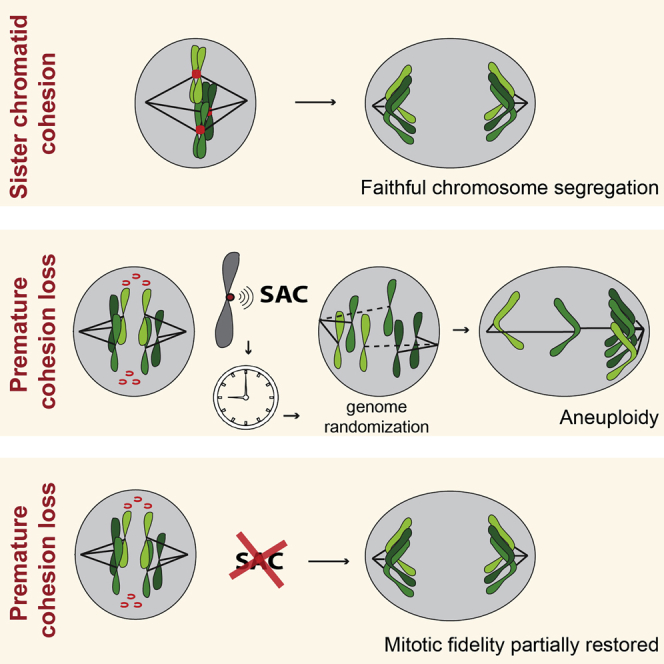
Summary
The fidelity of mitosis depends on cohesive forces that keep sister chromatids together. This is mediated by cohesin that embraces sister chromatid fibers from the time of their replication until the subsequent mitosis [1, 2, 3]. Cleavage of cohesin marks anaphase onset, where single chromatids are dragged to the poles by the mitotic spindle [4, 5, 6]. Cohesin cleavage should only occur when all chromosomes are properly bio-oriented to ensure equal genome distribution and prevent random chromosome segregation. Unscheduled loss of sister chromatid cohesion is prevented by a safeguard mechanism known as the spindle assembly checkpoint (SAC) [7, 8]. To identify specific conditions capable of restoring defects associated with cohesion loss, we screened for genes whose depletion modulates Drosophila wing development when sister chromatid cohesion is impaired. Cohesion deficiency was induced by knockdown of the acetyltransferase separation anxiety (San)/Naa50, a cohesin complex stabilizer [9, 10, 11, 12]. Several genes whose function impacts wing development upon cohesion loss were identified. Surprisingly, knockdown of key SAC proteins, Mad2 and Mps1, suppressed developmental defects associated with San depletion. SAC impairment upon cohesin removal, triggered by San depletion or artificial removal of the cohesin complex, prevented extensive genome shuffling, reduced segregation defects, and restored cell survival. This counterintuitive phenotypic suppression was caused by an intrinsic bias for efficient chromosome biorientation at mitotic entry, coupled with slow engagement of error-correction reactions. Thus, in contrast to SAC’s role as a safeguard mechanism for mitotic fidelity, removal of this checkpoint alleviates mitotic errors when sister chromatid cohesion is compromised.
Keywords: mitosis, sister chromatid cohesion, cohesin, spindle assembly checkpoint, aneuploidy, separation anxiety, SAN, Drosophila
Graphical Abstract
Highlights
-
•
A Drosophila screen identifies SAC genes as suppressors of cohesion-related defects
-
•
SAC removal enhances mitotic fidelity upon premature cohesion loss
-
•
SAC inactivation enhances cell survival and tissue homeostasis upon cohesion loss
The spindle assembly checkpoint (SAC) works as a safeguard mechanism ensuring mitotic fidelity. Here, Silva et al. describe that, in contrast to this safeguard role, a functional SAC aggravates the defects associated with premature loss of sister chromatid cohesion during mitosis.
Results
Depletion of SAC Genes Suppresses Developmental Defects Associated with Loss of Sister Chromatid Cohesion
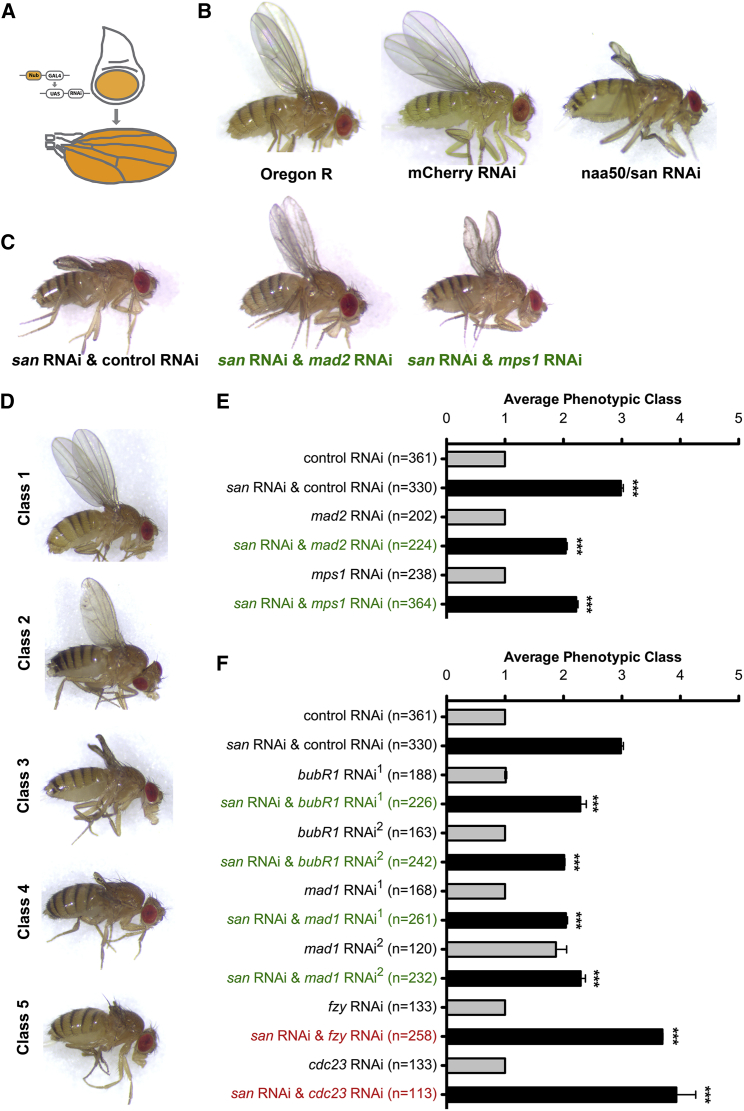
To probe for conditions that enhance or suppress cellular and tissue responses to cohesion defects, we performed a modifier screen in the adult Drosophila wing. We focused our analysis on the N-terminal acetyltransferase separation anxiety (San), required for establishment and/or maintenance of sister chromatid cohesion [9, 10, 11, 12] through regulation of the interaction between two cohesin subunits (Rad21/Smc3) [10]. Knockdown of San during development results in intermediate adult wing phenotype that is sensitive to phenotypic modulation (Figures 1A and 1B). Defects associated with San knockdown can be suppressed by several conditions that enhance cohesin stability on chromatin [10]. To search for modifiers of adult wing phenotype induced by San depletion, we co-expressed the san RNAi with 2,955 RNAis, theoretically depleting 2,920 gene products (21% of all gene products), specifically in larvae wing discs (Figure 1A). The resulting wings were scored in 5 categories, according to phenotype severity (Figure 1D) [10]. Co-expression of san RNAi with control RNAi transgene did not modify the wing phenotype when compared to san RNAi alone (Figures 1B and 1C) [10]. Any isolated enhancer gene whose depletion alone resulted in adult wings phenotype was discarded (Figure S1A). All tested RNAi lines and scored wing phenotypes are shown in Data S1.
Figure 1.
SAC Inhibition Modifies san RNAi-Induced Adult Wing Developmental Defects
(A) Tissue-specific RNAi in the pouch of the larvae wing imaginal using the nubbin-Gal4 driver and the upstream activating sequence (UAS)/Gal4 system.
(B) Adult wings of wild-type Drosophila (Oregon R) or Drosophila expressing a control RNAi (mCherry RNAi) or expressing RNAi for san in the larvae wing imaginal discs.
(C) Representative adult Drosophila wing phenotypes co-expressing san RNAi with mCherry RNAi, mad2 RNAi, or mps1 RNAi in the larvae wing imaginal discs.
(D) Adult wing phenotypic classes scored during the screen: class 1 (wild-type wings); class 2 (weak wing developmental defects); class 3 (san RNAi-like wing phenotype); class 4 (highly abnormal wings); and class 5 (absence or vestigial adult wings). Additional examples of the scored phenotypic classes are shown in [10].
(E) Quantification of Drosophila wing phenotypes expressing individual RNAi transgenes for control (mCherry), mad2, or mps1 (gray bars) or co-expressing san RNAi with control (mCherry) RNAi, mad2 RNAi, or mps1 RNAi (black bars) in the larvae wing imaginal discs, from the described screen.
(F) Candidate gene analysis for enhancers/supressors of san RNAi. Quantification of Drosophila wing phenotypes expressing individual RNAi transgenes for control (mCherry), bubR1, mad1, fzy, and cdc23 (gray bars) or co-expressing san RNAi with control (mCherry) RNAi, bubR1 RNAis, mad1 RNAis, fzy RNAi, and cdc23 RNAi (black bars) in the larvae wing imaginal discs. bubR1 RNAi1, bubR1 RNAi2, mad1 RNAi1, and mad1 RNAi2 correspond to the TRiP RNAis GL00236, GLV21065, GLV21088, and HMC03671, respectively.
The average phenotypic class of control and san RNAi and control RNAi (E and F) is the same. Phenotypic quantification of adult wings is mean ± SD of three independent experiments and is based on the classes described in (D) (∗∗∗p < 0.0001; one-way ANOVA with Bonferroni’s multiple comparison test; n represents the total number of scored flies). See also Figure S1, Table S1, and Data S1.
We identified 19 suppressors and 10 enhancers whose depletion modified san RNAi adult wing phenotype (Figure S1B). Given the known regenerative capacity of wing discs [13], we expected to isolate genes involved in cohesin maintenance, mitotic fidelity, and tissue response to mitotic damage. As expected, the screen revealed components previously implicated in cohesin dynamics (Mau2 and eco), validating its accuracy at isolating modifiers of cohesion state (Figure S1B) [10]. Most of the 29 genes identified in the screen were already characterized in Drosophila and/or in other species (Table S1). About half of the identified genes were either related with mitosis (Claspin, asp, Mps1, Eb1, eco, Mau2, γTub23C, and mad2) or with gene expression (CG5589, JMJD7, Pabp2, His3, and jumu). Other identified genes are important for maintaining apicobasal cell polarity and for actin cytoskeleton organization (capu, cno, and Cad99C). We identified additional suppressors or enhancer genes related with different metabolic processes (Sfxn1-3, CG3842, Dhap-at, and MFS18), protein glycosylation (CG11388), synaptic adhesion (Nlg4), a paralog of Naa20 N-terminal acetyltransferase (CG31730), and DNA repair or transcription (Parp). Surprisingly, two of the strongest suppressors were proteins that participate in the SAC, Mps1 and Mad2, whose depletion suppressed san RNAi adult wing phenotypes (Figures 1C and 1E). We thus hypothesized that impairment of SAC could rescue mitotic defects caused by cohesin deficiency. We tested this notion by candidate gene approach and probed for genetic interactions with other SAC genes. Among four additional SAC components probed (Bub3, Bub1, BubR1, and Mad1; Data S1), RNAi for Mad1 and BubR1 suppressed the morphological defects associated with San depletion (Figures 1F and S1B).
SAC Inactivation Rescues Chromosome Segregation Defects Associated with Loss of Cohesion
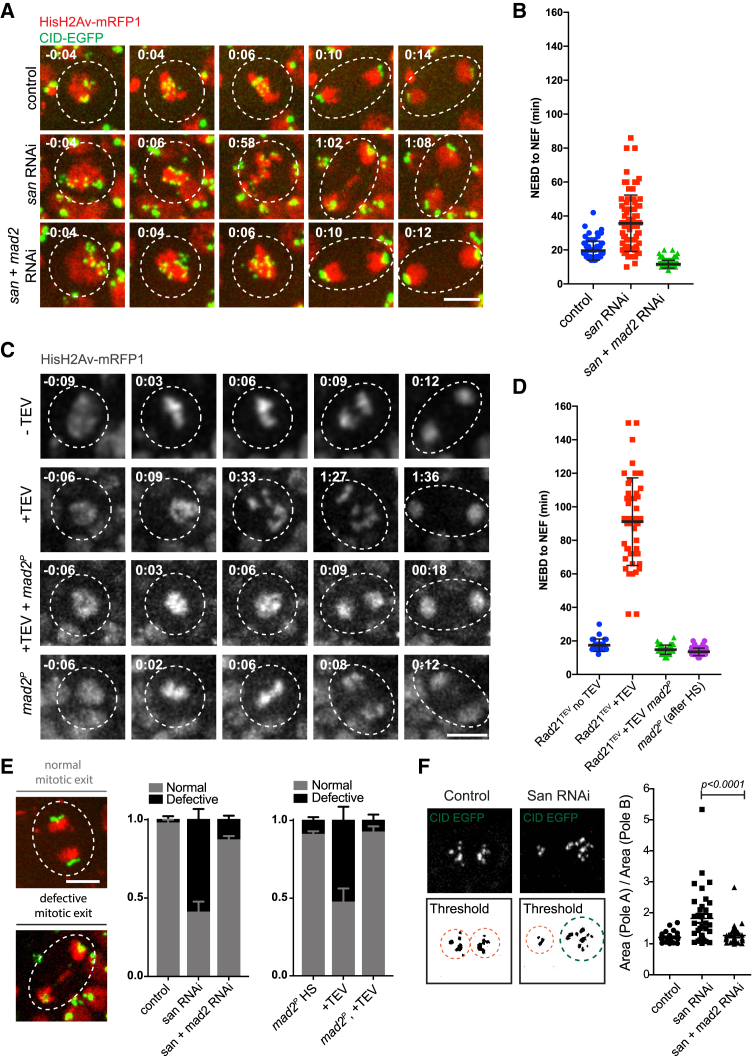
To elucidate whether SAC inactivation could rescue cohesion defects, we examined mitotic fidelity in various experimental conditions. Live imaging of wing discs revealed that, upon san RNAi, cells exhibit various degrees of cohesion defects. Whereas in controls, all cells underwent mitosis with normal metaphase morphology, upon san RNAi, only 13% ± 10% displayed normal mitosis and most cells underwent partial or full sister chromatid separation (17% ± 6% and 70% ± 13%, respectively), resulting in SAC activation and extended mitosis (Figures 2A, 2B, and S2; Video S1). More severe defects were obtained when cohesion loss was induced by acute cleavage of cohesin Rad21 subunit, using a previously established tobacco etch virus (TEV) protease-mediated system [14]. Heat-shock-induced TEV expression results in disappearance of Rad21 in cells expressing exclusively TEV-sensitive Rad21 (Figures S2C and S2D), leading to full cohesion loss in all cells (Figure S2B), an extended mitosis and chromatid shuffling between the poles (Figures 2C and 2D; Video S2).
Figure 2.
Inhibition of the SAC in Wing Imaginal Discs Alleviates Mitotic Errors Caused by Premature Loss of Cohesin
(A) Images from movies of the wing disc pouch in the control, san RNAi, and san and mad2 RNAi strains. Strains contained HisH2Av-RFP (red) and Cid-EGFP (green). Times are relative to NEBD. The scale bar represents 5 μm.
(B) Quantification of mitotic duration in control, san RNAi, or san and mad2 RNAi strains. The duration of mitosis was measured from nuclear envelope breakdown (NEBD) to nuclear envelope formation (NEF) using H2Av-RFP channel. Images were taken every 2 min. Each dot represents an individual cell and lines represent mean ± SD (n = 71/5 for control, 77/5 for san RNAi, and 124/5 for san+mad2 RNAi; n = number of cells and number of independent discs).
(C) Images from movies of the wing disc from strains surviving solely on TEV-cleavable Rad21 (Rad21TEV) with and without heat-shock-induced TEV protease cleavage, in strains wild-type or homozygous mutant for the mad2 gene. Strains also expressed HisH2Av-RFP (red) for visualization of mitotic duration and phenotype. Times are relative to NEBD. The scale bar represents 5 μm.
(D) Quantification of mitotic duration of the no TEV control upon TEV-protease-mediated cleavage of Rad21TEV, upon TEV-protease-mediated cleavage of Rad21TEV in a mad2 mutant background, and in a mad2 mutant without cohesin cleavage but after heat shock. The duration of mitosis was measured from NEBD to NEF using H2AvD-mRFP1. Images were taken every 2 or 3 min. Each dot represents an individual cell, and lines represent mean ± SD (n = 27/4 for Rad21TEV − TEV [no heat shock (HS)], 46/8 for Rad21TEV + TEV, 46/4 for Rad21TEV+TEV in a mad2P background, and 60/4 for mad2P after HS; n = number of cells and number of independent discs).
(E) Representative images of mitotic cells from san RNAi undergoing mitosis with normal and defective mitotic exit. The scale bar represents 5 μm and applies to all images. Graph represents the quantification of mitotic defects observed in the different experimental conditions as mean ± SEM of errors of individual discs (n ≥ 4 independent discs corresponding to over 50 cells analyzed per experimental condition).
(F) Quantification of centromere (cid-EGFP-labeled) segregation assymetry in the different experimental conditions; graph represents segregation symmetry index per cell calculated as the area of pole A (with higher area) divided by area of pole B (lower area), as illustrated on the left (n ≥ 4 independent discs corresponding to over 50 cells analyzed per experimental condition). Statistical analysis was performed using one-way ANOVA test.
DNA is labeled with H2Av-mRFP1 (red) and centromeres with cid-EGFP (green). Times are relative to NEBD.
DNA is labeled with H2Av-mRFP1 (gray). Times are relative to NEBD.
In order to inhibit the SAC, we focused on genetic conditions that remove Mad2, a key component of this checkpoint. Mad2 mutant flies were previously shown to be viable [15], and its depletion in the larvae wing disc did not compromise wing development (Figure 1E). Removal of Mad2 by RNAi or the mad2P-null allele abolished the mitotic delay in both experimental conditions for cohesion loss, san RNAi, and TEV-mediated Rad21 cleavage (Figures 2A–2D; Videos S1 and S2). Surprisingly, shortening of mitotic timing drastically reduced the frequency of abnormal anaphase figures (Figure 2E). Whereas, upon premature loss of cohesin, mitotic exit often displays lagging chromatids or chromatin bridges, these defects were reduced when SAC was removed (Figure 2E).
To further evaluate segregation defects, we quantified numerical errors in chromosome segregation. We measured the area occupied by centromeres in the vicinity of each pole during mitotic exit to calculate segregation symmetry as the ratio between the areas occupied by each centromere cluster (Centromere identifier [Cid]-EGFP-labeled; Figure 2F). This value was close to one in control, and San depletion caused a high degree of asymmetry between centromeric signals (Figure 2F). Importantly, segregation symmetry was significantly restored when San was co-depleted with Mad2 (Figure 2F).
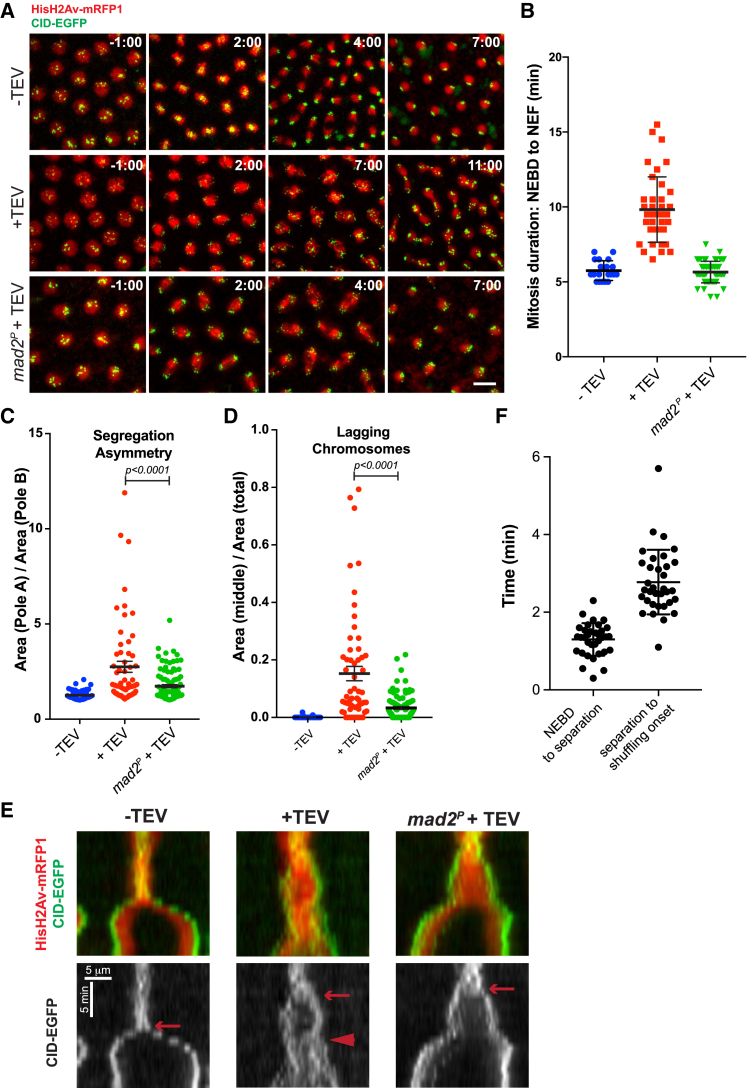
To test whether these results were restricted to larval wing discs, a parallel evaluation of chromosome segregation was performed in early syncytial blastoderm embryos. Cohesin cleavage in Drosophila syncytial embryos was induced by microinjection of TEV protease during interphase [16]. This led to full separation of sister chromatids after nuclear envelope breakdown (NEBD) and a short mitotic delay (Figures 3A and 3B; Video S3). SAC surveillance is responsible for the delay in mitotic progression upon premature loss of sister chromatid cohesion, given that mitotic duration in a mad2 mutant background was indistinguishable from controls (Figures 3A and 3B; Video S3).
Figure 3.
Inhibition of the SAC in Syncytial Blastoderm Embryos Alleviates Mitotic Errors Caused by Premature Loss of Cohesin
(A) Embryos surviving solely on Rad21TEV either non-injected (up) or injected with 5 mg/mL TEV protease (middle and bottom panels). Embryos are derived from females that are wild-type or homozygous mutant for mad2 gene and express HisH2Av-RFP (red) and Cid-EGFP (green). Images were taken every 30 s, and times are relative to NEBD. The scale bar represents 10 μm.
(B) Quantification of mitotic duration in un-injected embryos and embryos injected with TEV protease in strains containing solely Rad21TEV and wild-type or mutant for mad2. The duration of mitosis was measured from NEBD to NEF using HisH2Av-RFP. Images were taken every 30 s. Each dot represents a single mitosis, and lines represent mean ± SD (n = 20/4 for Rad21TEV no TEV, 40/8 for Rad21TEV + TEV, and 55/11 for Rad21TEV+TEV in a mad2P background; n = number of mitosis and number of independent embryos).
(C) Quantification of segregation asymmetry in control, cohesin cleavage, and cohesin cleavage in mad2 mutant background. Each value was quantified by normalizing the area of pole A (with higher area) and the area of pole B (lower area); (n = 46/5 for Rad21TEV no TEV, 60/6 for Rad21TEV + TEV, and 60/6 for Rad21TEV+TEV in a mad2P background; n = number of telophases and number of independent embryos); statistical analysis was performed using one-way ANOVA test.
(D) Relative area of lagging centromeres in control, Rad21TEV + TEV protease, and Rad21TEV + TEV protease in a mad2 mutant background; statistical analysis was performed using one-way ANOVA test.
(E) Kymographs of HisH2Av-RFP and Cid-EGFP of cells entering mitosis in control, cohesin cleavage, and cohesin cleavage in mad2 mutant background. Arrow points to centromere separation and arrowhead to the shuffling onset. The scale bars represent 5 min and 5 μm.
(F) Quantification of time for chromosome shuffling onset upon TEV-mediated cohesin cleavage, relative to NEBD. Each dot represents a single dividing nuclei from >10 independent embryos.
See also Figures S3 and S4 and Videos S3 and S4.
DNA is labeled with H2Av-mRFP1 (red) and centromeres with id-EGFP (green). Times are relative to NEBD.
Analysis of chromosome distribution revealed strong asymmetry upon cohesin cleavage (Figure 3C). We also quantified the frequency of chromosomes that lag behind the segregation plane during mitotic exit (most likely due to merotelic attachments; Figure 3D). Consistent with our previous results (Figure 2F), loss of SAC led to reduction of segregation errors after TEV cleavage, as evidenced by the significant recovery in centromere distribution symmetry and lagging centromere frequency decrease (Figures 3C and 3D). Altogether, these results demonstrate that SAC inactivation rescues chromosome segregation defects upon cohesion loss.
SAC Inactivation Suppresses Chromosome Shuffling after Loss of Cohesion
Premature sister-chromatid separation results in extensive genome randomization. Upon premature cohesin loss, single chromatids lack the opposing forces to ensure proper tension across and/or between kinetochores, leading to unstable microtubule-kinetochore interactions and error correction [17, 18]. These reactions are mediated by Aurora B kinase that destabilizes erroneous kinetochore-microtubule interactions, resulting in extensive shuffling of isolated sister chromatids between spindle poles [16, 17, 18, 19]. Consequently, mitosis in absence of cohesion results in random chromosome segregation, with high probability of generating aneuploidy.
We postulated that mitosis shortening due to SAC loss limits chromosome shuffling, therefore enhancing mitotic fidelity. To test this, we probed for genetic interactions between san RNAi and genes whose depletion should prolong mitosis (Data S1). As predicted, RNAi for the anaphase-promoting complex/cyclosome (APC/C) subunit cdc23 and the APC/C activator cdc20/Fzy aggravated morphological defects associated with san RNAi (Figures 1F and S1B; Data S1).
Aneuploidy levels should be proportional to the number of isolated chromatids crossing the middle of the segregation plane. We quantified the frequency of shuffling events, defined as each time a chromatid close to one pole undergoes erratic motion toward the opposite pole. In embryos, the SAC-dependent mitotic delay observed upon cohesin cleavage, albeit short (∼4 min; Figure 3B), was enough to allow a high degree of chromosome shuffling (Video S3; Figures S3A and S3B). In the absence of SAC, however, despite the full premature loss of cohesion, there was a decrease in chromosome-shuffling events (Figure S3B; Video S3). Thus, SAC abolishment substantially decreases frequency of shuffling by shortening mitosis.
These results suggest that, despite cohesin loss, error correction does not take place during early mitotic stages. To test this possibility, we measured the kinetics of chromosome shuffling onset upon loss of cohesion. Analysis in embryos and wing discs revealed that, despite cohesin removal, chromosomes retain a pseudo-metaphase configuration for an extended period of time (Figures 2A, t = 6 min, and 2C, t = 9 min; Figure 3A, t = 2 min; Videos S1, S2, and S3). During this pseudo-metaphase stage, sister centromeres were found fully disjoined, confirming loss of sister-chromatid cohesion. However, separation of chromatin itself was only initiated several minutes later.
We monitored the timing of error-correction engagement using kymographs that plot the positioning of centromeres over time. Time of initial centromere separation can be detected by the split in centromere signals and onset of chromosome shuffling by the time centromeres start crossing the segregation plane (Figure 3E, arrow and arrowheads, respectively). This analysis revealed that, upon cohesin cleavage, chromosome shuffling was only initiated 4.07 ± 0.96 min after NEBD (1.3 ± 0.4 min for NEBD to centromere separation and 2.8 ± 0.8 min from centromere separation to initiation of shuffling; Figure 3F). A similar, yet extended behavior was observed in the wing disc. Upon NEBD, chromosomes retained a prolonged pseudo-metaphase configuration despite sister chromatid separation (as judged by centromere distances), and chromosome shuffling was only observed much later (11.4 ± 2.9 min after NEBD; Figures S3C and S3D). These analyses reveal a significant delay in the initiation of major error-correction events. Such delay is similar to mitotic timing in absence of functional SAC (Figures 2B, 2D, and 3B). Thus, SAC inhibition counteracts genome shuffling in the absence of cohesin by shortening mitosis duration and thereby preventing extensive error correction.
A key prediction from this observation is that initial kinetochore-microtubule interactions are quite accurate and that inhibition of error correction should restore mitotic fidelity to a similar extent as SAC inactivation. Knowing that Aurora B has multiple roles during mitosis [20], we titrated the levels of an Aurora B inhibitor, Binucleine 2, to a concentration that does not impair chromosome condensation, mitotic timing, SAC competency, or separation of daughter nuclei in the wing disc (Figures S4A–S4D). Using such concentration (5 μM), we show that mild Aurora B inhibition shortened the mitotic delay induced by TEV-mediated Rad21 cleavage (Figure S4E). Furthermore, this treatment completely abolished chromosome shuffling and motion after the initial separation of single chromatids to the poles (Figure S4F; Video S4). Importantly, decrease in Aurora B activity is sufficient to restore centromere segregation symmetry upon premature cohesion loss and eliminate the frequency of lagging chromosomes during mitotic exit (Figures S4G and S4H). We therefore conclude that initial capture of kinetochores by the microtubules has a strong bias for symmetry, even in the complete absence of cohesin. Major asymmetry in chromosome distribution, in turn, depends on error-correction events.
DNA is labeled with H2Av-mRFP1 (gray). Times are relative to NEBD.
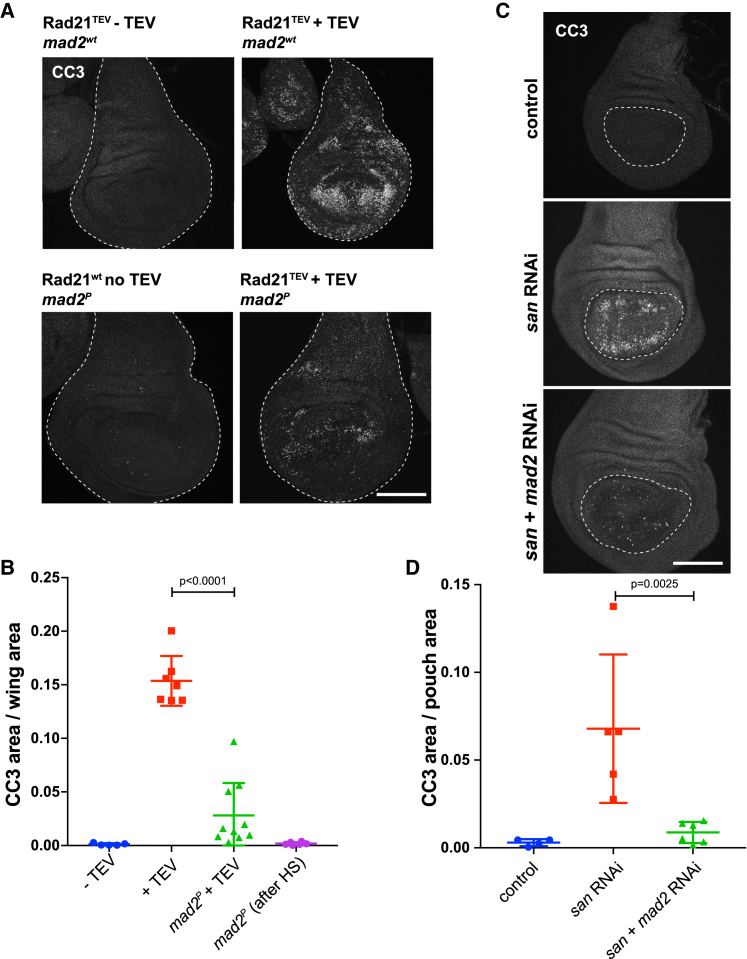
SAC Inactivation Restores Cell Survival after Loss of Cohesion
Our results indicate that mitotic defects upon loss of cohesion are less detrimental in absence of the SAC. If so, the degree of aneuploidy should follow a similar trend. Larvae wing discs are well known to eliminate cells with erroneous DNA content by apoptosis [13]. Therefore, SAC inactivation should reduce levels of apoptosis after cohesion loss. Virtually no apoptosis was detected (cleaved caspase-3 staining [CC3]) within the control wing discs (Rad21TEV without TEV; Figures 4A and 4B). In contrast, TEV-mediated cohesin cleavage induced high levels of apoptosis 24 hr after cohesin cleavage, extending to over 15% ± 2% of the entire wing disc area (Figures 4A and 4B). Remarkably, levels of apoptosis were significantly reduced when cohesin loss was induced in the absence of the SAC (3% ± 3%; Figures 4A and 4B). Similar results were obtained upon depletion of San, where apoptosis covered approximately 7% ± 4% of the wing disc pouch area, compared to only approximately 0.9% ± 0.6% of the pouch area after co-depletion of San and Mad2 (Figures 4C and 4D). These results show that inactivation of the SAC in proliferating tissues increases cell survival upon loss of cohesion.
Figure 4.
Inhibition of the SAC Suppresses Imaginal Wing Disc Apoptosis Caused by Premature Loss of Cohesin
(A) Images of cleaved caspase-3 (CC3) immunofluorescence in controls (Rad21TEV without TEV and mad2P after HS), Rad21TEV + TEV protease, and Rad21TEV + TEV protease in mad2 mutant background after HS. The scale bar represents 100 μm.
(B) Quantification of CC3-positive area of the entire wing disc, in the indicated experimental conditions; n ≥ 5 independent discs per experimental condition; statistical analysis was performed using one-way ANOVA test.
(C) Representative images of CC3 immunofluorescence in control, san RNAi, and san and mad2 double RNAi. Scale bar indicates 100 μm.
(D) Quantification of CC3-positive area of the wing disc pouch, in control, san, and san and mad2 RNAi; n ≥ 4 independent discs per experimental condition; multiple comparison analysis was perform using a one-way ANOVA test.
Discussion
In agreement with its “safeguard” function, mitotic errors are often exacerbated by impairment of the SAC. These include defects associated with multiple centrosomes, defective microtubule assembly, or kinetochore structure [21, 22, 23, 24]. Here, we demonstrate that the opposite happens with cohesion defects. Absence of the SAC alleviated mitotic errors and improved mitotic fidelity after cohesion loss. Cells with a functional SAC undergo extensive chromosome shuffling and consequent genome randomization, whereas virtually no shuffling is observed in absence of the SAC. The detrimental nature of the SAC in the absence of cohesin is likely related to irreversibility of cohesion loss. Most mitotic defects can be corrected over time, but premature cohesin loss is an irreversible error and prolonging mitosis further enhances genome randomization.
Improved mitotic fidelity after cohesion loss in absence of the SAC is likely a consequence of slow kinetics of error-correction engagement coupled with a bias toward correct chromosome alignment. Several mechanisms are known to bias chromosome segregation toward the right orientation, including chromosome positioning [25], centromere geometry [26], bias on microtubule growth toward the kinetochores [27, 28], and/or kinetochore-mediated microtubule nucleation [29, 30]. Among these, chromosome geometry is believed to facilitate bipolar attachment by facing one kinetochore to the opposite pole upon attachment. If so, what ensures geometric arrangement during the initial mitotic stages, even in the absence of cohesin? A possible mechanism enabling transient orientation of chromatids toward opposing poles is the incomplete resolution of sister chromatid intertwines. Yet residual catenation in metaphase chromosomes is unable to confer functional cohesion, as removal of cohesin is sufficient to induce immediate chromatid separation [4, 16]. Additional mechanisms may thus impair resolution of sister chromatids during early mitosis, in contrast to metaphase chromosomes. Spindle forces enhance decatenation [31, 32], and thus resolution of DNA intertwines may only be achieved upon chromosome capture. Recent findings propose that efficient decatenation requires constant “guiding action” from condensin I [33], whose maximal levels are observed on chromosomes in late metaphase or anaphase [34, 35]. This gradual condensin loading could limit full decatenation to later stages of mitosis, making residual catenation sufficient to allow transient pseudo-metaphase alignment that biases initial chromosome attachment toward biorientation. Although error prone, this process would be more accurate than total genome randomization resulting from chromosome shuffling.
Why cohesion loss is insufficient at triggering error correction during early mitosis remains to be addressed. This could be related to a partial tension state facilitated by pseudo-metaphase chromosomal configuration. Additionally, an intrinsic delayed action of error-correction machinery may further account for observed late shuffling onset. Indeed, slow kinetics or a lag time of Aurora-B-mediated chromosome detachment has been hypothesized in several theoretical studies [36, 37, 38] with little experimental validation. Such intrinsic delay would solve the “problem of initiation of biorientation” whereby initial low-tension interactions could survive such a tension-sensitive error correction [18, 36].
Interestingly, the interplay between mitotic timing and sister chromatid cohesion has been previously reported in mammalian cells whereby extension of mitosis predisposes to sister chromatid cohesion defects. Cells arrested in mitosis for long periods were shown to display sister chromatid separation (referred as “cohesion fatigue”) [39]. Moreover, defective sister chromatid cohesion was described to be synthetically lethal with impaired APC/C function in Warsaw breakage syndrome (WABS) patient-derived cells as well as several cancer cell lines with cohesion defects [40]. Our observations demonstrate how reduction of mitotic timing is sufficient to rescue segregation defects associated with premature cohesin loss. Importantly, these experiments highlight the detrimental effect of the SAC upon cohesion defects. When sister chromatid cohesion is compromised and mitotic fidelity irreversibly affected, the SAC exacerbates mitotic errors in contrast to its canonical protective function.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Guinea Pig α-Rad21 | [41] | Christian Lehner |
| Mouse α-tubulin (Cat#T9026) | Sigma-Aldrich | RRID:AB_477593 |
| Rabbit α-Cleaved Caspase 3 (Cat#9661) | Cell Signaling Technology | RRID:AB_2341188 |
| Rabbit α-Alexa Fluor 488 (Cat#A21206) | Life Technologies | RRID:AB_141708 |
| Donkey α-Guinea Pig IRDye 800CW 0.5mg (Cat# 926-32411) | Li-Cor | RRID:AB_1850024 |
| Goat α-Mouse IRDye 680RD 0.5 mg (Cat# 926-68070) | Li-Cor | RRID:AB_10956588 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Schneider medium | Biowest | Cat#L0207 |
| Fetal Bovine Serum | Life Technologies | Cat#16000-044 |
| Colchicine | Sigma-Aldrich | Cat#C9754 |
| Binucleine 2 | Sigma-Aldrich | Cat#B1186 |
| Halocarbon oil 700 | Sigma-Aldrich | Cat#H8898 |
| Heptane | Sigma-Aldrich | Cat#246654 |
| Formaldehyde 37% solution | Acros | Cat#BP531-500 |
| 10% Tween 20 | BioRad | Cat#1610781 |
| Triton X-100 | Sigma-Aldrich | Cat#X100-500ML |
| VECTASHIELD Mounting Medium with DAPI | Vector | Cat#H-1200 |
| TEV protease | In-house production | N/A |
| Experimental Models: Organisms/Strains | ||
| y, w1118; P{w+, nubbin-GAL4} | BDSC | RRID:FlyBase_FBst1001883 |
| w1118; nubbin-Gal4, UAS-san RNAi(P{KK101696}VIE-260B)/CyO | [10] | N/A |
| Control (mCherry) RNAi: y1 sc∗ v1; P{y+t7.7 v+t1.8 = VALIUM20-mCherry}attP2 | BDSC | RRID:BDSC_35785 |
| RNAi for mad2: y1 sc∗ v1; P{y+t7.7 v+t1.8 = TRiP.GLC01381}attP2 | BDSC | RRID:BDSC_44430 |
| RNAi for Mps1: y1 sc∗ v1; P{y+t7.7 v+t1.8 = TRiP.GL00184}attP2 | BDSC | RRID:BDSC_35283 |
| RNAi for BubR1: y1 sc∗ v1; P{y+t7.7 v+t1.8 = TRiP.GL00236}attP2 | BDSC | RRID:BDSC_35329 |
| RNAi for BubR1: y1 sc∗ v1; P{y+t7.7 v+t1.8 = TRiP.GLV21065}attP2 | BDSC | RRID:BDSC_35700 |
| RNAi for Mad1: y1 sc∗ v1; P{y+t7.7 v+t1.8 = TRiP.GLV21088}attP2 | BDSC | RRID:BDSC_35723 |
| RNAi for Mad1: y1 sc∗ v1; P{y+t7.7 v+t1.8 = TRiP.HMC03671}attP40 | BDSC | RRID:BDSC_52930 |
| RNAi for fzy: y1 sc∗ v1; P{y+t7.7 v+t1.8 = TRiP.HMS00964}attP2 | BDSC | RRID:BDSC_34001 |
| RNAi for Cdc23: y1 v1; P{y+t7.7 v+t1.8 = TRiP.HMJ23608}attP40 | BDSC | RRID:BDSC_61982 |
| All other trip RNAi lines used for the screen are detailed in Data S1 | BDSC | N/A |
| nubbin-Gal4/CyO; His2AvD-mRFP1 III.1, Cid-EGFP III.1/TM6B | This study | N/A |
| UAS-san RNAi(P{KK101696}VIE-260B)/CyO; UAS-Luciferase RNAi(P{TRiP.JF01355}attP2)/TM6B | This study | N/A |
| UAS-san RNAi(P{KK101696}VIE-260B)/CyO; UAS-Mad2 RNAi(P{TRiP.GLC01381}attP2)/TM6B | This study | N/A |
| yw; y+CyO/Sp; mad2P | [15] | Roger Karess |
| w;;His2Av-mRFP1 III.1, Cid-EGFP III.1 | [42] | Stefan Heidmann |
| w; 2x Rad21271-3TEV-myc; rad21ex3, mad2P | [43] | Christian Lehner |
| w; 2x Rad21271-3TEV-myc; rad21ex3 | [43] | Christian Lehner |
| w;; polyubiq-H2B-RFP, Rad21550TEV-EGFP, rad21ex3 | [44] | N/A |
| w; HisH2Av- mRFP1 II.2; Rad21ex15, Rad21550-3TEV –myc, Cid-EGFP III.1 | [16] | N/A |
| w; hspr-NLSv5TEV; Rad21ex3/TM6B ubiGFP | [14] | N/A |
| w; hsTEV, His2Av-mRFP, Cid-EGFP/CyO; rad21ex, mad2P/TM6B | This study | N/A |
| Software and Algorithms | ||
| FIJI | [45] | RRID:SCR_002285https://fiji.sc/ |
| Prism 7 | GraphPad | RRID:SCR_002798https://www.graphpad.com/ |
| Photoshop CS6 | Adobe | RRID:SCR_014199https://www.adobe.com/ |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Raquel A. Oliveira (rcoliveira@igc.gulbenkian.pt).
Experimental Model and Subject Details
Drosophila strains and rearing conditions
All RNAi lines used in the screen are from the Transgenic RNAi project (TRiP), are available in the Bloomington Drosophila Stock Center and are listed in Data S1. Other Drosophila stocks used in this study are indicated in the Key Resources Table. For the reported screen, 2-4 days old adult flies were used (both male and female). All cell biology analysis was performed in third instar larvae without gender selection. To induce full cohesin cleavage in a temporally controlled manner, by TEV protease cleavage, Drosophila strains were used with TEV-cleavable Rad21 (Rad21TEV) in a Rad21 null background (rad21ex15, Rad21271-3TEV-myc or rad21ex15, Rad21550-3TEV-EGFP) [14, 44], in strains mutant or wild-type for the Mad2 gene [15, 43]. TEV expression was induced by heat-shocking 3rd instar larvae at 37°C for 45 minutes. Larvae were then left to recover at room temperature. For live cell imaging, fly strains also expressed His2AvD-mRFP1 and Cid-EGFP [42] fluorescent markers. Drosophila melanogaster flies were raised at 25°C or 18°C for hs-TEV containing crosses in polypropylene vials (51 mm diameter) containing enriched medium (cornmeal, molasses, yeast, soya flour and beetroot syrup).
Method Details
Drosophila screen Details
2955 RNAi lines were analysed in the screen, which theoretically corresponds to depletion of approximately 21% of all protein coding genes annotated in Flybase (Flybase versionFB2017_06). The tested RNAi lines were ordered mostly alphabetically, according to gene name or CG number, from the TRiP website. Used RNAi lines were mostly constructed with Vallium20 or Valium22 vectors. In the screen, females carrying the nubbin-Gal4, UAS-san RNAi were crossed with males of different candidate RNAi lines from TRiP (see diagram in Figure S1A). The progeny of these flies was scored blind and classified into different classes according to the adult wing phenotypes: class 1 - wild-type wings; class 2 – flies with wings that present only mild morphological defects; class 3- flies whose wing morphological defects are intermediate (similar san RNAi); class 4 – flies whose wings show strong morphological defects; class 5 – flies without wings or vestigial wings (Figure 1C) [46]. The average adult wing class for each condition was always calculated using more than 50 adult flies (n ≥ 50). If the average class for a given genetic interaction was equal or below 2.6 than the RNAi line tested was classified as suppressor, if the average class was equal or above 3.5 than the RNAi line was classified as enhancer (Figure S1A). To exclude RNAi lines whose expression by itself led to wing morphological defects, in otherwise wild-type imaginal discs, all RNAi lines identified in the primary screen were crossed with nubbin-Gal4. All RNAi lines identified as enhancers but that produced significant adult wing phenotypes by themselves were discarded (Figure S1A). All identified enhancer/suppressor RNAi lines were confirmed by three independent replicas (see Data S1).
Live imaging
For imaging of wing discs, larval imaginal wing discs were dissected in Schneider medium with 10% FBS. Dissected discs were placed and oriented in a 200 μL drop of medium at the bottom of a glass-bottom petridish (MakTek). For Aurora B inhibition experiments (Figure S4), discs were incubated with Binuclein 2 (Sigma-Aldrich), at the indicated concentrations, throughout imaging acquisition. Time lapse imaging analysis was performed on a spinning disc microscope using either a Revolution XD microscope (Andor, UK), equipped with immersion a 60x (water) and 100x (oil) objectives (Nikon, Japan) and a iXon +512 EMCCD camera (Andor, UK), or a Revolution XD microscope (Andor, UK) equipped with immersion a 60x glycerol-immersion 1.30 NA objective (Leica Microsystems, Germany) and a 100x oil-immersion 1.4 NA objective (Leica Microsystems, Germany) and a iXon Ultra 888 1024∗1024 EMCCD (Andor, UK). Stacks of 20-30 frames 0,5 μm apart were taken every 1 to 3 minutes. For syncytial embryo imaging, embryos were aligned on coverslips and covered with Series 700 halocarbon oil (Sigma-Aldrich). Time-lapse microscopy was performed with an inverted wide-field DeltaVision microscope (Applied Precision, Issaquah, WA) in a temperature-controlled room (18–20°C). One stack of 15 frames (0.8 mm apart) was acquired every 30 s with a 100x 1.4 oil immersion objective (Olympus, Japan) and captured by an EMCCD camera (Roper Cascade 1024, Roper Technologies). Movies were assembled using FIJI software [45] and selected stills were processed with Photoshop CS6 (Adobe).
Microinjections
Microinjections were performed as previously described [16, 33]. 1-1.5 hr old embryos were collected and processed according to standard protocols, and were injected at the posterior pole. Injections were performed using a Burleigh Thorlabs Micromanipulator, a Femtojet microinjection system (Eppendorf, Germany), and pre-pulled Femtotip I needles (Eppendorf). TEV protease was injected at 5 mg/ml TEV protease in 20 mM Tris-HCl at pH 8.0, 1 mM EDTA, 50 mM NaCl and 2 mM DTT.
Immunofluorescence
Third instar wing imaginal disc fixation and staining was performed using standard procedures [47]. Briefly, third instar larvae wing disc tissue (still attached to the larva body) was fixed on ice for 30 min. The fixative consisted of 4% formaldehyde (Polysciences) in 1X PEM buffer solution. Following, tissues were washed by gentle agitation three times for 20 min in PBS-T (1x PBS + 0.1% Triton X-100). Primary antibodies incubation was performed overnight at 4 °C in PBS-T supplemented with 1% BSA and 1% donkey serum. The following day, the tissues were washed again and incubated for 2h at room temperature with the appropriate secondary antibodies diluted in PBS-T solution. Finally, after the wash of the secondary antibodies, wing discs were mounted in Vectashield (Vector Laboratories). Fluorescence images were acquired with a × 40 HCX PL APO CS oil immersion objective (numerical aperture: 1.25–0.75) on a Leica SP5 confocal microscope. Rabbit anti-cleaved caspase 3 at 1:300 (Cell Signaling, RRID:AB_2341188) and anti-Rabbit Alexa Fluor 488 at 1:1000 (Molecular Probes).
Western-blot
To analyze Rad21 protein amounts, Drosophila tissues were dissected in PBS and homogenized with a pestle in Sample buffer. Samples were centrifuged, and boiled for 5 minutes in 2x Sample Buffer. Samples were loaded on a 13% SDS-gel for electrophoresis and transferred to nitrocellulose membranes. Western-blot analysis was performed according to standard protocols using the following antibodies: anti-α-tubulin (1:50.000, DM1A, RRID:AB_477593), guinea pig anti-Rad21 (1:5.000, [41]).
Quantification and Statistical Analysis
Imaging analysis was performed using FIJI software [45]. Statistical analysis and graphic representations were performed using Prism 7 software. Multiple comparisons were performed using one-way ANOVA, using the Bonferroni’s multiple comparison test. Graphs depict mean ± standard deviations (SD) or mean ± standard error of the mean (SEM), as indicated. Sample size details are included in the respective figure legends.
Data and Software Availability
Detailed screen strains and results are summarized in Data S1.
Acknowledgments
We thank S. Heidmann, C. Lehner, R. Karess, and the Bloomington Stock Center for fly strains and antibodies; the Instituto Gulbenkian de Ciência’s (IGC) Fly Facility; all the members of the Oliveira and Martinho laboratories for discussions; Ricardo Matos for assistance with graphic design; and Bárbara Kellen for technical assistance in the pilot screen. We acknowledge the TRiP at Harvard Medical School (NIH/NIGMS R01-GM084947) for providing several transgenic RNAi fly stocks used in this study. We thank the technical support of IGC’s Advanced Imaging Facility, supported by the national Portuguese funding ref no. PPBI-POCI-01-0145-FEDER-022122, and the research infrastructure Congento: project LISBOA-01-0145-FEDER-022170. These two programs are co-financed by Lisboa Regional Operational Programme (Lisboa 2020), under the Portugal 2020 Partnership Agreement, through the European Regional Development Fund (FEDER) and Fundação para a Ciência e a Tecnologia (FCT; Portugal). The following authors were supported by FCT fellowships: R.D.S. (SFRH/BPD/87482/2012), M.M. (SFRH /BD/52438/2013), and O.S.R. (PD/BD/52428/2013, within the scope of the ProRegeM PhD program ref. PD/00117/2012, CRM: 0027030). R.G.M. is supported by funding from the Association for International Cancer Research (AICR 10–0553) and the following FCT grants: PTDC/BEX-BID/0395/2014 and UID/BIM/04773/2013 CBMR 1334. R.A.O. is supported by the following grants: FCT Investigator grant (IF/00851/2012/CP0185/CT0004), EMBO Installation Grant (IG2778), and European Research Council Starting Grant (ERC-2014-STG-638917). The following ORCID URLs apply to the authors: R.D.S. (orcid.org/0000-0003-2168-8599), M.M. (orcid.org/0000-0003-0802-7200), R.G.M. (orcid.org/0000-0002-1641-3403), and R.A.O. (orcid.org/0000-0002-8293-8603).
Author Contributions
Conceptualization, R.D.S., M.M., R.G.M., and R.A.O.; Investigation, R.D.S., M.M., L.G.G., O.S.R., and R.A.O.; Writing—Original Draft, R.G.M. and R.A.O.; Writing—Review & Editing, R.D.S., M.M., R.G.M., and R.A.O.; Supervision, R.G.M. and R.A.O.; Funding Acquisition, R.G.M. and R.A.O.
Declaration of Interests
The authors declare no competing interests.
Published: August 16, 2018
Footnotes
Supplemental Information includes four figures, one table, four videos, and one data file and can be found with this article online at https://doi.org/10.1016/j.cub.2018.06.062.
Contributor Information
Rui Gonçalo Martinho, Email: rgmartinho@ualg.pt.
Raquel A. Oliveira, Email: rcoliveira@igc.gulbenkian.pt.
Supplemental Information
Sheet 1: List of RNAi lines used in the Drosophila wing modifier screen, downloaded from the Transgenic RNAi Project (TRiP). Sheet 2: Adult wing phenotypes of Drosophila carrying the nubbin-Gal4 driver, the UAS-Naa50/san RNAi and the UAS-RNAi indicated at the table (first biological replica); RNAis were classified as suppressors or enhancers according to the criteria described in methods. When no adult carrying both RNAis was found, we classified the interaction as lethal. Empty cells indicate that adult flies were not scored. RNAi lines that genetically interacted with the Naa50/san adult wing phenotype (either by suppressing or enhancing it) were confirmed with a second independent biological replica (Sheet 3). Sheet 3: Adult wing phenotypes of Drosophila carrying the nubbin-Gal4 driver, the UAS-Naa50/san RNAi and the UAS-RNAi indicated at the table (second biological replica); RNAis were classified as suppressors or enhancers according to the criteria described in methods. Enhancer RNAi lines that alone with Nubbin-Gal4 gave rise to an adult wing phenotype were discarded. RNAi lines that reproducibly genetically interacted with the Naa50/san adult wing phenotype (either by suppressing or enhancing it) were further confirmed with a third independent biological replica (Sheet 4). Sheet 4: Adult wing phenotypes of Drosophila carrying the nubbin-Gal4 driver, the UAS-Naa50/san RNAi and the UAS-RNAi indicated at the table (third biological replica); RNAis were classified as suppressors or enhancers according to the criteria described in methods.
References
- 1.Michaelis C., Ciosk R., Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- 2.Guacci V., Koshland D., Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haering C.H., Farcas A.M., Arumugam P., Metson J., Nasmyth K. The cohesin ring concatenates sister DNA molecules. Nature. 2008;454:297–301. doi: 10.1038/nature07098. [DOI] [PubMed] [Google Scholar]
- 4.Uhlmann F., Wernic D., Poupart M.A., Koonin E.V., Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103:375–386. doi: 10.1016/s0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- 5.Uhlmann F., Lottspeich F., Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- 6.Mirkovic M., Oliveira R.A. Centromeric Cohesin: Molecular Glue and Much More. Prog. Mol. Subcell. Biol. 2017;56:485–513. doi: 10.1007/978-3-319-58592-5_20. [DOI] [PubMed] [Google Scholar]
- 7.Musacchio A., Salmon E.D. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 8.Foley E.A., Kapoor T.M. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat. Rev. Mol. Cell Biol. 2013;14:25–37. doi: 10.1038/nrm3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams B.C., Garrett-Engele C.M., Li Z., Williams E.V., Rosenman E.D., Goldberg M.L. Two putative acetyltransferases, san and deco, are required for establishing sister chromatid cohesion in Drosophila. Curr. Biol. 2003;13:2025–2036. doi: 10.1016/j.cub.2003.11.018. [DOI] [PubMed] [Google Scholar]
- 10.Ribeiro A.L., Silva R.D., Foyn H., Tiago M.N., Rathore O.S., Arnesen T., Martinho R.G. Naa50/San-dependent N-terminal acetylation of Scc1 is potentially important for sister chromatid cohesion. Sci. Rep. 2016;6:39118. doi: 10.1038/srep39118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hou F., Chu C.W., Kong X., Yokomori K., Zou H. The acetyltransferase activity of San stabilizes the mitotic cohesin at the centromeres in a shugoshin-independent manner. J. Cell Biol. 2007;177:587–597. doi: 10.1083/jcb.200701043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rong Z., Ouyang Z., Magin R.S., Marmorstein R., Yu H. Opposing Functions of the N-terminal Acetyltransferases Naa50 and NatA in Sister-chromatid Cohesion. J. Biol. Chem. 2016;291:19079–19091. doi: 10.1074/jbc.M116.737585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ryoo H.D., Gorenc T., Steller H. Apoptotic cells can induce compensatory cell proliferation through the JNK and the Wingless signaling pathways. Dev. Cell. 2004;7:491–501. doi: 10.1016/j.devcel.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 14.Pauli A., Althoff F., Oliveira R.A., Heidmann S., Schuldiner O., Lehner C.F., Dickson B.J., Nasmyth K. Cell-type-specific TEV protease cleavage reveals cohesin functions in Drosophila neurons. Dev. Cell. 2008;14:239–251. doi: 10.1016/j.devcel.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buffin E., Emre D., Karess R.E. Flies without a spindle checkpoint. Nat. Cell Biol. 2007;9:565–572. doi: 10.1038/ncb1570. [DOI] [PubMed] [Google Scholar]
- 16.Oliveira R.A., Hamilton R.S., Pauli A., Davis I., Nasmyth K. Cohesin cleavage and Cdk inhibition trigger formation of daughter nuclei. Nat. Cell Biol. 2010;12:185–192. doi: 10.1038/ncb2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nezi L., Musacchio A. Sister chromatid tension and the spindle assembly checkpoint. Curr. Opin. Cell Biol. 2009;21:785–795. doi: 10.1016/j.ceb.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 18.Khodjakov A., Pines J. Centromere tension: a divisive issue. Nat. Cell Biol. 2010;12:919–923. doi: 10.1038/ncb1010-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mirkovic M., Hutter L.H., Novák B., Oliveira R.A. Premature Sister Chromatid Separation Is Poorly Detected by the Spindle Assembly Checkpoint as a Result of System-Level Feedback. Cell Rep. 2015;13:469–478. doi: 10.1016/j.celrep.2015.09.020. [DOI] [PubMed] [Google Scholar]
- 20.Carmena M., Wheelock M., Funabiki H., Earnshaw W.C. The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell Biol. 2012;13:789–803. doi: 10.1038/nrm3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee M.S., Spencer F.A. Bipolar orientation of chromosomes in Saccharomyces cerevisiae is monitored by Mad1 and Mad2, but not by Mad3. Proc. Natl. Acad. Sci. USA. 2004;101:10655–10660. doi: 10.1073/pnas.0404102101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tarailo M., Tarailo S., Rose A.M. Synthetic lethal interactions identify phenotypic “interologs” of the spindle assembly checkpoint components. Genetics. 2007;177:2525–2530. doi: 10.1534/genetics.107.080408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daniel J.A., Keyes B.E., Ng Y.P., Freeman C.O., Burke D.J. Diverse functions of spindle assembly checkpoint genes in Saccharomyces cerevisiae. Genetics. 2006;172:53–65. doi: 10.1534/genetics.105.046441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Basto R., Brunk K., Vinadogrova T., Peel N., Franz A., Khodjakov A., Raff J.W. Centrosome amplification can initiate tumorigenesis in flies. Cell. 2008;133:1032–1042. doi: 10.1016/j.cell.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magidson V., O’Connell C.B., Lončarek J., Paul R., Mogilner A., Khodjakov A. The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly. Cell. 2011;146:555–567. doi: 10.1016/j.cell.2011.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanaka T., Fuchs J., Loidl J., Nasmyth K. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nat. Cell Biol. 2000;2:492–499. doi: 10.1038/35019529. [DOI] [PubMed] [Google Scholar]
- 27.Carazo-Salas R.E., Guarguaglini G., Gruss O.J., Segref A., Karsenti E., Mattaj I.W. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature. 1999;400:178–181. doi: 10.1038/22133. [DOI] [PubMed] [Google Scholar]
- 28.Wollman R., Cytrynbaum E.N., Jones J.T., Meyer T., Scholey J.M., Mogilner A. Efficient chromosome capture requires a bias in the ‘search-and-capture’ process during mitotic-spindle assembly. Curr. Biol. 2005;15:828–832. doi: 10.1016/j.cub.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 29.Maiato H., Rieder C.L., Khodjakov A. Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J. Cell Biol. 2004;167:831–840. doi: 10.1083/jcb.200407090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kitamura E., Tanaka K., Komoto S., Kitamura Y., Antony C., Tanaka T.U. Kinetochores generate microtubules with distal plus ends: their roles and limited lifetime in mitosis. Dev. Cell. 2010;18:248–259. doi: 10.1016/j.devcel.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baxter J., Sen N., Martínez V.L., De Carandini M.E., Schvartzman J.B., Diffley J.F., Aragón L. Positive supercoiling of mitotic DNA drives decatenation by topoisomerase II in eukaryotes. Science. 2011;331:1328–1332. doi: 10.1126/science.1201538. [DOI] [PubMed] [Google Scholar]
- 32.Charbin A., Bouchoux C., Uhlmann F. Condensin aids sister chromatid decatenation by topoisomerase II. Nucleic Acids Res. 2014;42:340–348. doi: 10.1093/nar/gkt882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piskadlo E., Tavares A., Oliveira R.A. Metaphase chromosome structure is dynamically maintained by condensin I-directed DNA (de)catenation. eLife. 2017;6:e26120. doi: 10.7554/eLife.26120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oliveira R.A., Heidmann S., Sunkel C.E. Condensin I binds chromatin early in prophase and displays a highly dynamic association with Drosophila mitotic chromosomes. Chromosoma. 2007;116:259–274. doi: 10.1007/s00412-007-0097-5. [DOI] [PubMed] [Google Scholar]
- 35.Gerlich D., Hirota T., Koch B., Peters J.M., Ellenberg J. Condensin I stabilizes chromosomes mechanically through a dynamic interaction in live cells. Curr. Biol. 2006;16:333–344. doi: 10.1016/j.cub.2005.12.040. [DOI] [PubMed] [Google Scholar]
- 36.Tubman E.S., Biggins S., Odde D.J. Stochastic Modeling Yields a Mechanistic Framework for Spindle Attachment Error Correction in Budding Yeast Mitosis. Cell Syst. 2017;4:645–650.e5. doi: 10.1016/j.cels.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang T., Oliveira R.A., Schmierer B., Novák B. Dynamical scenarios for chromosome bi-orientation. Biophys. J. 2013;104:2595–2606. doi: 10.1016/j.bpj.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalantzaki M., Kitamura E., Zhang T., Mino A., Novák B., Tanaka T.U. Kinetochore-microtubule error correction is driven by differentially regulated interaction modes. Nat. Cell Biol. 2015;17:530. doi: 10.1038/ncb3153. [DOI] [PubMed] [Google Scholar]
- 39.Daum J.R., Potapova T.A., Sivakumar S., Daniel J.J., Flynn J.N., Rankin S., Gorbsky G.J. Cohesion fatigue induces chromatid separation in cells delayed at metaphase. Curr. Biol. 2011;21:1018–1024. doi: 10.1016/j.cub.2011.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Lange J., Faramarz A., Oostra A.B., de Menezes R.X., van der Meulen I.H., Rooimans M.A., Rockx D.A., Brakenhoff R.H., van Beusechem V.W., King R.W. Defective sister chromatid cohesion is synthetically lethal with impaired APC/C function. Nat. Commun. 2015;6:8399. doi: 10.1038/ncomms9399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heidmann D., Horn S., Heidmann S., Schleiffer A., Nasmyth K., Lehner C.F. The Drosophila meiotic kleisin C(2)M functions before the meiotic divisions. Chromosoma. 2004;113:177–187. doi: 10.1007/s00412-004-0305-5. [DOI] [PubMed] [Google Scholar]
- 42.Schuh M., Lehner C.F., Heidmann S. Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr. Biol. 2007;17:237–243. doi: 10.1016/j.cub.2006.11.051. [DOI] [PubMed] [Google Scholar]
- 43.Althoff F., Karess R.E., Lehner C.F. Spindle checkpoint-independent inhibition of mitotic chromosome segregation by Drosophila Mps1. Mol. Biol. Cell. 2012;23:2275–2291. doi: 10.1091/mbc.E12-02-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oliveira R.A., Kotadia S., Tavares A., Mirkovic M., Bowlin K., Eichinger C.S., Nasmyth K., Sullivan W. Centromere-independent accumulation of cohesin at ectopic heterochromatin sites induces chromosome stretching during anaphase. PLoS Biol. 2014;12:e1001962. doi: 10.1371/journal.pbio.1001962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu J., Cohen S.M. Repression of Teashirt marks the initiation of wing development. Development. 2002;129:2411–2418. doi: 10.1242/dev.129.10.2411. [DOI] [PubMed] [Google Scholar]
- 47.Lee J.D., Treisman J.E. The role of Wingless signaling in establishing the anteroposterior and dorsoventral axes of the eye disc. Development. 2001;128:1519–1529. doi: 10.1242/dev.128.9.1519. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DNA is labeled with H2Av-mRFP1 (red) and centromeres with cid-EGFP (green). Times are relative to NEBD.
DNA is labeled with H2Av-mRFP1 (gray). Times are relative to NEBD.
DNA is labeled with H2Av-mRFP1 (red) and centromeres with id-EGFP (green). Times are relative to NEBD.
DNA is labeled with H2Av-mRFP1 (gray). Times are relative to NEBD.
Sheet 1: List of RNAi lines used in the Drosophila wing modifier screen, downloaded from the Transgenic RNAi Project (TRiP). Sheet 2: Adult wing phenotypes of Drosophila carrying the nubbin-Gal4 driver, the UAS-Naa50/san RNAi and the UAS-RNAi indicated at the table (first biological replica); RNAis were classified as suppressors or enhancers according to the criteria described in methods. When no adult carrying both RNAis was found, we classified the interaction as lethal. Empty cells indicate that adult flies were not scored. RNAi lines that genetically interacted with the Naa50/san adult wing phenotype (either by suppressing or enhancing it) were confirmed with a second independent biological replica (Sheet 3). Sheet 3: Adult wing phenotypes of Drosophila carrying the nubbin-Gal4 driver, the UAS-Naa50/san RNAi and the UAS-RNAi indicated at the table (second biological replica); RNAis were classified as suppressors or enhancers according to the criteria described in methods. Enhancer RNAi lines that alone with Nubbin-Gal4 gave rise to an adult wing phenotype were discarded. RNAi lines that reproducibly genetically interacted with the Naa50/san adult wing phenotype (either by suppressing or enhancing it) were further confirmed with a third independent biological replica (Sheet 4). Sheet 4: Adult wing phenotypes of Drosophila carrying the nubbin-Gal4 driver, the UAS-Naa50/san RNAi and the UAS-RNAi indicated at the table (third biological replica); RNAis were classified as suppressors or enhancers according to the criteria described in methods.