Abstract
Studying HIV-1 replication in the presence of functionally related proteins from different species has helped define host determinants of HIV-1 infection. Humans and owl monkeys, but not macaques, encode a CD4 receptor that permits entry of transmissible HIV-1 variants due to a single residue difference. However, little is known about whether divergent CCR5 receptor proteins act as determinants of host-range. Here we show that both owl monkey (Aotus vociferans) CD4 and CCR5 receptors are functional for the entry of transmitted HIV-1 when paired with human versions of the other receptor. By contrast, the owl monkey CD4/CCR5 pair is generally a suboptimal receptor combination, although there is virus-specific variation in infection with owl monkey receptors. Introduction of the human residues 15Y and 16T within a sulfation motif into owl monkey CCR5 resulted in a gain of function. These findings suggest there is cross-talk between CD4 and CCR5 involving the sulfation motif.
Keywords: HIV-1, receptor, entry, CD4, CCR5, owl monkey, species differences
Introduction
HIV-1 utilizes the CD4 receptor to gain entry into host cells. A second cell surface receptor that functions as a coreceptor is also required, and for early-stage HIV-1 variants this coreceptor is CCR5 (1). In later stages of infection, some HIV-1 variants can use CXCR4 as an alternate coreceptor (1). The HIV-1 surface glycoprotein, Envelope (Env), undergoes a conformational change upon engagement of the CD4 receptor; this new conformational state then allows Env to engage CCR5 (2). It is known that the extracellular N-terminus of both CD4 and CCR5 mediate interactions with HIV-1 Envelope. Changes in the amino acid sequences in either CD4 and/or CCR5 are known to modulate the ability of HIV-1 to enter target cells. For instance, sulfated tyrosines in the N-terminus of CCR5 at amino acid positions 3, 10, 14, and 15 have been shown to be most important for the interaction between Env gp120 and CCR5 (3, 4, 5, 6, 7, 8). It is commonly thought that CD4 and CCR5 do not interact with one another (9), although there is some data that suggests that they do (10, 11, 12, 13).
Nonhuman primates are integrally important to the study of HIV-1. Not only did HIV-1 emerge from nonhuman primate reservoirs, but nonhuman primates are also used as infection models in which to study HIV-1 pathogenesis. The CD4 and CCR5 molecules encoded by different primate species differ in sequence, occasionally affecting HIV-1 entry. For instance, we have previously shown that the macaque CD4 protein is a much less efficient receptor for HIV-1 than the human CD4 protein due to a single amino difference at position 39 in its first extracellular, D1 domain (14, 15). We found this to be particularly true for the variants of HIV-1 that are known to seed new infections in humans (transmissible variants). In addition, we have recently reported that several alleles of CD4 are circulating in a population of captive owl monkeys (Aotus vociferans) (16). Some of these alleles encode an asparagine (N) at position 39 in the D1 domain of the CD4 protein, while others encode an isoleucine (I). Owl monkey CD4-39N alleles are functional as receptors for transmitted HIV-1, while CD4-39I alleles are not. Less is known about the function of the HIV-1 coreceptors in nonhuman primate species.
Given that owl monkeys encode a CD4 that is functional as a receptor for HIV-1, at least when paired with human CCR5, this raises the question of whether the owl monkey CCR5 is a functional coreceptor for HIV-1. Here, we examined the function of owl monkey CCR5 with regards to HIV-1 entry. We found that this owl monkey CCR5 functions for HIV-1 entry when paired with human, but is suboptimal for HIV-1 entry when paired with owl monkey CD4. No transmissible variant of HIV-1 was able to use owl monkey CD4/CCR5 as efficiently as the corresponding human receptor pair. Once mutations were introduced into owl monkey CCR5 at positions 15 and 16 (the sulfation site), the owl monkey receptor pair functioned for HIV-1 entry as efficiently as the human receptor pair. However, viral Envelope-specific differences were observed in receptor function.
Results
Owl monkey CCR5 is a functional coreceptor for HIV-1 in some contexts
We first cloned the CCR5 from Spix’s owl monkey (Aotus vociferans; herein referred to as “owl monkeys”) (GenBank accession number MF632281). Cell lines were constructed to express CD4/CCR5 combinations of interest. Three CD4 receptors were explored: human (RefSeq number MN_000616), which functions as a receptor for HIV-1 and served as a positive control, rhesus macaque (GenBank accession number MF632287), which is not a functional receptor for transmitted and early stage HIV-1 (14) and the owl monkey allele of CD4 that encodes an N at position 39 and permits HIV-1 entry (14, 16) (GenBank accession number KR902344.3). These were paired with CCR5 from either human (AIDS reagent catalog number 3331) or owl monkey, in various combinations indicated in figure 1. Expression of CD4 and CCR5 was verified to be generally similar between each cell line stably expressing the CD4 and CCR5 receptors (Figure 1A).
Figure 1.
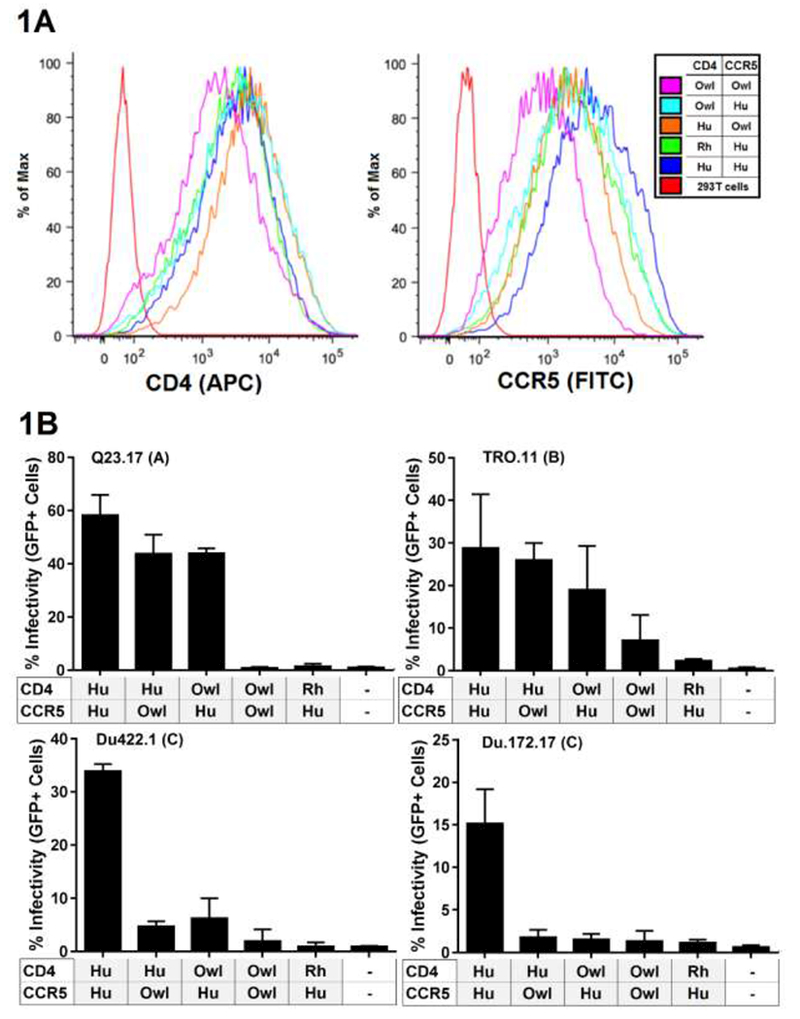
Characterization and infection of 293T cells stably expressing various CD4 and CCR5 combinations. (A) Flow cytometry fluorescence intensity plots of CD4 and CCR5 expression (B) Infection of cells from panel A were infected with virus encoding diverse Env variants, which are indicated above each bar graph, along with their subtype designation in parenthesis. Error bars represent the standard deviations between replicate wells from 2 separate experiments. indicates 293T cells that do not express CD4 or CCR5 and were infected as a negative control. Hu=Human, Rh=Rhesus macaque, Owl= owl monkey.
We infected each of these cell lines with single-cycle HIV-1 pseudotyped with four different Envs representative of transmitted/early stage HIV-1 from subtypes A, B and C (Table 1). All of the tested viruses infected cells expressing human CD4/CCR5 (positive control), and all of the viruses showed limited infection of cells expressing rhesus CD4/human CCR5 (negative control) (Figure 1B). None of the viruses infected cells expressing the owl monkey CD4/CCR5 at levels similar to those of the corresponding human receptor/coreceptor pair (Figure 1B). One Env clone, TRO.11, did mediate entry through the owl monkey receptors, but only at a level that was 25% relative to the level of entry into cells bearing human CD4/CCR5.
Table 1 –
Description of Env clones used in this study. Q23.ENV.17 (27), BG505.W6M.B1 (21), TRO.11 (28), Du172.17 and Du422.14 (20).
| Env clone | Subtype | Mode of transmission* | Source † | GenBank accession no. | Time postinfection (wks) |
|---|---|---|---|---|---|
| Q23.ENV.17 | A | M-F | PBMC | AF004885 | 11 |
| BG505.W6M.B1 | A | M-C | PBMC | DQ208457 | 6 |
| TRO.11 | B | M-M | ccPBMC | AY835445 | 4 |
| Du172.17 | C | M-F | ccPBMC | DQ411853 | 12 |
| Du422.14 | C | M-F | ccPBMC | DQ411854 | 8 |
M-F, male to female. M-M, male to male. M-C, mother to child.
PBMC - Env was cloned from uncultured PBMCs isolated directly from patient
ccPBMC - patient PBMCs (or virus from these PBMCs) underwent short-term coculture with PBMCs from HIV-1-negative donors in order to amplify virus before cloning.
Because the owl monkey CD4 allele used here was previously found to be functional for HIV-1 entry when paired with human CCR5 (16), our finding initially suggested that owl monkey CCR5 is not a functional coreceptor for HIV-1. However, columns 2 and 3 in each graph in Figure 1B reveal something unexpected. When either CD4 or CCR5 is derived from human, and the other from owl monkey, this receptor pair can function for the entry of HIV-1 and the levels of infection approach those of the paired human receptors (e.g. Q23-17 virus infection). Further, this phenomenon seems to be HIV-1 Envelope specific, as it is observed in the case of Envs Q23.17 and TRO.11 (top row in Figure 1B), but not in the case of Du422.1 and Du172.17 (bottom row in Figure 1B). Therefore, we conclude that both owl monkey CD4 and CCR5 are functional receptors for HIV-1, but only in certain contexts. This depends on the sequence of the other receptor with which each is paired, and the Env on the virus surface.
Mutations at owl monkey CCR5 positions 15 and 16 are sufficient to confer function as a coreceptor for viral entry.
We have observed that in certain contexts human CCR5 is a functional receptor when paired with owl monkey CD4, but that owl monkey CCR5 is not. To determine the basis for differences in receptor function of owl monkey versus human CCR5, the amino acid sequences of the two sequences were compared (Figure. 2A). Human and owl monkey CCR5 sequences are highly conserved (91.76% identity). Of note, there are 2 residue differences at positions 15 and 16 which have been implicated in human CCR5 coreceptor function (18); Hu CCR5 encodes Y15 and T16 while owl monkey CCR5 encodes D15 and A16 (Fig 2). Position 15 is of particular interest because it encodes a tyrosine that is thought to be sulfated, a modification that aids viral entry (3, 4).
Figure 2.
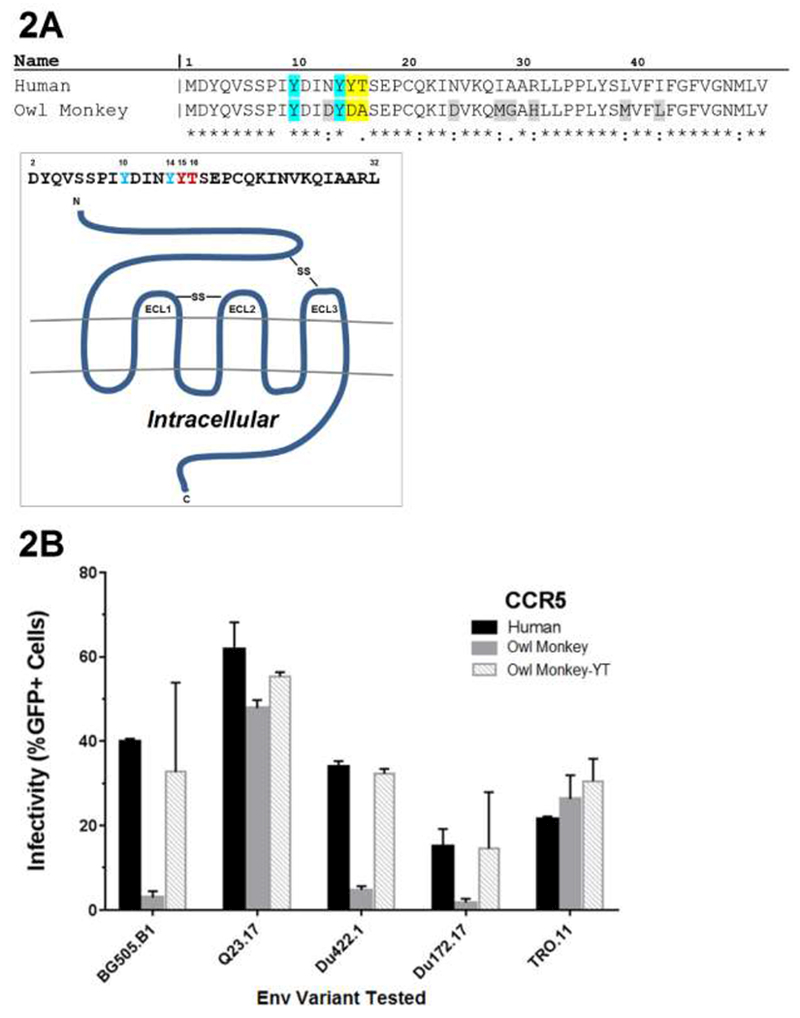
Amino acid sequences of a portion of CCR5. (A) Comparison of the human and owl monkey CCR5s with differences highlighted in grey and yellow. Sulfated tyrosines positions 10 and 14 shown to be important for gp120-CCR5 interaction are highlighted in blue. Figure adapted from Huang et al. 2007. Schematic representation of human CCR5. Sequence corresponds to the extracellular N-terminus of CCR5 (17). Residues of interest are depicted in red. Extracellular loops and disulfide linkages (-SS-) are labeled. (B) Effect of amino acids Y15 and T16 on the ability of owl monkey CCR5 to function with human CD4 as a coreceptor for different HIV-1 variants. 293T cells stably expressing the permissive human CD4 receptor and indicated CCR5 coreceptor were infected with pseudovirus with the Envs indicated on the x-axis. Infection was measured by flow cytometry as percent GFP-positive cells 48hrs postinfection. Error bars represent the standard deviations between replicate wells from 2 separate experiments.
To test whether these residues determine HIV-1 coreceptor function, we generated an owl monkey CCR5 protein encoding the human CCR5 residues at these two positions (Y15 and T16, yielding “CCR5-YT”) and tested it in infectivity assays. In this experiment, all cells expressed human CD4, which was paired with either human CCR5, owl monkey CCR5, or owl monkey CCR5-YT. There was little effect of the Y15 and T16 change on viruses that already showed high level infection with the human CD4 and owl monkey CCR5 receptor pair (Q23-17 and TRO.11). However, infections with pseudovirus bearing Env from BG505.W6M.B1, Du422.1 and Du172.17, which showed limited infection in cells expressing the combination of human CD4 and owl monkey CCR5, showed higher infection of cells expressing the owl monkey CCR5-YT protein compared to the wild type owl monkey CCR5 (11.0 fold for BG505.W6M.B1,6.8 fold for Du422.1, and 8.5 fold for Du172.17, Figure. 2B). These data suggest the two amino acids found at position 15 and 16 in human CCR5 are sufficient to confer HIV-1 coreceptor function to the owl monkey CCR5 when paired with human CD4.
Mutations at position 15 and 16 in owl monkey CCR5 make owl monkey CD4/CCR5 a functional receptor pair
The flow cytometry assay to measure infection using a GFP reporter virus is potentially subject to saturation at higher MOIs since it measures the number of infected cells, thus we developed an assay that measures total infection, using a secreted luciferase reporter. We used these cells to further examine the ability of the wildtype and the Y15 and T16 CCR5 coreceptors to permit entry when paired with human and owl monkey CD4s. We found that in these assays BG505.B1 infections of cell lines that expressed human CD4 paired with various CCR5s was lowest in the cell line expressing human CD4 and the wildtype owl monkey CCR5 (Figure. 3). This is in contrast to the cell line that expressed both human receptors or the cell line that expressed human CD4 paired with the mutated owl monkey CCR5 (Figure. 2A, Figure. 3) where there were relatively higher levels of infection. This experiment also clearly shows that the TRO.11 isolate can infect cells expressing owl monkey CD4/CCR5. In this case, the levels of infection were 2%-5% of the human receptor combination across the MOIs tested. This assay, which has a relatively large dynamic range, also suggests there is low-level infection of cells expressing owl monkey CCR5 by viruses pseudotyped with BG505.B1, Q23-17 and Du422.1 Envs (Figure. 2B). For these viruses, infection of cells expressing the owl monkey receptor pairs was higher than in control cells that were not expressing exogenous receptors. However, while low levels of viral entry in cells expressing owl monkey CCR5 could be detected with this assay, in all cases cells expressing human CCR5 were more permissive than those expressing owl monkey CCR5 (e.g. 18 fold lower for BG505 at and MOI of 1.25).
Figure 3.
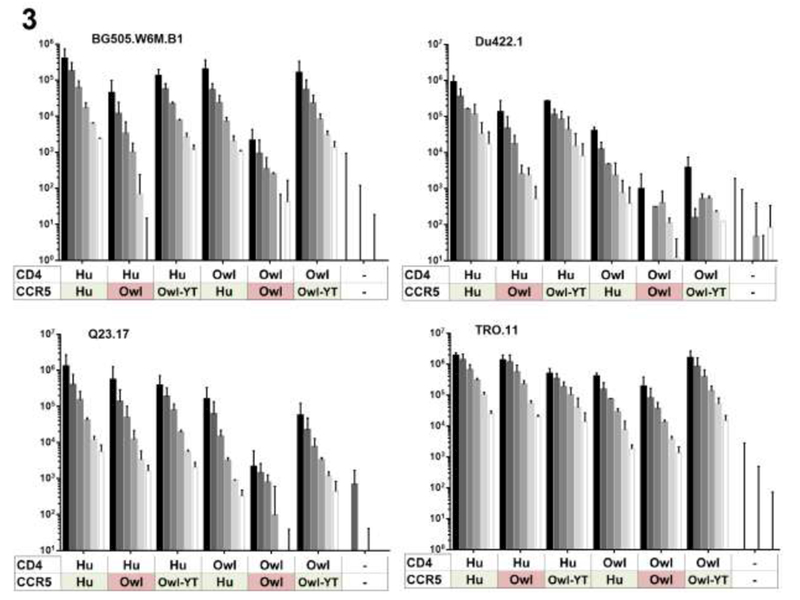
Infection of cells expressing various receptor combinations using a range of MOIs and a luciferase-based detection assay. 293T-CD4/CCR5 cell lines (x-axis) were infected with the viruses pseudotyped with indicated Envs (BG505.W6M.B1, Du.422.1, Q23.17, TRO.11), in this case with a proviral clone encoding luciferase. Infection was measured as luciferase expression 48hrs postinfection. Colors correspond to residues of interest encoded in the CCR5 allele. Results are representative of one independent experiment each. Shade of grey is indicative of MOI. “-” indicates 293T cells that do not express CD4 or CCR5 and were infected as a negative control. Hu=Human, Owl=owl monkey, Owl-YT=Owl monkey CCR5 encoding changes Y15 and T16.
Discussion
HIV-1 entry requires CD4 and a coreceptor – CCR5 in the case of transmissible viruses – and these receptors govern both host cell specificity and host range. Our previous studies revealed that the macaque CD4 is a suboptimal receptor for HIV-1 due to a single amino acid differences at position 39 compared to the human receptor, particularly for transmissible/early stage HIV-1 variants crucial to the HIV-1 epidemic (14). By contrast, owl monkeys encode a functional CD4 receptor with the same amino acid at position 39 as the human CD4, although there is some allelic variation with regards to viral entry (16). These findings are based on CD4s paired with a human CCR5 coreceptor, raising the questions of whether the CCR5 from owl monkeys is a functional coreceptor for HIV-1. Here we show that the owl monkey CD4 and CCR5 receptor combinations are suboptimal receptors for HIV-1 entry, permitting only low level of infection compared to the corresponding human receptors. However, the combination of human and owl monkey receptors can function for entry by the same viruses, suggesting there is an important interaction between the HIV receptor and coreceptor that governs entry.
The HIV-1 receptor function of owl monkey CCR5 is determined by two residues in the N terminus at positions 15 and 16. Previous studies have shown that sulfated tyrosines at position Y10 and Y14 in this N-terminal extracellular region of human CCR5 to interact with Env gp120 during HIV-1 entry and are critical for successful CCR5 usage as a coreceptor (3, 4, 5, 6). It has been suggested that Y15 and T16 contribute to the rigidity of the sulfated tyrosine (Y10, Y14) interactions with gp120 and are therefore important for successful CCR5 usage as a coreceptor for entry (17, 18). Indeed, we observed that the Y15, T16 mutant of owl monkey CCR5 functions as an HIV-1 coreceptor when combined with owl money CD4 and is more permissive to infection compared to the wild type-owl monkey CCR5 for all the viruses tested. These observations suggest residues at positions 15 and 16 are important for CCR5 coreceptor function.
Interestingly, the function of the owl monkey CCR5, paired with either the corresponding owl monkey CD4 or with human CD4 is Env dependent. In the case of one subtype A Env, Q23-17 and the subtype B Env TRO-11, human CD4 and owl monkey CCR5 were an effective receptor combination, but this was not true for the other Envs tested, which included two subtype C Env variants (20) and a subtype A Env (21). Moreover, only TRO-11 showed appreciable infection using the combination of owl monkey receptors; Q23-17 only infected the human CD4/owl monkey CCR5 cells but not the owl monkey CD4/owl monkey CCR5 cells despite the fact that both CD4 receptors encode the N at position 39. Of note, these differences cannot be explained by differences in expression level as the level of the receptors were generally similar across cell lines tested. When we compared the pattern of inhibition of the various Env variants used in this study to CD4 binding site antibodies or soluble CD4 (22), both of which can provide an indirect measure of the conformational states of the Env, there is no discernible pattern that predicts the ability of these Envs to use these receptors and coreceptors (not shown). However, given the small number of Envs tested, we cannot rule out a relationship between the ability of an Env to use the owl monkey CCR5 coreceptor for entry and whether the Env conformation is more open, potentially better exposing the CCR5 binding site. Overall, these findings, show selective entry is dependent on both viral strain and the combination of CD4 and CCR5 on the cells and highlights the complexity of the functional interactions that drive viral entry.
These findings also reveal the unexpected complexity of receptor function that is dependent on CD4 and CCR5 pairing and suggest that for transmissible HIV-1 variants owl monkey CCR5 may limit entry into cells of this host. Given that there is considerable within-species polymorphism in many genes relevant to viral replication (23), it is possible that there are additional alleles of owl monkey CCR5 that demonstrate improved function as a HIV-1 receptor. Moreover, the finding that TRO-11 can infect cells expressing the owl monkey receptors raise the possibility that this Env variant of HIV-1 may infect owl monkey lymphocytes if other post entry-restrictions are antagonized. Such viruses may hold clues to the viral and host determinants that define successful HIV-1 envelope/receptor interactions.
Methods
Generation of pBru-ΔEnv-Gaussia-luciferase Plasmid
pBru-ΔEnv-Gaussia-luciferase (pBruΔenv-Luciferase) was a gift from Dara Lehman and Mark Pankau. Gaussia luciferase was PCR amplified from pMCS-Gaussia-Dura-Luc vector (ThermoFisher) using the bamGlu forward (5’ GCAGGATCCATGGGAGTCAAAGTT) and Gluxho reverse (5’ AGCACTCGAGTTAGTCACCACCGG) primers. The PCR product was cut with BamHI and Xhol and the digested insert was ligated into pBru3oriΔenv-luc2 in the nef gene.
Pseudovirus production
HEK293T cells were seeded 16hrs prior to transfection at a density of 2×105 cells per ml in tissue culture plates in Dulbecco’s modified Eagle medium (DMEM) (Invitrogen) with 10% heat-inactivated fetal bovine serum (FBS), 0.5% penicillin-streptomycin-fungizone, 0.5% 2mM L-glutamine (complete DMEM). Cells were then transfected using Roche FuGene6 transfection reagent and manufacturer’s specifications with an env-deficient proviral plasmid (Q23ΔEnvGFP (15) or pBruΔenv-Luciferase and env gene of interest (Table 1). Forty-eight hours later, cell supernatants were harvested, centrifuged to remove cell debris (1,200g, room-temp, 5min) and concentrated using Amicon Ultra – 15 Centrifugal Filters (Ultracel – 100K) by centrifugation at 3,000g for 15min (room-temp) and then stored at −80°C. The viral titer of each transfection supernatant was determined by infecting TZM-bl cells (NIH AIDS Reagent Program) as previously described (24).
CCR5 mutagenesis
A CCR5 mutant of the owl monkey CCR5 encoding 2 amino acid substitutions (D15Y, A16T) was generated using the QuickChange site directed mutagenesis kit (Stratagene) with the appropriate mutagenic primers. Sanger sequencing was used to ensure the correct mutations and that the rest of the sequence remained identical to the owl monkey CCR5.
Generation of stable cell lines
HEK293T were cultured in complete DMEM at 37°C and 5% C02. For generation of CD4 and CCR5 expressing cell lines, retroviral virus-like particles (VLPs) were generated in HEK293T cells by cotransfecting pLPCX (retroviral vector encoding the CD4 or CCR5 of interest), pJK3 (MLV-based packaging plasmid) (25), and pMD.G (vesicular stomatitis virus glycoprotein [VSV-G] envelope plasmid) (26) 41 at a ratio of 1:1:0.5 using Fugene 6 (Roche) transfection reagent according to the manufacturer’s protocol. Forty-eight hrs post-transfection, the supernatants containing VLPs were collected, filtered through 0.22-micrometer filters, and concentrated using Amicon Ultracel 100K filters (Millipore). The concentrated VLPs (200μl) were used immediately to transduce HEK293T cells that had been plated 24hrs prior at a density of 105 cells/well in a 6-well plate in 2 ml of drug-free complete DMEM. The cells were transduced in the presence of 10pg/ml of DEAE-dextran by spinoculation at 1,200g for 90 min. The following day, cells were split and transferred in new T75 flasks in 10 ml of drug-free complete medium and cultured for 48 h. The cells were then passaged and maintained in complete medium supplemented with 2pg/ml of puromycin (to select for CD4 and CCR5 expression). The transduced cells with high levels of CD4 and CCR5 expression were obtained by sorting the cells on a FACSAria II cell sorter using an APC-conjugated CD4 antibody (BD Biosciences) and FITC-conjugated CCR5 antibody (BD Biosciences) as described previously (14).
CCR5 stable cell line GFP infectivity assay
Stable cell lines were seeded in 12 well plates in duplicate in 1ml complete DMEM at a density of 8*104 cells per well. Sixteen hours after plating, cells were infected by spinoculation (1200g for 90 min.) with HIV-1 pseudoviruses at an MOI of 1 in the presence of 10ug/ml of DEAE-dextran per well. All pseudoviruses used were Q23ΔEnvGFP (15) with env gene of interest. After 48hrs, the cells were washed once with 200μl of 1XPBS, harvested using 200μl of 0.05% trypsin-EDTA (Gibco), and fixed in 200μl of 2% PFA. The fixed cells were washed twice with 500μl FACS buffer. The cells were resuspended in 450μl of FACS buffer, filtered through a 35um pore size nylon mesh cap (BD Falcon), and analyzed for GFP expression using BD FACSCanto II flow cytometer. The data from 104 cells were analyzed using FlowJo version 9.7.5.
CCR5 stable cell line luciferase infectivity assay
Stable cell lines were seeded in 96 well plates in 125μl of complete DMEM at a density of 104 cells per well. Sixteen hours after plating, cells were infected by spinoculation (1200g for 90 min.) with HIV-1 pseudoviruses at a starting MOI of 5 with five subsequent two-fold dilutions in the presence of 10ug/ml of DEAE-dextran per well. All pseudoviruses used were pBruΔenv-Luciferase with env gene of interest. Four-six hours after spinoculation, media was aspirated and replaced with 125μl complete DMEM. Forty- Eight hrs after infection, 20μl of cell supernatants were collected into a separate 96 well plate, mixed with 50μl of Pierce™ Gaussia Luciferase Glow Assay Kit (Thermo Scientific) working stock (made according to kit recommendations), and then incubated for 10 min at room temperature. The samples were then immediately analyzed for luminosity using a luminometer. Data were gathered using Tecan© Magellan data analysis software and then plotted and analyzed further with Prism version 6.07 (GraphPad Software).
Highlights:
-
-
Owl monkey CCR5 acts as a receptor for HIV-1 when paired with human CD4.
-
-
The combination of owl monkey CD4 and CCR5 are suboptimal as receptors for HIV-1 compared to the corresponding human proteins.
-
-
Differences in the sulfation motif in CCR5 determine species differences in coreceptor function.
Acknowledgements
We thank Vanessa Bauer and Stephanie Rainwater for technical assistance and Mark Pankau and Dara Lehman for generating the pBruΔenv-Luciferase plasmid. Human CCR5 was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: pBABE.CCR5 (Cat #3331) from Dr. Nathaniel Landau. This work was supported by grants from the National Institutes of Health (DP1-DA039543 to J.O., R01-GM-093086 to S.L.S.) and an Investigator in the Pathogenesis of Infectious Disease award from the Burroughs Wellcome Fund (to S.L.S.). M.K. is supported by a National Science Foundation graduate research fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Ronen K, Sharma A, Overbaugh J. HIV Transmission Biology: Translation for HIV Prevention. AIDS (London, England). 2015;29(17):2219–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wilen CB, Tilton JC, Dorns RW. HIV: Cell Binding and Entry. Cold Spring Harbor Perspectives in Medicine. 2012;2(8):a006866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Dragic T, Trkola A, Lin SW, Nagashima KA, Kajumo F, Zhao L, Olson WC, Wu L, Mackay CR, Allaway GP, Sakmar TP, Moore JP, Maddon PJ. Amino-terminal substitutions in the CCR5 coreceptor impair gp120 binding and human immunodeficiency virus type 1 entry. J Virol. 1998;72:279–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Farzan M, Choe H, Vaca L, Martin K, Sun Y, Desjardins E, Ruffing N, Wu L, Wyatt R, Gerard N, Gerard C, Sodroski J. A tyrosinerich region in the N terminus of CCR5 is important for human immunodeficiency virus type 1 entry and mediates an association between gp120 and CCR5. J Virol. 1998;72:1160–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Farzan M, Mirzabekov T, Kolchinsky P, Wyatt R, Cayabyab M, Gerard N, Gerard C, Sodroski J, Choe H. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell. 1999;96:667–676. [DOI] [PubMed] [Google Scholar]
- [6].Kuhmann S, Platt E, Kozak S, Kabat D. Polymorphisms in the CCR5 genes of African green monkeys and mice implicate specific amino acids in infections by simian and human immunodeficiency viruses. J Virol. 1997;71:8642–8656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Platt E, Gomes MM, Kabat D. Reversible and efficient activation of HIV-1 cell entry by a tyrosine-sulfated peptide dissects endocytic entry and inhibitor mechanisms. J Virol. 2014;88:4304–4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Farzan M, Vasilieva N, Schnitzler CE, Chung S, Robinson J, Gerard NP, Gerard C, Choe H, Sodroski J. 2000. A tyrosine-sulfated peptide based on the N terminus of CCR5 interacts with a CD4-enhanced epitope of the HIV-1 gp120 envelope glycoprotein and inhibits HIV-1 entry. J Biol Chem. 2000;275:33516–33521 [DOI] [PubMed] [Google Scholar]
- [9].Steffens CM, Hope TJ. Localization of CD4 and CCR5 in Living Cells. J Virol. 2003;77:4985–4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Achour L, Scott MGH, Shirvani H, Thuret A, Bismuth G, Labbe-Jullie C, Marullo S. CD4-CCR5 interaction in intracellular compartments contributes to receptor expression at the cell surface. Blood. 2009;113:1938–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Xiao X, Wu L, Stantchev TS, Feng YR, Ugolini S, Chen H, Shen Z, Riley JL, Broder CC, Sattentau QJ, Dimitrov DS. Constitutive cell surface association between CD4 and CCR5. Proc Natl Acad Sci USA. 1999;96:7496–7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Gaibelet G, Planchenault T, Mazeres S, Dumas F, Arenzana-Seisdedos F, Lopez A, Lagane B, Bachelerie F. CD4 and CCR5 constitutively interact at the plasma membrane of living cells: a confocal fluorescence resonance energy transfer-based approach. J Biol Chem. 2006;281:37921–37929. [DOI] [PubMed] [Google Scholar]
- [13].Lapham CK, Zaitseva MB, Lee S, Romanstseva T, Golding H. Fusion of monocytes and macrophages with HIV-1 correlates with biochemical properties of CXCR4 and CCR5. Nat Med. 1999;5:303–308. [DOI] [PubMed] [Google Scholar]
- [14].Humes D, Emery S, Laws E, Overbaugh J. A species-specific amino acid difference in the macaque CD4 receptor restricts replication by global circulating HIV-1 variants representing viruses from recent infection. J Virol. 2012;86:12472–12483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Humes D, Overbaugh J. Adaptation of subtype a human immunodeficiency virus type 1 envelope to pig-tailed macaque cells. J Virol. 2011;85:4409–4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Meyerson NR, Sharma A, Wilkerson GK, Overbaugh J, Sawyer SL. Identification of owl monkey CD4 receptors broadly compatible with early-stage HIV-1 isolates. J Virol. 2015;89:8611–8622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Huang C, Lam SN, Acharya P, Tang M, Xiang S-H, Hussan SS, … Kwong PD. Structures of the CCR5 N Terminus and of a Tyrosine-Sulfated Antibody with HIV-1 gp120 and CD4. Science. 2007;317(5846): 1930–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hill CM, Kwon D, Jones M, Davis CB, Marmon S, Daugherty BL, DeMartino JA, Springer MS, Unutmaz D, Littman DR. The Amino Terminus of Human CCR5 Is Required for Its Function as a Receptor for Diverse Human and Simian Immunodeficiency Virus Envelope Glycoproteins. Virology. 1998;248(2):357–371 [DOI] [PubMed] [Google Scholar]
- [19].LaBonte JA, Babcock GJ, Patel T, & Sodroski J Blockade of HIV-1 Infection of New World Monkey Cells Occurs Primarily at the Stage of Virus Entry. The J of Exp Med. 2002;196:431–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Li M, Salazar-Gonzalez JF, Derdeyn CA, et al. Genetic and Neutralization Properties of Subtype C Human Immunodeficiency Virus Type 1 Molecular env Clones from Acute and Early Heterosexually Acquired Infections in Southern Africa. J Virol. 2006;80(23):11776–11790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Li M, Gao F, Mascola JR, et al. Human Immunodeficiency Virus Type 1 env Clones from Acute and Early Subtype B Infections for Standardized Assessments of Vaccine-Elicited Neutralizing Antibodies. J Virol. 2005;79(16):10108–10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wu X, Yang Z-Y, Li Y, et al. Rational Design of Envelope Identifies Broadly Neutralizing Human Monoclonal Antibodies to HIV-1. Science. 2010;329(5993):856–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Compton AA, Malik HS, Emerman M. Host gene evolution traces the evolutionary history of ancient primate lentiviruses. Philosophical Transactions of the Royal Society B: Biological Sciences. 2013;368(1626):20120496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wu X, Parast AB, Richardson BA, Nduati R, John-Stewart G, Mbori-Ngacha D, Rainwater SMJ, Overbaugh J. Neutralization escape variants of human immunodeficiency virus type 1 are transmitted from mother to infant. J Virol. 2006;80:835–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Bartz SR, Vodicka MA. Production of high-titer human immunodeficiency virus type 1 pseudotyped with vesicular stomatitis virus glycoprotein. Methods. 1997;12:337–42 [DOI] [PubMed] [Google Scholar]
- [26].Naldini L, Blomer U, Gage FH, Trono D, Verma IM. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc of the Natl Acad of Sci of the USA. 1996;93:11382–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Poss M, Overbaugh J. Variants from the Diverse Virus Population Identified at Seroconversion of a Clade A Human Immunodeficiency Virus Type 1-lnfected Woman Have Distinct Biological Properties. J Virol. 1999;73(7):5255–5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Li M, Gao F, Mascola JR, et al. Human Immunodeficiency Virus Type 1 env Clones from Acute and Early Subtype B Infections for Standardized Assessments of Vaccine-Elicited Neutralizing Antibodies. J Virol. 2005;79(16):10108–10125. [DOI] [PMC free article] [PubMed] [Google Scholar]


