Abstract
Background
Though the mechanisms of skeletal muscle regeneration are deeply understood, those involved in muscle contusion, one of the most common muscle injuries in sports medicine clinics, are not. The objective of this study is to explore the mechanisms involved in muscle regeneration after contusion injury.
Methods
In this study, a total of 72 mice were used. Eight of them were randomly chosen for the control group, while the rest were subjected to muscle contusion. Subsequently, their gastrocnemius muscles were harvested at different time points. The changes in muscle morphology were assessed by hematoxylin and eosin (HE) stain. In addition, the gene expression was analyzed by real-time polymerase chain reaction.
Results
The data showed that the expression of many genes, i.e., specific markers of immune cells and satellite cells, regulatory factors for muscle regeneration, cytokines, and chemokines, increased in the early stages of recovery, especially in the first 3 days. Furthermore, there were strict rules in the expression of these genes. However, almost all the genes returned to normal at 14 days post-injury.
Conclusion
The sequence of immune cells invaded after muscle contusion was neutrophils, M1 macrophages and M2 macrophages. Some CC (CCL2, CCL3, and CCL4) and CXC (CXCL10) chemokines may be involved in the chemotaxis of these immune cells. HGF may be the primary factor to activate the satellite cells after muscle contusion. Moreover, 2 weeks are needed to recover when acute contusion happens as used in this study.
Keywords: Chemokines, Contusion, Cytokines, Gene, Macrophages, Satellite cells, Skeletal muscle
1. Introduction
Muscle injuries are common musculoskeletal problems. The mechanisms of skeletal muscle recovery are becoming clearer recently. The healing process of injured skeletal muscle consists of three distinct phases: degeneration and inflammation, regeneration, and fibrosis.1, 2, 3 The first phase is characterized by local swelling at the injury site, formation of hematoma, necrosis of muscle tissue, degeneration, and inflammatory response. The second phase includes phagocytosis of the damaged tissue and regeneration of the injured muscle. And the final phase is characterized by scar tissue formation if the injury is serious.
The first phase usually consists of the infiltration of activated macrophages and neutrophils into the injured tissue. Many chemokines may play important roles in the chemotaxis of these immune cells.4, 5 In the regeneration phase, many growth factors can regulate the activation, proliferation, and differentiation of satellite cells, which are necessary for the regeneration of injured skeletal muscles.6
As we mentioned above, there is an appreciable understanding of the mechanisms of skeletal muscle regeneration among researchers. However, our understanding in this domain has been limited to the injury models of toxicant injection, freeze-induced injury, burn, disuse muscle atrophy, mdx mice, etc.7, 8, 9, 10 As a result, the mechanisms involved in muscle contusion, one of the most common muscle injuries in sports medicine clinics, are still not fully understood. Therefore, in the present study, we construct a model of skeletal muscle contusion with the objective of exploring the mechanisms involved in muscle regeneration. At different time points post-injury, we studied many genes expression such as the specific markers of neutrophils (MPO),11 M1 macrophages (CD68) and M2 macrophages (CD163),3, 12 proliferation (MyoD), and differentiation (myogenin)12, 13, 14 of satellite cells. Moreover, we examined the mRNA levels of inflammatory cytokines (i.e., TNF-α, IL-1β, IL-6, and IL-10) and chemokines (i.e., CCL2, CCL3, CCL4, and CCL8; CXCL9, CXCL10, and CXCL12).3, 5 We also analyzed the transcript levels of some regulatory factors which play important roles in satellite cell activation and muscle regeneration (i.e., HGF, uPA, IGF-I, MGF, and myostatin).15, 16, 17, 18, 19
2. Methods
2.1. Mice
Seventy-two C57BL/6 male mice (weighing 18.2–22.9 g, purchased from Shanghai Lab. Animal Research Center, Shanghai, China) were provided food and water ad libitum and maintained on a 12 h:12 h light–dark cycle. Eight mice were randomly chosen for the uninjured control group (n = 8), while the rest were subjected to muscle contusion (n = 64). In preparing the mice for muscle injury induction, they were anesthetized with 400 mg/kg chloral hydrate administered intraperitoneally. The study was approved by the Ethics Review Committee for Animal Experimentation of Shanghai University of Sport.
2.2. Contusion model
A simple and reproducible muscle contusion model in mice was used.20, 21 The animals' hind limbs were positioned on a board, dorsiflexing the ankle to 90°. A 16.8 g (diameter 15.9 mm) stainless steel ball was dropped from the height of 100 cm through a tube (interior diameter of tube:16 mm) onto an impactor20 resting with a surface of 28.26 mm2 on the middle of the gastrocnemius muscle of the mouse. The instantaneous force delivered by a falling object with these characteristics was calculated to equal 0.58 N·m/cm2, where 1 N·m is equal to the force of an object weighing 100 g falling over a distance of 1 m.22 The muscle contusion created by this method was a high-energy blunt injury that created a large hematoma and was followed by massive muscle regeneration,20, 23 healing processes that are very similar to those seen in humans.24 The mice that had bone fracture (fracture rate of 2.7%) were foreclosed. The injured mice in this study had signs of unrelieved pain such as piloerection of fur, reluctance to ambulate, overgrooming of the injured limb, and abnormal gait or posture.25 At different time points (6 h, 12 h, 1, 3, 5, 7, 14, and 21 days) post-injury, the mice were killed by cervical dislocation while under anesthesia and then gastrocnemius muscles were harvested.
2.3. Histology
At the time points of 1, 3, 7, 14, and 21 days post-injury, the right gastrocnemius muscles were collected and embedded in paraffin. Cross sections were cut 8 µm from the midbelly of each gastrocnemius muscle and were stained with hematoxylin and eosin (HE) for morphological analysis. Using a 40 lens objective, images were captured for each muscle section (Labphot-2; Nikon, New York, NY, USA).
2.4. RNA extraction and cDNA synthesis
Approximately 60 mg of tissue (from the middle of the left gastrocnemius muscle) was homogenized using an Ultra-Turrax homogenizer (IKA, Staufen, Germany) in a solution of TRIzol Reagent (Invitrogen, Carlsbad, CA, USA). Total RNA was isolated using a modified guanidiniumisothiocyanate-CsCl method,26, 27 and the concentration and purity were determined by measuring the absorbance at 260 nm and 280 nm in a spectrophotometer (NanoDrop 2000; Thermo Scientific, Wilmington, MA, USA). Total RNA was reverse transcribed into cDNA using the RevertaidTM First Strand cDNA Synthesis Kit from Fermentas (Fermentas, Vilnius, Lithuania). cDNA was synthesized using 2 µg of total RNA, 0.2 µg of random primers, 20 mmol/L dNTP mix, 5 × reaction buffer (Fermentas), 20U RiboLockTM RNase Inhibitor and 200 U of RevertaidTM M-MuLVreverse transcriptase in a total volume of 20 µL. The reaction was carried out at 25°C for 5 min followed by another 60 min at 42°C and was terminated by the deactivation of the enzyme at 70°C for 5 min. Control reactions lacking either reverse transcriptase or template were included to assess carryover of genomic DNA and non-specific contamination.28, 29
2.5. Real-time polymerase chain reaction (PCR)
Quantitative PCR was carried out in triplicate in reactions consisting of 12.5 µL 2 × Maxima SYBR Green/ROX qPCR Master mix (Fermentas), 1 µL cDNA, nuclease-free water and 300 nmol/L of each primer. Primer specifications are listed in Table 1. Amplifications were performed on a Rotor-Gene 3000 thermal cycler (Corbett, Sydney, Australia) with the following parameters: activation at 95°C for 10 min, 40 cycles of denaturation at 95°C for 15 s, and annealing/extension at 60°C for 1 min. The threshold cycle (CT, the number of cycles to reach threshold of detection) was determined for each reaction, and the levels of the target mRNAs were quantified relatively to the level of the housekeeping gene GAPDH using 2−△△CT method.30
Table 1.
Primers used for real-time polymerase chain reaction.
| Target gene | Forward primer sequence | Reverse primer sequence |
|---|---|---|
| MyoD | 5'-GAGCGCATCTCCACAGACAG-3' | 5'-AAATCGCATTGGGGTTTGAG-3' |
| myogenin | 5'-CCAGTACATTGAGCGCCTAC-3' | 5'-ACCGAACTCCAGTGCATTGC-3' |
| MPO | 5'-CTGAATCTGTTGTCCGTGTCA-3' | 5'-GTGATGGTGCGATACTTGTCAT-3' |
| CD68 | 5'-CAAAGCTTCTGCTGTGGAAAT-3' | 5'-GACTGGTCACGGTTGCAAG-3' |
| CD163 | 5'-GCAAAAACTGGCAGTGGG-3' | 5'-GTCAAAATCACAGACGGAGC-3' |
| HGF | 5'-AGGAACAGGGGCTTTACGTT-3' | 5'-GCTGCCTCCTTTACCAATGA-3' |
| uPA | 5'-AGTGTGGCCAGAAGGCTCTA-3' | 5'-GCTGCTCCACCTCAAACTTC-3' |
| IGF-I | 5'-GCTTGCTCACCTTTACCAGC-3' | 5'-AAATGTACTTCCTTCTGGGTCT-3' |
| MGF | 5'-GCTTGCTCACCTTTACCAGC-3' | 5'-AAATGTACTTCCTTTCCTTCTC-3' |
| myostatin | 5'-TGCAAAATTGGCTCAAA-CAG-3' | 5'-GCAGTCAAGCCCAAAGTCTC-3' |
| TNF-α | 5'-CTTCTGTCTACTGAACTTCGGG-3' | 5'-CACTTGGTGGTTTGCTACGAC-3' |
| IL-1β | 5'-TGACGTTCCCATTAGACAACTG -3' | 5'-CCGTCTTTCATTACACAGGACA-3' |
| IL-6 | 5'-GAACAACGATGATGCACTTGC-3' | 5'-CTTCATGTACTCCAGGTAGCTATGGT-3' |
| IL10 | 5'-CAAGGAGCATTTGAATTCCC-3' | 5'-GGCCTTGTAGACACCTTGGTC-3' |
| CCL2 | 5'-GCTCAGCCAGATGCAGTTAAC-3' | 5'-CTCTCTCTTGAGCTTGGTGAC-3' |
| CCL3 | 5'-ACCATGACACTCTGCAACCA-3' | 5'-CCCAGGTCTCTTTGGAGTCA-3' |
| CCL4 | 5'-CCACTTCCTGCTGTTTCTCTTA-3' | 5'-CTGTCTGCCTCTTTTGGTCAG-3' |
| CCL8 | 5'-CTTCTTTGCCTGCTGCTCATAG-3' | 5'-CACTTCTGTGTGGGGTCTACA-3' |
| CXCL9 | 5'-CTCCTTGCTTGCTTACCACTTT-3' | 5'-CCAGCCTTGTCTACTTTGAGAG-3' |
| CXCL10 | 5'-CCTCATCCTGCTGGGTCTG-3' | 5'-GTGGCAATGATCTCAACACG-3' |
| CXCL12 | 5'-ACGGAAGAACCAAAGAGAAAGA-3' | 5'-CTCAGACAGCGAGGCACAT-3' |
| GAPDH | 5'-ACTCCACTCACGGCAAATTC-3' | 5'-TCTCCATGGTGGTGAAGACA-3' |
2.6. Statistical analysis
All values are expressed as mean ± SD, and statistical significance was set at p < 0.05. Mean values were compared between groups by one-way ANOVA with the Bonferroni method as a post hoc test, or non-parametric Kruskal–Wallis test. Data were analyzed using SPSS 19.0 for Windows (SPSS Inc., Chicago, IL, USA).
3. Results
3.1. Muscle morphology
HE stain was used to assess changes in muscle morphology after injury. On Day 1 post-injury, cross sections of gastrocnemius muscles showed substantial fiber damage and edema, and a large number of inflammatory cell infiltration. On Day 3 post-injury, a small quantity of centronucleated myofibers were observed. On Days 7 and 14 post-injury, central nucleation phenomenon became more pronounced. On Day 21 post-injury, central nucleation almost disappeared. Since centrally nucleated myofibers are a sign of regeneration in injured muscle,19 it means that muscle regeneration was substantially completed on Day 21 (Fig. 1).
Fig. 1.
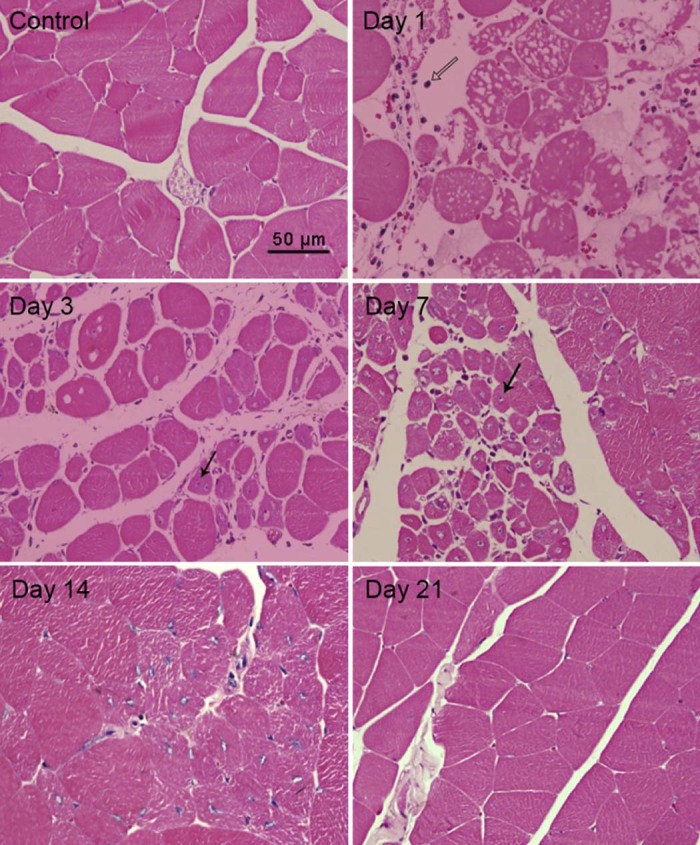
Histological evaluation of the muscle healing process in gastrocnemius muscle at five time points (1, 3, 7, 14, and 21 days post-injury).  inflammatory cells;
inflammatory cells;  central nucleation.
central nucleation.
3.2. Specific markers of satellite cells in muscle
MyoD and myogenin are the specific markers of satellite cells proliferation and differentiation, respectively. The data showed that MyoD mRNA increased significantly at the time points: 6 h (4.87 folds, p = 0.000), 12 h (p = 0.002), Day 1 (p = 0.009) and Day 3 (p = 0.002) post-injury (Fig. 2A). Unlike MyoD, myogenin did not increase significantly at 6 h, 12 h, and 1 day after injury (p > 0.05). However, a high expression of myogenin mRNA was observed at Day 3 (5.80 folds, p = 0.000), Day 5 (3.56 folds, p = 0.007), and Day 7 (3.93 folds, p = 0.003) post-injury (Fig. 2B).
Fig. 2.
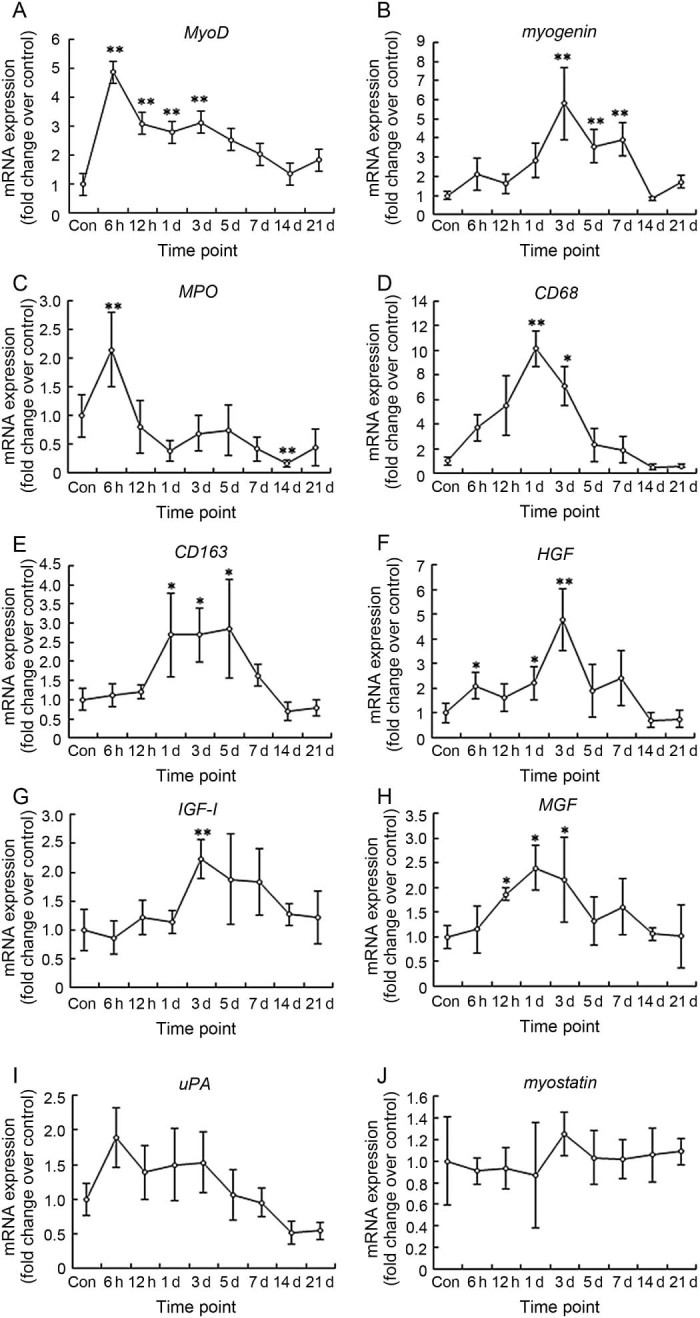
RNA levels of specific markers of satellite (A–B), immune (C–E) cells. And regeneration regulatory factors (F–J) in mice gastrocnemius muscle post-injury (mean ± SD, n = 8). Con = control; d = day. *p < 0.05, **p < 0.01; compared with Con.
3.3. Specific markers of immune cells in muscle
MPO, CD68, and CD163 are the specific markers of neutrophils, M1 macrophages and M2 macrophages, respectively. The data showed that MPO mRNA increased significantly at 6 h post-injury (2.15 folds, p = 0.000), and then declined quickly (Fig. 2C). CD68, the molecule marker of M1 macrophages, increased at 6 h and 12 h, peaked at 1 day (10.15 folds, p = 0.001), and then declined dramatically (Fig. 2D). Unlike CD68, CD163, the molecule marker of M2 macrophages almost did not change at 6 h and 12 h post-injury. However, CD163 mRNA level at 1 day (2.69 folds, p = 0.036), 3 days (2.69 folds, p = 0.023), and 5 days (2.86 folds, p = 0.028) post-injury was significantly higher than that of the control group (Fig. 2E).
3.4. Regulatory factors for muscle regeneration
We studied many regulatory factors which are involved in muscle regeneration, i.e., HGF, uPA, IGF-1, MGF (positive regulatory factors) and myostatin (negative regulatory factor). The data showed that HGF mRNA in muscle increased significantly at 6 h (2.11 folds, p = 0.015), 1 day (2.22 folds, p = 0.033), and 3 days (4.77 folds, p = 0.002) post-injury (Fig. 2F). IGF-1 mRNA increased significantly at 3 days (2.23 folds, p = 0.002) (Fig. 2G). MGF, the splicing isoform of IGF-1, increased significantly at 12 h (1.86 folds, p = 0.041), Day 1 (2.04 folds, p = 0.023), and Day 3 (2.15 folds, p = 0.038) (Fig. 2H). However, uPA (Fig. 2I) and myostatin (Fig. 2J) mRNA did not change significantly from the control group at all time points post-injury (p > 0.05).
3.5. Inflammatory cytokine levels
We studied the expression of pro-inflammatory cytokines (i.e., TNF-α, IL-1β, and IL-6). The data showed that TNF-α mRNA in muscle increased at 6 h and 12 h, and a high expression at 1 day (5.29 folds, p = 0.006) and 3 days (5.09 folds, p = 0.009) post-injury (Fig. 3A). IL-1β mRNA increased dramatically at 6 h (489.52 folds, p = 0.000), 12 h (p = 0.000), Day 1 (p = 0.002), and Day 3 (p = 0.043) (Fig. 3B). The expression of IL-6 was similar to IL-1β, but the extent of change was much smaller (Fig. 3C). On the other hand, a test on the mRNA level of IL-10 (anti-inflammatory cytokine) showed that IL-10 mRNA increased significantly at 6 h (8.56 folds, p = 0.041), Day 1 (12.95 folds, p = 0.001), and Day 3 (10.58 folds, p = 0.004) (Fig. 3D).
Fig. 3.
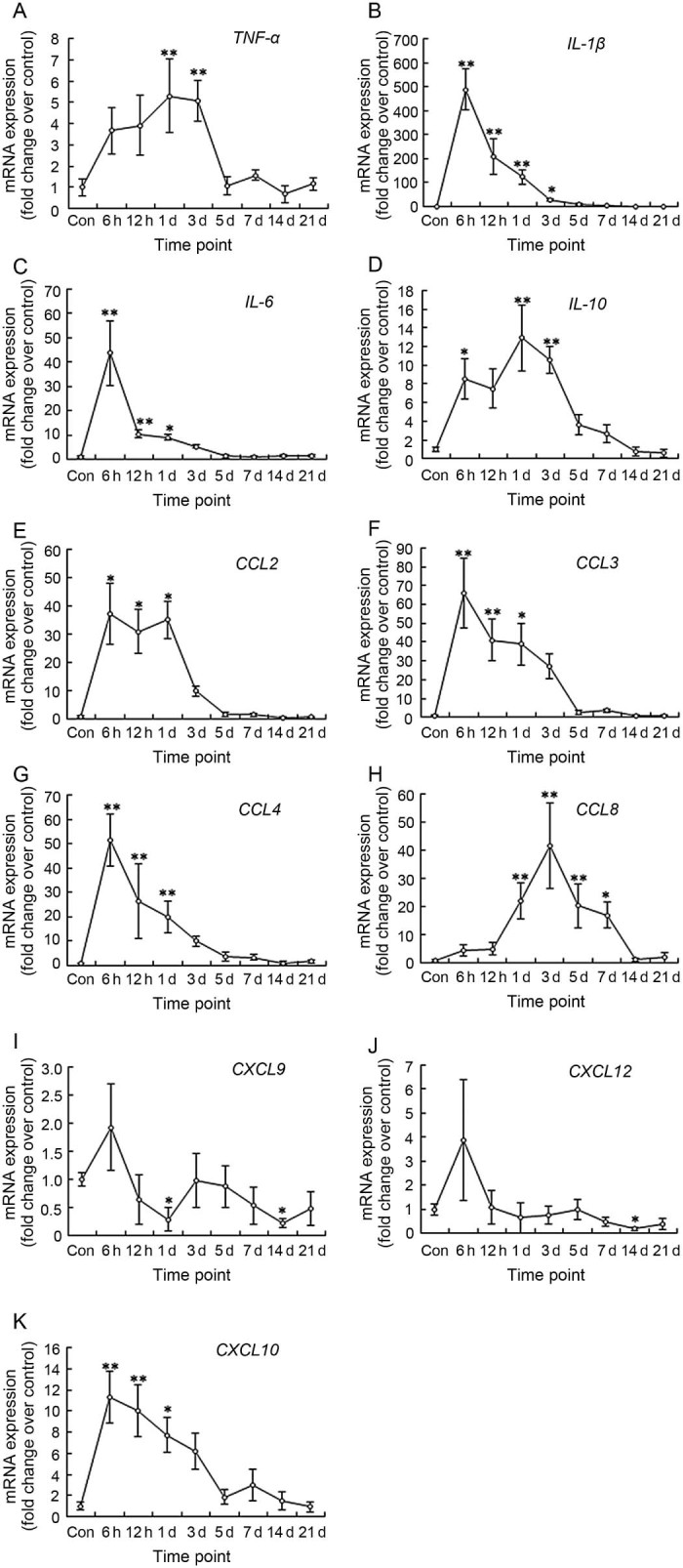
mRNA levels of inflammatory cytokines (A–D), CC chemokines (E–H), and CXC chemokines (I–K) in mice gastrocnemius muscle post-injury (mean ± SD, n = 8). Con = control; d = day. *p < 0.05, **p < 0.01; compared with Con.
3.6. Chemokine levels
We tested the CC chemokines which were involved in the chemotaxis of immune cells. The data showed that the expression patterns of CCL2, CCL3, and CCL4 were very similar. All of them peaked at 6 h (37.19 folds, p = 0.010; 65.86 folds, p = 0.000; 51.64 folds, p = 0.000; respectively) and then declined, but the mRNA levels at 12 h and 1 day were still much higher than the control group (Fig. 3E-G). However, CCL8 had different gene expression patterns. CCL8 mRNA increased significantly at Day 1 (p = 0.003), Day 3 (p = 0.000), Day 5 (p = 0.009), and Day 7 (p = 0.032) (Fig. 3H). Furthermore, we investigated the CXC chemokines. The data showed that CXCL9 and CXCL12 mRNA increased slightly only at 6 h (p > 0.05). At other time points, there was the tendency for the mRNA levels to decline, especially at 1 day (CXCL9, p < 0.05) and 14 days post-injury (CXCL9, CXCL12, p < 0.05) (Fig. 3I and J). However, different from CXCL9 and CXCL12, the expression of CXCL10 was similar to some CC chemokines (i.e., CCL3 and CCL4) (Fig. 3K).
4. Discussion
4.1. The sequence of immune cells invade after muscle contusion
Acute muscle injuries initiate a series of responses by specific myeloid cell populations. Studies have shown that type I macrophages are associated with muscle necrosis, whereas type II macrophages are associated with regenerative myofibers.3 In this study, we tested the specific markers of neutrophils (MPO),11 M1 macrophages (CD68), and M2 macrophages (CD163).3, 12 The data showed that neutrophils (MPO) were the first to respond and peak in concentration at 6 h post-injury before declining rapidly. Following the onset of neutrophil invasion, M1 macrophages also began to invade until it reached significant elevated concentrations at about 24 h post-injury and then declined sharply. Their invasion preceded the elevation of a population of M2 macrophages that remained significantly elevated for many days (from 1 day to 5 days post-injury). The fact that the invasion of M1 macrophages preceded that of M2 macrophages conformed to their functions. Indeed while type I macrophages enhance the proliferation of local myogenic precursor cells, type II macrophages stimulate their fusion and differentiation.31 Though our study was only based on genetic levels, we acknowledge that the results would have been more convincing had protein levels been tested too.
The sequence of immune cells invasion after muscle contusion, as described above, was similar to other injury models.3 Meanwhile, it coincided with the expression of the inflammatory cytokines. The high-energy blunt injury used in this study induced intense inflammatory response. Pro-inflammatory cytokines (i.e., TNF-α, IL-1β, and IL-6) were highly expressive in the first 3 days. Moreover, we studied the anti-inflammatory cytokine (IL-10) which plays a central role in regulating the switch of muscle macrophages from M1 to M2 phenotype in injured muscle.32 The study showed that ablation of IL-10 prevented a subsequent increase in CD163 post-injury. Furthermore, muscle regeneration and growth were greatly slowed by the loss of IL-10. In addition, in vitro assays showed that coculturing muscle cells with macrophages activated with IL-10 to the M2 phenotype increased myoblast proliferation.32 In this study, we found that the expression of IL-10 was faster than the specific marker of M2 macrophages (CD163). We speculate that IL-10 may be involved in the transition of macrophage phenotype after muscle contusion; however, this needs further study.
4.2. CC and CXC chemokines may be involved in the chemotaxis of immune cells after muscle contusion
Chemokines are small cytokines with chemoattractant properties, belonging to one of the four families: C, CC, CXC, and CX3C.33 These families are differentiated by the number and order of the amino-terminal cysteine residues and vary in their specific actions by binding to specific seven transmembrane-domain G protein-coupled receptors of the same family.34, 35 The C and CX3C families are chemotactic for lymphocytes,36 while CC chemokines and their receptors are primarily involved in the migration and activation of monocytes, macrophages and lymphocytes.34 The CXC chemokines that contain three residue motifs (ELR) have neutrophil specificity.36 Our study showed a rapid and coordinated elevation in CCL2, CCL3, and CCL4 in muscle post-injury, which is similar to the study of Warren et al.37, 38 CC chemokines increased dramatically post-injury. This has very important physiological significance. There were enough evidence to indicate that CC chemokines play a significant role in attracting immune cells to the injury site.3, 37, 38 Disruption of CCL2/CCR2 signaling in injured muscle may have multiple impacts on tissue response to injury that can lead to slower regeneration. Apart from attracting immune cells, CC chemokines were involved in the proliferation of myoblast. The study of Yahiaoui et al.39 showed that myoblasts constitutively expressed receptors for CCL2 (CCR2), CCL3 (CCR1 and CCR5), and CCL4 (CCR5); and that stimulation with either CCL2 or CCL4 was sufficient to promote myoblast proliferation. However, unlike CCL2, CCL3 and CCL4, CCL8 had different expression pattern. CCL8 did not increase significantly at 6 h and 12 h after injury; it did after 1 day. Our results are very similar to the study of Nicholas et al.,40 who found that CCL2 and CCL3 mRNA peaked at 8 h after muscle contusion, while CCL8 mRNA peaked at 48 h after injury. Immune cells invasion preceded the elevation of CCL8, meaning that CCL2, CCL3, and CCL4, but not CCL8, may be involved in the chemotaxis of immune cells after muscle contusion.
In addition, according to the literature,41 we studied three CXC chemokines which may be involved in the chemotaxis of immune cells after muscle injury. The data showed that CXCL9 and CXCL12 did not change significantly in the whole process of muscle recovery. However, CXCL10 increased rapidly after muscle injury, similar to CCL2, CCL3, and CCL4. In the study of Koh and Pizza,41 CXCL10 produced skeletal muscle cells or immune cells that can influence the chemotaxis of inflammatory cells. Therefore, CXCL10, but not CXCL9 and CXCL12, may be involved in the chemotaxis of immune cells after muscle contusion.
4.3. HGF may be the primary factor to activate muscle satellite cells after muscle contusion
Muscle regeneration consists of activation, proliferation, and differentiation of satellite cells into myotubes, which ultimately fuse with other existing myofibers or fuse together to form new myofibers.42, 43 The activation and differentiation of satellite cells are respectively indicated by the up-regulation of myogenic regulatory factor (MRF) MyoD and myogenin.12, 13, 14 In our study, we found that MyoD is expressed early in proliferating satellite cells, and myogenin appeared after the decline in MyoD expression (Fig. 2A and B). These phenomena also existed in other injury models in some previous studies.44, 45 MyoD acts as an early MRF that is mainly involved in satellite cell activation and proliferation, whereas myogenin is a late-acting MRF expressed during differentiation.13
Satellite cells can be activated by some growth factors after muscle injury. However, there still exists a controversy as to which specific factor is involved after muscle contusion. Most studies have chosen the 1 day post-injury as the first time point. They pointed out that IGF-1, MGF, uPA, or HGF may be involved in the activation of satellite cells.15, 16, 18 Researchers of early studies opined that IGF-1 was the activate factor of satellite cells;46 as a result, MGF, the isoform of IGF-1, was deemed to be involved.15 On the other hand, some studies found that uPA may play an important role in satellite cells activation.47, 48
In this study, we added two time points, 6 h and 12 h, before the 1 day post-injury. We speculated that the negative regulatory factor of muscle regeneration, myostatin, would increase after muscle contusion, as in other injury models.49 However, interestingly, myostatin did not change in the whole process of recovery. On the other hand, when we tested some positive regulatory factors such as HGF, IGF-1, MGF, and uPA, the data showed that the expression of IGF-1 and MGF increased significantly after muscle injury, though they trailed the activation of satellite cell (MyoD). In addition, uPA, an activate factor, as described in some studies, did not change after muscle contusion. Nonetheless, due to the unavailability of protein data, we cannot certainly conclude that these factors (IGF-1, MGF, and uPA) are not involved in the activation of satellite cells. However, HGF mRNA increased significantly at 6 h post-injury, which is synchronous with the activation of satellite cell. We speculate that HGF may be the primary factor to activate muscle satellite cells after muscle contusion. HGF is a mesenchyme-derived heparin-binding glycoprotein that regulates cell proliferation, cell survival, cell motility, and morphogenesis.50 The evidence from early experiments showed that HGF can activate quiescent satellite cells in vivo or in vitro. The activation of satellite cells by HGF can undergo both paracrine and autocrine signaling.51 HGF is essential in inducing the migration of myogenic precursor cells in embryonic myogenesis.51 Furthermore, HGF is the only growth factor that has been established to have the ability to stimulate quiescent satellite cells to enter the cell cycle early in a culture assay and in vivo.18, 52
4.4. Two weeks is needed to recover when acute muscle contusion happens
In order to investigate the process of muscle recovery, we did the morphological analysis. The high-energy blunt injury induced a large hematoma and edema, widened the interstitial spaces between the muscles fibers, infiltrated the mononuclear cell into the interstitial spaces, and disorganized the muscle architecture on the first day of post-injury53 (Fig. 1). On Day 3 of post-injury, central nucleation started and became more obviously on Days 7 and 14. However, on Day 21 post-injury, central nucleation almost disappeared. Since central nucleated myofibers are a sign of regeneration in injured muscle,47 it means that muscle regeneration was substantially completed on Day 21 post-injury.
Furthermore, we studied many genes expression at different times post-injury. The data showed that many genes, e.g., specific markers of immune cells (MPO and CD68), regeneration regulatory factors (HGF, IGF-1, MGF), inflammatory cytokines (TNF-α, IL-1β, IL-6, IL-10), and chemokines (CCL2, CCL3, CCL4, CXCL10), increased in the first phase of the recovery, especially in the first 3 days. In the second phase of regeneration, specific markers of satellite cells differentiation (myogenin) and M2 macrophages (CD163) increased significantly. In addition, we found that intense inflammatory response and muscle regeneration existed simultaneously (6 h after injury), which suggested that there were no strict boundaries between the first and the second stages of regeneration. However, almost all the genes returned to normal at 14 days post-injury. Some genes such as MPO, CXCL9, and CXCL12 even decreased significantly at 14 days. Though we are currently not sure about the reasons why these inflammatory genes inhibited in the later stage of recovery, we speculate that it may be related to the severe inflammatory reaction of the early stage of recovery. Accordingly, these data suggest that skeletal muscle needs no less than 2 weeks to recover after acute contusion, as used in our study. However, it is unclear whether or not 2 weeks are sufficient for the recovery of muscle function since our study only focused on morphology and genes. This needs further study.
5. Conclusion
From the genetic perspective, we can conclude that the sequence of immune cells invaded after muscle contusion was neutrophils, M1 macrophages and M2 macrophages. Some CC (CCL2, CCL3, and CCL4) and CXC (CXCL10) chemokines may be involved in the chemotaxis of these immune cells. HGF may be the primary factor to activate the satellite cells. In addition, intense inflammatory response and regeneration existed simultaneously, which suggested that there were no strict boundaries between the first and second stages of regeneration. Almost all index returned to normal at 14 days. This means that 2 weeks are needed to recover when the acute contusion, as used in the study, happens.
Authors' contributions
WX, BL, LZ, and XL carried out the genetic studies. YL and ZZ carried out the morphological analysis. WX performed the statistical analysis and drafted the manuscript; PC conceived the study, and participated in its design and coordination and helped to draft the manuscript. All authors have read and approved the final version of the manuscript, and agree with the order of presentation of the authors.
Competing interests
None of the authors declare competing financial interests.
Acknowledgment
This work was supported by the grants from National Natural Science Foundation of China (No. 31271273, No. 31300975), the Doctoral Fund of Ministry of Education of China (No. 20133156120004), and the Key Lab of Exercise and Health Sciences of Ministry of Education (Shanghai University of Sport).
Footnotes
Peer review under responsibility of Shanghai University of Sport.
References
- 1.Carosio S., Berardinelli M.G., Aucello M., Musarò A. Impact of ageing on muscle cell regeneration. Ageing Res Rev. 2011;10:35–42. doi: 10.1016/j.arr.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 2.Fukushima K., Badlani N., Usas A., Riano F., Fu F., Huard J. The use of an antifibrosis agent to improve muscle recovery after laceration. Am J Sports Med. 2001;29:394–402. doi: 10.1177/03635465010290040201. [DOI] [PubMed] [Google Scholar]
- 3.Tidball J.G., Villalta S.A. Regulatory interactions between muscle and the immune system during muscle regeneration. Am J Physiol Regul Integr Comp Physiol. 2010;298:R1173–87. doi: 10.1152/ajpregu.00735.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griffin C.A., Apponi L.H., Long K.K., Pavlath G.K. Chemokine expression and control of muscle cell migration during myogenesis. J Cell Sci. 2010;123:3052–3060. doi: 10.1242/jcs.066241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tidball J.G. Inflammatory processes in muscle injury and repair. Am J Physiol Regul Integr Comp Physiol. 2005;288:R345–53. doi: 10.1152/ajpregu.00454.2004. [DOI] [PubMed] [Google Scholar]
- 6.Ten Broek R.W., Grefte S., Von den Hoff J.W. Regulatory factors and cell populations involved in skeletal muscle regeneration. J Cell Physiol. 2010;224:7–16. doi: 10.1002/jcp.22127. [DOI] [PubMed] [Google Scholar]
- 7.Lesault P.F., Theret M., Magnan M., Cuvellier S., Niu Y., Gherardi R.K. Macrophages improve survival, proliferation and migration of engrafted myogenic precursor cells into MDX skeletal muscle. PLoS One. 2012;7:e46698. doi: 10.1371/journal.pone.0046698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu H., Huang D., Saederup N., Charo I.F., Ransohoff R.M., Zhou L. Macrophages recruited via CCR2 produce insulin-like growth factor-1 to repair acute skeletal muscle injury. FASEB J. 2011;25:358–369. doi: 10.1096/fj.10-171579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Powers S.K., Kavazis A.N., DeRuisseau K.C. Mechanisms of disuse muscle atrophy: role of oxidative stress. Am J Physiol Regul Integr Comp Physiol. 2005;288:R337–44. doi: 10.1152/ajpregu.00469.2004. [DOI] [PubMed] [Google Scholar]
- 10.Warren G.L., Summan M., Gao X., Chapman R., Hulderman T., Simeonova P.P. Mechanisms of skeletal muscle injury and repair revealed by gene expression studies in mouse models. J Physiol. 2007;582:825–841. doi: 10.1113/jphysiol.2007.132373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bradley P.P., Priebat D.A., Christensen R.D., Rothstein G. Measurement of cutaneous inflammation: estimation of neutrophil content with an enzyme marker. J Invest Dermatol. 1982;78:206–209. doi: 10.1111/1523-1747.ep12506462. [DOI] [PubMed] [Google Scholar]
- 12.Xiao W., Chen P., Dong J. Effects of overtraining on skeletal muscle growth and gene expression. Int J Sports Med. 2012;33:846–853. doi: 10.1055/s-0032-1311585. [DOI] [PubMed] [Google Scholar]
- 13.Chargé S.B., Rudnicki M.A. Cellular and molecular regulation of muscle regeneration. Physiol Rev. 2004;84:209–238. doi: 10.1152/physrev.00019.2003. [DOI] [PubMed] [Google Scholar]
- 14.Koishi K., Zhang M., McLennan I.S., Harris A.J. MyoD protein accumulates in satellite cells and is neurally regulated in regenerating myotubes and skeletal muscle fibers. Dev Dyn. 1995;202:244–254. doi: 10.1002/aja.1002020304. [DOI] [PubMed] [Google Scholar]
- 15.Hill M., Goldspink G. Expression and splicing of the insulin-like growth factor gene in rodent muscle is associated with muscle satellite (stem) cell activation following local tissue damage. J Physiol. 2003;549:409–418. doi: 10.1113/jphysiol.2002.035832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lluıs F., Roma J., Suelves M., Parra M., Aniorte G., Gallardo E. Urokinase-dependent plasminogen activation is required for efficient skeletal muscle regeneration in vivo. Blood. 2001;97:1703–1711. doi: 10.1182/blood.v97.6.1703. [DOI] [PubMed] [Google Scholar]
- 17.Nozaki M., Li Y., Zhu J., Ambrosio F., Uehara K., Fu F.H. Improved muscle healing after contusion injury by the inhibitory effect of suramin on myostatin, a negative regulator of muscle growth. Am J Sports Med. 2008;36:2354–2362. doi: 10.1177/0363546508322886. [DOI] [PubMed] [Google Scholar]
- 18.Tatsumi R., Anderson J.E., Nevoret C.J., Halevy O., Allen R.E. HGF/SF is present in normal adult skeletal muscle and is capable of activating satellite cells. Dev Biol. 1998;194:114–128. doi: 10.1006/dbio.1997.8803. [DOI] [PubMed] [Google Scholar]
- 19.Whittemore L.A., Song K., Li X., Aghajanian J., Davies M., Girgenrath S. Inhibition of myostatin in adult mice increases skeletal muscle mass and strength. Biochem Biophys Res Commun. 2003;300:965–971. doi: 10.1016/s0006-291x(02)02953-4. [DOI] [PubMed] [Google Scholar]
- 20.Kasemkijwattana C., Menetrey J., Somogyi G., Moreland M.S., Fu F.H., Buranapanitkit B. Development of approaches to improve the healing following muscle contusion. Cell Transplant. 1998;7:585–598. doi: 10.1177/096368979800700609. [DOI] [PubMed] [Google Scholar]
- 21.Wright-Carpenter T., Opolon P., Appell H.J., Meijer H., Wehling P., Mir L.M. Treatment of muscle injuries by local administration of autologous conditioned serum: animal experiments using a muscle contusion model. Int J Sports Med. 2004;25:582–587. doi: 10.1055/s-2004-821303. [DOI] [PubMed] [Google Scholar]
- 22.Fisher B.D., Baracos V.E., Shnitka T.K., Mendryk S.W., Reid D.C. Ultrastructural events following acute muscle trauma. Med Sci Sports Exerc. 1990;22:185–193. [PubMed] [Google Scholar]
- 23.Crisco J.J., Jokl P., Heinen G.T., Connell M.D., Panjabi M.M. A muscle contusion injury model biomechanics, physiology, and histology. Am J Sports Med. 1994;22:702–710. doi: 10.1177/036354659402200521. [DOI] [PubMed] [Google Scholar]
- 24.Diaz J.A., Fischer D.A., Rettig A.C., Davis T.J., Shelbourne K.D. Severe quadriceps muscle contusions in athletes a report of three cases. Am J Sports Med. 2003;31:289–293. doi: 10.1177/03635465030310022201. [DOI] [PubMed] [Google Scholar]
- 25.Dobek G.L., Fulkerson N.D., Nicholas J., Schneider B.S. Mouse model of muscle crush injury of the legs. Comp Med. 2013;63:227–232. [PMC free article] [PubMed] [Google Scholar]
- 26.Chirgwin J.M., Przybyla A.E., MacDonald R.J., Rutter W.J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 27.Xiao W., Chen P., Wang R., Dong J. Overload training inhibits phagocytosis and ROS generation of peritoneal macrophages: role of IGF-1 and MGF. Eur J Appl Physiol. 2013;113:117–125. doi: 10.1007/s00421-012-2418-5. [DOI] [PubMed] [Google Scholar]
- 28.Fuxjager M.J., Barske J., Du S., Day L.B., Schlinger B.A. Androgens regulate gene expression in avian skeletal muscles. PLoS One. 2012;7:e51482. doi: 10.1371/journal.pone.0051482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moreno-Navarrete J.M., Ortega F., Gómez-Serrano M., Garcia-Santos E., Ricart W., Tinahones F. The MRC1/CD68 ratio is positively associated with adipose tissue lipogenesis and with muscle mitochondrial gene expression in humans. PLoS One. 2013;8:e70810. doi: 10.1371/journal.pone.0070810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Arnold L., Henry A., Poron F., Baba-Amer Y., Van Rooijen N., Plonquet A. Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J Exp Med. 2007;204:1057–1069. doi: 10.1084/jem.20070075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng B., Wehling-Henricks M., Villalta S.A., Wang Y., Tidball J.G. IL-10 triggers changes in macrophage phenotype that promote muscle growth and regeneration. J Immunol. 2012;189:3669–3680. doi: 10.4049/jimmunol.1103180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baggiolini M. Chemokines in pathology and medicine. J Intern Med. 2001;250:91–104. doi: 10.1046/j.1365-2796.2001.00867.x. [DOI] [PubMed] [Google Scholar]
- 34.Zlotnik A., Yoshie O. Chemokines: a new classification system and their role in immunity. Immunity. 2000;12:121–127. doi: 10.1016/s1074-7613(00)80165-x. [DOI] [PubMed] [Google Scholar]
- 35.Baggiolini M. Chemokines and leukocyte traffic. Nature. 1998;392:565–568. doi: 10.1038/33340. [DOI] [PubMed] [Google Scholar]
- 36.Laing K.J., Secombes C.J. Chemokines. Dev Comp Immunol. 2004;28:443–460. doi: 10.1016/j.dci.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 37.Warren G.L., Hulderman T., Mishra D., Gao X., Millecchia L., O'Farrell L. Chemokine receptor CCR2 involvement in skeletal muscle regeneration. FASEB J. 2005;19:413–415. doi: 10.1096/fj.04-2421fje. [DOI] [PubMed] [Google Scholar]
- 38.Warren G.L., O'Farrell L., Summan M., Hulderman T., Mishra D., Luster M.I. Role of CC chemokines in skeletal muscle functional restoration after injury. Am J Physiol Cell Physiol. 2004;286:C1031–6. doi: 10.1152/ajpcell.00467.2003. [DOI] [PubMed] [Google Scholar]
- 39.Yahiaoui L., Gvozdic D., Danialou G., Mack M., Petrof B.J. CC family chemokines directly regulate myoblast responses to skeletal muscle injury. J Physiol. 2008;586:3991–4004. doi: 10.1113/jphysiol.2008.152090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nicholas J., Voss J.G., Tsuji J., Fulkerson N.D., Soulakova J., Schneider B.S. Time course of chemokine expression and leukocyte infiltration after acute skeletal muscle injury in mice. Innate Immun. 2015;21:266–274. doi: 10.1177/1753425914527326. [DOI] [PubMed] [Google Scholar]
- 41.Koh T.J., Pizza F.X. Do inflammatory cells influence skeletal muscle hypertrophy? Front Biosci (Elite Ed) 2009;1:60–71. doi: 10.2741/E7. [DOI] [PubMed] [Google Scholar]
- 42.Hawke T.J., Garry D.J. Myogenic satellite cells: physiology to molecular biology. J Appl Physiol. 2001;91:534–551. doi: 10.1152/jappl.2001.91.2.534. [DOI] [PubMed] [Google Scholar]
- 43.Seale P., Rudnicki M.A. A new look at the origin, function, and “stem-cell” status of muscle satellite cells. Dev Biol. 2000;218:115–124. doi: 10.1006/dbio.1999.9565. [DOI] [PubMed] [Google Scholar]
- 44.Cornelison D.D., Wold B.J. Single-cell analysis of regulatory gene expression in quiescent and activated mouse skeletal muscle satellite cells. Dev Biol. 1997;191:270–283. doi: 10.1006/dbio.1997.8721. [DOI] [PubMed] [Google Scholar]
- 45.Yablonka-Reuveni Z., Rivera A.J. Temporal expression of regulatory and structural muscle proteins during myogenesis of satellite cells on isolated adult rat fibers. Dev Biol. 1994;164:588–603. doi: 10.1006/dbio.1994.1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chakravarthy M.V., Davis B.S., Booth F.W. IGF-I restores satellite cell proliferative potential in immobilized old skeletal muscle. J Appl Physiol. 2000;89:1365–1379. doi: 10.1152/jappl.2000.89.4.1365. [DOI] [PubMed] [Google Scholar]
- 47.Koh T.J., Bryer S.C., Pucci A.M., Sisson T.H. Mice deficient in plasminogen activator inhibitor-1 have improved skeletal muscle regeneration. Am J Physiol Cell Physiol. 2005;289:C217–23. doi: 10.1152/ajpcell.00555.2004. [DOI] [PubMed] [Google Scholar]
- 48.Sisson T.H., Nguyen M.H., Yu B., Novak M.L., Simon R.H., Koh T.J. Urokinase-type plasminogen activator increases hepatocyte growth factor activity required for skeletal muscle regeneration. Blood. 2009;114:5052–5061. doi: 10.1182/blood-2008-12-196212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kirk S., Oldham J., Kambadur R., Sharma M., Dobbie P., Bass J. Myostatin regulation during skeletal muscle regeneration. J Cell Physiol. 2000;184:356–363. doi: 10.1002/1097-4652(200009)184:3<356::AID-JCP10>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 50.Miyazawa K., Shimomura T., Naka D., Kitamura N. Proteolytic activation of hepatocyte growth factor in response to tissue injury. J Biol Chem. 1994;269:8966–8970. [PubMed] [Google Scholar]
- 51.Fu X., Wang H., Hu P. Stem cell activation in skeletal muscle regeneration. Cell Mol Life Sci. 2015;72:1663–1677. doi: 10.1007/s00018-014-1819-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Allen R.E., Temm-Grove C.J., Sheehan S.M., Rice G. Skeletal muscle satellite cell cultures. Methods Cell Biol. 1997;52:155–176. doi: 10.1016/s0091-679x(08)60378-7. [DOI] [PubMed] [Google Scholar]
- 53.Farges M.C., Balcerzak D., Fisher B.D., Attaix D., Béchet D., Ferrara M. Increased muscle proteolysis after local trauma mainly reflects macrophage-associated lysosomal proteolysis. Am J Physiol Endocrinol Metab. 2002;282:E326–35. doi: 10.1152/ajpendo.00345.2001. [DOI] [PubMed] [Google Scholar]


