Abstract
Following synthesis, RNA can be modified with over 100 chemically distinct modifications. Recently, two studies—one by our group—developed conceptually similar approaches to globally map N1-methyladenosine (m1A) at single nucleotide resolution. Surprisingly, the studies diverged quite substantially in their estimates of the abundance, whereabouts, and stoichiometry of m1A within internal sites in cytosolic mRNAs: One study reported it to be a very rare modification, present at very low stoichiometries, and invariably catalyzed by TRMT6/61A. The other found it to be present at >470 sites, often at high levels, and suggested that the vast majority were highly unlikely to be TRMT6/61A substrates. Here we reanalyze the data from the latter study, and demonstrate that the vast majority of the detected sites originate from duplications, misannotations, mismapping, SNPs, sequencing errors, and a set of sites from the very first transcribed base that appear to originate from nontemplated incorporations by reverse transcriptase. Only 53 of the sites detected in the latter study likely reflect bona-fide internal modifications of cytoplasmically encoded mRNA molecules, nearly all of which are likely TRMT6/TRMT61A substrates and typically modified at low to undetectable levels. The experimental data sets from both studies thus consistently demonstrate that within cytosolic mRNAs, m1A is a rare internal modification where it is typically catalyzed at very low stoichiometries via a single complex. Our findings offer a clear and consistent view on the abundance and whereabouts of m1A, and lay out directions for future studies.
Keywords: m1A, RNA modifications, epitranscriptome, post-transcriptional regulation
INTRODUCTION
Post-transcriptional modifications of RNA form an emerging layer of regulation of gene expression, analogous in potential importance to post-translational modifications of proteins. Over 100 modifications exist throughout the three domains of life. While these modifications were traditionally studied in the highly abundant—and hence biochemically tractable—tRNA and rRNA molecules, in recent years high-throughput sequencing approaches have allowed to generate transcriptome-wide maps of RNA modifications. These maps have revealed that some modifications are widespread also in other classes of RNA, most notably in mRNA (Dominissini et al. 2012; Meyer et al. 2012; Carlile et al. 2014; Schwartz et al. 2013, 2014a; Li et al. 2015), where they can impact mRNA processing, localization, stability, and translational efficiency (Schwartz et al. 2014b; Wang et al. 2014, 2015; Meyer et al. 2015; Zhou et al. 2015; Haussmann et al. 2016; Lence et al. 2016; Shi et al. 2017).
Nearly two years ago, two reports—by the groups of He and Yi—coupled the use of an anti-m1A antibody with RNA-sequencing, collectively reported the identification of >7000 putative m1A harboring regions in one study (Dominissini et al. 2016) and nearly 1000 regions in the other (Li et al. 2016). Both studies found m1A to originate primarily from 5′ UTRs, and particularly near start codons or first exon–exon junctions (Dominissini et al. 2016; Li et al. 2016). One study found the putative m1A sites to be associated within a degenerate GC rich consensus (Dominissini et al. 2016); the other observed an enrichment for a degenerate purine-rich motif (Li et al. 2016). Both studies lacked the ability to detect m1A at single nucleotide resolution, and did not identify enzymes catalyzing formation of m1A on mRNA, and hence could not directly validate these sites, nor assay their functions and mechanisms of action.
Recently, two studies—one by our group, the other again by the Yi group—developed conceptually similar approaches for mapping m1A at single nucleotide resolution (Li et al. 2017; Safra et al. 2017). Rather than relying on enrichment of m1A-containing fragments upon m1A-IP, these two studies relied on misincorporation patterns introduced upon reverse transcription of m1A-containing RNA (Hauenschild et al. 2015). The studies used additional measures to ensure that the misincorporation patterns were specific. Both studies measured misincorporation levels in the input RNA and upon pulldown using the anti-m1A antibody, and sought to identify misincorporation signals enriched in the latter. To achieve additional stringency, both studies further measured misincorporation levels in an immunoprecipitated sample subjected to a treatment that eliminates, or reduces, m1A levels: In one case the samples were subjected to Dimroth rearrangement, a chemical treatment that converts m1A residues into m6A residues and thereby reduces misincorporation levels (Safra et al. 2017). In the other case, elimination of m1A was achieved via the employment and careful calibration of an RNA demethylating enzyme, leading to almost complete elimination of m1A levels (Li et al. 2017). The Safra et al. (2017) study further developed an approach relying on reverse transcription using an enzyme that predominantly leads to premature truncation of reverse transcription, as an additional control (Safra et al. 2017).
The two studies converged on some of their findings. Both identified sites in the cytoplasm sharing an identical sequence and structural features, and found them to be modified by the TRMT6/TRMT61A complex at an identical sequence and structural motif as found in position 58 of tRNA, the well-characterized substrate of this complex. Both studies further found that m1A was present at a number of sites within mitochondria, leading both of them to focus in particular on the same site in ND5, a mitochondrially encoded gene forming part of complex I of the respiratory complex. In addition, both studies found that m1A within internal positions of mRNA represses translation. Finally, the single nucleotide resolution sites identified by both groups show negligible overlap with the previously identified “m6A peaks” (see analysis below).
Nonetheless, the two studies diverged substantially in terms of their estimates of the abundance, whereabouts, and stoichiometry of m1A. Safra et al. (2017) reported the identification of eight m1A sites in cytosolic mRNAs and lncRNAs, most of which were estimated to be modified at very low levels (most were undetectable in the Input samples), and all of them catalyzed via TRMT6/TRM61A. In addition, this study reported that m1A-IP enriches for the 5′ end of genes, but that this enrichment does not originate from 5′ UTRs, the start codon, or the first splice junction as previously reported (Dominissini et al. 2016; Li et al. 2016), but rather from the very first transcribed base (“TSS sites”). Finally, this study did not detect evidence for m1A presence at the TSS sites on the basis of analysis of premature truncation or misincorporation patterns, and hence left open the question as to whether these sites originate from m1A (or an m1A-derivative) at the TSS or from antibody promiscuity. In contrast, Li et al. (2017) reported a total of 474 sites, of which only 53 harbored a TRMT6/61A motif. They further classified a total of 277 sites as originating from the 5′ UTR, only 24 of which mapping to the first transcribed nucleotide. The findings of Li et al. (2017) suggest that (i) m1A at internal positions on mRNA is substantially more prevalent than reported by the Safra study, (ii) TRMT6/61A only has a minor role in shaping the m1A landscape, suggesting that other m1A methyltransferases remain to be discovered, (iii) m1A at internal sites can be present at considerable stoichiometry: 76 of the mRNA sites reported by Li et al. (2017) have mean misincorporation rates >20% within the Input—unenriched—fractions, and the authors further emphasize that misincorporation rates likely substantially underestimate the true m1A levels. Although the actual sites identified by Li et al. (2017) are nearly completely distinct from the originally identified “m1A peaks,” they were interpreted to provide support to the two original publications on m1A, which characterized this modification to be widespread at internal sites within mRNAs, primarily within 5′ UTRs, and to be present at a stoichiometry of ∼20% (Dominissini et al. 2016; Li et al. 2016).
Here, we reconcile the points of divergence between the two studies, primarily by reanalyzing and reannotating the set of sites identified by Li et al. (2017). We find that 53 of the 474 sites likely reflect bona fide internal modifications of cytoplasmically encoded mRNA molecules. All of these sites harbor TRMT6/TRMT61A consensus motifs and are modified at low to undetectable levels, in the absence of IP, fully consistent with the observations made in the Safra et al. (2017) study. The remaining sites correspond to (i) sites appearing redundantly within this data set, (ii) sites originating from tRNAs which were misannotated as mRNA, (iii) sites originating from mitochondrial RNA that were mismapped to mRNA, (iv) Genomic SNPs, (v) sequencing errors, or (vi) sites originating from the very first transcribed base (“TSS sites”), which we suggest may have originated from nontemplated activity of reverse transcriptase. The differences in the conclusions of the two studies are thus not due to differences in the experimental approaches used by the two groups, but to critical differences in the analytical pipeline used by the two groups, which led to an inflated estimate of the number and stoichiometry of m1A sites by the Li et al. (2017) study.
RESULTS AND DISCUSSION
Reclassification of putative m1A sites
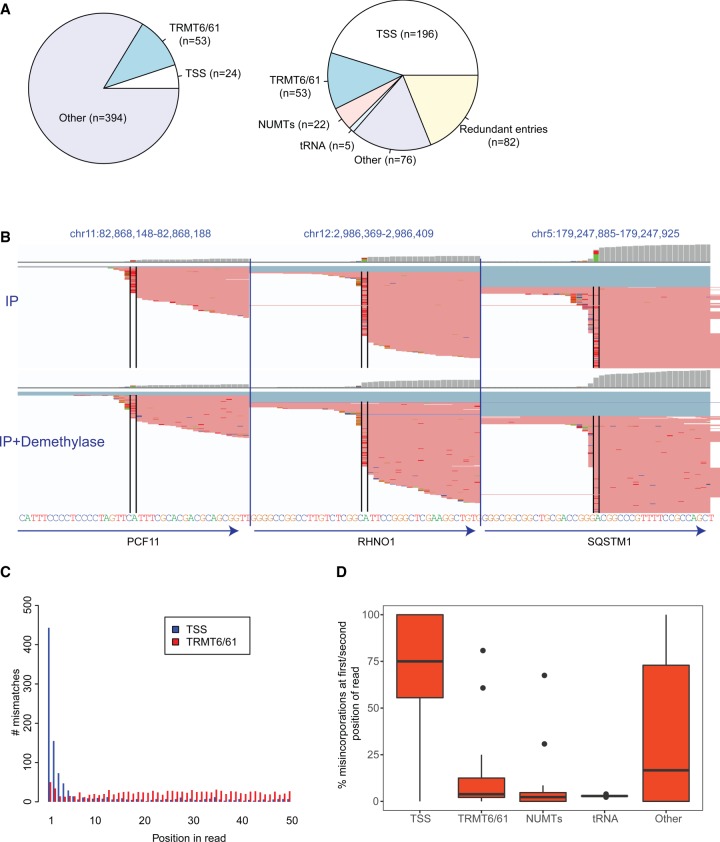
Li et al. (2017) aligned the sequencing reads to a transcriptomic reference, rather than to the human genome, and identified 474 putative m1A sites. Fifty-three of these sites were classified as putative TRMT6/61A substrates, 24 as originating from the first transcribed nucleotide, and the remaining sites were unclassified (Fig. 1A, left panel). To allow comparison of these sites to other data sets, we first converted the transcriptomic coordinates to genomic coordinates. In this context we eliminated from this data set three sites that did not form part of the current RefSeq database and 37 sites that did not have an “A” at the reported site in this annotation. Importantly, given that 37 of the 474 sites did not harbor an A at the reported position, we estimate a minimal “false conversion rate” on our end of ∼7.8%, and thus estimate that a further ∼39 of the remaining sites analyzed here do not reflect the original sites identified by Li et al. (2017) (these discrepancies are most likely due to updates of the RefSeq database; the Li et al. (2017) study does not specify the precise RefSeq version used, and these discrepancies thus reflect changes made in the RefSeq annotation between their downloading of it and our own).
FIGURE 1.
Reannotation and characterization of putative m1A sites identified by Li et al. (2017) (A) Pie charts depicting the original classification of the putative m1A sites (left) and the reclassified sites (right). (B) Genome browser snapshots of three genomic loci, harboring putative m1A peaks originally classified by Li and coworkers as originating from the 5′ UTR. The genomic reference is depicted at the bottom. The red lines overlapping each locus are reads, with bases diverging from the reference encoded by different colors (green - A, blue - C, orange - G, red - T). The black bars enclose the putative m1A sites. Although the gene annotations (depicted by arrows on the bottom) all indicate that the “official” gene start site is upstream of the modified site, it is apparent that all the misincorporations occur at the first base of the sequencing read. The top panel presents the mismatches upon m1A-IP, the bottom upon IP and demethylase treatment. (C) Barplots depicting the distribution of misincorporation events along the first 50 positions of the sequencing reads. Misincorporation in “TSS sites” (in blue) are highly skewed to occur in the first two positions, whereas misincorporation in TRMT6/61 sites (in red) are relatively uniformly distributed. (D) Boxplots depicting the percentage of misincorporation events occurring within the first or second base (from among all misincorporation events) at sites across the five classes.
We then reclassified and filtered the 434 remaining sites (Fig. 1A, right panel), as follows:
Redundant entries: Eighty-two of the remaining sites were redundant entries, i.e., entries appearing at least twice in the reported data set (in one extreme case the same site appears eight times in the table). Although Li et al. (2017) eliminated transcripts harboring identical RefSeq IDs, this was not sufficient to eliminate such redundancies, as transcripts with distinct RefSeq IDs can also overlap the same genomic locus, hence resulting in an artificial inflation of the number of detected sites.
Mitochondrial pseudogenes and tRNAs: Twenty-two of the remaining sites mapped to mitochondrial pseudogenes, also known as nuclear mitochondrial DNA segments (NUMTS), and five sites originated from tRNAs in regions that had overlapping annotations also with mRNAs. In both cases, the identified sites likely arise from the mitochondrial RNA and from tRNAs (both of which have well documented m1A sites), respectively, and not from the mRNA annotated as overlapping them. Indeed, visual inspection of the alignment patterns at these sites further confirmed that the mapping patterns within these genes were inconsistent with the annotation of the mRNAs annotated at this site, and hence likely stem from reads originating from the mitochondria, that underwent mismapping to the NUMTs, and reads originating from tRNAs that were misannotated as mRNAs.
TRMT6/TRMT61A substrates: Li et al. (2017) required a strictly defined consensus sequence (GTTCRA) in order to classify a site as TRMT6/61A dependent, with no structural constraints. As we previously reported, TRMT6/61A substrates require both a consensus sequence and a structure, although there is considerable flexibility with regard to both (Safra et al. 2017). We thus classified sites as harboring a TRMT6/TRM61A motif if they harbored both a slightly relaxed motif (G[ATC]TCNA) and a stem of at least 3 bp, including base-pairing between position −5 and +3 with respect to the m1A site, −6 and +4; these two base-pairings are the most critical for the formation of m1A (Safra et al. 2017). Sites with only one of these two criteria were still considered TRMT6/TRMT61A substrates, unless they were within the first 200 bp, in which case they were annotated as TSS sites (see below). Fifty-three sites matched these criteria (note that although this number is identical to the one reported by Li and coworkers, the catalog of sites is distinct and both include sites that were not originally included by Li and coworkers due to our relaxed consensus requirements, and eliminate sites that were reported by Li and coworkers, but that originate from tRNAs). Of note, 35 of these sites (66%) formed part of the data set of 495 sites that we reported to acquire m1A upon overexpression of TRMT6/TRMT61A (Safra et al. 2017), lending strong evidence to the notion that these sites are highly enriched in TRMT6/TRMT61A substrates.
TSS sites: Li et al. (2017) identified many sites mapping to 5′ UTRs, but classified a site as originating from the first transcribed nucleotide only if it mapped to the very first base of the “annotated” transcript. This approach results in a substantial underestimate of the number of sites originating from the first transcribed base, as transcription typically does not begin at a single nucleotide but across a region comprising several dozens of nucleotides, of which the RefSeq annotation chooses a representative site (Carninci et al. 2006; Plessy et al. 2010; Takahashi et al. 2012). Given that the library preparation procedure used by the Li et al. (2017) study allows capturing the first transcribed base at single nucleotide precision, we first inspected the reads at the identified sites. This analysis revealed that the mutations, when present in the sites annotated as 5′ UTR, almost invariably occurred at the first base of the read (Fig. 1B), strongly suggesting that they occur at the first transcribed nucleotide. Consistently, a systematic analysis of 196 sites residing within the first 200 bp revealed that misincorporations in support of these sites were massively enriched to originate from the first one or two bases of the reads (Fig. 1C,D), in stark contrast to the above-defined TRMT6/61 sites for which mutations were relatively uniformly spread across the entire read (Fig. 1C,D). Of note, the first bases of the reads are particularly prone to various experimental artifacts (see below). Finally, to gain further support that these sites serve as TSSs, we examined CAGE data, a technique dedicated to obtaining single nucleotide resolution mapping of TSS. ENCODE CAGE data from A549 cells (ENCODE CAGE data for HEK293 does not exist) reveals that 79% of the non-TRMT6/61A sites that occur within 200 bp of the annotated start site have at least one CAGE tag associated with them, indicating that they serve at least occasionally as the first transcribed nucleotide (in contrast, only 3% of sites classified as TRMT6/61A sites had one such tag). We thus classified 196 sites residing within the first 200 bp as putative TSS sites.
- Other sites: Seventy-six (of the 434) sites do not readily fall into any of the above-defined criteria. Forty-three of these sites had misincorporation ratios in the Input samples exceeding 10%, and we individually inspected each of them. We found only two cases of convincing misincorporation patterns, consistent with m1A (i.e., high in IP, lower in Input, low upon demethylase treatment). Both sites are with very high likelihood TRMT6/61A substrates, but have minor variations in the motif/structure causing them not to be classified as such. Of the remaining sites:
- Eleven originate from the first transcribed nucleotide (with misincorporations present only at the first base of the read), but are not annotated as such by the RefSeq annotation (and were hence not classified by us as TSS sites). This is consistent with the fact that this group is also enriched for misincorporations occurring at the first 2 bp of the reads (Fig. 1D).
- Four sites have a known SNP at the modified sites, and the identified misincorporations merely reflect the presence of two alleles.
- Six sites occurred within poly(C) stretches, and the misincorporation patterns in them were always A → C, which is highly atypical of the m1A signature (which is typically A → T and A → G). These sites also generally exhibited very poor enrichment upon m1A-IP. Moreover, mutations to C are in some of the cases also observed in adjacent positions that do not harbor an A, but are also embedded within poly(C) stretches. These sites thus likely reflect Illumina errors at homopolymeric C runs, where the basecaller appears to call the homopolymer base also in the cycle following the homopolymer. Such a phenomenon has been previously attributed to limited handling of (pre-) phasing in homopolymeric stretches and to signals accumulating over cycles due to incomplete cleavage of fluorophore (Whiteford et al. 2009; Ledergerber and Dessimoz 2011; Chang et al. 2014).
- Three sites had “noisy” alignment patterns, i.e., there were many “misincorporations” either in the reads supporting them or in the adjacent region. In two of these cases the sites are within an Alu repeat, and hence the noisy alignments likely reflect reads originating from elsewhere.
- Seventeen sites either harbored no reads or no misincorporations when aligned to the human genome. The discrepancy between these patterns and the reported ones are likely a result of differences between our annotation and the one used by Li et al. (2017), resulting in the above-described “false conversion rate.” It is furthermore possible that misincorporations reported at some of these sites are due to reads that were mapped to a region in the transcriptome, despite originating from a different region in the genome. This is a key limitation when mapping reads to the transcriptome, as it is heavily dependent on the annotation and can force alignments even in the presence of better genomic alignments.
The reannotated and reclassified table of 352 nonredundant sites is available as Supplemental Table S1.
TRMT6/TRMT61A sites are typically modified at very low levels
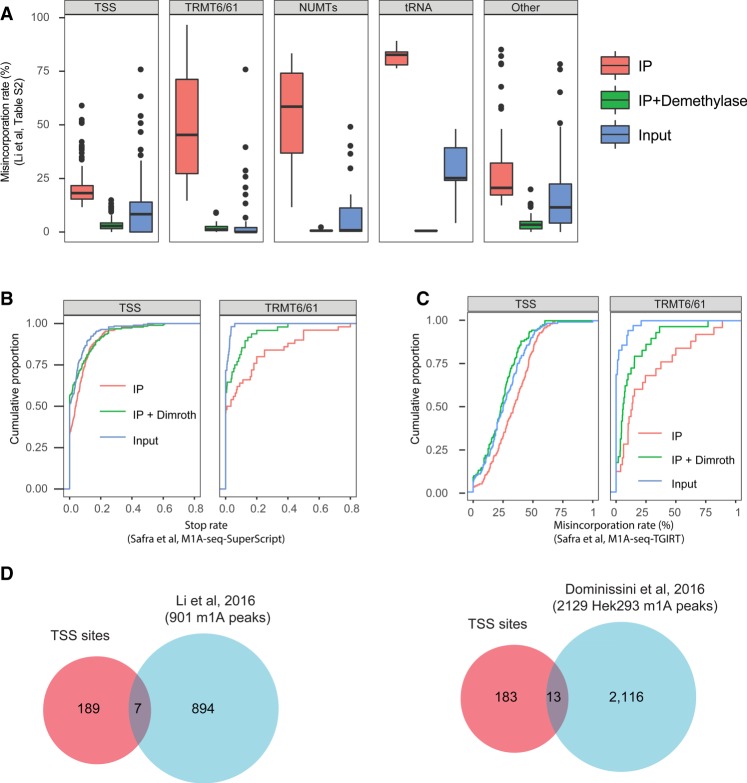
To evaluate the potential physiological relevance of a modification, it is critical to not only interrogate its whereabouts but its stoichiometry. To obtain a lower boundary on the stoichiometry of m1A at the identified sites, we next examined the misincorporation profile at the identified TRMT6/TRMT61A sites in Input, IP, and IP + Demethylated samples. For each site we refer to the mean misincorporation ratio, based on two experimental replicates reported in Supplementary Table S2 in the manuscript by Li et al. (2017). This analysis reveals that the median misincorporation rate for TRMT6/TRMT61A sites in the Input samples was 0% (but 45% after IP); only eight sites had misincorporation rates exceeding 5% (Fig. 2A). Moreover, inspection of each of these eight sites revealed only a single site, in the PRUNE gene 5′ UTR, that had convincing misincorporation rates (16%) in the Input sample. Note that this is also the one cytoplasmic site identified and followed up on in the Safra and coworkers manuscript, due to the higher misincorporation levels detected there as well (Safra et al. 2017). In all other cases, upon genomic mapping of the Li et al. (2017) reads in the input samples, we either observed very low read counts at these sites or no reads at all. The paucity or lack of coverage is likely due to false conversion rates or to issues with alignment to transcriptomes instead of genomes, as above, and in a few cases may also be due to the lower depth to which the Input samples were sequenced. While it thus remains possible that some of them are modified to a low extent, which could be quantifiable via targeted sequencing, overall the data does not provide evidence for m1A being present at substantial levels.
FIGURE 2.
Characterization of TSS sites. (A) Distribution of misincorporation rates across Input, IP, and IP + Demethylase treated samples across the five different classes into which the Li et al. (2017) sites were reclassified, on the basis of the misincorporation ratios reported in Supplementary Table S2. These quantifications highlight strong enrichment of TRMT6/61A sites, but much reduced enrichment of TSS sites. (B) Analysis of the “stop rate,” defined as the fraction of reads beginning immediately after a site (indicating reverse transcriptase termination) divided by the number of reads overlapping it, on the basis of the m1A-seq-SuperScript data set released by Safra et al. (2017). SuperScript leads to a high termination rate of reverse transcription (and lower misincorporation rate) and hence serves as an orthogonal methodology for detecting m1A. The rate of termination does not differ significantly between IP and IP + Dimroth across TSS sites (left panel) in stark contrast to TRMT6/61A sites (right panel) used as positive control. (C) Analysis of the misincorporation rate at the Li et al. (2017) sites on the basis of the Safra and coworkers’ data set, aligned with the enforcement of an “end-to-end” (global) alignment parameter. Low differences are observed for TSS sites (left panel) in contrast to the TRMT6/61A sites (right panel). (D) Venn diagram depicting the overlap between the TSS sites, as classified here, and the sets of “m1A peaks” identified as enriched in Li et al. (2016) (left panel) and Dominissini et al. (2016) (right panel), respectively.
Are misincorporation rates an accurate proxy of m1A stoichiometries? Given that misincorporation rates at m1A are sequence dependent (Hauenschild et al. 2015), we assessed the “maximal” incorporation rate that can be expected on the basis of the TRMT6/TRMT61A consensus sequence. For this we utilized the misincorporation rates within tRNAs at position 58—which harbor the identical consensus sequence as the positions identified in mRNA, obtained from Supplementary Table 1 released by Li et al. (2017). We found that the median misincorporation rates at this site upon m1A-IP was 83%. Even under the conservative assumption that the IP was 100% efficient, and that it isolated only m1A-containing molecules, this would imply that misincorporation rates provide a mild underestimate of m1A stoichiometry, at the order of ∼20% of the methylation levels. Thus, the fact that the vast majority of sites harbor virtually undetectable misincorporation levels in the input samples strongly suggests that the stoichiometry of m1A at these sites is extremely low.
Misincorporations at the TSS—nontemplated incorporations of reverse transcriptase?
Although the m1A antibody clearly enriches for TSS as supported by all studies applying it (Dominissini et al. 2016; Li et al. 2016, 2017; Safra et al. 2017), several considerations raise important concerns as to whether the TSS sites truly harbor m1A, or whether the enrichment may be attributed to an m1A derivative or potentially also to promiscuous binding by the antibody: (i) Although the TSS sites have substantially higher levels of misincorporations in the Input samples (median: 8.3%), the sites reported by Li et al. (2017) are only poorly enriched upon IP (median: 18%). This is in stark contrast to the TRMT6/61A substrates, which we consider as positive controls, which increase from median values of 0% to 45% (Fig. 2A). (ii) Bona fide m1A sites are expected to lead to premature truncation of reverse transcription when reverse transcribed using SuperScript; the rates of such truncations are expected to be decreased upon Dimroth rearrangement, which converts m1A to m6A, and which no longer induces the stop (Safra et al. 2017). Using our published m1A-seq-SuperScript data set, we found no significant change in stop rates at the detected sites between samples subjected to IP and samples subjected to IP + Dimroth treatment (paired T-test, P = 0.12) (Fig. 2B, left panel). In contrast, there was a highly significant drop in stop rates when performing this analysis for sites in the TRMT6/61A group as a control (Paired T-test, P = 0.006) (Fig. 2B, right panel). (iii) Similarly, when quantifying misincorporation rates in the m1A-Seq-TGIRT data set generated in Safra et al. (2017) at the set of putative sites identified in Li et al. (2017) (using an end-to-end alignment mode that allows identification of misincorporations at the end), the magnitude of the differences between Input, IP, and IP + Dimroth were very low in stark contrast to the TRMT6/61A sites (Fig. 2C, right panel). (iv) There is a very poor overlap between the set of TSS sites reported here and the set of m1A peaks identified by the two original studies. Of the 901 “m1A peaks” reported by the authors in their previous study (Li et al. 2016), only seven overlap with the TSS peaks; and of 2129 “m1A peaks” reported in Hek293 cells (Dominissini et al. 2016), only 13 overlap with the TSS sites (Fig. 2D). Thus, for the overwhelming majority of the previously reported peak regions that were enriched by the anti-m1A antibody, no evidence of misincorporation is observed. Lack of overlap can, to some extent, potentially be due to limited sensitivity of peak-calling, which may lead to some peaks failing to be identified. Nonetheless, given that the methodology developed by Li et al. (2017) is highly sensitive, and—as exemplified by the TRMT6/61A substrates—is able to detect sites that are modified at levels close to 0%, and given that the peak regions typically have extremely deep coverage by virtue of being enriched upon m1A-IP, the lack of detected misincorporations across the vast majority of the peaks that were detected thus strongly indicates that such misincorporations that are reversible upon demethylase treatment typically do not occur at these sites.
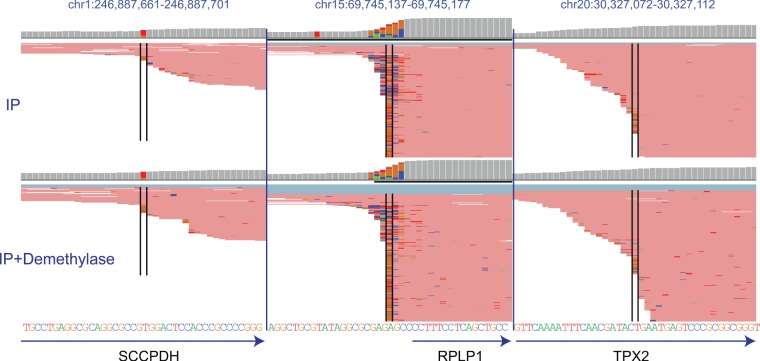
One source for the observed misincorporations at the very first position is the potential for nontemplated nucleotide incorporations in the context of reverse transcription: It is a well characterized phenomenon that reverse transcriptases can include such nontemplated additions (Chen and Patton 2001). Indeed, inspection of the misincorporations patterns across many TSS, both enriched and not enriched via the anti-m1A antibody, reveals myriad “misincorporations” occurring at the very first position at nearly every gene (Fig. 2D). Moreover, such misincorporations are not exclusive to first bases harboring an “A” (Fig. 3, middle panel) but also occur at transcripts beginning with other bases (Fig. 3, left and right panels). While these misincorporations are typically not enriched upon pulldown nor eliminated upon treatment with the demethylase, by explicitly filtering for such sites from across the entire genome, such sites may potentially be spuriously detected. Why, then, were none of these misincorporations flagged as a putative m1A site by the Safra et al. (2017) study? Due to these widespread misincorporations at the very first position, the Safra et al. (2017) manuscript utilizes soft-clipping (Dobin et al. 2013), which enforces local, rather than global alignment. Thus, this option trims bases from their beginning (and ends), to maximize the alignment score. Therefore, misincorporations occurring at the very first base of the read are clipped, and not taken into account in the context of misincorporation detection.
FIGURE 3.
Genome browser snapshots (as in Fig. 1B) of three arbitrarily selected regions around the transcription start sites of genes, in which misincorporations enriched upon IP with respect to IP + Dimroth were not identified. In all cases, misincorporations can be viewed both at the position marked by the black lines, but often also in nearby positions. In the left and right panels the highlighted position does not contain an A. In all cases, the mismatches with respect to the genome occur at the very first base of the read (in some cases they encompass the first two or three bases of the read). The misincorporations are present both in IP and IP + Dimroth treated samples.
Of note, the need to soft-clip reads from the very beginning of reads when searching for misincorporation signals was highlighted also in the past in the context of reanalysis of RNA:DNA mismatch patterns. Such patterns were originally reported to be widespread at over 10,000 sites (Li et al. 2011) and thought to reflect widespread editing of an uncharacterized nature, but subsequent reanalysis of the data revealed that the original study had performed end-to-end alignment, rather than allowing for soft-clipping, and that the majority of RNA:DNA differences originated from the first positions within the reads (Kleinman and Majewski 2012; Lin et al. 2012; Pickrell et al. 2012), and hence likely originate from technical artifacts.
High overlap in detection of TRMT6/61A sites—but not of TSS sites—between computational approaches
Why did the Li et al. (2017) approach result in identification of 53 TRMT6/61 substrates, and the Safra et al. (2017) approach only in eight? A trivial explanation would be that this is a consequence of the increased coverage in the Li et al. (2017) study. Due to increased sequencing depth, increased read length, and improved depletion of rRNA sequences, the median coverage obtained for coding (and noncoding) genes in Li et al. (2017) was 10.2-fold higher in the two IP samples in comparison to the Safra and coworkers data set, and such enrichment should allow detection of sites in more poorly expressed genes. However, the differences in the computational approaches used by the two groups could also lead to differential detection. To directly assess whether the approach used by Safra and coworkers has reduced sensitivity in detecting m1A sites, we applied the computational approach developed in the Safra et al. (2017) manuscript to the raw data obtained from the Li et al. (2017) studies. This approach uncovered a total of 65 sites within mRNAs. Of these, 51 (78%) underwent classification as TRMT6/61A sites on the basis of the above-defined criteria; visual examination of the sequences/potential structures of the remaining ones revealed that at least eight of them are likely to be TRMT6/61A candidates as well, but with atypical secondary structures causing them to undergo misclassification. Among the putative TRMT6/61A substrates, 32/53 (∼60%) of the TRMT6/61A sites identified by Li and coworkers were also identified by the Safra et al. (2017) approach. Of note, only five of the 196 (2.5%) sites within the TSS class were identified using the Safra et al. (2017) approach, and inspection of all of these revealed that they are all likely to be TRMT6/61A substrates (four of these five sites formed part of the sites previously identified by Safra and coworkers upon TRMT6/61A overexpression). Thus, overall the computational approaches by Safra et al. (2017) and Li et al. (2017) yield a similar number of putative TRMT6/61A substrates when applied to the same data set, and with a reasonable extent of overlap between them. In stark contrast, despite the fact that approximately fourfold more sites in the Li and coworkers' data set are classified as TSS sites than TRMT6/61A sites, essentially none of these sites is detected when applying the Safra et al. (2017) computational approach.
Conclusion
Reanalysis of the Li et al. (2017) data reveals a view highly similar to the one identified in Safra et al: Both data sets demonstrate that within internal positions in cytosolic mRNA molecules, m1A is nearly exclusively found within a typical sequence and secondary structure, where it is catalyzed by TRMT6/TRMT61A, in the vast majority of cases at very low stoichiometries. Thus, there does not seem to be evidence in support of an additional m1A methyltransferase, acting on a distinct subset of internal sites on mRNA. While the higher coverage in the Li et al. (2017) study facilitated the identification of more sites compared to the Safra et al. (2017) study, the characteristics of these sites in terms of their sequence/structural requirements, their stoichiometries, and the enzymatic complex catalyzing their formation are essentially identical.
It is likely that, in immunoprecipitated samples sequenced to even greater depth, it will be possible to find evidence for the existence of even more TRMT6/61A sites. In fact, it is possible that it will be possible to find evidence for m1A at very minor levels for all ∼400 sites identified in the Safra et al. (2017) study upon overexpression of TRMT6/61A. An interesting analogy is to A → I editing, where—based on ultradeep sequencing of selected loci—it was estimated that over 100 million editing residues may exist in the human genome, albeit the vast majority at levels <<1% (Bazak et al. 2014). The fact that m1A levels within mRNA TRMT6/61A sites are—in the overwhelming majority of cases—present at very low stoichiometries, suggests that m1A at internal positions is unlikely to play a major role in regulation of gene expression. This, however, does not preclude a potentially more dramatic role played by this modification in mitochondrial mRNAs, where this modification is found, at least in one case, at a higher stoichiometry (Li et al. 2017; Safra et al. 2017).
The differences in the conclusion between the two studies are thus not due to differences in experimental approaches, the source of the HEK293T cell line, or differences in library prep strategies as has been speculated (Dominissini and Rechavi 2017; Xiong et al. 2018). They are due to critical differences in the analytic pipeline used by the two groups.
An important future direction to better understand is the nature of the signal at the transcription start site. Our reanalysis of the Li et al. (2017) data set strongly suggests that the enrichment previously reported to originate from the 5′ UTR, or from the first exon–exon junction (Dominissini et al. 2016; Li et al. 2016, 2017), in fact originates from the first transcribed base, consistent with our previous report (Safra et al. 2017). The low-level enrichment observed for TSS sites, the absence of RT-induced truncations, and the fact that such sites can only be detected at a very low fraction of the previously reported m1A-peaks raise concerns as to whether these sites truly reflect m1A occurring at the first transcribed nucleotide. These concerns are compounded by the fact that detection of these sites relies on mismatches occurring at the first position of the read, which is particularly prone to nontemplated nucleotide incorporations occurring as part of reverse transcription. This notwithstanding, none of these observations provide an answer as to why certain TSSs are enriched with the anti-m1A antibody, nor do they definitely rule out that m1A—or an m1A derivative—is present at the TSS. The reports that the presence of such enrichment—whatever its nature—correlates positively with translation (Dominissini et al. 2016; Li et al. 2017) are intriguing. We anticipate that a definitive response to these questions will arise from biochemical approaches.
MATERIALS AND METHODS
The reanalysis of this study primarily relies on Supplementary Table S2 in the Li et al. (2017) study. Transcriptomic coordinates were mapped to genomic coordinates using an in-house script on the basis of an updated RefSeq annotation (downloaded on Dec. 12, 2017 from the UCSC Table Browser). The “tRNA Genes,” “NumtS,” and “Common SNPs (147)” tables providing the coordinates of tRNAs, mitochondrial pseudogenes, and common SNPs were downloaded from the UCSC Table Browser. CAGE tags in A549 cells, generated by the ENCODE project, were obtained from http://ccg.vital-it.ch/mga/hg19/encode/GSE34448/A549_cell_longPolyA_rep1.sga.gz. Intersections between genomic coordinates were performed using BEDTools (Quinlan 2014).
To assess the stop rates across the various classes of detected sites, we utilized the m1A-seq-SuperScript data sets generated in Safra et al. (2017). The stop rate is calculated as the fraction of reads beginning at each site, divided by the number of reads overlapping it (Safra et al. 2017).
To visually inspect and reanalyze the Li et al. (2017) data, raw data were downloaded from GEO: GSE102040. The reads from Input, IP, and IP + demethylase samples (two replicates each) were aligned to the human genome (assembly: hg19) using STAR aligner (Dobin et al. 2013). To clip the 10 nt barcode at the 5′ end and the adapter at the 3′ end, we used the following criteria “--clip3pAdapterSeq AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGT --clip5pNbases 10”. The data sets were analyzed using the approach described in Safra et al. (2017).
The analyses of the relative position of misincorporations within reads presented in Figure 1C and D were done on the basis of one of the m1A-IP data sets (GSM2722295) in the above GEO. This data was aligned to the genome with the above parameters but also enforcing an end-to-end alignment (“--alignEndsType EndToEnd”), to allow detection of misincorporations occurring at the very end of the read. For Figure 2C we used this m1A-IP data set, along with input and demethylase treatment samples (GSM2722297, GSM2722299), also aligned using EndToEnd mode.
The Venn diagrams comparing the putative m1A sites with previously defined peaks were done on the basis of an intersection of the transcriptome level coordinates provided for the “m1A peaks,” downloaded as a Supplementary Table from Li et al. (2016). For the comparison with the Dominissini et al. paper, we defined a window of 200 nt centered around the center of the 2129 “m1A peaks” that were reported for HEK293 cells, downloaded from the Supplementary Tables in Dominissini et al. (2016).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This project has received funding from the Israel Science Foundation (543165), the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program (grant agreement no. 714023), by research grants from the Abramson Family Center for Young Scientists, the David and Fela Shapell Family Foundation INCPM Fund for Preclinical Studies, the Estate of David Turner, and the Berlin Family Foundation New Scientist Fund.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.067348.118.
REFERENCES
- Bazak L, Haviv A, Barak M, Jacob-Hirsch J, Deng P, Zhang R, Isaacs FJ, Rechavi G, Li JB, Eisenberg E, et al. 2014. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res 24: 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV. 2014. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515: 143–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engström PG, Frith MC, et al. 2006. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet 38: 626–635. [DOI] [PubMed] [Google Scholar]
- Chang H, Lim J, Ha M, Kim VN. 2014. TAIL-seq: genome-wide determination of poly(A) tail length and 3′ end modifications. Mol Cell 53: 1044–1052. [DOI] [PubMed] [Google Scholar]
- Chen D, Patton JT. 2001. Reverse transcriptase adds nontemplated nucleotides to cDNAs during 5′-RACE and primer extension. Biotechniques 30: 574–580, 582. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D, Rechavi G. 2017. Loud and clear epitranscriptomic m1A signals: now in single-base resolution. Mol Cell 68: 825–826. [DOI] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. 2012. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485: 201–206. [DOI] [PubMed] [Google Scholar]
- Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, Dai Q, Di Segni A, Salmon-Divon M, Clark WC, et al. 2016. The dynamic N1-methyladenosine methylome in eukaryotic messenger RNA. Nature 530: 441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauenschild R, Tserovski L, Schmid K, Thüring K, Winz M-L, Sharma S, Entian K-D, Wacheul L, Lafontaine DLJ, Anderson J, et al. 2015. The reverse transcription signature of N1-methyladenosine in RNA-seq is sequence dependent. Nucleic Acids Res 43: 9950–9964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussmann IU, Bodi Z, Sanchez-Moran E, Mongan NP, Archer N, Fray RG, Soller M. 2016. m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature 540: 301–304. [DOI] [PubMed] [Google Scholar]
- Kleinman CL, Majewski J. 2012. Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”. Science 335: 1302; author reply 1302. [DOI] [PubMed] [Google Scholar]
- Ledergerber C, Dessimoz C. 2011. Base-calling for next-generation sequencing platforms. Brief Bioinform 12: 489–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lence T, Akhtar J, Bayer M, Schmid K, Spindler L, Ho CH, Kreim N, Andrade-Navarro MA, Poeck B, Helm M, et al. 2016. m6A modulates neuronal functions and sex determination in Drosophila. Nature 540: 242–247. [DOI] [PubMed] [Google Scholar]
- Li M, Wang IX, Li Y, Bruzel A, Richards AL, Toung JM, Cheung VG. 2011. Widespread RNA and DNA sequence differences in the human transcriptome. Science 333: 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhu P, Ma S, Song J, Bai J, Sun F, Yi C. 2015. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat Chem Biol 11: 592–597. [DOI] [PubMed] [Google Scholar]
- Li X, Xiong X, Wang K, Wang L, Shu X, Ma S, Yi C. 2016. Transcriptome-wide mapping reveals reversible and dynamic N1-methyladenosine methylome. Nat Chem Biol 12: 311–316. [DOI] [PubMed] [Google Scholar]
- Li X, Xiong X, Zhang M, Wang K, Chen Y, Zhou J, Mao Y, Lv J, Yi D, Chen X-W, et al. 2017. Base-resolution mapping reveals distinct m1A methylome in nuclear- and mitochondrial-encoded transcripts. Mol Cell 68: 993–1005.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Piskol R, Tan MH, Li JB. 2012. Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”. Science 335: 1302; author reply 1302. [DOI] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. 2012. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149: 1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian S-B, Jaffrey SR. 2015. 5′ UTR m6A promotes cap-independent translation. Cell 163: 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Gilad Y, Pritchard JK. 2012. Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”. Science 335: 1302; author reply 1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plessy C, Bertin N, Takahashi H, Simone R, Salimullah M, Lassmann T, Vitezic M, Severin J, Olivarius S, Lazarevic D, et al. 2010. Linking promoters to functional transcripts in small samples with nanoCAGE and CAGEscan. Nat Methods 7: 528–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR. 2014. BEDTools: the Swiss-army tool for genome feature analysis. Curr Protoc Bioinformatics 47: 11.12.1–11.12.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safra M, Sas-Chen A, Nir R, Winkler R, Nachshon A, Bar-Yaacov D, Erlacher M, Rossmanith W, Stern-Ginossar N, Schwartz S. 2017. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature 551: 251–255. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Agarwala SD, Mumbach MR, Jovanovic M, Mertins P, Shishkin A, Tabach Y, Mikkelsen TS, Satija R, Ruvkun G, et al. 2013. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell 155: 1409–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, León-Ricardo BX, Engreitz JM, Guttman M, Satija R, Lander ES, et al. 2014a. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159: 148–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, Mertins P, Ter-Ovanesyan D, Habib N, Cacchiarelli D, et al. 2014b. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep 8: 284–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, He C. 2017. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res 27: 315–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Kato S, Murata M, Carninci P. 2012. CAGE (cap analysis of gene expression): a protocol for the detection of promoter and transcriptional networks. Methods Mol Biol 786: 181–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. 2014. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505: 117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. 2015. N6-methyladenosine modulates messenger RNA translation efficiency. Cell 161: 1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteford N, Skelly T, Curtis C, Ritchie ME, Löhr A, Zaranek AW, Abnizova I, Brown C. 2009. Swift: primary data analysis for the Illumina Solexa sequencing platform. Bioinformatics 25: 2194–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Li X, Yi C. 2018. N1-methyladenosine methylome in messenger RNA and non-coding RNA. Curr Opin Chem Biol 45: 179–186. [DOI] [PubMed] [Google Scholar]
- Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian S-B. 2015. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 526: 591–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.