Abstract
How signaling molecules achieve signal diversity and specificity is a long-standing cell biology question. Here, we report the development of a targeted delivery method that permits specific expression of homologous Ras family small GTPases (i.e., Ras, Rap2 and Rap1) in different subcellular microdomains, including the endoplasmic reticulum, lipid rafts, bulk membrane, lysosome and Golgi complex, in rodent hippocampal CA1 neurons. The microdomain-targeted delivery, combined with multicolor fluorescence protein-tagging and high-resolution dual-quintuple simultaneous patch-clamp recordings, allows a systematic analysis of microdomain-specific signaling. The analysis shows that Ras signals long-term potentiation via endoplasmic reticulum PI3K and lipid rafts ERK, while Rap2 and Rap1 signal depotentiation and long-term depression via bulk membrane JNK and lysosome p38MAPK, respectively. These results establish an effective subcellular microdomain-specific targeted delivery method, and unveil subcellular microdomain-specific signaling as the mechanism for homologous Ras and Rap to achieve signal diversity and specificity to control multiple forms of synaptic plasticity.
eTOC Blurb
Zhang et al. develop an effective subcellular microdomain-specific targeted delivery method and demonstrate that homologous proteins (e.g., Ras and Rap) confine their signaling within distinct subcellular microdomains to achieve signal transduction diversity and specificity.
Different forms of synaptic plasticity, including the long-term potentiation (LTP), depotentiation and long-term depression (LTD), control synaptic transmission and thereby contribute to higher brain functions such as learning and memory, yet how the multiple forms of synaptic plasticity are independently regulated at individual synapses remains unclear (Henley and Wilkinson, 2016; Huganir and Nicoll, 2013; Nishiyama and Yasuda, 2015; Roth et al., 2017). Previous studies have identified Ras family small GTPases (i.e., Ras, Rap2 and Rap1) as molecular switches of multiple signal transduction cascades that control synaptic plasticity, and linked genetic defects of molecules in the signal transduction cascades to a large number of mental, neurological and psychiatric disorders (Costa and Silva, 2003; Stornetta and Zhu, 2011; Volk et al., 2015). In particular, Ras initiates PI3K and ERK signaling to drive synaptic delivery of AMPA-Rs containing subunits with long cytoplasmic termini (e.g., GluA1- and GluA2L-containing AMPA-Rs) during LTP (Kielland et al., 2009; Kim et al., 2005; Man et al., 2003; Qin et al., 2005; Zhu et al., 2002), and Rap2 activates JNK signaling to induce synaptic removal of AMPA-Rs containing subunits with long cytoplasmic termini during depotentiation (Kielland et al., 2009; Yang et al., 2011; Zhu et al., 2005), whereas Rap1 stimulates p38MAPK signaling to trigger synaptic removal of AMPA-Rs containing only subunits with short cytoplasmic termini (i.e., GluA2/3 AMPA-Rs) during LTD (Hsieh et al., 2006; Kielland et al., 2009; Nabavi et al., 2013; Zhu et al., 2002). However, Ras and Rap proteins, which share a high degree of sequence and structure homology, can be activated by the same signals and/or stimulate the same effectors; these signals and effectors introduce abundant opportunities for cross-talk (Bos et al., 2001; Reuther and Der, 2000). It is thus perplexing how Ras and Rap proteins may independently regulate different forms of synaptic plasticity at the same synapses.
As a general biology phenomenon, homologous signaling molecules may initiate an astonishing number of complex cellular responses to diverse environmental stimuli (Gloerich and Bos, 2011; Simanshu et al., 2017). Although the mechanism underlying this intriguing phenomenon remains elusive, the prevailing hypothesis favors the idea that signaling molecules achieve signal diversity and specificity via selective participation into different signaling platforms confined in distinct subcellular microdomains (Ahearn et al., 2011; Rocks et al., 2006; Zhou and Hancock, 2015). Indeed, custom-recombinated signaling platforms can create diversified synthetic signaling pathways (Chau et al., 2012; Peisajovich et al., 2010), and trafficking and localization of signaling molecules, including members in Ras family small GTPases, are precisely regulated (Casar et al., 2009; Mochizuki et al., 2001; Prior et al., 2003; Zhou et al., 2017). The remaining question is whether the microdomain-confined signaling cascades do exist and mediate diverse, specific signaling responses under physiological conditions. In this study, we systematically investigated synaptic signaling of three general members of Ras family small GTPases, Ras, Rap2 and Rap1. Adapting a subcellular microdomain-targeting strategy, we successfully made targeted expression of Ras and Rap mutants in the five major subcellular microdomains, including the endoplasmic reticulum, lipid rafts, bulk membrane, lysosome and Golgi complex. Using this approach, we examined the endogenous Ras, Rap2 and Rap1 signaling in these microdomains, in both in vitro and in vivo preparations. We found that endogenous Ras preferentially signaled synaptic potentiation via the endoplasmic reticulum PI3K and lipid rafts ERK pathways, while endogenous Rap2 and Rap1 predominantly signaled synaptic depression via the bulk membrane JNK and lysosome p38MAPK pathways, respectively. These results provide the first evidence indicating that under physiological conditions, homologous Ras and Rap proteins use distinct subcellular microdomains to create multiple specific signaling responses to regulate different forms of synaptic plasticity.
RESULTS
Microdomain-specific sequences achieve targeted delivery
To study how Ras family small GTPases may signal synaptic transmission at synapses, we fused the specific membrane targeting signal sequences, including M1, LCK, CD8, LAMP1 and KDELr (see the methods), to Ras and Rap to selectively deliver these signaling molecules into the endoplasmic reticulum, lipid rafts, bulk membrane, lysosome and Golgi complex, respectively. To verify the delivery specificity, we fused YFP to C termini of various microdomain-targeting Ras constructs and expressed them in cultured rat hippocampal neurons (Fig 1A). M1-, LCK-, CD8-, LAMP1- and KDELr-Ras-YFP, appeared in the expected subcellular domains as revealed by co-immunostaining of the specific domain markers (Fig 1B). To independently confirm these findings, we micro-fractionated cultured hippocampal slices (Fig 1C). Western blots showed that Ras, Rap1 and Rap2 were present in the endoplasmic reticulum, lipid rafts, bulk membrane, lysosome and Golgi complex fractions (Fig 1D–E), as validated by the specific domain markers (Fig S1). Next, we acutely overexpressed M1-, LCK-, CD8-, LAMP1- or KDELr-Ras-YFP in the cultured rat hippocampal slices for ~16 hours using the established Sindbis viral expression system (Fig 1C; see (Lim et al., 2014; Wang et al., 2015a) for the methods), and then micro-fractionated the endoplasmic reticulum, lipid rafts, bulk membrane, lysosome and Golgi complex from the tissues. Western blot analysis verified that M1-, LCK-, CD8-, LAMP1- and KDELr-Ras-YFP were predominantly expressed in the endoplasmic reticulum, lipid rafts, bulk membrane, lysosome and Golgi complex, respectively (Fig 1F–G). Together, these results indicate that M1, LCK, CD8, LAMP1 and KDELr signal sequences are effective in targeting Ras expression into five distinct subcellular domains.
Figure 1. Microdomain-targeting sequences deliver Ras to distinct subcellular compartments.
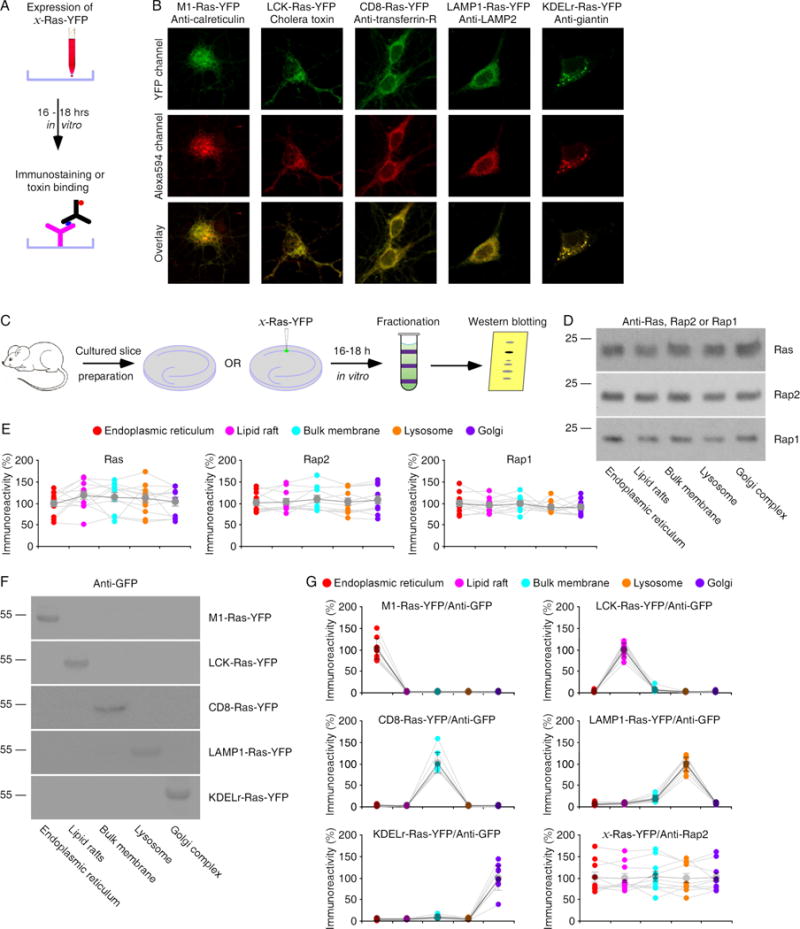
(A) Schematic drawing outlines in vitro immunostaining and toxin binding experimental design.
(B) Upper images show YFP fluorescence of expressed M1-, LCK-, CD8-, LAMP1- and KDELr-Ras-YFP in cultured hippocampal neurons. Middle images show red fluorescence of Alexa 594-conjugated anti-calreticulin, -transferring-R, -LAMP2 or –giantin, or Alexa 594-conjugated cholera toxin. Lower images show the overlays.
(C) Schematic drawing outlines micro-fractionation experimental design.
(D) Blots of endogenous Ras, Rap2 and Rap1 in the endoplasmic reticulum, lipid rafts, bulk membrane, lysosome and Golgi complex fractionated from cultured rat hippocampal slices.
(E) Relative levels of Ras, Rap2 and Rap1 in all microdomains. See Table S1 for values.
(F) Blots of recombinated Ras-YFP in the endoplasmic reticulum, lipid rafts, bulk membrane, lysosome and Golgi complex fractionated from cultured rat hippocampal slices after 16–18 h expression of M1-, LCK, CD8-, LAMP1- and KDELr-Ras-YFP.
(G) Relative levels of Ras-YFP and Rap2 in all microdomains in tissues expressing M1-, LCK, CD8-, LAMP1-and KDELr-Ras-YFP. See Table S1 for values.
Ras and Rap signal synaptic transmission in separate microdomains
To determine in which subcellular domain(s) Ras signals synaptic transmission, we first made Sindbis viral expression of constitutively active (ca), dead (dd) and dominant negative (dn) mutant forms of M1-Ras in CA1 pyramidal neurons in cultured rat hippocampal slices for ~10 hours (Fig 2A). To identify the expression of different Ras mutant constructs, M1-Ras(ca), -Ras(dd) and -Ras(dn) were co-expressed with different fluorescence proteins using an internal ribosome entry site sequence (IRES). Electrophysiological recordings were then obtained from nearby M1-Ras(ca)-IRES-GFP, M1-Ras(dd)-IRES-CFP, M1-Ras(dn)-IRES-RFP expressing (identified by the green, cyan or red fluorescence) and control non-expressing neuron quadruplets (Fig 2A image insets). Afferent fibers were stimulated and AMPA-R- and NMDA-R-mediated excitatory postsynaptic currents were recorded. Neurons expressing M1-Ras(ca)-IRES-GFP had enhanced AMPA responses, while neurons expressing M1-Ras(dd)-IRES-CFP or M1-Ras(dn)-IRES-RFP had the same AMPA responses compared to nearby control non-expressing neurons (Fig 2B–C and Table S2). There was no difference in NMDA responses between M1-Ras(ca)-IRES-GFP, M1-Ras(dd)-IRES-CFP, M1-Ras(dn)-IRES-RFP expressing and non-expressing neurons (Fig 2B–C and Table S2). These results indicate that constitutively active Ras signaling in the endoplasmic reticulum potentiates AMPA transmission.
Figure 2. Ras signaling in the endoplasmic reticulum and lipid rafts potentiates transmission.
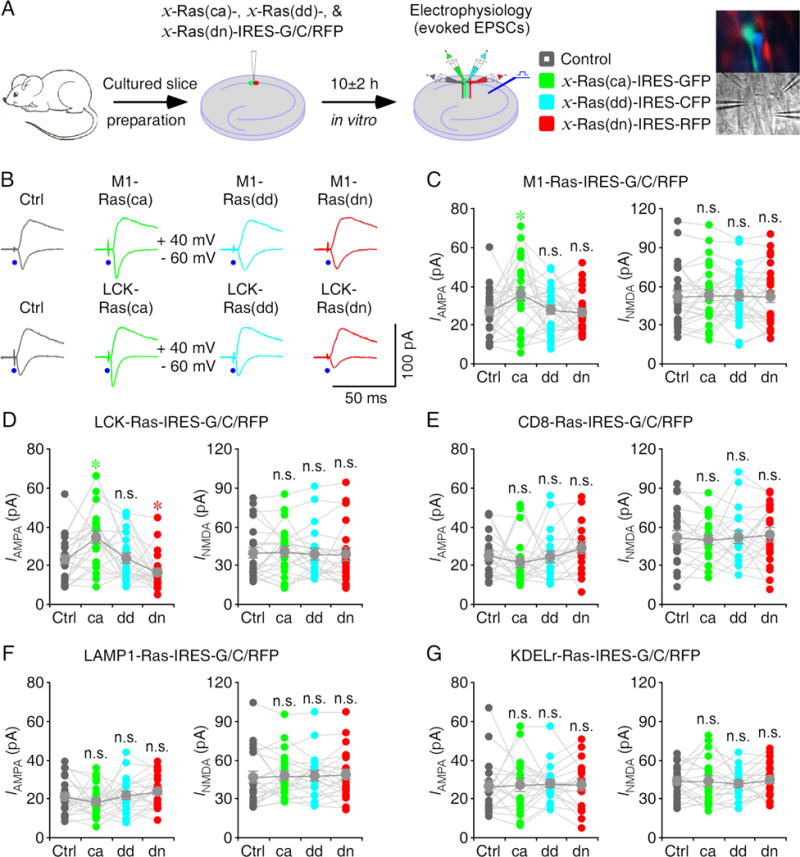
(A) Schematic drawing outlines in vitro experimental design. The right images show simultaneous whole-cell recordings from CA1 pyramidal neuron quadruplets under fluorescence (upper) and transmitted light (lower) microscopy.
(B) Upper, evoked AMPA-R- (-60 mV) and NMDA-R- (+40 mV) mediated responses recorded from control non-expressing (Ctrl), M1-Ras(ca)-IRES-GFP, M1-Ras(dd)-IRES-CFP and M1-Ras(dn)-IRES-RFP expressing CA1 cells after 10±2 h expression. Lower, evoked AMPA and NMDA responses recorded from non-expressing, LCK-Ras(ca)-IRES-GFP, LCK-Ras(dd)-IRES-CFP and LCK-Ras(dn)-IRES-RFP expressing cells after 10±2 h expression.
(C–G) AMPA and NMDA responses in M1-Ras (C), LCK-Ras (D), CD8-Ras (E), LAMP1-Ras (F) and KDELr-Ras (G) mutant expressing neurons relative to non-expressing control cells. See Table S2 for values. Asterisks indicate p<0.05 (Wilcoxon tests).
We then acutely overexpressed mutant forms of LCK-Ras in CA1 neurons in cultured slices for ~10 hours. Neurons expressing LCK-Ras(ca)-IRES-GFP had enhanced AMPA responses, whereas neurons expressing LCK-Ras(dn)-IRES-RFP had reduced AMPA responses compared to nearby control non-expressing neurons (Fig 2B and D). Neurons expressing LCK-Ras(dd)-IRES-CFP had the same AMPA responses compared to nearby control non-expressing neurons (Fig 2B, 2D and Table S2). In addition, there was no difference in NMDA responses between expressing and control non-expressing neurons (Fig 2B, 2D and Table S2). These results suggest that constitutively active Ras signaling in the lipid rafts potentiates AMPA transmission and endogenous Ras activity in the lipid rafts contributes to a tonic potentiation of AMPA transmission.
Similarly, we acutely overexpressed mutant forms of CD8-, LAMP1- and KDELr-Ras in CA1 neurons in cultured slices for ~10 hours. Neurons expressing CD8-, LAMP1- and KDELr-Ras(ca)-IRES-GFP, CD8-, LAMP1- and KDELr-Ras(dd)-IRES-CFP, and CD8-, LAMP1- and KDELr-Ras(dn)-IRES-RFP, all had the same AMPA and NMDA responses compared to nearby control non-expressing neurons (Fig 2E–G and Table S2). Collectively, these results suggest that constitutively active Ras signaling in both the endoplasmic reticulum and lipid rafts potentiates AMPA transmission though only endogenous Ras activity in the lipid rafts tonically potentiates AMPA transmission.
Using the same approach, we examined subcellular domain(s) in which Rap2 may signal synaptic transmission by acutely overexpressing mutant forms of M1-, LCK-, CD8-, LAMP1- and KDELr-Rap2 in CA1 neurons in cultured rat hippocampal slices for ~10 hours (Fig 3A). Neurons expressing CD8-Rap2(ca)-IRES-GFP had reduced AMPA responses and neurons expressing CD8-Rap2(dd)-IRES-CFP had the same AMPA responses, whereas neurons expressing CD8-Rap2(dn)-IRES-RFP had enhanced AMPA responses compared to nearby control non-expressing neurons (Fig 3B, 3E and Table S3). Neurons expressing M1-, LCK-, LAMP1- and KDELr-Rap2(ca)-IRES-GFP, M1-, LCK-, LAMP1- and KDELr-Rap2(dd)-IRES-CFP, and M1-, LCK-, LAMP1-, and KDELr-Rap2(dn)-IRES-RFP, all had the same AMPA and NMDA responses compared to nearby control non-expressing neurons (Fig 3C–D, 3F–G and Table S3). Together, these results suggest that constitutively active Rap2 signaling in the bulk membrane depresses AMPA transmission while endogenous Rap2 activity in the bulk membrane contributes to a tonic depression of AMPA transmission.
Figure 3. Rap2 signaling in the bulk membrane depresses transmission.
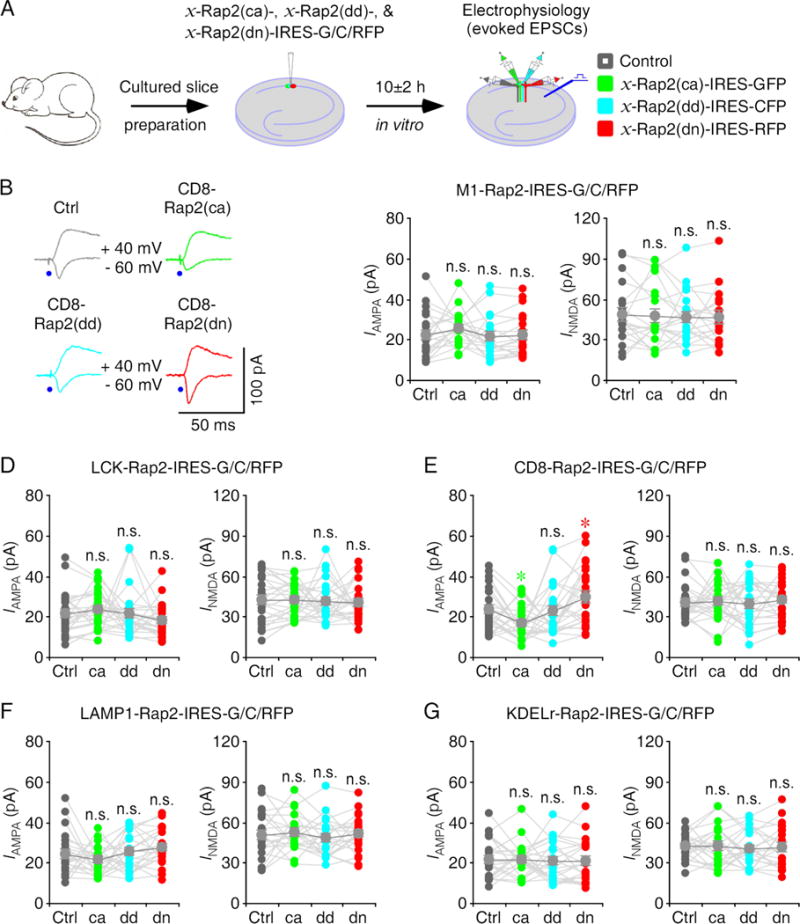
(A) Schematic drawing outlines in vitro experimental design.
(B) Evoked AMPA-R- (−60 mV) and NMDA-R- (+40 mV) mediated responses recorded from control non-expressing (Ctrl), CD8-Rap2(ca)-IRES-GFP, CD8-Rap2(dd)-IRES-CFP and CD8-Rap2(dn)-IRES-RFP expressing CA1 cells after 10±2 h expression.
(C–G) AMPA and NMDA responses in M1-Rap2 (C), LCK-Rap2 (D), CD8-Rap2 (E), LAMP1-Rap2 (F) and KDELr-Rap2 (G) mutant expressing neurons relative to non-expressing control cells. See Table S3 for values. Asterisks indicate p<0.05 (Wilcoxon tests).
We next investigated subcellular domain(s) in which Rap1 may signal synaptic transmission by acutely overexpressing mutant forms of M1-, LCK-, CD8-, LAMP1- and KDELr-Rap1 in CA1 neurons in cultured rat hippocampal slices for ~10 hours (Fig 4A). In these experiments, neurons expressing LAMP1-Rap1(ca)-IRES-GFP had reduced AMPA responses and neurons expressing LAMP1-Rap1(dd)-IRES-CFP had the same AMPA responses, whereas neurons expressing LAMP1-Rap1(dn)-IRES-RFP had enhanced AMPA responses compared to nearby control non-expressing neurons (Fig 4B, 4F and Table S4). Neurons expressing M1-, LCK-, CD8- and KDELr-Rap1(ca)-IRES-GFP, M1-, LCK-, CD8- and KDELr-Rap1(dd)-IRES-CFP, and M1-, LCK-, CD8-, and KDELr-, all had the same AMPA and NMDA responses compared to nearby control non-expressing neurons (Fig 4C–E, 3G and Table S4). Collectively, these results suggest that constitutively active Rap1 signaling in the lysosome depresses AMPA transmission whereas endogenous Rap1 activity in the lysosome contributes to a tonic depression of AMPA transmission.
Figure 4. Rap1 signaling in the lysosome depresses transmission.
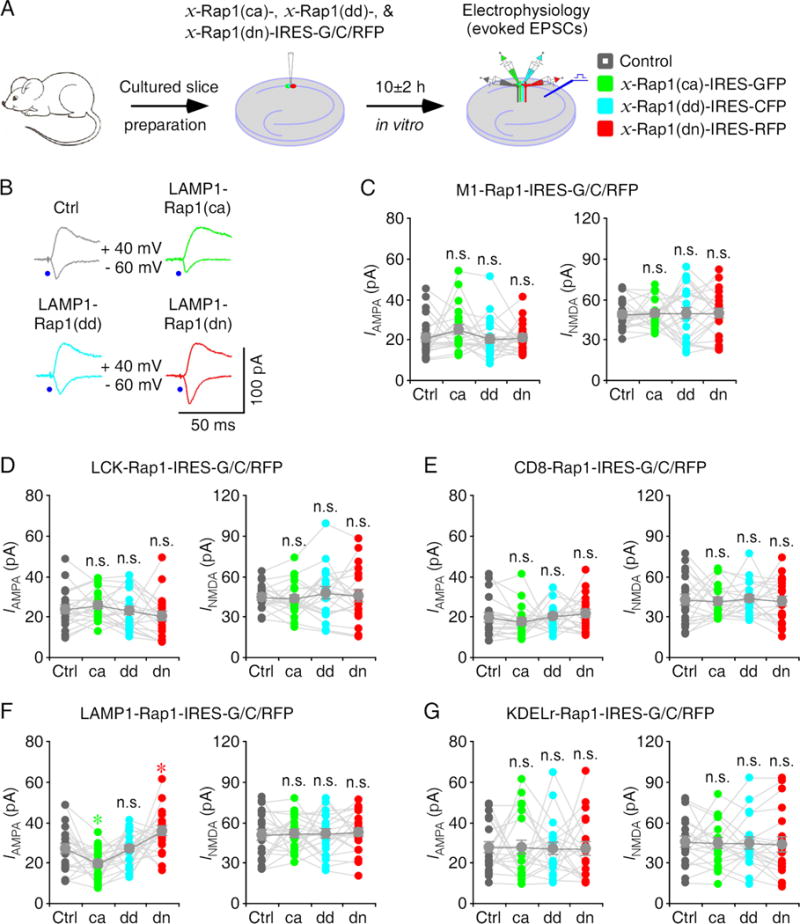
(A) Schematic drawing outlines in vitro experimental design.
(B) Evoked AMPA-R- (−60 mV) and NMDA-R- (+40 mV) mediated responses recorded from control non-expressing (Ctrl), LAMP1-Rap1(ca)-IRES-GFP, LAMP1-Rap1(dd)-IRES-CFP and LAMP1-Rap1(dn)-IRES-RFP expressing CA1 cells after 10±2 h expression.
(C–G) AMPA and NMDA responses in M1-Rap1 (C), LCK-Rap1 (D), CD8-Rap1 (E), LAMP1-Rap1 (F) and KDELr-Rap1 (G) mutant expressing neurons relative to non-expressing control cells. See Table S4 for values. Asterisks indicate p<0.05 (Wilcoxon tests).
We noted that neurons expressing CD8- and LAMP1-Ras(ca)-IRES-GFP had somewhat decreased AMPA responses, whereas neurons expressing CD8- and LAMP1-Ras(dn)-IRES-RFP had slightly increased AMPA responses, although the changes were insignificant (Fig 2E–F). Because Ras shares a high degree (>~50%) of sequence and structure homology with Rap proteins (Bos et al., 2001), we speculated that the small changes in AMPA responses resulted from the less effective Ras mutants on Rap signaling in the bulk membrane and lysosome microdomains. To test this idea, we increased the expression level of Ras mutants by overexpressing CD8-Ras(ca)-IRES-CFP, LAMP1-Ras(ca)-IRES-GFP, CD8-Ras(dn)-IRES-OFP and LAMP1-Ras(dn)-IRES-RFP in CA1 pyramidal neurons in cultured rat hippocampal slices for a longer time, ~16 hours (Fig S2A), and then compared the evoked synaptic responses in nearby expressing and control non-expressing neuron quintuplets (Fig S2A image insets). After 16-hour expression, neurons expressing CD8-Ras(ca)-IRES-CFP and LAMP1-Ras(ca)-IRES-GFP had significantly reduced AMPA responses, whereas neurons expressing CD8-Ras(dn)-IRES-OFP and LAMP1-Ras(dn)-IRES-RFP had significantly enhanced AMPA responses compared to nearby control non-expressing neurons (Fig S2B–C and Table S5). NMDA responses in expressing and non-expressing neurons were the same (Fig S2B–C and Table S5). These results indicate that with sufficient overexpression, the target-delivered constitutively active Ras can stimulate and dominant negative Ras can suppress Rap signaling in the bulk membrane and lysosome.
In contrast, neurons expressing M1- and LCK-Rap2(ca)-IRES-GFP had somewhat increased AMPA responses, whereas neurons expressing LCK-Rap2(dn)-IRES-RFP had slightly decreased AMPA responses (Fig 3C–D). We verified the idea that targeted delivery of Rap2 mutants may interfere with Ras signaling in the endoplasmic reticulum and lipid rafts by overexpressing M1-Rap2(ca)-IRES-CFP, LCK-Rap2(ca)-IRES-GFP, M1-Rap2(dn)-IRES-OFP and LCK-Rap2(dn)-IRES-RFP in cultured rat hippocampal slices for ~16 hours, and comparing the evoked synaptic responses in nearby expressing and control non-expressing CA1 pyramidal neuron quintuplets (Fig S3A). After 16-hour expression, neurons expressing M1-Rap2(ca)-IRES-CFP and LCK-Rap2(ca)-IRES-GFP had significantly enhanced AMPA responses, whereas neurons expressing M1-Rap2(dn)-IRES-OFP has the same, and neurons expressing LCK-Rap2(dn)-IRES-RFP had significantly reduced AMPA responses compared to nearby control non-expressing neurons (Fig S3B–C and Table S5). NMDA responses in expressing and non-expressing neurons were the same (Fig S3B–C and Table S5). These results indicate that with sufficient overexpression, the target-delivered constitutively active Rap2 can stimulate Ras signaling in the endoplasmic reticulum and lipid rafts and dominant negative Rap2 can suppress Ras signaling in the lipid rafts.
Likewise, neurons expressing M1- and LCK-Rap1(ca)-IRES-GFP had somewhat increased AMPA responses, whereas neurons expressing LCK-Rap1(dn)-IRES-RFP had slightly decreased AMPA responses (Fig 4C–D). We validated the notion that targeted delivery of Rap1 may interfere with Ras signaling in the endoplasmic reticulum and lipid rafts by overexpressing M1-Rap1(ca)-IRES-CFP, LCK-Rap1(ca)-IRES-GFP, M1-Rap1(dn)-IRES-OFP and LCK-Rap1(dn)-IRES-RFP in cultured rat hippocampal slices for ~16 hours, and comparing the evoked synaptic responses in nearby expressing and control non-expressing CA1 pyramidal neuron quintuplets (Fig S3A). After 16-hour expression, neurons expressing M1-Rap1(ca)-IRES-CFP and LCK-Rap1(ca)-IRES-GFP had significantly enhanced AMPA responses, whereas neurons expressing M1-Rap1(dn)-IRES-OFP has the same, and neurons expressing LCK-Rap1(dn)-IRES-RFP had significantly reduced AMPA responses compared to nearby control non-expressing neurons (Fig S3D–E and Table S5). NMDA responses in expressing and non-expressing neurons were the same (Fig S3D–E and Table S5). These results indicate that with sufficient overexpression, the target-delivered constitutively active Rap1 can stimulate Ras signaling in the endoplasmic reticulum and lipid rafts and dominant negative Rap1 can suppress Ras signaling in the lipid rafts.
Microdomain-specific Ras and Rap signaling control transmission via different pathways
Activation of Ras-ERK signaling is required to potentiate GluA2L-mediated AMPA transmission whereas activation of both Ras-ERK and -PI3K signaling is required to potentiate GluA1-mediated AMPA transmission, although in the cultured hippocampal slices, spontaneous activity is only sufficient to stimulate Ras-ERK signaling, but not Ras-PI3K signaling (Kolleker et al., 2003; Qin et al., 2005; Zhu et al., 2002). Thus, our results that stimulating Ras in the endoplasmic reticulum and lipid rafts potentiates AMPA transmission while blocking Ras in the lipid rafts but not that in the endoplasmic reticulum suppresses AMPA transmission are suggestive of existence of Ras-PI3K signaling complex in the endoplasmic reticulum and Ras-ERK signaling complex in the lipid rafts. To directly examine this possibility, we first performed pharmacology experiments (Fig 5A). Including LY294002, an inhibitor of PI3K, in culture media blocked the potentiation of AMPA transmission in M1-Ras(ca)-IRES-GFP expressing neurons, but had no effect on the potentiation of AMPA transmission in LCK-Ras(ca)-IRES-RFP expressing neurons, the normal AMPA transmission in M1-Ras(dn)-IRES-GFP expressing neurons and the depression of AMPA transmission in LCK-Ras(dn)-IRES-RFP expressing neurons. Conversely, including PD98059, an inhibitor of ERK activator MEK, in culture media blocked the potentiation of AMPA transmissions in M1-Ras(ca)-IRES-GFP and LCK-Ras(ca)-IRES-RFP expressing neurons, and occluded the depression of AMPA transmission in LCK-Ras(dn)-IRES-RFP-stimulated, but had no effect on the normal AMPA transmission in M1-Ras(dn)-IRES-GFP expressing neurons (Fig 5B–F and Table S6). To confirm the idea, we examined the synaptic transmission in cultured slices prepared from GluA1 and GluA2 knockout, and GluA1(S831A/S845A) transgenic mice. We found that M1-Ras(ca)-IRES-GFP-, but not LCK-Ras(ca)-IRES-RFP-stimulated potentiation of AMPA transmission was barred in GluA1 knockout CA1 neurons, as well as in GluA1(S831A/S845A) transgenic CA1 neurons (Fig 5G–I and Table S7), in which GluA1 phosphorylation and GluA1-mediated synaptic potentiation are impaired (Lee et al., 2003). In contrast, LCK-Ras(ca)-IRES-GFP-, but not M1-Ras(ca)-IRES-RFP-stimulated potentiation of AMPA transmission was barred in GluA2 knockout CA1 neurons (Fig 5G, 5J and Table S7). Consistently, GluA1 knockout had no effect on the normal AMPA transmission in M1-Ras(dn)-IRES-GFP expressing neurons, and the depression of AMPA transmission in LCK-Ras(dn)-IRES-RFP expressing neurons, while GluA2 knockout had no effect on the normal AMPA transmission in M1-Ras(dn)-IRES-GFP-expressing neurons, but prevented the depression of AMPA transmission in LCK-Ras(dn)-IRES-RFP expressing neurons (Fig 5K–L and Table S7). Together, these results suggest that Ras controls PI3K signaling in the endoplasmic reticulum and ERK signaling in the lipid rafts.
Figure 5. Ras signals PI3K and ERK in the endoplasmic reticulum and lipid rafts.
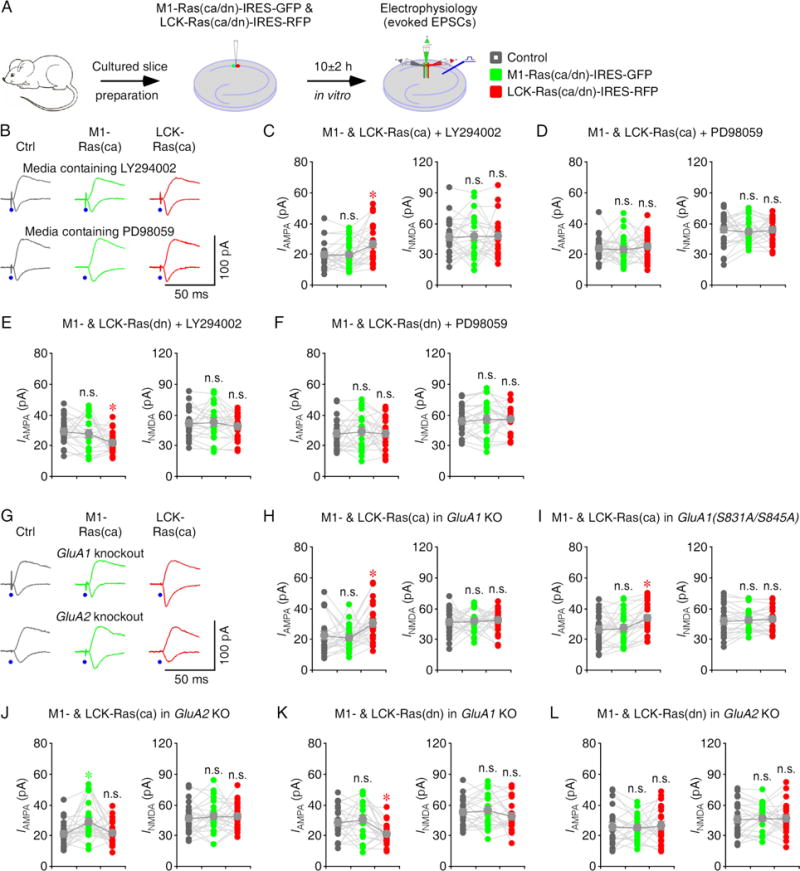
(A) Schematic drawing outlines in vitro experimental design.
(B) Evoked AMPA-R- (−60 mV) and NMDA-R- (+40 mV) mediated responses recorded from control non-expressing (Ctrl), M1-Ras(ca)-IRES-GFP and LCK-Ras(ca)-IRES-RFP expressing CA1 cells cultured in media containing LY294002 or PD98059.
(C–D) AMPA and NMDA responses in M1-Ras(ca) and LCK-Ras(ca) expressing neurons relative to non-expressing control CA1 cells cultured in media containing 10 μM LY294002 (C) or 25 μM PD98059 (D). See Table S6 for values.
(E–F) AMPA and NMDA responses in M1-Ras(dn) and LCK-Ras(dn) expressing neurons relative to non-expressing control CA1 cells cultured in media containing 10 μM LY294002 (E) or 25 μM PD98059 (F). See Table S6 for values.
(G) Evoked AMPA-R- (−60 mV) and NMDA-R- (+40 mV) mediated responses recorded from non-expressing (Ctrl), M1-Ras(ca)-IRES-GFP and LCK-Ras(ca)-IRES-RFP expressing CA1 cells in cultured slices prepared from GluA1 knockout and GluA2 knockout mice.
(H–J) AMPA and NMDA responses in M1-Ras(ca) and LCK-Ras(ca) expressing neurons relative to non-expressing control CA1 cells in cultured slices prepared from GluA1 knockout (H), GluA1(S831A/S845A) transgenic (I) and GluA2 knockout (J) mice. See Table S7 for values.
(K–L) AMPA and NMDA responses in M1-Ras(dn) and LCK-Ras(dn) expressing neurons relative to non-expressing control CA1 cells in cultured slices prepared from GluA1 knockout (K) and GluA2 knockout (L) mice. See Table S7 for values. Asterisks indicate p<0.05 (Wilcoxon tests).
To verify that Ras signals the PI3K pathway in the endoplasmic reticulum, we made subcellular domain-specific expression of dominant negative PTEN, PTEN(dn) (Fig S4A), which stimulates PI3K signaling (Hu et al., 2008). Specifically, we overexpressed M1-, LCK-, CD8- and LAMP1-PTEN(dn) in CA1 neurons in cultured rat slices for ~10 hours. We found that neurons expressing M1-PTEN(dn)-IRES-GFP had increased AMPA responses, whereas neurons expressing LCK-, CD8- and LAMP1-PTEN(dn)-IRES-GFP had the same AMPA responses compared to nearby control non-expressing neurons (Fig S4B and Table S8). Similarly, to confirm that Ras signals the ERK pathway in the lipid rafts, we acutely overexpressed M1-, LCK-, CD8- and LAMP1-MEK(dn)-IRES-GFP in CA1 neurons in cultured rat slices for ~10 hours to make subcellular domain-specific suppression of MEK-ERK signaling (Fig S4A). Neurons expressing LCK-MEK(dn)-IRES-GFP had decreased AMPA responses, whereas neurons expressing M1-, CD8- and LAMP1-MEK(dn)-IRES-GFP had the same AMPA responses compared to nearby control non-expressing neurons (Fig S4C and Table S8). Collectively, these results consistently support the idea that Ras signals via the PI3K pathway in the endoplasmic reticulum and the ERK pathway in the lipid rafts.
While Rap2 signals via JNK to depotentiate GluA1- and GluA2L-mediated AMPA transmission, Rap1 signals via p38MAPK to depress GluA2-mediated AMPA transmission (Kielland et al., 2009; Myers et al., 2012; Sheng et al., 2016; Zhu et al., 2005). Thus, our results that activating and blocking Rap2 activity in the bulk membrane suppresses and potentiates AMPA transmission, respectively, are indicative of existence of the Rap2-JNK signaling complex in the bulk membrane. Similarly, our results that activating and blocking Rap1 in the lysosome suppresses and potentiates AMPA transmission, respectively, are indicative of existence of the Rap1-p38MAPK signaling complex in the lysosome. We evaluated these deductions with pharmacology experiments, GluA1 and GluA2 knockout mice, and GluA2(K882A) transgenic mice (Fig 6B–F and Table S6). Including SP600125, an inhibitor of JNK, in culture media blocked the depression of AMPA transmission in CD8-Rap2(ca)-IRES-GFP expressing neurons and occluded the potentiation of AMPA transmission in CD8-Rap2(dn)-IRES-GFP expressing neurons, but had no effect on the depression of AMPA transmission in LAMP1-Rap1(ca)-IRES-RFP expressing neurons and the potentiation of AMPA transmission in LAMP1- Rap1(dn)-IRES-RFP expressing neurons. In contrast, including SB203580, an inhibitor of p38MAPK, in culture media blocked the depression of AMPA transmission in LAMP1-Rap1(ca)-IRES-RFP expressing neurons and occluded the potentiation AMPA transmission in LAMP1-Rap1(dn)-IRES-RFP expressing neurons, but had no effect on the depression of AMPA transmission in CD8-Rap2(ca)-IRES-GFP expressing neurons and the potentiation of AMPA transmission in CD8-Rap2(dn)-IRES-GFP expressing neurons. Moreover, CD8-Rap2(ca)-IRES-GFP- and LAMP1-Rap1(ca)-IRES-RFP-stimulated depressions of AMPA transmission, and CD8-Rap2(ca)-IRES-GFP- and LAMP1-Rap1(ca)-IRES-RFP-stimulated potentiations of AMPA transmission were preserved in GluA1 knockout CA1 neurons, but all were prevented in GluA2 knockout CA1 neurons (Fig 6G–L and Table S9). Finally, LAMP1-Rap1(ca)-IRES-GFP-, but not CD8-Rap2(ca)-IRES-GFP-stimulated depression of AMPA transmission was prevented in GluA2(K882A) transgenic CA1 neurons (Fig 6G, 6J and Table S9), in which GluA2-mediated synaptic depression is impaired (Steinberg et al., 2006). Together, these results suggest that Rap2 controls JNK signaling in the bulk membrane and Rap1 controls p38MAPK signaling in the lysosome.
Figure 6. Rap signals JNK and p38MAPK in the bulk membrane and lysosome.
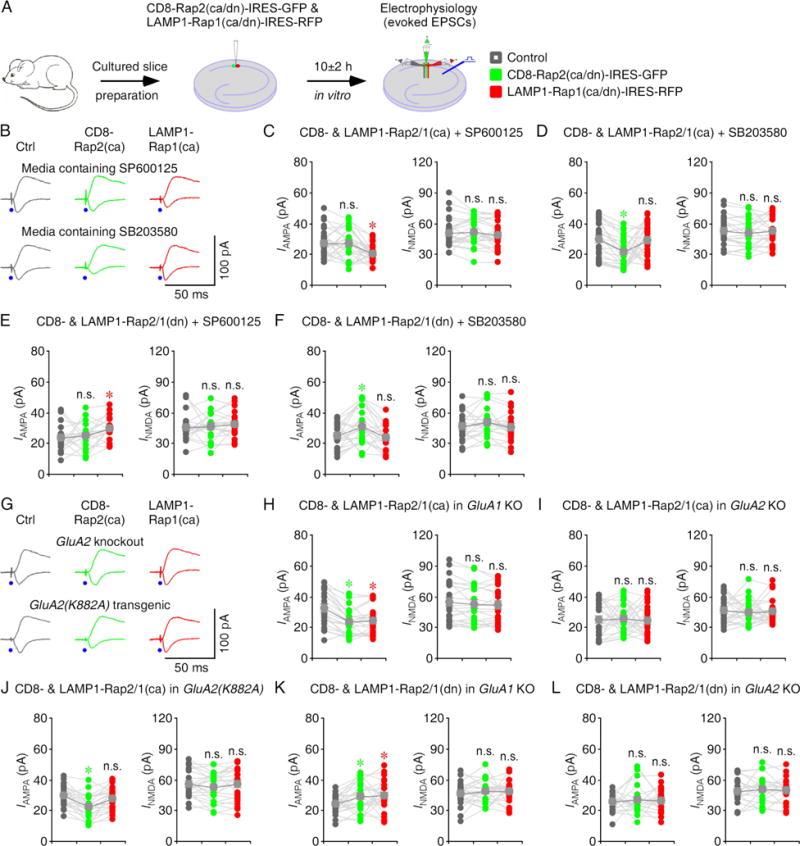
(A) Schematic drawing outlines in vitro experimental design.
(B) Evoked AMPA-R- (−60 mV) and NMDA-R- (+40 mV) mediated responses recorded from control non-expressing (Ctrl), CD8-Rap2(ca)-IRES-GFP and LAMP1-Rap1(ca)-IRES-RFP expressing CA1 cells cultured in media containing SP600125 or SB203580.
(C–D) AMPA and NMDA responses in CD8-Rap2(ca) and LAMP1-Rap1(ca) expressing neurons relative to non-expressing CA1 cells cultured in media containing 5 μM SP600125 (C) or 2 μM SB203580 (D). See Table S6 for values.
(E–F) AMPA and NMDA responses in CD8-Rap2(dn) and LAMP1-Rap1(dn) expressing neurons relative to non-expressing CA1 cells cultured in media containing 5 μM SP600125 (E) or 2 μM SB203580 (F). See Table S6 for values.
(G) Evoked AMPA and NMDA responses recorded from non-expressing (Ctrl), CD8-Rap2(ca)-IRES-GFP and LAMP1-Rap1(ca)-IRES-RFP expressing CA1 cells in cultured slices prepared from GluA2 knockout and GluA2(K882A) transgenic mice.
(H–J) AMPA and NMDA responses in CD8-Rap2(ca) and LAMP1-Rap1(ca) expressing neurons relative to non-expressing CA1 cells in cultured slices prepared from GluA1 knockout (H), GluA2 knockout (I) and GluA2(K882A) transgenic (J) mice. See Table S9 for values.
(K–L) AMPA and NMDA responses in CD8-Rap2(dn) and LAMP1-Rap1(dn) expressing neurons relative to non-expressing CA1 cells in cultured slices prepared from GluA1 knockout (K) and GluA2 knockout (L) mice. See Table S9 for values. Asterisks indicate p<0.05 (Wilcoxon tests).
To verify that Rap2 signals the JNK pathway in the bulk membrane, we acutely overexpressed M1-, LCK-, CD8- and LAMP1-TNIK(dn)-IRES-GFP in CA1 neurons in cultured rat slices for ~10 hours to make subcellular domain-specific suppression of TNIK signaling (Fig S4A), which relays Rap2 signaling to JNK (Zhu et al., 2005). Neurons expressing CD8-TNIK(dn)-IRES-GFP had increased AMPA responses, whereas neurons expressing M1-, LCK- and LAMP1-TNIK(dn)-IRES-GFP had the same AMPA responses compared to nearby control non-expressing neurons (Fig S4D and Table S8). Similarly, to confirm that Rap1 signals the p38MAPK pathway in the lysosome, we acutely overexpressed M1-, LCK-, CD8- and LAMP1-p38MAPK(dn)-IRES-GFP in CA1 neurons in cultured rat slices for ~10 hours to make subcellular domain-specific suppression of p38MAPK signaling (Fig S4A). Neurons expressing LAMP1-p38MAPK(dn)-IRES-GFP had increased AMPA responses, whereas neurons expressing M1-, LCK- and CD8-p38MAPK(dn)-IRES-GFP had the same AMPA responses compared to nearby control non-expressing neurons (Fig S4E and Table S8). Collectively, these results consistently support the notion that Rap2 signals the JNK pathway in the bulk membrane and Rap1 signals the p38MAPK pathway in the lysosome.
Because LTP depends on Ras-ERK (required by both GluA2L-and GluA1-mediated LTP) and -PI3K (required by GluA1-mediated LTP) signaling, depotentiation depends on Rap2-JNK signaling, and LTD depends on Rap1-p38MAPK signaling (Kielland et al., 2009; Kolleker et al., 2003; Qin et al., 2005; Zhu et al., 2002; Zhu et al., 2005), we investigated whether blocking subcelluar microdomain-specific Ras/Rap signaling selectively affects LTP, depotentiation and LTD by overexpressing M1-Ras(dn)-, LCK-Ras(dn)-, CD8-Rap2(dn)- and LAMP1-Rap1(dn)-IRES-GFP in CA1 neurons in cultured rat slices for ~10 hours (Fig 7A). Subsequently examining LTP revealed that M1-Ras(dn)-IRES-GFP expressing neurons had reduced LTP (by ~50%), LCK-Ras(dn)-IRES-GFP expressing neurons had abolished LTP, while CD8-Rap2(dn)- and LAMP1-Rap1(dn)-IRES-GFP expressing neurons had the same LTP compared to nearby control non-expressing neurons (Fig 7B–E and Table S10). Depotentiating the newly formed LTP in the same neurons showed that M1-Ras(dn)-IRES-GFP expressing neurons had reduced depotentiation (by ~50%), CD8-Rap2(dn)-IRES-GFP expressing neurons had no depotentiation, and LAMP1-Rap1(dn)-IRES-GFP expressing neurons had the same depotentiation compared to nearby control non-expressing neurons (Fig 7B–E and Table S10). LTD experiments showed that M1-Ras(dn)-, LCK-Ras(dn)- and CD8-Rap2(dn)-IRES-GFP expressing neurons had the same LTD, whereas LAMP1-Rap1(dn)-IRES-GFP expressing neurons had the eliminated LTD compared to nearby control non-expressing neurons (Fig 7F–I and Table S10). These results suggest that the endoplasmic reticulum- and lipid rafts-specific Ras signals LTP, the bulk membrane-specific Rap2 signals depotentiation and the lysosome-specific Rap1 signals LTD.
Figure 7. Microdomain-specific Ras and Rap signal distinct forms of synaptic plasticity.
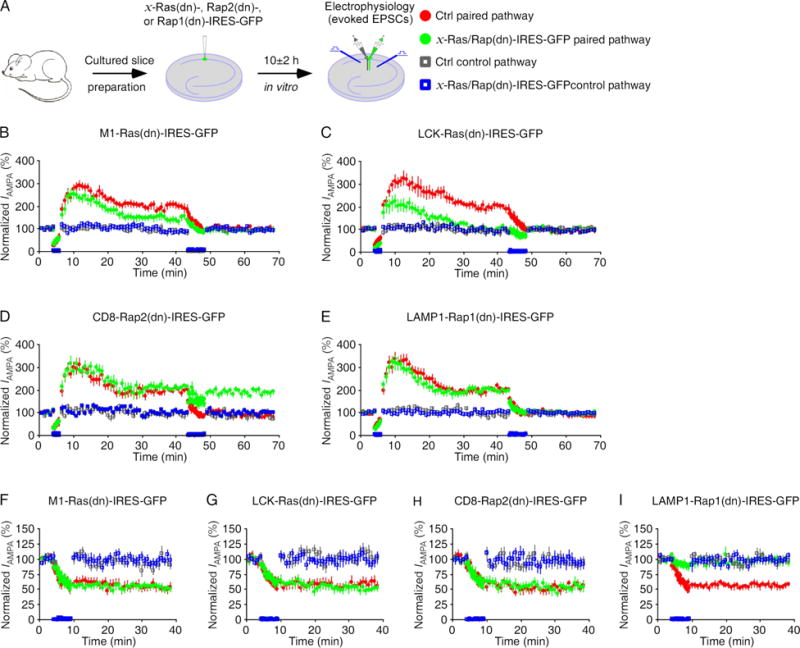
(A) Schematic drawing outlines in vitro experimental design.
(B–E) Relative LTP and depotentiation in M1-Ras(dn)-, LCK-Ras(dn)-, CD8-Rap2(dn) and LAMP1-Rap1(dn)-IRES-GFP expressing neurons compared to control non-expressing neurons. See Table S10 for values.
(F–I) Relative LTD in M1-Ras(dn)-, LCK-Ras(dn)-, CD8-Rap2(dn) and LAMP1-Rap1(dn)-IRES-GFP expressing neurons compared to control non-expressing neurons. See Table S10 for values.
Endogenous Ras and Rap signal synaptic transmission in distinct microdomains in vivo
To determine whether endogenous Ras signals transmission in the endoplasmic reticulum and lipid rafts in intact brains, we virally expressed M1-Ras(dn)-IRES-GFP and LCK-Ras(dn)-IRES-RFP in the hippocampal CA1 regions of intact rats. After 10 hours in vivo expression, we made simultaneous recordings from M1- Ras(dn)-IRES-GFP and LCK-Ras(dn)-IRES-RFP expressing and nearby control non-expressing CA1 pyramidal neuron triplets in the acutely prepared hippocampal slices (Fig 8A). Neurons expressing M1-Ras(dn)-IRES-GFP and LCK-Ras(dn)-IRES-RFP had reduced AMPA responses compared to non-expressing neurons (Fig 8B–C and Table S11), suggesting that the endogenous endoplasmic reticulum and lipid rafts Ras activity tonically potentiates AMPA transmission. To examine whether the potentiations depend on activity, we infused TTX with the viral solutions during the expression (Fig 8A), which blocks the local neuronal activity during the expression (McCormack et al., 2006). Hippocampal infusion of TTX blocked M1-Ras(dn)-IRES-GFP- and LCK-Ras(dn)-IRES-RFP-mediated suppressions of AMPA responses (Fig 8B, 8D and Table S11). As expected, neurons expressing M1-Ras(ca)-IRES-GFP and LCK-Ras(ca)-IRES-RFP had enhanced AMPA responses compared to non-expressing neurons (Fig 8E and Table S11). Together, these results suggest that endogenous Ras signaling in the endoplasmic reticulum and lipid rafts potentiates synaptic AMPA transmission in an activity-dependent manner in intact brains.
Figure 8. Microdomain-specific Ras, Rap2 and Rap1 signal synaptic transmission in intact brains.
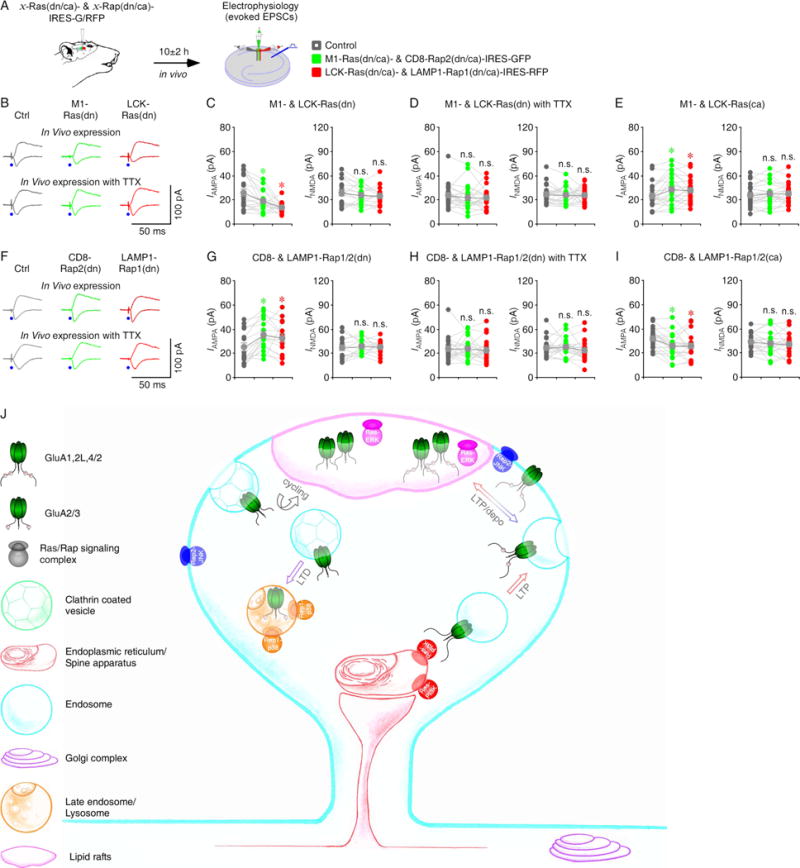
(A) Schematic drawing outlines in vivo experimental design.
(B) Evoked AMPA-R- (−60 mV) and NMDA-R- (+40 mV) mediated responses recorded from control non-expressing (Ctrl), M1-Ras(dn)-IRES-GFP and LCK-Ras(dn)-IRES-RFP expressing CA1 cells after 10±2 h expression in vivo without or with 100 μM TTX.
(C–E) AMPA and NMDA responses in M1-Ras(dn), LCK-Ras(dn), M1-Ras(ca) and LCK-Ras(ca) expressing neurons relative to non-expressing control cells. See Table S11 for values.
(F) Evoked AMPA and NMDA responses recorded from non-expressing (Ctrl), CD8-Rap2(dn)-IRES-GFP and LAMP1-Rap1(dn)-IRES-RFP expressing CA1 cells after 10±2 h expression in vivo without or with 100 μM TTX.
(G–I) AMPA and NMDA responses in CD8-Rap2(dn), LAMP1-Rap1(dn), CD8-Rap2(ca) and LAMP1-Rap1(ca) expressing neurons relative to non-expressing control cells. See Table S11 for values. Asterisks indicate p<0.05 (Wilcoxon tests).
(J) Schematic model for microdomain-specific Ras and Rap signaling controlling LTP, depotentiation and LTD. See text for detail.
We next investigate endogenous Rap signaling in the bulk membrane and lysosome in intact brains by virally expressing CD8-Rap2(dn)-IRES-GFP and LAMP1-Rap1(dn)-IRES-RFP in the hippocampal CA1 regions of intact rats. After 10 hours of in vivo expression, we made simultaneous recordings from CD8-Rap2(dn)-IRES-GFP and LAMP1-Rap1(dn)-IRES-RFP expressing and nearby control non-expressing CA1 pyramidal neuron triplets in the acutely prepared hippocampal slices (Fig 8A). Neurons expressing CD8-Rap2(dn)-IRES-GFP and LAMP1-Rap1(dn)-IRES-RFP had enhanced AMPA responses compared to non-expressing neurons (Fig 8F–G and Table S11), suggesting that endogenous Rap2 signaling in the bulk membrane and endogenous Rap1 signaling in the lysosome tonically depress AMPA transmission. Hippocampal infusion of TTX blocked CD8-Rap2(dn)-IRES-GFP and LAMP1-Rap1(dn)-IRES-RFP-mediated potentiations of AMPA responses (Fig 8F, 8H and Table S11). As expected, neurons expressing CD8-Rap2(ca)-IRES-GFP and LAMP1-Rap1(ca)-IRES-RFP had depressed AMPA responses compared to non-expressing neurons (Fig 8I and Table S11). Collectively, these results suggest that endogenous Rap2 signaling in the bulk membrane and endogenous Rap1 signaling in the lysosome depress synaptic AMPA transmission in an activity-dependent manner in intact brains.
DISCUSSION
In this study, we have created and validated a targeted delivery method that permits selective expression of Ras and Rap proteins into distinct subcellular microdomains. Using this method, we reveal that endogenous Ras preferentially signals synaptic potentiation via the endoplasmic reticulum PI3K and lipid rafts ERK pathways, while endogenous Rap2 and Rap1 predominantly signal synaptic depression via the bulk membrane JNK and lysosome p38MAPK pathways, respectively (Fig 8J).
Microdomain-dependent signal diversity and specificity
We show here that Ras, Rap2 and Rap1 utilize the different subcellular microdomains to signal multiple different forms of synaptic plasticity (Fig 8J). Signaling molecules generate a plethora of specific cellular outputs, even though many of them have a high degree of sequence and structural identity in functional domains (Gloerich and Bos, 2011; Simanshu et al., 2017). How the signal diversity and specificity are achieved is a long-standing cell biology question. To determine whether Ras and Rap utilize the microdomain-specific signaling to create signal diversity and specificity to control synaptic plasticity, we first adapted or engineered multiple domain-targeting sequences, including M1-, LCK-, CD8-, LAMP1- and KDELr-targeting sequences, to drive expression of recombinant proteins predominantly into the desired subcellular domains. We verified the delivery specificity with microdomain-specific markers and Western blotting after micro-fractionation. Importantly, electrophysiology and pharmacology experiments in wild type, knockout and/or transgenic animals showed no effect on synaptic responses resulted from any potential non-specific expressions in undesired microdomains, suggesting no significant non-specific expression in undesired microdomains. Moreover, expression of Ras and Rap in their preferred, as well as in all non-preferred signaling microdomains for 10±2 hours and 16±2 hours resulted in similar changes in synaptic responses that were suggestive of approximately equivalent levels of construct expressions in these microdomains (cf. (Zhu et al., 2002; Zhu et al., 2005)). Finally, expressions of Ras and Rap dead mutants with these five targeting sequences had no effect on synaptic transmission. Collectively, these results validate the method as an effective approach to deliver recombinant proteins into multiple distinct subcellular microdomains.
Using the microdomain-specific delivery method, we systematically interrogated endogenous Ras and Rap signaling in the endoplasmic reticulum, lipid rafts, bulk membrane, lysosome and Golgi complex. Although endogenous Ras and Rap are present in all five microdomains, Ras and Rap only use the endoplasmic reticulum-, lipid rafts-, bulk membrane- and lysosome-specific signaling to achieve their perspective physiological functions at synapses. For example, targeting Ras into the endoplasmic reticulum and lipid rafts microdomains results in the independent PI3K and ERK signal transductions that control synaptic potentiation during LTP, while targeting Rap2 and Rap1 (which share ~70% homology) into the bulk membrane and lysosome microdomains initiates the specific JNK and p38MAPK signaling cascades that control synaptic depressions during depotentiation and LTD, respectively. Therefore, confining endogenous, homologous Ras and Rap within distinct microdomains, as seen here in CA1 neurons, diversifies signaling responses. Moreover, forcing Ras into the functional Rap microdomains and forcing Rap into in the functional Ras microdomains induce non-specific signaling responses, underscoring the importance of restraining Ras and Rap within the correct microdomains in maintaining synaptic signal specificity. Together, our data provide the first evidence indicating that the microdomain-specific Ras and Rap signaling mechanisms play key roles in diversifying and specifying signals at single synapses.
Microdomain-specific signaling in regulation of synaptic plasticity
Unveiling the microdomain-specific Ras and Rap signaling at synapses is central to the effort of deciphering the entire nanoscale molecular regulatory machinery governing synaptic plasticity, crucial for understanding the molecular and cellular basis of behavior, as well as a number of mental, neurological and psychiatric disorders (Costa and Silva, 2003; Henley and Wilkinson, 2016; Huganir and Nicoll, 2013; Nishiyama and Yasuda, 2015; Roth et al., 2017; Stornetta and Zhu, 2011). It is established that activity-dependent exocytosis and endocytosis bring AMPA-Rs into and out of the cell plasma membrane to regulate transmission (Lledo et al., 1998; Luscher et al., 1999). In particular, synaptic activity drives exocytosis of AMPA-Rs during LTP, with majority of AMPA-Rs being inserted into extrasynaptic membrane first and then trapped into synapses by activity (Ehlers et al., 2007; Esteves da Silva et al., 2015; Kennedy et al., 2010; Makino and Malinow, 2009; Patterson et al., 2010; Penn et al., 2017; Zhang et al., 2015). The existing evidence also supports the idea that some AMPA-Rs may be directly inserted into synapses via exocytosis (Esteves da Silva et al., 2015; Kennedy et al., 2010; Patterson et al., 2010; Penn et al., 2017). Here we show that Ras-PI3K signaling in the endoplasmic reticulum potentiates transmission when activated together with Ras-ERK signaling in the lipid rafts, whereas Ras-ERK signaling in the lipid rafts alone is sufficient to potentiate transmission, shedding the first light on the subcellular signaling mechanisms that control the trafficking events. Synaptic activity activates Ras-ERK signaling that stimulates phosphorylation of GluA1 at S845, GluA4 at S842 and GluA2L at S841, and this single phosphorylation is sufficient to drive synaptic delivery of GluA2L- and GluA4-, but not GluA1-containing AMPA-Rs (Hu et al., 2008; Kolleker et al., 2003; Qin et al., 2005). On the other hand, strong synaptic activity, experience-dependent activity and/or the presence of neuromodulatory factors stimulate additional Ras signaling and activate Ras-PI3K signaling that stimulates phosphorylation of GluA1 at S831, and this phosphorylation, together with the phosphorylation of GluA1 at S845, is required for driving GluA1-containing AMPA-Rs into synapses (Hu et al., 2008; Kolleker et al., 2003; Lim et al., 2014; Qin et al., 2005). Thus, it is tempting to speculate that Ras-PI3K signaling at the endoplasmic reticulum (or spine apparatus) may lead to phosphorylation of intracellular GluA1 at S831 and drive exocytosis of GluA1-containing AMPA-Rs at extrasynaptic membrane (Fig 8J), and that at the membrane surface the single-site phosphorylated GluA1-containing AMPA-Rs remain to be highly mobile (cf. (Ehlers et al., 2007)). Synaptic activity-stimulated Ras-ERK signaling at the lipid rafts may further phosphorylate GluA1 at S845, which can reduce the AMPA-Rs’ mobility and restrict them locally at synapses. Indeed, our pilot immunoelectron microscopic analysis of subcellular locations of phosphorylated GluA1 at synapses supports this notion (Fig S5; our unpublished observation). It is likely that lipid rafts Ras-ERK signaling may also entrap GluA2L- and GluA4-containing AMPA-Rs into synapses by phosphorylation, via the lateral trafficking and/or direct exocytosis routes.
We here report that Rap2-JNK signaling in the bulk membrane depotentiates transmission whereas Rap1-p38MAPAK signaling in the lysosome depresses transmission, inspiring a few new thoughts of how synaptic signaling regulates AMPA-R trafficking during depressions. Rap2-JNK signaling stimulates dephosphorylation and synaptic removal of GluA1-, GluA2L- and GluA4-containing AMPA-Rs during depotentiation (Kielland et al., 2009; Myers et al., 2012; Sheng et al., 2016; Zhu et al., 2005). Thereby, we envisage that depotentiating activity-stimulated Ras-JNK signaling at the bulk membrane might induce dephosphorylation of GluA1, GluA2L and GluA4 in the lipid raft islands surrounded by the bulk membrane to enhance their mobility to escape from synapses (Fig 8J). Undepotentiated GluA1-, GluA2L- and GluA4-containing AMPA-Rs will eventually be exchanged with GluR2/3 AMPA-Rs via an activity-independent process at a slow rate time constants of ~16 h (Adesnik et al., 2005; Kolleker et al., 2003; McCormack et al., 2006; Zhu, 2009; Zhu et al., 2000). GluA2/3 AMPA-Rs, on the other hand, are continuously cycling between synaptic and non-synaptic sites in an activity-independent manner at a fast rate time constant of ~20 min (Lee et al., 2002; Luscher et al., 1999; Nishimune et al., 1998; Osten et al., 1998; Song et al., 1998). Therefore, we imagine that the cycling could give the synaptic activity-stimulated Rap1-JNK signaling at lysosome/late endosome an opportunity to phosphorylate GluA2/3 AMPA-Rs during LTD (Fig 8J) (Kielland et al., 2009; Zhu et al., 2002). Phosphorylation of GluA2 at S880 disrupts its interaction with glutamate receptor-interacting protein/AMPA-binding protein (GRIP/ABP) and favors its binding with PKC-interacting protein 1 (PICK1) (Chung et al., 2000; Matsuda et al., 2000; Perez et al., 2001), which reduces the recycling of GluA2/3 AMPA-Rs and depresses synaptic transmission (Braithwaite et al., 2002; Daw et al., 2000; Lin and Huganir, 2007; Seidenman et al., 2003; Zhou et al., 2018). As with LTP, it remains in debate whether synaptic AMPA-Rs are removed via direct endocytosis at synapses and/or through lateral diffusion followed by endocytosis at extrasynaptic sites during depotentiation and LTD (Ashby et al., 2004; Nadif Kasri et al., 2011; Tardin et al., 2003). Combining the microdomain-specific signaling manipulation technique that we report here with immunoelectron microscopy (Kielland et al., 2009; Racz et al., 2004), two-photon fluorescence lifetime imaging (Nishiyama and Yasuda, 2015) and super-resolution live-cell imaging (MacGillavry et al., 2013; Roth et al., 2017) of phosphorylation and trafficking of AMPA-Rs should allow direct visualization of many elusive nanoscale signaling and trafficking events at synapses during different forms of plasticity. We expect that these experiments will directly test the above hypotheses of signaling regulations of LTP, depotentiation and LTD.
Understanding microdomain-specific signaling is essential to intervene in the aberrant signaling-driven diseases (Ahearn et al., 2011; Simanshu et al., 2017). Lack of knowledge of specificities of disease-involved signaling, that prevents identification of precise drugable targets, is the primary obstacle that hinders development of effective treatments for cancers and cognitive diseases. For example, although Ras and Rap are known to interact with a large number of effector proteins and control many signal responses, so far only a very few specific Ras and Rap signaling pathways have been identified to be responsible for particular tumors and cognitive disorders (Aoki et al., 2008; Lim et al., 2014; Lim et al., 2017b). This study provides a precision regulation toolbox and an effective strategy that may inspire more systematic and high-throughput investigations of subcellular microdomain-specific signaling pathways, which should promote development of precision medications for treating a variety of diseases.
STAR☼METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER | |
|---|---|---|---|
| Antibodies | |||
| mouse anti-calreticulin monoclonal antibody | BD Biosciences | Cat No: 612136; RRID:AB_399507 |
|
| Goat anti-caveolin-1 polyclonal antibody | Sigma-Aldrich | Cat No: SAB2500204; RRID:AB_10605605 |
|
| mouse anti-transferrin receptor monoclonal antibody | Innovative Research | Cat No: 13-6800; RRID:AB_86623 |
|
| rabbit anti-giantin polyclonal antibody | Covance | Cat No: PRB-114C-200; RRID:AB_291560 |
|
| rabbit anti-LAMP2 polyclonal antibody | Thermo Fisher Scientific | Cat No: PA-1655; RRID:AB_2134625 |
|
| Alexa 594-conjugated goat anti-mouse antibody | Thermo Fisher Scientific | Cat No: A-11005; RRID:AB_2534073 |
|
| Alexa 594-conjugated goat anti-rabbit antibody | Thermo Fisher Scientific | Cat No: A-11012; RRID:AB_2534079 |
|
| rabbit anti-GFP monoclonal antibody | Cell Signaling Technology | Cat No: 2956S; RRID:AB_1196615 |
|
| mouse anti-Ras monoclonal antibody | BD Biosciences | Cat No: 610001; RRID:AB_397424 |
|
| mouse anti-Rap1 monoclonal antibody | BD Biosciences | Cat No: 610195; RRID:AB_397594 |
|
| mouse anti-Rap2 monoclonal antibody | BD Biosciences | Cat No: 610215; RRID:AB_397612 |
|
| Bacterial and Virus Strains | |||
| Sindbis virus | Invitrogen Corporation | Cat No: K750-01 | |
| Chemicals, Peptides, and Recombinant Proteins | |||
| Alexa 594-conjugated cholera toxin | Thermo Fisher Scientific |
|
|
| LY294002 | Calbiochem | A15147 | |
| PD98059 | Calbiochem | A10705 | |
| SP600125 | Calbiochem | A10860 | |
| SB203580 | Calbiochem | A10824 | |
| Tetrodotoxin citrate | Calbiochem | 554412 | |
| Critical Commercial Assays | |||
| Sigma endoplasmic reticulum isolation kit | Sigma-Aldrich | ER0100 | |
| Sigma caveolae/rafts isolation kit | Sigma-Aldrich | CS0750 | |
| Sigma lysosome isolation kit | Sigma-Aldrich | LYSIS01 | |
| Sigma Golgi isolation kit | Sigma-Aldrich | GL0010 | |
| Experimental Models: Organisms/Strains | |||
| Rat: Sprague-Dawley | Charles River | Strain Code: 400 | |
| Mouse: C57BL/6 | Jackson Laboratory | Stock No: 005304; RRID: IMSR_JAX:005304 |
|
| Mouse: GluA2 knockout | Jackson Laboratory | Stock No: 003143 | |
| Mouse: GluA1 knockout | Jackson Laboratory | Stock No: 019011 | |
| Mouse: GluA1(S831A/S845A) transgenic | Jackson Laboratory | Stock No: 008892 | |
| Mouse: GluA1(K882A) transgenic | Jackson Laboratory | Stock No: 024419 | |
| Recombinant DNA | |||
| Ras(G12V), Ras(S17N) and Ras(T35A/D38A) | Home-made; (Zhu et al., 2002; Zhu et al., 2005) | N/A | |
| Rap2(G12E), Rap2(S17N) and Rap2(F39S) | Home-made; (Zhu et al., 2002; Zhu et al., 2005) | N/A | |
| Rap1(Q63E), Rap1(S17N) and Rap1(F39S) | Home-made; (Zhu et al., 2002; Zhu et al., 2005) | N/A | |
| M1-, LCK-, CD8-, LAMP1- and KDELr-Ras(G12V)/Ras(S17N)/Ras(T35A/D38A)-YFP | Home-made; this paper, see STAR Methods for detail | N/A | |
| M1-, LCK-, CD8-, LAMP1- and KDELr-Ras(G12V)/Ras(S17N)/Ras(T35A/D38A)-IRES-G/R/C/OFP | Home-made; this paper, see methods for detail | N/A | |
| M1-, LCK-, CD8-, LAMP1- and KDELr-Rap2(G12E)/Rap2(S17N)/Rap2(F39S)-IRES-G/R/C/OFP | Home-made; this paper, see STAR Methods for detail | N/A | |
| M1-, LCK-, CD8-, LAMP1- and KDELr-Rap1(Q63E)/Rap1(S17N)/Rap1(F39S)-IRES-G/R/C/OFP | Home-made; this paper, see STAR Methods for detail | N/A | |
| Software and Algorithms | |||
| Igor Pro 6 | Wavemetrics | http://www.wavemetrics.com/ | |
| PEPOI; an Igor-based operation and analysis program for simultaneous Electrophysiology, Optogenetics & Imaging experiments | Home-made; (Wang et al., 2015b) | Contact UVA Patent Foundation (https://lvg.virginia.edu/) for the end user license | |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents can be directed to and will be fulfilled by the Lead Contact Dr. Li Lin (linliwz@163.com). The requests may also be sent to and will be fulfilled by Dr. J. Julius Zhu (jjzhu@virginia.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
Male and female Sprague Dawley rats (Charles River), knockout and/or transgenic mice bred congenically on a C57BL/6 background (Jackson Laboratory, Bar Harbor, MA) at embryonic day 18 (E18 pregnant females) and/or postnatal 6–7 day (P6–7) and P14–21 were used to prepare cultured neurons, cultured slices and intact brain experiments in this study. Both male and female animals were used in the study given no difference detected in Ras- and Rap-regulated synaptic transmission and plasticity between the animal groups (Zhu et al., 2002; Zhu et al., 2005). Animals were maintained in the animal facility at the University of Virginia, and family or pair housed in the temperature-controlled animal room with 12 h/12 h light/dark cycle. Food and water were available ad libitum. All procedures for animal surgery and maintenance were performed following protocols approved by the Animal Care & Use Committee of the University of Virginia and in accordance with US National Institutes of Health guidelines.
METHOD DETAILS
Neuronal culture preparation
Cultured hippocampal neurons were prepared from rat embryos at 18 as previously described (Lim et al., 2017a; Zhang and Macara, 2006). Cells were dissociated mechanically after trypsin treatment, and plated onto poly-D-lysine (Sigma-Aldrich; P1149)-coated plastic culture dishes at the density of ~5×104 cells/cm2. Plated cells were recovered in the plating media (DMEM with 2 mM glutamine, 10% FBS, 0.45% glucose and 0.11 mg/mL sodium pyruvate) for 3–4 h, and then maintained in the Neurobasal media (Invitrogen, 21103-049) supplemented with B27 (Invitrogen, 17504-044) and Glutamax (Invitrogen, 35050-061) before use. Neurons were infected using Sindbis virus on DIV4–5.
Cultured slice preparation
Cultured slices were prepared from postnatal 6–7 day old rats, knockout and transgenic mice following the previous studies (Lim et al., 2017a; Lim et al., 2017b; Wang et al., 2015a). In brief, the hippocampi were dissected out in ice-cold HEPES-buffered Hanks’ solution (pH 7.35) under sterile conditions, sectioned into 400 μm slices on a tissue chopper, and explanted onto a 0.4-mm pore size Millicell-CM culture membrane insert (Millipore; PICM03050). The membranes were then placed in 750 μl of MEM culture medium, contained (in mM): HEPES 30, heat-inactivated horse serum 20%, glutamine 1.4, D-glucose 16.25, NaHCO3 5, CaCl2 1, MgSO4 2, insulin 1 mg/ml, ascorbic acid 0.012% at pH 7.28 and osmolarity 320. Cultured slices were maintained at 35°C, in a humidified incubator (ambi ent air enriched with 5% CO2).
Subcellular microdomain-targeting constructs
Ras and Rap constructs were made as previously described (Zhu et al., 2002; Zhu et al., 2005). Briefly, Ras mutant constructs, including constitutively active mutant (G12→V), dominant negative mutant (S17→N) and null/dead mutant (T35→A, D38→A), Rap2 mutant constructs, including the constitutively active mutant (G12→E), dominant negative mutant (S17→N) and null/dead mutant (F39→S), and Rap1 mutant constructs, including constitutively active mutant (Q63→E), dominant negative mutant (S17→N) and null/dead mutant (T35→A, D38→A), were generated from wild type Ras (NCBI Reference: NM_001130442.1), Rap2 (NCBI Reference: NM_021033) and Rap1 (NCBI Reference: NM_001291896) sequences using Quick Change Site-Directed mutagenesis kit (Stratagene, La Jolla, CA). Subcellular microdomain-targeting Ras and Rap constructs were generated by first mutating putative palmitoylation-relevant cysteines at C-termini of H-Ras and Rap2 to serines to make them palmitoylation-deficient. The palmitoylation-deficient H-Ras and Rap2, and wild type Rap1 were then fused to specific tethering signals at their N-termini, with the sequence encoding amino acids 1–66 of the avian infectious bronchitis virus M protein (M1-Ras/Rap) to restrict their expression in the endoplasmic reticulum, with the sequence encoding N-terminal palmitoylated myristoylation LCK signal (LCK-Ras/Rap) to restrict their expression in the lipid rafts, with the sequence encoding the targeting signal and transmembrane domain of CD8α receptor (CD8-Ras/Rap) to restrict their expression in the bulk membrane, with the sequence encoding N-terminal and transmembrane domain of LAMP1 (LAMP1-Ras/Rap) to restrict their expression in the lysosome, or with the sequence encoding the resident Golgi protein KDELr receptor with N193D mutation (KDELr-Ras/Rap) to restrict their expression in the Golgi complex (cf. (Chiu et al., 2002; Honing et al., 1996; Matallanas et al., 2006; Rohrer et al., 1996)). Given the critical role of C-terminal YXXI signal of LAMP1 in its lysosome trafficking (Honing et al., 1996; Rohrer et al., 1996), the sequence encoding C-terminal 11 amino acids of LAMP1 was linked to the C-termini of Ras and Rap constructs to improve their lysosome targeting. In some experiments, YFP was fused at the C-termini of Ras constructs to follow their expression distributions. In the other experiments, Ras and Rap constructs and GFP, CFP, OFP and RFP (mCherry) (gifts from Dr Roger Tsien) were subcloned before and after an internal ribosomal entry site (IRES) sequence of the pCITE vector (Novagen) to achieve co-expression. These Ras and Rap constructs were then subcloned into a Sindbis viral construct and viral particles were produced following the supplier manual (http://tools.thermofisher.com/content/sfs/manuals/sindbis_man.pdf).
Construct expression
Recombinant proteins were expressed using the Sindbis viral expression system, which does not infect individual neurons in intact brain slices/tissues more than once (Malinow et al., 2010). For in vitro expression, CA1 pyramidal neurons in hippocampal cultured rat and mouse slices were infected after 7–14 days in vitro with Sindbis virus, and then incubated on culture media and 5% CO2 for 10±2 h before experiments unless stated otherwise. For in vivo expression, P14–21 rats were initially anesthetized by an intraperitoneal injection of ketamine and xylazine (10 and 2 mg/kg, respectively). Animals were then placed in a stereotaxic frame and one ~1×1 mm hole was opened above the right side of the somatosensory cortex. A glass pipette was used to make pressure injections of ~100 nl diluted Sindbis viral solution in the hippocampal CA1 region according to stereotaxic coordinates. After injection, animals were allowed to recover from the anesthesia and returned to their cages. When expression time is properly controlled, dominant negative proteins serve as an effective way to study endogenous Ras and Rap signaling (Zhu et al., 2002; Zhu et al., 2005). Therefore, experiments were typically performed within 10±2 h or 16±2 h after Sindbis viral infection.
Immunocytochemistry
For M1-, KDELr- and LAMP1-Ras constructs, neurons were fixed in 4% paraformaldehyde with 4% sucrose in PBS for 15 min at room temperature. They were then permeabilized with 0.2% Triton X-100 in PBS for 5 min at room temperature. For CD8- and LCK-Ras constructs, neurons were fixed in ice-cold 4% paraformaldehyde with 4% sucrose for 15 minutes without permeabilization. Paraformaldehyde-fixed neurons were blocked with 20% goat serum in PBS for one hour at room temperature, and then incubated with primary antibodies diluted in 5% goat serum in PBS overnight at 4°C. Primary a ntibodies used include mouse monoclonal calreticulin antibody (1:100, BD Biosciences, San Jose, CA; 612136, RRID:AB_399507), mouse monoclonal transferrin receptor antibody (1:100, Innovative Research, Novi, MI; 13-6800, RRID:AB_86623), rabbit polyclonal giantin antibody (1:500, Covance, Princeton, NJ; PRB-114C-200, RRID:AB_291560), rabbit polyclonal LAMP2 antibody (1:500, Thermo Fisher Scientific, Waltham, MA; PA-1655, RRID:AB_2134625). Following washes in PBS, Alexa 594-conjugated goat anti-mouse (1:500, Thermo Fisher Scientific, Waltham, MA; A-11005, RRID:AB_2534073) or goat anti-rabbit (1:500, Thermo Fisher Scientific, Waltham, MA; A-11012, RRID:AB_2534079) secondary antibodies diluted in 5% goat serum were incubated with the neurons at room temperature for one hour. For LCK-Ras construct, neurons were incubated with Alexa 594-conjugated cholera toxin (Thermo Fisher Scientific, Waltham, MA; C34777) for one hour at room temperature. After incubation, neurons were washed with PBS and mounted using Vectashield (Vector Laboratories, Burlingame, CA). Fluorescence images were acquired with an Olympus FV1000 confocal microscope or an custom-made two-photon microscope (Wang et al., 2015b) with a 60× water-immersion lens (NA 1.00, Olympus, Center Valley, PA).
Micro-fractionation and Western analysis
The endoplasmic reticulum, lipid rafts, lysosome and Golgi complex fractionations were isolated with the Sigma endoplasmic reticulum (Sigma-Aldrich, St. Louis, MO; ER0100), caveolae/rafts (Sigma-Aldrich; CS0750), lysosome (Sigma-Aldrich; LYSIS01) and Golgi (Sigma-Aldrich; GL0010) isolation kits, respectively. The bulk membrane fraction was isolated following a previous protocol (Agudo-Ibanez et al., 2017). Briefly, the hippocampal tissues were homogenized in 0.25 M sucrose, 10 mM Tris (pH 8), 1 mM MgCl2, 1 mM EDTA, and 0.5 mM DTT with protease inhibitors. The lysates were then centrifuged at 800 × g for 10 min at 4°C to remove the nuclear fraction, and the supernatant was further centrifuged at 5,000 × g for 10 min at 4°C to remove the mitochondrial fraction, and at 100,000 × g for 60 min at 4°C to isolate the membrane fraction. Homogenization was made for every 48 cultured slices, and the lysates were micro-fractionated for either endoplasmic reticulum, lipid rafts, lysosome or Golgi complex fractions using 0.8 ml ultra-clear tubes (Beckman Coulter, Brea, CA; 344090). Recombinant Ras-YFP, endogenous calreticulin, caveolin-1, transferrin receptor, LAMP2, giantin, Ras and Rap in 5–10% of the isolated fractions were loaded and separated with SDS-PAGE, blotted with anti-GFP (1:1000; Cell Signaling Technology, Beverly, MA; 2956S, RRID:AB_1196615), anti-calreticulin (1:1000, BD Biosciences; 612136, RRID:AB_399507), anti-caveolin-1 (1:1000, Sigma-Aldrich; SAB2500204, RRID:AB_10605605), anti-transferrin receptor (1:1000, Innovative Research; 13-6800, RRID:AB_86623), anti-giantin (1:2000, Covance; PRB-114C-200, RRID:AB_291560), anti-Ras (1:2000; BD Biosciences; 610001, RRID:AB_397424), anti-Rap1 (1:2000; BD Biosciences; 610195, RRID:AB_397594) or anti-Rap2 (1:2000; BD Biosciences; 610215, RRID:AB_397612). Chemiluminescent Western blots were captured by a digital camera under linear exposure conditions.
Electrophysiology
Multiple patch-clamp recordings were obtained simultaneously and/or sequentially from nearby infected and non-infected CA1 neuron doublets, triplets, quadruplets or quintuplets (Lim et al., 2017a; Wang et al., 2015b), under visual guidance using fluorescence and transmitted light illumination, using up to five Axopatch-200B amplifiers (Molecular Devices, Sunnyvale, CA). The operation and analysis of multiple patch-clamp recordings (and imaging) were made with a single custom-written IGOR Pro 6 program (WaveMetrics, Lake Oswego, OR)-based software PEPOI (Wang et al., 2015b). Bath solution (29±1.5°C), unless otherwise stated, cont ained (in mM): NaCl 119, KCl 2.5, CaCl2 4, MgCl2 4, NaHCO3 26, NaH2PO4 1, glucose 11, picrotoxin (PTX) 0.1, bicuculline 0.01, and 2-chloroadenosine 0.002, at pH 7.4 and gassed with 5% CO2/95% O2. 2-chloroadenosine was included to prevent bursting. For acutely prepared slices, 2-mM CaCl2- and 1-mM MgCl2-containing bath solution was used instead and 2-chloroadenosine was excluded. For experiments in which slices were maintained in culture media with additional 10 μM LY294002 (Calbiochem, La Jolla, CA; A15147), 25 M PD98059 (Calbiochem; A10705), 5 M SP600125 (Calbiochem; A10860) and 2 μM SB203580 (Calbiochem; A10824), these inhibitors were included at the time of viral infection and removed during recordings. For some intact brain experiments, 100 μM TTX (Calbiochem; 554412) was infused together with viral solutions during the expression and TTX was removed during the recordings. Patch recording pipettes (3–6 MΩ) for current (voltage-clamp) recordings contained (in mM): cesium methanesulfonate 115, CsCl 20, HEPES 10, MgCl2 2.5, Na2ATP 4, Na3GTP 0.4, sodium phosphocreatine 10, EGTA 0.6, and spermine 0.1, at pH 7.25. Synaptic responses were evoked by bipolar electrodes with single voltage pulses (200 μs, up to 20 V) placed in the stratum radiatum ~300–500 μm from the CA1 cells. Synaptic AMPA and NMDA responses at −60 mV and +40 mV were averaged over 90 trials. To minimize the effect from AMPA responses, the peak NMDA responses at +40 mV were measured after digital subtraction of estimated AMPA responses at +40 mV. Synaptic plasticity experiments followed our previous reports (McCormack et al., 2006; Zhu et al., 2005). Briefly, LTP was induced by a pairing protocol using 200 pulses at 2 Hz at −5 mV within 5 min after formation of whole-cell configuration. Depotentiation was induced by a pairing protocol using 300 pulses at 1 Hz at −45 mV ~38 min after induction of LTP in the presence of SB203580, which blocks LTD. LTD was induced by pairing 300 pulses at 1 Hz at −45 mV 15 min after formation of whole-cell configuration. The cultured slices used for synaptic plasticity experiments were cultured in high Mg2+ media during the expression to avoid change in the basal transmission (McCormack et al., 2006; Zhu et al., 2005).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical results were reported as mean±s.e.m. Animals or cells were randomly assigned into control or experimental groups and investigators were blinded to experiment treatments in cultured slices and animals. Given the negative correlation between the variation and square root of sample number, n, the group sample size was typically set to be ~16–36 to optimize the efficiency and power of statistical tests. All the sample numbers, including animals, cells and/or experimental replications, were summarized in the supplemental tables S1–11. Because the evoked synaptic responses depend on the arbitrarily applied stimulation intensity, the Wilcoxon non-parametric test, that is independent of means and requires no pre-assumption, was used to determine the statistical significance of the means (p<0.05; two sides).
Supplementary Material
Highlights.
Microdomain-targeted delivery method reveals signal diversity and specificity
Endoplasmic reticulum Ras-PI3K and lipid rafts Ras-ERK signal long-term potentiation
Bulk membrane Rap2-JNK signals depotentiation
Lysosome Rap1-p38MAPK signals long-term depression
Acknowledgments
We thank members of the Zhu laboratory for comments and technical assistance, and Drs Ian Macara and Roger Tsien for reagents and/or use of lab resources. This study is supported in part by the Virginia Center on Aging postdoctoral ARDRAF Award (LZ) and Epilepsy Foundation postdoctoral fellowship No. 310443 (GW), MINECO/FEDER-UE grant SAF-2015-63638R, RTICC grant RD-12-0036-0033 and AECC grant GCB141423113 (PC), National Natural Science Foundation of China grants NSFC81722028 (JX) and NSFC81771284 (LL), HHMI (RLH), NIH grants NS036715 (RLH), NS065183 and NS089578 (HZ), NS078792 (JWH), and NS053570, NS091452, NS094980, NS092548 and NS104670 (JJZ). JJZ is the Radboud Professor and Sir Yue-Kong Pao Chair Professor.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
JJZ conceived the concept and led the project with input and assistance from LL; LZ, PZ, GW and HZ carried out the key experiments; YZ, YY, MZ and JX performed the other experiments and/or assisted on the experiments; PC, JWH and RLH provided additional supervision and/or input on the design of experiments; JJZ wrote the manuscript with input from all coauthors.
DECLARATION OF INTERESTS
The authors declare no competing interests.
References
- Adesnik H, Nicoll RA, England PM. Photoinactivation of native AMPA receptors reveals their real-time trafficking. Neuron. 2005;48:977–985. doi: 10.1016/j.neuron.2005.11.030. [DOI] [PubMed] [Google Scholar]
- Agudo-Ibanez L, Crespo P, Casar B. Analysis of Ras/ERK compartmentalization by subcellular fractionation. Methods Mol Biol. 2017;1487:151–162. doi: 10.1007/978-1-4939-6424-6_11. [DOI] [PubMed] [Google Scholar]
- Ahearn IM, Haigis K, Bar-Sagi D, Philips MR. Regulating the regulator: post-translational modification of RAS. Nature reviews. Molecular cell biology. 2011;13:39–51. doi: 10.1038/nrm3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki Y, Niihori T, Narumi Y, Kure S, Matsubara Y. The RAS/MAPK syndromes: novel roles of the RAS pathway in human genetic disorders. Human mutation. 2008;29:992–1006. doi: 10.1002/humu.20748. [DOI] [PubMed] [Google Scholar]
- Ashby MC, De La Rue SA, Ralph GS, Uney J, Collingridge GL, Henley JM. Removal of AMPA receptors (AMPARs) from synapses is preceded by transient endocytosis of extrasynaptic AMPARs. J Neurosci. 2004;24:5172–5176. doi: 10.1523/JNEUROSCI.1042-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos JL, de Rooij J, Reedquist KA. Rap1 signalling: adhering to new models. Nature reviews. Molecular cell biology. 2001;2:369–377. doi: 10.1038/35073073. [DOI] [PubMed] [Google Scholar]
- Braithwaite SP, Xia H, Malenka RC. Differential roles for NSF and GRIP/ABP in AMPA receptor cycling. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:7096–7101. doi: 10.1073/pnas.102156099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casar B, Arozarena I, Sanz-Moreno V, Pinto A, Agudo-Ibanez L, Marais R, Lewis RE, Berciano MT, Crespo P. Ras subcellular localization defines extracellular signal-regulated kinase 1 and 2 substrate specificity through distinct utilization of scaffold proteins. Molecular and cellular biology. 2009;29:1338–1353. doi: 10.1128/MCB.01359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau AH, Walter JM, Gerardin J, Tang C, Lim WA. Designing synthetic regulatory networks capable of self-organizing cell polarization. Cell. 2012;151:320–332. doi: 10.1016/j.cell.2012.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu VK, Bivona T, Hach A, Sajous JB, Silletti J, Wiener H, Johnson RL, 2nd, Cox AD, Philips MR. Ras signalling on the endoplasmic reticulum and the Golgi. Nature cell biology. 2002;4:343–350. doi: 10.1038/ncb783. [DOI] [PubMed] [Google Scholar]
- Chung HJ, Xia J, Scannevin RH, Zhang X, Huganir RL. Phosphorylation of the AMPA receptor subunit GluR2 differentially regulates its interaction with PDZ domain-containing proteins. J Neurosci. 2000;20:7258–7267. doi: 10.1523/JNEUROSCI.20-19-07258.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa RM, Silva AJ. Mouse models of neurofibromatosis type I: bridging the GAP. Trends in molecular medicine. 2003;9:19–23. doi: 10.1016/s1471-4914(02)00008-4. [DOI] [PubMed] [Google Scholar]
- Daw MI, Chittajallu R, Bortolotto ZA, Dev KK, Duprat F, Henley JM, Collingridge GL, Isaac JT. PDZ proteins interacting with C-terminal GluR2/3 are involved in a PKC-dependent regulation of AMPA receptors at hippocampal synapses. Neuron. 2000;28:873–886. doi: 10.1016/s0896-6273(00)00160-4. [DOI] [PubMed] [Google Scholar]
- Ehlers MD, Heine M, Groc L, Lee MC, Choquet D. Diffusional trapping of GluR1 AMPA receptors by input-specific synaptic activity. Neuron. 2007;54:447–460. doi: 10.1016/j.neuron.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteves da Silva M, Adrian M, Schatzle P, Lipka J, Watanabe T, Cho S, Futai K, Wierenga CJ, Kapitein LC, Hoogenraad CC. Positioning of AMPA receptor-containing endosomes regulates synapse architecture. Cell reports. 2015;13:933–943. doi: 10.1016/j.celrep.2015.09.062. [DOI] [PubMed] [Google Scholar]
- Gloerich M, Bos JL. Regulating Rap small G-proteins in time and space. Trends in cell biology. 2011;21:615–623. doi: 10.1016/j.tcb.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Henley JM, Wilkinson KA. Synaptic AMPA receptor composition in development, plasticity and disease. Nature reviews. Neuroscience. 2016;17:337–350. doi: 10.1038/nrn.2016.37. [DOI] [PubMed] [Google Scholar]
- Honing S, Griffith J, Geuze HJ, Hunziker W. The tyrosine-based lysosomal targeting signal in lamp-1 mediates sorting into Golgi-derived clathrin-coated vesicles. The EMBO journal. 1996;15:5230–5239. [PMC free article] [PubMed] [Google Scholar]
- Hsieh H, Boehm J, Sato C, Iwatsubo T, Tomita T, Sisodia S, Malinow R. AMPAR removal underlies Abeta-induced synaptic depression and dendritic spine loss. Neuron. 2006;52:831–843. doi: 10.1016/j.neuron.2006.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Qin Y, Bochorishvili G, Zhu Y, van Aelst L, Zhu JJ. Ras signaling mechanisms underlying impaired GluR1-dependent plasticity associated with fragile X syndrome. J Neurosci. 2008;28:7847–7862. doi: 10.1523/JNEUROSCI.1496-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huganir RL, Nicoll RA. AMPARs and synaptic plasticity: the last 25 years. Neuron. 2013;80:704–717. doi: 10.1016/j.neuron.2013.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy MJ, Davison IG, Robinson CG, Ehlers MD. Syntaxin-4 defines a domain for activity-dependent exocytosis in dendritic spines. Cell. 2010;141:524–535. doi: 10.1016/j.cell.2010.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielland A, Bochorishvili G, Corson J, Zhang L, Rosin DL, Heggelund P, Zhu JJ. Activity patterns govern synapse-specific AMPA-R trafficking between deliverable and synaptic pools. Neuron. 2009;62:84–101. doi: 10.1016/j.neuron.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Dunah AW, Wang YT, Sheng M. Differential roles of NR2A- and NR2B-containing NMDA receptors in Ras-ERK signaling and AMPA receptor trafficking. Neuron. 2005;46:745–760. doi: 10.1016/j.neuron.2005.04.031. [DOI] [PubMed] [Google Scholar]
- Kolleker A, Zhu JJ, Schupp BJ, Qin Y, Mack V, Borchardt T, Kohr G, Malinow R, Seeburg PH, Osten P. Glutamatergic plasticity by synaptic delivery of GluR-B(long)-containing AMPA receptors. Neuron. 2003;40:1199–1212. doi: 10.1016/s0896-6273(03)00722-0. [DOI] [PubMed] [Google Scholar]
- Lee HK, Takamiya K, Han JS, Man H, Kim CH, Rumbaugh G, Yu S, Ding L, He C, Petralia RS, et al. Phosphorylation of the AMPA receptor GluR1 subunit is required for synaptic plasticity and retention of spatial memory. Cell. 2003;112:631–643. doi: 10.1016/s0092-8674(03)00122-3. [DOI] [PubMed] [Google Scholar]
- Lee SH, Liu L, Wang YT, Sheng M. Clathrin adaptor AP2 and NSF interact with overlapping sites of GluR2 and play distinct roles in AMPA receptor trafficking and hippocampal LTD. Neuron. 2002;36:661–674. doi: 10.1016/s0896-6273(02)01024-3. [DOI] [PubMed] [Google Scholar]
- Lim CS, Hoang ET, Viar KE, Stornetta RL, Scott MM, Zhu JJ. Pharmacological rescue of Ras signaling, GluA1-dependent synaptic plasticity, and learning deficits in a fragile X model. Genes & development. 2014;28:273–289. doi: 10.1101/gad.232470.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CS, Kang X, Mirabella V, Zhang H, Bu Q, Araki Y, Hoang ET, Wang S, Shen Y, Choi S, et al. BRaf signaling principles unveiled by large-scale human mutation analysis with a rapid lentivirus-based gene replacement method. Genes & development. 2017a;31:537–552. doi: 10.1101/gad.294413.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CS, Wen C, Sheng Y, Wang G, Zhou Z, Wang S, Zhang H, Ye A, Zhu JJ. Piconewton-scale analysis of Ras-BRaf signal transduction with single-molecule force spectroscopy. Small. 2017b;13 doi: 10.1002/smll.201701972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DT, Huganir RL. PICK1 and phosphorylation of the glutamate receptor 2 (GluR2) AMPA receptor subunit regulates GluR2 recycling after NMDA receptor-induced internalization. J Neurosci. 2007;27:13903–13908. doi: 10.1523/JNEUROSCI.1750-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lledo PM, Zhang X, Sudhof TC, Malenka RC, Nicoll RA. Postsynaptic membrane fusion and long-term potentiation. Science. 1998;279:399–403. doi: 10.1126/science.279.5349.399. [DOI] [PubMed] [Google Scholar]
- Luscher C, Xia H, Beattie EC, Carroll RC, von Zastrow M, Malenka RC, Nicoll RA. Role of AMPA receptor cycling in synaptic transmission and plasticity. Neuron. 1999;24:649–658. doi: 10.1016/s0896-6273(00)81119-8. [DOI] [PubMed] [Google Scholar]
- MacGillavry HD, Song Y, Raghavachari S, Blanpied TA. Nanoscale scaffolding domains within the postsynaptic density concentrate synaptic AMPA receptors. Neuron. 2013;78:615–622. doi: 10.1016/j.neuron.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino H, Malinow R. AMPA receptor incorporation into synapses during LTP: the role of lateral movement and exocytosis. Neuron. 2009;64:381–390. doi: 10.1016/j.neuron.2009.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinow R, Hayashi Y, Maletic-Savatic M, Zaman SH, Poncer JC, Shi SH, Esteban JA, Osten P, Seidenman K. Introduction of green fluorescent protein (GFP) into hippocampal neurons through viral infection. Cold Spring Harbor; 2010. (protocols 2010). pdb prot5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man HY, Wang Q, Lu WY, Ju W, Ahmadian G, Liu L, D’Souza S, Wong TP, Taghibiglou C, Lu J, et al. Activation of PI3-kinase is required for AMPA receptor insertion during LTP of mEPSCs in cultured hippocampal neurons. Neuron. 2003;38:611–624. doi: 10.1016/s0896-6273(03)00228-9. [DOI] [PubMed] [Google Scholar]
- Matallanas D, Sanz-Moreno V, Arozarena I, Calvo F, Agudo-Ibanez L, Santos E, Berciano MT, Crespo P. Distinct utilization of effectors and biological outcomes resulting from site-specific Ras activation: Ras functions in lipid rafts and Golgi complex are dispensable for proliferation and transformation. Molecular and cellular biology. 2006;26:100–116. doi: 10.1128/MCB.26.1.100-116.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda S, Launey T, Mikawa S, Hirai H. Disruption of AMPA receptor GluR2 clusters following long-term depression induction in cerebellar Purkinje neurons. The EMBO journal. 2000;19:2765–2774. doi: 10.1093/emboj/19.12.2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack SG, Stornetta RL, Zhu JJ. Synaptic AMPA receptor exchange maintains bidirectional plasticity. Neuron. 2006;50:75–88. doi: 10.1016/j.neuron.2006.02.027. [DOI] [PubMed] [Google Scholar]
- Mochizuki N, Yamashita S, Kurokawa K, Ohba Y, Nagai T, Miyawaki A, Matsuda M. Spatio-temporal images of growth-factor-induced activation of Ras and Rap1. Nature. 2001;411:1065–1068. doi: 10.1038/35082594. [DOI] [PubMed] [Google Scholar]
- Myers KR, Wang G, Sheng Y, Conger KK, Casanova JE, Zhu JJ. Arf6-GEF BRAG1 regulates JNK-mediated synaptic removal of GluA1-containing AMPA receptors: a new mechanism for nonsyndromic X-linked mental disorder. J Neurosci. 2012;32:11716–11726. doi: 10.1523/JNEUROSCI.1942-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabavi S, Kessels HW, Alfonso S, Aow J, Fox R, Malinow R. Metabotropic NMDA receptor function is required for NMDA receptor-dependent long-term depression. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:4027–4032. doi: 10.1073/pnas.1219454110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadif Kasri N, Nakano-Kobayashi A, Van Aelst L. Rapid synthesis of the X-linked mental retardation protein OPHN1 mediates mGluR-dependent LTD through interaction with the endocytic machinery. Neuron. 2011;72:300–315. doi: 10.1016/j.neuron.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimune A, Isaac JT, Molnar E, Noel J, Nash SR, Tagaya M, Collingridge GL, Nakanishi S, Henley JM. NSF binding to GluR2 regulates synaptic transmission. Neuron. 1998;21:87–97. doi: 10.1016/s0896-6273(00)80517-6. [DOI] [PubMed] [Google Scholar]
- Nishiyama J, Yasuda R. Biochemical computation for spine ctructural plasticity. Neuron. 2015;87:63–75. doi: 10.1016/j.neuron.2015.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osten P, Srivastava S, Inman GJ, Vilim FS, Khatri L, Lee LM, States BA, Einheber S, Milner TA, Hanson PI, Ziff EB. The AMPA receptor GluR2 C terminus can mediate a reversible, ATP-dependent interaction with NSF and alpha- and beta-SNAPs. Neuron. 1998;21:99–110. doi: 10.1016/s0896-6273(00)80518-8. [DOI] [PubMed] [Google Scholar]
- Patterson MA, Szatmari EM, Yasuda R. AMPA receptors are exocytosed in stimulated spines and adjacent dendrites in a Ras-ERK-dependent manner during long-term potentiation. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:15951–15956. doi: 10.1073/pnas.0913875107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisajovich SG, Garbarino JE, Wei P, Lim WA. Rapid diversification of cell signaling phenotypes by modular domain recombination. Science. 2010;328:368–372. doi: 10.1126/science.1182376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn AC, Zhang CL, Georges F, Royer L, Breillat C, Hosy E, Petersen JD, Humeau Y, Choquet D. Hippocampal LTP and contextual learning require surface diffusion of AMPA receptors. Nature. 2017;549:384–388. doi: 10.1038/nature23658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez JL, Khatri L, Chang C, Srivastava S, Osten P, Ziff EB. PICK1 targets activated protein kinase Calpha to AMPA receptor clusters in spines of hippocampal neurons and reduces surface levels of the AMPA-type glutamate receptor subunit 2. J Neurosci. 2001;21:5417–5428. doi: 10.1523/JNEUROSCI.21-15-05417.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior IA, Muncke C, Parton RG, Hancock JF. Direct visualization of Ras proteins in spatially distinct cell surface microdomains. The Journal of cell biology. 2003;160:165–170. doi: 10.1083/jcb.200209091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Zhu Y, Baumgart JP, Stornetta RL, Seidenman K, Mack V, van Aelst L, Zhu JJ. State-dependent Ras signaling and AMPA receptor trafficking. Genes & development. 2005;19:2000–2015. doi: 10.1101/gad.342205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racz B, Blanpied TA, Ehlers MD, Weinberg RJ. Lateral organization of endocytic machinery in dendritic spines. Nature neuroscience. 2004;7:917–918. doi: 10.1038/nn1303. [DOI] [PubMed] [Google Scholar]
- Reuther GW, Der CJ. The Ras branch of small GTPases: Ras family members don’t fall far from the tree. Current opinion in cell biology. 2000;12:157–165. doi: 10.1016/s0955-0674(99)00071-x. [DOI] [PubMed] [Google Scholar]
- Rocks O, Peyker A, Bastiaens PI. Spatio-temporal segregation of Ras signals: one ship, three anchors, many harbors. Current opinion in cell biology. 2006;18:351–357. doi: 10.1016/j.ceb.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Rohrer J, Schweizer A, Russell D, Kornfeld S. The targeting of Lamp1 to lysosomes is dependent on the spacing of its cytoplasmic tail tyrosine sorting motif relative to the membrane. The Journal of cell biology. 1996;132:565–576. doi: 10.1083/jcb.132.4.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth RH, Zhang Y, Huganir RL. Dynamic imaging of AMPA receptor trafficking in vitro and in vivo. Current opinion in neurobiology. 2017;45:51–58. doi: 10.1016/j.conb.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidenman KJ, Steinberg JP, Huganir R, Malinow R. Glutamate receptor subunit 2 Serine 880 phosphorylation modulates synaptic transmission and mediates plasticity in CA1 pyramidal cells. J Neurosci. 2003;23:9220–9228. doi: 10.1523/JNEUROSCI.23-27-09220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Y, Zhang L, Su SC, Tsai LH, Zhu JJ. Cdk5 is a new rapid synaptic homeostasis regulator capable of initiating the early alzheimer-like pathology. Cereb Cortex. 2016;26:2937–2951. doi: 10.1093/cercor/bhv032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simanshu DK, Nissley DV, McCormick F. RAS proteins and their regulators in human disease. Cell. 2017;170:17–33. doi: 10.1016/j.cell.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song I, Kamboj S, Xia J, Dong H, Liao D, Huganir RL. Interaction of the N-ethylmaleimide-sensitive factor with AMPA receptors. Neuron. 1998;21:393–400. doi: 10.1016/s0896-6273(00)80548-6. [DOI] [PubMed] [Google Scholar]
- Steinberg JP, Takamiya K, Shen Y, Xia J, Rubio ME, Yu S, Jin W, Thomas GM, Linden DJ, Huganir RL. Targeted in vivo mutations of the AMPA receptor subunit GluR2 and its interacting protein PICK1 eliminate cerebellar long-term depression. Neuron. 2006;49:845–860. doi: 10.1016/j.neuron.2006.02.025. [DOI] [PubMed] [Google Scholar]
- Stornetta RL, Zhu JJ. Ras and Rap signaling in synaptic plasticity and mental disorders. The Neuroscientist: a review journal bringing neurobiology, neurology and psychiatry. 2011;17:54–78. doi: 10.1177/1073858410365562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardin C, Cognet L, Bats C, Lounis B, Choquet D. Direct imaging of lateral movements of AMPA receptors inside synapses. The EMBO journal. 2003;22:4656–4665. doi: 10.1093/emboj/cdg463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk L, Chiu SL, Sharma K, Huganir RL. Glutamate synapses in human cognitive disorders. Annual review of neuroscience. 2015;38:127–149. doi: 10.1146/annurev-neuro-071714-033821. [DOI] [PubMed] [Google Scholar]
- Wang G, Bochorishvili G, Chen Y, Salvati KA, Zhang P, Dubel SJ, Perez-Reyes E, Snutch TP, Stornetta RL, Deisseroth K, et al. CaV3.2 calcium channels control NMDA receptor-mediated transmission: a new mechanism for absence epilepsy. Genes & development. 2015a;29:1535–1551. doi: 10.1101/gad.260869.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Wyskiel DR, Yang W, Wang Y, Milbern LC, Lalanne T, Jiang X, Shen Y, Sun QQ, Zhu JJ. An optogenetics- and imaging-assisted simultaneous multiple patch-clamp recordings system for decoding complex neural circuits. Nature protocols. 2015b;10:397–412. doi: 10.1038/nprot.2015.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Courtney MJ, Martinsson P, Manahan-Vaughan D. Hippocampal long-term depression is enhanced, depotentiation is inhibited and long-term potentiation is unaffected by the application of a selective c-Jun N-terminal kinase inhibitor to freely behaving rats. The European journal of neuroscience. 2011;33:1647–1655. doi: 10.1111/j.1460-9568.2011.07661.x. [DOI] [PubMed] [Google Scholar]
- Zhang H, Macara IG. The polarity protein PAR-3 and TIAM1 cooperate in dendritic spine morphogenesis. Nature cell biology. 2006;8:227–237. doi: 10.1038/ncb1368. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Cudmore RH, Lin DT, Linden DJ, Huganir RL. Visualization of NMDA receptordependent AMPA receptor synaptic plasticity in vivo. Nature neuroscience. 2015;18:402–407. doi: 10.1038/nn.3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Hancock JF. Ras nanoclusters: Versatile lipid-based signaling platforms. Biochimica et biophysica acta. 2015;1853:841–849. doi: 10.1016/j.bbamcr.2014.09.008. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Prakash P, Liang H, Cho KJ, Gorfe AA, Hancock JF. Lipid-sorting specificity encoded in K-Ras membrane anchor regulates signal output. Cell. 2017;168:239–251 e216. doi: 10.1016/j.cell.2016.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Liu A, Xia S, Leung C, Qi J, Meng Y, Xie W, Park P, Collingridge GL, Jia Z. The C-terminal tails of endogenous GluA1 and GluA2 differentially contribute to hippocampal synaptic plasticity and learning. Nature neuroscience. 2018;21:50–62. doi: 10.1038/s41593-017-0030-z. [DOI] [PubMed] [Google Scholar]
- Zhu JJ. Activity level-dependent synapse-specific AMPA receptor trafficking regulates transmission kinetics. J Neurosci. 2009;29:6320–6335. doi: 10.1523/JNEUROSCI.4630-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JJ, Esteban JA, Hayashi Y, Malinow R. Postnatal synaptic potentiation: delivery of GluR4-containing AMPA receptors by spontaneous activity. Nature neuroscience. 2000;3:1098–1106. doi: 10.1038/80614. [DOI] [PubMed] [Google Scholar]
- Zhu JJ, Qin Y, Zhao M, Van Aelst L, Malinow R. Ras and Rap control AMPA receptor trafficking during synaptic plasticity. Cell. 2002;110:443–455. doi: 10.1016/s0092-8674(02)00897-8. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Pak D, Qin Y, McCormack SG, Kim MJ, Baumgart JP, Velamoor V, Auberson YP, Osten P, van Aelst L, et al. Rap2-JNK removes synaptic AMPA receptors during depotentiation. Neuron. 2005;46:905–916. doi: 10.1016/j.neuron.2005.04.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


