Abstract
Ochrobactrum anthropi, a rare human pathogen, has been isolated predominantly from patients with catheter-related bacteraemia and rarely from other infections. In 2016, six cases of pseudo-bacteraemia caused by carbapenem-resistant O. anthropi isolates were recovered from an Argentinian hospital. The resistant phenotype exposed by the isolates caught our attention and led to an extensive epidemiologic investigation. Here we describe the characterization of a carbapenem-resistant O. anthropi outbreak whose probable cause was by contaminated collection tubes. The genome analysis of one strain revealed the presence of various resistant determinants. Among them, a metal-dependent hydrolase of the β-lactamase superfamily I, phnP, was found. Lately the recovery of unusual multidrug-resistant pathogens in the clinical setting has increased, thus emphasizing the need to implement standardized infection control practice and epidemiologic investigation to identify the real cause of hospital outbreaks.
Keywords: Carbapenem resistant, Ochrobactrum anthropi, outbreak, pseudo-bacteraemia
Introduction
In recent years the use of technologies in the microbiology diagnostic laboratory has contributed to the identification of pathogens not previously recognized. These pathogens are often environmental microorganisms with a high level of resistance to several antibiotics, converting them into potential reservoirs of antibiotic resistance traits. Ochrobactrum anthropi is an emerging Gram-negative, motile, nonfermentative, oxidase-negative and urease-positive aerobic bacillus [1]. It has been found to be widely distributed in soil, environmental and water sources, including antiseptic solutions and dialysis fluid [1], [2], [3], [4]. Several reports recognize it as a causative agent of central catheter–related infections in severely ill, immunocompromised patients, as well as patients with osteochondritis, necrotizing fasciitis, endophthalmitis, meningitis and peritonitis [5], [6], [7]. Few outbreaks related to O. anthropi have been described associated with endophthalmitis after cataract surgery [8] and bacteraemia in organ transplant recipients [9], as well as with pseudo-outbreak related to cross-contamination from erythrocyte sedimentation tubes [10].
Here we describe six cases of pseudo-bacteraemia caused by carbapenem-resistant (CR) Ochrobactrum anthropi isolates recovered from an Argentinian hospital in 2016. Notably, the isolates involved in the pseudo-bacteraemia were carbapenem resistant, which makes our findings unique. Our aim was to characterize the strains involved in the outbreak, to characterize the resistant determinants that can explain the multidrug-resistant phenotype observed and to identify the source.
Materials and methods
Setting, sample collection and outbreak investigation
The Hospital Interzonal de Agudos Eva Perón (San Martin, Buenos Aires, Argentina) has 280 beds and processes more than 500 blood cultures monthly. It has an emergency unit, a paediatric unit, a medical clinic unit, a trauma unit and a maternity unit. At our institution, blood cultures are analysed via an automated instrument (BacT/ALERT Microbial Identification System; bioMérieux, Marcy l’Etoile, France). The recovered isolates were characterized to the species level by conventional biochemical tests, the VITEK 2 System (bioMérieux) and 16S rDNA amplification. PCR products of the gyrB, rpoB and 16S rDNA gene, using specific primers [11], were obtained with the Apex Master Mix according to the manufacturer's directions (Genesee Scientific, San Diego, CA, USA). Sequencing was performed on both DNA strands at the Eton Bioscience sequencing facility (San Diego, CA, USA). The sequences were analysed by BLAST 2.0 software (National Center for Biotechnology Information, Bethesda, MD, USA; https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Six CR O. anthropi isolates were recovered from blood cultures from six patients (Table 1) admitted to the hospital between July 2016 and February 2017. Because of the rarity of isolation of CR O. anthropi, the institution decided to begin an investigation to identify the occurrence and source of the outbreak. A case patient was defined as any patient from whom O. anthropi was isolated in blood culture from July 2016 to February 2017.
Table 1.
Clinical and microbiologic data of Ochrobactrum anthropi isolates resistant to carbapenems
| Isolate | Age/sex | Unit | Diagnosis | IMP MIC (μg/mL) | MEM MIC (μg/mL) | Rapid Blue-Carba test | MEM-DA | bla PCR reactionsa |
|---|---|---|---|---|---|---|---|---|
| 1 | 2 months/F | Paediatrics | Suspected sepsis | 8 | >16 | + | + | — |
| 2 | 2 years/F | Paediatrics | Pneumonia | 8 | >16 | + | + | — |
| 3 | 6 years/F | Paediatrics | Pneumonia | 4 | >16 | + | + | — |
| 4 | 59 years/M | Trauma | Chronic osteomyelitis | 8 | >16 | + | + | — |
| 5 | 18 years/F | Maternity | Puerperal fever | 8 | >16 | + | + | — |
| 6 | 6 months/F | Paediatrics | Pneumonia | 8 | >16 | + | + | — |
| BCT isolate | NA | NA | NA | 8 | >16 | + | + | — |
| Unrelated strain | NA | NA | NA | 0.06 | 0.25 | — | — | — |
BCT, blood culture tube; MEM-DA, meropenem and dipicolinic acid synergy test; NA, not applicable.
bla PCR reactions: IMP, VIM, KPC, NDM-1.
All samples were processed in according to the Clinical Microbiology Procedures Handbook [12]. Samples were subcultured on sheep's blood agar and Levine eosin methylene blue agar plates. The blood tube collections were inoculated with thioglycollate broth.
Considering the high frequency of pseudo-bacteraemia associated with environmental sources, environmental samples from the hospital units involved in the outbreak were screened. Sinks used for handwashing and countertop surfaces in each unit were swabbed. In addition, samples from povidone–iodine and 70% alcohol solutions, blood collection tubes with EDTA solution, sodium citrate solution and dry tubes and gloves were also taken and cultured.
A total of 61 environmental samples from the paediatric, gynecology and orthopedics units were cultured to locate the O. anthropi reservoir.
Susceptibility assay
Susceptibility assay was performed by the VITEK 2 (AST-082 panel). The MIC results were interpreted according to the Clinical and Laboratory Standards Institute [13]. For the rapid detection of carbapenemases, a colourimetric method (Rapid Blue-Carba test) was also performed [14]. To study the presence of metallo-β-lactamases (MBL), a double-disc diffusion test using meropenem and imipenem (10 μg; Oxoid, Basingstoke, UK) was performed on Müller-Hinton agar using an 1900/750 μg EDTA/SMA disc (Laboratorios Britania, Buenos Aires, Argentina) [15].
The screening method for the detection of carbapenemase enzymes using the KPC-MBL Kit Confirm ID Pack (Rosco Taastrup, Taastrup, Denmark) containing meropenem (10 μg), meropenem 10 μg + dipicolinic acid (metallo-β-lactamase inhibitor), meropenem 10 μg + cloxacillin (AmpC inhibitor) and meropenem 10 μg + phenylboronic acid (KPC and AmpC inhibitor) was carried out.
General molecular techniques
Total DNA extraction was carried with the Wizard Genomic DNA Purification Kit according to the manufacturer's instructions (Promega, Madison, WI, USA). PCR reactions using Apex Master Mix (Genesee Scientific) were carried out for the most common carbapenemase genes (blaIMP, blaVIM, blaKPC and blaNDM).
Degenerate oligonucleotide PCR using a specific primer (5′-GGTCGACYTTNGYNGGRTC) and a low-stringency amplification protocol (5 minutes of denaturation at 95°C, 40 cycles for 1 minute at 93°C and 1.5 minutes at 36°C, 2 minutes at 72°C and 10 minutes at 72°C) [16] was used to determine the genetic relationship between the isolates.
Whole-genome sequence of OA 5 clinical strain
Genomic DNA was extracted using a MasterPure DNA Purification kit from Epicentre Biotechnologies (Madison, WI, USA). Whole-genome shotgun sequencing of one of the strains was performed using the Illumina MiSeq-I and Nextera XT DNA library (Illumina, San Diego, CA, USA). De novo assembly was performed by SPADES assembler 3.1.0 [17] using a preassembly approach with VELVET assembler 1.2 [18]. The RAST web server was used for gene prediction and functional annotation [19]. Predictions were confirmed, whenever necessary, by BLASTp 2.0 [20] of the GenBank protein database. Further genomic analysis was carried out using average ARG-ANNOT, ISFinder, PHAST and PathogenFinder, among others [20], [21], [22], [23], [24] Assembly and annotation of OA 5 were deposited at a local server; the deposit is freely available online (http://www.higiene.edu.uy/ddbp/Andres/montana_et_al_2018_data.html).
Phylogenetic analysis and distribution of phnP gene
Genome assemblies and available annotation of the genus Ochrobactrum were downloaded from the National Center for Biotechnology Information ftp site (ftp://ftp.ncbi.nlm.nih.gov/genomes/) (February 2018). When not available, annotation was done in the RAST web server [19] as described above. The final data set comprised the genome reported here plus 61 genomes of the genus Ochrobactrum and two genomes of the order Rhizobiales, also downloaded from the GenBank database. Agrobacterium tumefaciens Ach5 and Mesorhizobium loti NZP2037 strains were used as the outgroup in a phylogenetic analysis (Supplementary Table S1).
A total of 556 putative orthologous sequences were identified among the studied genomes by the OrthoMCL method [25], as implemented in get_homologous 1.3 software [26]. BLASTp searches were performed with a minimal identity value of 30%, minimal query coverage of 75% and a maximum E value of 1e-05. Each orthologous protein cluster was aligned using ClustalO 1.2.0 [27]. The resulting 556 alignments were concatenated by means of local script, and subsequently a phylogenetic tree was inferred. The maximum-likelihood method with an amino acid LG+G model [28] was used as implemented in the programme RAxML 8.2.9 [29]. The RELL bootstrapping technique was used to evaluate branch supports [30].
The phylogenetic distribution of phnP in the genus was studied by tblastn [20]. The protein sequence of this gene in O. anthropi ATCC 49188 strain was used as initial query for BLAST search (accession no. WP_012091991.1). Minimum query coverage of 75% and an identity value of 50% were taken as thresholds for presence definition. The pairwise sequence identity of the identified proteins with the PhnP protein of O. anthropi OA 5 was calculated.
Results and discussion
Blood collection tubes as source of CR O. anthropi outbreak
Six isolates identified as Ochrobactrum sp. by conventional biochemical tests were recovered from patients' blood cultures. All were identified as O. anthropi (biocodes 6000041320601001, 6001041320501001 and 6000041320001001) by the VITEK 2 system with a probability of 99%. This result was confirmed with the gyrB, rpoB and 16S rDNA sequence analysis, showing 98%, 99% and 99% of identity with O. anthropi OAB (GenBank accession no. CP008820), respectively, for each gene.
The first set of four CR O. anthropi isolates occurred in the paediatric unit, and the later two CR O. anthropi isolates were recovered in the trauma and maternity units (Table 1). There was no previous record of CR O. anthropi strains at our institution. This led to the suspicion of a CR O. anthropi outbreak. In all 6 cases, O. anthropi was recovered in one of two bottles of blood culture. Considering this, as well as the fact that patients had no demonstrable clinical signs of septicaemia, the isolates were not clinically hierarchized. An epidemiologic investigation screening the potential reservoirs took place to locate the source of the outbreak, and 69 environmental samples from the paediatric (n = 53), gynecology (n = 8) and orthopedic (n = 8) units were obtained, as follows: antiseptic solutions (n = 2), handwashing solutions (n = 2), swabbing surfaces (n = 8), blood collection tubes with EDTA solution (n = 6), blood collection tubes with sodium citrate solution (n = 14), dry tubes (n = 6), gloves (n = 4 pairs), sterile needles and syringes used for blood culture sampling (n = 4) and gauze (n = 4). Additionally, enrichment broth media from unopened bottles and bottle caps were also controlled. Culture results from antiseptic solutions, handwashing solutions, swabbing surfaces, blood collection tubes with EDTA solution, dry tubes, gloves, sterile needles, syringes, gauze, broth media from unopened bottles and bottle caps were negative. However, in the blood collection tubes with sodium citrate solution, used for the erythrocyte sedimentation rate determination, an organism that biochemically and phenotypically resembled those recovered from the initial blood cultures was obtained.
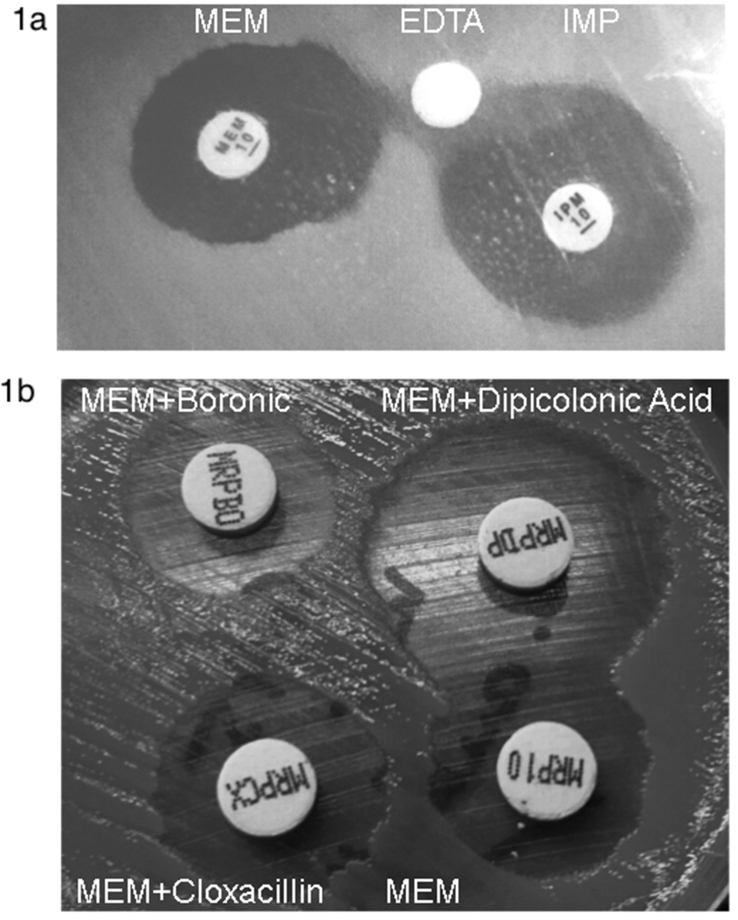
All isolates showed an unusual resistance profile harbouring resistance to carbapenems (Table 1). Taking this into account, the Rapid Blue-Carba test was performed, and a weakly positive result (light green colour) was obtained for the six O. anthropi isolates at between 90 and 120 minutes of incubation. Moreover, synergy between carbapenems and EDTA was observed for the six O. anthropi isolates (Fig. 1(a)). Additionally, the KPC-MBL Kit Confirm ID Pack showed the presence of MBL-type carbapenemase in all O. anthropi isolates. A halo difference of ≥5 mm in the case of meropenem + dipicolinic acid with respect to the meropenem 10 μg disc was observed in OA 5 (Fig. 1(b)). PCR reaction for blaIMP, blaVIM, blaKPC and blaNDM was negative.
Fig. 1.
(a) Synergy test between carbapenems (left, meropenem; right, imipenem) and EDTA (middle) observed in Ochrobactrum anthropi isolate OA 5. (b) Picture of KPC-MBL Kit Confirm ID Pack Representative results showing presence of MBL-type carbapenemase in O. anthropi isolate OA 5 are shown. MBL, metallo-β-lactamase; MRP10, meropenem; MRPBO, meropenem + boronic; MRPCX, meropenem + cloxacillin; MRPDP, meropenem + dipicolinic acid.
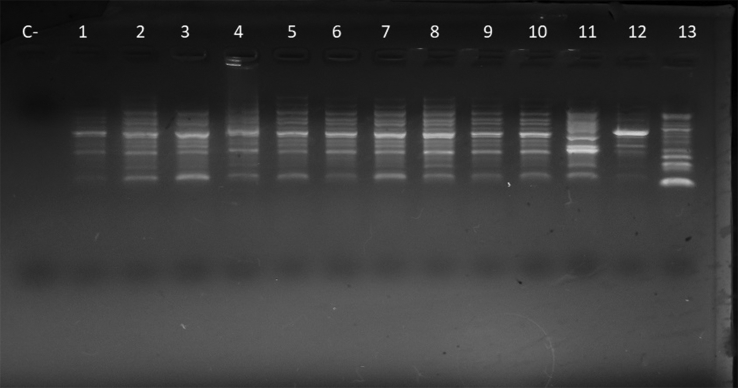
Oligonucleotide PCR results exhibited the same pattern of bands in the six patient isolates; in addition, the pattern obtained from the blood extraction tube with sodium citrate solution was the same, suggesting an epidemiologic relationship among the isolates (Fig. 2). These results, along with the fact that only CR O. anthropi was recovered from collection tubes, indicate this to be the probable origin of the outbreak. As mentioned above, an outbreak related to cross-contamination from erythrocyte sedimentation tubes due to O. anthropi has been described in the literature [10]. However, our case represents the first outbreak due to this microorganism exhibiting carbapenem resistance.
Fig. 2.
Clonal relationship among Ochrobactrum anthropi (OA) isolates. Oligonucleotide PCR of OA isolates included in study: C−, control negative; lanes 1–6, OA carbapenem-resistant clinical isolates; lanes 7–10, OA carbapenem-resistant isolates from blood collection tubes; lane 11, OA carbapenem-sensitive isolates from blood collection tubes; lanes 12 and 13, outgroup strains unrelated to pseudo-outbreak.
Whole-genome analysis of CR O. anthropi (OA 5) representative strains
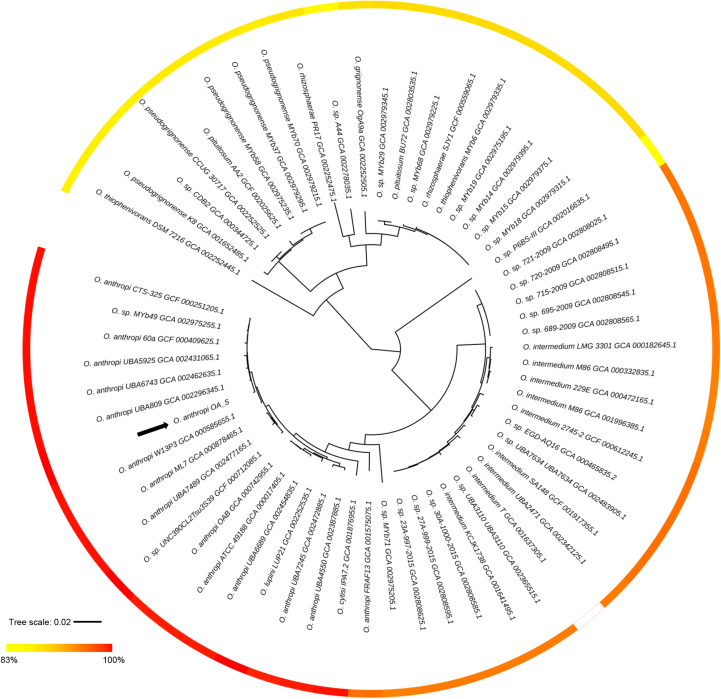
To further characterize the strains involved in the outbreak, one representative isolate (OA 5) was chosen, and whole-genome sequencing was performed. The draft genome of OA 5 consists of 4 792 719 bp sequences, distributed in 163 contigs, with an N50 size of 378 864 bp. The observed coverage for this sequenced genome was 312×, and the G+C content was 56%. By means of the RAST web server, 4554 protein-coding genes were predicted. The ARG-ANNOT database was queried to identify resistance genes within the OA 5 genome. One class C extended-spectrum β-lactamase (blaOCH-4), other three class C β-lactamases and, most importantly, a metal-dependent hydrolase of the β-lactamase superfamily I, phnP, were identified. The analysis of the genetic context of phnP gene revealed presence of a deoxyribonuclease TatD family gene, a methionyl-tRNA synthetase ligase (metG) upstream and a glucan exporter ATP-binding protein downstream. These genes were also associated with phnP in a previous description of O. anthropi ATCC 49188 (GenBank accession no. CP000758) and O. anthropi strain OAB (GenBank accession no. CP008820). The analysis of the phylogenetic distribution of this gene among sequenced genomes also supports the ubiquity of this gene in the genus (Fig. 3). The amino acid sequence identity among identified genes ranged from 80% to 100%, suggesting that is relatively well conserved at the sequence level. The analysis of the more divergent sequences and the functional relevance of the observed substitutions in this gene in different lineages deserve further study, although this falls outside the scope of our study here.
Fig. 3.
Maximum likelihood phylogenetic tree of genus Ochrobactrum. Calculated amino acid sequence identity of each identified PhnP against annotated OA 5 PhnP is shown in coloured gradient scale that ranges from 83% to 100%. Phylogenetic analysis was done based on concatenation of protein sequence of 556 putative orthologous genes and was inferred using RAxML 8.2.9. RELL test was used to evaluate branch supports with 100 replicates. All nodes showed support between 98% and 100% (data not shown). Identification of PhnP in studied genomes was done by tblastn [20]. Black arrow indicates position of O. anthropi OA 5 strain.
A variety of genes coding for efflux pumps were identified. Among them, genes coding for the macrolide efflux pump family (MacAB-TolC) and genes belonging to the efflux pumps of resistance nodulation cell division (RND) were present, including the membrane fusion protein, the inner membrane transporter and the transcriptional regulator. In addition, genes coding for multidrug and toxin extrusion (MATE) family efflux pump and genes of the AcrAB efflux pump system were present. This result highlights the importance of efflux pump systems in Gram-negative bacteria, which can contribute to the resistant phenotype development.
Among mobile elements, two intact phages were also found in OA 5 genome; one complete (ISRle4) and one incomplete (IS426) insertion sequence were also found.
In addition to being described in O. anthropi genomes, ISRle4 was also found in other species, such as Sinorhizobium sp. CCBAU 05631 (GenBank accession no. CP023065) and Rhizobium sp. N941 (GenBank accession no. CP013643). IS426 was described mainly in Agrobacterium tumefaciens, some of which were associated with plasmids, such as in A. tumefaciens C58 (GenBank accession no. FJ004947) as well as in O. anthropi OAB (GenBank accession no. CP008818).
Using PathogenFinder, 195 predicted genes known to be associated with pathogenicity were found in the OA 5 genome. Genes related with the bacterial flagellum were found, such as fliL and fliG, which are the flagellar basal body–associated protein and the flagellar motor switch protein, respectively. In addition, a flagellar hook length control protein and a flagellar hook-associated protein were present. Genes related to the type IV pilus, which is known to be important for adhesion and surface motility, were found, such as pilZ.
Six peptidases, 18 proteins with unknown function and 78 conserved hypothetical proteins were also identified in this genome. Finally, we report a robust phylogenetic tree for the genus based on more than 500 orthologs, comprising 146 453 aa–aligned sites. As expected, OA 5 appears clustered with other O. anthropi strains as a monophyletic group, but it is closely related to strains UBA809, UBA6743, UBA5925, 60a, MYb49 and CTS-325, which are all associated with environmental niches.
Conclusion
We have described an outbreak and provided a detailed whole-genome analysis of a representative strains of O. anthropi causing pseudo-bacteraemia. After an intense epidemiologic investigation, the isolates recovered from the patients' sample were linked to the source of the outbreak.
The uncommon resistant profile, not previously reported in the literature, exhibited in the strains involved in this outbreak, led us to sequence the whole genome of one strain. Different β-lactamase genes in the genome of O. anthropi were found that might contribute to the exposed phenotype. In addition, genes coding for efflux pumps belonging to the macrolide efflux pump family, multidrug RND family transporters, MATE efflux pump and AcrAB efflux pump system were also found, and can also in part explain the resistance profile of the isolates. Further analysis to reveal the mechanisms responsible for the observed phenotype is needed.
This report highlights the importance of correct practices on a daily basis and the implementation of exhaustive epidemiologic surveillance upon the occurrence of uncommon multidrug-resistant pathogens in a clinical setting. This could help to find and identify the routes of transmission and allow the implementation of infection control prevention practices to stop or control the spread of the pathogen.
Acknowledgement
Part of the authors’ work was supported by NIH 1SC3GM125556-01 to MSR. JF has a Strengthening Opportunity, Access and Resources (SOAR)-ELEVAR Scholar Fellowship from Latina/o Graduate Students, Title V Grant from the U.S. Department of Education. SM has a Doctoral Fellowship from CONICET. AI is member of Sistema Nacional de Investigadores (SNI) and PEDECIBA, Uruguay.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nmni.2018.09.002.
Contributor Information
M.S. Ramirez, Email: msramirez@fullerton.edu.
M. Almuzara, Email: marisaalmuzara@gmail.com.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Holmes B., Popoff M., Kiredjian M., Kersters K. Ochrobactrum anthropi gen. nov., sp. nov. from human clinical specimens and previously known as group Vd. Int J Syst Evol Microbiol. 1988;38:406–416. [Google Scholar]
- 2.Mudshingkar S.S., Choure A.C., Palewar M.S., Dohe V.B., Kagal A.S. Ochrobactrum anthropi: an unusual pathogen: are we missing them? Indian J Med Microbiol. 2013;31:306–308. doi: 10.4103/0255-0857.115664. [DOI] [PubMed] [Google Scholar]
- 3.Cieslak T.J., Robb M.L., Drabick C.J., Fischer G.W. Catheter-associated sepsis caused by Ochrobactrum anthropi: report of a case and review of related nonfermentative bacteria. Clin Infect Dis. 1992;14:902–907. doi: 10.1093/clinids/14.4.902. [DOI] [PubMed] [Google Scholar]
- 4.Alnor D., Frimodt-Moller N., Espersen F., Frederiksen W. Infections with the unusual human pathogens Agrobacterium species and Ochrobactrum anthropi. Clin Infect Dis. 1994;18:914–920. doi: 10.1093/clinids/18.6.914. [DOI] [PubMed] [Google Scholar]
- 5.Khasawneh W., Yusef D. Ochrobactrum anthropi fulminant early-onset neonatal sepsis: a case report and review of literature. Pediatr Infect Dis J. 2017;36:1167–1168. doi: 10.1097/INF.0000000000001660. [DOI] [PubMed] [Google Scholar]
- 6.Gigi R., Flusser G., Kadar A., Salai M., Elias S. Ochrobactrum anthropi–caused osteomyelitis in the foot mimicking a bone tumor: case report and review of the literature. J Foot Ankle Surg. 2017;56:851–853. doi: 10.1053/j.jfas.2017.02.008. [DOI] [PubMed] [Google Scholar]
- 7.Alparslan C., Yavascan O., Kose E., Sanlioglu P., Aksu N. An opportunistic pathogen in a peritoneal dialysis patient: Ochrobactrum anthropi. Indian J Pediatr. 2013;80:72–74. doi: 10.1007/s12098-012-0800-2. [DOI] [PubMed] [Google Scholar]
- 8.Mattos F.B., Saraiva F.P., Angotti-Neto H., Passos A.F. Outbreak of Ochrobactrum anthropi endophthalmitis following cataract surgery. J Hosp Infect. 2013;83:337–340. doi: 10.1016/j.jhin.2012.11.027. [DOI] [PubMed] [Google Scholar]
- 9.Ezzedine H., Mourad M., Van Ossel C., Logghe C., Squifflet J.P., Renault F. An outbreak of Ochrobactrum anthropi bacteraemia in five organ transplant patients. J Hosp Infect. 1994;27:35–42. doi: 10.1016/0195-6701(94)90066-3. [DOI] [PubMed] [Google Scholar]
- 10.Labarca J.A., Garcia P., Balcells M.E., de la Cerda G., Salinas A.M., Roman J.C. Pseudo-outbreak of Ochrobactrum anthropi bacteremia related to cross-contamination from erythrocyte sedimentation tubes. Infect Control Hosp Epidemiol. 2007;28:763–765. doi: 10.1086/517979. [DOI] [PubMed] [Google Scholar]
- 11.Weisburg W.G., Barns S.M., Pelletier D.A., Lane D.J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Isenberg H.D., American Society for Microbiology . ASM Press; Washington, DC: 2004. Clinical microbiology procedures handbook. [Google Scholar]
- 13.Clinical Laboratory Standards Institute . Clinical and Laboratory Standards Institute; Wayne, PA: 2017. Performance standards for antimicrobial susceptibility testing. Twenty-seventh informational supplement. Document M100-S27. [Google Scholar]
- 14.Pires J., Novais A., Peixe L. Blue-Carba, an easy biochemical test for detection of diverse carbapenemase producers directly from bacterial cultures. J Clin Microbiol. 2013;51:4281–4283. doi: 10.1128/JCM.01634-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee K., Lim Y.S., Yong D., Yum J.H., Chong Y. Evaluation of the Hodge test and the imipenem-EDTA double-disk synergy test for differentiating metallo-beta-lactamase–producing isolates of Pseudomonas spp. and Acinetobacter spp. J Clin Microbiol. 2003;41:4623–4629. doi: 10.1128/JCM.41.10.4623-4629.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Limansky A.S., Viale A.M. Can composition and structural features of oligonucleotides contribute to their wide-scale applicability as random PCR primers in mapping bacterial genome diversity. J Microbiol Methods. 2002;50:291–297. doi: 10.1016/s0167-7012(02)00040-4. [DOI] [PubMed] [Google Scholar]
- 17.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zerbino D.R., Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aziz R.K., Bartels D., Best A.A., DeJongh M., Disz T., Edwards R.A. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 21.Cosentino S., Voldby Larsen M., Moller Aarestrup F., Lund O. PathogenFinder—distinguishing friend from foe using bacterial whole genome sequence data. PLoS One. 2013;8 doi: 10.1371/journal.pone.0077302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupta S.K., Padmanabhan B.R., Diene S.M., Lopez-Rojas R., Kempf M., Landraud L. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother. 2014;58:212–220. doi: 10.1128/AAC.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siguier P., Perochon J., Lestrade L., Mahillon J., Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Y., Liang Y., Lynch K.H., Dennis J.J., Wishart D.S. PHAST: a fast phage search tool. Nucleic Acids Res. 2011;39:W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L., Stoeckert C.J., Jr., Roos D.S. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Contreras-Moreira B., Vinuesa P. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl Environ Microbiol. 2013;79:7696–7701. doi: 10.1128/AEM.02411-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le S.Q., Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 29.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minh B.Q., Nguyen M.A.T., von Haeseler A. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 2013;30:1188–1195. doi: 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.