Proc. R. Soc. B 284, 20170799. (Published Online 9 August 2017). (doi:10.1098/rspb.2017.0799)
We recently found an error in our calculation of the probability of a wild-type allele moving from s to j susceptible sites (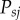 ), and acquiring the gene drive (
), and acquiring the gene drive (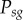 ), during gene-drive homing, under the assumption that multiplexed gRNAs are expressed simultaneously. In the R code provided (function GeneDriveSimRec, appendix S1), these probabilities are calculated recursively and the multiplier
), during gene-drive homing, under the assumption that multiplexed gRNAs are expressed simultaneously. In the R code provided (function GeneDriveSimRec, appendix S1), these probabilities are calculated recursively and the multiplier 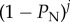 was missing (from line 11), where j is the number of cut target sites along an inter-site NHEJ-mediated deletion at which NHEJ did not occur. We have assessed the impact of this omission on our results, and it is negligible for the baseline of
was missing (from line 11), where j is the number of cut target sites along an inter-site NHEJ-mediated deletion at which NHEJ did not occur. We have assessed the impact of this omission on our results, and it is negligible for the baseline of 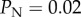 used in our study. However, the impact of this error increases as
used in our study. However, the impact of this error increases as 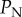 increases. Therefore, we have redone the sensitivity analysis in which we tested
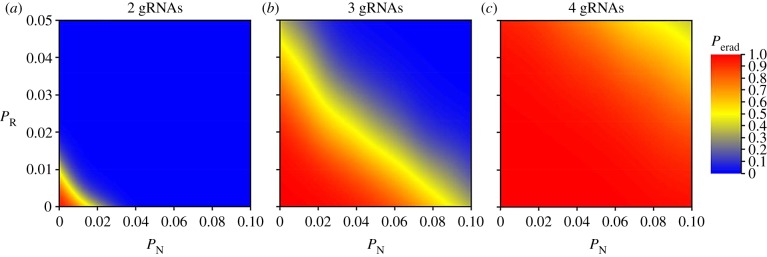
increases. Therefore, we have redone the sensitivity analysis in which we tested 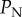 ranging up to 0.1. The amended figure 6 below is essentially identical to the original figure, with the exception that probabilities of eradication are slightly lower for
ranging up to 0.1. The amended figure 6 below is essentially identical to the original figure, with the exception that probabilities of eradication are slightly lower for 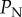 close to 0.1 in panel (c) of the corrected version.
close to 0.1 in panel (c) of the corrected version.
Figure 6 (corrected).
The impact of the probability of non-homologous end-joining (PN) and existing polymorphic resistance (PR) on the probability of successful mouse eradication (Perad) under the homozygotic XX sterility gene-drive strategy. The results shown assume an island carrying capacity of 50 000 mice, 100 gene-drive carriers used for inoculation, simultaneous gRNA expression and (a–c) 2, 3 or 4 guide RNAs. The plotted probabilities are derived from a binomial spatial spline fitted to the sensitivity-analysis output separately for each panel.
This same error invalidates the expression provided in the paper for the probability of successful homing (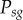 ) for a wild-type allele with s susceptible sites, assuming multiplexed gRNAs are expressed simultaneously. The original expression published was:
) for a wild-type allele with s susceptible sites, assuming multiplexed gRNAs are expressed simultaneously. The original expression published was:
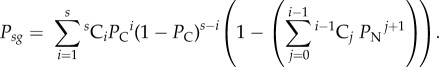 |
We correct this here to:
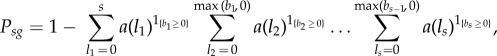 |
where
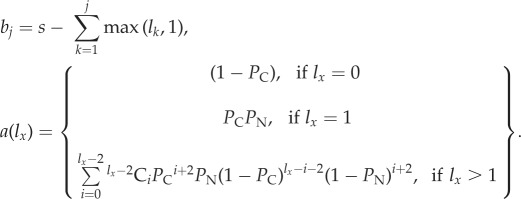 |
The figure above illustrates that this correction has little impact on the probability of successful homing (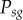 ) for the baseline of
) for the baseline of 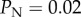 used in our study (panel (a)), but also that the impact of this error increases as
used in our study (panel (a)), but also that the impact of this error increases as 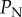 increases (panel (b)).
increases (panel (b)).
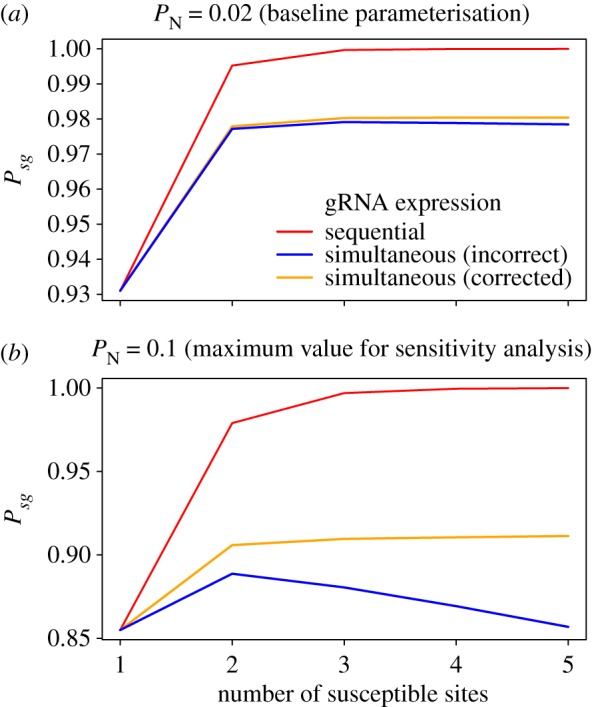
For completeness, we also provide here a corrected version of figure S3 from the electronic supplementary material. Note that in this figure, the maximum number of susceptible sites exceeds that which is tested by modelling in the paper (which was limited to a maximum of five cutting sites targeted by five gRNAs).
Figure S3 (corrected).
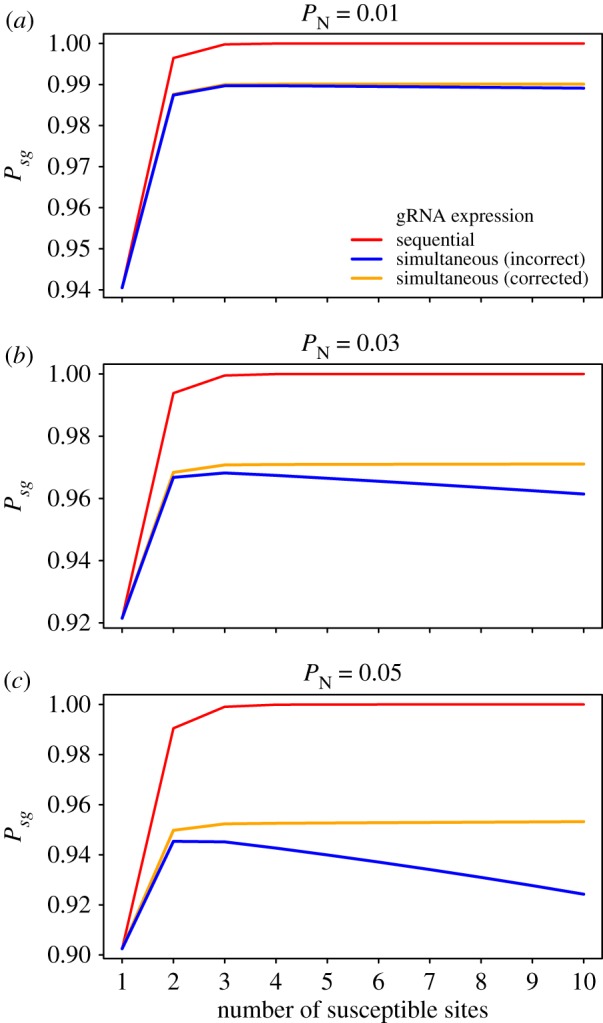
The probability of successful homing (Psg) as a function of the number of susceptible DNA recognition sites, and assuming either sequential or simultaneous expression of guide RNAs. Analytical results are shown for different probabilities of non-homologous end-joining (PN): (a) PN = 0.01, (b) PN = 0.03 and (c) PN = 0.05.