Abstract
BACKGROUND
The aim of this study was to comprehensively test the associations of genetic variants of nicotinamide adenine dinucleotide phosphate (NADPH) oxidase-related genes with blood pressure (BP) responses to dietary sodium intervention in a Chinese population.
METHODS
We conducted a 7-day low-sodium intervention followed by a 7-day high-sodium intervention among 1,906 participants in rural China. BP measurements were obtained at baseline and each dietary intervention using a random-zero sphygmomanometer. Linear mixed-effect models were used to assess the additive associations of 63 tag single-nucleotide polymorphisms in 11 NADPH oxidase-related genes with BP responses to dietary sodium intervention. Gene-based analyses were conducted using the truncated product method. The Bonferroni method was used to adjust for multiple testing in all analyses.
RESULTS
Systolic BP (SBP) response to high-sodium intervention significantly decreased with the number of minor T allele of marker rs6967221 in RAC1 (P = 4.51 × 10−4). SBP responses (95% confidence interval) for genotypes CC, CT, and TT were 5.03 (4.71, 5.36), 4.20 (3.54, 4.85), and 0.56 (−1.08, 2.20) mm Hg, respectively, during the high-sodium intervention. Gene-based analyses revealed that RAC1 was significantly associated with SBP response to high-sodium intervention (P = 1.00 × 10−6) and diastolic BP response to low-sodium intervention (P = 9.80 × 10−4).
CONCLUSIONS
These findings suggested that genetic variants of NADPH oxidase-related genes may contribute to the variation of BP responses to sodium intervention in Chinese population. Further replication of these findings is warranted.
Keywords: blood pressure, genetic association, hypertension, NADPH oxidase, salt sensitivity
Essential hypertension is a major risk factor of cardiovascular disease and has become a serious public health problem.1 An excess of dietary salt is one of the established environmental factors of hypertension.2,3 Blood pressure (BP) response to dietary sodium intake varies considerably among individuals, termed salt sensitivity of BP (SSBP).4 The Genetic Epidemiology Network of Salt-Sensitivity (GenSalt) study indicates that the heritability of SSBP ranges from 20% to 33% among people from northern rural area in China.3 However, the genomic mechanisms of SSBP remain to be elucidated.
Reactive oxygen species, including superoxide anion (O2−), hydrogen peroxide (H2O2), and hydroxyl anion (OH−), play an important role in hypertension.5,6 Increased renal and vascular oxidative stress is implicated in SSBP.7 Nicotinamide adenine dinucleotide phosphate (NADPH) oxidases (Noxs) are major sources to increase the level of reactive oxygen species.8 NADPH oxidase is a multicomponent enzyme consisting of at least one membrane-bound NOX subunit. The classical NADPH oxidase is comprised of 2 integral membrane proteins, the catalytic subunits gp91phox (now referred to as NOX2) and p22phox (CYBA), and the cytosolic components p47phox (NCF1), p67phox (NCF2), p40phox (NCF4), and Rac1 (RAC1) or 2 (RAC2).8 In mammalian, 7 distinct NOX genes (NOX1 to 5 and DUOX1 and 2) have been identified.9 Among them, only NOX1, NOX2, NOX4, and NOX5 have been identified in the cardiovascular–renal systems and have been implicated in the pathophysiology of cardiovascular and renal disease.10,11
Recently, evidence from Dahl salt-sensitive (SS) rats revealed that NOX4, NCF2, and RAC1 contributed to the development of salt-induced hypertension.12–14 For instance, SSNox4−/− rats had reduction of renal injury and attenuation of BP response to high salt compared with SS rats.12 In comparison to a salt-resistant strain, the higher expression of NCF2 was associated with higher NADPH oxidase activity and salt sensitivity in SS rats.13 In Dahl SS rats, high-salt status activates renal RAC1 in SS hypertension, leading to high BP and renal damage.14 Previous candidate gene studies have reported associations between common variants of the CYBA gene and BP, hypertension, and SSBP.15–17 However, the comprehensive relationship of NADPH oxidase-related genes polymorphisms with SSBP had not been investigated. Thus, we systematically selected 11 NADPH oxidase-related genes (Table 1) and conducted single-marker and gene-based analyses to examine the associations of 11 NADPH oxidase-related genes with BP response to sodium intervention among 1,906 Chinese participants in the GenSalt study.
Table 1.
Characteristics of 11 NADPH oxidase-related genes
| Gene symbol | Physical position | |||
|---|---|---|---|---|
| Chr | ±5 kb | Tag SNPs | Encoded protein | |
| NCF2 | 1 | (183519697, 183565056) | 5 | Neutrophil cytosolic factor 2, p67phox |
| RAC1 | 7 | (6409126, 6448598) | 14 | RAS-related C3 botulinum toxin substrate 1 |
| NOXA1 | 9 | (140312847, 140333858) | 1 | NADPH oxidase activator 1 |
| NOX4 | 11 | (89052522, 89229653) | 17 | NADPH oxidase 4 |
| NOX5 | 15 | (69302034, 69354501) | 3 | NADPH oxidase 5 |
| CYBA | 16 | (88704697, 88722492) | 1 | Cytochrome b-245, alpha polypeptide, p22phox |
| NOXO1 | 16 | (2023918, 2036550) | 3 | NADPH oxidase organizer 1 |
| NCF4 | 22 | (37252030, 37279059) | 5 | Neutrophil cytosolic factor 4, p40phox |
| RAC2 | 22 | (37616310, 37645305) | 9 | RAS-related C3 botulinum toxin substrate 2 |
| NOX1 | X | (100093313, 100134334) | 2 | NADPH oxidase 1 |
| NOX2 | X | (37634270, 37677714) | 3 | NADPH oxidase 2, p91phox |
Abbreviations: NADPH, nicotinamide adenine dinucleotide phosphate; SNP, single-nucleotide polymorphism.
METHODS
Study population
The GenSalt study was conducted in a Han Chinese population from rural areas in northern China. A community-based BP screening was carried out among residents aged 18–60 years in the study villages to identify potential probands and their families. Those with a mean systolic BP (SBP) between 130–160 mm Hg and/or a diastolic BP (DBP) between 85–100 mm Hg and no use of antihypertensive medications as well as their spouses, siblings, and offspring were recruited as volunteers for the dietary intervention study. Individuals with stage 2 hypertension, current use of antihypertensive medications, secondary hypertension, history of clinical cardiovascular disease, diabetes, chronic kidney disease, along with pregnant women, heavy alcohol users, and those currently on a low-sodium diet were excluded from the study. The detailed eligibility criteria for the probands and their family members are presented elsewhere.18 Institutional Review Boards at all of the participating institutions approved the GenSalt study. Written informed consents for the baseline observation and for the intervention program were obtained from each participant.
Dietary intervention
The study participants received a 7-day low-sodium intervention (3 g of sodium chloride or 51.3 mmol of sodium per day) followed by a 7-day high-sodium intervention (18 g of sodium chloride or 307.8 mmol of sodium per day). Total energy intake was varied according to each participant’s baseline energy intake. All of the foods were cooked without salt, and prepackaged salt was added to the individual study participant’s meal when it was served by the study staff. All participants were required to have their breakfast, lunch, and dinner at the study kitchen under supervision of the study staff during the entire study period and they were instructed to avoid consuming any foods that were not provided by the study. To ensure study participants’ compliance to the dietary sodium intervention, 3 timed urinary specimens (one 24-hour and 2 overnight) were collected at baseline and at the end of each phase of intervention (days 5, 6, and 7). The overnight urinary excretion of sodium was converted to 24-hour values on the basis of formula developed from a random sample of 238 participants.19 The mean (SD) 24-hour urinary excretions of sodium was 242.4 (66.7) mmol at baseline, 47.5 (16.0) mmol during the low-sodium intervention, and 244.3 (37.7) mmol during the high-sodium intervention, respectively, which showed excellent compliance with the study diet. Among the 1,906 eligible participants, 1,871 (98.2%) and 1,860 (97.6%) completed the low-sodium and high-sodium interventions, respectively, and were included in the current analysis.
Phenotype measurements
During the 3 days of baseline examination, trained staff collected information on family structure, demographic characteristics, personal and family medical history, and lifestyle risk factors using a standard questionnaire. Three morning BP measurements were obtained according to a standard protocol during each day of baseline observation and on days 5, 6, and 7 of each intervention period. All BP readings were measured by trained and certified observers using random-zero sphygmomanometer.20 BP was measured with the participants in the sitting position after 5 minutes of rest. In addition, participants were advised to avoid alcohol, cigarette smoking, coffee/tea, and exercise for at least 30 minutes prior to their BP measurements. In addition, body weight, height, and waist circumference were measured twice in light indoor clothing without shoes during the baseline examination. Body mass index (BMI) was calculated as kilograms per meters squared (kg/m2).
BP responses were defined continuously as the absolute changes in SBP, DBP, and mean arterial pressure when switching from baseline to low-sodium intervention and from low-sodium to high-sodium intervention. Mean BP responses to low-sodium intake were calculated as the mean of 9 measurements on days 5, 6, and 7 during the low-sodium intervention minus the mean of 9 measurements at baseline, and responses to high-sodium intake were calculated as the mean of 9 measurements on days 5, 6, and 7 during the high-sodium intervention minus the mean of 9 measurements on days 5, 6, and 7 during the low-sodium intervention.
Genotype data and quality control
Eleven NADPH oxidase-related genes were selected based on their potential biological effects on BP regulation (Table 1). Within the 11 candidate genes, 124 single-nucleotide polymorphisms (SNPs) were genotyped on the Affymetrix 6.0 platform (Affymetrix, Santa Clara, CA). Quality control excluded 26 SNPs based on low minor allele frequency <1%, low genotyping call rate (<95%), or deviation from the Hardy–Weinberg equilibrium after using the false discovery rate procedure (Q value with the significance level of 0.05 was used) to correct for multiple testing. Among the remaining 98 SNPs, 63 tag SNPs were selected using Haploview software (version 4.2, http://www.broad.mit.edu/mpg/haploview) with r2 <0.8 for inclusion in the statistical analysis.21 Quality control information on the tagged 63 SNPs was listed in Supplementary Table 1.
Statistical analysis
Quality control, including checks of Mendelian consistency, genotyping call rate, minor allele frequency, and Hardy–Weinberg equilibrium was performed using PLINK software (version 1.07; Dr Sean Purcell, http://pngu.mgh.harvard.edu/~purcell/plink/).22
The means or percentages of phenotypic and genotypic data were calculated for the 1,906 GenSalt study participants. Additive associations between SNPs and BP responses to each dietary sodium intervention were assessed by a mixed-effect linear regression model using the PROC MIXED procedure in SAS (version 9.3; SAS Institute, Cary, NC).23 The mixed-effect model we used was as follows:
In the formula, γij represents the BP responses to each dietary sodium intervention for the ith individual in the jth family. β0 was the mean BP responses after accounting for covariates and genetic effects. The terms ageij, genderij, BMIij, and 24_hour_urinary_sodiumij represented baseline age, gender, BMI, and 24-hour urinary sodium excretion of the ith individual in the jth family, respectively. SNPij modeled the genetic effect, where the genotype was coded under an additive model. The random effects term aj accounts for the correlation among individuals in the same family. The last term eij stands for residual. P values for β5 were used to evaluate the significance of the association of each SNP with BP responses to each dietary sodium intervention.
A sandwich estimator was used to account for the nonindependence of family members. To account for the sex-specific structure genes in X chromosome, models were calculated that assumed inactivation as well as not assuming inactivation. A similar, gender-stratified analysis was conducted for those SNPs located on the X-chromosome. For significant SNPs, the mean effect size and 95% confidence interval was estimated for each genotype using a mixed-linear regression model. Comparatively, to evaluate our results from the mixed-effect models, we also used the packages kinship2 and GWAF to account for the kinships between individuals in R software (version 3.2.4; http://www.r-project.org).
In the gene-based analysis, the truncated product method (TPM) was used to determine the overall association of each NADPH oxidase-related genes, in which at least 2 SNPs were genotyped, with BP responses to dietary sodium intervention.24 The truncation point was set as τ = 0.10, and the P value for TPM was estimated by 1,000,000 simulations. Sensitivity analyses were conducted using the TPM after removing the lead SNP within a gene to examine their influence on the gene-based association. Additionally, to evaluate the robustness of findings from the TPM, the Versatile Gene-based Association Study (VEGAS) was also employed.25,26
Bonferroni correction was used to adjust for multiple comparisons. The thresholds for the single SNP-based and gene-based analyses were α = 0.05/63 = 7.94 × 10−4 and α = 0.05/9 = 5.56 × 10−3, respectively. Single-SNP-based analysis was conducted using SAS (version 9.3; SAS Institute). TPM was performed using R software (version 3.2.4; http://www.r-project.org).
RESULTS
Characteristics of the GenSalt participants and BP responses to sodium interventions were shown in Table 2. On average, study participants were 38.7 years old and had a mean BMI of 23.3 kg/m2, mean SBP of 116.9 mm Hg, and mean DBP of 73.7 mm Hg. Approximately 53% of participants were male. BP levels were similar during baseline and the high-sodium intervention, and BP responses to sodium intervention were significantly different from zero.
Table 2.
Characteristics of 1,906 GenSalt dietary intervention participants
| Variable | Mean ± SD or percentage | Median (interquartile range) |
|---|---|---|
| Age, years | 38.7 ± 9.6 | 39.0 (33.0, 46.0) |
| Men, % | 53.0 | |
| BMI, kg/m2 | 23.3 ± 3.2 | 22.9 (21.1, 25.2) |
| SBP, mm Hg | ||
| Baseline | 116.9 ± 14.2 | 115.8 (106.4, 127.1) |
| Response to low-sodium intake | −5.5 ± 7.0* | −4.4 (−8.9, −1.3) |
| Response to high-sodium intake | 4.9 ± 6.0* | 4.7 (0.6, 8.2) |
| DBP, mm Hg | ||
| Baseline | 73.7 ± 10.3 | 73.3 (66.7, 80.7) |
| Response to low-sodium intake | −2.8 ± 5.5* | −2.7 (−5.6, 0.4) |
| Response to high-sodium intake | 1.9 ± 5.4* | 1.8 (−1.6, 5.3) |
| MAP, mm Hg | ||
| Baseline | 88.1 ± 10.9 | 87.7 (80.0, 95.4) |
| Response to low-sodium intake | −3.7 ± 5.3* | −3.3 (−6.6, −0.6) |
| Response to high-sodium intake | 2.9 ± 5.0* | 2.7 (−0.4, 5.9) |
Abbreviations: BMI, body mass index; DBP, diastolic blood pressure; MAP, mean arterial pressure; SBP, systolic blood pressure. *P value <0.0001 when compared to no BP change during sodium interventions.
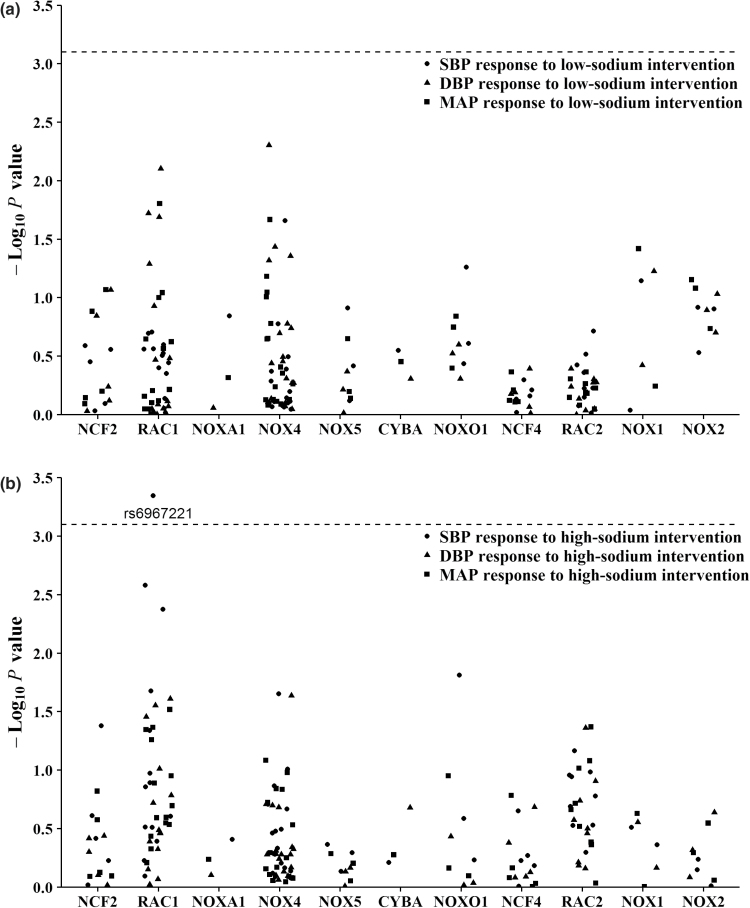
Figure 1 presents the association of each SNP with absolute SBP, DBP, and mean arterial pressure responses to sodium intervention. After adjustment for multiple testing, RAC1 marker rs6967221 was significantly associated with SBP response to high-sodium intervention (P = 4.51 × 10−4). Mean SBP responses (95% confidence interval) were 5.03 (4.71, 5.36), 4.20 (3.54, 4.85), and 0.56 (−1.08, 2.20) mm Hg for genotypes CC, CT, and TT, respectively, during the high-sodium intervention (Table 3). Exact P values for all SNPs association tests were shown in Supplementary Table 2. Similar results were obtained after including kinships between individuals.
Figure 1.
-Log10P values for the associations of 63 tag SNPs in NADPH oxidase-related genes with blood pressure responses to low-sodium (a) and high-sodium (b) interventions (points were jittered to reduce overlap). Labeled SNP was significant after Bonferroni correction. The horizontal dashed lines indicated the Bonferroni corrected significant level (P = 7.94 × 10−4). Abbreviations: NADPH, nicotinamide adenine dinucleotide phosphate; SNP, single-nucleotide polymorphism.
Table 3.
BP response to dietary sodium intervention according to rs6967221 genotype
| Low-sodium intervention | High-sodium intervention | ||||
|---|---|---|---|---|---|
| BP response | Genotype | Absolute change (95% CI) | P value for additive model | Absolute change (95% CI) | P value for additive model |
| SBP | CC | −5.60 (−5.97, −5.22) | 0.27 | 5.03 (4.71, 5.36) | 4.51 × 10−4 |
| CT | −5.07 (−5.85, −4.29) | 4.20 (3.54, 4.85) | |||
| TT | −5.85 (−7.78, −3.92) | 0.56 (−1.08, 2.20) | |||
| DBP | CC | −2.81 (−3.12, −2.50) | 0.89 | 2.00 (1.68, 2.32) | 0.41 |
| CT | −2.93 (−3.63, −2.24) | 1.85 (1.21, 2.49) | |||
| TT | −2.27 (−4.92, 0.38) | 0.60 (−2.11, 3.32) | |||
| MAP | CC | −3.74 (−4.03, −3.44) | 0.75 | 3.01 (2.72, 3.30) | 5.50 × 10−2 |
| CT | −3.65 (−4.31, −2.99) | 2.64 (2.07, 3.21) | |||
| TT | −3.46 (−5.59, −1.34) | 0.60 (−1.50, 2.69) | |||
Abbreviations: CI, confidence interval; DBP, diastolic blood pressure; MAP, mean arterial pressure; SBP, systolic blood pressure.
Table 4 presents the results of gene-based analyses. CYBA and NOXA1 could not be examined since only one SNP was available within each of these genes. Among the remaining 9 genes, RAC1 was significantly associated with DBP response to low-sodium intervention (P = 9.80 × 10−4) and SBP response to high-sodium intervention (P = 1.00 × 10−6) even after adjustment for multiple testing. In sensitivity analyses, the overall associaitons of RAC1 with SBP response to high-sodium intervention (P = 6.20 × 10−5) remained significant even after removing the lead marker rs6967221 from gene-based analyses. DBP responses to low-sodium intervention (P = 0.02) remained nominal significant after the removal of the lead SNP rs2689420. NOX4 showed nominal significance with DBP response to low-sodium intervention (P = 0.01). Similar results were obtained using the VEGAS analyses (Supplementary Table 3).
Table 4.
Associations of 9 NADPH oxidase-related genes with BP response to dietary sodium intervention (truncated product method)
| Genea | Number of SNPs | Absolute blood pressure response | |||||
|---|---|---|---|---|---|---|---|
| Low-sodium intervention | High-sodium intervention | ||||||
| SBP | DBP | MAP | SBP | DBP | MAP | ||
| NCF2 | 5 | 0.39 | 0.40 | 0.37 | 0.22 | 0.37 | 0.37 |
| RAC1 | 14 | 0.67 | 9.80 × 10−4b | 0.09 | 1.00 × 10−6b | 0.01 | 0.12 |
| NOX4 | 17 | 0.44 | 0.01 | 0.13 | 0.32 | 0.37 | 0.66 |
| NOX5 | 3 | 0.26 | 0.28 | 0.22 | 0.17 | 0.25 | 0.20 |
| NOXO1 | 3 | 0.13 | 0.15 | 0.17 | 0.05 | 0.29 | 0.29 |
| NCF4 | 5 | 0.28 | 0.33 | 0.20 | 0.44 | 0.32 | 0.46 |
| RAC2 | 9 | 0.47 | 0.60 | 0.26 | 0.21 | 0.28 | 0.09 |
| NOX1 | 2 | 0.14 | 0.11 | 0.08 | 0.12 | 0.17 | 0.20 |
| NOX2 | 3 | 0.16 | 0.13 | 0.09 | 0.26 | 0.24 | 0.25 |
Abbreviations: DBP, diastolic blood pressure; MAP, mean arterial pressure; NADPH, nicotinamide adenine dinucleotide phosphate; SBP, systolic blood pressure; SNP, single-nucleotide polymorphism.
aGenes containing at least 2 SNPs.
bSignificant after adjustment for multiple testing using Bonferroni correction.
DISCUSSION
In the present study, we examined the association between the NADPH oxidase-related genes and SSBP among a large sample of Han Chinese population. We identified that the minor T allele of rs6967221 in RAC1 was associated with the decreased SBP response to high-sodium intervention. Consistent with these findings, gene-based analyses also revealed a significant association of the RAC1 gene with DBP response to low-sodium intervention and SBP response to high-sodium intervention. Besides, NOX4 was nominally associated with DBP response to low-sodium intervention. In aggregate, these findings contribute genetic evidence for a role of NADPH oxidase-related genes in SSBP.
Rac1 is a member of the Rho family of GTPases, and it cycles between an inactive GDP-bound and an active GTP-bound state.27 It is part of the NADPH oxidase complex, which induces the generation of reactive oxygen species leading to increased oxidative stress, alterations in the cell membrane, and endothelial damage.28 Several studies have provided important evidence of the link between RAC1 and BP control and SSBP.29–31 In Dahl SS rats, it is reported that enhanced NaCl–Rac1–NADPH oxidase–reactive oxygen species–Na reabsorption cascade might be an important mechanism of SS hypertension.32 Besides, several studies reported that Rac1-mineralocorticoid receptor pathway plays a key role in the development of SS hypertension.14,33,34 However, few studies have implicated the association between RAC1 gene variants and SSBP. To the best of our knowledge, this is the first investigation to examine the association of genetic variants in RAC1 with SSBP. The marker rs6967221 locates in the intronic region of the RAC1 gene. Although we used the web tools SNPinfo and RegulomeDB to speculate the functional implication of the significant SNP,35,36 little evidence showed that rs6967221 and its highly correlated SNPs are causally associated with the regulation of RAC1 expression. Thus, the function of rs6967221 needs to be further investigated. A previous study in a Chilean pediatric population demonstrated that polymorphism rs836478 and rs10951982 of the RAC1 gene was associated with hypertension and DBP.37 SNP rs10951982 was not genotyped in our study. Due to weak correlation between rs10951982 and rs6967221 (r2 = 0.012) in HapMap Chinese Han in Beijing data, rs6967221 could be a novel marker associated with BP response to diet salt intervention in Chinese. Besides, the present study showed that rs836478 was nominally associated with DBP and mean arterial pressure responses to low-sodium (P values = 0.001 and 0.043, respectively) and high-sodium interventions (P values = 0.014 and 0.023, respectively). Furthermore, finding from gene-based analysis supported that RAC1 influenced the BP responses to dietary sodium intake even after removing the prominent marker rs6967221. Further genetic and functional research are still needed to delineate the role of RAC1 in SSBP.
Several studies indicated that CYBA variants influenced the indices of oxidative stress and were associated with hypertension and SSBP.15–17,38 For instance, the CYBA gene C242T (rs4673) polymorphism was associated with SSBP in Hispanics.16 With only rs12709102 available in the current study, we could not replicate these findings. Several possibilities may explain the discrepancies among studies. Our study was conducted in Han Chinese population, whose linkage disequilibrium structure may be different from other populations. Furthermore, the same genetic variants may have different BP effects considering gene–environment interaction. Moreover, other potential factors, such as different characteristics of subjects, sodium intervention design, and lifestyle factors, might contribute to the inconsistency of results.
Strengths and limitations
Several strengths of this study should be noted. First of all, it is a comprehensive investigation to examine associations of NADPH oxidase-related genes with BP responses to dietary sodium intervention. Study attributes, such as homogeneity of the population, should make the analysis robust to population stratification. In addition, measurement of BP for 9 times during each period had ensured the accuracy of BP and stringent quality control procedures were also conducted for genotyping data, data collection, and dietary intervention. Moreover, Bonferroni correction procedures were performed accounting for multiple testing. However, this study also had several limitations. Our research was conducted in a Han Chinese population. These novel associations reported here need be replicated in other populations with different genetic background. Thus, the generalizability of our results to other populations with different genetic and environmental background was unknown. Furthermore, although the Affymetrix 6.0 platform generally provides good genomic coverage of common polymorphisms in the Han Chinese population (approximately 75%),39 limited genotype data were available for the CYBA and NOXA1 genes. Therefore, future researches to examine the associations between common variants in these genes and SSBP are still needed.
In conclusion, our study suggested that common variants of NADPH oxidase-related genes were associated with BP responses to dietary sodium intervention in Han Chinese. These findings may contribute to a better understanding of the genetic mechanism of underlying BP regulation. However, replications of these results in other populations with different genetic background are needed. Furthermore, functional studies are warranted to pinpoint the underlying mechanism.
SUPPLEMENTARY MATERIAL
Supplementary data are available at American Journal of Hypertension online.
Supplementary Material
ACKNOWLEDGMENTS
This study is supported by the National Natural Science Foundation of China (grant no. 81570386). The Genetic Epidemiology Network of Salt Sensitivity (GenSalt) is supported by a cooperative agreement project grant (U01HL072507, R01HL087263, and R01HL090682) from the National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, MD, USA.
DISCLOSURE
The authors declared no conflict of interest.
References
- 1. Yusuf S, Rangarajan S, Teo K, Islam S, Li W, Liu L, Bo J, Lou Q, Lu F, Liu T, Yu L, Zhang S, Mony P, Swaminathan S, Mohan V, Gupta R, Kumar R, Vijayakumar K, Lear S, Anand S, Wielgosz A, Diaz R, Avezum A, Lopez-Jaramillo P, Lanas F, Yusoff K, Ismail N, Iqbal R, Rahman O, Rosengren A, Yusufali A, Kelishadi R, Kruger A, Puoane T, Szuba A, Chifamba J, Oguz A, McQueen M, McKee M, Dagenais G; PURE Investigators Cardiovascular risk and events in 17 low-, middle-, and high-income countries. N Engl J Med 2014; 371:818–827. [DOI] [PubMed] [Google Scholar]
- 2. Weinberger MH. Salt sensitivity of blood pressure in humans. Hypertension 1996; 27:481–490. [DOI] [PubMed] [Google Scholar]
- 3. Gu D, Rice T, Wang S, Yang W, Gu C, Chen CS, Hixson JE, Jaquish CE, Yao ZJ, Liu DP, Rao DC, He J. Heritability of blood pressure responses to dietary sodium and potassium intake in a Chinese population. Hypertension 2007; 50:116–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Elijovich F, Weinberger MH, Anderson CA, Appel LJ, Bursztyn M, Cook NR, Dart RA, Newton-Cheh CH, Sacks FM, Laffer CL; American Heart Association Professional and Public Education Committee of the Council on Hypertension; Council on Functional Genomics and Translational Biology; and Stroke Council Salt sensitivity of blood pressure: a scientific statement from the American Heart Association. Hypertension 2016; 68:e7–e46. [DOI] [PubMed] [Google Scholar]
- 5. Crowley SD. The cooperative roles of inflammation and oxidative stress in the pathogenesis of hypertension. Antioxid Redox Signal 2014; 20:102–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Montezano AC, Touyz RM. Reactive oxygen species, vascular Noxs, and hypertension: focus on translational and clinical research. Antioxid Redox Signal 2014; 20:164–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Manning RD, Jr, Meng S, Tian N. Renal and vascular oxidative stress and salt-sensitivity of arterial pressure. Acta Physiol Scand 2003; 179:243–250. [DOI] [PubMed] [Google Scholar]
- 8. Santillo M, Colantuoni A, Mondola P, Guida B, Damiano S. NOX signaling in molecular cardiovascular mechanisms involved in the blood pressure homeostasis. Front Physiol 2015; 6:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bedard K, Krause KH. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol Rev 2007; 87:245–313. [DOI] [PubMed] [Google Scholar]
- 10. Sedeek M, Hébert RL, Kennedy CR, Burns KD, Touyz RM. Molecular mechanisms of hypertension: role of Nox family NADPH oxidases. Curr Opin Nephrol Hypertens 2009; 18:122–127. [DOI] [PubMed] [Google Scholar]
- 11. Lassègue B, San Martín A, Griendling KK. Biochemistry, physiology, and pathophysiology of NADPH oxidases in the cardiovascular system. Circ Res 2012; 110:1364–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cowley AW, Jr, Yang C, Zheleznova NN, Staruschenko A, Kurth T, Rein L, Kumar V, Sadovnikov K, Dayton A, Hoffman M, Ryan RP, Skelton MM, Salehpour F, Ranji M, Geurts A. Evidence of the importance of Nox4 in production of hypertension in Dahl salt-sensitive rats. Hypertension 2016; 67:440–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Feng D, Yang C, Geurts AM, Kurth T, Liang M, Lazar J, Mattson DL, O’Connor PM, Cowley AW., Jr Increased expression of NAD(P)H oxidase subunit p67(phox) in the renal medulla contributes to excess oxidative stress and salt-sensitive hypertension. Cell Metab 2012; 15:201–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shibata S, Mu S, Kawarazaki H, Muraoka K, Ishizawa K, Yoshida S, Kawarazaki W, Takeuchi M, Ayuzawa N, Miyoshi J, Takai Y, Ishikawa A, Shimosawa T, Ando K, Nagase M, Fujita T. Rac1 GTPase in rodent kidneys is essential for salt-sensitive hypertension via a mineralocorticoid receptor-dependent pathway. J Clin Invest 2011; 121:3233–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zalba G, San José G, Moreno MU, Fortuño A, Díez J. NADPH oxidase-mediated oxidative stress: genetic studies of the p22(phox) gene in hypertension. Antioxid Redox Signal 2005; 7:1327–1336. [DOI] [PubMed] [Google Scholar]
- 16. Castejon AM, Bracero J, Hoffmann IS, Alfieri AB, Cubeddu LX. NAD(P)H oxidase p22phox gene C242T polymorphism, nitric oxide production, salt sensitivity and cardiovascular risk factors in Hispanics. J Hum Hypertens 2006; 20:772–779. [DOI] [PubMed] [Google Scholar]
- 17. Kumar R, Kohli S, Ali Z, Duhan K, Ram R, Gupta M, Tyagi S, Mohammad G, Pasha MQ. CYBA (p22phox) variants associate with blood pressure and oxidative stress markers in hypertension: a replication study in populations of diverse altitudes. Hypertens Res 2015; 38:498–506. [DOI] [PubMed] [Google Scholar]
- 18. GenSalt Collaborative Research G. GenSalt: rationale, design, methods and baseline characteristics of study participants. J Hum Hypertens 2007; 21:639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. He J, Gu D, Chen J, Jaquish CE, Rao DC, Hixson JE, Chen JC, Duan X, Huang JF, Chen CS, Kelly TN, Bazzano LA, Whelton PK; GenSalt Collaborative Research Group Gender difference in blood pressure responses to dietary sodium intervention in the GenSalt study. J Hypertens 2009; 27:48–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Perloff D, Grim C, Flack J, Frohlich ED, Hill M, McDonald M, Morgenstern BZ. Human blood pressure determination by sphygmomanometry. Circulation 1993; 88:2460–2470. [DOI] [PubMed] [Google Scholar]
- 21. de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet 2005; 37:1217–1223. [DOI] [PubMed] [Google Scholar]
- 22. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Singer JD. Using SAS PROC MIXED to fit multilevel models, hierarchical models, and individual growth models. J Educ Behav Stat 1998; 23:323–355. [Google Scholar]
- 24. Zaykin DV, Zhivotovsky LA, Westfall PH, Weir BS. Truncated product method for combining P-values. Genet Epidemiol 2002; 22:170–185. [DOI] [PubMed] [Google Scholar]
- 25. Liu JZ, McRae AF, Nyholt DR, Medland SE, Wray NR, Brown KM, Hayward NK, Montgomery GW, Visscher PM, Martin NG, Macgregor S; AMFS Investigators A versatile gene-based test for genome-wide association studies. Am J Hum Genet 2010; 87:139–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mishra A, Macgregor S. VEGAS2: software for more flexible gene-based testing. Twin Res Hum Genet 2015; 18:86–91. [DOI] [PubMed] [Google Scholar]
- 27. Matos P, Skaug J, Marques B, Beck S, Veríssimo F, Gespach C, Boavida MG, Scherer SW, Jordan P. Small GTPase Rac1: structure, localization, and expression of the human gene. Biochem Biophys Res Commun 2000; 277:741–751. [DOI] [PubMed] [Google Scholar]
- 28. Hordijk PL. Regulation of NADPH oxidases: the role of Rac proteins. Circ Res 2006; 98:453–462. [DOI] [PubMed] [Google Scholar]
- 29. Hassanain HH, Gregg D, Marcelo ML, Zweier JL, Souza HP, Selvakumar B, Ma Q, Moustafa-Bayoumi M, Binkley PF, Flavahan NA, Morris M, Dong C, Goldschmidt-Clermont PJ. Hypertension caused by transgenic overexpression of Rac1. Antioxid Redox Signal 2007; 9:91–100. [DOI] [PubMed] [Google Scholar]
- 30. Loirand G, Pacaud P. The role of Rho protein signaling in hypertension. Nat Rev Cardiol 2010; 7:637–647. [DOI] [PubMed] [Google Scholar]
- 31. Nagase M. Role of Rac1 GTPase in salt-sensitive hypertension. Curr Opin Nephrol Hypertens 2013; 22:148–155. [DOI] [PubMed] [Google Scholar]
- 32. Kirchner KA. Greater loop chloride uptake contributes to blunted pressure natriuresis in Dahl salt sensitive rats. J Am Soc Nephrol 1990; 1:180–186. [DOI] [PubMed] [Google Scholar]
- 33. Kawarazaki W, Fujita T. Aberrant Rac1-mineralocorticoid receptor pathways in salt-sensitive hypertension. Clin Exp Pharmacol Physiol 2013; 40:929–936. [DOI] [PubMed] [Google Scholar]
- 34. Kawarazaki H, Ando K, Shibata S, Muraoka K, Fujita M, Kawarasaki C, Fujita T. Mineralocorticoid receptor–Rac1 activation and oxidative stress play major roles in salt-induced hypertension and kidney injury in prepubertal rats. J Hypertens 2012; 30:1977–1985. [DOI] [PubMed] [Google Scholar]
- 35. Xu Z, Taylor JA. SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res 2009; 37:W600–W605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng S, Cherry JM, Snyder M. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 2012; 22:1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tapia-Castillo A, Carvajal CA, Campino C, Vecchiola A, Allende F, Solari S, García L, Lavanderos S, Valdivia C, Fuentes C, Lagos CF, Martínez-Aguayo A, Baudrand R, Aglony M, García H, Fardella CE. Polymorphisms in the RAC1 gene are associated with hypertension risk factors in a Chilean pediatric population. Am J Hypertens 2014; 27:299–307. [DOI] [PubMed] [Google Scholar]
- 38. Meijles DN, Fan LM, Ghazaly MM, Howlin B, Krönke M, Brooks G, Li JM. p22phox C242T single-nucleotide polymorphism inhibits inflammatory oxidative damage to endothelial cells and vessels. Circulation 2016; 133:2391–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kato N, Takeuchi F, Tabara Y, Kelly TN, Go MJ, Sim X, Tay WT, Chen CH, Zhang Y, Yamamoto K, Katsuya T, Yokota M, Kim YJ, Ong RT, Nabika T, Gu D, Chang LC, Kokubo Y, Huang W, Ohnaka K, Yamori Y, Nakashima E, Jaquish CE, Lee JY, Seielstad M, Isono M, Hixson JE, Chen YT, Miki T, Zhou X, Sugiyama T, Jeon JP, Liu JJ, Takayanagi R, Kim SS, Aung T, Sung YJ, Zhang X, Wong TY, Han BG, Kobayashi S, Ogihara T, Zhu D, Iwai N, Wu JY, Teo YY, Tai ES, Cho YS, He J. Meta-analysis of genome-wide association studies identifies common variants associated with blood pressure variation in east Asians. Nat Genet 2011; 43:531–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.