Abstract
Freeze-thaw cycles impact the amount of aggregation observed in antibodies and Fc-fusion proteins. Various formulation strategies are used to mitigate the amount of aggregation that occurs upon putting a protein solution through a freeze-thaw cycle. Additionally, low pH solutions cause native antibodies to unfold, which are prone to aggregate upon pH neutralization. There is great interest in the mechanism that causes therapeutic proteins to aggregate since aggregate species can cause unwanted immunogenicity in patients. Herein, an increase in aggregation is reported when the pH is adjusted from pH 3 up to a pH ranging from pH 4 to pH 7 during the thaw process of a frozen antibody solution. Raising the pH during the thaw process caused a significant increase in the percent aggregation observed. Two antibodies and one Fc-fusion protein were evaluated during the pH jump thaw process and similar effects were observed. The results provide a new tool to study the kinetics of therapeutic protein aggregation upon pH increase.
Keywords: Monoclonal antibodies, Freeze–thaw, Protein aggregation, Protein stability
1. Introduction
Aggregation is a critical quality attribute of therapeutic proteins such as monoclonal antibodies (mAbs). It is important to understand the mechanism of protein aggregation since aggregate species often elicit an unwanted immunogenic response in patients. Therefore, reproducibility of manufactured therapeutic proteins with very low levels of aggregates is highly desired in the pharmaceutical industry so that safe products can be delivered. Aggregate formation during the therapeutic protein production process can potentially be the result of any one of many different steps in a typical commercial process [1]. The impact of freeze-thaw on aggregate formation and stability of antibodies has been studied extensively. The pH [2], [3], [4], excipients [5], [6], [7], ionic strength, containers [2], and heating and cooling rates [8], [9] have been shown to impact the stability of antibodies during the freeze-thaw process.
In this work, we describe how an increase in aggregation occurs upon thawing an antibody solution, stored at pH 3 and −80 °C, with a solution that is at a higher pH. The impacts of pH, antibody concentration, buffer, rate of thaw and stabilizing molecules were investigated. In order to ensure this phenomenon was not specific to one particular antibody, key experiments were repeated with another antibody and an Fc-fusion protein. All three of the proteins contained the Fc region of IgG1, which is implicated in aggregate formation when the pH is increased during thaw. The results are explained by a model for kinetically trapped aggregation that is promoted under these conditions.
2. Materials and methods
2.1. Samples
All protein samples were either produced at MilliporeSigma from CHOZN® cell lines grown in chemically defined media (mAb 1 and mAb 2) or purchased commercially (Fc-fusion protein). Unless otherwise noted, all reagents were purchased from Sigma-Aldrich (USA). Buffered solutions were prepared using deionized water (18.2 MΩ·cm) and were filtered (0.22 µm) prior to use. Frozen protein samples were stored in 0.2 mL Greiner Sapphire PCR vials at −80 °C. For protein purifications, 200 µL per sample of Poros® MAbCapture™ Protein A resin (Thermo) was used and rinsed with pH 7 wash buffer (20 mM citrate, 150 mM NaCl) prior to use. Protein samples produced at MilliporeSigma were purified by mixing the centrifuged supernatant (900 µL) with protein A resin for 10 min. The samples were washed twice with 900 µL of pH 7 wash buffer and eluted with 150 µL of pH 3 elution buffer (25 mM citrate).
Following purification, mAb 1 was frozen at −80 °C in 40 µL aliquots, unless otherwise noted in the text. The second protein, mAb 2, was further processed following the standard purification procedure at MilliporeSigma. Namely, it was adjusted to pH 5.5 using 50 µL of additive buffer (2 M L-arginine, 400 mM acetate, pH 8.12) and subsequently frozen at −80 °C. For the purpose of this study, the mAb 2 solution was thawed and subsequently exchanged into pH 3 elution buffer using Amicon® 30 K molecular weight cutoff filters for buffer exchange, and then frozen in either 20 µL or 40 µL aliquots at −80 °C. The Fc-fusion protein was received as a solid lyophilized formulation. The lyophilized Fc-fusion powder was hydrated with water, exchanged into 25 mM citrate buffer at pH 3 and subsequently frozen in either 20 µL or 40 µL aliquots at −80 °C. Both mAb 1 and mAb 2 are IgG1 antibodies and the Fc-fusion protein contains the Fc region of human IgG1. The samples were analyzed for identification and purity using SDS-PAGE analysis (Fig. S1), and the major impurities observed were typically fragments of fusion proteins and mAbs (missing heavy or light chain, Table S1).
2.2. Thaw process
The samples were either allowed to thaw completely at room temperature (RT) and then adjusted with buffers at the specified conditions, or were mixed immediately with buffers at the specified conditions during thaw. Unless otherwise noted, all dilution buffers contained 100 mM citrate. The dilution factor was kept at 7 to 1 for all samples. Unless otherwise noted, 40 µL samples were used with 240 µL dilution buffer. For experiments where the samples were mixed after the thaw process, the frozen aliquots were allowed to completely thaw at RT (3 min) and then mixed with buffer at RT using continuous aspiration (10 aspiration cycles with micro-pipette set at 240 µL for 10 s, 1 aspiration cycle per second). For experiments where the samples were mixed during the thaw process, the frozen aliquots were immediately mixed with buffer at RT using continuous aspiration until the sample completely dissolved (10 aspiration cycles with micro-pipette set at 240 µL for 10 s, 1 aspiration cycle per second). For samples that were frozen in 20 µL aliquots, 120 µL dilution buffer was used (10 aspiration cycles with micro-pipette set at 120 µL for 10 s, 1 aspiration cycle per second). Care was taken to avoid bubble formation. All samples were filtered through 0.45 µm GHP filters (Pall) and immediately analyzed by size exclusion chromatography (SEC) for aggregation levels.
2.3. Aggregation assessment
Aggregation was assessed with a Waters Acquity H-Class UPLC system. A Waters UPLC BEH200 SEC column (200 Å, 1.7 µm, 4.6 mm × 150 mm) was used for analysis. Peak resolution and aggregate recoveries were increased by adding arginine to the mobile phase in accordance with previous work by Ejima et al. [10]. After further optimization, reproducible recoveries of the aggregates were maximized when the arginine concentration was increased to 500 mM in the mobile phase. The column temperature was also optimized for peak resolution and a temperature of 35 °C gave ideal peak separation while showing no effect on aggregate recoveries. All of the samples were analyzed using an isocratic mobile phase of pH 7.6, 500 mM arginine, 100 mM sodium phosphate, and 200 mM NaCl. The flow rate was 0.4 mL/min. The column temperature was 35 °C, and the samples were stored at 10 °C during analysis. The injection volume was 15 µL, and the runtime of the method was 7.5 min. The chromatograms were extracted at a wavelength of 280 nm.
3. Results
3.1. Initial observations of mAb 1 aggregation during thaw
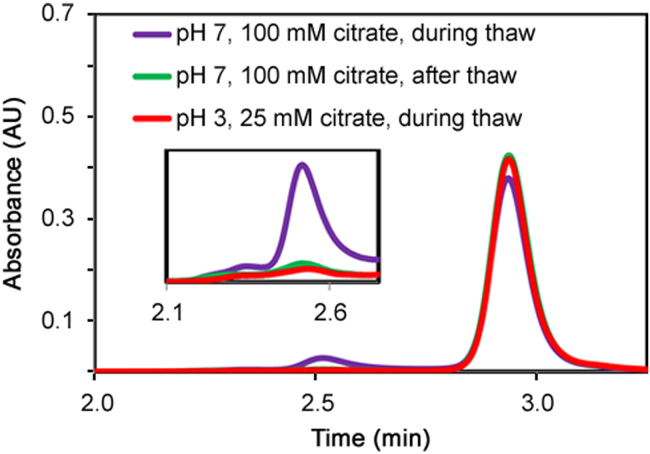
When an mAb 1 sample, stored frozen at pH 3, was allowed to completely thaw before being mixed with a higher pH buffer (pH 7), there was no increase in aggregation (Fig. 1). However, when the same sample was immediately mixed with pH 7 buffer during the thaw process, aggregation levels were significantly increased. In order to test whether the induced aggregation was due to a rapid increase in pH or temperature, a control experiment was performed where the mAb 1 sample was mixed with the same storage buffer (pH 3) during the thaw process. The results in Fig. 1 confirm that a rapid increase in temperature alone, was not responsible for causing aggregation since mixing the protein sample with buffer at pH 3 while the sample was still thawing did not result in aggregation. Note that when the frozen protein sample was allowed to completely thaw prior to addition of pH 7 buffer there was no increase in aggregation, which suggests that a rapid increase in pH alone was not responsible for aggregation since the pH change was also relatively fast in this case. There was clearly no difference in aggregation levels when the sample was mixed with either pH 3 buffer during thaw, or pH 7 buffer after the sample was allowed to completely thaw, indicating there was no effect of pH on aggregate formation when the samples were allowed to completely thaw prior to pH adjustment. Taken together, these results show the impact on mAb aggregation was both pH- and thaw-dependent, and not attributable to just a rapid pH increase or the thaw process alone.
Fig. 1.
Size exclusion chromatograms of three mAb 1 samples thawed in different conditions (absorbance at 280 nm). For each sample, the peak observed at 2.8 min was the monomer peak of mAb 1, and anything eluting earlier was considered aggregates. Inset: Zoom-in on the chromatogram of the aggregate species.
3.2. Effect of dilution process on aggregation
3.2.1. Rate of thaw
The temperature of the thawing buffer during the pH jump with mAb 1 was evaluated to see how big of an effect the rate of thaw would have on aggregation levels. The following temperature experiments were performed to investigate this effect on aggregation levels. Frozen samples of mAb 1 were thawed with pH 7 buffer at different rates (with or without aspiration) or with buffer kept at different temperatures (RT or 4 °C). The mAb 1 samples were also allowed to completely thaw before being mixed with pH 7 buffer at RT or 4 °C (Fig. 2).
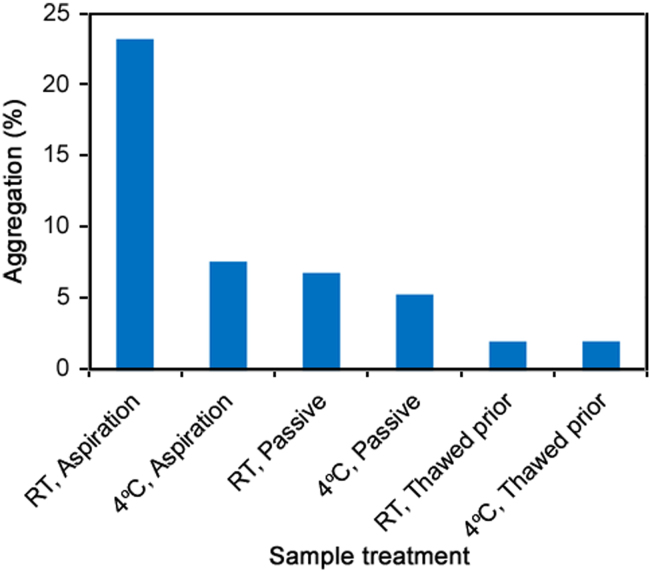
Fig. 2.
The percent of aggregation observed when frozen mAb 1 samples were thawed in six different conditions with pH 7, 100 mM citrate (dilution buffer). In the first two conditions, samples were aspirated with dilution buffer stored at the specified temperature. In the third and fourth conditions, the samples were allowed to thaw passively with dilution buffer stored at RT or 4 °C and without aspiration. In the last two conditions, the samples were allowed to completely thaw and then mixed with the dilution buffer at RT or 4 °C.
Different aggregation levels were observed with mAb 1 depending on both the temperature of the dilution buffer and the thawing rate of the sample. In the sample aspirated immediately with RT buffer, the aggregation level was significantly higher than that of the sample aspirated with buffer at 4 °C. When the buffer was added without aspiration (passive), aggregation still occurred, but there was no difference in aggregation levels when the dilution buffer was at 4 °C or RT. The amount of time required for complete thaw of the mAb 1 samples was in the following order: RT Aspiration < 4 °C Aspiration < RT Passive < 4 °C Passive. When the samples were allowed to completely thaw prior to buffer addition, aggregation levels were even lower than that of passive addition, but the temperature of the dilution buffer had no impact on aggregation levels in this case. The results illustrate how the kinetics of protein thawing has an impact on induced aggregation levels when the protein is exposed to higher pH.
3.2.2. pH of dilution buffer
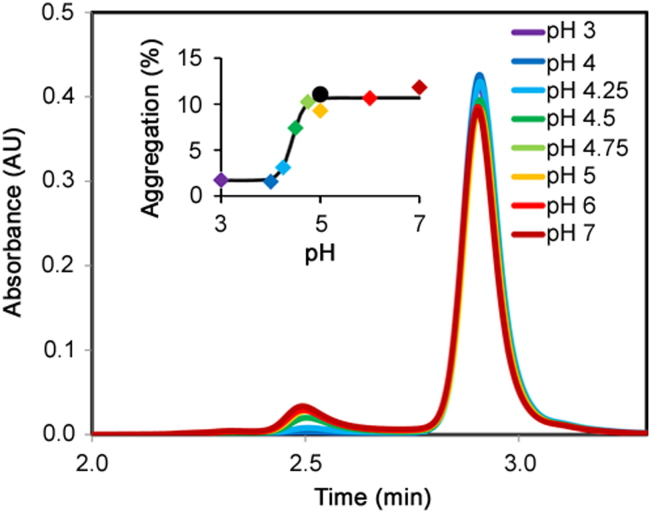
Knowing that the rate of thaw has an effect on aggregation levels, attention was shifted to the role of pH on aggregate formation. In order to further investigate the impact of pH on aggregate formation during the thaw process, pH titration experiments were performed on mAb 1. The pH titration was performed by thawing mAb 1 samples (stored at pH 3) in dilution buffers containing 100 mM citrate at a pH ranging from pH 3 to pH 7. The frozen samples were immediately aspirated with the corresponding dilution buffer at RT. A clear sigmoidal relationship exists between the amount of aggregate formed and the solution pH (Fig. 3). This sigmoidal fit suggests that a titratable group with a pKa of 4.5 is involved in the aggregation process (Fig. 3 inset). Aspiration of the frozen sample with an alternative dilution buffer consisting of 100 mM acetate at pH 5 gave similar results to the corresponding citrate buffer at pH 5.
Fig. 3.
Size exclusion chromatograms (SEC) of mAb 1 thawed in dilution buffers (100 mM citrate) of varying pH (pH 3 to pH 7). Inset: The percent aggregation observed when SEC are plotted vs. solution pH. The colors of the data points coordinate with the colors in the chromatograms. ● represents the measurement when pH 5, 100 mM acetate was used.
3.3. Effect of storage buffer on aggregation
3.3.1. pH of storage buffer
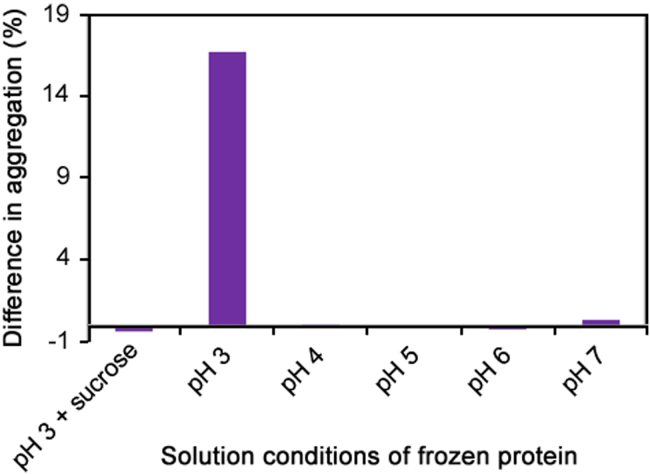
In an effort to determine whether the temperature and pH jump on the thaw-induced aggregation process was also dependent on the pH of the frozen solution, samples of mAb 1 were frozen in other solution at pH values of 4–7 (each with 25 mM citrate) and again adjusted with pH 7 dilution buffer. All of the samples were mixed with dilution buffer at RT using continuous aspiration either during or after the protein thawed. The results (Fig. 4) are shown as a difference in percent aggregation (difference between when the sample was allowed to thaw completely and when the sample was mixed during thaw). The largest increase in aggregation was observed when the pH 3, protein sample was thawed (aspirated) with pH 7, 100 mM citrate buffer, as previously shown. When the pH of the mAb storage solution was adjusted to a higher pH (pH 4–7) prior to freezing, no increase in aggregation was observed upon thawing with pH 7 buffer. This shows that storing the samples at pH 3 is significant upon thawing at higher pH.
Fig. 4.
Difference in percent aggregation when samples were mixed with pH 7 buffer during and after the thaw process. The mAb 1 samples were stored frozen at the pH indicated (with 25 mM citrate). In the experiments with sucrose, the protein sample was frozen with 25 mM citrate and 500 mM sucrose. All of the frozen samples (stored at 2 mg/mL) were adjusted to pH 7 with 7:1 dilution into pH 7, 100 mM citrate solution.
3.3.2. Effect of sucrose
Sucrose is known to protect frozen proteins by serving as a cryoprotectant [6], [7] and plays a role in protecting mAb 1 from induced aggregation upon thawing with a higher pH buffer. To test this hypothesis, sucrose was added at a final concentration of 500 mM to the mAb 1 sample and frozen at pH 3 (25 mM citrate). Upon aspirated thawing with pH 7 buffer, no increase in aggregation was observed (Fig. 4), showing that sucrose protects mAb 1 from induced aggregation when samples stored at pH 3 were thawed with a higher pH buffer.
3.4. Generality of protein aggregation during thaw
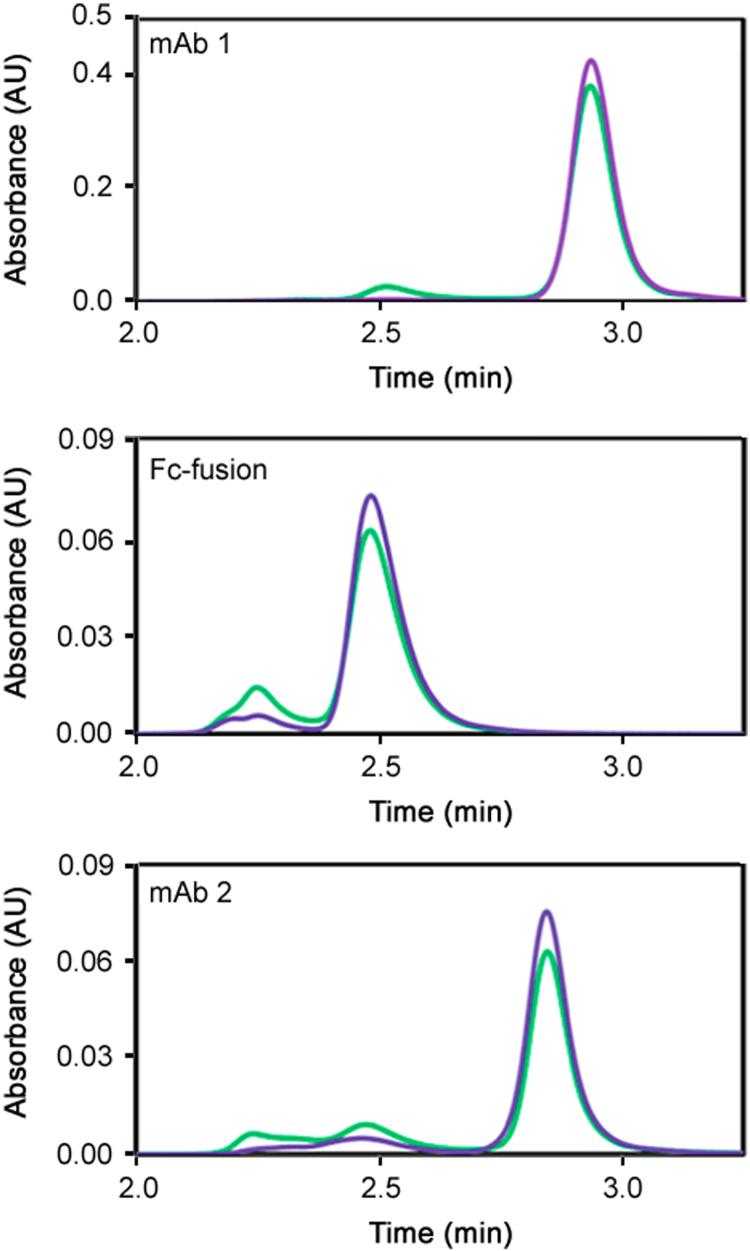
In order to ensure the aggregation induced phenomenon was not isolated to a single mAb, two other therapeutic proteins were evaluated. An Fc-fusion protein and mAb 2 were exchanged into pH 3, 25 mM citrate buffer and then frozen at −80 °C in 20 µL aliquots. The samples were either thawed with mixing in the presence of pH 7 buffer, or allowed to completely thaw before being mixed with the pH 7 buffer. The results (Fig. 5) show that the phenomenon of induced aggregation upon thawing low pH protein samples with higher pH buffers at RT is not isolated to mAb 1 alone. Both the Fc-fusion protein and mAb 2 exhibited an increase in aggregate levels when the samples were subjected to a rapid elevation in pH and temperature. However, in order to fully generalize the kinetic model observed for mAb 1 with other Fc-containing proteins, additional experiments are needed with the Fc-fusion protein and mAb 2, including the effect of thaw rates on aggregation levels.
Fig. 5.
Absorbance chromatograms of mAb 1, an Fc-fusion protein and mAb 2 when the pH was adjusted during thaw (green) and after thaw (purple). The pH was adjusted from pH 3 in the frozen sample to pH 7 in the final solution. An increase in aggregation is observed for all three proteins. The Fc-fusion and mAb 2 proteins were both at 2 mg/mL in the frozen samples, and the mAb 1 protein was at 7 mg/mL in the frozen sample.
4. Discussion
4.1. Proposed model for pH jump induced aggregation during thaw
4.1.1. CH2 unfolding at low pH
Numerous studies have reported in recent years the mechanism of therapeutic antibody aggregation at low pH [3], [4], [11], [12], [13], [14], [15]. Previous work by Latypov et al. [4] showed that IgG1 and IgG2 Fc fragments are largely unfolded at low pH (2.5) and become more ordered in secondary and tertiary structure as the pH is increased to pH 4.7. They discovered unfolding of the CH2 domain precluded unfolding in the CH3 domain as the pH is lowered. From pH 3.1 to 3.5, the CH2 domain is completely unfolded, while the CH3 domain is partially unfolded. The driving force for unfolding at low pH is largely influenced by protonation of side residues that are involved in stabilizing secondary and tertiary structures in the native protein at neutral pH [16]. In the native conformation, these residues are oppositely charged and held together through electrostatic attractions, with the native protein having a net neutral charge. At low pH, the non-native protein has an increased net charge due to multiple, positively charged residues [12], [17]. The electrostatic repulsions between intramolecular charged residues cause the protein to unfold, exposing hydrophobic segments to the solvent that is usually buried in the core of the folded, native protein. As a result, hydrophobic attractions between adjacent monomers become possible and are strong enough to compete with the energetically unfavorable intermolecular repulsions that exist between positively charged residues on the opposite binding partner [16]. The association of unfolded monomers to form larger oligomers in this manner represents the foundation for non-native protein aggregation, which is a plausible mechanism for explaining the aggregation results observed in this study.
The requirement that the solution pH must be as low as pH 3 to achieve complete unfolding of the native CH2 region can explain the results in Fig. 4. The non-native conformation of the monomer that is capable of self-association will only be present when enough side residues have become protonated, which is only possible when the pH is at pH 3. When the pH is at pH 3.5 or above, the CH2 domain is partially folded and incapable of binding neighboring monomers. This explains why the frozen proteins must be at pH 3 in order to achieve aggregation upon a pH jump to neutral pH. The pH limit for achieving adequate neutralization of net charge in the non-native monomer appears to be pH 4.5 based on the aggregation results shown in Fig. 3. This result fits the net charge calculations of an IgG1-derived Fc fragment reported by Wu et al. [12], in which the net charge was positive at pH 4 and neutral at pH 5.
4.1.2. Kinetically trapped aggregates
In current models for non-native protein aggregation, reversible pathways are used to explain aggregate formation starting from the native monomer [16], [18]. Unfolding of the monomer at pH 3 is not considered rate-limiting. Once unfolded, the monomer can associate with adjacent monomers, as mentioned earlier, to form dimer and trimer clusters, which are reversible. However, as the pH is increased towards the isoelectric point of the protein, the dispersed charges that are repelled between monomers in close proximity are neutralized and the hydrophobic attractions become significant. At this point, the protein will either re-fold into the native conformation (thermodynamically reversible formation) or form strong hydrophobic interactions with another monomer in the form of β–sheets, leading to aggregate species which are irreversible [19], [20]. The unfolded, neutralized proteins are transiently formed upon exposure to increased pH. The population of monomers that overcome the energy barrier for self-association will reach a local energy minimum and will be kinetically trapped as an aggregate [13], [21]. Although the corresponding aggregates are considered energetically stable with high energy barriers to dissociation, the trapped aggregates formed in this study were found to be slowly reversible (Fig. S2).
4.1.3. Concentration influence on aggregation
As discussed in the previous section, an increase in pH from pH 3 to above pH 4.5 affected the amount of aggregates observed in mAb 1 and was attributed to the stability of the CH2 domain in the unfolded protein. However, aggregation levels were not influenced by a pH jump alone. As shown in Fig. 2, the rate at which the frozen sample thawed showed an influence on aggregation. When the sample, frozen at pH 3, was thawed with neutral buffer at pH 7, aggregation levels were appreciably higher when the samples were thawed faster. Cryoconcentration, in which proteins are concentrated into localized sections of a frozen sample, can occur upon freezing [8]. Higher concentrated protein when exposed to a rapid pH shift during the thaw process would lead to unfolded CH2 domains being neutralized in closer proximity to other proteins, thus leading to higher aggregation. This model can be used to explain the results in Fig. 2, in which frozen samples that were thawed with aspiration showed higher aggregation levels compared to samples that were passively thawed. The samples that were thawed prior to addition of neutral buffer would have been diluted by the time the pH was increased and expected to show less aggregation, as was the case in this study.
The hypothesis for cryoconcentration as the cause of increased aggregation with increased thaw rate is supported by the observation that sucrose was able to decrease aggregation during rapid thaw with neutral buffer, when it was present in the frozen mAb 1 sample at pH 3 (Fig. 4). Sucrose has the ability to act as a cryoprotectant when added to frozen protein samples [6], [7] and as a result, local protein concentrations would change less during the freezing process. Additional experiments were performed to test concentration dependence, in which samples of mAb 1 were frozen in pH 3 buffer at different protein concentrations (Fig. S3). Upon aspirated dilution with neutral buffer, increased aggregation was observed that was proportional to the concentration of frozen protein, further supporting this hypothesis.
5. Conclusions
Frozen samples of three different therapeutic proteins stored at pH 3 showed appreciable amounts of aggregation upon a pH jump and rapid thaw with neutral buffer. The Fc region, common to all three proteins, was implicated in the aggregation process. The stability of the CH2 domain within the Fc region was used to explain the kinetic results obtained with mAb 1 and compliments the findings of recent reports on the mechanism of aggregation by non-native therapeutic proteins. In particular, aggregates formed by the pH jump thaw process are attributed to a kinetically trapped intermediate that is formed. Various factors promoted the aggregation process such as the rate of thaw and the pH of the frozen protein sample and dilution buffer. Cryoconcentration and pH increase are the proposed root causes to explain the observed phenomenon. Altogether, the findings can be used as a research tool to further study the aggregation kinetics of therapeutic proteins.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Peer review under responsibility of Xi'an Jiaotong University.
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.jpha.2017.09.002.
Appendix A. Supplementary material
Supplementary material Analytical data for samples subjected to the pH jump thaw process, with respect to aggregate stability, protein concentration, and SDS-PAGE analysis, are located in the supporting information.
References
- 1.Vázquez-Ray M., Lang D.A. Aggregates in monoclonal antibody manufacturing processes. Biotech. Bioeng. 2011;108:1494–1508. doi: 10.1002/bit.23155. [DOI] [PubMed] [Google Scholar]
- 2.Kueltzo L.A., Wang W., Randolph T.W. Effects of solution conditions, processing parameters, and container materials on aggregation of a monoclonal antibody during freeze-thawing. J. Pharm. Sci. 2008;97:1801–1812. doi: 10.1002/jps.21110. [DOI] [PubMed] [Google Scholar]
- 3.Hari S.B., Lau H., Razinkov V.I. Acid-induced aggregation of human monoclonal IgG1 and IgG2: molecular mechanism and the effect of solution composition. Biochemistry. 2010;49:9328–9338. doi: 10.1021/bi100841u. [DOI] [PubMed] [Google Scholar]
- 4.Latypov R.F., Hogan S., Lau H. Elucidation of acid-induced unfolding and aggregation of human immunogolbulin IgG1 and IgG2 Fc. J. Biol. Chem. 2012;287:1381–1396. doi: 10.1074/jbc.M111.297697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kreilgaard L., Jones L.S., Randolph T.W. Effect of Tween 20 on freeze-thawing- and agitation-induced aggregation of recombinant human factor XIII. J. Pharm. Sci. 1998;87:1597–1603. doi: 10.1021/js980126i. [DOI] [PubMed] [Google Scholar]
- 6.Kendrick B.S., Chang B.S., Arakawa T. Preferential exclusion of sucrose from recombinant interleukin-1 receptor antagonist: role in restricted conformational mobility and compaction of native state. Proc. Natl. Acad. Sci. USA. 1997;94:11917–11922. doi: 10.1073/pnas.94.22.11917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee J.C., Timasheff S.N. The stabilization of proteins by sucrose. J. Biol. Chem. 1981;256:7193–7201. [PubMed] [Google Scholar]
- 8.Kohle P., Badkar A. Protein and solute distribution in drug substance containers during frozen storage and post-thawing: a tool to understand and define freezing-thawing parameters in biotechnology process development. Biotechnol. Prog. 2011;27:494–504. doi: 10.1002/btpr.530. [DOI] [PubMed] [Google Scholar]
- 9.Chi E.Y., Krishnan S., Randolph T.W. Physical stability of proteins in aqueous solution: mechanism and driving forces in nonnative protein aggregation. Pharm. Res. 2003;20:1325–1336. doi: 10.1023/a:1025771421906. [DOI] [PubMed] [Google Scholar]
- 10.Ejima D., Yumioka R., Arakawa T. Arginine as an effective additive in gel permeation chromatography. J. Chromatogr. A. 2005;1094:49–55. doi: 10.1016/j.chroma.2005.07.086. [DOI] [PubMed] [Google Scholar]
- 11.Arosio P., Rima S., Morbidelli M. Aggregation mechanism of an IgG2 and two IgG1 monoclonal antibodies at low pH: from oligomers to larger aggregates. Pharm. Res. 2013;30:641–654. doi: 10.1007/s11095-012-0885-3. [DOI] [PubMed] [Google Scholar]
- 12.Wu H., Truncali K., Ritchie J. Weak protein interactions and pH- and temperature-dependent aggregation of human Fc1. mAbs. 2015;7:1072–1083. doi: 10.1080/19420862.2015.1079678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imamura H., Honda S. Kinetics of antibody aggregation at neutral pH and ambient temperatures triggered by temporal exposure to acid. J. Phys. Chem. B. 2016;120:9581–9589. doi: 10.1021/acs.jpcb.6b05473. [DOI] [PubMed] [Google Scholar]
- 14.Liu B., Guo H., Xu J. Acid-induced aggregation propensity of nivolumab is dependent on the Fc. mAbs. 2016;8:1107–1117. doi: 10.1080/19420862.2016.1197443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skamris T., Tian X., Thorolfsson M. Monoclonal antibodies follow distinct aggregation pathways during production-relevant acidic incubation and neutralization. Pharm. Res. 2016;33:716–728. doi: 10.1007/s11095-015-1821-0. [DOI] [PubMed] [Google Scholar]
- 16.Roberts C.J. Therapeutic protein aggregation: mechanisms, design, and control. Trends Biotechnol. 2014;32:372–380. doi: 10.1016/j.tibtech.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu H., Kroe-Barrett R., Singh S. Competing aggregation pathways for monoclonal antibodies. FEBS Lett. 2014;588:936–941. doi: 10.1016/j.febslet.2014.01.051. [DOI] [PubMed] [Google Scholar]
- 18.Saha S., Deep S. Protein aggregation: elucidation of the mechanism and determination of associated thermodynamic and kinetic parameters. Curr. Phys. Chem. 2014;4:114–136. [Google Scholar]
- 19.Baker D., Agard D.A. Kinetics versus thermodynamics in protein folding. Biochemistry. 1994;33:7505–7509. doi: 10.1021/bi00190a002. [DOI] [PubMed] [Google Scholar]
- 20.Fink A.L. Protein aggregation: folding aggregates, inclusion bodies and amyloid. Fold. Des. 1998;3:R9–R23. doi: 10.1016/S1359-0278(98)00002-9. [DOI] [PubMed] [Google Scholar]
- 21.Filipe V., Kukrer B., Hawe A. Transient molten globules and metastable aggregates induced by brief exposure of a monoclonal IgG to low pH. J. Pharm. Sci. 2012;101:2327–2339. doi: 10.1002/jps.23157. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material Analytical data for samples subjected to the pH jump thaw process, with respect to aggregate stability, protein concentration, and SDS-PAGE analysis, are located in the supporting information.