Abstract
Heterogeneous patterns of mutations and RNA expression have been well documented in invasive cancers. However, technological challenges have limited the ability to study heterogeneity of protein expression. This is particularly true for pre-invasive lesions such as ductal carcinoma in situ of the breast. Cell level heterogeneity in ductal carcinoma in situ was analyzed in a single 5um tissue section using a multiplexed immunofluorescence analysis of 11 disease-related markers (EGFR, HER2, HER4, S6, pmTOR, CD44v6, SLC7A5 and CD10, CD4, CD8 and CD20, plus pan-cytokeratin, pan-cadherin, DAPI, Na+K+ATPase for cell segmentation). Expression was quantified at cell level using a single cell segmentation algorithm. K-means clustering was used to determine co-expression patterns of epithelial cell markers and immune markers. We document for the first time the presence of epithelial cell heterogeneity within ducts, between ducts and between patients with ductal carcinoma in situ. There was moderate heterogeneity in a distribution of 8 clusters within each duct (average Shannon Index 0.76; ranged 0 – 1.61). Furthermore, within each patient, the average Shannon Index across all ducts ranged from 0.33 to 1.02 (S.D. 0.09 to 0.38). As the distribution of clusters within ducts was uneven, the analysis of 8 ducts might be sufficient to represent all the clusters i.e. within- and between-duct heterogeneity. The pattern of epithelial cell clustering was associated with the presence and type of immune infiltrates, indicating a complex interaction between the epithelial tumor and immune system for each patient. This analysis also provides the first evidence that simultaneous analysis of both the epithelial and immune/stromal components might be necessary to understand the complex milieu in ductal carcinoma in situ lesions.
Keywords: Breast cancer, Ductal carcinoma in situ, tumor heterogeneity, multiplex protein analysis
Introduction:
Cellular heterogeneity presents a major problem in treating cancer. Tumor heterogeneity has been elucidated in detail using next generation sequencing (1–4). Studies have identified substantial tumor heterogeneity with mutations in spatially distinct parts of the tumor in several cancer types. More recently, next generation sequencing techniques using single tumor cells from invasive cancers have increased our ability to understand cellular heterogeneity (5). Understanding heterogeneity in protein expression is still a major challenge. This is even more problematic in small pre-invasive lesions such as ductal carcinoma in situ.
Ductal carcinoma in situ, the most common type of pre-invasive lesion, is being detected with increasing frequency with the advent of mammographic screening. Surgery is the mainstay for the treatment of ductal carcinoma in situ and on the clinic-pathological features, this may be followed by radiotherapy and/ or endocrine therapy (6). The qualitative assessment of histological grade, expression of single protein biomarkers and more recently, mRNA analysis (Oncotype DX Breast DCIS Score®) (7–9) have been used to make these decisions. However, these approaches do not fully reflect the degree of cellular heterogeneity of ductal carcinoma in situ, cellular interactions with surrounding microenvironment and other cell types. Using quantitative multiplexed immunofluorescence of a single formalin fixed paraffin embedded tissue section (10), we have for the first time investigated the degree of cellular heterogeneity by analyzing the expression and co-expression of biologically relevant proteins at a single cell and subcellular levels in tissues in ductal carcinoma in situ and correlations with immune response. In total, 14 protein markers were multiplexed on single formalin fixed paraffin embedded section per patient, including EGFR, HER2, HER4, pmTOR, CD44v6, SLC7A5, CD10, CD4, CD8 and CD20. Pan-cytokeratin, pan-cadherin, DAPI, Na+K+ATPase were used for epithelial and stromal cell segmentation. Markers were selected based on previously reported expression in ductal carcinoma in situ lesions and association with recurrence and included members of the ErbB family (EGFR, HER2, HER4) (11–17) ); pmTOR, which has been associated with autophagy pathway in ductal carcinoma in situ and breast cancer (18). CD44v6 expression has been reported in expressed in ductal carcinoma in situ, invasive cancers, but not in benign tissue (19) and is also reported to be a putative stem cell marker (20). It may also facilitate receptor tyrosine kinase signaling via binding to its heparin sulfate side chain (21). SLC7A5/LAT1 is an amino acid exchange transporter that has not been studied in ductal carcinoma in situ, but is upregulated in breast cancers at both primary and metastatic sites (22). Increased expression correlates with poor prognosis and disruption leads to the inhibition of mTOR (23). CD10 is expressed on myoepithelial cells and loss of expression may be associated with invasive disease (24). Lymphocyte infiltration in triple negative and HER2 positive subtypes have been associated with prognosis in ductal carcinoma in situ and breast cancer, with CD4+ T-helper cells responsible for facilitating antigen presentation; FOXP3+CD4+ regulatory T-cells supporting proliferation of B-cells and CD8+ cytotoxic T-cells required for tumor destruction (25–28). The expression pattern of individual markers (both epithelial and stromal) in the current study conformed to that described in prior publications and hence is not described in detail herein.
Materials and Methods:
Antibody Screening and Selection-
Antibody validation processes have been previously described (10). Briefly, for each protein target examined in the study, a minimum of 3 commercial initial antibodies were compared in parallel using Cy3 or Cy5 conjugated secondary antibodies (Jackson Immunologicals). Antibodies were assessed based on staining specificity and sensitivity characteristics using a multi-tissue array that contained breast and other tissue types (Pantomics #MNT241) and compared to patterns reported in literature and or the Human Protein Atlas (http://www.proteinatlas.org/). Where possible, antibodies used in clinical immunohistochemistry lab were included in the screenings. Slides were cleared and processed for antigen retrieval as previously reported (10) and all antibodies used at manufacturer recommended concentrations with 1 hour room temperature incubation. Specificity was determined based on localization pattern and comparison to non-specific antibodies of the same isotype. The strongest staining and most specific antibodies were further tested to ensure that the antigen was not altered by the dye inactivation process by comparing staining on samples that were untreated or treated 1, 5, or 10 times with the dye inactivation process (used to multiplex) and subsequent washing in PBS prior to antibody staining. The 14 markers were: pan-cytokeratin, pan-cadherin, NaKATPase, SLC7A5, CD10, EGFR, HER2, HER4, S6, phospho-mTOR, CD44v6, CD4, CD8, and CD20.
Antibody Conjugation-
After selection, each antibody was conjugated with either Cy3 or Cy5 bis-NHS-ester dyes using standard protocols as previously described (10). Where needed, antibodies were purified from BSA, glycerol, or other stabilizing agents by protein A/G purification. Antibodies were initially conjugated at 2 different loading concentrations (2 and 4 dyes/antibody) and staining performance was compared to the secondary-detected antibodies used in the screening stage.
Clinical Cohort:
13 de-identified cases of ductal carcinoma in situ were selected from the archives at Singapore General Hospital. The criteria for the selection were as follows 1) they were excision specimens 2) patients did not have invasive cancer 3) patients had additional blocks of ductal carcinoma in situ 4) multiple foci of ductal carcinoma in situ were present in the sections examined. Clinical information on the samples included age at diagnosis, size (mm) of ductal carcinoma in situ. An initial histopathological review was performed to confirm the diagnosis of ductal carcinoma in situ. The diagnosis of ductal carcinoma in situ was confirmed independently by two pathologists. A second tier of histological analysis was performed using pseudo-H&E digital images of the stained slides to study the relationships between the epithelial and stromal components and to quantify the different elements (see below) within any given field. Multiple fields were quantified from each case. Data for the expression of estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor (HER2) was obtained from the files of Singapore General Hospital. These stains were performed as a part of standard of care using ASCO-CAP guidelines to define positivity (29, 30).
Multiplexed Immunofluorescence Staining-
Multiplexed immunofluorescence staining was performed as previously described (10). Slides were initially baked at 60°C for 1 hour prior to clearing with xylene and rehydration through a graded series of ethanol. Two-step antigen retrieval was performed as previously described (10) and slides were blocked overnight in 4% BSA, 10% donkey serum, in PBS at 4°C. After blocking, slides were stained with DAPI (1ug/ml), cover-slipped with anti-fade mounting media, and background images collected in DAPI, eGFP, Cy3, and Cy5 filter channels. For initial imaging, a 10X objective was used, and the whole tissue imaged, followed by image stitching to create a composite image of the sample. The large representation of the whole tissue was then converted to a pseudo-H&E image and used for mapping specific fields for further interrogation for each slide. A minimum of 30 fields were chosen for each histological section and included ductal carcinoma in situ, and surrounding stromal areas. After selection, microscope coordinates were determined and exported for use during imaging calibrated to a 20X objective. Slides were then sequentially stained, imaged with a 20X objective, dyes inactivated, and re-stained iteratively for a total of 14 markers under analysis (Table 1). All dye conjugated antibody stains were for 1 hour at room temperature, and slides were processed for staining and washing using a Biocare autostainer. A summary table of the specific staining sequence and exposure times are provided in Supplemental data. All fields of view acquired were subsequently re-evaluated by a pathologist for % tumor, % normal epithelial, % immune along with pathological grading. The % tumor pertains to the % of any given field of view that is represented by ductal carcinoma in situ. Similarly, the amount in each field of view was quantified for normal epithelium and immune cells. The complete image set was then reviewed for tissue quality (tissue loss or damage) and image analysis segmentation output.
Image processing and statistical analyses
Data preprocessing
After data collection, images from all rounds are registered to the initial round using the DAPI channel to determine any offsets. Images are then processed for field flattening and autofluorescence is subtracted. Several quality control steps were applied to ensure usable cell level intensities. In addition, manual scoring of tissue quality and segmentation for every “Field Of View” was performed; this resulted in exclusion of 35 field of views. For total 13 Slides, 416 field of views were remaining for further analysis.
Single cell analysis algorithm segments cells in the epithelial compartments using DAPI, pan-cytokeratin, S6, and NaKATPase, and just DAPI in the stromal compartment, as previously described (10). Expression of SLC7A5, CD10, EGFR, HER2, HER4, S6, phospho-mTOR, CD44v6 in the epithelial masked area was quantified at single cell level and metrics included mean intensity, maximum intensity, and standard deviation in the membrane, cytoplasm and nucleus. Quality checks and filtering at single cell level were also performed. For example, epithelial cells were required to have the following characteristics: 1) maximum 1–2 nuclei per cell; 2) Each sub-cellular compartment (nucleus, membrane, cytoplasm) area had to have > 10 pixels; and 3) nuclei in each round of staining have to have perfect alignment to the first round of staining (automatic tissue quality index=1 at each round, which is the correlation between DAPI in each image). After the quality control steps, exposure time was re-set at 200 milli-second for cy3, 500 millisecond for cy5, 50 millisecond for eGFP and 20 millisecond for DAPI across all slides and markers. Finally, biomarker intensities were log2 transformed. After all the pre-processing and quality control steps (and exclusion of the control slides), we have 379,408 epithelium cells of 13 cases for all the markers with available valid cell level intensities.
For immune cell analysis (CD4, CD8 and CD20), support vector machine classification was used to classify immune cells. Model training was carried out by manual annotations using a tool developed in “FIJI” program to annotate CD4, CD8 and CD20 positive cells for ground truth and model training. These features were used to assign each cell in the image a probability of being a member of the CD4+, CD8+, or CD20+ classes. Ultimately, each cell in the image was assigned to one of the following classes: negative for all immune markers; cytotoxic T-cells (CD8+); T-helper cells (CD4+) and B-cells (CD20+).
Cellular Heterogeneity and Shannon Index methods
Shannon diversity index was applied to the cell data to measures the diversity of cells within the dataset using the proportion of each cell types. It is a quantitative index that was originally developed by Shannon (31) to quantify uncertainty in strings of text. It has been applied to ecology applications to quantify to proportion of individuals belonging to a species in a given dataset, and quantifies the uncertainty in predicting the species identity of an individual taken at random from the dataset. It is calculated as the following:
where R is the total number of clusters and pi is the proportion of the cluster i. The higher H is, the more heterogeneity is present in the dataset. For example, in the case of this study, if all the cells from the same duct are assigned to one K-mean cluster, the H value approaches to zero. However, if all the cells come from multiple clusters evenly, the H value is high and approaches to lnR= ln(8)= 2.08 (R=8 clusters in our dataset). A Shannon index was computed for each duct, then for a single patient, the average and standard deviation of Shannon index from all their imaged ducts was calculated to describe duct-to-duct diversity.
K-means clustering algorithm was applied to all cellular data (379,408 cells for all 13 patients) in R for K=2,3, …,15 and repeat 10 runs with randomly selected initial K centroids for each K. To help determine the best K, we used Consensus Clustering (R library ConsensusClusterPlus) and based on the Delta area metric. The best results were obtained with a cluster solution using the following 7 epithelial markers: EGFR, HER2, HER4, SLC7A5, pmTOR, CD44v6 and CD10. A heatmap was plotted to show cluster definition. Using cell IDs and spatial coordinates, the epithelial cell clusters were then overlaid with the tissue images using a color-coding schema.
Once the clusters were defined at cell level, the compositions of clusters for each patient and tumor field of views was calculated using level enrichment statistics. Each patient was represented by a vector of 8 elements with each element being the percentage of cells falling in that cluster and all the elements adding up to 100 percent. A pie chart was also generated to visually assess the cell type variation across all patients. The same statistic was also calculated for each field of view and the relationship between enrichment and the histological scores at patient level was assessed.
For the immune markers (CD4, CD8, CD20) we limited analysis to the stromal region (543,355 cells in total) as described above. The patient level immune profiles were then summarized by the proportion of CD4 positive cells relative to stromal cells, the proportion of CD8 positive cells, the proportion of CD20 positive cells, the total T cells (CD4+CD8), the CD4 to CD8 ratios, and the T cell to B cell ratios. The relationship between immune profiles and traditional histological scores at patient level was analyzed. The correlation between epithelium cell clusters and stromal area immune positivity was also analyzed.
Results:
Heterogeneity within ductal carcinoma in situ – single duct analyses within same patients
To evaluate expression heterogeneity of marker expression in ductal carcinoma in situ, we analyzed 416 individual ducts involving ductal carcinoma in situ on 13 patient tissue sections (average 29 ducts per patient; range: 20–40; Figure 1 and Supplementary Table S1) containing a total of 379,408 epithelial cells from 13 cases (average 29,185 cells per patient) (Supplementary Table S2). Similarly, 543,355 stromal cells (average 41,796 cells per patient) were studied for expression of the three immune markers CD4, CD8, and CD20. Using this cellular data, cluster analysis for epithelial markers was performed within ducts, between ducts, and between patients. Additional correlation analysis was performed to investigate relationships between these epithelial clusters and immune cell infiltration.
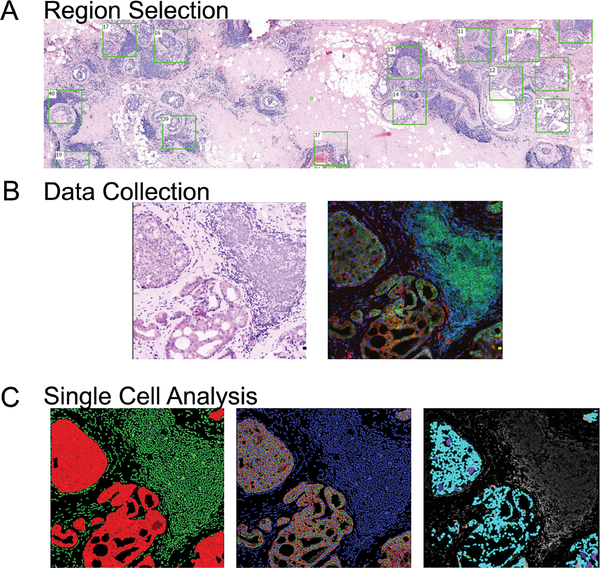
Figure 1: Data collection and analysis workflow.
1A. Tissue sections are scanned for auto-fluorescence and DAPI, and an H&E like image is generated allowing for specific regions to be analyzed by multiplex IF.
1B. Enlarged region from H&E scan and a companion composite image of multiplex staining for epithelial/ tumor cells (anti-pan cytokeratins AE1 and pck26), CD3 T-Cells, and CD20 B-cells are shown.
1C. Output images from single cell analysis workflow showing (left) separation of tumor (red) and stroma (green), (middle) single cell segmentation (blue-nuclei, red-epithelial membranes, green- epithelial cytoplasm), and (right) mapping of cell expression patterns based on K-means clustering back on original images.
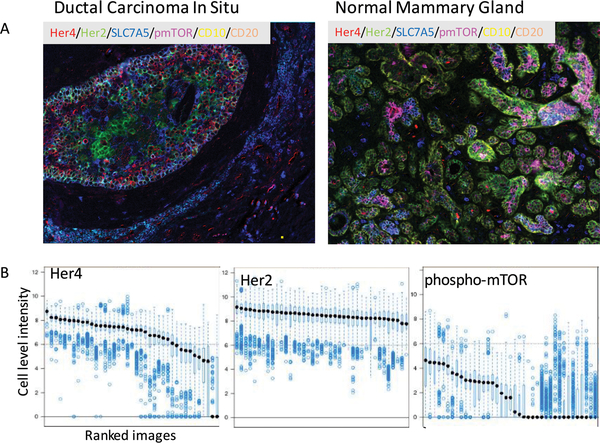
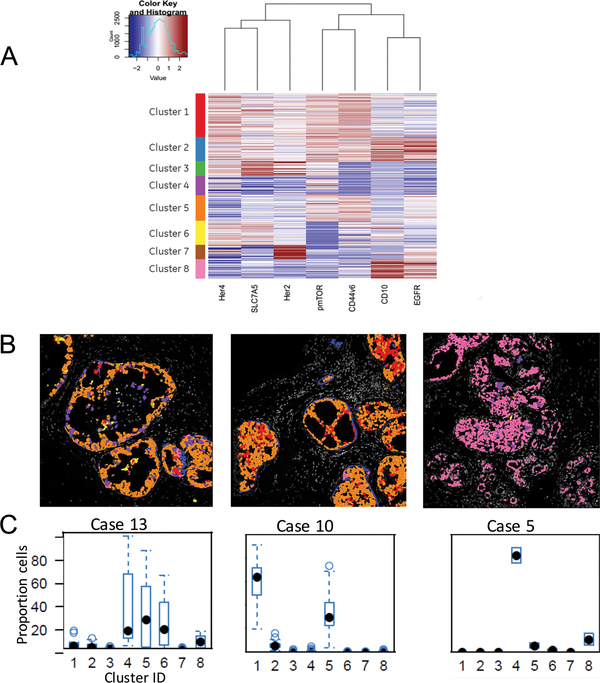
Analysis of the markers in the epithelial compartment showed heterogeneity of expression within single individual ducts from the same patient (Fig 2A & B; Supplementary Figure S1). Expression pattern for markers such as HER4 and HER2 was relatively stable across multiple cells/ducts. On the other hand, pmTOR, CD44v6, and CD10 were infrequently expressed (5%, 2% and 2% respectively) and varied significantly among cells from the same duct (Supplementary Figure S2). To better understand marker co-expression and quantify heterogeneity, a clustering algorithm (K-means) was applied to the epithelial cell biomarker data. Eight different clusters were identified based on expression of 7 epithelial markers (Figure 3A); these cluster IDs were mapped back to the cells as illustrated as in Figure 3B (Supplementary Table S3). The proportion of each cluster was quantified and heterogeneity was assessed using Shannon Diversity Index. Briefly, Clusters 2 and 8 both showed high expression of CD10 and EGFR, but cluster 8 showed low expression of other markers. HER2 expression was high in clusters 3 and 7; pmTOR expression was higher in clusters 1–3 and 5. There was moderate heterogeneity in distribution of 8 clusters within each duct (average Shannon Index 0.76; ranged 0 −1.61).
Figure 2: Evaluation of heterogeneity in antibody staining for individual markers.
2A. Representative composite images of staining from a typical ductal carcinoma in situ lesion (left), and a region of normal ducts from the same patient sample.
2B: Boxplot of HER4, HER2 and pmTOR intensity for all the cells of one example patient. X-axis is the ducts of the lesion. Y-axis is the log2(protein intensity). Within each duct, each point is a cell, the black dot is the median cell intensity and the boxplot around the black dot is the 25th and 75th quantiles. This figure shows the cell to cell and duct to duct variabilities for HER2, HER4 and pmTOR.
Figure 3: Determining complex patterns of protein expression in ductal carcinoma in situ.
3A. K-means clustering was applied to single cell data to determine patterns of expression for 7 markers. Each cluster group represents a pattern of unique cellular expression. Scale set with blue representing low/negative expression and red is highest expression.
3B. Cluster IDs mapped back onto images to confirm accuracy of segmentation demonstrates variable levels of heterogeneity. Shown are 3 representative cases with high, intermediate and low heterogeneity. Colors of cells correspond to color code given to cluster groups in figure 3A.
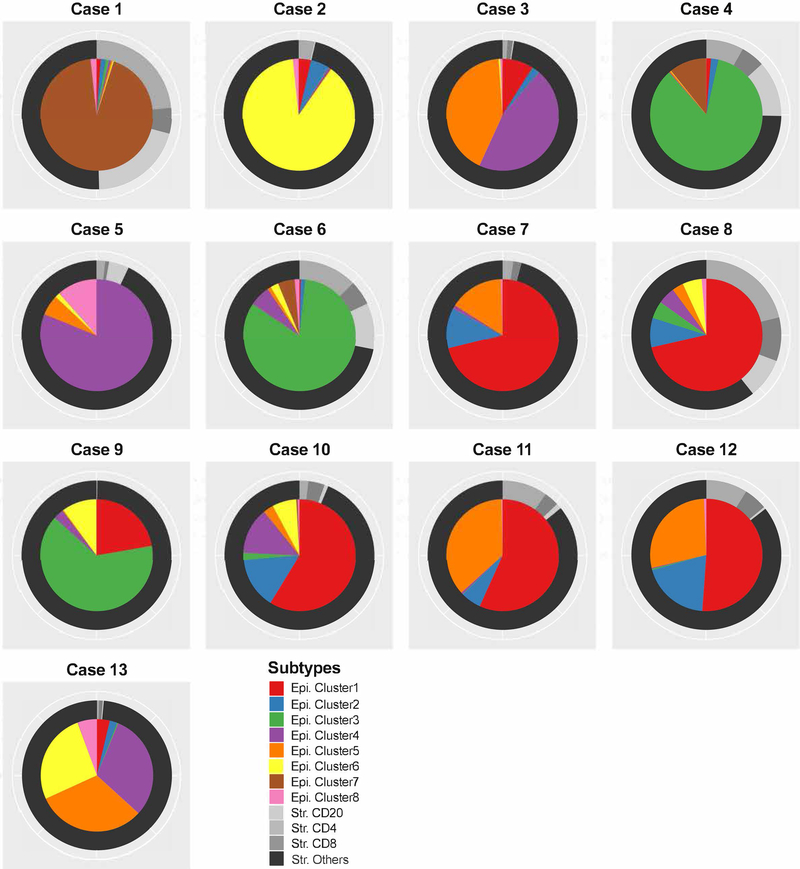
3C. Boxplot of cellular heterogeneity of patients. X-axis is the 8 cell level clusters. Y-axis is the percentage of cells within each cluster. Each point is a duct. Most of the ducts for case 5 has more than 80% cells in cluster 4. Case 10 has majorly cluster 1 and 5. However, case 13 has 3 main clusters and the percent of cells within each duct can vary from 0–100% for cluster 4, 0–90% for cluster 5 and 0–60% for cluster 6.
Heterogeneity within ductal carcinoma in situ – single patient analyses
To further address the issue of the heterogeneity of ductal carcinoma in situ lesions within the same patient, the distribution of the clusters was analyzed in different ducts from the same patient (Figure 3C and Supplementary Figure S3). Four patients had a dominant cellular phenotype (>80%) represented by cluster groups 3 and 7 (high HER2), cluster 6 (High HER4 and SLC7A5 and low HER2 and pmTOR), or cluster 4 (no dominant markers define this group). In 5 patients, the pattern was more heterogeneous with 50–70% of the cells belonging to cluster 1 (moderate to high levels for all markers except EGFR and CD10). The remaining patients had a strong representation of clusters 4 and 5 (CD44v6 and pmTOR) cells. Within each patient, the average Shannon Index across all ducts ranged from 0.33 to 1.02 (S.D. 0.09 to 0.38). This data documents the presence of high degree of heterogeneity at the protein level in patients with ductal carcinoma in situ. Additional analyses were performed to estimate the minimal number of ducts required to represent all 8 clusters. A cluster was deemed to be represented, if more than 5% of the duct cells belong to the particular cluster. On average, the number of clusters represented within a single duct was 2.5 (range 106), which corresponds to Shannon Index 0.76. As the distribution of clusters within ducts is uneven, the analysis of 8 ducts would be sufficient to represent all 8 clusters i.e. within and between duct heterogeneity.
Correlation of Stromal/Immune Analysis with epithelial expression pattern-
To analyze the impact of heterogeneity on immune infiltration, immune cells were quantified in both the stromal and epithelial compartments (representative data in Figure 4A & 4B). The stromal tumor infiltrating lymphocytes were classified as (CD20+) B-cells or (CD4+ or CD8+) T- cells. The distribution of stromal tumor infiltrating lymphocytes in ductal carcinoma in situ cases (Supplementary Table S4) was either a mixed B-cell (CD20+) or T-cell response (n=10), or one dominated by CD4 T-cells (n=1). The relationship between histology and immune markers was assessed in each field of view for all patients (Figure 5A-C). Although the sample size is small, larger lesion size was positively correlated with proportion of cluster 7 (high HER2) cells and CD20 infiltration (P= 0.002 and 0.0027 from t-test, respectively). Higher nuclear grade was associated with proportion of epithelial cluster 5 (high CD44v6 and pmTOR) cells (P value 0.008) and CD20 positivity (P = 0.019; F test). The cases were further analyzed as per four subtypes defined by expression of ER, PR and HER2 (ER+/PR-/HER2-, ER+/PR+/HER2-, ER+ HER2+, and HER2+) accounting for 3, 7, 1 and 2 cases, respectively. Proportion of cluster 3 cells was lower in all ER+PR+ cases (P= 0.05 from F-test). The ER+/HER2- cases had lower representation of cluster 7 cells (P= 0.08 from F=test). The HER2+ patients had higher B cells irrespective of ER status (F-test; P = 0.003).
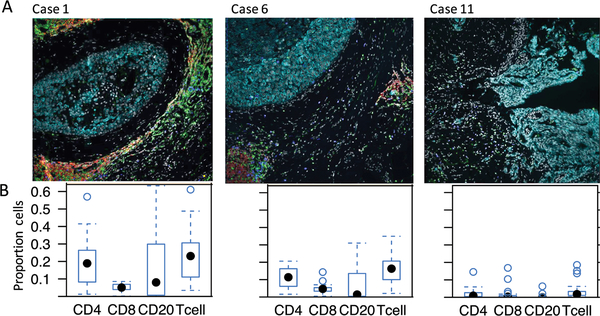
Figure 4: Immune quantification in ductal carcinoma in situ samples.
4A. Representative images from 3 cases of ductal carcinoma in situ with differing levels of local immune response. Tumor cells (turquoise), CD20 (red), CD4 (green) and CD8 (blue).
4B. Boxplot of immune marker enrichment of patients. X-axis is the 4 main immune types: CD4, CD8, CD20 and total T cells (CD4+CD8). Y-axis is the proportion of positive immune cells. Each point is a duct. Most of the ducts for case 11 have very few immune cells. Case 6 has moderate level of immune cells. However, case 1 has relatively higher immune proportions for both T cells and B cells.
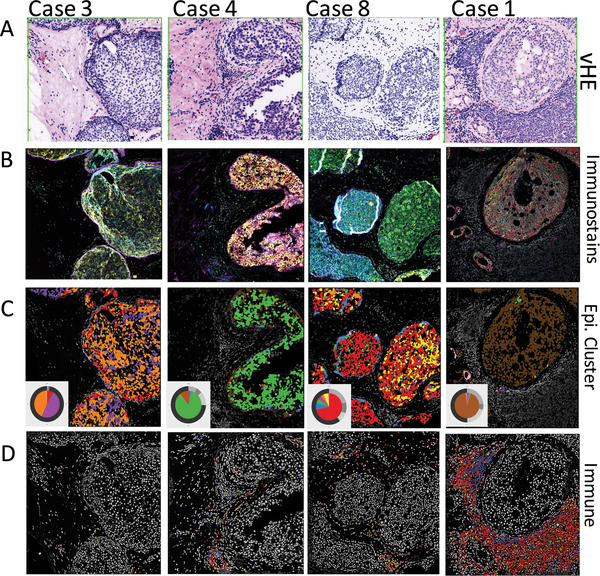
Figure 5: Mapping of epithelial and immune components in ductal carcinoma in situ.
5A. Representative pseudo-H&E images showing 4 cases of ductal carcinoma in situ with local immune responses.
5B. Composite immunofluorescence images for fields shown in A. Markers include HER4, SLC7A5, HER2, pmTOR, CD44v6, CD10 and EGFR.
5C. Cluster IDs mapped back to images allow clear visualization of cells with different patterns of expression. Colors correspond to cluster IDs in Figure 3, and pie charts shown in inset are for the specific field of view - the inner pie shows relative abundance of the 8 epithelial groups, and the outer ring show proportions of different immune populations.
5D. Verification of immune patterns after semi-automated quantification. CD20 positive cells in red, CD4 in green, and CD8 in blue.
A composite image representing the relationship of the epithelial clusters with immune markers is illustrated in Figure 6. This clearly highlights the degree of heterogeneity in protein expression in ductal carcinoma in situ. Cluster 2 (high EGFR and CD10 expression) was associated with a high T-cell to B-cell ratio (Spearman correlation coefficient = 0.83, P = 0.0004), while Cluster 7 (high HER2 expression) was associated with a B-cell response (Spearman correlation coefficient = 0.68, P = 0.009). The relative scarcity of intra-epithelial tumor infiltrating lymphocytes made correlations difficult.
Figure 6: Summary of epithelial and associated immune patterns in ductal carcinoma in situ.
Pie charts for cluster distribution and immune profile for each patient. The inner pie is composed of the 8 epithelial cell clusters. The outer ring is composed of the immune cell types as a proportion of the stromal space.
Discussion:
Tumors being monoclonal in origin were believed to lack significant cellular diversity. This was particularly true for ductal carcinoma in situ, which is thought to arise in/ from terminal duct units and then spread outwards to involve the ductal system in a segmental manner. The currently used CAP classification system is based on the finding of uniformity of nuclear (grade) morphology in ductal carcinoma in situ lesions (32). These factors feed in to the notion that ductal carcinoma in situ is a homogeneous lesion. Recent studies have documented intra-tumor heterogeneity in several cancers as an important determinant for tumor recurrence and resistance to therapy (33). The extent of intra- and inter-ductal heterogeneity in ductal carcinoma in situ has not been yet well documented.
The traditional methods of assessing heterogeneity involve studying different regions from the same lesion, extracting DNA and studying the mutational profile. This approach is excellent for larger lesions and fresh tumors but has limited utility in ductal carcinoma in situ, where the lesions are small, and admixed with normal tissues. Furthermore, these methods only permit regional assessment of heterogeneity and do not permit single cell analysis of lesion diagnosed only after histological examination. Multiplexed immunofluorescent staining and single cell analysis of cancer tissue provides a potentially powerful approach for measuring tumor cell expression and heterogeneity in intact formalin fixed paraffin embedded tissue, avoiding the “grind and find” approach required for genomic analyses and potential loss of spatial information(10). Previous work using this multiplexed immunofluorescence measured 27 proteins at single cell level in 26 patients with untreated stage II/III breast cancer and found 8 clusters of proteins that were co-expressed within the tumor region. The ER-negative cluster phenotype was significantly correlated with uptake of radiotracer 18F-fluorodeoxyglucose (34). Quantitative intra-tumor spatial heterogeneity algorithms have been demonstrated in multiplexed invasive breast cancer (35) and 9 multi-marker analysis of Reed Sternberg cells has been used for diagnosis of Hodgkin Lymphoma (36). Other reports have showed how multiplexing could decipher the complexity of the tumor-immune relationships and patient outcomes. Multispectral imaging of CD3, CD8, FoxP3, CD163, PD-L1 was used to analyze the tumor microenvironment in melanoma patients and predicted ability to generate tumor-infiltrating lymphocytes for T-cell therapy (37).
We have therefore applied a multiplex immunofluorescence based approach to study the heterogeneity of ductal carcinoma in situ of protein expression pattern in formalin fixed paraffin embedded tissues. This analysis is performed on a single 5-micron section and represents a paradigm shift in that it permits multiplex in situ protein analysis at the single cell level. Furthermore, co-expression analysis, application of clustering algorithms, and annotation of the expression data to single cells are possible, while retaining the tissue structural context.
Co-expression and correlation analysis of 7 different proteins of epithelial and 3 immune origin showed marked differences in expression of proteins in the cells. A single duct was found to contain cells located adjacent to one another expressing different proteins. The application of K-means clustering identified distinct co-expression profiles within individual cells of the ductal carcinoma in situ lesions. The use of these algorithms, although typical for gene expression studies, has not been commonly applied for tissue based protein analysis. This analysis identified moderate to marked degree of heterogeneity of protein expression by cells within a single duct (average Shannon Index 0.76; range 0 to 1.61). This highlights the degree of heterogeneity that is not appreciated by mutational analyses. More importantly, it raises the possibility of co-operative action, as a colony, in which single cells could have different roles in propagating the growth and progression of the disease.
Further analysis of multiple ducts from the same patient showed moderate degree of heterogeneity with individual ducts being composed of one or 2 (co-) dominant clusters/ clones or showing a scattered mix of phenotypes. Similar moderate degree of heterogeneity was seen in ducts from different patients. Higher expression of cluster 5 cells was associated in nuclear grade 2 (P= 0.008; F-test), particularly in ER+ patients. There was also a trend for lower cluster 3 and 7 expression in ER+ patients. Grade and ER/PR expression have prognostic value in determining the likelihood of recurrence and/or progression to invasive cancer. One could postulate that the observed tumor heterogeneity could have a bearing on the propensity to recur or progress.
In the current study, specific protein expression profiles were associated with TIL responses. In invasive breast cancer, immune cells have been documented to contribute to prognosis even in cases treated with chemotherapy (27, 28, 38–40). The amount of tumor infiltrating lymphocytes has been correlated with mutational burden in gastrointestinal cancers (41). In breast cancer, specific mutational signatures have been associated with tumor infiltrating lymphocytes (42). Tantalizingly, an association of epithelial clustering pattern with the presence and type of immune infiltrates was observed. This analysis provides the first evidence of that simultaneous analysis of both the epithelial and immune/stromal components might be necessary to understand the complex milieu in ductal carcinoma in situ lesions.
The current study is based on the analysis of a larger number of cells and ducts albeit from a limited number of patients with no outcome data (work is ongoing to expand sample size with outcomes and will include spatial cell analysis for deeper analysis of cell-to-cell interactions and relationships). In spite of these limitations, the study raises several issues that are fundamental to understanding of the biology of cancer, particularly breast cancer. The study, for the first time, documents the significant heterogeneity in protein expression in pre-invasive breast cancer. The documentation of moderate tumor heterogeneity within cells of a single duct has profound implications. It seems to suggest that even at the pre-invasive stage, cancer is a heterogeneous disease, and perhaps different clones fighting for dominance. This is consistent with the stochastic model of cancer evolution (43, 44). On the other hand, the finding of single dominant clone in some cases would support the “Big Bang” model (45). Does this indicate one should not focus on analyzing mass lesions for mutational and protein expression and focus on single cells? Is this the future of single cell sequencing? If so, how many cells need to be sequenced is an important question. From the patient perspective, “Is heterogeneity or lack of it good?” Homogeneous lesions could indicate the establishment of clonal dominance as a sign of aggressiveness. On the other hand, as cases with marked heterogeneity would have different clones, one or more of them could survive/ give rise to invasive disease. Understanding the biological impact is complicated by the fact that it is rare to see direct progression of ductal carcinoma in situ to invasive carcinoma and the low recurrence rates in treated patients. The only way forward is to analyze a large series of well annotated cases with established follow-up or a case-control design study. The current study provides the scientific basis for the design and performance of analysis of well-annotated ductal carcinoma in situ patient cohorts using multiplexed immunofluorescence technique and clustering algorithm described herein. These will lead to identification of novel clustering patterns that are related with outcomes
Supplementary Material
Acknowledgments:
Funding is through NCI - R01CA194600 to S. Badve and M. Gerdes and internal funds from General Electric to F. Ginty and M. Gerdes. In addition, S. Badve is supported by 100 Voices of Hope and Joshua Edwards Endowed Chair.
Funding: This study is supported by the National Cancer Institute of the National Institutes of Health under Award Number R01CA194600 to S Badve and M Gerdes. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References:
- 1.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012:883–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cancer Govindan R.. Attack of the clones. Science. 2014:169–70. [DOI] [PubMed] [Google Scholar]
- 3.de Bruin EC, McGranahan N, Mitter R, Salm M, Wedge DC, Yates L, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 2014:251–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J, Fujimoto J, Zhang J, Wedge DC, Song X, Zhang J, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014:256–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Navin NE. Cancer genomics: one cell at a time. Genome Biol. 2014:452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith BD. When is good enough really good enough? Defining the role of radiation in low-risk ductal carcinoma in situ. J Clin Oncol. 2015:686–91. [DOI] [PubMed] [Google Scholar]
- 7.Solin LJ, Gray R, Hughes LL, Wood WC, Lowen MA, Badve SS, et al. Surgical Excision Without Radiation for Ductal Carcinoma in Situ of the Breast: 12-Year Results From the ECOG-ACRIN E5194 Study. J Clin Oncol. 2015:3938–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Solin LJ, Gray R, Baehner FL, Butler SM, Hughes LL, Yoshizawa C, et al. A multigene expression assay to predict local recurrence risk for ductal carcinoma in situ of the breast. J Natl Cancer Inst. 2013:701–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Badve S, A’Hern RP, Ward AM, Millis RR, Pinder SE, Ellis IO, et al. Prediction of local recurrence of ductal carcinoma in situ of the breast using five histological classifications: a comparative study with long follow-up. Hum Pathol. 1998:915–23. [DOI] [PubMed] [Google Scholar]
- 10.Gerdes MJ, Sevinsky CJ, Sood A, Adak S, Bello MO, Bordwell A, et al. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc Natl Acad Sci U S A. 2013:11982–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou W, Johansson C, Jirstrom K, Ringberg A, Blomqvist C, Amini RM, et al. A Comparison of Tumor Biology in Primary Ductal Carcinoma In Situ Recurring as Invasive Carcinoma versus a New In Situ. Int J Breast Cancer. 2013:582134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pape-Zambito D, Jiang Z, Wu H, Devarajan K, Slater CM, Cai KQ, et al. Identifying a highly-aggressive DCIS subgroup by studying intra-individual DCIS heterogeneity among invasive breast cancer patients. PLoS One. 2014:e100488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lebeau A, Unholzer A, Amann G, Kronawitter M, Bauerfeind I, Sendelhofert A, et al. EGFR, HER-2/neu, cyclin D1, p21 and p53 in correlation to cell proliferation and steroid hormone receptor status in ductal carcinoma in situ of the breast. Breast Cancer Res Treat. 2003:187–98. [DOI] [PubMed] [Google Scholar]
- 14.Lari SA, Kuerer HM. Biological Markers in DCIS and Risk of Breast Recurrence: A Systematic Review. J Cancer. 2011:232–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collins LC, Schnitt SJ. HER2 protein overexpression in estrogen receptor-positive ductal carcinoma in situ of the breast: frequency and implications for tamoxifen therapy. Mod Pathol. 2005:615–20. [DOI] [PubMed] [Google Scholar]
- 16.Barnes NL, Khavari S, Boland GP, Cramer A, Knox WF, Bundred NJ. Absence of HER4 expression predicts recurrence of ductal carcinoma in situ of the breast. Clin Cancer Res. 2005:2163–8. [DOI] [PubMed] [Google Scholar]
- 17.Altintas S, Lambein K, Huizing MT, Braems G, Asjoe FT, Hellemans H, et al. Prognostic significance of oncogenic markers in ductal carcinoma in situ of the breast: a clinicopathologic study. Breast J. 2009:120–32. [DOI] [PubMed] [Google Scholar]
- 18.Buckley N, Boyle D, McArt D, Irwin G, Harkin DP, Lioe T, et al. Molecular classification of non-invasive breast lesions for personalised therapy and chemoprevention. Oncotarget. 2015:43244–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Afify A, McNiel MA, Braggin J, Bailey H, Paulino AF. Expression of CD44s, CD44v6, and hyaluronan across the spectrum of normal-hyperplasia-carcinoma in breast. Appl Immunohistochem Mol Morphol. 2008:121–7. [DOI] [PubMed] [Google Scholar]
- 20.Miller TJ, McCoy MJ, Hemmings C, Bulsara MK, Iacopetta B, Platell CF. Objective analysis of cancer stem cell marker expression using immunohistochemistry. Pathology. 2017:24–9. [DOI] [PubMed] [Google Scholar]
- 21.Vermeulen JF, van Brussel AS, Adams A, Mali WP, van der Wall E, van Diest PJ, et al. Near-infrared fluorescence molecular imaging of ductal carcinoma in situ with CD44v6-specific antibodies in mice: a preclinical study. Mol Imaging Biol. 2013:290–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Juhasz C, Nahleh Z, Zitron I, Chugani DC, Janabi MZ, Bandyopadhyay S, et al. Tryptophan metabolism in breast cancers: molecular imaging and immunohistochemistry studies. Nucl Med Biol. 2012:926–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayashi K, Anzai N. Novel therapeutic approaches targeting L-type amino acid transporters for cancer treatment. World J Gastrointest Oncol. 2017:21–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moritani S, Kushima R, Sugihara H, Bamba M, Kobayashi TK, Hattori T. Availability of CD10 immunohistochemistry as a marker of breast myoepithelial cells on paraffin sections. Mod Pathol. 2002:397–405. [DOI] [PubMed] [Google Scholar]
- 25.Stanton SE, Disis ML. Clinical significance of tumor-infiltrating lymphocytes in breast cancer. J Immunother Cancer. 2016:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Denkert C, Loibl S, Noske A, Roller M, Muller BM, Komor M, et al. Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J Clin Oncol. 2010:105–13. [DOI] [PubMed] [Google Scholar]
- 27.Perez EA, Ballman KV, Tenner KS, Thompson EA, Badve SS, Bailey H, et al. Association of Stromal Tumor-Infiltrating Lymphocytes With Recurrence-Free Survival in the N9831 Adjuvant Trial in Patients With Early-Stage HER2-Positive Breast Cancer. JAMA Oncol. 2016:56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adams S, Gray RJ, Demaria S, Goldstein L, Perez EA, Shulman LN, et al. Prognostic value of tumor-infiltrating lymphocytes in triple-negative breast cancers from two phase III randomized adjuvant breast cancer trials: ECOG 2197 and ECOG 1199. J Clin Oncol. 2014:2959–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. Arch Pathol Lab Med. 2010:907–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013:3997–4013. [DOI] [PubMed] [Google Scholar]
- 31.Shannon CE. A Mathematical Theory of Communication. Bell System Technical Journal. 1948:379–423. [Google Scholar]
- 32.Lester SC, Connolly JL, Amin MB. College of American Pathologists protocol for the reporting of ductal carcinoma in situ. Arch Pathol Lab Med. 2009:13–4. [DOI] [PubMed] [Google Scholar]
- 33.Badve S, Gokmen-Polar Y. Tumor Heterogeneity in Breast Cancer. Adv Anat Pathol. 2015:294–302. [DOI] [PubMed] [Google Scholar]
- 34.Sood A, Miller AM, Brogi E, Sui Y, Armenia J, McDonough E, et al. Multiplexed immunofluorescence delineates proteomic cancer cell states associated with metabolism. JCI Insight. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spagnolo DM, Gyanchandani R, Al-Kofahi Y, Stern AM, Lezon TR, Gough A, et al. Pointwise mutual information quantifies intratumor heterogeneity in tissue sections labeled with multiple fluorescent biomarkers. J Pathol Inform. 2016:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hollman-Hewgley D, Lazare M, Bordwell A, Zebadua E, Tripathi P, Ross AS, et al. A single slide multiplex assay for the evaluation of classical Hodgkin lymphoma. Am J Surg Pathol. 2014:1193–202. [DOI] [PubMed] [Google Scholar]
- 37.Tsujikawa T, Kumar S, Borkar RN, Azimi V, Thibault G, Chang YH, et al. Quantitative Multiplex Immunohistochemistry Reveals Myeloid-Inflamed Tumor-Immune Complexity Associated with Poor Prognosis. Cell Rep. 2017:203–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams S, Goldstein LJ, Sparano JA, Demaria S, Badve SS. Tumor infiltrating lymphocytes (TILs) improve prognosis in patients with triple negative breast cancer (TNBC). Oncoimmunology. 2015:e985930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salgado R, Denkert C, Demaria S, Sirtaine N, Klauschen F, Pruneri G, et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann Oncol. 2015:259–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loi S, Michiels S, Salgado R, Sirtaine N, Jose V, Fumagalli D, et al. Tumor infiltrating lymphocytes are prognostic in triple negative breast cancer and predictive for trastuzumab benefit in early breast cancer: results from the FinHER trial. Ann Oncol. 2014:1544–50. [DOI] [PubMed] [Google Scholar]
- 41.Giannakis M, Mu XJ, Shukla SA, Qian ZR, Cohen O, Nishihara R, et al. Genomic Correlates of Immune-Cell Infiltrates in Colorectal Carcinoma. Cell Rep. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smid M, Rodriguez-Gonzalez FG, Sieuwerts AM, Salgado R, Prager-Van der Smissen WJ, Vlugt-Daane MV, et al. Breast cancer genome and transcriptome integration implicates specific mutational signatures with immune cell infiltration. Nat Commun. 2016:12910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yates LR, Campbell PJ. Evolution of the cancer genome. Nat Rev Genet. 2012:795–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sidow A, Spies N. Concepts in solid tumor evolution. Trends Genet. 2015:208–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sottoriva A, Kang H, Ma Z, Graham TA, Salomon MP, Zhao J, et al. A Big Bang model of human colorectal tumor growth. Nat Genet. 2015:209–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.