Abstract
MRP1/2 is a heteromeric protein complex that functions in the trypanosomatid mitochondrion as part of the RNA editing machinery, which facilitates multiple targeted insertions and deletions of uridines. MRP1/2 was shown to interact with MRB8170, which initiates RNA editing by marking pre-edited mRNAs, while TbRGG2 is required for its efficient progression on pan-edited mRNAs. Both MRP1/2 and TbRGG2 are capable of modulating RNA–RNA interactions in vitro. As determined by using iCLIP and RIP-qPCR, RNAs bound to MRP1/2 are characterized and compared with those associated with MRB8170 and TbRGG2. We provide evidence that MRP1 and MRB8170 have correlated binding and similar RNA crosslinking peak profiles over minimally and never-edited mRNAs. Our results suggest that MRP1 assists MRB8170 in RNA editing on minimally edited mRNAs.
Keywords: RNA editing, RNA binding proteins, ribonuclear protein, mitochondrion, trypanosome, iCLIP
INTRODUCTION
The trypanosomatid flagellates belong to excavate protists and constitute a major clade within the Kinetoplastea group. Their best-studied representative is Trypanosoma brucei, the causative agent of African sleeping sickness in humans and livestock. In the single mitochondrion of trypanosomatids, complex molecular machinery executes a unique type of RNA editing: insertions and deletions of uridines (Us) into most of the mitochondrial mRNA transcripts that are essential for the generation of a correct open reading frame. The unusually organized mitochondrial DNA on which these “cryptogenes” are encoded, termed kinetoplast (k)DNA, is composed of dozens of large maxicircles and thousands of small minicircles, mutually catenated into a single densely packed network (Jensen and Englund 2012; Verner et al. 2015).
The ∼27-kb-long maxicircle carries 18 protein-coding genes and/or cryptogenes, most of which constitute subunits of the respiratory complexes (Verner et al. 2015; Read et al. 2016). The ∼1-kb-long minicircles encode 50- to 70-bp-long noncoding RNAs termed guide (g) RNAs, which represent trans-acting templates to specify the precise insertions/deletions of Us. In T. brucei, nine out of 12 mRNAs, edited throughout their entire length and using multiple gRNAs, are classified as “pan-edited” transcripts. The remaining three mRNAs are subject to limited insertions and/or deletions requiring only one or two gRNAs and are termed “minimally edited.” The multi-round editing is carried out from 3′ to 5′ in pre-edited mRNAs. Finally, the six mRNAs that do not undergo editing constitute “never-edited” transcripts.
Editing enzymatic activities are provided by the RNA editing core complex (RECC or editosome) (Schnaufer et al. 2001, 2003), which lacks RNAs and processivity in vitro. Therefore, the RECC requires several additional protein complexes for execution of editing (Rusché et al. 1997; Li et al. 2009; Suematsu et al. 2016). Two of these extensively characterized factors are RBP16 and the mitochondrial RNA binding protein (RBP) complex (MRP1/2). RBP16 carries an RNA binding domain and belongs to the Y-box protein family (Hayman and Read 1999), and its depletion affects editing of CYb mRNA and decreases the levels of two never-edited transcripts (ND4 and COX1) (Pelletier and Read 2003). MRP1/2 is a heterotetrametric complex consisting of two molecules each of MRP1 and MRP2 proteins (Allen et al. 1998; Müller and Göringer 2002; Aphasizhev et al. 2003; Vondrušková et al. 2005). RNAi-mediated ablation of MRP1/2 results in a phenotype that has a limited overlap with that of RBP16 (Fisk et al. 2009). Interestingly, both RBP16 and MRP1/2 promote the annealing of gRNAs to respective cognate pre-mRNAs (Vondrušková et al. 2005; Miller et al. 2006; Schumacher et al. 2006; Zíková et al. 2008). Simultaneous depletion of both proteins leads to the destabilization and/or decrease of 3′ to 5′ editing progression of several mRNAs and results in a strong growth phenotype (Fisk et al. 2009). However, the exact function of MRP1/2 in RNA editing and processing remains elusive and is thus often ignored in current models of trypanosome RNA editing (Aphasizhev and Aphasizheva 2011; Aphasizheva and Aphasizhev 2016; Read et al. 2016).
Both RECC and MRP1/2 associate with mitochondrial RNA binding protein complex 1 (MRB1) (Weng et al. 2008) that itself comprises of two subcomplexes. The first, termed gRNA-binding complex (GRBC), is composed of six proteins involved in gRNA stability and availability, while the other, RNA editing mediator complex (REMC) (Aphasizheva and Aphasizhev 2016), is more loosely defined, likely due to many transient interactions (see Read et al. 2016 for a guide to MRB1-associated proteins alternative nomenclature). REMC functions have been largely defined by investigating the activities of individual protein subunits (Hashimi et al. 2013; Simpson et al. 2017). For instance, the functionally redundant MRB8170 and MRB4160 subunits of REMC (Kafková et al. 2012) act as editing initiators by selectively marking pre-edited mRNAs (Dixit et al. 2017; Simpson et al. 2017). However, for editing to progress, another REMC subunit, TbRGG2, is needed, which exhibits both RNA melting and annealing properties (Fisk et al. 2008; Ammerman et al. 2010).
MRB8170 is involved in editing of both pan- and minimally edited mRNAs (Kafková et al. 2012; Dixit et al. 2017), while TbRGG2 is nonessential for editing of the latter category (Fisk et al. 2008; Acestor et al. 2009; Aphasizheva et al. 2014). Hence, it is plausible that an unidentified TbRGG2-like protein assists MRB8170 over minimally edited mRNAs. Previous biochemical analyses showed that MRP1/2 and TbRGG2 possess similar functional attributes, such as the ability to modulate RNA–RNA interactions (Müller et al. 2001; Müller and Göringer 2002; Schumacher et al. 2006; Ammerman et al. 2010). However, despite these functional similarities, the ablations of TbRGG2 and MRP1/2 affect different sets of transcripts (Vondrušková et al. 2005; Fisk et al. 2008, 2009; Acestor et al. 2009).
This study aims to characterize in vivo transcriptome-wide binding of MRP1/2 and compare it with RNA molecules bound by MRB8170 and TbRGG2. For this we have used individual nucleotide resolution UV cross-linking and immuno- or affinity-precipitation (iCLIP/iCLAP). Our results demonstrate higher binding of MRP1 to minimally and never-edited mRNAs than to pan-edited mRNAs. We observed a strong correlation and similar peak profile of minimally and never-edited mRNAs crosslinked to MRP1 and MRB8170.
RESULTS
MRP1 binds more to minimally and never-edited, and less to pan-edited mRNAs
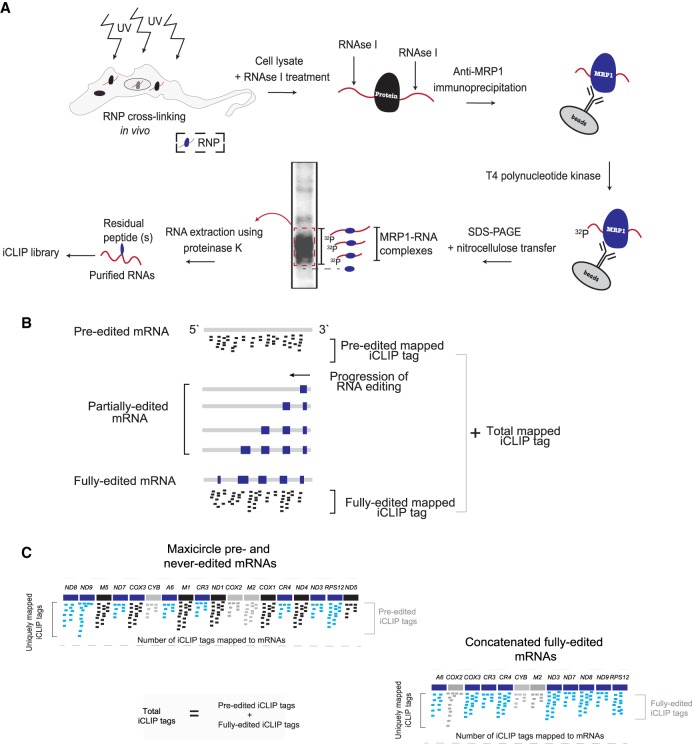
The abundances and identities of RNAs bound to an RNA editing factor can provide clues to its function. Therefore, we aimed to identify and quantify relative populations of RNAs bound by MRP1 in vivo with the highly stringent iCLIP protocol (Fig. 1A; Huppertz et al. 2014). Immunoprecipitated ribonucleoprotein complexes from UV-crosslinked cells showed a strong signal of radiolabeled RNAs at the expected size of ∼21–50 kDa (Supplemental Fig. S1A, lane 3). As expected, a mock immunoprecipitation (IP) and a non-UV-crosslink control experiment yielded no radiolabeled signal (Supplemental Fig. S1A, lanes 1 and 2). The high RNase-treated UV-crosslinked parental cell lysate was used to control for possible presence of contaminating RBPs (Supplemental Fig. S1B, lanes 1 and 2). Hence, if another RBP was present in the eluate, due to high RNase treatment a sharp radiolabeled band would appear in the autoradiogram as compared to a smear that is usually present in the low RNase-treated cell lysates. In our iCLIP pull-down, we found three additional RBPs migrating at ∼55, 75 and 150 kDa (Supplemental Fig. S1B, lanes 1 and 2). To avoid contaminating RBPs and to co-IP exclusively the MRP1-bound RNAs, the labeled ∼21–50 kDa smears from the non-UV and UV-crosslinked cells were excised and used to generate iCLIP libraries (Supplemental Fig. S1A, lanes 2 and 3). Finally, the size-selected iCLIP libraries have been verified by gel electrophoresis (Supplemental Fig. S1C).
FIGURE 1.
MRP1 iCLIP analysis. (A) Schematic depiction of the MRP1 iCLIP workflow to purify UV-crosslinked RNA-MRP1 complexes and sequence the mRNAs. (B) Schematic representation of the problem of iCLIP mapping assignment ambiguity due to the presence of editing intermediates in the transcriptome. Many of the approximately 30 to 50 nt iCLIP tags can be attributed to a partially edited or to a pre/fully edited version of a transcript. Therefore, the editing state of a transcript from which an iCLIP tag was derived cannot be unambiguously defined. Small blue boxes depict editing events. (C) Illustration of the MRP1 iCLIP read mapping strategy. Preprocessed iCLIP reads were mapped against maxicircle-encoded mRNAs. (Top) iCLIP reads mapped to reference sequences existing on the maxicircle; namely, never-edited and pre-edited mRNA sequences. (Bottom) iCLIP reads mapped to fully edited sequences of edited mRNAs. Pan-, minimally, and never-edited mRNAs are marked in blue, gray, and black, respectively.
It would be very informative to know exactly at what time during the editing process MRP1 is present on the edited mRNAs. The current iCLIP protocol generates ∼30- to 50-nt-long iCLIP tags that represent only a short segment of the transcript from which they are derived. The editing process proceeds in a 3′–5′ direction and generates a high number of editing intermediates (Fig. 1B; David et al. 2015; Simpson et al. 2016, 2017; Gerasimov et al. 2018). However, with the available iCLIP technology, it is impossible to unambiguously determine whether an iCLIP tag arose from a partially or fully edited transcript. As this ambiguity could lead to incorrect or inaccurate conclusions, we used alternative approaches to define the set of mRNAs bound by MRP1.
iCLIP libraries were mapped to both genomic and, where relevant, fully edited maxicircle sequences (Fig. 1C). The uniquely mapped reads from two biological replicates of this experiment, termed iCLIP tags, were compared. A strong correlation (r = 0.99) between them (Supplemental Fig. S2A) allowed their unification into a single data set. The MRP1 replicates yielded a total of 6,137,846 iCLIP tags, compared to 12,856 acquired from the non-UV-crosslink experiments. Due to the ∼500-fold difference in iCLIP tags, it is highly unlikely that the MRP1-IP RNAs were nonspecific. To further verify stringency of the protocol, we generated correlation plots between MRP1, MRB8170, and MRB4160 (Dixit et al. 2017). The three iCLIP libraries varied considerably in total amount of iCLIP tags, which might be due to the two-step affinity purification in the case of MRB8170 and MRB4160 (iCLAP), and the single-step IP in the case of MRP1 (iCLIP), yet it may also reflect different in vivo features of these proteins. The iCLIP libraries were normalized using the DEseq2 package and used in subsequent analysis (Love et al. 2014). As expected, a high correlation is observed between the two functional paralogs MRB8170 and MRB4160 (r = 0.94) (Supplemental Fig. S2B; Kafková et al. 2012), while MRP1 demonstrated no such correlation with MRB8170 or MRB4160 (r = 0.19) (Supplemental Fig. S2B). Therefore, the differences between the MRP1 iCLIP library and the MRB8170 and MRB4160 iCLIP libraries appear legitimate.
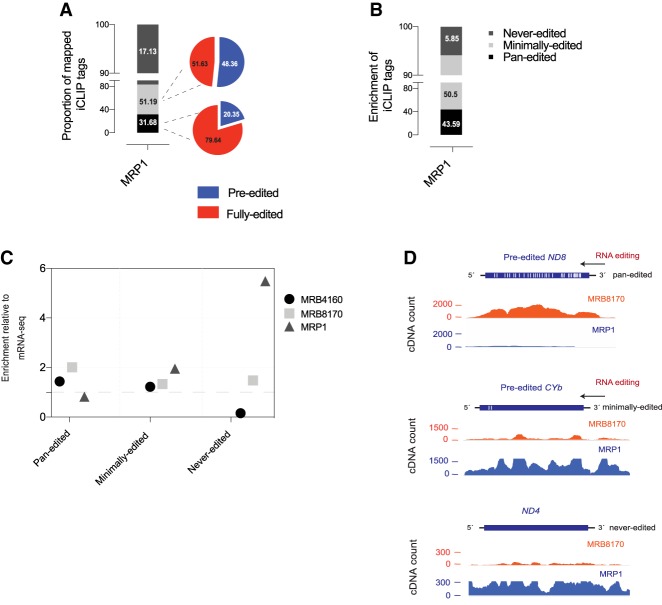
To examine MRP1 binding across mRNAs, we organized the iCLIP tags from the UV-crosslinked data set into three categories: nine pan-edited, three minimally edited, and six never-edited mRNAs (Fig. 1C). Including both pre-edited and fully edited mapped reads, 31.6% and 51.1% of iCLIP tags belonged to the pan- and minimally edited categories, respectively. The remaining 17.1% belonged to the never-edited category (Fig. 2A). In pan-edited mRNAs, the majority of mapped MRP1 iCLIP tags arise from the fully edited region (79.64%) as compared to the pre-edited one (20.35%) (Fig. 2A). For the minimally edited mRNAs, the similar breakdown resulted in 48.36% and 51.63% iCLIP tags mapped to pre- and fully edited regions, respectively. In MRB8170 the similar analysis mainly differed with MRP1only in the case of pan-edited mRNAs with the majority of iCLIP tags mapped to pre-edited mRNAs (Dixit et al. 2017). After normalization to account for varied gene lengths, these numbers were 43.5%, 50.5%, and 5.85% in the pan-, minimally, and never-edited categories, respectively (Fig. 2B).
FIGURE 2.
MRP1 iCLIP results. (A) Percentage of total MRP1 iCLIP tags obtained from each of the three mRNA categories. (B) Transcript length-normalized percentage of total MRB1 iCLIP tags obtained from each of the three mRNA categories. (C) Fold enrichment of MRB8170, MRB4160, and MRP1 iCLIP tags relative to mapped mRNA-seq read counts for three mRNA categories. (D) Genomic browser snapshots of MRP1 and MRB8170 presence on the pre-edited forms of ND8 (pan-edited) and CYB (minimally edited), and never-edited ND4 mRNAs. White lines show the approximate distribution of editing sites on ND8 and CYB. See also Supplemental Figures S1–S5.
Both normalized and non-normalized percentages of MRP1 IP-derived iCLIP tags show that binding of MRP1 to minimally and never-edited mRNAs is increased compared to its interacting partner MRB8170, for which ∼90% of iCLIP tags originate from pan-edited mRNAs (Dixit et al. 2017). To quantify and compare MRP1, MRB4160, and MRB8170 binding across the three mRNA categories, we plotted the mapped normalized iCLIP tags of these proteins relative to the published mRNA-seq mapped read counts (Lott et al. 2015) and compared the plots (Fig. 2C).
In the case of MRP1, iCLIP tags were enriched approximately fivefold in the never-edited category and approximately twofold for the minimally edited category relative to what is observed in the transcriptome steady-state population. In contrast, pan-edited mRNAs were slightly underrepresented (Fig. 2C). The situation appears to be reversed in MRB8170 and MRB4160, where iCLIP tags are most enriched in the pan-edited category compared to the other two categories. As expected, the iCLIP pre-edited mRNAs genomic browser snapshots when visualized with the same cDNA count cutoff on individual mRNAs revealed an enrichment of MRP1 iCLIP tags relative to those of MRB8170 on minimally and never-edited mRNAs (Fig. 2D; Supplemental Figs. S3–S5). Combined, these results reveal that MRP1 binds more minimally and never-edited mRNAs, and less pan-edited mRNAs compared to the same categories bound by MRB8170 or MRB4160. In short, MRP1, MRB8170 are not aligned in their RNA binding preferences.
MRP1 binding of pan-edited mRNAs correlates neither with MRB8170 nor with the extent of editing
We wanted to further dissect the binding of MRP1 to pan-edited mRNAs relative to those of its interacting partner MRB8170 (or MRB4160; the subsequent result and discussion will be limited to the functionally more active paralog MRB8170). For this comparison, mapped MRP1 iCLIP tags on pre- and fully edited sequences of individual pan-edited mRNA were merged as a single “total” category for each examined mRNA (Supplemental Fig. S6A–C).
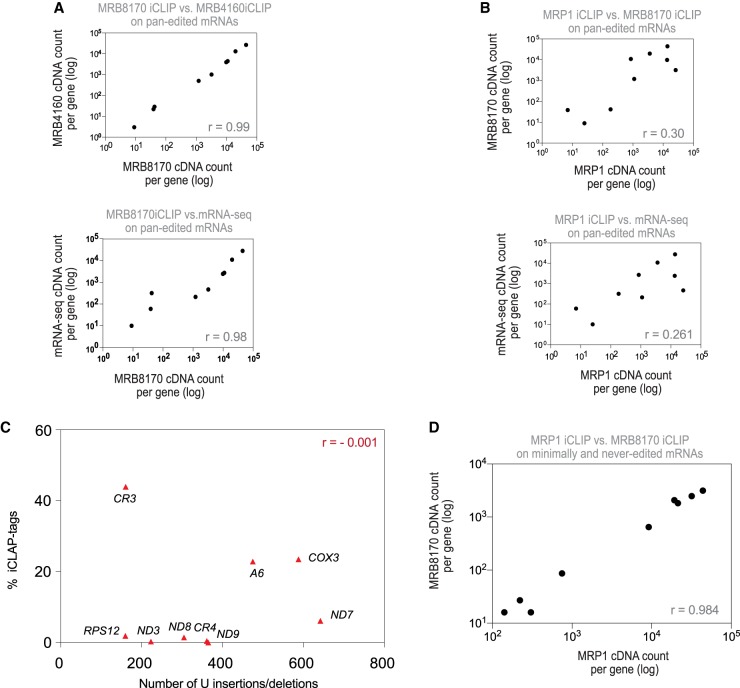
To indirectly analyze the binding relationship between the fraction of MRP1 and MRB8170 associated with pan-edited mRNAs, we calculated Pearson's correlation coefficient of iCLIP tag counts for each of the nine pan-edited mRNAs between the MRP1 and MRB8170 (Fig. 3A,B). As expected for functional paralogs (Kafková et al. 2012), there was a strong correlation (r = 0.99) between MRB8170 and MRB4160 distribution of iCLIP tags among pan-edited transcripts and this served as a positive control (Fig. 3A). MRB8170 is essential for the editing of pan-edited mRNAs (Kafková et al. 2012), so we additionally expected the abundance of each bound RNA to be correlated with that of mRNA's relative abundance as determined in RNA-seq analysis. In our data, we found this correlation to be strong (r = 0.98) (Fig. 3A). In contrast, the amount of bound MRP1 and MRB8170 among individual pan-edited transcripts was uncorrelated (r = 0.30) (Fig. 3B). Similarly, no such correlation was seen when examining pan-edited mRNA read and tag abundances from MRP1 iCLIP and mRNA-seq data (r = 0.261).
FIGURE 3.
Comparison of MRP1 and MRB8170 association with mRNAs. (A) Plots comparing the total number of iCLIP tags of each of nine pan-edited mRNAs from MRB8170 IPs to those of MRB4160 IPs, and MRB8170 iCLIP tags to total mRNA-seq data. Pearson correlation coefficients are (r) indicated. (B) As in B, from MRP1 iCLIP, MRB8170 iCLIP, and total mRNA-seq data. (C) Correlation between total MRP1 iCLIP tags (y-axis) of each pan-edited transcript and its respective number of U insertions and deletions required to generate the fully edited, translatable sequence (x-axis). Pearson correlation coefficients (r) are indicated. (D) Plots comparing the total number of iCLIP tags of the combined minimally and never-edited mRNAs from MRB8170 and MRP1 iCLIP data. The Pearson correlation coefficient (r) is indicated. See also Supplemental Figure S6.
MRP1 binding on pan-edited mRNAs appears unrelated to that of MRB8170, so we asked whether MRP1 binding instead correlates with the number of U insertions/deletions in a given transcript. In contrast to the previously observed high correlation of this parameter and iCLIP tags per transcript for MRB8170 and MRB4160 (r = 0.72, r = 0.75, respectively) (Dixit et al. 2017), MRP1 iCLIP data showed no correlation (r = −0.001) (Fig. 3C). Taken together, these analyses indicate that the extent of MRP1 binding to each pan-edited mRNA is neither governed by the degree of editing required for that transcript, nor its binding or association with MRB8170.
MRP1 association with minimally and never-edited mRNAs correlates with MRB8170
Since the binding of MRP1 to pan-edited mRNAs appears unrelated to MRB8170 binding, we explored another potential associative relationship for these proteins by examining their footprints on the three minimally edited and six never-edited mRNAs. We used the above approaches to assess MRP1 binding on minimally and never-edited mRNAs, and generated bar-plots of the results, which showed highly variable binding of MRP1 to the various minimally and never-edited mRNAs (Supplemental Fig. S6B,C).
Interestingly, unlike the case with pan-edited mRNAs, the iCLIP data revealed a high correlation of relative binding between MRP1 and MRB8170 to each of the minimally edited mRNAs (r = 0.99) (Supplemental Fig. S6D). When we performed the same MRP1 and MRB8170 binding correlations on the never-edited mRNAs, the outcome was again a high correlation (r = 0.99) (Supplemental Fig. S6E). Of course, these values are obtained with only three and six data points, respectively. To overcome this limitation, we combined minimally and never-edited mRNAs that revealed a high correlation of greater confidence between MRB8170 and MRP1 (Fig. 3D).
The ability of iCLIP to mark the RNA cross-linking sites allowed us to analyze whether the observed cDNA count-based correlation analyses between MRP1 and MRB8170 also translate into their similar mRNA binding sites. To overcome the difficulty in resolving the RNA cross-linking sites at nucleotide resolution for proteins involved in RNA editing, we relied on comparing the peak characteristics at similar nucleotide positions in two iCLIP data sets. Both MRP1 and MRB8170 iCLIP libraries vary considerably in the total amount of sequenced iCLIP tags (Dixit et al. 2017). To ensure that the highest cDNA count peaks in both MRP1 and MRB8170 data sets are visible in the genomic browser snapshots, we used different cDNA count cutoffs.
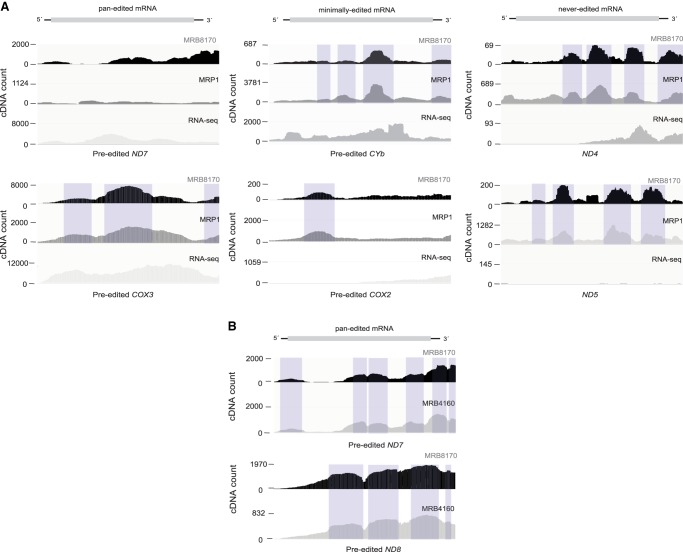
As expected from the correlation analyses, MRP1 and MRB8170 have similar peak profiles at corresponding nucleotide positions over minimally and never-edited mRNAs (Fig. 4A), whereas in the case of most pan-edited mRNAs, peak profiles differ considerably. In all three mRNA classes the iCLIP and mRNA-seq peak profiles differ markedly at the same nucleotide positions. Hence, the iCLIP peaks likely correspond to the cross-linked RNA and do not merely reflect the transcript's steady-state abundance. The MRB8170 and MRB4160 peak profiles that are highly similar over pan-edited mRNAs (Fig. 4B) served as a positive control. Altogether, the iCLIP data reflect a substantial similarity between MRB8170 and MRP1 when binding to minimally and never-edited mRNAs is concerned.
FIGURE 4.
RNA crosslinked peak profile comparison between MRP1- and MRB8170- associated mRNAs. (A) Genomic browser snapshots of MRP1 and MRB8170, as well as RNA-seq on the pre-edited forms of ND7 and COX3 (pan-edited), CYb and COX2 (minimally edited), and ND4 and ND5 (never-edited) mRNAs. The shaded peaks show the clustering of RNA crosslinked peaks at similar positions over mRNAs in MRB8170 and MRP1. (B) Genomic browser snapshots of MRB8170 and MRB4160, as well as RNA-seq on the pre-edited forms of ND7 and ND8 (pan-edited). The shaded peaks show the clustering of RNA crosslinked peaks at similar positions over mRNAs in MRB8170 and MRB4160.
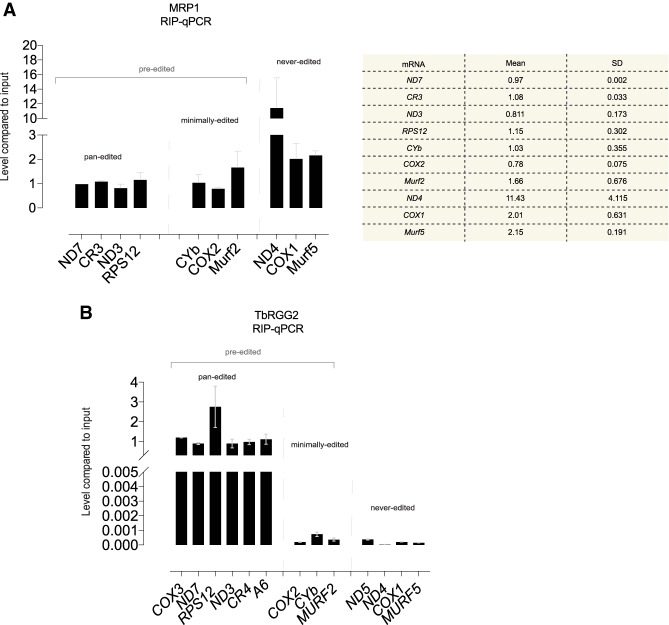
We performed RNA immunoprecipitation (RIP) followed by reverse transcription and quantitative (q) PCR on the MRP1-tagged cells to validate these results (Fig. 5A). This revealed a strong association of MRP1 with never-edited mRNAs, as these coprecipitated with MRP1 in high amounts. When comparing MRP1 binding of representative mRNAs of minimally and pan-edited categories, the binding to minimally edited Murf2 mRNAs was higher, while CYb and Cox2 showed similar enrichment for pan-edited mRNAs (Fig. 5A). Taken together, both the MRP1 qPCR and iCLIP data shows its enrichment on never-edited mRNAs relative to pan-edited ones. The discrepancy in the enrichment of minimally edited mRNAs between the MRP1 iCLIP and RIP-qPCR experiments might be due to UV-crosslinking and RNAse-I treatment in the former technique, which allow higher accuracy in quantifying the protein-bound RNA footprints as compared to RIP-qPCR. In summary, in the never-edited and minimally edited categories, MRB8170 and MRP1 mRNA binding exhibit a correlative relationship that is not present when analyzed within the pan-edited mRNA category.
FIGURE 5.
RIP-qPCR for MRP1 and TbRGG2. (A) MRP1 RIP-qPCR. Bar plots show the relative amount of representative pre- and never-edited mRNAs coimmunoprecipitated with MRP1. Abundance of each mRNA is presented relative to the abundance of the same mRNA recovered from 1% of input lysate. Two biological replicates (collected on different days) were performed in triplicate. Error bars show standard deviation. Table shows the averages and standard deviation of the tested mRNAs. (B) TbRGG2 RIP-qPCR. As in A.
Differential recruitment of TbRGG2, MRP1, and MRB8170
MRB8170 interacts with both MRP1 and the editing processivity factor TbRGG2 vital for pan-editing (Fisk et al. 2008; Ammerman et al. 2010; Kafková et al. 2012; Aphasizheva et al. 2014; Dixit et al. 2017). Therefore, after probing mRNA binding preferences of MRB8170 and MRP1, we also explored those of TbRGG2. The latter protein was suspected to preferentially bind pan-edited mRNAs, since its depletion does not impact minimally and never-edited transcripts (Fisk et al. 2008; Acestor et al. 2009; Aphasizheva et al. 2014). Indeed, TbRGG2 binds almost exclusively pan-edited mRNAs (Fig. 5B).
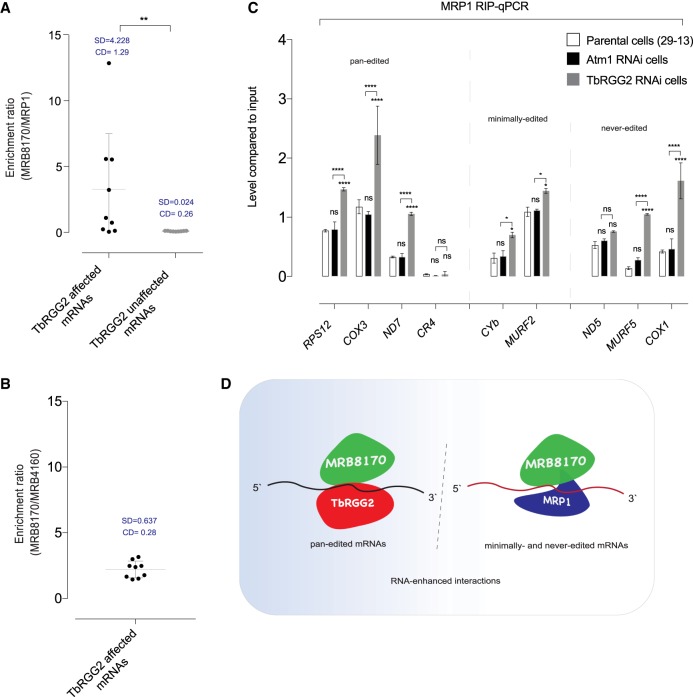
In the iCLIP peak profile analysis, RNA cross-linked peaks between MRP1 and MRB8170 over certain pan-edited mRNAs raised a prospect of a sub-fraction of the MRP1/2 complex being associated with MRB8170 on pan-edited mRNAs, which would not appear in the analysis of the entire bound pan-edited mRNA populations. To explore this indirectly, we separately tested the variability of the protein enrichment ratios over nine pan-edited mRNAs (TbRGG2-affected mRNAs) and minimally and never-edited mRNAs (TbRGG2-unaffected mRNAs).
The collective enrichment ratios of transcripts associated with MRB8170 versus MRP1 significantly differ, as assessed by iCLIP tag abundances (Fig. 6A). This was expected since MRB8170 is predominantly associated with pan-edited mRNAs and MRP1 with the other classes. Individual transcripts affected by the depletion of TbRGG2 show a large variation in the enrichment ratio. However, the same pan-edited transcript analysis comparing MRB8170 to its functional homolog MRB4160 instead of MRP1 revealed negligible variability and served as a negative control (Fig. 6B). This result is consistent with MRP1 being associated with MRB8170-bound transcripts to a different degree for each pan-edited mRNA, presumably allowing TbRGG2 differential access to MRB8170 on these transcripts.
FIGURE 6.
MRP1 and TbRGG2 recruited to 8170 on mRNAs in a class-specific manner. (A) Enrichment ratios of bound MRB8170 relative to bound MRP1 (y-axis) of each pan-edited, minimally edited, and never-edited mRNA. The first column includes all pan-edited mRNAs; these are affected by TbRGG2 depletion. The second column includes minimally and never-edited mRNAs; these are unaffected by TbRGG2 depletion (Mann–Whitney test; [**] P < 0.005; SD, standard deviation; CV, coefficient of variation). (B) As in A, except that the enrichment ratio compares bound MRB8170 relative to bound MRB4160. MRB4160 does not bind to never-edited mRNAs. (C) Relative amounts of MRP1 coimmunoprecipitated mRNAs in three different conditions using RIP-qPCR (Tukey's multiple comparison test, [*] P < 0.05, [***] P < 0.001, [****] P < 0.0001, biological replicates n = 2–3). Each mRNA abundance is presented relative to the abundance of the same mRNA recovered from 1% of input lysate. (D) Combinatorial interplay involving three RNA-binding proteins. In pan-edited transcripts, MRB8170 marks the pre-edited mRNAs and is followed by TbRGG2 to allow efficient progression of editing. Since TbRGG2 is nonessential for minimally and never-edited mRNAs, MRP1 with similar functional capabilities to modulate RNA–RNA interactions operates on these transcripts.
Next, we measured the relative abundances of mRNAs coimmunoprecipitated by MRP1 in cells with down-regulated TbRGG2 and compared them to abundances when TbRGG2 was present (Fig. 6C). Cells depleted for Atm1, an inner mitochondrial membrane protein that neither affects gene expression (Horáková et al. 2015), nor presumably competes with MRP1, were used as a control. In the Atm1-depleted cells, MRP1 coimmunoprecipitated similar levels of mitochondrial mRNAs as in the parental cells. In the absence of TbRGG2, we found an increased coimmunoprecipitation of RPS12, COX3, ND7, MURF5, and COX1 with MRP1 (Fig. 6C). The increase was modest for minimally edited transcripts (CYb and MURF2), while pan-edited CR4 and never-edited ND5 mRNAs showed no significant increase. Taken together, our results favor a scenario, in which MRP1 and TbRGG2 exhibit complimentary behavior with each other to bind with MRB8170 in a transcript-specific manner (Fig. 6D).
DISCUSSION
The functional paralogs MRB8170 and MRB4160 from the REMC act as initiation factors and their depletion impacts RNA editing of both pan- and minimally edited mRNAs (Kafková et al. 2012; Dixit et al. 2017; Simpson et al. 2017). Another REMC-associated protein, TbRGG2, assists in the progression of editing past intrinsic pause sites, yet it is not essential for minimally and never-edited mRNAs (Fisk et al. 2008; Acestor et al. 2009; Aphasizheva et al. 2014). Hence, it is plausible that an as yet unidentified protein acting in a way reminiscent of TbRGG2 assists the MRB1 complex in editing of minimally edited mRNAs.
MRP1/2 exhibit similar functional capabilities to modulate RNA–RNA interactions as TbRGG2 (Müller et al. 2001; Müller and Göringer 2002; Schumacher et al. 2006; Ammerman et al. 2010), although different subsets of mRNAs are affected following depletion of these proteins (Vondrušková et al. 2005; Fisk et al. 2008, 2009; Acestor et al. 2009), which makes it an ideal protein complex to assist MRB8170 over minimally edited mRNAs. Therefore, this study aims to identify and quantify transcripts associated with MRP1 and compare them with those bound to MRB8170 and TbRGG2. A tight mutual dependence between MRP1 and MRP2 makes their phenotypic effects highly similar (Vondrušková et al. 2005; Zíková et al. 2008), so we have extrapolated MRP1-specific results to be true for the entire MRP1/2 complex. However, this should eventually be confirmed by performing MRP2 iCLIP analysis.
We report efficient binding of MRP1 to minimally and never-edited mRNAs. The lower levels of MRP1 associated with pan-edited mRNAs suggest the possibility that TbRGG2 binds a substantial fraction of MRB8170 on this category of transcripts. In support of this conclusion, we showed an almost exclusive association of TbRGG2 with pan-edited mRNAs. Furthermore, in spite of both MRB8170 and MRP1 being associated with pan-edited mRNAs, there is no apparent correlation between the two proteins in the binding of individual mRNAs to greater or lesser extent from the pan-edited category. If we assume that most MRB8170 on pan-edited mRNAs binds either MRP1/2 or TbRGG2 but not both at the same time, then the most reasonable explanation for this lack of MRB8170-MRP1 correlation is that a substantial fraction of the former protein is bound to TbRGG2. The iCLIP data support this interpretation, since they revealed just a few RNA cross-linked peaks shared between MRB8170 and MRP1 over pan-edited mRNAs. However, the COX3 transcript is an outlier in this respect, as both proteins exhibit a similar RNA cross-linking peak profile, suggesting their involvement in COX3 editing. The iCLIP data derived from COX3 fits nicely with the previously described effect of MRP1/2 depletion on the editing and/or stability of COX3 (Fisk et al. 2009).
In contrast to pan-edited mRNAs, we detected a strong correlation between MRP1 and MRB8170 binding of minimally and never-edited mRNAs. It seems that MRB8170 is associated with MRP1 on minimally and never-edited mRNAs, while TbRGG2, a protein essential for the progression of editing of pan-edited mRNAs (Ammerman et al. 2010; Simpson et al. 2017), is a major binding partner of MRB8170 on this class of transcripts (Foda et al. 2012). Moreover, MRB8170 and MRP1 have very similar peak profiles over minimally and never-edited mRNAs as compared to pan-edited ones, nicely supporting the above assumption. Similar to COX3, we observed coordinated clustering of cross-linked RNA peaks by MRB8170 and MRP1 over CYb, which is another transcript with its editing disrupted in cells depleted for these two proteins (Vondrušková et al. 2005; Fisk et al. 2009). Overall, the iCLIP data suggest that a similar cross-linked peak profile of MRB8170 and MRP1 over specific mRNAs may reflect their function in editing. Moreover, binding of both proteins beyond the regions undergoing editing is likely associated with their involvement in other post-transcriptional events such as gRNA–mRNA annealing and RNA stability.
Since the enrichment ratios of MRP1 with respect to MRB8170 varies greatly among pan-edited mRNAs, it is possible that a high ratio of bound MRP1/2 with respect to MRB8170 on an mRNA will inversely affect the TbRGG2 versus MRB8170 ratio, interfering with efficient processing. Conversely, a low MRP1/2 to MRB8170 ratio on a given transcript likely means higher TbRGG2 presence, in return positively impacting editing processing efficiency. Therefore, MRP1 may influence editing in either a positive or negative way for pan-edited transcripts in regions where MRB8170 and MRP1 share just a few RNA cross-linking peaks. The MRP1/2 knockdown phenotypes in both the procyclic and bloodstream stages of T. brucei support this assertion. In the former stage, the depletion of MRP1/2 substantially reduced the level of edited RPS12, while edited ND7 increased approximately fivefold. In the bloodstream stage, editing of A6 was decreased, yet the efficiency of ND7 editing was increased (Vondrušková et al. 2005). To test, we measured the amount of RNA bound by MRP1 in cells depleted of TbRGG2. TbRGG2 silencing resulted in a significant increase in abundance of the majority of tested pan-edited mRNAs, consistent with this model.
Interestingly, we have also found a higher amount of bound minimally and never-edited mRNAs to MRP1 in TbRGG2-depleted cells. The iCLIP data is not informative as to the abundance of non-RNA-containing protein complexes, some of which likely contain MRB8170 and TbRGG2 (Kafková et al. 2012; Aphasizheva et al. 2014). MRB8170 shares an RNA-enhanced interaction with both TbRGG2 and MRP1 (Kafková et al. 2012; Aphasizheva et al. 2014; Dixit et al. 2017). Therefore, the RNase-resistant reservoir of these proteins or TbRGG2 effects on other proteins could explain the increased abundance of never- and minimally edited mRNAs coimmunoprecipitated with MRP1.
An alternative explanation of the obtained results is that the role of MRB8170 bound to MRP1/2 is to stabilize most of the translation-competent mRNAs, while the fraction bound to TbRGG2 is essential for editing. Most minimally and never-edited mRNAs are likely translationally competent, while in the case of pan-edited mRNAs, their fully edited versions represent, in a transcript-dependent fashion, different fractions of the total reads (Zimmer et al. 2018). Therefore, a smaller fraction of TbRGG2 would along with MRB8170 occupy minimally and never-edited RNAs, while a bigger fraction of this protein would bind pan-edited mRNAs. In contrast, we predict that most MRP1 associated with MRB8170 binds minimally and never-edited mRNAs.
Results from our study are consistent with or add clarity to observations from previous studies, such as TbRGG2 depletion resulting in dramatic decreases in pan-edited mRNAs and a moderate increase of never-, minimally, and pre-edited mRNAs (Fisk et al. 2008; Acestor et al. 2009). According to MRP1 RIP-qPCR in TbRGG2 depleted cells, the lack of TbRGG2 will cause an increase in majority of MRP1 associated mRNAs. MRP1/2's implied role in mRNA stability (Vondrušková et al. 2005; Zíková et al. 2008) may provide a rationale for the observed moderate stabilization of never-, minimally and pre-edited mRNAs in TbRGG2-depleted cells (Fisk et al. 2008; Acestor et al. 2009). Simultaneously, the absence of TbRGG2 will halt editing at pause sites leading to diminished levels of fully edited pan-edited mRNAs observed in the same studies (Fisk et al. 2008; Ammerman et al. 2010; Simpson et al. 2017). The unimpeded editing of minimally edited mRNAs observed in TbRGG2-depleted cells (Fisk et al. 2008; Foda et al. 2012) is to be expected considering that we found almost no TbRGG2 associated with this category of transcripts; thus, TbRGG2 plays no role in their editing.
Intriguingly, both MRP1/2 and TbRGG2 exhibit gRNA/mRNA annealing activity and share similar RNA modulated interactions with the GRBC sub-complex of MRB1 (Schumacher et al. 2006; Zíková et al. 2008; Aphasizheva et al. 2014). Therefore, we speculate that MRP1/2 can replace TbRGG2 bound to MRB8170 as editing progresses, narrowing the indispensability of TbRGG2 exclusively to the editing pause sites. If correct, it further explains why TbRGG2 mostly affects editing of pan-edited mRNAs and much less that of minimally edited ones (Fisk et al. 2008), as pan-edited mRNAs harbor more editing sites. Additionally, the highly correlated binding of MRP1 and MRB8170 on minimally edited transcripts might be important for some yet unidentified aspect of their proper editing, as involvement of both MRP1/2 and MRB8170 in Murf2 and CYb has previously been identified (Vondrušková et al. 2005; Fisk et al. 2009; Kafková et al. 2012). The same reasoning might explain the observed lack of overlap in positions where editing gets stalled on mRNAs in MRB8170- compared to TbRGG2-depleted cells (Simpson et al. 2017). MRB8170 is present throughout editing (Dixit et al. 2017), although bound to different partners, while the TbRGG2 essential role might be limited to editing pause sites. Future TbRGG2 iCLIP analysis would be highly useful to test this model, particularly if performed in both the presence and absence of MRP1/2. Similarly, analysis of MRP1 iCLIP tags from a TbRGG2-depleted environment could be compared to results obtained here. Such experiments would allow higher resolution in tracking and quantitating the amount of cross-linked RNA to the protein of investigation in absence of the other.
The RBP16 similar function in RNA stability and annealing as that of MRP1/2 adds another layer of complexity to these factors overlapping roles (Fisk et al. 2009). Functional redundancy between MRP1/2 and RBP16 was invoked to explain the limited effects of their individual depletions and dramatic effects of co-depletion that leads to destabilization of several transcripts from all three mRNA classes (Fisk et al. 2009). Specifically, two of the three strongly down-regulated never-edited mRNAs (ND4, ND5) in the MRP1/2-depleted cells (Vondrušková et al. 2005) showed significant enrichment in the MRP1 iCLIP data (Supplemental Fig. S6B). A future study involving RBP16 iCLIP could help in better understanding of RBP16 and MRP1/2 unique and shared roles over individual mRNAs. Additionally, we will pursue MRP1/2 role(s) in RNA stability of all three classes of mRNAs in future studies. In conclusion, we provide evidence supporting the differential recruitment of MRP1 and TbRGG2 to MRB8170 in a transcript-specific manner aided by an RNA-enhanced interaction (Fig. 6D).
MATERIALS AND METHODS
T. brucei culture conditions and generation of cell lines
Previously constructed procyclic form (PF) T. brucei strain 29–13 cell lines harboring MRB8170/MRB4160 (Kafková et al. 2012), MRP1 with TAP tags (Zíková et al. 2008), plus those for simultaneous inducible RNAi silencing of MRB8170 and MRB4160 (Kafková et al. 2012), Atm1 (Horáková et al. 2015), or TbRGG2 (Fisk et al. 2008) were used as previously described. RNAi was induced by the addition of 1 µg/mL tetracycline to culture media. Cell densities were measured with a Z2 cell counter (Beckman Coulter) and were maintained in the exponential mid-log growth phase at 27°C with constant shaking.
In vivo UV-cross linking and immunoprecipitation (iCLIP)
Parental 29–13 cell line was irradiated once with 800 J/cm2 UV light (254 nm) to covalently crosslink protein–RNA in vivo (Dixit et al. 2017). The UV-crosslinked cell lysates treated with low concentration of RNase I were used to immunopurify RNA bound to MRP1 using protein G Dynabeads coupled with anti-MRP1 antibodies (Vondrušková et al. 2005). Two replicates of IPs, one each for MRP1 (UV-crosslinked 29–13 cells) and negative control (non-UV crosslinked 29–13 cells), were performed. Immunoblots confirmed the specificity and efficiency of IP. Crosslinked immunopurified RNA was digested to size range of 60–150 nt and reverse transcribed to generate iCLIP libraries. All the buffers, reagents, adapter oligonucleotides, and reverse transcription primers for amplification were as described elsewhere (Huppertz et al. 2014). The iCLIP libraries were sequenced using Illumina Hi-Seq 2000 (single-end sequencing, 75 nt length). The MRP1 and MRB8170/MRB4160 iCLIP protocol differed at the step of protein–RNA complex purification. In the case of MRB8170/MRB4160, two-step purification was used, which involves IgG sepharose and His-tag isolation Dynabeads to purify the TAP-his-MRB8170 or MRB4160-RNA complex (Dixit et al. 2017), while anti-MRP1 coated protein-G Dynabeads were used to isolate MRP1-bound RNA. The downstream iCLIP library preparation steps were the same in both protocols.
Computational analysis of next-generation sequencing data
The processing of iCLIP tags was done as previously described (Dixit et al. 2017). For all subsequent analyses, replicates were merged into one iCLIP data set. In total, we obtained 6,137,846 uniquely aligned iCLIP tags for MRP1 (1,238,207 and 5,899,639 for the individual replicates). The control library from non-UV-crosslinked parental cells contained 12,856 uniquely mapping iCLIP tags (7588 and 5268 for the individual replicates). Already published MRB8170, MRB4160, and mRNA-seq libraries were used in this study (Lott et al. 2015; Dixit et al. 2017). Mapping of iCLIP data was done using Bowtie 2 (Langmead et al. 2009). For normalization and comparison of different iCLIP libraries, the Bioconductor package DESeq2 was used (Love et al. 2014), which provides a statistical method based on negative binomial linear model testing differential expression (for a detailed workflow see: http://bioconductor.org/packages/devel/bioc/vignettes/DESeq2/inst/doc/DESeq2.html). The visualization of iCLIP and RNA-seq data was performed with the Integrative Genomics Viewer (https://www.broadinstitute.org/igv/) (Thorvaldsdottir et al. 2013).
RNA immunoprecipitation and reverse transcription-quantitative PCR
TbRGG2 and Atm1 knockdown cells harvested after 24 h of RNAi induction were resuspended in RIP lysis buffer (50 mM Tris pH 7.5, 250 mM NaCl, 1 mM DTT, 10% glycerol, 0.5% NP-40) containing cOmplete EDTA-free Protease Inhibitor Cocktail (Roche) and RNaseOUT (100 units; Life Technologies). The supernatant was Turbo DNase treated (Life Technologies) and subjected to IP using Protein G Dynabeads coated with anti-MRP1 antibodies (Vondrušková et al. 2005) for 2 h at 4°C. Ten percent of the supernatant was reserved as input to generate cDNA for normalizing the respective IP samples. After IP, the beads were washed with RIP wash buffer (50 mM Tris pH7.5, 1 M NaCl, 0.5% NP40, and 0.1% SDS) three times before phenol extraction of RNA. RNA obtained from the supernatant (input) and eluate (output) was transcribed into cDNA using the QuantiTect Reverse Transcription Kit (Qiagen) and further analyzed by qPCR (Light Cycler, Roche). We used primers described previously (Carnes et al. 2005) that anneal to the specified mRNAs. The PCR conditions were 95°C for 10 min, followed by 40 cycles (95°C for 15 sec, 60°C for 1 min). Relative ratios were calculated for each mRNA from the immunoprecipitated samples normalized against its respective input. A separate IP strategy was used for MRP1 TAP tag RIP-qPCR by using IgG-Sepharose beads (GE Healthcare) with the downstream steps as described above. The TbRGG2 RIP-qPCR was performed as above except that TbRGG2 antibodies coated the Protein G Dynabeads. The relative abundances were determined using LightCycler480 instrumentation and software version 1.5.1.62 (Roche) with relative quantification settings.
Statistical analysis
All plots, graphs and statistical analyses were done using GraphPad Prism 5.00 (GraphPad).
DATA DEPOSITION
All the iCLIP sequences are available at ArrayExpress with accession number E-MTAB-4934 (http://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-4934/).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sara Zimmer (University of Minnesota Medical School, Duluth, USA) for suggestions and critical reading of the manuscript and Andreas Dahl (Technische Universitat Dresden, Germany) for sequencing. Funding for this study was provided by the Czech Grant Agency (15-21974S and 16-18699S), the European Research Council CZ (LL1601), and the European Regional Development Fund, project OPVVV 16_019/0000759.
Author contributions: S.D. performed the experiments. Both authors designed experiments, analyzed data, and wrote the manuscript.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.066233.118.
REFERENCES
- Acestor N, Panigrahi AK, Carnes J, Zíková A, Stuart KD. 2009. The MRB1 complex functions in kinetoplastid RNA processing. RNA 15: 277–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen TE, Heidmann S, Reed R, Myler PJ, Göringer HU, Stuart KD. 1998. Association of guide RNA binding protein gBP21 with active RNA editing complexes in Trypanosoma brucei. Mol Cell Biol 18: 6014–6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammerman ML, Presnyak V, Fisk JC, Foda BM, Read LK. 2010. TbRGG2 facilitates kinetoplastid RNA editing initiation and progression past intrinsic pause sites. RNA 16: 2239–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R, Aphasizheva I. 2011. Uridine insertion/deletion editing in trypanosomes: a playground for RNA-guided information transfer. Wiley Interdiscip Rev RNA 2: 669–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R, Aphasizheva I, Nelson RE, Simpson L. 2003. A 100-kD complex of two RNA-binding proteins from mitochondria of Leishmania tarentolae catalyzes RNA annealing and interacts with several RNA editing components. RNA 9: 62–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizheva I, Aphasizhev R. 2016. U-insertion/deletion mRNA-editing holoenzyme: definition in sight. Trends Parasitol 32: 144–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizheva I, Zhang L, Wang X, Kaake RM, Huang L, Monti S, Aphasizhev R. 2014. RNA binding and core complexes constitute the U-insertion/deletion editosome. Mol Cell Biol 34: 4329–4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnes J, Trotter JR, Ernst NL, Steinberg A, Stuart K. 2005. An essential RNase III insertion editing endonuclease in Trypanosoma brucei. Proc Natl Acad Sci 102: 16614–16619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David V, Flegontov P, Gerasimov E, Tanifuji G, Hashimi H, Logacheva MD, Maruyama S, Onodera NT, Gray MW, Archibald JM, et al. 2015. Gene loss and error-prone RNA editing in the mitochondrion of Perkinsela, an endosymbiotic kinetoplastid. mBio 6: e01498-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit S, Müller-Mcnicoll M, David V, Zarnack K, Ule J, Hashimi H, Lukeš J. 2017. Differential binding of mitochondrial transcripts by MRB8170 and MRB4160 regulates distinct editing fates of mitochondrial MRNA in trypanosomes. mBio 8: e02288-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk JC, Ammerman ML, Presnyak V, Read LK. 2008. TbRGG2, an essential RNA editing accessory factor in two trypanosoma brucei life cycle stages. J Biol Chem 283: 23016–23025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk JC, Presnyak V, Ammerman ML, Read LK. 2009. Distinct and overlapping functions of MRP1/2 and RBP16 in mitochondrial RNA metabolism. Mol Cell Biol 29: 5214–5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foda BM, Downey KM, Fisk JC, Read LK. 2012. Multifunctional G-rich and RRM-containing domains of TbRGG2 perform separate yet essential functions in trypanosome RNA editing. Eukaryot Cell 11: 1119–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimov ES, Gasparyan AA, Kaurov I, Tichý B, Logacheva MD, Kolesnikov AA, Lukeš J, Yurchenko V, Zimmer SL, Flegontov P. 2018. Trypanosomatid mitochondrial RNA editing: dramatically complex transcript repertoires revealed with a dedicated mapping tool. Nucleic Acids Res 46: 765–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimi H, Zimmer SL, Ammerman ML, Read LK, Lukeš J. 2013. Dual core processing: MRB1 is an emerging kinetoplast RNA editing complex. Trends Parasitol 29: 91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayman ML, Read LK. 1999. Trypanosoma brucei RBP16 is a mitochondrial Y-box family protein with guide RNA binding activity. J Biol Chem 274: 12067–12074. [DOI] [PubMed] [Google Scholar]
- Horáková E, Changmai P, Paris Z, Salmon D, Lukeš J. 2015. Simultaneous depletion of Atm and Mdl rebalances cytosolic Fe-S cluster assembly but not heme import into the mitochondrion of Trypanosoma brucei. FEBS J 282: 4157–4175. [DOI] [PubMed] [Google Scholar]
- Huppertz I, Attig J, D'Ambrogio A, Easton LE, Sibley CR, Sugimoto Y, Tajnik M, König J, Ule J. 2014. iCLIP: protein–RNA interactions at nucleotide resolution. Methods 65: 274–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RE, Englund PT. 2012. Network news: the replication of kinetoplast DNA. Annu Rev Microbiol 66: 473–491. [DOI] [PubMed] [Google Scholar]
- Kafková L, Ammerman ML, Faktorová D, Fisk JC, Zimmer SL, Sobotka R, Read LK, Lukeš J, Hashimi H. 2012. Functional characterization of two paralogs that are novel RNA binding proteins influencing mitochondrial transcripts of Trypanosoma brucei. RNA 18: 1846–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Ge P, Hui WH, Atanasov I, Rogers K, Guo Q, Osato D, Falick AM, Zhou ZH, Simpson L. 2009. Structure of the core editing complex (L-complex) involved in uridine insertion/deletion RNA editing in trypanosomatid mitochondria. Proc Natl Acad Sci 106: 12306–12310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lott K, Mukhopadhyay S, Li J, Wang J, Yao J, Sun Y, Qu J, Read LK. 2015. Arginine methylation of DRBD18 differentially impacts its opposing effects on the trypanosome transcriptome. Nucleic Acids Res 43: 5501–5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MM, Halbig K, Cruz-Reyes J, Read LK. 2006. RBP16 stimulates trypanosome RNA editing in vitro at an early step in the editing reaction. RNA 12: 1292–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller UF, Göringer HU. 2002. Mechanism of the gBP21-mediated RNA/RNA annealing reaction: matchmaking and charge reduction. Nucleic Acids Res 30: 447–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller UF, Lambert L, Göringer HU. 2001. Annealing of RNA editing substrates facilitated by guide RNA-binding protein gBP21. EMBO J 20: 1394–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier M, Read LK. 2003. RBP16 is a multifunctional gene regulatory protein involved in editing and stabilization of specific mitochondrial mRNAs in Trypanosoma brucei. RNA 9: 457–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read LK, Lukeš J, Hashimi H. 2016. Trypanosome RNA editing: the complexity of getting U in and taking U out. Wiley Interdiscip Rev RNA 7: 33–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusché LN, Cruz-Reyes J, Piller KJ, Sollner-Webb B. 1997. Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J 16: 4069–4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaufer A, Panigrahi AK, Panicucci B, Igo RP, Salavati R, Stuart K. 2001. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science 291: 2159–2162. [DOI] [PubMed] [Google Scholar]
- Schnaufer A, Ernst NL, Palazzo SS, O'Rear J, Salavati R, Stuart K. 2003. Separate insertion and deletion subcomplexes of the Trypanosoma brucei RNA editing complex. Mol Cell 12: 307–319. [DOI] [PubMed] [Google Scholar]
- Schumacher MA, Karamooz E, Zíková A, Trantírek L, Lukeš J. 2006. Crystal structures of T. brucei MRP1/MRP2 guide-RNA binding complex reveal RNA matchmaking mechanism. Cell 126: 701–711. [DOI] [PubMed] [Google Scholar]
- Simpson RM, Bruno AE, Bard JE, Buck MJ, Read LK. 2016. High-throughput sequencing of partially edited trypanosome mRNAs reveals barriers to editing progression and evidence for alternative editing. RNA 22: 677–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson RM, Bruno AE, Chen R, Lott K, Tylec BL, Bard JE, Sun Y, Buck MJ, Read LK. 2017. Trypanosome RNA editing mediator complex proteins have distinct functions in gRNA utilization. Nucleic Acids Res 45: 7965–7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suematsu T, Zhang L, Aphasizheva I, Monti S, Huang L, Wang Q, Costello CE, Aphasizhev R. 2016. Antisense transcripts delimit exonucleolytic activity of the mitochondrial 3′ processome to generate guide RNAs. Mol Cell 61: 364–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14: 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verner Z, Basu S, Benz C, Dixit S, Dobáková E, Faktorová D, Hashimi H, Horáková E, Huang Z, Paris Z, et al. 2015. Malleable mitochondrion of Trypanosoma brucei. Int Rev Cell Mol Biol 315: 73–151. [DOI] [PubMed] [Google Scholar]
- Vondrušková E, van den Burg J, Zíková A, Ernst NL, Stuart K, Benne R, Lukeš J. 2005. RNA interference analyses suggest a transcript-specific regulatory role for mitochondrial RNA-binding proteins MRP1 and MRP2 in RNA editing and other RNA processing in Trypanosoma brucei. J Biol Chem 280: 2429–2438. [DOI] [PubMed] [Google Scholar]
- Weng J, Aphasizheva I, Etheridge RD, Huang L, Wang X, Falick AM, Aphasizhev R. 2008. Guide RNA-binding complex from mitochondria of trypanosomatids. Mol Cell 32: 198–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zíková A, Kopečná J, Schumacher MA, Stuart K, Trantírek L, Lukeš J. 2008. Structure and function of the native and recombinant mitochondrial MRP1/MRP2 complex from Trypanosoma brucei. Int J Parasitol 38: 901–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer SL, Simpson RM, Read LK. 2018. High throughput sequencing revolution reveals conserved fundamentals of U-indel editing. WIREs RNA 10.1002/wrna.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.