Capsid (CA) plays multiple roles in the HIV-1 replication cycle. CA-CA domain interactions are responsible for multimerization of the Gag polyprotein at virus assembly sites, and in the mature virion, CA monomers assemble into a conical core that encapsidates the viral RNA genome. Multiple CA regions that contribute to the assembly and release of HIV-1 particles have been mapped and investigated. Here, we identified and characterized a Pro-rich loop in CA that is important for the formation of the immature Gag lattice. Changes in this region disrupt viral production and abrogate the formation of infectious, mature virions. Propagation of the mutants in culture led to the selection of second-site compensatory mutations within CA. These results expand our knowledge of the assembly and maturation steps in the viral replication cycle and may be relevant for development of antiviral drugs targeting CA.
KEYWORDS: Cryo-EM, HIV-1, assembly, capsid, maturation, virus replication
ABSTRACT
Late in the HIV-1 replication cycle, the viral structural protein Gag is targeted to virus assembly sites at the plasma membrane of infected cells. The capsid (CA) domain of Gag plays a critical role in the formation of the hexameric Gag lattice in the immature virion, and, during particle release, CA is cleaved from the Gag precursor by the viral protease and forms the conical core of the mature virion. A highly conserved Pro-Pro-Ile-Pro (PPIP) motif (CA residues 122 to 125) [PPIP(122–125)] in a loop connecting CA helices 6 and 7 resides at a 3-fold axis formed by neighboring hexamers in the immature Gag lattice. In this study, we characterized the role of this PPIP(122–125) loop in HIV-1 assembly and maturation. While mutations P123A and P125A were relatively well tolerated, mutation of P122 and I124 significantly impaired virus release, caused Gag processing defects, and abolished infectivity. X-ray crystallography indicated that the P122A and I124A mutations induce subtle changes in the structure of the mature CA lattice which were permissive for in vitro assembly of CA tubes. Transmission electron microscopy and cryo-electron tomography demonstrated that the P122A and I124A mutations induce severe structural defects in the immature Gag lattice and abrogate conical core formation. Propagation of the P122A and I124A mutants in T-cell lines led to the selection of compensatory mutations within CA. Our findings demonstrate that the CA PPIP(122–125) loop comprises a structural element critical for the formation of the immature Gag lattice.
INTRODUCTION
HIV-1 particles assemble predominantly at the plasma membrane (PM) of infected cells. Assembly is driven by the main viral structural polyprotein, Gag, which consists of four domains, namely, matrix (MA), capsid (CA), nucleocapsid (NC), and p6, and of two short spacer peptides, SP1 and SP2 (1–3). The mechanism and pathways that direct Gag molecules to the site of assembly are not fully understood. It has been shown that a patch of basic amino acids near the N terminus of the MA domain and a myristic acid moiety attached to the N-terminal Gly of MA mediate Gag targeting and binding to the PM (4, 5). After membrane binding, Gag molecules multimerize to form a hexameric lattice on the inner leaflet of the PM, leading to the assembly and release of immature virions. Particle release is driven by direct interactions between the p6 domain of Gag and the cellular endosomal sorting complex required for transport (ESCRT) machinery (6, 7).
The immature Gag lattice, both before and after particle release, is hexameric in its organization, with gaps in the lattice that accommodate the strain of curvature (8). During or shortly after release of the immature particle, the viral protease (PR) cleaves the Gag polyprotein into four mature Gag proteins—MA, CA, NC, and p6—and SP1 and SP2. PR-mediated Gag cleavage results in extensive structural rearrangements of the particle interior. The most significant rearrangement is the formation of a cone-shaped core, the shell of which is composed predominantly of CA hexamers and 12 pentamers that close off the ends of the capsid (9). The structural rearrangements that occur during virus maturation lead to the formation of the mature, infectious virion. Because the two copies of the single-stranded viral genomic RNA are packaged into the core, along with the enzymes reverse transcriptase (RT) and integrase (IN), HIV-1 capsids provide a suitable environment for reverse transcription early after entry into the host cell. Thus, capsid formation is an essential step in the generation of an infectious virion. The mechanism of conical core formation is still under investigation. Reports from several groups have supported an assembly-disassembly model in which CA monomers released by PR-mediated Gag processing reassemble to form the capsid core (10, 11). In contrast, other models invoke a nondiffusional transition of the cleaved CA layer without full disassembly of the hexameric Gag lattice (12, 13) or a combination of the two mechanisms for the core maturation pathway (14).
In addition to their role in capsid assembly, CA-CA contacts are also indispensable for the assembly of the immature Gag lattice. The CA domain of Gag consists of N-terminal and C-terminal subdomains—referred to as the CA-NTD and CA-CTD, respectively—connected by a short, flexible linker (Fig. 1A). The CA-NTD (amino acids 1 to ∼145) is composed of an N-terminal β-hairpin and seven α-helices and a loop (residues 85 to 93) that binds the cellular protein cyclophilin A (CypA). The CA-CTD (residues ∼150 to 231) consists of the major homology region (MHR) and four α-helixes (15). Extensive mutational analyses have identified a number of CA residues that are critical for the assembly of the immature particle and the mature capsid (3). For example, using a library of HIV-1 mutants bearing single amino acid substitutions in CA, it was observed that most mutations in CA disrupt virion assembly (16). Key residues are located in several regions, including helices 4, 5, and 6 of the CA-NTD; helix 9 of the CA-CTD; and the part of the CA-CTD that includes the MHR as well as helices 10 and 11 (17). Interestingly, deletion of the CA-NTD results in only moderate impairment of the formation of virus-like particles (VLPs), pointing to a major role for CA-CTD in virion assembly. However, CA-NTD-deleted Gag forms VLPs with a wide size distribution, indicating that the CA-NTD is involved in determining particle morphology (18).
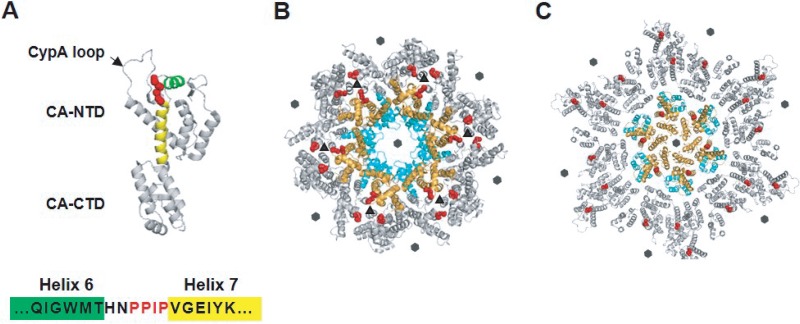
FIG 1.
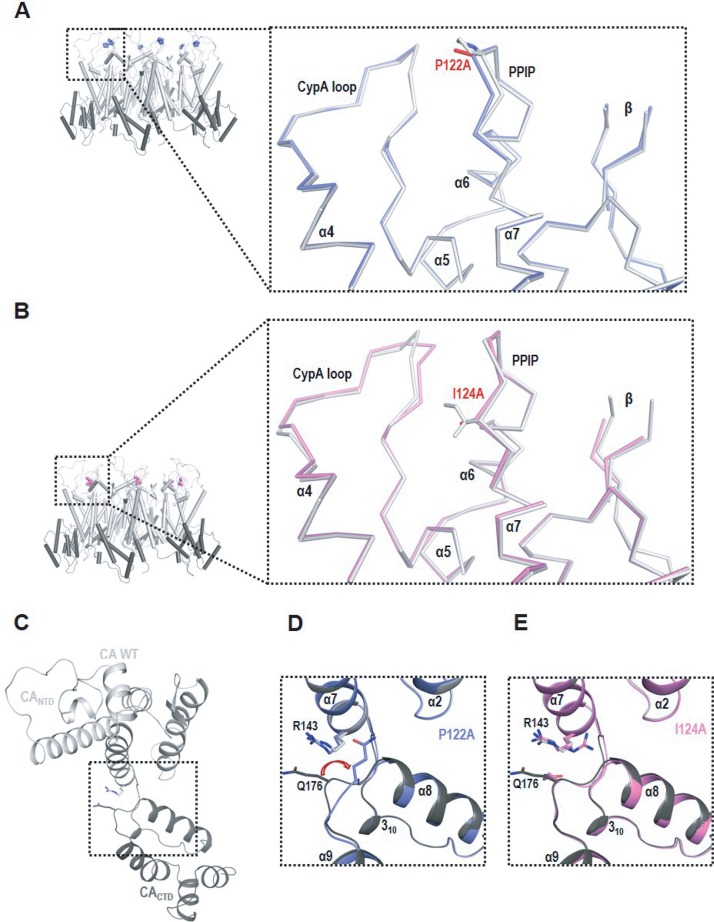
Location of the PPIP(122–125) motif in the HIV-1 CA monomer, immature Gag lattice, and mature capsid. (A) HIV-1 CA is composed of N-terminal and C-terminal domains (CA-NTD and CA-CTD, respectively) connected by a flexible linker. The loop that binds the cellular protein CypA is indicated (CypA loop; arrow). The PPIP motif (red) is located between helices 6 (green) and 7 (yellow) in the CA-NTD (PDB ID: 5MCX [27]). (B) The CA domain is arranged in a hexameric fashion in the immature Gag lattice (in the central hexamer CA-NTDs in orange and CA-CTDs in cyan; neighboring hexamers in gray) (PDB ID: 4USN [21]). The PPIP motifs (red) of neighboring hexamers meet in the 3-fold interhexamer interface (triangles). Sixfold symmetry axes are indicated by hexagons. (C) A mature hexameric lattice in the intact virion (in the central hexamer, CA-NTDs in orange, CA-CTDs in cyan, neighboring hexamers in gray). Conformational shifts during maturation move the PPIP motif (red) away from the interhexamer interface in the immature Gag lattice to a more central position in the mature CA lattice (PDB ID: 5MCX [27]). Sixfold symmetry axes are indicated by hexagons.
Numerous structural studies have provided insights into the folding and conformation of CA in both the immature Gag lattice and the mature conical capsid (19, 20). More recently, Briggs and colleagues used cryo-electron tomography (cryo-ET) to solve the structure of the CA domain in the immature particle and to define more precisely the roles of individual CA domains in the formation of the immature Gag lattice (Fig. 1B) (21). Each CA-NTD forms multiple contacts with CA-NTDs from the same or neighboring hexamers. Multiple contacts between CA-CTDs also maintain the integrity of the immature Gag lattice, and MHR residues interact within a hexamer. Residues within helix 9, in particular, two hydrophobic amino acids, W184 and M185, form interhexameric contacts across a CA-CA dimer interface. The C terminus of CA-CTD and the N-terminal eight residues of SP1 form a six-helix bundle in the immature Gag lattice (22, 23). The CA-CTD, including a flexible hinge formed by a Val-Gly-Gly motif (residues 221 to 223), and the six-helix bundle together form an assembly unit that plays a central role in stabilizing the immature Gag lattice (22). Although the tertiary structure of the CA monomer is highly conserved, the arrangements of the two CA domains in the immature Gag shell differ significantly between retroviruses (21).
The CA arrangement in the mature core has been established based on analysis of in vitro-assembled hexameric wild-type (WT) and mutant lattices by X-ray crystallography and cryo-electron microscopy (24–26). Most recently, the structure of the HIV-1 capsid derived from intact virions has been examined by using cryo-ET (Fig. 1C) (27). Comparisons of the orientations of CA in the immature and mature hexameric lattices revealed the presence of large-scale CA domain rotations and rearrangements during the maturation process (13, 20, 21). The integrity of one mature hexamer is supported by CA-NTD–CA-NTD and CA-NTD–CA-CTD contacts between adjacent CA monomers and by CA-NTD–CA-CTD intrasubunit contacts. In contrast to the immature Gag lattice, interhexameric contacts in the mature lattice are formed exclusively by the CA-CTD (26). Subtle changes in CA structure, including movement of CA-NTD relative to CA-CTD and rotations around CA-CTD at the 2-fold and 3-fold interfaces of CA hexamers, together with the presence of CA pentamers, allow formation of the variably curved mature core (27).
In this study, we determined the role in HIV-1 assembly of a highly-conserved, Pro-rich sequence, Pro-Pro-Ile-Pro (PPIP), spanning CA residues 122 to 125 [PPIP(122–125)], in a short loop connecting helices 6 and 7 (H6-H7) (Fig. 1A). These loops surround the trimer interface between neighboring hexamers in the immature Gag lattice (Fig. 1B), while this region is exposed in the mature core (Fig. 1C). It has been reported that mutation of residue P122 or residue I124 drastically decreases the production and infectivity of HIV-1 particles (16, 28–30); however, the basis for these defects has not been investigated. To analyze the contribution of this loop to assembly of HIV-1 particles, we substituted alanine at each position in the PPIP motif and characterized the mutant viruses by a combination of virological, biochemical, and structural approaches. Substitutions at two positions, P122 and I124, resulted in impaired release of noninfectious particles containing a defective immature Gag lattice. Propagation of the P122 and I124 mutants in T-cell line cultures led to the generation of revertant viruses containing substitutions at several positions within the CA-NTD. These compensatory mutations restored particle assembly and infectivity. Our results demonstrate that the PPIP(122–125) motif plays a key role in assembly of the Gag lattice of the immature virion and in subsequent particle maturation.
RESULTS
Mutations in the PPIP(122–125) motif cause defects in HIV-1 particle assembly and maturation.
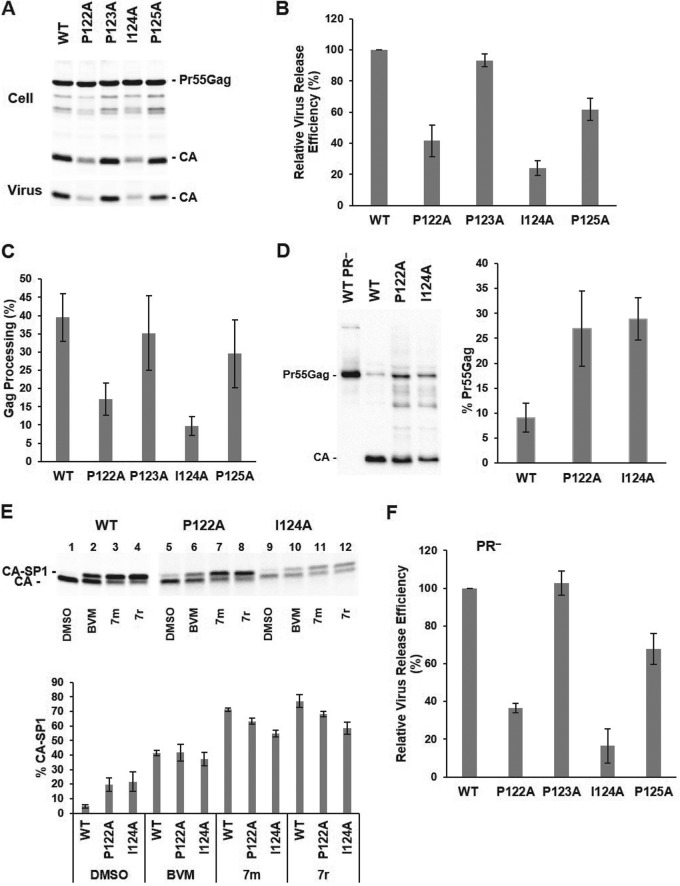
To characterize the role of the PPIP motif spanning HIV-1 CA residues 122 to 125, we introduced Ala mutations into each position of this motif in the pNL4-3 full-length infectious molecular clone and analyzed the resulting effects on HIV-1 particle production, Gag processing efficiency, infectivity, replication, and maturation. To probe the role of the PPIP(122–125) motif in virus particle production, HeLa cells were transfected in parallel with WT and mutant molecular clones and the relative levels of virus release efficiency were calculated based on the amount of radiolabeled viral proteins in cell and virion lysates. We found that while mutation P123A did not affect virus production, mutations P122A, I124A, and P125A reduced virus particle production to ∼40%, 25%, and 60% of WT levels, respectively (Fig. 2A and B). The virus release defects exhibited by the P122A and I124A mutants were associated with impaired Gag processing, as measured by the ratio of cell-associated CA to the total amount of Pr55Gag and CA in cell lysates (Fig. 2C). To determine the levels of unprocessed Gag in virions, we collected viral particles from transfected cells and measured the ratio of Pr55Gag to the total amount of Gag (Pr55Gag plus CA) in particles by western blotting. The loading volumes were adjusted to account for the fact that the P122A and I124A mutants release less virus than the WT. The levels of Pr55Gag were about three times higher in the two mutants than in the WT virions (Fig. 2D). The efficiency of the last step in Gag processing—release of CA from CA-SP1—was measured in radiolabeled viruses produced from transfected cells (Fig. 2E). We observed that CA-SP1 cleavage was around four times less efficient in P122A and I124 particles than in the WT particles (Fig. 2E, lanes 1, 5, and 9 and the leftmost three bars on the graph). These data demonstrate that P122A and I124A mutants exhibit Gag processing defects not only in producer cells but also in released virions.
FIG 2.
P122A and I124A mutations impair virus particle production and Gag processing. HeLa cells were transfected with WT and PPIP mutant proviral clones. At 24 h posttransfection, the cells were metabolically labeled with [35S]Met/Cys. Viral particles were collected by ultracentrifugation. Cell lysates were immunoprecipitated with anti-HIV immunoglobulin (HIV-Ig). Viral proteins were separated by SDS-PAGE and quantified by phosphorimager analysis. (A) A representative gel. (B) Relative virus release efficiency was calculated as [virus-associated CA]/[total (cell plus virus) Gag + CA], with the WT value set at 100; error bars indicate standard deviations (SD); n = 3 independent experiments. (C) Gag processing efficiency in cell lysates was calculated as CA/(CA + Pr55Gag). Error bars indicate SD; n = 3 independent experiments. (D) The level of unprocessed Pr55Gag in virions collected from 293T cells was assessed by western blotting and calculated as Pr55Gag/(Pr55Gag + CA). Sample loading was adjusted to reflect the decreased particle production of the P122A and I124A mutants (a representative gel is shown on the left; quantitation indicated on the right). Error bars = SD; n = 3 independent experiments. (E) Percentages of CA-SP1 calculated as CA-SP1/(CA-SP1 + CA). 293T cells transfected with WT and mutant clones were incubated in the presence of dimethyl sulfoxide (DMSO) or 100 nM maturation inhibitors (BVM or the 7m or 7r analogs) and were metabolically labeled in [35S]Met/Cys. Radiolabeled virions were collected and viral proteins separated by SDS-PAGE. WT protein bands were exposed to a phosphorimager screen for 1 day; mutant viral samples were exposed for 4 to 5 days. Error bars indicate SD; n = 3 independent experiments. (F) HeLa cells were transfected with PR− molecular clones. Virus release efficiency was calculated as virion Pr55Gag/total (cell plus virus) Pr55Gag. Error bars indicate SD; n = 3 independent experiments.
To evaluate whether the defects in virus particle production are linked to dysregulation of PR-mediated Gag processing, we examined virus release efficiency in the context of PR-negative (PR−) molecular clones. Consistent with the results of virus release assays performed with PR+ mutants, production of immature particles was impaired by the P122A and I124A mutations (Fig. 2F). These data suggest that defects in virus particle production by P122A and I124A viruses are not caused by aberrant Gag processing.
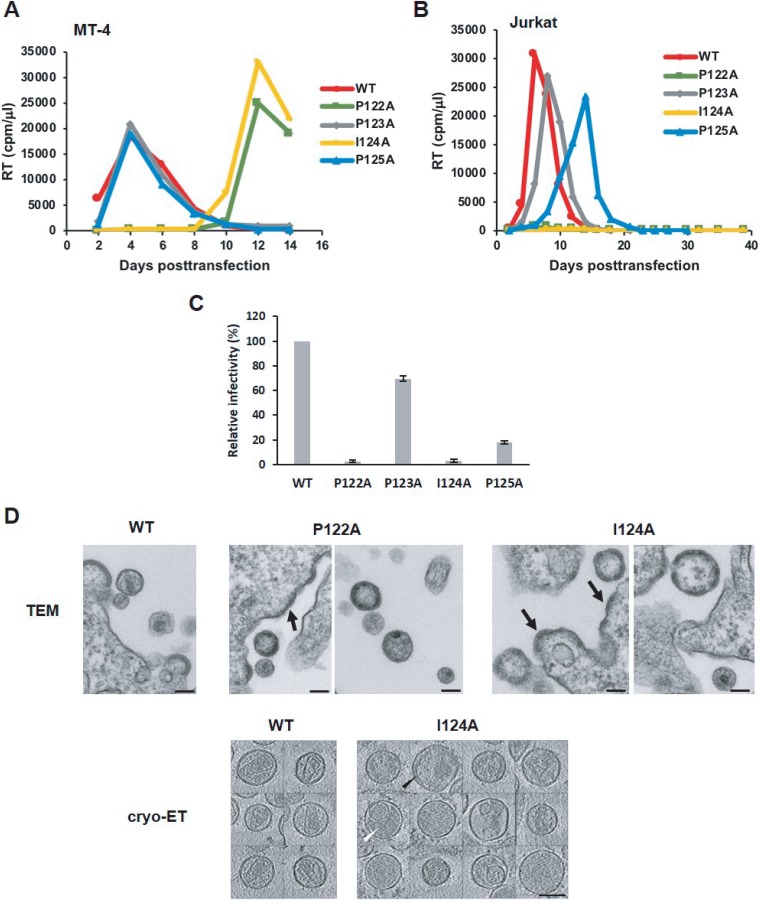
To analyze the effects of the PPIP mutations on virus replication kinetics, we transfected highly permissive MT-4 (Fig. 3A) and less-permessive Jurkat (Fig. 3B) T-cell lines with WT or mutant molecular clones and monitored virus replication kinetics by RT assay. We observed that the P123A and P125A viruses were replication competent in MT-4 and Jurkat cells. In contrast, and consistent with their defects in virus assembly and release, the P122A and I124A mutants replicated with a significant delay relative to the WT in MT-4 cells and were not able to replicate in Jurkat cells (Fig. 3A and B).
FIG 3.
P122A and I124A mutants exhibit defects in virus replication kinetics, particle infectivity, and virion morphology. (A and B) MT-4 (A) or Jurkat (B) cells were transfected with the indicated viral clones. Aliquots of supernatants were collected every other day, and RT activity was measured. (C) TZM-bl cells were incubated with HeLa-derived viral stocks in the presence of DEAE-dextran. After 2 to 3 h, the medium was replaced, and at 48 h postinfection, luciferase activity was analyzed and normalized based on the levels of virus-associated RT. Error bars indicate SD; n = 3 independent experiments. (D) (Upper panels) HeLa cells were transfected with WT, P122A, or I124 molecular clones. At 24 h posttransfection, the medium was removed, and the cells were fixed with 2% glutaraldehyde–0.1 M sodium cacodylate buffer and analyzed by TEM. Arrows indicate thick-walled, patchy Gag layers along the PM of virus-producing cells. (Lower panels) Cryo-ET analysis of WT and I124A virions. HeLa cells were transfected with the indicated viral clones. At 48 h posttransfection, the supernatants were filtered, viral particles were pelleted by ultracentrifugation, resuspended in 1× Tris-buffered saline, fixed in 3% to 4% glutaraldehyde, and imaged by cryo-ET. Shown are central slices through I124A virions that have a disordered Gag layer (black arrowhead) or are at later stages of maturation, with the protein density, presumably representing processed Gag, distributed throughout the interior of the particles (white arrowhead). No I124A virions containing conical, WT-like cores were observed. Scale bars, 100 nm.
To examine the effects of these mutations on particle infectivity, HeLa cells were transfected with WT and mutant molecular clones, and virus-containing supernatants were analyzed for infectivity of specific particles in the TZM-bl indicator cell line. Relative to WT viruses, the P123A and P125A mutants showed 70% and 20% infectivity, respectively, whereas P122A and I124A viruses were noninfectious (Fig. 3C).
We next examined the morphology of mutant particles by thin-section transmission electron microscopy (TEM) and cryo-ET. The electron micrographs demonstrated that P123A and P125A mutant particles were morphologically similar to the WT particles (data not shown). In contrast, P122A and I124A viral particles lacked the characteristic conical core observed with WT HIV-1 virions (Fig. 3D, upper panels). The P122A and I124A mutants also showed an accumulation of Gag patches at the PM of virus-producing cells, suggesting a structural role for these residues early in the assembly process. As similar defects in virion morphology were observed for the two mutants, we performed cryo-ET analysis only for the I124A particles (Fig. 3D, lower panels). We verified that I124A virions lacked the conical core seen in WT particles and also lacked any other higher-order structure made of the same (mature) lattice; we infer that these particles represent maturation-defective virions at various stages of Gag processing. Some virions contained a poorly ordered Gag-like layer lining the envelope/MA layer, while others contained a density distributed throughout the particle; at least part of that density is likely to have been derived from unassembled CA. Overall, these data demonstrate that mutations of P122 and I124 in the highly conserved PPIP(122–125) motif have similar phenotypes and that the motif plays a critical role in HIV-1 particle assembly, maturation, and infectivity.
Protease-deficient P122A and I124A virions have a morphologically abnormal Gag lattice.
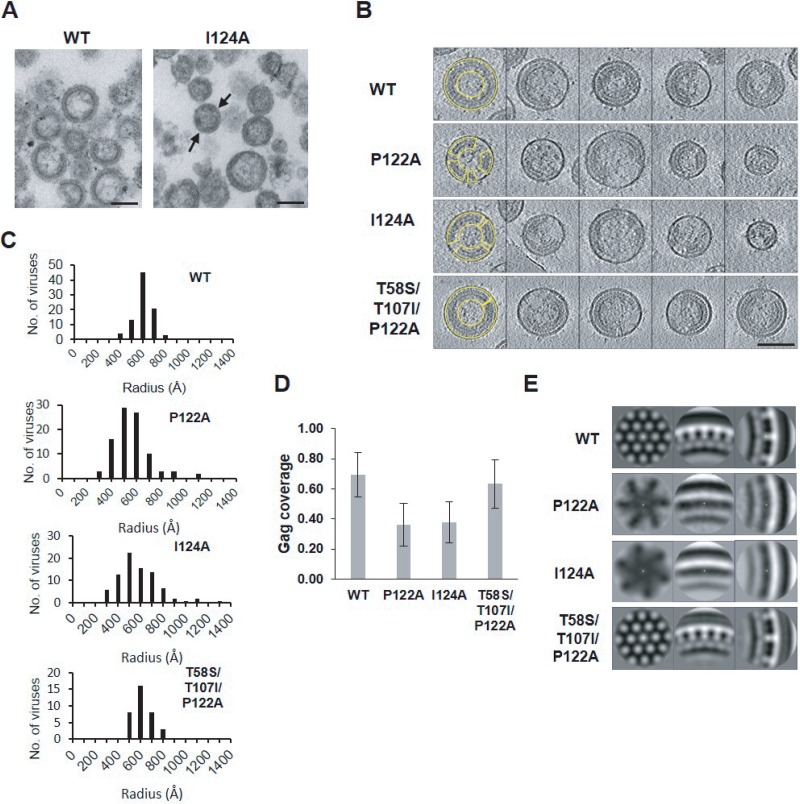
To further define the role of the PPIP loop in virus assembly, we characterized PR− derivatives of the P122A and I124A molecular clones. TEM analysis of those virus particles revealed significant gaps or discontinuities in the Gag-like lattice for both mutants (I124A, Fig. 4A; P122A, data not shown).
FIG 4.
In PR-deficient virions, the P122A and I124A mutations cause defects in the Gag lattice. HeLa cells were transfected with WT or I124A HIV-1 PR− clones. At 48 h posttransfection, the supernatants were filtered and viral particles were collected by ultracentrifugation. The pelleted virions were resuspended with 2% glutaraldehyde–0.1 M sodium cacodylate buffer and then concentrated by ultracentrifugation. (A) TEM analysis revealed discontinuities (black arrows) in the Gag lattices of I124A virions compared to the WT virions. Scale bars, 100 nm. (B to D) Cryo-ET analysis of PR− derivatives of the two mutants, a WT control, and a revertant. HeLa cells were transfected with the indicated clones. At 48 h posttransfection, the supernatants were filtered and released virions were pelleted by ultracentrifugation followed by fixation in 3% to 4% glutaraldehyde and visualized by cryo-ET. P122A and I124A particles exhibited broader size distributions that peaked at a smaller radius than those of the WT or the revertant (C). The Gag layers of the P122A and I124A mutants are seen to be distributed sparsely and discontinuously along the viral envelope compared to the WT or the T58S/T107I/P122A revertant (examples delineated in yellow) (B). (D) Gag coverage was estimated for 87, 92, 84, and 35 virions for WT, P122A, I124A, and T58S/T107I/P122A samples, respectively. Error bars indicate SD. Scale bar, 100 nm. (E) Subtomogram averaging of data from the immature Gag lattice demonstrated that the P122A and I124A mutations disrupted the organization of the Gag lattice, whereas the T58S/T107I/P122A revertant regained WT-like Gag lattice organization.
To further characterize the defects in the structure of the immature Gag lattice caused by the P122A and I124A mutations, protease-deficient WT and mutant particles were visualized by cryo-ET (Fig. 4B). Analysis of these data revealed broader size distributions for the P122A PR− and I124A PR− particles than for the WT PR− particles, and the size range peaked at a smaller diameter (Fig. 4C). Moreover, Gag was distributed more sparsely and in discontinuous patches along the viral envelope in these mutants (Fig. 4B and D). To discern the local structure of this region, subtomogram averaging was performed. For WT PR− particles, the honeycomb pattern characteristic of the immature Gag lattice was clearly seen in the top-down view (Fig. 4E, left panel), and the trilaminar nature of the cross-sectional view (8, 31) was also well defined (Fig. 4E, middle and right panels). In contrast, the subtomogram average data determined for the P122A PR− particles showed no evidence of the WT immature Gag lattice, and the CA layer was thinner and nonstriated in the cross-sectional view. The I124A PR− particles were of essentially the same morphotype (Fig. 4E).
To determine whether mutations in the PPIP(122–125) motif of the CA-NTD affect the arrangement of downstream domains in Gag in the assembled immature lattice, we used maturation inhibitors (MIs) to probe the formation of the six-helix bundle that spans the junction between the CA-CTD and SP1. The first-in-class MI bevirimat (BVM) and BVM analogs inhibit the final cleavage step in the Gag processing cascade—the separation of CA from CA-SP1—thereby preventing the formation of mature, infectious particles (32–34). Recent studies suggest that BVM binds at the center of the CA-SP1 bundle and stabilizes its structure, thereby impeding the access of PR to the cleavage site (22, 23). We thus examined whether BVM and two potent BVM analogs (7m and 7r [34]) are able to block CA-SP1 processing of the P122A and I124A mutants. Indeed, we observed that BVM and the two BVM analogs significantly inhibited P122A and I124A CA-SP1 processing, albeit not to the same level as observed with the WT (Fig 2E). Because the six-helix CA-SP1 bundle is the target of MI activity, these data suggest that bundle formation is not blocked by the P122A and I124A mutations, despite their marked impact on particle assembly.
P122A and I124A subtly affect formation of the CA hexagonal lattice.
As demonstrated above, the P122A and I124A mutants are unable to produce virions containing mature cone-shaped cores, thereby impeding investigation of the role of the motif in assembling the mature CA lattice in the context of virus particles. To address the effect of the PPIP mutations on the formation of the mature CA hexamer, full-length CA proteins bearing P122A or I124A mutations were crystallized. The structures were solved in the P6 space group with one molecule/asymmetric unit (see Table S1 in the supplemental material). Overall, the mutant structures are similar to those of the WT CA (root mean square deviation [RMSD], 0.394 Å for P122A and 0.400 Å for I124A), indicating that the mutations did not disrupt the overall folding of the CA protein. Rather, the mutations primarily resulted in local structural rearrangements at or near the site of the mutations. Specifically, the P122A and I124A mutations resulted in alterations in the PPIP motif itself and induced subtle repositioning of residues 92 to 96 in the CypA loop (Fig. 5A and B). Moreover, residue Q176 in the P122A mutant assumed a conformation different from that seen with CA WT (Fig. 5C and D), while Q176 adopted the WT conformation in the P124A mutant, despite some flexibility (Fig. 5E).
FIG 5.
X-ray crystallography demonstrated that P122A and I124A mutations subtly affect the PPIP motif, CypA-binding loop, and CANTD-CACTD intraprotomer interface. (A and B) CA WT hexamers (CANTD in white, CACTD in gray) with mutation sites (in spheres) are shown in cartoon form. Small boxes show closeup views of changes induced by (A) P122A (in blue) and (B) I124A (in pink). The mutant proteins (shown as a Cα tracing) are superposed with CA WT (in white). (C) The CA WT protomer (CANTD in white, CACTD in gray) is shown in ribbon. Side chains of R143 and Q176 are shown as sticks. (D and E) Enlarged views of the boxed region in panel C. P122A (in blue) (D) or I124A (in pink) (E) is shown superposed with the CA WT. The red arrow in panel D highlights repositioning of Q176 in P122A mutant. CypA loop, CypA binding loop.
Summary of data collection and refinement statistics. Download Table S1, PDF file, 0.1 MB (90.5KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
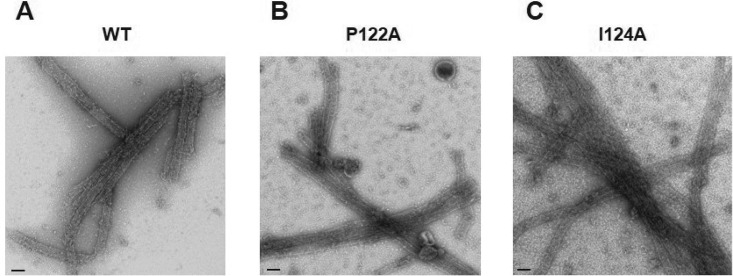
To characterize in more detail the ability of the PPIP mutants to assemble into a mature CA lattice, we investigated their in vitro assembly properties. Under high-salt conditions, WT CA monomers assemble into tubular structures composed of mature CA hexamers (35, 36). The efficiency of tube formation can be monitored either by direct EM observation or by measuring the increase in optical density (turbidity) of the reaction as the CA protein assembles in solution. We performed in vitro assembly reactions with WT, P122A, and I124A CA proteins and examined the assembly products by EM (Fig. 6). The results indicated that both the P122A and I124A CA proteins were able to assemble into tubes in vitro. However, some differences in assembly efficiency were observed; P122A CA protein assembled with kinetics that were even faster than those of the WT, whereas I124A CA assembled more slowly than the WT and, unlike the WT and P122A CA protein, did not assemble at concentrations of <6 mg/ml (data not shown). Overall, these data suggest that the PPIP(122–125) loop is a structural element critical for the formation of the immature Gag lattice but plays a less significant role in formation of the mature CA lattice.
FIG 6.
P122A and I124 CA mutants assemble into tubes in vitro. Wild-type and the two mutant CA proteins were incubated in the presence of 1M NaCl, then stained and analyzed by EM. Tubular structures were observed for WT CA protein (A) as well as for the P122A (B) and I124A (C) mutants. Scale bars, 100 nm.
Compensatory mutations obtained by in vitro selection rescue the replication defects imposed by PPIP(122–125) mutations.
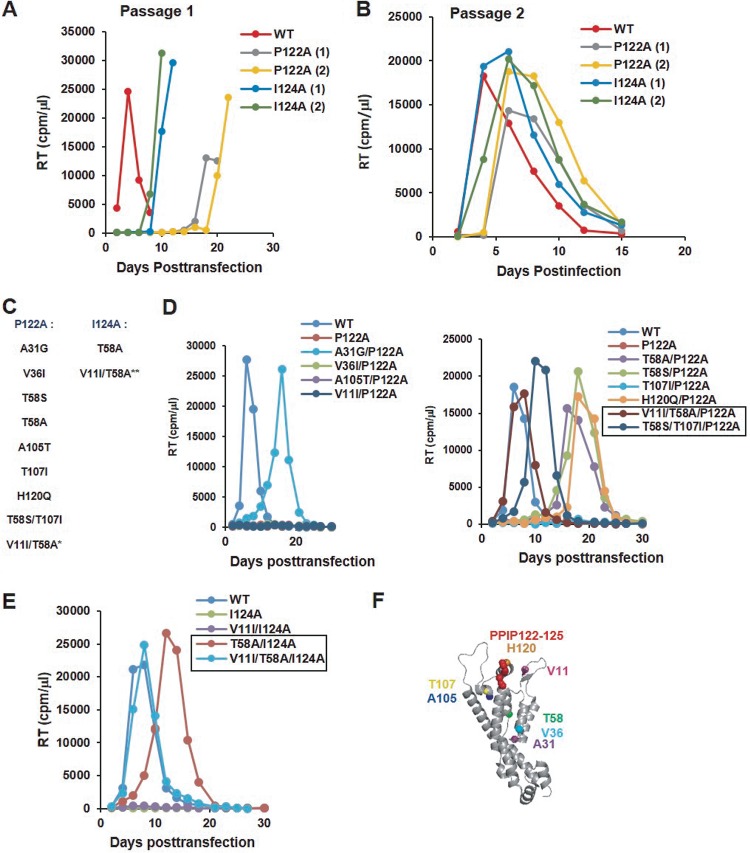
To elucidate the interplay between the PPIP loop and the remainder of CA during virus assembly and maturation, we selected for viruses with compensatory mutations that rescue the defects imposed by P122A and I124A. As a starting point, the highly permissive MT-4 T-cell line was transfected with WT, P122A, and I124A pNL4-3 molecular clones, and virus replication was monitored by RT assay. As shown in Fig. 3A, virus replication was markedly delayed in the cultures transfected with the P122A and I124A mutants (Fig. 7A). Supernatants were harvested from the peaks of virus replication, normalized for RT activity, and used to reinfect fresh MT-4 cells. The repassaged virus, derived from multiple independent flasks, replicated with near-WT kinetics (Fig. 7B), demonstrating that the viruses had reverted and/or acquired second-site compensatory mutations that rescued the defects imposed by the original PPIP motif substitutions. The Gag coding region from ∼20 putative revertant viruses was sequenced, and a number of second-site mutations were identified (Fig. 7C). In some cases, the I124A mutant reverted via a primary-site change (I124V or I124T). To analyze the ability of the second-site mutations acquired during passaging to rescue the defects conferred by the P122A and I124A substitutions, we introduced these mutations into the P122A or I124A clones. All of the viruses containing second-site changes, except for the A105T/P122A mutant, were replication competent in MT-4 cells (data not shown).
FIG 7.
P122A and I124A mutants acquire compensatory substitutions upon propagation in cell culture. (A) Multiple flasks of MT-4 cells were transfected with either P122A or I124A molecular clones (data from two flasks are shown for each mutant, indicated as “1” and “2”). Aliquots of the supernatants were collected every other day and analyzed for RT activity. (B) Supernatants collected at the peak of the RT level analyzed as described for panel A were used to infect fresh MT-4 cells. (C) At the RT peak in infected cultures, infected cells were pelleted, genomic DNA was isolated, and gag regions of the proviral DNA were amplified by PCR. Sequencing of these PCR-amplified fragments identified a set of second-site mutations in the CA-NTD. All revertant mutations were selected in MT-4 cells, except for V11I (*), which arose upon propagation of T58A/P122A in Jurkat cells. V11I was also engineered into the T58A/I124A clone (**). (D, E) Jurkat cells were transfected with P122A-containing (D) or I124A-containing (E) viral clones. Supernatants were collected every other day and analyzed for RT activity. Revertant viruses that were selected for further analysis are indicated in boxes. (F) Location of the second-site mutations in the structure of the mature CA monomer (PDB ID: 5MCX [27]).
As mentioned above, the MT-4 T-cell line is highly permissive for HIV-1 replication. To investigate further the ability of the selected second-site mutations to rescue P122A- and I124A-imposed defects, we performed replication assays in the less permissive Jurkat T-cell line. In contrast to the results obtained in MT-4 cells, we observed in the Jurkat cells a wider range of replicative capacity for the mutants (Fig. 7D to E). Among the P122A-containing viruses, three mutants—T107I/P122A, V36I/P122A, and A105T/P122A—did not replicate during 2 months in culture in three independent experiments (Fig. 7D and data not shown). All other mutants exhibited modest to marked delays in replication, with RT levels peaking between 10 and 30 days. We collected several viral samples at the RT peaks and repassaged them in Jurkat cells to identify additional mutations (data not shown). Analysis of Gag sequences was performed for the viruses that demonstrated improved replication kinetics relative to the first passage. We found that T58A/P122A acquired an additional change, V11I. We introduced V11I in the context of P122A and T58A/P122A and observed that, while V11I alone was not able to rescue the replication defects exhibited by P122A, the V11I/T58A/P122A triple mutant displayed WT-like replication kinetics in three independent experiments (Fig. 7D and data not shown). As the T58A mutation also rescued the replication capacity of I124A in MT-4 cells (data not shown) and Jurkat cells (Fig. 7E), V11I was introduced in the I124A molecular clone as well. In contrast to P122A mutant viruses, T58A significantly improved replication of the I124A mutant, with T58A/I124A showing a delay of only 4 to 6 days relative to the WT (Fig. 7E). However, again, WT-like replication kinetics were consistently observed only for the triple mutant, V11I/T58A/I124A (Fig. 7E). Similarly to P122A-containing viruses, V11I alone was unable to compensate for the replication defects conferred by I124A. All of the compensatory mutations identified in this analysis were located in the CA-NTD and in most cases at residues distal to the PPIP motif (Fig. 7F). Overall, experiments performed to select revertant viruses revealed that the combination of two mutations, V11I and T58A, completely rescues the replication defects imposed by both P122A and I124A in Jurkat cells and that a variety of other mutations are able to confer partial rescue.
Compensatory mutations restore WT-like virus particle production, infectivity, and virion morphology.
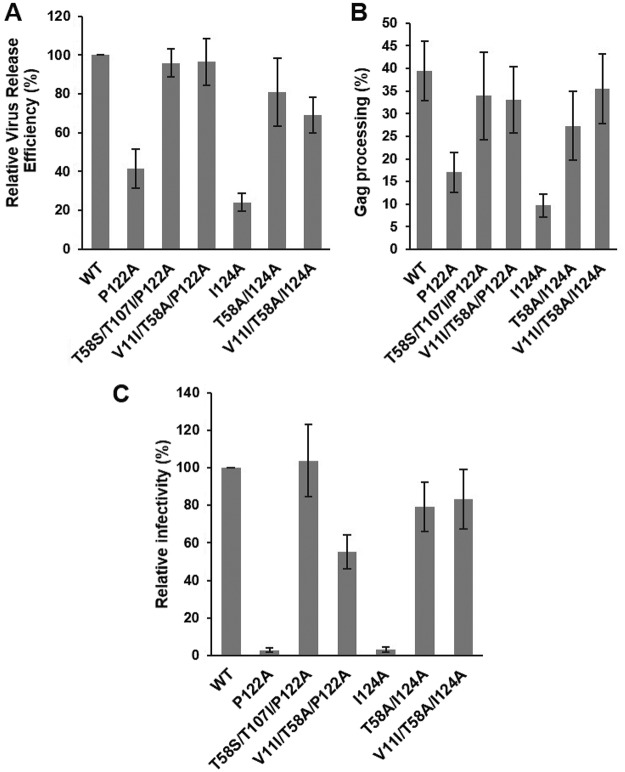
To elucidate the mechanism by which the identified CA-NTD mutations corrected the replication defects imposed by the P122A and I124A substitutions, we focused on the four viral revertants—T58S/T107I/P122A, V11I/T58A/P122A, T58A/I124A, and V11I/T58A/I124A—that exhibited the most improved replication kinetics in Jurkat cells (Fig. 7D and E). Virus particle production, Gag processing, virion infectivity, and particle morphology were examined. For all of the mutants, the efficiency of virus assembly and release was restored to near-WT levels (Fig. 8A). Similarly, the Gag processing defects observed with the original P122A and I124A mutants were corrected in the revertants (Fig. 8B). Infectivity of the mutant viruses was also significantly improved (Fig. 8C). Virions with WT morphology were detected in all samples by TEM (data not shown). Analyzing the T58S/T107I/P122A PR− revertant particles further by cryo-ET, we found that the compensatory mutations restored immature Gag lattice formation and particle size distribution to WT-like levels (Fig. 4B, C, D, and E). Overall, a combination of two mutations—V11I/T58A or T58S/T107I—was able to compensate for the original defect caused by the P122A substitution, leading to production of infectious virions (Fig. 8C); the former combination was also capable of rescuing the replication competency of I124A.
FIG 8.
Compensatory mutations reverse the P122A- and I124A-imposed defects in virus particle production, Gag processing, and infectivity. (A and B) HeLa cells were transfected with the indicated HIV-1 molecular clones. At 24 h posttransfection, cells were metabolically labeled with [35S]Met/Cys, and released virus particles were collected by ultracentrifugation. Cell lysates were immunoprecipitated with HIV-Ig. Cell-associated and virion-associated proteins were separated by SDS-PAGE and quantified by phosphorimager analysis. Relative virus release efficiency was calculated as the ratio of virus-associated CA to the total (cell plus virus) Gag, with the WT value set at 100; error bars indicate SD; n = 3 independent experiments. Gag processing efficiency in cell lysates was calculated as CA/(CA + Pr55Gag). Error bars indicate SD; n = 3 independent experiments. (C) TZM-bl cells were incubated with HeLa-derived viral stocks in the presence of DEAE-dextran. After 2 to 3 h, the medium was replaced, and luciferase activity was analyzed at 48 h postinfection. Luciferase signals were normalized based on levels of virus-associated RT. Error bars indicate SD; n = 3 independent experiments.
DISCUSSION
Analysis of CA sequences across HIV-1 subtypes revealed the presence of a highly conserved, Pro-rich motif in a short, exposed loop between helices 6 and 7 in the CA-NTD. In the present study, we determined the role of this PPIP(122–125) motif in the HIV-1 replication cycle by biochemical, virological, and structural approaches. We demonstrated that substitutions in the PPIP motif result in severe defects in the assembly of the immature Gag lattice and subsequent virion maturation, leading to reduced production of infectious HIV-1 particles. Compensatory mutations within the CA-NTD that rescue these defects were identified.
In the immature Gag lattice, the H6-H7 loop is located in close proximity to the same loop from two neighboring hexamers (Fig. 1B). In contrast, in the mature hexameric lattice, this region does not appear to be directly involved in forming CA-CA contacts (Fig. 1C). The three Pro residues of the PPIP(122–125) motif are conserved in >95% of HIV-1 isolates; Ile124 is found in 67% of the viral isolates in the HIV-1 database, while Val is present at that position in 32% of the HIV-1 sequences (http://hivmut.org/) (37). Among other primate lentiviruses (e.g., HIV-2 Gag-Pol [UniProt entry P04584], SIVmac Gag-Pol [UniProt entry P05897], and SIVagm Gag-Pol [UniProt entry Q02836]), an Ile or Val and at least one Pro are present, presumably forming a conserved structural element in CA—a turn between two helices. Despite the highly conserved nature of the HIV-1 PPIP(122–125) motif, a mutation of either P123 or P125 to Ala is relatively well tolerated in terms of virus assembly, although P125A exhibits a severe infectivity defect. In contrast, mutation of either P122 or I124 markedly impairs virus assembly; aberrant Gag lattices are also observed at sites of P122A and I124A assembly at the PM of virus-producing cells, indicating severe defects at early stages of assembly. The P122A and I124A mutants both produce virions with a disordered Gag lattice and a wider size distribution than the WT. Together, these data demonstrate that the PPIP(122–125) loop plays a critical role in the assembly of an ordered, immature Gag lattice.
Although our data demonstrate that mutation of the PPIP(122–125) motif blocks the formation of an ordered, immature Gag lattice, EM analysis of P122A and I124A virions revealed heterogeneous particles with diverse structural defects, i.e., particles with patches of Gag-like lattices and condensed or dispersed viral material and a complete absence of mature, conical cores. Several explanations could account for the failure of these mutants to complete conical core formation. (i) Although X-ray crystallography analysis did not reveal any major structural perturbations resulting from these mutations, we did observe conformational changes in several regions of CA, including the intraprotomer interface. In addition, EM analysis of in vitro CA assembly products revealed that both mutant proteins are able to form tubular structures, albeit with some differences in kinetics relative to the WT protein. (ii) P122A and I124A mutants exhibit a defect in Gag processing which results in reduced levels of mature CA in mutant virions relative to the WT. It is well established that incomplete Gag processing impairs conical core formation. Complete capsid cores consist of approximately 1,500 copies of mature CA, and similar numbers of CA subunits remain unassembled in WT virions (38). It is possible that the requisite number of CA subunits is not generated in P122A or I124A particles. (iii) P122A and I124A mutant virions contain an elevated level of the CA-SP1 processing intermediate relative to the WT virions. It has been demonstrated that the presence of Gag processing intermediates, including CA-SP1, exerts a dominant-negative effect on core condensation and that dysregulation of the sequential Gag processing pathway is detrimental to proper virion maturation (39–43). These three models are not mutually exclusive; the combination of conformational changes in CA, lower levels of mature CA, and higher levels of CA-SP1 in P122A and I124A virions could act in concert to prevent conical core formation.
HIV-1 MIs are thought to block CA-SP1 processing by binding within a six-helix bundle that forms at the CA-SP1 junction in the immature Gag lattice (22, 23). Despite the severe defects in immature lattice assembly induced by the P122A and I124A mutations, these CA-NTD substitutions do not prevent MIs from blocking CA-SP1 processing. This result implies that changes in the PPIP(122–125) motif do not prevent the formation of the six-helix bundle at the CA-SP1 junction and thus do not abrogate hexamer assembly. Interestingly, there does appear to be some cross talk between the PPIP(122–125) motif and SP1, as a mutation at residue 8 of SP1 (SP1-T8I [44, 45]) results in conformational changes in the PPIP(122–125) loop (46).
Sequential propagation of the P122A and I124A mutants in the MT-4 and Jurkat T-cell lines led to the emergence of escape mutants containing second-site changes within the CA-NTD. Most of the compensatory mutations were located at sites distal to the original substitutions and were distributed throughout the CA-NTD. Among the changes identified, only a combination of V11I and T58A mutations led to nearly complete recovery of virus particle production, virion infectivity, and multicycle replication. V11 is located in the unfolded flexible region that connects MA and CA in the immature Gag lattice, and V11 is a part of the β-hairpin that forms between Pro1 and D51 of CA in mature CA (47). In both immature and mature structures, T58 is located in the hydrophobic core formed by CA-NTD helices. V11I alone did not rescue the replication ability of either P122A or I124A. T58A improved replication capacity only for I124A virus, but the addition of V11I was required to fully rescue replication kinetics of both T58A/P122A and T58A/I124A mutants. Interestingly, a previous study demonstrated that T58I enhances CA-NC assembly in vitro (48).
In this study, we characterized the role of the PPIP(122–125) motif in the CA-NTD in HIV-1 Gag assembly and particle maturation. Our functional data, consistent with recent structural data on CA in the immature lattice (21), suggest that this exposed loop plays a key role in the formation of the interhexameric contacts required for assembly of the immature HIV-1 Gag lattice. Future studies will examine in more detail the mechanism by which second-site changes within the CA-NTD, distal from the location of the original mutations, are able to rescue particle assembly and infectivity.
MATERIALS AND METHODS
Cell lines.
HeLa, 293T, and HeLa-derived TZM-bl cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM), supplemented with 10% (vol/vol) fetal bovine serum (FBS), 2 mM l-glutamine (Gibco), 100 U/ml penicillin, and 100 μg/ml streptomycin, at 37°C and 5% CO2. The TZM-bl indicator cell line expresses luciferase upon infection by HIV (49). MT-4 and Jurkat CD4+ T-cell lines were cultured in RPMI 1640 medium, supplemented with 10% (vol/vol) FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine, at 37°C and 5% CO2.
Plasmids.
Full-length proviral clone pNL4-3 (50) was used for production of HIV-1 particles. pNL4-3 clones bearing CA mutations were generated either by site-directed mutagenesis using a pGEM-based subclone containing a nucleotide (nt) 588 to 2295 gag fragment or by direct amplification of gag regions bearing CA mutations from genomic DNA of infected cells followed by ligation of BssHII-ApaI or BssHII-SpeI fragments into pNL4-3. For collection of immature HIV-1 WT and PPIP mutant particles, molecular clones bearing the Asp25Asn mutation that inactivates PR were used (51). For expression and purification of CA proteins, P122A and I124A mutations were introduced into the pET11a HIV-1 CA Escherichia coli expression plasmid kindly provided by C. Tang (52).
Virus release assay.
HeLa cells were plated in a 12-well plate and transfected with viral clones using Lipofectamine 2000 according to the manufacturer’s instructions. At 24 h posttransfection, the medium was replaced with 0.5 ml of Met/Cys-free medium for 30 min and then metabolically labeled with a [35S]Met/Cys-Pro mix (PerkinElmer). After incubation for 3 h, viral particles were collected by filtering supernatants through 0.45-μm-pore-size membrane filters and were pelleted by ultracentrifugation. Virion-containing pellets were resuspended in lysis buffer. Cells were solubilized in 250 μl of lysis buffer, preheated with 15 μl of 2 × sample buffer (125 mM Tris-HCl [pH 6.8] containing 6% SDS, 10% 2-mercaptoethanol, and 20% glycerol) for 5 min at 95°C to disrupt Gag multimers and were immunoprecipitated with anti-HIV-1 IgG, a pooled patient serum obtained from the NIH AIDS Reagent Program. Cell and virus protein samples were separated on a 13% SDS-polyacrylamide gel, exposed to a phosphorimager cassette, and quantified by the use of Quantity One software (Bio-Rad). Relative virus release efficiency values were calculated as the amount of CA in virions divided by the sum of viral CA, cellular Gag, and cellular CA; the numbers were subsequently normalized to WT sample values. For calculation of standard deviations and standard errors of the means, at least three independent experiments were performed.
Analysis of Gag processing products in virions.
To analyze Pr55Gag levels, 293T cells were transfected with the indicated viral clones using Lipofectamine 2000 (Invitrogen) following the manufacturer’s protocol. At 48 h posttransfection, 3.5 ml of the supernatants was filtered through 0.45-μm-pore-size membrane filters, pelleted, and resuspended in lysis buffer (50 mM Tris-HCl [pH 7.5], 300 mM NaCl, 0.5% Triton X-100, 10 mM iodoacetamide, and protease inhibitor cocktail tablets [Roche]). The samples were subjected to 12% SDS-PAGE and then transferred to a polyvinylidene fluoride (PVDF) membrane (Immobilon, Millipore) by wet electroblotting. The membrane was blocked for 30 min with 5% nonfat milk–Tris-buffered saline–0.05% Tween 20 detergent and incubated overnight at 4°C with anti-HIV-1 IgG. The membrane was washed, incubated for 45 min with anti-human horseradish peroxidase-conjugated secondary antibody, and washed again. Supersignal West Pico reagent (Thermo Scientific) was used to reveal protein bands followed by analysis performed with ImageLab software (Bio-Rad). To analyze CA-SP1 levels in virions produced in the absence or presence of MIs, CA-SP1 accumulation assays were performed with [35S]Met/Cys-containing media as described previously using 293T cells as virus producer cells (34, 53). To obtain a clear separation between CA and CA-SP1, samples were resolved on a 15% SDS-polyacrylamide gel. The effect of BVM, 7m, or 7r was tested at 100 nM.
Virus replication kinetics.
For transfection, 2.5 × 106 MT-4 cells and Jurkat cells were mixed with 1 μg and 2.5 μg plasmid DNA, respectively, in 500 μl of 0.8 mg/ml DEAE-dextran solution. After 15 min of incubation at 37°C, cells were washed with 4 ml of 1× STBS (25 mM Tris-HCl [pH 7.4], 0.6 mM Na2HPO4, 5 mM KCl, 140 mM NaCl, 0.7 mM CaCl2, 0.5 mM MgCl2) and pelleted by centrifugation. The cell pellets were resuspended in 2.5 ml RPMI medium and transferred to tissue culture flasks. Aliquots of supernatants were collected to monitor RT activity, and cells were split every other day. For reinfection, 1.5 × 106 MT-4 cells or 2.5 × 106 Jurkat cells were mixed with RT-normalized viral stocks, incubated for 2 h at 37°C, and pelleted by centrifugation. Cell pellets were resuspended in 2.5 ml RPMI medium and transferred to tissue culture flasks. On the days of peak RT activity, the genomic DNA was extracted using a QIAamp genomic DNA extraction kit (Qiagen), and CA-coding regions were amplified and sequenced (Macrogen).
Single-cycle infectivity assay.
TZM-bl cells (2 × 104) were plated into each well of a 96-well plate. After 24 h, serial dilutions of viral stocks collected from HeLa cells were added to the cells in the presence of 10 μg/ml DEAE-dextran, and the medium was changed after incubation for 2 h to 4 h. At 48 h postinfection, the culture medium was removed and infected cells were washed once in 1× PBS and lysed in 30 μl of britelite plus reagent (Perkin-Elmer). Detected luminescent signals were normalized to the RT activity of viral samples measured as described previously (54). WT values were set at 100%. At least three independent experiments were performed.
Cryo-ET.
WT and I124A virions and PR− derivatives of WT, P122A, I124A, and T58S/T107I/P122A virions were imaged by cryo-ET as described previously (55). Briefly, purified fixed virus was mixed (2:1) with a suspension of colloidal gold particles (Electron Microscopy Sciences), applied to Quantifoil R2/2 200 mesh holey carbon grids (Structure Probe, Inc.), and plunge-frozen in ethane using a Leica EM GP (Leica Microsystems). For data acquisition, grids were transferred to a cryo-holder (type 626 or 914; Gatan), and single-axis tilt series were recorded at 120 keV on a Tecnai-12 electron microscope through a zero-loss energy filter (GIF 2002; Gatan) with a slit width of 20 eV. Images were acquired on a 2,048-by-2,048-pixel charge-coupled-device (CCD) camera (Gatan) using SerialEM (56). Tilt-series projections were typically acquired at 1.5° or 2° increments from −60° to +60° and at ×38,500 magnification (7.8 Å/pixel) and −3.5-μm defocus. The electron dose per projection was ∼1.2 e−/Å2, giving a total cumulative dose of ∼75 e−/Å2. Tilt series were reconstructed using the Bsoft package (57), and virions were extracted and denoised by 20 iterations of anisotropic nonlinear diffusion (58).
Morphological analysis.
Size and Gag layer coverage were evaluated for each PR− particle in the cryo-ET data set. The central section was located for each particle. The diameter was determined at the longest axis of each central section using the “measure” tool in Bshow. To assess Gag layer coverage for a given particle, the angular measure for each visible Gag layer arc in the central slice was determined manually. The angular measures of all Gag layer arcs were summed, and this sum was divided by 360° to yield the overall Gag layer coverage for that particle.
Subtomogram averaging.
Subtomogram averaging was performed as previously described (45). Briefly, subtomograms containing patches of the Gag layer were manually selected in the denoised tomograms and extracted from the corresponding raw tomograms. Initial orientations of the subtomograms were defined by vectors from the virion centers directed radially outward. The average of all selected subtomograms was used as an initial reference for alignment. Subtomogram alignment, taking into account the missing wedge of information, was performed using routines from Bsoft (57) wrapped into Python scripts. Initial alignment steps involved 2 iterations of only translational alignment. As a result of this process, subtomograms were not rotationally aligned, and therefore the Gag-related lattices were not in register. For the next steps, the viral membrane and MA layers were masked off. One subtomogram was selected, C6 symmetry was applied, and this subtomogram was used as a reference for a full rotational and translational alignment. This alignment procedure was iterated five times, using as a reference the average of the top 50% of the particles (as ranked by correlation coefficients) from the previous iteration. Classification and averaging procedures were then performed by maximum likelihood analysis as implemented in the Xmipp package (59). Approximately, the top 75% of the initially selected subtomograms were used to calculate the final averages.
Crystallization of CA mutants, data collection, and structure determination.
P122A and I124A CA were cloned without fusion tags in the pET11a plasmid. Mutant proteins were expressed and purified as previously described (26). Crystals of the P122A and I124A CA grew at 18°C in hanging drops containing 5 mg/ml of protein, 6% to 9% polyethylene glycol (PEG) 3350, 2% to 6% glycerol, sodium iodide, and sodium cacodylate. Hexagonal plate-like crystals appeared after 5 days, and crystal growth was completed over 2 weeks. Crystals were briefly soaked in 20% glycerol or paraffin oil before they were subjected to cryo-cooling in liquid nitrogen. Data were collected on a complementary metal-oxide semiconductor (CMOS) detector at Advanced Light Source (ALS) beamline 4.2.2 (Lawrence Berkeley National Laboratory) or a Dectris Eiger-16M detector at Advanced Photon Source (APS) beamline 23 ID-B (Argonne National Laboratory). Datasets were collected and processed using XDS (60). The data were examined for the presence of systematic absences; however, no characteristic patterns were observed. Thus, the crystals were indexed in hexagonal space group P6 with one CA molecule in the asymmetric unit. No twinning was present, as determined by either POINTLESS (61) or XTRIAGE (62). Space group and twinning data were also verified in ZANUDA (63). The CA phase problem was solved using molecular replacement, with the native CA (PDB identifier [ID]: 4XFX) as a starting model. Initial phases were solved via the use of PHASER (63). Several rounds of iterative model building and refinement were carried out using Coot (64) and PHENIX (62), REFMAC (65), or PDBREDO (http://www.cmbi.ru.nl/pdb_redo/). Structure validation of final models was performed with MOLPROBITY (http://molprobity.biochem.duke.edu/). Accessible and buried surface area data were calculated using PISA (http://www.ccp4.ac.uk/pisa/). The figures showing structural information were generated in PyMOL (http://www.pymol.org/).
In vitro assembly of HIV-1 CA proteins.
In vitro assembly reactions were performed by using methods developed in other studies (35, 47, 66) with some modifications. Briefly, CA protein, purified as described previously (26), was incubated at a concentration of 6 mg/ml in 50 mM Tris-HCl (pH 7.5 to 8)–1 M NaCl for 15 to 30 min at room temperature and then overnight at 4°C. For negatively stained EM images, aliqouts of in vitro assembly reactions were placed on Formvar carbon-coated grids, incubated for 30 s, and then rinsed with water three times and stained with 2% uranyl formate for 30 s. Excess material was removed from the grids with Whatman filter paper. The images were collected using a Hitachi 7600 transmission electron microscope.
Structural modeling.
Models of the CA monomer, immature Gag lattice, and mature core were generated using PyMOL software.
Accession numbers.
ACKNOWLEDGMENTS
We thank members of the Freed laboratory for helpful discussion and critical review of the manuscript.
This work was supported by the Intramural Research Programs of the Center for Cancer Research, National Cancer Institute, NIH (E.O.F.), and the National Institute of Arthritis and Musculoskeletal and Skin Diseases, NIH (A.C.S.); by the Intramural AIDS Targeted Antiviral Program (E.O.F. and A.C.S.), National Institute of Allergy and Infectious Diseases (R01-AI120860, R21-AI112417, and U54 GM103368), NIH (S.G.S.); and by the University of Leeds (University Academic Fellow scheme; J.F.). This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN26120080001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Citation Novikova M, Adams LJ, Fontana J, Gres AT, Balasubramaniam M, Winkler DC, Kudchodkar SB, Soheilian F, Sarafianos SG, Steven AC, Freed EO. 2018. Identification of a structural element in HIV-1 Gag required for virus particle assembly and maturation. mBio 9:e01567-18. https://doi.org/10.1128/mBio.01567-18.
REFERENCES
- 1.Freed EO. 2015. HIV-1 assembly, release and maturation. Nat Rev Microbiol 13:484–496. doi: 10.1038/nrmicro3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sundquist WI, Kräusslich HG. 2012. HIV-1 assembly, budding, and maturation. Cold Spring Harb Perspect Med 2:a006924. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lingappa JR, Reed JC, Tanaka M, Chutiraka K, Robinson BA. 2014. How HIV-1 Gag assembles in cells: putting together pieces of the puzzle. Virus Res 193:89–107. doi: 10.1016/j.virusres.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freed EO, Orenstein JM, Buckler-White AJ, Martin MA. 1994. Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. J Virol 68:5311–5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saad JS, Miller J, Tai J, Kim A, Ghanam RH, Summers MF. 2006. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc Natl Acad Sci U S A 103:11364–11369. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Votteler J, Sundquist WI. 2013. Virus budding and the ESCRT pathway. Cell Host Microbe 14:232–241. doi: 10.1016/j.chom.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lippincott-Schwartz J, Freed EO, van Engelenburg SB. 2017. A consensus view of ESCRT-mediated human immunodeficiency virus type 1 abscission. Annu Rev Virol 4:309–325. doi: 10.1146/annurev-virology-101416-041840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wright ER, Schooler JB, Ding HJ, Kieffer C, Fillmore C, Sundquist WI, Jensen GJ. 2007. Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J 26:2218–2226. doi: 10.1038/sj.emboj.7601664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pornillos O, Ganser-Pornillos BK, Yeager M. 2011. Atomic-level modelling of the HIV capsid. Nature 469:424–427. doi: 10.1038/nature09640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Briggs JA, Grünewald K, Glass B, Forster F, Krausslich HG, Fuller SD. 2006. The mechanism of HIV-1 core assembly: insights from three-dimensional reconstructions of authentic virions. Structure 14:15–20. doi: 10.1016/j.str.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 11.Keller PW, Huang RK, England MR, Waki K, Cheng N, Heymann JB, Craven RC, Freed EO, Steven AC. 2013. A two-pronged structural analysis of retroviral maturation indicates that core formation proceeds by a disassembly-reassembly pathway rather than a displacive transition. J Virol 87:13655–13664. doi: 10.1128/JVI.01408-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frank GA, Narayan K, Bess JW Jr, Del Prete GQ, Wu X, Moran A, Hartnell LM, Earl LA, Lifson JD, Subramaniam S. 2015. Maturation of the HIV-1 core by a non-diffusional phase transition. Nat Commun 6:5854. doi: 10.1038/ncomms6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meng X, Zhao G, Yufenyuy E, Ke D, Ning J, Delucia M, Ahn J, Gronenborn AM, Aiken C, Zhang P. 2012. Protease cleavage leads to formation of mature trimer interface in HIV-1 capsid. PLoS Pathog 8:e1002886. doi: 10.1371/journal.ppat.1002886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ning J, Erdemci-Tandogan G, Yufenyuy EL, Wagner J, Himes BA, Zhao G, Aiken C, Zandi R, Zhang P. 2016. In vitro protease cleavage and computer simulations reveal the HIV-1 capsid maturation pathway. Nat Commun 7:13689. doi: 10.1038/ncomms13689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gamble TR, Yoo S, Vajdos FF, von Schwedler UK, Worthylake DK, Wang H, McCutcheon JP, Sundquist WI, Hill CP. 1997. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science 278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 16.Rihn SJ, Wilson SJ, Loman NJ, Alim M, Bakker SE, Bhella D, Gifford RJ, Rixon FJ, Bieniasz PD. 2013. Extreme genetic fragility of the HIV-1 capsid. PLoS Pathog 9:e1003461. doi: 10.1371/journal.ppat.1003461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.von Schwedler UK, Stray KM, Garrus JE, Sundquist WI. 2003. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J Virol 77:5439–5450. doi: 10.1128/JVI.77.9.5439-5450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borsetti A, Ohagen A, Gottlinger HG. 1998. The C-terminal half of the human immunodeficiency virus type 1 Gag precursor is sufficient for efficient particle assembly. J Virol 72:9313–9317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ganser-Pornillos BK, Yeager M, Sundquist WI. 2008. The structural biology of HIV assembly. Curr Opin Struct Biol 18:203–217. doi: 10.1016/j.sbi.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mattei S, Schur FK, Briggs JA. 2016. Retrovirus maturation — an extraordinary structural transformation. Curr Opin Virol 18:27–35. doi: 10.1016/j.coviro.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 21.Schur FK, Hagen WJ, Rumlová M, Ruml T, Müller B, Kräusslich H-G, Briggs JA. 2015. Structure of the immature HIV-1 capsid in intact virus particles at 8.8 Å resolution. Nature 517:505–508. doi: 10.1038/nature13838. [DOI] [PubMed] [Google Scholar]
- 22.Schur FK, Obr M, Hagen WJ, Wan W, Jakobi AJ, Kirkpatrick JM, Sachse C, Kräusslich H-G, Briggs JA. 2016. An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation. Science 353:506–508. doi: 10.1126/science.aaf9620. [DOI] [PubMed] [Google Scholar]
- 23.Wagner JM, Zadrozny KK, Chrustowicz J, Purdy MD, Yeager M, Ganser-Pornillos BK, Pornillos O. 2016. Crystal structure of an HIV assembly and maturation switch. Elife 5:e17063. doi: 10.7554/eLife.17063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pornillos O, Ganser-Pornillos BK, Kelly BN, Hua Y, Whitby FG, Stout CD, Sundquist WI, Hill CP, Yeager M. 2009. X-ray structures of the hexameric building block of the HIV capsid. Cell 137:1282–1292. doi: 10.1016/j.cell.2009.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao G, Perilla JR, Yufenyuy EL, Meng X, Chen B, Ning J, Ahn J, Gronenborn AM, Schulten K, Aiken C, Zhang P. 2013. Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature 497:643–646. doi: 10.1038/nature12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gres AT, Kirby KA, Kewalramani VN, Tanner JJ, Pornillos O, Sarafianos SG. 2015. X-ray structures of native HIV-1 capsid protein reveal conformational variability. Science 349:99–103. doi: 10.1126/science.aaa5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mattei S, Glass B, Hagen WJ, Kräusslich H-G, Briggs JA. 2016. The structure and flexibility of conical HIV-1 capsids determined within intact virions. Science 354:1434–1437. doi: 10.1126/science.aah4972. [DOI] [PubMed] [Google Scholar]
- 28.Hatziioannou T, Cowan S, von Schwedler UK, Sundquist WI, Bieniasz PD. 2004. Species-specific tropism determinants in the human immunodeficiency virus type 1 capsid. J Virol 78:6005–6012. doi: 10.1128/JVI.78.11.6005-6012.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maillard PV, Zoete V, Michielin O, Trono D. 2011. Homology-based identification of capsid determinants that protect HIV1 from human TRIM5a restriction. J Biol Chem 286:8128–8140. doi: 10.1074/jbc.M110.187609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rahm N, Gfeller D, Snoeck J, Martinez R, Mclaren PJ, Ortiz M, Ciuffi A, Telenti A. 2013. Susceptibility and adaptation to human TRIM5α alleles at positive selected sites in HIV-1 capsid. Virology 441:162–170. doi: 10.1016/j.virol.2013.03.021. [DOI] [PubMed] [Google Scholar]
- 31.Keller PW, Adamson CS, Heymann JB, Freed EO, Steven AC. 2011. HIV-1 maturation inhibitor bevirimat stabilizes the immature Gag lattice. J Virol 85:1420–1428. doi: 10.1128/JVI.01926-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li F, Goila-Gaur R, Salzwedel K, Kilgore NR, Reddick M, Matallana C, Castillo A, Zoumplis D, Martin DE, Orenstein JM, Allaway GP, Freed EO, Wild CT. 2003. PA-457: a potent HIV inhibitor that disrupts core condensation by targeting a late step in Gag processing. Proc Natl Acad Sci U S A 100:13555–13560. doi: 10.1073/pnas.2234683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou J, Yuan X, Dismuke D, Forshey BM, Lundquist C, Lee K-H, Aiken C, Chen CH. 2004. Small-molecule inhibition of human immunodeficiency virus type 1 replication by specific targeting of the final step of virion maturation. J Virol 78:922–929. doi: 10.1128/JVI.78.2.922-929.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Urano E, Ablan SD, Mandt R, Pauly GT, Sigano DM, Schneider JP, Martin DE, Nitz TJ, Wild CT, Freed EO. 2016. Alkyl amine bevirimat derivatives are potent and broadly active HIV-1 maturation inhibitors. Antimicrob Agents Chemother 60:190–197. doi: 10.1128/AAC.02121-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gross I, Hohenberg H, Kräusslich HG. 1997. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur J Biochem 249:592–600. doi: 10.1111/j.1432-1033.1997.t01-1-00592.x. [DOI] [PubMed] [Google Scholar]
- 36.Li S, Hill CP, Sundquist WI, Finch JT. 2000. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 407:409–413. doi: 10.1038/35030177. [DOI] [PubMed] [Google Scholar]
- 37.Davey NE, Satagopam VP, Santiago-Mozos S, Villacorta-Martin C, Bharat TA, Schneider R, Briggs JA. 2014. The HIV mutation browser: a resource for human immunodeficiency virus mutagenesis and polymorphism data. PLoS Comput Biol 10:e1003951. doi: 10.1371/journal.pcbi.1003951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. 1999. Assembly and analysis of conical models for the HIV-1 core. Science 283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 39.Checkley MA, Luttge BG, Soheilian F, Nagashima K, Freed EO. 2010. The capsid-spacer peptide 1 Gag processing intermediate is a dominant-negative inhibitor of HIV-1 maturation. Virology 400:137–144. doi: 10.1016/j.virol.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee SK, Harris J, Swanstrom R. 2009. A strongly transdominant mutation in the human immunodeficiency virus type 1 gag gene defines an Achilles heel in the virus life cycle. J Virol 83:8536–8543. doi: 10.1128/JVI.00317-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Müller B, Anders M, Akiyama H, Welsch S, Glass B, Nikovics K, Clavel F, Tervo H-M, Keppler OT, Kräusslich H-G. 2009. HIV-1 Gag processing intermediates trans-dominantly interfere with HIV-1 infectivity. J Biol Chem 284:29692–29703. doi: 10.1074/jbc.M109.027144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaplan AH, Zack JA, Knigge M, Paul DA, Kempf DJ, Norbeck DW, Swanstrom R. 1993. Partial inhibition of the human immunodeficiency virus type 1 protease results in aberrant virus assembly and the formation of noninfectious particles. J Virol 67:4050–4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moore MD, Fu W, Soheilian F, Nagashima K, Ptak RG, Pathak VK, Hu W-S. 2008. Suboptimal inhibition of protease activity in human immunodeficiency virus type 1: effects on virion morphogenesis and RNA maturation. Virology 379:152–160. doi: 10.1016/j.virol.2008.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waki K, Durell SR, Soheilian F, Nagashima K, Butler SL, Freed EO, Krausslich H-G. 2012. Structural and functional insights into the HIV-1 maturation inhibitor binding pocket. PLoS Pathog 8:e1002997. doi: 10.1371/journal.ppat.1002997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fontana J, Keller PW, Urano E, Ablan SD, Steven AC, Freed EO. 2016. Identification of an HIV-1 mutation in spacer peptide 1 that stabilizes the immature CA-SP1 lattice. J Virol 90:972–978. doi: 10.1128/JVI.02204-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang M, Quinn CM, Perilla JR, Zhang H, Shirra R, Hou G, Byeon I-J, Suiter CL, Ablan S, Urano E, Nitz TJ, Aiken C, Freed EO, Zhang P, Schulten K, Gronenborn AM, Polenova T. 2017. Quenching protein dynamics interferes with HIV capsid maturation. Nat Commun 8:1779. doi: 10.1038/s41467-017-01856-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.von Schwedler UK, Stemmler TL, Klishko VY, Li S, Albertine KH, Davis DR, Sundquist WI. 1998. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J 17:1555–1568. doi: 10.1093/emboj/17.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lemke CT, Titolo S, von Schwedler UK, Goudreau N, Mercier J-F, Wardrop E, Faucher A-M, Coulombe R, Banik SS, Fader L, Gagnon A, Kawai SH, Rancourt J, Tremblay M, Yoakim C, Simoneau B, Archambault J, Sundquist WI, Mason SW. 2012. Distinct effects of two HIV-1 capsid assembly inhibitor families that bind the same site within the N-terminal domain of the viral CA protein. J Virol 86:6643–6655. doi: 10.1128/JVI.00493-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol 72:2855–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol 59:284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang M, Orenstein JM, Martin MA, Freed EO. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol 69:6810–6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang C, Loeliger E, Kinde I, Kyere S, Mayo K, Barklis E, Sun Y, Huang M, Summers MF. 2003. Antiviral inhibition of the HIV-1 capsid protein. J Mol Biol 327:1013–1020. doi: 10.1016/S0022-2836(03)00289-4. [DOI] [PubMed] [Google Scholar]
- 53.Adamson CS, Ablan SD, Boeras I, Goila-Gaur R, Soheilian F, Nagashima K, Li F, Salzwedel K, Sakalian M, Wild CT, Freed EO. 2006. In vitro resistance to the human immunodeficiency virus type 1 maturation inhibitor PA-457 (bevirimat). J Virol 80:10957–10971. doi: 10.1128/JVI.01369-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Willey RL, Smith DH, Lasky LA, Theodore TS, Earl PL, Moss B, Capon DJ, Martin MA. 1988. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J Virol 62:139–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fontana J, Cardone G, Heymann JB, Winkler DC, Steven AC. 2012. Structural changes in influenza virus at low pH characterized by cryo-electron tomography. J Virol 86:2919–2929. doi: 10.1128/JVI.06698-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mastronarde DN. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 57.Heymann JB, Cardone G, Winkler DC, Steven AC. 2008. Computational resources for cryo-electron tomography in Bsoft. J Struct Biol 161:232–242. doi: 10.1016/j.jsb.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Frangakis AS, Hegerl R. 2001. Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J Struct Biol 135:239–250. doi: 10.1006/jsbi.2001.4406. [DOI] [PubMed] [Google Scholar]
- 59.Scheres SH, Melero R, Valle M, Carazo JM. 2009. Averaging of electron subtomograms and random conical tilt reconstructions through likelihood optimization. Structure 17:1563–1572. doi: 10.1016/j.str.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kabsch W. 2010. XDS. Acta Crystallogr D Biol Crystallogr 66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Evans P. 2006. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr 62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 62.Adams PD, Grosse-Kunstleve RW, Hung L-W, Ioerger TR, Mccoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58:1948–1954. doi: 10.1107/S0907444902016657. [DOI] [PubMed] [Google Scholar]
- 63.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 65.Murshudov GN, Skubák P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. 2011. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.López CS, Tsagli SM, Sloan R, Eccles J, Barklis E. 2013. Second site reversion of a mutation near the amino terminus of the HIV-1 capsid protein. Virology 447:95–103. doi: 10.1016/j.virol.2013.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary of data collection and refinement statistics. Download Table S1, PDF file, 0.1 MB (90.5KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.