Abstract
For most marine organisms, species richness peaks in the Central Indo-Pacific region and declines longitudinally, a striking pattern that remains poorly understood. Here, we used phylogenetic approaches to address the causes of richness patterns among global marine regions, comparing the relative importance of colonization time, number of colonization events, and diversification rates (speciation minus extinction). We estimated regional richness using distributional data for almost all percomorph fishes (17 435 species total, including approximately 72% of all marine fishes and approximately 33% of all freshwater fishes). The high diversity of the Central Indo-Pacific was explained by its colonization by many lineages 5.3–34 million years ago. These relatively old colonizations allowed more time for richness to build up through in situ diversification compared to other warm-marine regions. Surprisingly, diversification rates were decoupled from marine richness patterns, with clades in low-richness cold-marine habitats having the highest rates. Unlike marine richness, freshwater diversity was largely derived from a few ancient colonizations, coupled with high diversification rates. Our results are congruent with the geological history of the marine tropics, and thus may apply to many other organisms. Beyond marine biogeography, we add to the growing number of cases where colonization and time-for-speciation explain large-scale richness patterns instead of diversification rates.
Keywords: marine biodiversity, fish, time-for-speciation, diversification, freshwater, historical biogeography
1. Introduction
Why is the Central Indo-Pacific (CIP) so diverse? This region, including the coral reefs of Southeast Asia and Australia, contains more species than any other in the world's oceans, across diverse taxa [1]. At similar latitudes, richness declines dramatically with distance from this hotspot [1]. As a consequence, richness differences among marine regions overwhelm correlations between richness and present-day environmental variables [2–4]. Thus, explaining the remarkable richness of the CIP region is crucial for understanding the origins of marine diversity patterns in general.
The peak in marine diversity in the CIP has fascinated researchers for more than 60 years [5]. Traditionally, four major hypotheses have been proposed to explain this pattern (reviewed by [6,7]). First, the ‘centre-of-origin’ hypothesis states that more new species originated in the CIP than surrounding regions [8]. Second, the ‘centre-of-accumulation’ hypothesis states that lineages originating elsewhere preferentially colonized the CIP [9]. Third, the ‘centre-of-overlap’ hypothesis states that species have widespread ranges that overlap in the CIP due to its central position in the broader Indo-West Pacific [10]. Fourth, the ‘centre-of-survival’ hypothesis states that lineages in the CIP experienced less extinction than those in surrounding regions [11]. These four hypotheses differ in the biogeographic origin of species (within the CIP, elsewhere, or no prediction), and in the processes ultimately responsible for high richness in the CIP (colonization, speciation, or extinction).
Despite many important studies clarifying the origins of tropical marine species [11–13], it remains unclear why the CIP has more species than other regions. Based on first principles, species richness in a region can only change via three processes: in situ speciation, local extinction and colonization from other regions [14]. The four ‘centre-of’ hypotheses do invoke these core processes, but do not make specific predictions about their rates or timing. For example, a region's biota could be entirely generated by in situ speciation (i.e. the region is a centre-of-origin), but still have low richness if speciation rates were lower than in other regions, or if the region was colonized more recently (allowing less time to build up richness [15]). Moreover, recent analyses suggest that no single ‘centre-of’ hypothesis fully explains this pattern, and that these processes instead act in concert [6,11–13,16]. For example, Cowman & Bellwood [12] detected many colonizations of the CIP during the early history of reef fish clades (accumulation), followed by in situ diversification (origin and survival). However, it remains unclear whether the CIP has high richness because of older colonization(s), more colonizing lineages or faster diversification rates.
We propose a new direction in answering this 60-year-old mystery. Researchers have already suggested departing from the traditional ‘centre-of’ framework to focus on rates of diversification and colonization instead [7]. However, the timing of colonization may also explain richness patterns, irrespective of diversification rates. The time-for-speciation hypothesis predicts that richness will be higher in regions that were colonized earlier [15]. To our knowledge, no previous studies have compared the importance of the time-for-speciation versus diversification-rate hypotheses for explaining global marine richness patterns. Yet many studies have compared these hypotheses in the broader literature on species richness [17–19].
Here, we analyse the dominant group of marine fishes to test three non-mutually exclusive explanations for the high richness of the CIP: (i) lineages there have higher diversification rates (speciation minus extinction), (ii) colonizations of the CIP tend to be older, allowing more time for richness to build up through in situ speciation, or (iii) lineages have simply colonized the CIP more frequently, regardless of timing. The cosmopolitan clade Percomorpha contains approximately 90% of reef-associated fishes [20], including important focal groups in previous studies of tropical marine diversity (e.g. wrasses, damselfishes [11,12]). We assembled a biogeographic dataset for more than 17 000 percomorph species, encompassing approximately 75% of all marine and approximately 33% of freshwater ray-finned fishes [21], and 72% of marine fishes overall [22]. We included all percomorphs, including freshwater and cold-marine species. This allowed us to compare the processes generating diversity in all three habitats, as well as patterns within warm oceans. We then performed analyses of biogeography and diversification using an extensive time-calibrated phylogeny [23]. We found that differences in diversification rates among regions do not explain fish richness patterns in the oceans. Instead, we found that repeated, older colonizations of the CIP explain its exceptionally high diversity.
2. Material and methods
(a). Phylogeny and biogeographic data
We first extracted the clade Percomorpha (sensu [24]) from a larger phylogeny of actinopterygian fishes [23]. This was the most complete species-level phylogeny published at the time of analysis (but see the very recent [25]). We then obtained a list of all known percomorph species from FishBase.org (17 458 species as of August 2016 [22]). This allowed us to resolve discrepancies in taxonomy among data sources, and estimate richness in each region, including species not sampled in the phylogeny. After resolving taxonomic differences (electronic supplementary material, appendix S1), the resulting phylogeny had 4571 species (26% of Percomorpha).
We compared eight marine biogeographic regions (figure 1), delimited based on previous studies of coastal fish biogeography [26–28]. We divided the warm oceans (tropical + warm temperate) into six regions. To incorporate all percomorph species, we also included two cold-marine regions (cold temperate + polar: Northern and Southern Hemispheres). Additionally, we treated freshwater and brackish habitats (‘freshwater’ for brevity) as a ninth region, irrespective of geography. We also performed alternative analyses in which we combined the six warm-marine regions to reflect the three major biogeographic realms: the Atlantic (Western and Eastern Atlantic), Indo-West Pacific (Western Indian, CIP and Central Pacific) and Eastern Pacific [26–28].
Figure 1.
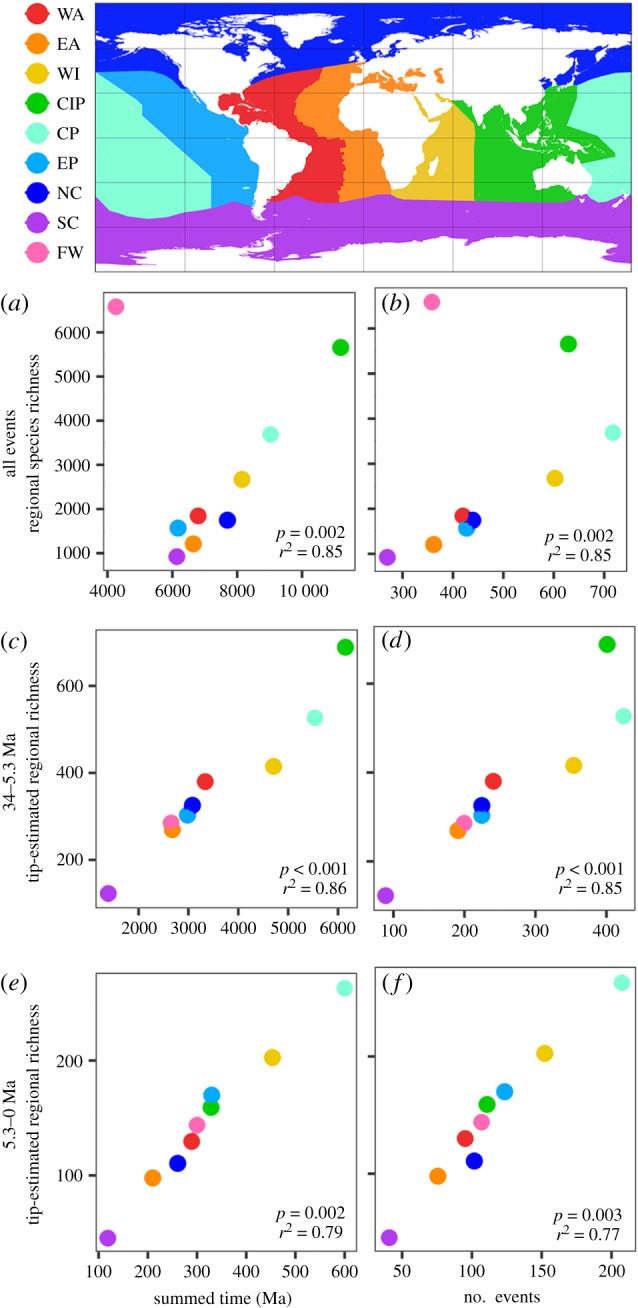
The time-for-speciation effect and the number of colonization events explain differences in species richness among global-scale marine regions. (a) When all events are included, the CIP leads in summed time-for-speciation, with freshwater (FW) an outlier removed prior to analysis. (b) The number of colonizations is also strongly related to regional richness, but the CP has more colonizations than the CIP. (c,d) From 34–5.3 Ma, summed time also best explains high richness in the CIP. The CIP leads in number of colonizations only if fossil constraints are used (see electronic supplementary material, appendix S2). (e,f) From 5.3 Ma to the present, the WI and CP have the greatest summed time, most colonizations, and highest richness descended from these colonizations. All values are means of 100 stochastic reconstructions [36] (confidence intervals too narrow to be visible). The ‘tip-estimated regional richness’ (c–f) is the extant richness descended from lineages that colonized each region during the focal period, estimated from the phylogeny. Richness was log10-transformed prior to performing regressions. WA, Western Atlantic; EA, Eastern Atlantic; WI, Western Indian; CIP, Central Indo-Pacific; CP, Central Pacific; EP, Eastern Pacific; NC, northern cold; SC, southern cold; FW, freshwater.
We estimated total richness in each marine region using georeferenced localities. Most data were from the Ocean Biogeographic Information System (OBIS) [29], augmented using the Global Biodiversity Information Facility (GBIF) [30] and FishBase [22] when needed. We were able to categorize 12 434 marine species (more than 99% of all marine percomorphs) as present or absent in each region. A total of 5019 species were restricted to freshwater (based on habitat data from [22]), and were thus absent from the 8 marine regions.
We systematically searched the occurrence data for errors (electronic supplementary material, appendix S1). Nevertheless, even at a coarse scale, a species's range can differ across repositories [31]. To assess uncertainty in area assignments, we constructed two alternative biogeographic datasets, one based on FishBase only [22] and the other based on IUCN only [32]. Regional richness was strongly related among the three datasets (electronic supplementary material, figure S1). Species ranges were identical between repositories in most cases. Details of obtaining and cleaning occurrence data are in the electronic supplementary material, appendix S1.
(b). Ancestral-range estimation and colonization history
We estimated the frequency and timing of colonization of each region using the Dispersal-Extinction-Cladogenesis framework (DEC) [33] implemented in the R package BioGeoBEARS v. 0.2.1 [34]. We restricted dispersal among marine regions across time and space based on changing ocean connectivity. Since these reconstructions were based on modern localities only, we also performed a second set of reconstructions incorporating fossil occurrences. In this second set, following [12], we constrained clade origins to the East Atlantic if they had Eocene fossil representatives in the Tethys Ocean (fossil occurrences from [13]). Additionally, we excluded the CIP and adjacent Central Pacific from these nodes, and disallowed colonization of the CIP until 34 Ma (million years ago). This conforms to the ‘hopping hotspots’ model [35], in which the CIP was potentially unsuitable for reef fishes prior to 34 Ma. Additional details of both reconstructions are in the electronic supplementary material, appendix S2.
A colonization to a new region was inferred when the estimated geographical range for a node contained a different region than its parent node. Under a DEC model, this can only be achieved through anagenetic range expansion. Since our goal is to understand effects of colonization on richness, we did not distinguish between forms of range inheritance that do not involve colonizing new regions (e.g. sympatry or vicariance [33]). After identifying colonizations, the age of each event was estimated as the crown-group age of the focal node. For single-species colonizations (which lack a crown age), we assumed that the colonization occurred at the midpoint of that species's terminal branch (as illustrated in electronic supplementary material, figure S2).
We performed linear regressions between log10-transformed regional species richness and each of three metrics of colonization history: (i) the earliest colonization of each region, (ii) the number of colonizations of a region, and (iii) the summed ages of all colonizations of a region (‘summed time-for-speciation’). Only the last two metrics reflect contributions of multiple colonizations, which may be important in highly connected marine systems [6]. The summed time-for-speciation metric [18] reflects both the amount of time each colonizing lineage has been present in the region, and the cumulative contribution of multiple lineages colonizing the region. This metric may be high for a region because of frequent colonizations, older colonizations or both. To incorporate uncertainty in ancestral-range estimations, we simulated 100 possible biogeographic histories given the phylogeny, species ranges and model of range inheritance [36]. For each of the 100 replicates, we calculated colonization metrics and performed regressions of these metrics with regional richness. We did not perform phylogenetic regression analyses since the units of analysis here were regions (which are not connected by a phylogeny).
Biogeographic models (i.e. DEC) do not implement a correction for missing species. To explore how our results might change as phylogenetic sampling increases, we compared biogeographic reconstructions of Labridae (wrasses and parrotfishes, a focal group in previous biogeographic studies [11,12]) between the 2013 phylogeny [23] and a newly published phylogeny with greater sampling [25]. Of 630 total labrid species, these phylogenies contained 244 and 339 species respectively [22]. Results were very similar between phylogenies (electronic supplementary material, appendix S2). Therefore, our reconstructions should capture the broad-scale differences in colonization influencing regional richness patterns, even with incomplete sampling.
(c). Biogeographic analyses of different time periods
To assess the relative contributions of older versus more recent colonizations to extant regional richness, we separately analysed the colonizations occurring during two well-established periods of reef-fish evolution. We first focused on the period from 34 to 5.3 Ma, when peak marine biodiversity shifted from the Tethys to the CIP, and when many coral reef fish genera originated [13]. We then analysed the period between 5.3 and 0 Ma, a period of increased diversification in reef fish species globally [13].
We performed regression analyses for each time period as described above. Unlike analyses across all Percomorpha, the regional richness in these regressions was the pool of extant species in the region descended from colonizations occurring during the time period of interest. For each colonization occurring in the focal time bin, we counted the number of terminal taxa sampled in the phylogeny descended from that colonization and still occurring in the same region. To obtain the regional richness for the time period, we summed this count across all colonizations for the region during that time bin (single-species events were simply added to this number). We used only species sampled in the phylogeny to count regional richness, because we could not assign phylogenetically unsampled species to colonizations from a specific time period. Phylogenetically sampled richness is strongly related to total richness for each region (p = 0.0001, r2 = 0.89; electronic supplementary material, table S2), so our conclusions should not be explained by incomplete sampling alone. These regional richness counts estimated from the tree were stable across 100 simulated histories (figure 1c–f).
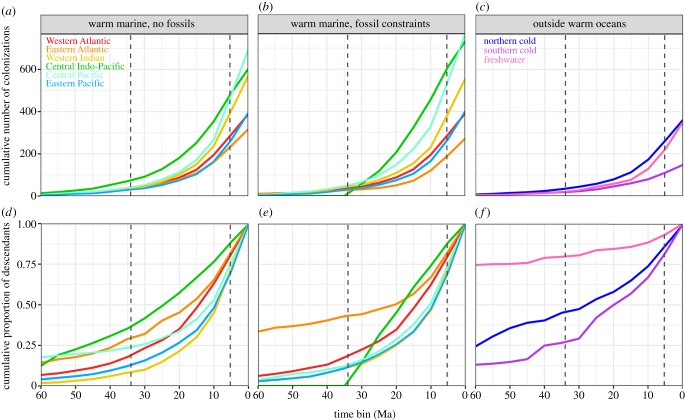
Finally, to visualize the temporal origins of extant richness in each region more generally, we counted the number of colonizations and their descendant richness within 5 Ma time bins from the root of the phylogeny to the present (figure 2). Descendant richness for each time bin was calculated as described above.
Figure 2.
The CIP is dominated by more frequent and older colonizations compared with other regions. (a–c) The cumulative number of colonizations across the Cenozoic, in 5 million-year time bins. (a) In reconstructions without fossil constraints, the CIP was colonized by more lineages than any other region until approximately 5 Ma, when it was surpassed by the CP. (b) Reconstructions with fossil constraints yield similar results. (c) Compared with warm oceans, there are fewer colonizations into freshwater and the cold oceans. (d–f) The proportion of living species in the region descended from colonizations that occurred during or prior to each time-bin. (d) Without fossil constraints, a greater proportion of species in the CIP are descended from older colonizations overall, compared with other warm marine regions. (e) With fossil constraints, the oldest colonizations (greater than 34 Ma) contribute more to Atlantic richness than other regions, but the Atlantic is ultimately dominated by more recent colonizations (approximately last 20 Ma). The CIP is still dominated by relatively older colonizations approximately over the last 30 million years. (f) A high proportion of living freshwater richness comes from older colonizations (greater than 34 Ma). Dashed lines indicate the start of the Oligocene (34 Ma) and Pliocene (5.3 Ma).
(d). Diversification rates
We used two approaches to test if richness patterns were explained by regional differences in diversification rates. First, we performed regressions of log10-transformed regional richness and each region's weighted mean net diversification rate, which is the average net diversification rate of clades weighted by their richness in each region [19]. This approach allowed us to directly include all known species of a clade in calculations of diversification rates (including undersampled groups [7,37]). Second, we used state-dependent speciation-extinction (SSE) models that allow for ‘hidden states’ [38] (see below).
To obtain a region's weighted mean net diversification rate, we first calculated net diversification rates of clades using the method-of-moments estimators [39]. These estimators only require clade age (crown or stem), known extant richness, and a correction for the failure to sample extinct clades (ɛ = extinction/speciation rate). Simulations show that this approach is relatively accurate, even given incomplete species sampling and heterogeneous rates within clades [40]. We performed separate analyses using families and genera, which differ systematically in age and geographical range. Non-monophyletic families were aggregated into clades with related families; non-monophyletic genera were excluded (electronic supplementary material, appendix S3). Following standard practice, we assumed three values of ε (0, 0.5, 0.9). Overall, we performed analyses including diversification rates for 200 family-level clades and 1131 monophyletic genera. The weighted mean rate was calculated for each region as the net diversification rate of each clade multiplied by the clade's richness in the region, summed across clades, and divided by the total richness of these clades in the region. We also performed alternative analyses in which we weighed rates by endemic richness (species restricted to each region).
In addition, we compared regional diversification rates using SSE-class models, which do not depend on a priori clade delimitation. We used the Hidden State Speciation and Extinction framework (HiSSE and GeoHiSSE) [38,41], which estimates speciation, extinction, and transition rates for a known character and an unknown (‘hidden’) character that overlaps with the character of interest. Inclusion of hidden states significantly reduces type-1 error by allowing diversification rates to vary with factors other than geography, thereby creating biologically meaningful null models [38].
Since HiSSE and GeoHiSSE models are currently limited to comparing two states, we performed binary comparisons by combining regions. We compared: (i) the combined Indo-West Pacific (Western Indian, CIP and Central Pacific) versus other warm oceans (East Pacific and Atlantic); (ii) warm versus cold oceans; and (iii) freshwater versus marine habitats. We pruned the phylogeny to only include species occurring in the relevant regions for each binary comparison (warm marine species, marine species and all species). Since one species cannot be assigned to two regions simultaneously in HiSSE, we performed alternative analyses assigning widespread species to one group or the other (e.g. only species restricted to the Indo-West Pacific assigned there versus all species occurring there). We fitted alternative models that partitioned variation in rates by geography, hidden states or both. GeoHiSSE [41] explicitly models geographical range evolution, and allows species to occur in both regions. We only include results from comparisons among warm oceans using GeoHiSSE, because parameter estimates were problematic for the other comparisons. We did not use GeoSSE (geographical model without hidden states [42]) because simulations suggested a type-1 error rate of 65% given our data. Details of HiSSE, GeoHiSSE and GeoSSE analyses are in the electronic supplementary material, appendix S3.
3. Results
Our analyses of biogeography and diversification revealed three major results. First, the hotspot of marine richness in the CIP is explained by many, relatively old colonizations that allowed greater time to accrue species richness (through in situ speciation) than in other regions. Second, diversification rates are similar among the warm oceans, but are higher in cold marine clades, despite the low richness of these habitats. Third, in contrast to marine diversity, high freshwater richness is derived largely from a few successful lineages, with ancient origins and high diversification rates.
(a). Regional richness
Based on occurrence data from all 17 453 species of percomorph fishes, regional richness was highest in freshwater (6584 species globally) and in the CIP (5659 species). Richness was moderate in the Central Pacific (3697) and Western Indian Oceans (2677), both flanking the CIP. Richness was lower in the Western Atlantic (1845), northern cold oceans (1749), Eastern Pacific (1570) and Eastern Atlantic (1210), and lowest in the southern cold oceans (929). The distribution of percomorph richness among regions reflects the longitudinal and latitudinal diversity gradients [1], as well as high richness in freshwater.
(b). Timing and frequency of colonization
Results are summarized as means across 100 simulated biogeographic histories [36]. We found that variation in richness among marine regions (excluding freshwater) was strongly related to both summed time-for-speciation (figure 1a; mean p = 0.002; mean r2 = 0.85) and the number of colonizations of each region (figure 1b; mean p = 0.002, mean r2 = 0.85). The CIP had the greatest summed time among all marine regions. However, there were more colonizations of the adjacent Central Pacific region than the CIP. Thus, the age of colonizations is needed to explain the peak in richness at the CIP, not their number alone. These results were similar using reconstructions with additional fossil constraints (electronic supplementary material, appendix S2). Results using the earliest colonization were not significant (electronic supplementary material, appendix S2), apparently because colonizations after 34 Ma generated present-day richness patterns among warm oceans (figures 1 and 2).
Our results from analyses including only colonizations from 34 to 5.3 Ma (figure 1c,d) were similar to those spanning the entire history of Percomorpha (figure 1a,b). The number of colonizations into each region and their summed time-for-speciation were both strongly related to the extant richness descended from those events (number of events: mean p < 0.001, mean r2 = 0.85; summed time: mean p < 0.001, mean r2 = 0.86). The CIP had greater summed time-for-speciation in reconstructions both with and without fossil constraints. However, in those with fossil constraints, the CIP also had the most colonizations from 34 Ma to 5.3 Ma (electronic supplementary material, appendix S2). Interestingly, even after preventing colonization of the CIP before 34 Ma, its high richness is still explained by many older colonizations compared to other regions (figure 2; electronic supplementary material, appendix S2).
From 5.3 Ma to the present, the relationships between regional richness and both metrics of colonization were again significant (figure 1e,f; number of events: mean p = 0.003; mean r2 = 0.77; summed time: mean p = 0.002, mean r2 = 0.79). In this period, however, the two regions adjacent to the CIP (Western Indian, Central Pacific) surpassed all other marine regions in regional richness derived from these more recent colonizations. Results from both sets of reconstructions were very similar.
To summarize, the high richness of the CIP is explained by both the number of colonizations and their age, as combined by the summed-time metric (figure 1). Specifically, there were many more colonizations of the CIP than other regions from 34 Ma to 5.3 Ma, allowing more time-for-speciation cumulatively across many lineages (figures 1 and 2).
After combining regions, we still found a strong relationship between summed time-for-speciation and regional richness (electronic supplementary material, appendix S2). The combined Indo-West Pacific region had higher colonization metrics in all analyses. This suggests the relationship described above is not an artefact of our delimitation of regions, nor of frequent interchange between contiguous regions (electronic supplementary material, appendix S2).
Freshwater habitats (containing 38% of all percomorphs) showed a very different pattern of colonization relative to marine regions. Although global freshwater richness is similar to that of the CIP, freshwater diversity is derived from relatively few habitat transitions. Thus, freshwater habitats appeared as an outlier in the relationships between regional richness, colonizations, and summed time-for-speciation (figure 1a,b). This pattern is driven by ancient transitions. An estimated 80% of extant freshwater richness was derived from only approximately 18 habitat transitions, all before 35 Ma (figure 2c,f). These include Cichlidae (1685 species; crown age = 71.4 Ma) and Cyprinodontiformes (1280 species; crown age = 93.5 Ma). In contrast, extant diversity in the CIP is predominately descended from approximately 530–700 colonizations since 35 Ma (depending on constraints used; figure 2). Thus, similar richness in freshwater and the CIP was achieved through very different colonization histories (figures 1 and 2).
(c). Diversification rates
Regional richness was not significantly related to weighted mean diversification rates of regions (for families: p = 0.22, r2 = 0.20, using the crown age and assuming ɛ = 0.5; figure 3a). When using families, the diversification-richness relationship was negative. Despite their relatively low richness, the two cold marine regions had the highest mean net diversification rates. Among 200 family-level clades, the 10 clades with the highest diversification rates all had peak richness in the northern or southern cold oceans (including Zoarcidae, Nototheniidae and members of Scorpaeniformes; electronic supplementary material, dataset S2). Weighted mean rates were similar among the six warm marine regions, despite dramatic differences in richness. The clades with the highest richness were shared among the six warm regions (i.e. gobies, blennies, wrasses; electronic supplementary material, figure S9, appendix S3). Analyses using genera, which are typically younger and more geographically restricted than families, produced similar rates among warm regions (figure 3a). Interestingly, mean freshwater rates were much higher using genera than families. Results were congruent with rates calculated using stem ages, alternative values of ɛ, and a weighting scheme based on endemic richness (electronic supplementary material, appendix S3).
Figure 3.
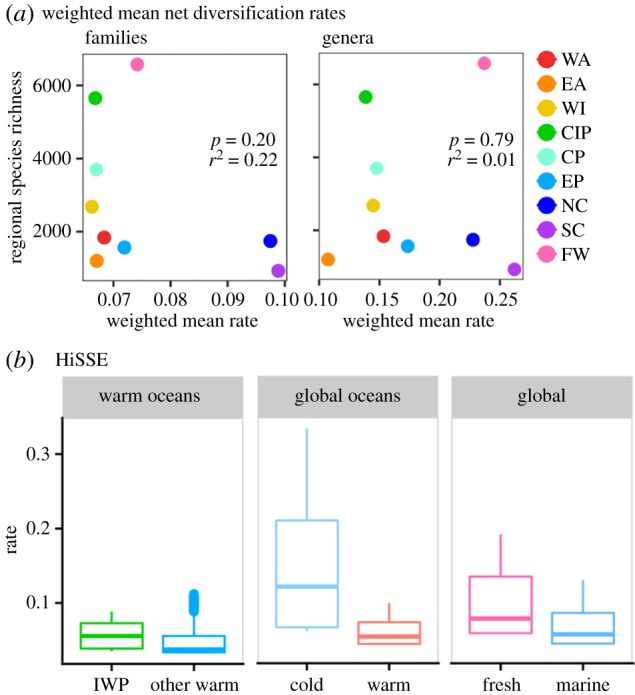
Net diversification rates are similar among warm marine regions, and highest in the cold oceans. (a) The relationship between weighted mean net diversification rates for each region, and the regions' total richness. Means for each region are the mean net diversification rates of families or genera present, weighted by each clade's richness in that region. Here, rates were calculated using the crown age and assuming ɛ = 0.5. Richness was log10-transformed prior to performing regressions. (b) Net diversification rates inferred by best-fit HiSSE models. We performed three binary comparisons by combining regions (Methods) and pruning the phylogeny to include only species in the regions of interest. For the analyses shown, only species restricted to the Indo-West Pacific (IWP), cold oceans, and freshwater habitats were assigned to each of these groups. Alternative HiSSE analyses assigned widespread species to these categories. Rates were averaged for each region across hidden states, based on the distribution of the hidden states among regions. Details of alternative analyses, hidden states and GeoHiSSE are given in the electronic supplementary material, appendix S3.
Hidden-state analyses were consistent with those based on clades. The combined Indo-West Pacific had similar net diversification rates to other warm oceans on average (figure 3b), and the hidden states had a stronger influence on diversification rates than geography. This was consistent using both SSE methods, and with alternative assignments of widespread species to either binary group using HiSSE. When only species restricted to cold oceans were assigned to the cold group, cold oceans had higher net diversification rates than warm oceans. Similarly, when only species restricted to freshwater were assigned to the freshwater group, freshwater clades had slightly higher inferred net diversification rates than marine clades. However, assigning species found in both habitats to these groups (cold and freshwater, respectively) eliminated these differences. Extended results can be found in the electronic supplementary material, appendix S3. Overall, these results were consistent with the idea that species richness patterns in the ocean are better explained by the frequency and timing of colonization than by strong differences in diversification rates.
4. Discussion
In this study, we explored the causes of the dramatic differences in species richness among major regions in the world's oceans, especially the high richness of the CIP. We show that the exceptional richness of the CIP is explained by the relatively older colonization of this region by many lineages, followed by in situ speciation. Surprisingly, the CIP hotspot was not explained by higher diversification rates relative to other warm marine regions. We discuss these drivers of global marine richness patterns below, as well as the origins of freshwater percomorph diversity.
(a). Colonization history among warm marine regions
Our results suggest that the dramatic differences in species richness among warm oceans are explained by differences in both the number and timing of colonizations among regions. These patterns appear to reflect the geological history of the global marine tropics. The shift in peak richness away from the Tethys (present-day East Atlantic) during the Oligocene and Miocene (34–5.3 Ma) was concomitant with the collision of the Australian and Pacific plates with continental Southeast Asia [35]. This collision created a wide platform of shallow ocean, allowing reef formation in the present-day CIP. Our biogeographic analyses revealed many colonizations of the CIP during this time, which gave rise to most of the extant diversity of the region (figures 1 and 2). Among the other five warm marine regions, richness differences are explained by fewer colonizing lineages altogether (Atlantic, East Pacific [12,43]), and only limited timespans with high colonization rates (Western Indian and Central Pacific; figures 1 and 2 [12]).
Most importantly, lineages descended from these relatively old colonizations of the CIP persisted in the region up to the present-day (figure 2). Historical extinction events likely contributed to the lower richness of other warm marine regions [3,43–45]. For example, a biodiversity hotspot also existed in the Western Indian Ocean from 23 to 16 Ma, during the formation of the present-day CIP hotspot [35]. However, the lineages that occurred there appear to have migrated or gone extinct [35]. Thus, most living species in the Western Indian Ocean are descended from more recent colonizations than those in the CIP (figure 2d,e). A similar pattern occurred in the East Atlantic (the former Tethys hotspot), although a greater proportion of its extant diversity can be traced to earlier colonizations than the Western Indian (figure 2d,e). Our results support the idea that biodiversity centres first accumulate lineages via colonization, followed by in situ diversification within these lineages [12,46]. In particular, our results demonstrate that the traditional centre-of-accumulation, centre-of-origin and centre-of-survival hypotheses may be synthesized via the time-for-speciation effect, which explains the high richness of the CIP. Specifically, lineages colonized the CIP earlier, were not subsequently eliminated by extinction, and thus were able to diversify longer than lineages in other regions.
Many marine organisms seem to have biogeographic histories similar to those found here in fishes, including benthic invertebrates, mangroves and planktonic eukaryotes [3,13,35,45,47]. In addition, recent analyses suggest that diversification rates in corals are not higher in the CIP than other regions (see below), and instead colonization rates explain the region's high coral richness [47]. Therefore, our proposed explanation for the exceptional diversity of the CIP (more colonizations and time, not faster diversification) may apply broadly across marine groups.
(b). Net diversification rates in the warm oceans
Our results suggest that marine richness patterns are explained by differences in colonization history, especially time-for-speciation, rather than differences in diversification rates per se. This explanation has important precedents in earlier studies on the biogeography and diversification of reef fishes [11,12]. These studies showed that rapid diversification began much earlier and was sustained for longer in the Indo-West Pacific than in other regions. However, it was previously unclear if high richness in the CIP was thus due to earlier diversification or higher diversification rates.
A ‘centre-of-origin’ model is sometimes interpreted to predict higher speciation rates in the CIP (e.g. [47]). Whereas higher speciation rates would indeed support this model, they are not necessary. Our results demonstrate that earlier colonization to a region may also result in relatively high richness via in situ speciation, independent of the speciation rate itself. In support of this idea, a recently published study showed that speciation rates are similar in ray-finned fishes across the warm oceans, despite differences in richness [25]. Our results concur, using net diversification rates (speciation minus extinction). Note that richness patterns could potentially be explained by differences in diversification rates even if speciation rates were constant, given variation in extinction rates.
Several studies have reported higher diversification rates in reef-inhabiting lineages relative to nonreef lineages [44,48] (but see [20]). The CIP supports greater area of reef habitat than other tropical oceans [2], including during times of extensive Pleistocene glaciation [3]. Therefore, one would expect higher diversification rates in reef-fish lineages within the CIP, perhaps due to reduced extinction rates [3,7,45]. In light of our results, instead of increasing diversification rates per se, larger reef area in the CIP may have attracted and maintained more colonists, and supported longer periods of sustained diversification, compared to the younger reefs of other tropical marine regions [43]. Extinction may play a different role by erasing the diversity of historical hotspots. These extinctions may reduce the time for extant lineages to rebuild richness via colonization and speciation [49]. Further, extinction rates estimated from extant clades may not reflect these historical extinction events [49]. This hypothesis should be further investigated in the context of marine richness, especially by using the fossil record.
The rate estimates used here [39] reflect the outcome of speciation and extinction over a given period of time (clade age). Further analysis may elucidate how diversification rates changed over time. For example, were diversification rates in the CIP higher in the past? The comparison of rates among regions is complicated by widespread species [16], limitations on the number of regions allowed by available methods [42], and limited phylogenetic sampling in key groups (e.g. cryptobenthic fishes [7,37]). In principle, the region-of-origin of widespread species could be inferred using ancestral-range reconstructions, and then temporal differences in rates among regions could be assessed (e.g. [50]). However, this approach may be limited when few species are sampled, because only splitting events in the phylogeny are used to record in situ speciation. Overall, assessments of diversification-rate differences among regions should become more precise in the future. However, our results suggest that colonization timing may be a more important driver of richness patterns than diversification rates alone, and should be prioritized in future studies. Future studies might also consider how diversification, colonization and time-for-speciation explain richness differences at smaller scales within regions (e.g. the richness of the Red and Mediterranean Seas [51]).
(c). Rapid diversification outside the warm oceans
Marine lineages in coldwater regions had the highest diversification rates across Percomorpha (see also [25,48]), yet these regions also had low richness (figure 3a). This decoupling of rates and richness could be attributed to the young ages of rapidly diversifying coldwater families (electronic supplementary material, dataset S2). In addition, the few successful colonizations of cold regions from warm oceans also contributes to their low richness (figure 1), and is consistent with the tropical conservatism hypothesis ([52], see also [48]). These observations also help resolve the richness paradox posed by [25]. Thus, coldwater lineages exemplify the problem with implicitly linking high species richness with high diversification rates, without testing the role of colonization.
Notably, rapid diversification in cold regions is a replicated pattern, both across clades and geographical space, occurring in both northern and southern cold waters (figure 3a; in agreement with [25] for speciation rates). This rapid diversification may be driven by ecological opportunity after repeated glaciation events drove high-latitude faunas locally extinct [53]. Note that the northern cold oceans have higher richness than the southern cold oceans, apparently due to more colonizing lineages in the north (figures 1 and 2).
Our results also shed light on the origins of freshwater fish diversity. We found that freshwater percomorph diversity, though similar in magnitude to the CIP, is dominated by a few ancient clades with exceptional species richness and high diversification rates (figures 1 and 3; electronic supplementary material, figure S6). Some authors have hypothesized that freshwater clades have higher rates of diversification than marine clades due to finer population fragmentation [48,54]. However, high diversification rates are not a universal feature of freshwater clades (electronic supplementary material, appendix S3). Thus, we speculate that rapid diversification might be better explained by ecological factors within freshwater (e.g. rivers versus lakes) rather than occupancy in freshwater alone.
5. Conclusion
The high species richness of the CIP is a striking biodiversity pattern seen in many groups of marine organisms [1]. The processes that explain this pattern have remained poorly understood. Here, we provide an explanation for these regional differences in marine species richness, a 60-year-old mystery [5]: in fishes, the CIP's high richness is explained by many, relatively old colonizations of the region, allowing more time for in situ speciation to build up richness. This explanation is consistent with the geologic history of the marine tropics, and may apply to many other marine organisms with similar biogeographic histories [35]. Beyond marine biogeography, our study adds to the growing number of cases where regional differences in richness are unrelated to diversification rates [55]. Instead, the time-for-speciation effect may be the predominant driver of spatial richness patterns formed over relatively short geological time scales (e.g. Cenozoic [17]).
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank S. Lambert, C. Román-Palacios, C. Hutter, B. Larsen, R. Ferriere, M. Donoghue and J. Chang for helpful discussion. We thank J. Pandolfi and four anonymous reviewers for constructive comments on the manuscript. We thank D. Caetano, J. Beaulieu and B. O'Meara for advice on HiSSE analyses. We thank P. Santoro for assistance with a pilot version of this project. Part of this research used computational resources and services at the Center for Computation and Visualization, Brown University.
Data accessibility
All data are available in the electronic supplementary material and the Dryad Digital Repository: https://doi.org/10.5061/dryad.gh162v1 [56].
Authors' contributions
E.C.M. conceived the study and designed analyses. E.C.M., K.T.H. and D.S. collected data and implemented analyses. All authors contributed to writing the manuscript and gave final approval for publication.
Competing interests
We declare we have no competing interests.
Funding
E.C.M. was supported by a Graduate Research Fellowship from the US National Science Foundation no. DGE-1143953. K.T.H. was funded by a 2016 BrownConnect LINK Award from Brown University. D.S. was funded by the National Top Talent Undergraduate Training Program in 2016. J.J.W. was funded by NSF grant no. DEB 1655690.
References
- 1.Tittensor DP, Mora C, Jetz W, Lotze HK, Ricard D, Berghe EV, Worm B. 2010. Global patterns and predictors of marine biodiversity across taxa. Nature 466, 1098–1101. ( 10.1038/nature09329) [DOI] [PubMed] [Google Scholar]
- 2.Parravicini V, et al. 2013. Global patterns and predictors of tropical reef fish species richness. Ecography 36, 1254–1262. ( 10.1111/j.1600-0587.2013.00291.x) [DOI] [Google Scholar]
- 3.Pellissier L, et al. 2014. Quaternary coral reef refugia preserved fish diversity. Science 344, 1016–1019. ( 10.1126/science.1249853) [DOI] [PubMed] [Google Scholar]
- 4.Jablonski D, Huang S, Roy K, Valentine JW. 2017. Shaping the latitudinal diversity gradient: new perspectives from a synthesis of paleobiology and biogeography. Am. Nat. 189, 1–12. ( 10.1086/689739) [DOI] [PubMed] [Google Scholar]
- 5.Hoeksema BW. 2007. Delineation of the Indo-Malayan centre of maximum marine biodiversity: the Coral Triangle. In Biogeography, time, and place: distributions, barriers, and islands (ed. Renema W.), pp. 117–178. Dordrecht, The Netherlands: Springer. [Google Scholar]
- 6.Bowen BW, et al. 2013. The origins of tropical marine biodiversity. Trends Ecol. Evol. 28, 359–366. ( 10.1016/j.tree.2013.01.018) [DOI] [PubMed] [Google Scholar]
- 7.Cowman PF. 2014. Historical factors that have shaped the evolution of tropical reef fishes: a review of phylogenies, biogeography, and remaining questions. Front. Genet. 5, 1–15. ( 10.3389/fgene.2014.00394) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Briggs JC. 1974. Marine zoogeography. New York, NY: McGraw-Hill Book Company. [Google Scholar]
- 9.Ladd HS. 1960. Origin of the Pacific Island molluscan fauna. Am. J. Sci. 258A, 137–150. [Google Scholar]
- 10.Woodland DJ. 1983. Zoogeography of the Siganidae (Pisces): an interpretation of distribution and richness patterns. Bull. Mar. Sci. 33, 713–717. [Google Scholar]
- 11.Barber PH, Bellwood DR. 2005. Biodiversity hotspots: evolutionary origins of biodiversity in wrasses (Labridae) in the Indo-Pacific and New World tropics. Mol. Phylogenet. Evol. 35, 235–253. ( 10.1016/j.ympev.2004.10.004) [DOI] [PubMed] [Google Scholar]
- 12.Cowman PF, Bellwood DR. 2013. The historical biogeography of coral reef fishes: global patterns of origination and dispersal. J. Biogeogr. 40, 209–224. ( 10.1111/jbi.12003) [DOI] [Google Scholar]
- 13.Bellwood DR, Goatley CHR, Bellwood O. 2017. The evolution of fishes and corals on reefs: form, function, and interdependence. Biol. Rev. 92, 878–901. ( 10.1111/brv.12259) [DOI] [PubMed] [Google Scholar]
- 14.Ricklefs RE. 1987. Community diversity: relative roles of local and regional processes. Science 235, 167–171. ( 10.1126/science.235.4785.167) [DOI] [PubMed] [Google Scholar]
- 15.Stephens PR, Wiens JJ. 2003. Explaining species richness from continents to communities: the time-for-speciation effect in emydid turtles. Am. Nat. 161, 112–128. [DOI] [PubMed] [Google Scholar]
- 16.Barber PH, Meyer CP. 2015. Pluralism explains diversity in the Coral Triangle. In Ecology of fishes on coral reefs (ed. Mora C.), pp. 258–263. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 17.Pontarp M, Wiens JJ. 2017. The origin of species richness patterns along environmental gradients: uniting explanations based on time, diversification rate, and carrying capacity. J. Biogeogr. 44, 722–735. ( 10.1111/jbi.12896) [DOI] [Google Scholar]
- 18.Kozak KH, Wiens JJ. 2012. Phylogeny, ecology, and the origins of climate-richness relationships. Ecology 93, S167–S181. ( 10.1890/11-0542.1) [DOI] [Google Scholar]
- 19.Wiens JJ, Pyron RA, Moen DC. 2011. Phylogenetic origins of local-scale diversity patterns and the causes of Amazonian megadiversity. Ecol. Lett. 14, 643–652. ( 10.1111/j.1461-0248.2011.01625.x) [DOI] [PubMed] [Google Scholar]
- 20.Price SA, Claverie T, Near TJ, Wainwright PC. 2015. Phylogenetic insights into the history and diversification of fishes on reefs. Coral Reefs 34, 997–1009. ( 10.1007/s00338-015-1326-7) [DOI] [Google Scholar]
- 21.Carrete Vega G, Wiens JJ. 2012. Why are there so few fish in the sea? Proc. R. Soc. B 279, 2323–2329. ( 10.1098/rspb.2012.0075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Froese R, Pauly D. 2017. FishBase version 2/2017. See http://www.fishbase.org/ .
- 23.Rabosky DL, Santini F, Eastman J, Smith SA, Sidlauskas B, Chang J, Alfaro ME. 2013. Rates of speciation and morphological evolution are correlated across the largest vertebrate radiation. Nat. Commun. 4, 1958 ( 10.1038/ncomms2958) [DOI] [PubMed] [Google Scholar]
- 24.Near TJ, et al. 2013. Phylogeny and tempo of diversification in the superradiation of spiny-rayed fishes. Proc. Natl Acad. Sci. USA 110, 12 738–12 743. ( 10.1073/pnas.1304661110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rabosky DL, et al. 2018. An inverse latitudinal gradient in speciation rate for marine fishes. Nature 559, 392–395. ( 10.1038/s41586-018-0273-1) [DOI] [PubMed] [Google Scholar]
- 26.Briggs JC, Bowen BW. 2012. A realignment of marine biogeographic provinces with particular reference to fish distributions. J. Biogeogr. 39, 12–30. ( 10.1111/j.1365-2699.2011.02613.x) [DOI] [Google Scholar]
- 27.Kulbicki M, et al. 2013. Global biogeography of reef fishes: a hierarchical quantitative delineation of regions. PLoS ONE 8, e81847 ( 10.1371/journal.pone.0081847) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cowman PF, Parravicini V, Kulbicki M, Floeter SR. 2017. The biogeography of tropical reef fishes: endemism and provinciality through time. Biol. Rev. 92, 2112–2130. ( 10.1111/brv.12323) [DOI] [PubMed] [Google Scholar]
- 29.OBIS. 2017. Data from the Ocean Biogeographic Information System. Intergovernmental Oceanographic Commission of UNESCO. See http://www.iobis.org/.
- 30.Global Biodiversity Information Facility. 2017. See http://www.gbif.org/.
- 31.Robertson DR. 2008. Global biogeographical data bases on marine fishes: caveat emptor. Divers. Distrib. 14, 891–892. ( 10.1111/j.1472-4642.2008.00519.x) [DOI] [Google Scholar]
- 32.International Union for the Conservation of Nature. 2017. IUCN Red List of Threatened Species Version 2017.1. See http://www.iucnredlist.org/.
- 33.Ree RH, Smith SA. 2008. Likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Syst. Biol. 57, 4–14. ( 10.1080/10635150701883881) [DOI] [PubMed] [Google Scholar]
- 34.Matzke NJ. 2014. Model selection in historical biogeography reveals that founder-event speciation is a crucial process in island clades. Syst. Biol. 63, 951–970. ( 10.1093/sysbio/syu056) [DOI] [PubMed] [Google Scholar]
- 35.Renema W, et al. 2008. Hopping hotspots: global shifts in marine biodiversity. Science 321, 654–657. ( 10.1126/science.1155674) [DOI] [PubMed] [Google Scholar]
- 36.Matzke NJ. 2015. Stochastic mapping under biogeographical models. See http://phylo.wikidot.com/biogeobears#stochastic_mapping/ .
- 37.Brandl SJ, Goatley CHR, Bellwood DR, Tornabene L. In press The hidden half: ecology and evolution of cryptobenthic fishes on coral reefs. Biol. Rev. ( 10.1111/brv.12423) [DOI] [PubMed] [Google Scholar]
- 38.Beaulieu JM, O'Meara BC. 2016. Detecting hidden diversification shifts in models of trait-dependent speciation and extinction. Syst. Biol. 65, 583–601. ( 10.1093/sysbio/syw022) [DOI] [PubMed] [Google Scholar]
- 39.Magallón S, Sanderson MJ. 2001. Absolute diversification rates in angiosperm clades. Evolution 55, 1762–1780. ( 10.1111/j.0014-3820.2001.tb00826.x) [DOI] [PubMed] [Google Scholar]
- 40.Meyer ALS, Romá-Palacios C, Wiens JJ. In press BAMM gives misleading rate estimates in simulated and empirical datasets. Evolution. ( 10.1111/evo.13574) [DOI] [PubMed] [Google Scholar]
- 41.Caetano D, O'Meara B, Beaulieu J. In press Hidden state models improve state-dependent diversification approaches, including biogeographical models. Evolution ( 10.1101/evo.13602) [DOI] [PubMed] [Google Scholar]
- 42.Goldberg EE, Lancaster LT, Ree RH. 2011. Phylogenetic inference of reciprocal effects between geographic range evolution and diversification. Syst. Biol. 60, 451–465. ( 10.1093/sysbio/syr046) [DOI] [PubMed] [Google Scholar]
- 43.Lessios HA. 2008. The Great American Schism: divergence of marine organisms after the rise of the Central American isthmus. Annu. Rev. Ecol. Evol. Syst. 39, 63–91. ( 10.1146/annurev.ecolsys.38.091206.095815) [DOI] [Google Scholar]
- 44.Cowman PF, Bellwood DR. 2011. Coral reefs as drivers of cladogenesis: expanding coral reefs, cryptic extinction events, and the development of biodiversity hotspots. J. Evol. Biol. 24, 2543–2562. ( 10.1111/j.1420-9101.2011.02391.x) [DOI] [PubMed] [Google Scholar]
- 45.Di Martino E, Jackson JBC, Taylor PD, Johnson KG. 2018. Differences in extinction rates drove modern biogeographic patterns of tropical marine biodiversity. Sci. Adv. 4, eaaq1508 ( 10.1126/sciadv.aaq1508) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Briggs JC, Bowen BW. 2013. Evolutionary patterns: marine shelf habitat. J. Biogeogr. 40, 1023–1035. ( 10.1111/jbi.12082) [DOI] [Google Scholar]
- 47.Huang D, Goldberg EE, Chou LM, Roy K. 2018. The origin and evolution of coral species richness in a marine biodiversity hotspot. Evolution 72, 288–302. ( 10.1111/evo.13402) [DOI] [PubMed] [Google Scholar]
- 48.Tedesco PA, Paradis E, Lévêque C, Hugueny B. 2017. Explaining global-scale diversification patterns in actinopterygian fishes. J. Biogeogr. 44, 773–783. ( 10.1111/jbi.12905) [DOI] [Google Scholar]
- 49.Miller EC, Wiens JJ. 2017. Extinction and time help drive the marine-terrestrial biodiversity gradient: is the ocean a deathtrap? Ecol. Lett. 20, 911–921. ( 10.1111/ele.12783) [DOI] [PubMed] [Google Scholar]
- 50.Xing Y, Ree RH. 2017. Uplift-driven diversification in the Hengduan Mountains, a temperate biodiversity hotspot. Proc. Natl Acad. Sci. USA 114, E3444–E3451. ( 10.1073/pnas.1616063114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DiBattista JD, Berumen ML, Gaither MR, Rocha LA, Eble JA, Choat JH, Craig MT, Skillings DJ, Bowen BW. 2013. After continents divide: comparative phylogeography of reef fishes from the Red Sea and Indian Ocean. J. Biogeogr. 40, 1170–1181. ( 10.1111/jbi.12068) [DOI] [Google Scholar]
- 52.Wiens JJ, Donoghue MJ. 2004. Historical biogeography, ecology, and species richness. Trends Ecol. Evol. 19, 639–644. ( 10.1016/j.tree.2004.09.011) [DOI] [PubMed] [Google Scholar]
- 53.Crame JA. 2018. Key stages in the evolution of the Antarctic marine fauna. J. Biogeogr. 45, 986–994. ( 10.1111/jbi.13208) [DOI] [Google Scholar]
- 54.Grosberg RK, Vermeij GJ, Wainwright PC. 2012. Biodiversity in water and on land. Curr. Biol. 22, R900–R903. ( 10.1016/j.cub.2012.09.050) [DOI] [PubMed] [Google Scholar]
- 55.Schluter D, Pennell MW. 2017. Speciation gradients and the distribution of biodiversity. Nature 546, 48–55. ( 10.1038/nature22897) [DOI] [PubMed] [Google Scholar]
- 56.Miller EC, Hayashi KT, Song D, Weins JJ. 2018. Data from: Explaining the ocean's richest biodiversity hotspot and global patterns of fish diversity Dryad Digital Repository. ( 10.5061/dryad.gh162v1) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Miller EC, Hayashi KT, Song D, Weins JJ. 2018. Data from: Explaining the ocean's richest biodiversity hotspot and global patterns of fish diversity Dryad Digital Repository. ( 10.5061/dryad.gh162v1) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
All data are available in the electronic supplementary material and the Dryad Digital Repository: https://doi.org/10.5061/dryad.gh162v1 [56].