Abstract
Monocarboxylate transporter-8 (MCT8) is a specific thyroid hormone transporter, essential for the uptake of thyroid hormone into target tissues. Mutations in the MCT8 gene have been identified as the cause of Allan-Herndon-Dudley syndrome (AHDS). It has been reported that soy isoflavones influence thyroid hormone system and can interact with thyroid hormone transporter proteins. Therefore, the present study aimed to find out whether soy isoflavones (genistein, daidzein and glycitein) can be used as a natural inhibitor to target MCT8 in AHDS. Docking studies were performed for soy isoflavones in order to evaluate their binding affinity to MCT8 protein using AutoDock4 (version 4.2.6) and AutoDock Vina. After docking, the ligands were ranked according to their binding energy and the best lead compound was selected based on the least binding energy. The docking results indicated that daidzein possesses the lowest binding energy against MCT8. Moreover, it was found that the residues PRO-338, HIS-341, and GLU-348 were involved in hydrogen bond interactions with genistein and daidzein. This study suggests that daidzein is a promising natural inhibitor to target MCT8 in AHDS.
Keywords: Molecular docking, MCT8, AHDS, Soy isoflavones, Daidzein, Genistein
1. Introduction
Thyroid hormone plays an important role in our body with widespread biological actions. Adequate levels of thyroid hormone are crucial for the development and proper function of multiple organs [1], [2]. Thyroid hormone exists in two forms: T4 (3,3’,5,5’-tetraiodothyronine) and T3 (3,3’,5-triodothyronine). The biological activity of thyroid hormone is related to the level of T3 within the cell. MCT8 (monocarboxylate transporter 8) protein is a transporter specific for T3 [1]. The MCT8 gene is located in Xq13 and mutations in MCT8 are responsible for an X-linked condition, known as the Allan–Herndon–Dudley syndrome (AHDS) showing high serum T3 levels in affected male patients [1], [2].
Currently, there is no effective treatment available for AHDS. Hence, there is an urgent need for the identification and validation of novel drug lead compounds for treating AHDS. Therefore, this study aimed to identify the natural potent inhibitors of MCT8 from soy isoflavones. While isoflavones occur in many types of legumes, soybean contains the highest concentration of isoflavones. Genistein, daidzein and glycitein are the soy isoflavones typically accounting for 50%, 40% and 10%, respectively [3]. Due to their chemical structure, the isoflavones can bind to estrogen receptors. As a result of this binding, isoflavones inhibit and promote the expression of estrogen-sensitive genes [4]. Previous researches have shown that the occurrence of breast cancer is lower in Asian individuals than in other populations because of the high soy consumption as part of their regular diet [3], [4].
Soy isoflavones influence thyroid hormone system and can interact with thyroid hormone transporter proteins [5], [6]. In vitro and in vivo studies have indicated that genistein and daidzein are the potent inhibitors of thyroid peroxidase [5]. Moreover, soy isoflavones are the potent ligands for transthyretin in serum and cerebrospinal fluid [5]. However, soy isoflavones have not been studied against MCT8. So this study made an attempt to find out whether soy isoflavones (genistein, daidzein and glycitein) can be used as a natural inhibitor to target MCT8 in AHDS.
2. Materials and methods
2.1. Drug-likeness of the ligands
Genistein, daidzein and glycitein were considered as ligands. SwissADME tool [7] was used to calculate the molecular properties of the ligands. The molecular properties were screened based on the “Lipinski's rule of five” [8], [9]. The total polar surface area (TPSA) and the number of rotatable bonds were also calculated using SwissADME [7].
2.2. Protein preparation for docking
The human MCT8 protein is not available in protein data bank (PDB) [10]. The protein used in the docking study was obtained through multi-template based homology modeling [11]. In the previous study [11], a good-quality 3D model of MCT8 was predicted based on multiple templates (PDB IDs: 1pw4A, 4u4tA, 4ikxA, 4j05A, and 4gbyA) using the advanced modeling feature of MODELLERv9.17 [12], [13]. The structure refinement of modeled protein was done by ModRefiner [14]. For docking, all water molecules were removed and polar hydrogen atoms were added to the refined model using AutoDock Tools (ADT) [15]. The prepared protein was saved in PDBQT format.
2.3. Ligand preparation for docking
The ligands were downloaded from Pubchem Database [16], [17] and converted to PDB [10] file format by using Openbabel software [18]. The ligands were prepared using ADT [15]. Gasteiger charge was assigned to the ligands. The prepared ligands were saved in PDBQT format.
2.4. Molecular docking
AutoDock4 (version 4.2.6) [15] and AutoDock Vina [19] were used for molecular docking studies. AutoGrid program supplied with AutoDock4 [15] was used for the preparation of grid maps. The grid box size was set at 76, 70, and 76 Å for x, y, and z, respectively. The spacing between the grid points was 1.0 Å. The grid centre was set at 30.375, 17.112, and −37.003 Å for x, y, and z, respectively. The Lamarckian Genetic Algorithm (LGA) was chosen to search for the best conformers. During the docking process, a maximum of 10 conformers was considered for each ligand. All the docking processes were performed with the default parameters of AutoDock 4 [15]. Population size was set to 150, maximum number of evaluations 2,500,000, maximum number of generations 27,000, maximum number of top individual that automatically survived 1, gene mutation rate 0.02 and crossover rate 0.8. AutoDock4 [15] and AutoDock Vina [19] were compiled and run under Windows 10 Operating System. All figures with structure representations were produced using Discovery Studio Visualizer [20].
3. Results and discussion
3.1. Drug-likeness of the ligands
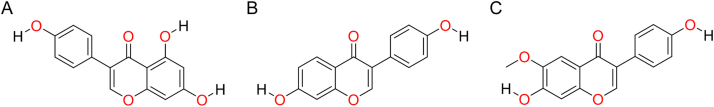
“Lipinski’s rule of five” [8], [9] is used to evaluate the drug-likeness of a chemical compound. The molecular properties of a chemical compound consist of molecular weight, hydrogen bond donor, hydrogen bond acceptor, and log P. “Lipinski’s rule of five” states that an orally active drug has no more than one violation of the following criteria: (1) less than 5 hydrogen-bond donors, (2) less than 10 hydrogen-bond acceptors, (3) a molecular mass less than 500 Da, and (4) log P not greater than 5. All ligands of the present study met the requirements of “Lipinski’s rule of five”. The other significant properties such as total polar surface area (TPSA) and the number of rotatable bonds were also calculated. TPSA of a compound should be less than 140 Å2 and the number of rotatable bonds should be less than 10 [21]. All the ligands had the above properties. The chemical structures of the ligands are shown in Fig. 1 and their molecular properties are shown in Table 1.
Fig. 1.
Chemical structures of (A) genistein, (B) daidzein and (C) glycitein.
Table 1.
Molecular properties of the ligands.
| Ligand | Num. H-bond acceptors | Num. H-bond donors | MLOGP | Number of rotatable bonds | Molecular weight (g/mol) | TPSA (Å2) |
|---|---|---|---|---|---|---|
| Genistein | 5 | 3 | 0.52 | 1 | 270.24 | 90.90 |
| Daidzein | 4 | 2 | 1.08 | 1 | 254.24 | 70.67 |
| Glycitein | 5 | 2 | 0.77 | 2 | 284.26 | 79.90 |
3.2. Molecular docking
Molecular docking is an important tool in pharmaceutical research [22]. The molecular docking approach can be used to model the interaction between a ligand and a protein at the atomic level [22], [23]. The docking process involves two basic steps: prediction of the ligand conformation and assessment of the binding affinity [22]. These two steps are related to sampling methods and scoring schemes, respectively [22].
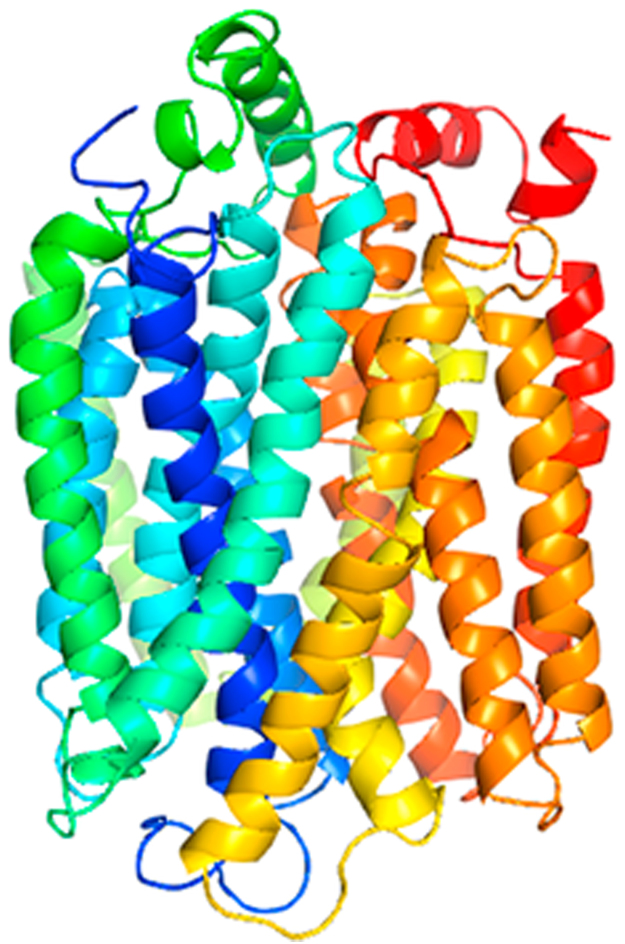
Docking studies were performed for 3 compounds in order to evaluate their binding affinity to MCT8 protein using AutoDock4 (version 4.2.6) [15] and AutoDock Vina [19]. The 3D structure of the MCT8 protein is shown in Fig. 2. The results were analyzed based on the binding energies of the docked complexes. AutoDock4 [15] and AutoDock Vina [19] generated 10 poses for each ligand. The selection of the best pose was done on the least binding energy between the ligand and the protein. After docking, the ligands were ranked according to their binding energy.
Fig. 2.
3D structure of MCT8 protein by multi-template homology modeling [11]. This 3D model of MCT8 was predicted based on multiple templates (PDB IDs: 1pw4A, 4u4tA, 4ikxA, 4j05A, and 4gbyA) using the advanced modeling feature of MODELLERv9.17 [12], [13].
3.2.1. Docked results with AutoDock4
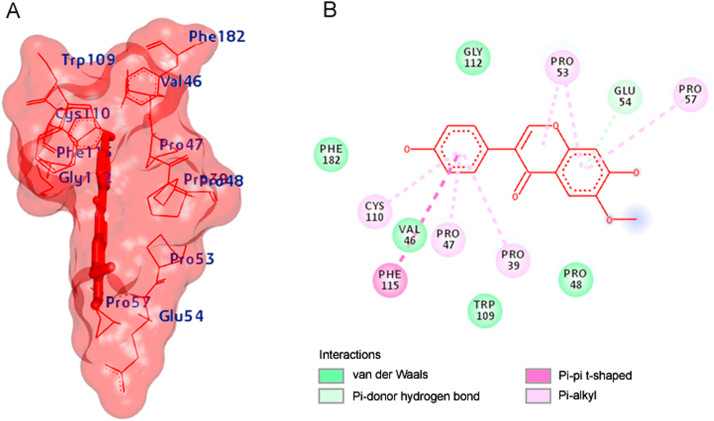
All the ligand molecules were docked against MCT8 using AutoDock4 [15]. The best selected pose of MCT8-genistein docked complex (binding energy −5.76 kcal/mol) with binding site residues is shown in Fig. 3A. The hydrogen bonds and the types of contacts involved in MCT8-genistein complex are shown in Fig. 3B. It was observed that PRO-338, HIS-341, LEU-342, MET-343 and GLU-348 were involved in hydrogen bond interactions.
Fig. 3.
(A) The output of AutoDock showing the binding site residues of MCT8 protein with the ligand genistein. The residues in the binding site are shown in red color. Genistein is shown in purple stick format. (B) 2D diagram showing the types of contacts formed between MCT8 and genistein. The green dotted lines indicate H‑bond interactions between MCT8 and genistein. The values adjacent to the green dotted lines indicate their distance.
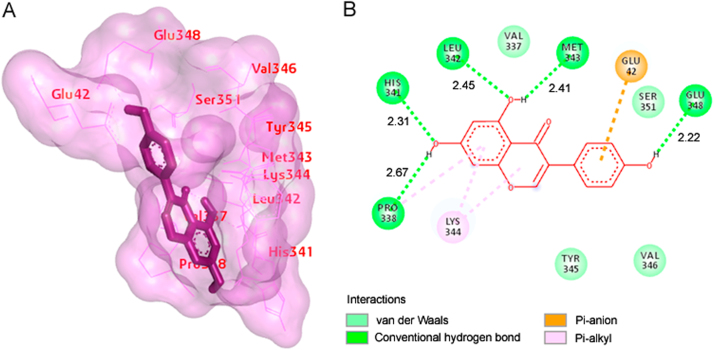
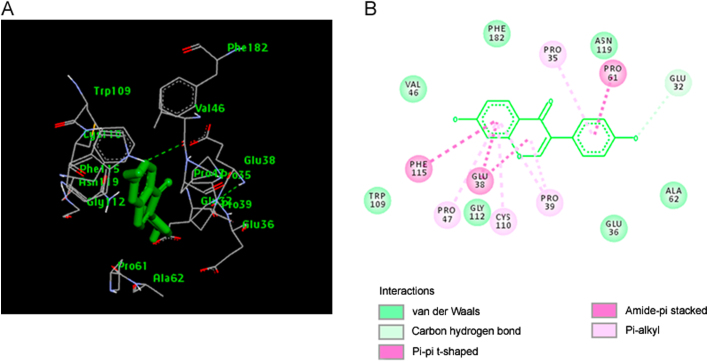
The best selected pose of MCT8-daidzein docked complex (binding energy −6.22 kcal/mol) with binding site residues is shown in Fig. 4A. The hydrogen bonds and the types of contacts involved in MCT8-daidzein complex are shown in Fig. 4B. It was observed that PRO-338, HIS-341, and GLU-348 were involved in hydrogen bond interactions.
Fig. 4.
(A) The output of AutoDock showing the binding site residues of MCT8 protein with the ligand daidzein. The residues in the binding site are shown in red color. Daidzein is shown in green stick format. (B) 2D diagram showing the types of contacts formed between MCT8 and daidzein. The green dotted lines indicate H‑bond interactions between MCT8 and daidzein. The values adjacent to the green dotted lines indicate their distance.
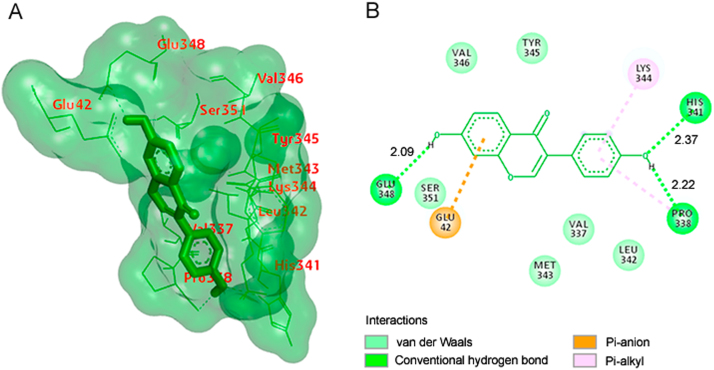
The best selected pose of MCT8-glycitein docked complex (binding energy −5.54 kcal/mol) with binding site residues is shown in Fig. 5A. The types of contacts involved in MCT8-glycitein complex are shown in Fig. 5B. The results showed that there were no hydrogen bonds formed between MCT8 and glycitein.
Fig. 5.
(A) The output of AutoDock showing the binding site residues of MCT8 protein with the ligand glycitein. The residues in the binding site are shown in blue color. Glycitein is shown in red stick format. (B) 2D diagram showing the types of contacts formed between MCT8 and glycitein.
The docking with AutoDock4 showed that daidzein was the best scored compound against MCT8 with the lowest binding energy.
3.2.2. Docked results with AutoDock Vina
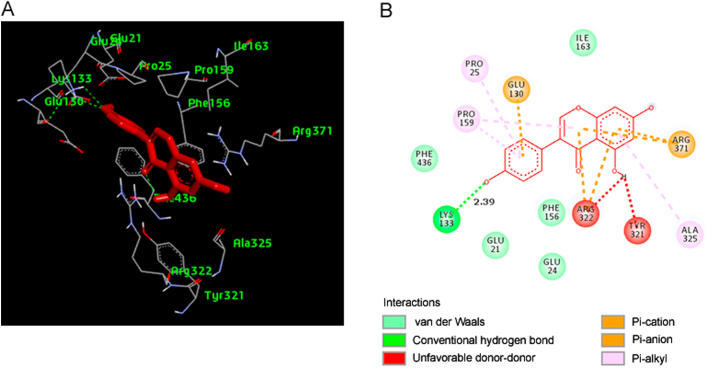
The ligands were docked against MCT8 using AutoDock Vina [19]. The best scored conformation was selected by considering the lowest binding energy between the protein and the ligand. The best selected pose of MCT8-genistein docked complex (binding energy −8.6 kcal/mol) predicted by AutoDock Vina is shown in Fig. 6A. The 2D diagram showing the hydrogen bonds and the types of contacts involved in MCT8-genistein complex is shown in Fig. 6B. The results showed that genistein interacted with MCT8 by forming hydrogen bonds with LYS-133.
Fig. 6.
(A) The output of AutoDock Vina showing the binding site residues of MCT8 protein with the ligand genistein. The residues in the binding site are shown in green color. Genistein is shown in red stick format. (B) 2D diagram showing the types of contacts formed between MCT8 and genistein. The green dotted lines indicate H‑bond interactions between MCT8 and genistein. The values adjacent to the green dotted lines indicate their distance.
The best docking pose of MCT8-daidzein docked complex (binding energy −8.6 kcal/mol) is shown in Fig. 7A. The types of contacts involved in MCT8-daidzein complex is shown in Fig. 7B. There were no hydrogen bonds formed between MCT8 and daidzein.
Fig. 7.
(A) The output of AutoDock Vina showing the binding site residues of MCT8 protein with the ligand daidzein. The residues in the binding site are shown in green color. Daidzein is shown in green stick format. (B) 2D diagram showing the types of contacts formed between MCT8 and daidzein.
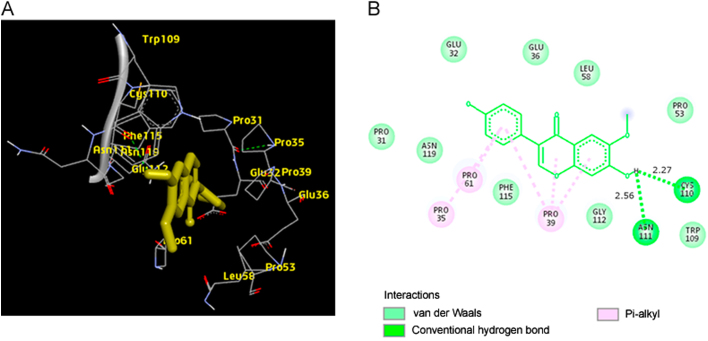
The best docking pose of MCT8-glycitein complex (binding energy −8.5 kcal/mol) is shown in Fig. 8A. The 2D diagram showing the hydrogen bonds and the types of contacts involved in MCT8-glycitein complex is shown in Fig. 8B. It was observed that CYS-110 and ASN-111 were involved in hydrogen bond interactions.
Fig. 8.
(A) The output of AutoDock Vina showing the binding site residues of MCT8 protein with the ligand glycitein. The residues in the binding site are shown in yellow color. Glycitein is shown in yellow stick format. (B) 2D diagram showing the types of contacts formed between MCT8 and glycitein. The green dotted lines indicate H‑bond interactions between MCT8 and glycitein. The values adjacent to the green dotted lines indicate their distance.
3.2.3. Analysis of the docked results
The docking results predicted by AutoDock4 [15] were compared to those of AutoDock Vina [19]. Docking analysis showed that there were hydrogen bonds formed between MCT8 and the inhibitors used. Opposite to the results of AutoDock4 [15], AutoDock Vina [19] generated hydrogen bonds in MCT8-glycitein interaction. Opposite to the results of AutoDock Vina, AutoDock generated hydrogen bonds in MCT8-daidzein interaction. Moreover, van der Waals interactions were also involved in addition to hydrogen bonds. It is important to point out that both compounds, genistein and daidzein, have hydrogen bonds with residues PRO-338, HIS-341 and GLU-348.
The best lead compound was selected in terms of binding energy. The binding energies of the ligands calculated by AutoDock4 [15] and AutoDock Vina [19] are shown in Table 2. Based on the analysis with AutoDock Vina [19], it was observed that binding energies of the three compounds were almost the same. Docking studies with AutoDock4 [15] and AutoDock Vina [19] showed that the natural compound Daidzein showed the lowest binding energy value of −6.22 and −8.6 kcal/mol, respectively (Table 2). Based on these findings, gaidzein can be used as a natural inhibitor to target MCT8 in AHDS. However, daidzein should be subjected to further investigation using in vitro studies.
Table 2.
Binding energies of the ligands.
| Ligand | Binding energy calculated by AutoDock4 (kcal/mol) | Binding energy calculated by AutoDock Vina (kcal/mol) |
|---|---|---|
| Genistein | − 5.76 | − 8.6 |
| Daidzein | − 6.22 | − 8.6 |
| Glycitein | − 5.54 | − 8.5 |
4. Conclusion
Recently, many researches have focused on the identification of inhibitors from natural sources. This study concludes that daidzein is an effective lead compound which will be useful for the design of novel less toxic and highly efficient drugs for the treatment of AHDS. Daidzein should be subjected to further experimental study in order to confirm this finding. This study also identified that PRO-338, HIS-341 and GLU-348 of MCT8 play an important role in hydrogen bonding with genistein and daidzein.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Peer review under responsibility of Xi'an Jiaotong University.
References
- 1.Schwartz C.E., Stevenson R.E. The MCT8 thyroid hormone transporter and Allan–Herndon–Dudley syndrome. Best. Pract. Res. Clin. Endocrinol. Metab. 2007;21:307–321. doi: 10.1016/j.beem.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vander Deure W.M., Peeters R.P., Visser T.J. Molecular aspects of thyroid hormone transporters, including MCT8, MCT10, and OATPs, and the effects of genetic variation in these transporters. J. Mol. Endocrinol. 2010;44:1–11. doi: 10.1677/JME-09-0042. [DOI] [PubMed] [Google Scholar]
- 3.He F.J., Chen J.Q. Consumption of soybean, soy foods, soy isoflavones and breast cancer incidence: differences between Chinese women and women in Western countries and possible mechanisms. Food Sci. Human. Wellness. 2013;2:146–161. [Google Scholar]
- 4.Ziaei S., Halaby R. Dietary isoflavones and breast cancer risk. Medicines. 2017;4:18. doi: 10.3390/medicines4020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Radović B., Mentrup B., Köhrle J. Genistein and other soya isoflavones are potent ligands for transthyretin in serum and cerebrospinal fluid. Br. J. Nutr. 2006;95:1171–1176. doi: 10.1079/bjn20061779. [DOI] [PubMed] [Google Scholar]
- 6.Lima S.T.C., Merrigan T.L., Rodrigues E.D. Synthetic and plant derived thyroid hormone analogs. Thyroid Parathyr. Dis.- New Insights into some Old. some New Issues. 2012:221–236. [Google Scholar]
- 7.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lipinski C.A. Drug-like properties and the causes of poor solubility and poor permeability. J. Pharmacol. Toxicol. Methods. 2000;44:235–249. doi: 10.1016/s1056-8719(00)00107-6. [DOI] [PubMed] [Google Scholar]
- 9.Lipinski C.A., Lombardo F., Dominy B.W. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997;23:3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 10.Berman H.M., Westbrook J., Feng Z. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaji D. Multi-template homology modeling of human MCT8 protein. Int. J. Adv. Res. 2017;5:1025–1036. [Google Scholar]
- 12.Šali A., Potterton L., Yuan F. Evaluation of comparative protein modeling by MODELLER. Protein. Struct. Funct. Bioinform. 1995;23:318–326. doi: 10.1002/prot.340230306. [DOI] [PubMed] [Google Scholar]
- 13.Eswar N., Webb B., Martin-Renom M.A. Comparative protein structure modeling using modeller. Curr. Protoc. Bioinform. 2006;15:5.6.1–5.6.32. [Google Scholar]
- 14.Xu D., Zhang Y. Improving the physical realism and structural accuracy of protein models by a two-step atomic-level energy minimization. Biophys. J. 2011;101:2525–2534. doi: 10.1016/j.bpj.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morris G.M., Huey R., Lindstrom W. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.NCBI-PubChem Compound database, 〈http://pubchem.ncbi.nlm.nih.gov〉.
- 17.Wheeler D.L., Barrett T., Benson D.A. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2007;36:D13–D21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Boyle N.M., Banck M., James C.A. Open Babel: an open chemical toolbox. J. Cheminf. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Visualizer A.D.S. Version 4. 5. Softw. Vis. Anal. Protein Struct. 2017 , <http://www.accelrys.com>. [Google Scholar]
- 21.Veber D.F., Johnson S.R., Cheng H.Y. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002;45:2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 22.Meng X.Y., Zhang H.X., Mezei M. Molecular docking: a powerful approach for structure-based drug discovery. Curr. Comput. Aided Drug Des. 2011;7:146–157. doi: 10.2174/157340911795677602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McConkey B.J., Sobolev V., Edelman M. The performance of current methods in ligand–protein docking. Curr. Sci. 2002;83:845–856. [Google Scholar]