, take over the cortex, thus suggesting “rostroventralization” of the dorsal telencephalon, in accordance with the high caudal to low rostral gradient of expression of Dmt3/5 genes. Conversely, in Dmrt5-overexpressing embryos, DP genes expand ventrally, although some PSB markers remain in place, and OBs are smaller than in WT mice. In addition, Dbx1 expression expands, supporting an expansion of the VP. These findings revealed a new early role for DMRT5 and DMRT3 in DV telencephalic patterning, which could not have been predicted from examining the phenotype of either single KO line alone. This role fits with the timing of activation in the developing telencephalon. Both Dmrt3 and Dmrt5 expression is detected from the open neural plate stage in the prospective dorsal telencephalon when patterning is initiated (data not shown).
As noted, WNT and BMP signaling molecules and downstream dorsal telencephalic transcription factors are dramatically reduced in the cortex of the double KO embryos. The reduction of these pathways is likely to be involved in the ventralization. In contrast, FGF signals appear unaffected, and SHH and downstream targets, such as Nkx2.1, appear normally restricted to the MGE (data not shown). Gli3R is another major player in telencephalic DV patterning (Theil et al., 1999; Tole et al., 2000a; Rallu et al., 2002; Kuschel et al., 2003). RNA-seq analysis of the cortex of E12.5 single and double Dmrt KO embryos did not detect a significant difference in Gli3 mRNA levels compared with WT controls. This unaltered expression of Gli3 in double Dmrt KO embryos has been confirmed by ISH and qRT-PCR experiments (data not shown). Given the similarity of the telencephalic patterning defects observed in Dmrt3;Dmrt5 and Gli3Pdn/Pdn mutants, whether a reduction of Gli3R activity occurs in the Dmrt double KO embryos is a hypothesis that remains to be tested. A more significant role in the ventralization of double Dmrt KO ermbryos is likely to be played by the transcription factor GSX2. In the ventral telencephalon, GSX2 is known to be required for the specification of LGE progenitors. GSX2, further, represses the expression of Dbx1 and other VP markers (Yun et al., 2001; Carney et al., 2009). Our data indicate that it also represses Dmrt5 expression. GSX2 acts in the specification of LGE progenitors upstream of the homeobox Dlx and proneural Ascl1 genes (Wang et al., 2009, 2013). Thus, the increase of Dlx2 and Ascl1 and the loss of dorsal and ventral pallium markers in Dmrt3;Dmrt5 double KO embryos may be a consequence of the dorsal expansion of Gsx2. The large expansion of Gsx2 and other subpallial markers in the cortical neuroepithelium when Dmrt3 and Dmrt5 are lost is intriguing because it occurs despite continued strong Pax6 expression in the cortex, which is reduced only close to the PSB. PAX6 has been shown to repress Gsx2 expression to position the PSB (Toresson et al., 2000; Yun et al., 2001; Carney et al., 2009), but PAX6 may be not sufficient for this role in the absence of DMRT3 and DMRT5.
Gsx2 is expressed at high level in dLGE progenitors that generate ER81- and Sp8-expressing OB interneurons, and at lower levels in vLGE progenitors giving rise to Isl1-expressing striatal projection neurons (Toresson and Campbell, 2001; Yun et al., 2001; Stenman et al., 2003; Waclaw et al., 2006; Ehrman et al., 2013). Gsx2 mutants have reduced vLGE and dLGE and exhibit a significant reduction of their derivatives, striatal projection, and OB interneurons (Corbin et al., 2000; Toresson et al., 2000; Yun et al., 2001, 2003). Conversely, in Gsx2 conditional transgenics, GSX2 sequentially favors striatal and OB fates (Waclaw et al., 2009). In Dmrt double KO embryos, we observed at E12.5 strong ectopic expression of Sp8 but no or low Isl1 indicating a predominant expansion of dLGE. Accordingly, at E18.5, TH-positive interneurons appear abundant in the OBLS structure and Gad67, expressed in OB interneurons, is increased. At the same stage in the double mutant cortex, we also observed a strong reduction of Math2, suggesting that some respecification of cortical neuroblasts is occurring as a consequence of the observed early patterning defects.
Upon ectopic expression of Dmrt5 throughout the telencephalon, we observed at E12.5–E13.5 a loss of Gsx2 expression and downstream dLGE gene expression markers, and an expansion of DP and some VP gene expression markers into the subpallium. Other PSB markers appeared less affected. SFrp2 expression shifted ventrally at E12.5 but recovered at E13.5. In Gsx2 mutants, a strong LGE phenotype at early stages goes away later, due to increased Gsx1 expression (Toresson and Campbell, 2001; Yun et al., 2003; Carney et al., 2009). Whether there is a compensatory increase in Gsx1 in the Dmrt5Tg/+;Foxg1-IRES-Cre that rescues the location of sFrp2 expression remains to be investigated. An alternative possibility is that the PSB repositions itself. The VP, however, does not, and instead expands in subpallial territory, thus dissociating the VP from the PSB. A similar dorsalizing phenotype was observed when Dmrt5 is selectively misexpressed in the ventral telencephalon. This indicates that the ventral expansion of DP and VP gene expression markers is indirect and caused by downregulation of ventral determinants by DMRT5. Gsx2 is again a good DMRT3/5 target candidate, as SHH and Nkx2.1 were found to be unaffected in Dmrt5Tg/Tg;Gsx2-CIE embryos (data not shown). A further suggestion is that dorsal identity in the telencephalon is a “ground state,” which must be suppressed by ventral determinants.
Finally, our findings indicate that Emx2, a target of Dmrt5 (Saulnier et al., 2013), cooperates with Dmrt3 and Dmrt5 in repressing Gsx2, and that these transcription factors bind to an evolutionarily conserved, PAX6-bound element, located downstream of Gsx2. This Emx2 involvement in Dmrt-dependent Gsx2 repression echoes a previous report that coablation of Emx2 and Pax6 results into generalized spreading of subpallial programs into dorsal telencephalon and suppression of pallial programs (Muzio et al., 2002a). Intriguingly, despite the prominent ventralization of dorsal telencephalon occurring in mid-neuronogenic Emx2−/−;Pax6−/− and Dmrt3−/−;Dmrt5−/− embryos, vestigial and abortive pallial specification can still be detected at earlier developmental stages in both double KO embryos.
In conclusion, our findings suggest that DMRT3/5 transcription factors and GSX2 mutually antagonize one another and that DMRT3/5 cooperate with EMX2 and PAX6 to establish and maintain DV patterning in the telencephalon. How DMRT3 and DMRT5 interact with EMX2, and possibly PAX6 and other dorsal regulators such as LHX2, to control the positioning of the PSB and, more globally, telencephalon patterning remains to be investigated.
Footnotes
This work was supported by Fonds National de la Recherche Scientifique Grants FRFC 6973823 and CDR 29148846, Walloon Region First International Project “CORTEX,” and Fonds pour la Recherche Médicale dans le Hainaut. Work in the laboratories of E.A.G., D.Z., and K.C. was supported by National Institutes of Health Grants RO1 MH103211, GM059152, and R01 NS044080, respectively. Work in the laboratory of T.T. was supported by Grant BB/P00122X/1 by the Biotechnology and Biological Sciences Research Council. Work in the Center for Microscopy and Molecular Imaging was supported by the Hainaut-Biomed FEDER program. E.D. is a Wallonie-Bruxelles International doctoral fellow from the Wallonia-Brussels Federation. M.K. was a First International and L.R. a BEWARE postdoctoral fellow from the Walloon Region. S.D.C. was a Fonds National de la Recherche Scientifique postdoctoral fellow. We thank Dr. Daichi Kawaguchi for the kind gift of the Foxg1-IRES-Cre line used in this study; Dr. François Guillemot, John Rubenstein, Setsuko Sahara, Denis O'Leary, Anne Chotteau-Lelièvre, and Y. Sun for gifts of plasmid DNA; Carlos Parras for the Ascl1 antibody; and Louis Delhaye for technical help.
The authors declare no competing financial interests.
References
- Allen ZJ 2nd, Waclaw RR, Colbert MC, Campbell K (2007) Molecular identity of olfactory bulb interneurons: transcriptional codes of periglomerular neuron subtypes. J Mol Histol 38:517–525. 10.1007/s10735-007-9115-4 [DOI] [PubMed] [Google Scholar]
- Anderson SA, Eisenstat DD, Shi L, Rubenstein JL (1997) Interneuron migration from basal forebrain to neocortex: dependence on dlx genes. Science 278:474–476. 10.1126/science.278.5337.474 [DOI] [PubMed] [Google Scholar]
- Aoto K, Nishimura T, Eto K, Motoyama J (2002) Mouse GLI3 regulates Fgf8 expression and apoptosis in the developing neural tube, face, and limb bud. Dev Biol 251:320–332. 10.1006/dbio.2002.0811 [DOI] [PubMed] [Google Scholar]
- Assimacopoulos S, Grove EA, Ragsdale CW (2003) Identification of a Pax6-dependent epidermal growth factor family signaling source at the lateral edge of the embryonic cerebral cortex. J Neurosci 23:6399–6403. 10.1523/JNEUROSCI.23-16-06399.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backman M, Machon O, Mygland L, van den Bout CJ, Zhong W, Taketo MM, Krauss S (2005) Effects of canonical wnt signaling on dorso-ventral specification of the mouse telencephalon. Dev Biol 279:155–168. 10.1016/j.ydbio.2004.12.010 [DOI] [PubMed] [Google Scholar]
- Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D (2004) Ultraconserved elements in the human genome. Science 304:1321–1325. 10.1126/science.1098119 [DOI] [PubMed] [Google Scholar]
- Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Peña-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, Khalid F, Zhang W, Newburger D, Jaeger SA, Morris QD, Bulyk ML, Hughes TR (2008) Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell 133:1266–1276. 10.1016/j.cell.2008.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besse L, Neti M, Anselme I, Gerhardt C, Rüther U, Laclef C, Schneider-Maunoury S (2011) Primary cilia control telencephalic patterning and morphogenesis via Gli3 proteolytic processing. Development 138:2079–2088. 10.1242/dev.059808 [DOI] [PubMed] [Google Scholar]
- Carney RS, Cocas LA, Hirata T, Mansfield K, Corbin JG (2009) Differential regulation of telencephalic pallial-subpallial boundary patterning by Pax6 and Gsh2. Cereb Cortex 19:745–759. 10.1093/cercor/bhn123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H, Beachy PA (1996) Cyclopia and defective axial patterning in mice lacking sonic hedgehog gene function. Nature 383:407–413. 10.1038/383407a0 [DOI] [PubMed] [Google Scholar]
- Chotteau-Lelièvre A, Desbiens X, Pelczar H, Defossez PA, de Launoit Y (1997) Differential expression patterns of the PEA3 group transcription factors through murine embryonic development. Oncogene 15:937–952. 10.1038/sj.onc.1201261 [DOI] [PubMed] [Google Scholar]
- Corbin JG, Gaiano N, Machold RP, Langston A, Fishell G (2000) The Gsh2 homeodomain gene controls multiple aspects of telencephalic development. Development 127:5007–5020. [DOI] [PubMed] [Google Scholar]
- De Clercq S, Keruzore M, Desmaris E, Pollart C, Assimacopoulos S, Preillon J, Ascenzo S, Matson CK, Lee M, Nan X, Li M, Nakagawa Y, Hochepied T, Zarkower D, Grove EA, Bellefroid EJ (2018) DMRT5 together with DMRT3 directly controls hippocampus development and neocortical area map formation. Cereb Cortex 28:493–509. 10.1093/cercor/bhw384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrman LA, Mu X, Waclaw RR, Yoshida Y, Vorhees CV, Klein WH, Campbell K (2013) The LIM homeobox gene Isl1 is required for the correct development of the striatonigral pathway in the mouse. Proc Natl Acad Sci U S A 110:E4026–E4035. 10.1073/pnas.1308275110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericson J, Muhr J, Placzek M, Lints T, Jessell TM, Edlund T (1995) Sonic hedgehog induces the differentiation of ventral forebrain neurons: a common signal for ventral patterning within the neural tube. Cell 81:747–756. 10.1016/0092-8674(95)90536-7 [DOI] [PubMed] [Google Scholar]
- Faedo A, Ficara F, Ghiani M, Aiuti A, Rubenstein JL, Bulfone A (2002) Developmental expression of the T-box transcription factor T-bet/Tbx21 during mouse embryogenesis. Mech Dev 116:157–160. 10.1016/S0925-4773(02)00114-4 [DOI] [PubMed] [Google Scholar]
- Fernandes M, Gutin G, Alcorn H, McConnell SK, Hébert JM (2007) Mutations in the BMP pathway in mice support the existence of two molecular classes of holoprosencephaly. Development 134:3789–3794. 10.1242/dev.004325 [DOI] [PubMed] [Google Scholar]
- Flames N, Pla R, Gelman DM, Rubenstein JL, Puelles L, Marín O (2007) Delineation of multiple subpallial progenitor domains by the combinatorial expression of transcriptional codes. J Neurosci 27:9682–9695. 10.1523/JNEUROSCI.2750-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fode C, Ma Q, Casarosa S, Ang SL, Anderson DJ, Guillemot F (2000) A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes Dev 14:67–80. 10.1101/gad.14.1.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaitanis JN, Walsh CA (2004) Genetics of disorders of cortical development. Neuroimaging Clin North Am 14:219–229. 10.1016/j.nic.2004.03.007 [DOI] [PubMed] [Google Scholar]
- Grove EA, Tole S, Limon J, Yip L, Ragsdale CW (1998) The hem of the embryonic cerebral cortex is defined by the expression of multiple wnt genes and is compromised in Gli3-deficient mice. Development 125:2315–2325. [DOI] [PubMed] [Google Scholar]
- Gutin G, Fernandes M, Palazzolo L, Paek H, Yu K, Ornitz DM, McConnell SK, Hébert JM (2006) FGF signalling generates ventral telencephalic cells independently of SHH. Development 133:2937–2946. 10.1242/dev.02465 [DOI] [PubMed] [Google Scholar]
- Hasenpusch-Theil K, Magnani D, Amaniti EM, Han L, Armstrong D, Theil T (2012) Transcriptional analysis of Gli3 mutants identifies wnt target genes in the developing hippocampus. Cereb Cortex 22:2878–2893. 10.1093/cercor/bhr365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasenpusch-Theil K, Watson JA, Theil T (2017) Direct interactions between Gli3, Wnt8b, and Fgfs underlie patterning of the dorsal telencephalon. Cereb Cortex 27:1137–1148. 10.1093/cercor/bhv291 [DOI] [PubMed] [Google Scholar]
- Hu WF, Chahrour MH, Walsh CA (2014) The diverse genetic landscape of neurodevelopmental disorders. Annu Rev Genomics Hum Genet 15:195–213. 10.1146/annurev-genom-090413-025600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber K, Narasimhan P, Shtukmaster S, Pfeifer D, Evans SM, Sun Y (2013) The LIM-homeodomain transcription factor islet-1 is required for the development of sympathetic neurons and adrenal chromaffin cells. Dev Biol 380:286–298. 10.1016/j.ydbio.2013.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahoud RJ, Elsen GE, Hevner RF, Hodge RD (2014) Conditional ablation of Tbr2 results in abnormal development of the olfactory bulbs and subventricular zone-rostral migratory stream. Dev Dyn 243:440–450. 10.1002/dvdy.24090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi D, Sahara S, Zembrzycki A, O'Leary DDM (2016) Generation and analysis of an improved Foxg1-IRES-cre driver mouse line. Dev Biol 412:139–147. 10.1016/j.ydbio.2016.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno D, Iwashita M, Satoh Y, Momiyama A, Abe T, Kiyonari H, Matsuzaki F (2012) The mammalian DM domain transcription factor Dmrta2 is required for early embryonic development of the cerebral cortex. PLoS One 7:e46577. 10.1371/journal.pone.0046577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll TT, O'Leary DD (2005) Ventralized dorsal telencephalic progenitors in Pax6 mutant mice generate GABA interneurons of a lateral ganglionic eminence fate. Proc Natl Acad Sci U S A 102:7374–7379. 10.1073/pnas.0500819102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuschel S, Rüther U, Theil T (2003) A disrupted balance between Bmp/Wnt and fgf signaling underlies the ventralization of the Gli3 mutant telencephalon. Dev Biol 260:484–495. 10.1016/S0012-1606(03)00252-5 [DOI] [PubMed] [Google Scholar]
- Murphy MW, Zarkower D, Bardwell VJ (2007) Vertebrate DM domain proteins bind similar DNA sequences and can heterodimerize on DNA. BMC Mol Biol 8:58. 10.1186/1471-2199-8-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzio L, DiBenedetto B, Stoykova A, Boncinelli E, Gruss P, Mallamaci A (2002a) Emx2 and Pax6 control regionalization of the pre-neuronogenic cortical primordium. Cereb Cortex 12:129–139. 10.1093/cercor/12.2.129 [DOI] [PubMed] [Google Scholar]
- Muzio L, DiBenedetto B, Stoykova A, Boncinelli E, Gruss P, Mallamaci A (2002b) Conversion of cerebral cortex into basal ganglia in Emx2(−/−) Pax6(Sey/Sey) double-mutant mice. Nat Neurosci 5:737–745. 10.1038/nn892 [DOI] [PubMed] [Google Scholar]
- Ohkubo Y, Chiang C, Rubenstein JL (2002) Coordinate regulation and synergistic actions of BMP4, SHH and FGF8 in the rostral prosencephalon regulate morphogenesis of the telencephalic and optic vesicles. Neuroscience 111:1–17. 10.1016/S0306-4522(01)00616-9 [DOI] [PubMed] [Google Scholar]
- Pellegrini M, Mansouri A, Simeone A, Boncinelli E, Gruss P (1996) Dentate gyrus formation requires Emx2. Development 122:3893–3898. [DOI] [PubMed] [Google Scholar]
- Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, Plajzer-Frick I, Akiyama J, De Val S, Afzal V, Black BL, Couronne O, Eisen MB, Visel A, Rubin EM (2006) In vivo enhancer analysis of human conserved non-coding sequences. Nature 444:499–502. 10.1038/nature05295 [DOI] [PubMed] [Google Scholar]
- Porteus MH, Bulfone A, Ciaranello RD, Rubenstein JL (1991) Isolation and characterization of a novel cDNA clone encoding a homeodomain that is developmentally regulated in the ventral forebrain. Neuron 7:221–229. 10.1016/0896-6273(91)90260-7 [DOI] [PubMed] [Google Scholar]
- Puelles L, Kuwana E, Puelles E, Bulfone A, Shimamura K, Keleher J, Smiga S, Rubenstein JL (2000) Pallial and subpallial derivatives in the embryonic chick and mouse telencephalon, traced by the expression of the genes dlx-2, Emx-1, Nkx-2.1, Pax-6, and Tbr-1. J Comp Neurol 424:409–438. [DOI] [PubMed] [Google Scholar]
- Qin S, Madhavan M, Waclaw RR, Nakafuku M, Campbell K (2016) Characterization of a new Gsx2-cre line in the developing mouse telencephalon. Genesis 54:542–549. 10.1002/dvg.22980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rallu M, Machold R, Gaiano N, Corbin JG, McMahon AP, Fishell G (2002) Dorsoventral patterning is established in the telencephalon of mutants lacking both Gli3 and hedgehog signaling. Development 129:4963–4974. [DOI] [PubMed] [Google Scholar]
- Rash BG, Grove EA (2007) Patterning the dorsal telencephalon: a role for sonic hedgehog? J Neurosci 27:11595–11603. 10.1523/JNEUROSCI.3204-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahara S, Kawakami Y, Izpisua Belmonte JC, O'Leary DD (2007) Sp8 exhibits reciprocal induction with Fgf8 but has an opposing effect on anterior–posterior cortical area patterning. Neural Dev 2:10. 10.1186/1749-8104-2-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saulnier A, Keruzore M, De Clercq S, Bar I, Moers V, Magnani D, Walcher T, Filippis C, Kricha S, Parlier D, Viviani L, Matson CK, Nakagawa Y, Theil T, Götz M, Mallamaci A, Marine JC, Zarkower D, Bellefroid EJ (2013) The doublesex homolog Dmrt5 is required for the development of the caudomedial cerebral cortex in mammals. Cereb Cortex 23:2552–2567. 10.1093/cercor/bhs234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimogori T, Banuchi V, Ng HY, Strauss JB, Grove EA (2004) Embryonic signaling centers expressing BMP, WNT and FGF proteins interact to pattern the cerebral cortex. Development 131:5639–5647. 10.1242/dev.01428 [DOI] [PubMed] [Google Scholar]
- Stenman J, Toresson H, Campbell K (2003) Identification of two distinct progenitor populations in the lateral ganglionic eminence: implications for striatal and olfactory bulb neurogenesis. J Neurosci 23:167–174. 10.1523/JNEUROSCI.23-01-00167.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm EE, Garel S, Borello U, Hebert JM, Martinez S, McConnell SK, Martin GR, Rubenstein JL (2006) Dose-dependent functions of Fgf8 in regulating telencephalic patterning centers. Development 133:1831–1844. 10.1242/dev.02324 [DOI] [PubMed] [Google Scholar]
- Stoykova A, Treichel D, Hallonet M, Gruss P (2000) Pax6 modulates the dorsoventral patterning of the mammalian telencephalon. J Neurosci 20:8042–8050. 10.1523/JNEUROSCI.20-21-08042.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Rockowitz S, Xie Q, Ashery-Padan R, Zheng D, Cvekl A (2015) Identification of in vivo DNA-binding mechanisms of Pax6 and reconstruction of Pax6-dependent gene regulatory networks during forebrain and lens development. Nucleic Acids Res 43:6827–6846. 10.1093/nar/gkv589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theil T, Alvarez-Bolado G, Walter A, Rüther U (1999) Gli3 is required for emx gene expression during dorsal telencephalon development. Development 126:3561–3571. [DOI] [PubMed] [Google Scholar]
- Tole S, Hébert J (2013) Telencephalon patterning. In: Patterning and cell type specification in the developing CNS and PNS: comprehensive developmental neuroscience, pp 3–24. New York: Elsevier. [Google Scholar]
- Tole S, Ragsdale CW, Grove EA (2000a) Dorsoventral patterning of the telencephalon is disrupted in the mouse mutant extra-toes(J). Dev Biol 217:254–265. 10.1006/dbio.1999.9509 [DOI] [PubMed] [Google Scholar]
- Tole S, Goudreau G, Assimacopoulos S, Grove EA (2000b) Emx2 is required for growth of the hippocampus but not for hippocampal field specification. J Neurosci 20:2618–2625. 10.1523/JNEUROSCI.20-07-02618.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tole S, Remedios R, Saha B, Stoykova A (2005) Selective requirement of Pax6, but not Emx2, in the specification and development of several nuclei of the amygdaloid complex. J Neurosci 25:2753–2760. 10.1523/JNEUROSCI.3014-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toresson H, Campbell K (2001) A role for Gsh1 in the developing striatum and olfactory bulb of Gsh2 mutant mice. Development 128:4769–4780. [DOI] [PubMed] [Google Scholar]
- Toresson H, Potter SS, Campbell K (2000) Genetic control of dorsal-ventral identity in the telencephalon: opposing roles for Pax6 and Gsh2. Development 127:4361–4371. [DOI] [PubMed] [Google Scholar]
- Urquhart JE, Beaman G, Byers H, Roberts NA, Chervinsky E, O'Sullivan J, Pilz D, Fry A, Williams SG, Bhaskar SS, Khayat M, Simanovsky N, Shachar IB, Shalev SA, Newman WG (2016) DMRTA2 (DMRT5) is mutated in a novel cortical brain malformation. Clin Genet 89:724–727. 10.1111/cge.12734 [DOI] [PubMed] [Google Scholar]
- Waclaw RR, Allen ZJ 2nd, Bell SM, Erdélyi F, Szabó G, Potter SS, Campbell K (2006) The zinc finger transcription factor Sp8 regulates the generation and diversity of olfactory bulb interneurons. Neuron 49:503–516. 10.1016/j.neuron.2006.01.018 [DOI] [PubMed] [Google Scholar]
- Waclaw RR, Wang B, Pei Z, Ehrman LA, Campbell K (2009) Distinct temporal requirements for the homeobox gene Gsx2 in specifying striatal and olfactory bulb neuronal fates. Neuron 63:451–465. 10.1016/j.neuron.2009.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waclaw RR, Ehrman LA, Pierani A, Campbell K (2010) Developmental origin of the neuronal subtypes that comprise the amygdalar fear circuit in the mouse. J Neurosci 30:6944–6953. 10.1523/JNEUROSCI.5772-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Waclaw RR, Allen ZJ 2nd, Guillemot F, Campbell K (2009) Ascl1 is a required downstream effector of gsx gene function in the embryonic mouse telencephalon. Neural Dev 4:5. 10.1186/1749-8104-4-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Long JE, Flandin P, Pla R, Waclaw RR, Campbell K, Rubenstein JL (2013) Loss of Gsx1 and Gsx2 function rescues distinct phenotypes in Dlx1/2 mutants. J Comp Neurol 521:1561–1584. 10.1002/cne.23242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee SP, Rigby PW (1993) The regulation of myogenin gene expression during the embryonic development of the mouse. Genes Dev 7:1277–1289. 10.1101/gad.7.7a.1277 [DOI] [PubMed] [Google Scholar]
- Yoshida M, Suda Y, Matsuo I, Miyamoto N, Takeda N, Kuratani S, Aizawa S (1997) Emx1 and Emx2 functions in development of dorsal telencephalon. Development 124:101–111. [DOI] [PubMed] [Google Scholar]
- Young FI, Keruzore M, Nan X, Gennet N, Bellefroid EJ, Li M (2017) The doublesex-related Dmrta2 safeguards neural progenitor maintenance involving transcriptional regulation of Hes1. Proc Natl Acad Sci U S A 114:E5599–E5607. 10.1073/pnas.1705186114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun K, Potter S, Rubenstein JL (2001) Gsh2 and Pax6 play complementary roles in dorsoventral patterning of the mammalian telencephalon. Development 128:193–205. [DOI] [PubMed] [Google Scholar]
- Yun K, Garel S, Fischman S, Rubenstein JL (2003) Patterning of the lateral ganglionic eminence by the Gsh1 and Gsh2 homeobox genes regulates striatal and olfactory bulb histogenesis and the growth of axons through the basal ganglia. J Comp Neurol 461:151–165. 10.1002/cne.10685 [DOI] [PubMed] [Google Scholar]