Abstract
Background
Mitochondrial dysfunction has been implicated in the pathologies of a number of retinal degenerative diseases in both the outer and inner retina. In the outer retina, photoreceptors are particularly vulnerable to mutations affecting mitochondrial function due to their high energy demand and sensitivity to oxidative stress. However, it is unclear how defective mitochondrial biogenesis affects neural development and contributes to neural degeneration. In this report, we investigated the in vivo function of nuclear respiratory factor 1 (Nrf1), a major transcriptional regulator of mitochondrial biogenesis in both proliferating retinal progenitor cells (RPCs) and postmitotic rod photoreceptor cells (PRs).
Methods
We used mouse genetic techniques to generate RPC-specific and rod PR-specific Nrf1 conditional knockout mouse models. We then applied a comprehensive set of tools, including histopathological and molecular analyses, RNA-seq, and electroretinography on these mouse lines to study Nrf1-regulated genes and Nrf1’s roles in both developing retinas and differentiated rod PRs. For all comparisons between genotypes, a two-tailed two-sample student’s t-test was used. Results were considered significant when P < 0.05.
Results
We uncovered essential roles of Nrf1 in cell proliferation in RPCs, cell migration and survival of newly specified retinal ganglion cells (RGCs), neurite outgrowth in retinal explants, reconfiguration of metabolic pathways in RPCs, and mitochondrial morphology, position, and function in rod PRs.
Conclusions
Our findings provide in vivo evidence that Nrf1 and Nrf1-mediated pathways have context-dependent and cell-state-specific functions during neural development, and disruption of Nrf1-mediated mitochondrial biogenesis in rod PRs results in impaired mitochondria and a slow, progressive degeneration of rod PRs. These results offer new insights into the roles of Nrf1 in retinal development and neuronal homeostasis and the differential sensitivities of diverse neuronal tissues and cell types of dysfunctional mitochondria. Moreover, the conditional Nrf1 allele we have generated provides the opportunity to develop novel mouse models to understand how defective mitochondrial biogenesis contributes to the pathologies and disease progression of several neurodegenerative diseases, including glaucoma, age-related macular degeneration, Parkinson’s diseases, and Huntington’s disease.
Keywords: Mitochondrial biogenesis, Nrf1, Retinal progenitor cell, Retinal ganglion cell, Optic atrophy, Photoreceptor degeneration
Background
Mitochondrial biogenesis is a dynamic subcellular process through which existing mitochondria continuously import and integrate new proteins and lipids, replicate mitochondrial DNA (mtDNA), and fuse and divide upon environment changes. This process is intricately regulated to maintain a healthy mitochondrial network, essential for energy homeostasis, metabolism, signaling, and apoptosis. The vast majority of the ~ 1500 proteins involved in mitochondrial structure and function are encoded by nuclear genes, which are regulated in concert with a set of transcriptional regulators, including peroxisome proliferative activated receptor gamma coactivator 1 (PGC-1) family members, nuclear respiratory factor 1 (Nrf1), and nuclear respiratory factor 2 (Nrf2/GABP) [1–4].
Nrf1 encodes an evolutionarily conserved transcription activator [5–9]. Nrf1 binds to GC-rich DNA elements in promoters of many nuclear genes required for mitochondrial biogenesis and respiratory function [9–11]. In primary cortical neurons, Nrf1 has been shown to co-regulate all cytochrome c oxidase (COX) subunits and several glutamatergic neurochemicals, implying that a Nrf1-mediated higher-order mechanism coordinately controls the expression of genes involved in neuronal activity and energy metabolism [12–15]. In muscle, Nrf1 has been shown to be a direct PGC-1 target, the master regulator of mitochondrial biogenesis, whose dysfunction has been implicated in several neurodegenerative diseases, such as Parkinson’s disease [1, 4]. In addition, Nrf1 plays a significant role in cell growth and proliferation. A recent study using chromatin immunoprecipitation sequencing (ChIP-seq) analysis identified 2470 potential Nrf1 targets in human neuroblastoma cells, indicating roles for Nrf1 in regulating genes for mitochondrial biogenesis and cell growth and in the pathogenesis of neurodegenerative diseases [16]. Interestingly, several genes involved in the glycolytic pathway, such as PFKB2, PGAM1, PGKM5, and ALDOA, were also found in this list, suggesting a possible Nrf1 role in reprogramming metabolic processes. Nrf1 also interacts with several proteins involved in different cellular functions. For example, it interacts directly with poly(ADP-ribose) polymerase 1 (PARP-1), and PARP-1 modulates Nrf1’s DNA-binding domain for transcriptional regulation [17]. Dynein light chain was also shown to interact with NRF-1, although the functional significance remains unknown [18].
Several in vivo studies have revealed distinct functions of Nrf1 in different developing organisms. In zebrafish, an insertional mutation in the Nrf1 locus caused a cell death phenotype in developing photoreceptors [7]. In Drosophila, the Nrf1 homolog gene erect wing (ewg) has been shown to regulate Hippo pathway activity in a neuronal subtype-specific manner to determine neuronal fate in developing retinas [19]. In mice, Nrf1-null embryos fail to maintain mtDNA and die between embryonic day 3.5 (E3.5) and 6.5 [20]. These studies offer insights into the understanding of Nrf1’s in vivo function in different developmental systems and cellular context, but how Nrf1-regulated pathways function in retinal development and how they contribute to defective mitochondrial biogenesis to affect neural development and contribute to neural degeneration is unknown.
In this report, we studied the function of Nrf1 during mouse retinal development. We show that Nrf1 is expressed in proliferating retinal progenitor cells (RPCs) in embryonic retinas and enriched in retinal ganglion cells (RGCs) and rod photoreceptors cells (PRs), both of which consume large amounts of energy. Using cell-type-specific Nrf1 knockout mice, we demonstrate that Nrf1 controls cell proliferation in RPCs and the extension of neurite processes in developing retinal neurons. Nrf1-deficient embryonic retinas exhibited affected expression of genes involved in multiple cellular processes. In differentiated rod PRs, deleting Nrf1 caused abnormal mitochondrial morphology, deteriorated mitochondrial functions, abnormal photoreceptor inner and outer segments, and reduced electroretinography (ERG) activities. Eventually, mutant rod PRs completely degenerated. Together, these results demonstrate the crucial role of Nrf1-mediated mitochondrial biogenesis in retinal development and homeostasis and provide new insights into Nrf1 function in neurite outgrowth and metabolic reprogramming.
Methods
Gene targeting and animal breeding
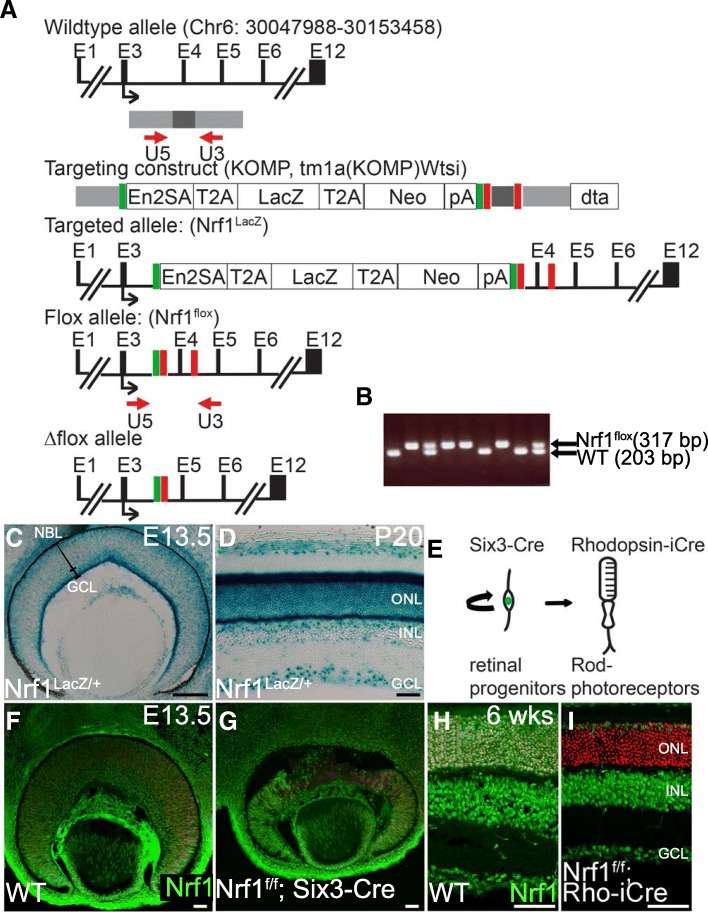
A Nrf1-targeted embryonic stem (ES) clone was obtained from the knockout mouse project repository (http://www.mousephenotype.org/data/alleles/MGI:1332235/tm1a(KOMP)Wtsi). This allele contains 2 loxP sites inserted into the third and fourth introns, and a FLP recombinase target (FRT)-site flanked T2A-LacZ-T2A-neomycin fusion cassette inserted into intron 3. Exon 4 in the floxed allele can be deleted by Cre-mediated recombination. ES cells were injected into B6(GC)-Tyrc-2 J/J blastocysts, and the injected blastocysts were transferred into C57/BL6 albino females. Chimeric males obtained by blastocyst injection were bred to wildtype B6(GC)-Tyrc-2 J/J females to generate the Nrf1LacZ/+ allele, which was subsequently bred with a 17T17TRosa26-FLPeR0T17T0T17T 0T0Ttransgene to remove a LacZ-neomycin fusion cassette to generate a Nrf1flox/+ allele (Fig. 1a). PCR primers used to distinguish the Nrf1flox allele from the wildtype allele were U5 (5’-CCAAGACTTGTATGCATTGGTCTCAG-3′) and U3 (5’-GCACTTCTGGCTCCATGGTCC-3′) (Fig. 1a, b). PCR primers for Six3-Cre were Cre1 (5′-AACGAGTGATGAGGTTCGCAAGAAC-3′) and Cre2 (5′-CGCTATTTTCCATGAGTGAACGAACC-3′); and for Rho-iCre were iCre1 (5’-GGATGCCACCTCTGATGAAG-3′) and iCre2 (5’-CACACCATTCTTTCTGACCCG-3′). Embryos were designated as E0.5 at noon on the day in which vaginal plugs were observed. Both male and female mice were used in this study, and no differences were observed according to sex.
Fig. 1.
Generation of Nrf1 expression and conditional Nfr1 alleles. (a) Genomic structure of Nrf1, the targeting construct, the targeted Nrf1LacZ and Nrf1flox alleles, and the deleted allele. Exons are indicated as E1-E12. The gray and black bars indicate the regions used in the targeting construct. A black arrow indicates the translational start site for the Nrf1 protein. Red arrows indicate PCR primers used for PCR genotyping of the wildtype and floxed alleles. Red boxes indicate loxP sites, and green boxes indicate FRT sites. (b) PCR genotyping using U5 and U3 primers for wildtype (203 bp) and floxed (317 bp) alleles. (c, d) Nrf1 expression during retinogenesis revealed by LacZ expression in Nrf1LacZ/+ retinas at E13.5 (c) and P20 (d). (e) Schematic representation of the developing retinal cells expressing Six3 and rhodopsin. Proliferative retinal progenitor cells expressing Six3 will give rise to all mature retinal cells. Rhodopsin is expressed in differentiated rod photoreceptor cells. (f–i) Nrf1 expression detected by immunofluorescent staining on E13.5 wildtype (f) and Nrf1f/f;Six3-Cre (g) retinal sections and on 6-week-old wildtype (h) and Nrf1f/f;Rho-iCre (i) retinal sections. Scale bars: 100 μm in C, 50 μm in d-i. ONL: outer nuclear layer. INL: inner nuclear layer. GCL: ganglion cell layer. WT: wildtype. NBL: neural blast layer
Histology, immunohistochemistry, X-gal staining, COX activity
Embryos or eyeballs dissected from mice were fixed in 4% paraformaldehyde at 4 °C for 2 h or overnight, embedded in paraffin or optimal cutting temperature (OCT) compound, and sectioned into 7 μm thickness for histological analysis. After dewaxing and rehydration, the sections were stained with Hematoxylin and Eosin.
For immunohistochemical analysis, cryo- or paraffin-embedded embryos or eyes were sectioned into 7 μm or 30 μm thickness. Sections were heat-treated in a microwave oven at 600 W in 10 mM sodium citrate for 15 min. The sections were blocked with 2% bovine serum albumin and 5% normal serum for 2 h at room temperature. The primary antibody was applied to the sections for 1–3 days at 4oC. The primary antibodies used were mouse anti-Nrf1 (1:300, catalog #PCRP-NFR1-3D4; DSHB, The University of Iowa, Iowa City, IA), mouse anti-Isl1 (1:200, catalog# 37.3F7; DSHB), goat anti-Brn3/Pou4f2 (1:150, catalog #sc-6026; Santa Cruz Biotechnology, Dallas, TX), mouse anti-Pax6 (1:200, catalog #MAB5552; Chemicon, Burlington, MA), sheep anti-Chx10 (1:300, catalog #X1180P; Exalpha, Shirley, MA), rabbit anti-cleaved caspase-3 (1:300, catalog #9579; Cell Signaling, Danvers, MA), mouse anti-BrdU (1:10, catalog #05–633; Millipore, Burlington, MA), mouse anti-Phospho-Histone H3/PH3 (1:700, catalog #9706; Cell Signaling), rabbit anti-Cyclin D1 (1:300, catalog #MA1–39546; Thermo Fisher Scientific, Waltham, MA), mouse anti-rhodopsin (1:20, catalog #MS-1233-R7; Thermo Fisher Scientific), rabbit anti-cone arrestin (1:2000, catalog #AB16282; Millipore), and rabbit anti-Tfam (1:500, catalog #ab131607; Abcam, Cambridge, MA). Secondary antibodies conjugated with Alexa-488, 555 or 633 (Thermo Fisher Scientific) were used in 1:800 dilution. For indirect immunofluorescence, a tyramide signal amplification kit was used (PerkinElmer, Waltham, MA). HRP-conjugated secondary antibodies were from Jackson ImmunoResearch Laboratories (West Grove, PA). DAPI (2.5 μg/ml, catalog #D1306; Thermo Fisher Scientific) was used to stain nuclei. Images were captured using Olympus (Tokyo, Japan) FluoView 1000 or Zeiss (Thornwood, NY) LSM 780 confocal microscopes. SimplePCI software (Hamamatsu Corporation, Sewickley, PA) was used to analyze the number of cells.
For X-gal staining, embryos or eyes were fixed in 10% formalin for 30 min, embedded in OCT compound, and sectioned into 30 μm thickness. Sections were dried at room temperature for 3 h, washed with wash buffer (0.1 M sodium phosphate containing 2 mM MgCl2, 0.01% deoxycholate, and 0.02% Nonidet P-40). LacZ color reaction was performed in wash buffer containing 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, and 1 mg/ml X-gal at 37oC overnight. Color reaction was terminated by incubation in 10% formalin for 10 min. Post-fixed sections were washed, dehydrated, and mounted with Cytoseal 60 (Thermo Fisher Scientific). Images were collected with a Canon EOS 10 digital camera (Melville, NY) mounted on an Olympus IX71 microscope.
Cytochrome c oxidase (COX) analysis was performed as described previously [21] with slight modifications. E13.5 embryonic heads from wildtype and Nrf1f/f;Six3-Cre embryos or 6 week-old adult eyeballs from wildtype and Nrf1f/f;Rho-iCre were fixed in 10% formalin for 20 min at room temperature. Samples were washed in phosphate buffered saline (PBS) 3 times and embedded in OCT; 14 μm cryo-sections were collected. Sections were dried at 4oC for 1 hour, rehydrated, and incubated in COX reacting solution (1× DAB, 100 μM cytochrome C, and 2 μg/ml bovine catalase in 0.1 M PBS, pH 7.0) at 37oC. Color reactions were terminated by incubating in 10% formalin for 10 min. Post-fixed sections were washed, dehydrated, and mounted with Cytoseal 60. Images were collected as described for X-gal staining.
BrdU labeling and TUNEL assays
Terminal deoxynucleotidyl transferase dUTP nick end label (TUNEL) assays were performed using an in situ cell death detection kit (Roche, Pleasanton, CA). For pulse labeling with BrdU, 0.1 mg per body gram of BrdU (Sigma, St. Louis, MO) was injected intraperitoneally into pregnant females 1 h before embryo collection.
RNA sequencing analysis
Eighteen retinas from wildtype and Nrf1f/f;Six3-Cre embryos at E13.5 of multiple littermates were pooled, and RNA was extracted using TRI reagent (Sigma) and purified with a Pure Link RNA mini kit (Thermo Fisher Scientific). RNA sequencing (RNA-seq) was performed in the Sequencing and Microarray Core Resource Facility at The University of Texas MD Anderson Cancer Center. RNAs were treated with DNase, and cDNAs were synthesized using a cDNA synthesis kit (NuGen, San Carlos, CA). One hundred nt paired-end reads were obtained using an Illumina HiSeq 3000 Next Generation Sequencing instrument (San Diego, CA). The RNA-seq experiment was duplicated, thus making statistical comparisons possible, although there is still a lack of statistical power. The RNA-seq reads were mapped to the mouse genome (mm10) via the Tophat 2.7.2 program. We performed QC and sum counts (reads) for each gene using HTseq. The differential expression analyses were performed by Cuffdiff software. Nrf1-dependent genes (fold change ≥ 1.4; adjusted P-value ≤ 0.2) were analyzed with DAVID Bioinformatics Resources 6.8 (https://david.ncifcrf.gov/home.jsp). The raw datasets and normalized count data for each gene have been deposited in NCBI (GSE101550).
Retinal explant culture
Retinal explant culture was described previously [22]. In brief, retinas were isolated from E13.5 wildtype and Nrf1f/f;Six3-Cre embryos and cut in 4 pieces, then placed on laminin-coated coverslips and cultured in Neurobasal media containing N2 supplement and penicillin-streptomycin (Thermo Fisher Science). Images were examined and collected using an Olympus IX-70 inverted microscope.
In situ hybridization
Embryo heads from wildtype and Nrf1f/f;Six3-Cre at E13.5 were dissected and fixed in fresh 10% neutral buffered formalin for 24 h. Samples were washed with PBS then dehydrated with serial ethanol and embedded in paraffin. Sections were cut to 7 μm or 10 μm in thickness. In situ hybridization was performed as described previously [23]. Antisense Idh1 (957 bp) and Ldha (950 bp) probes were cloned by reverse transcriptase PCR using Idh1 probe F (5’-AGGTTCTGTGGTGGAGATGC-3′), Idh1 probe R (5’-GACGTCTCTTGCCCTTTCTG-3′), Ldha probe F (5’-TCCGTTACCTGATGGGAGAG-3′) and Ldha probe R (5’-ACACTTGGGTGGTTGGTTCC-3′). RNAscope in situ hybridization (ISH) was performed using the RNAscope 2.5 HD Detection Reagents-Brown kit following manufacturer’s protocol (cat# 322310, Advanced Cell Diagnostics, Newark, CA). According to instructions, each mRNA molecule hybridized to a probe appears as a separate brown color dot. The probes used were mouse Cpt1a-C1 (cat# 443071) and mouse Slc16a1-C1 (cat# 423661).
Quantitative reverse transcriptase PCR (qRT-PCR)
Eight retinas from wildtype or Nrf1f/f;Six3-Cre embryos at E13.5 or one retina from wildtype or Nrf1f/f; Rho-iCre at 6 weeks old of multiple littermates were pooled, and RNAs were extracted using TRI reagent (Sigma). First-strand cDNA was synthesized using the Superscript III First-Strand Synthesis System (Thermo Fisher Scientific). Real-time PCR was performed using CFX Connect Real-Time System (BioRad, Hercules, CA) with SsoAdvanced Universal SYBR Green Supermix (BioRad). Relative RNA levels were normalized to that of β-actin. Sequences of PCR primers are listed in Table 1.
Table 1.
Primer sets used for qRT-PCR
| Gene | Position | 5′-sequence-3’ |
|---|---|---|
| b-actin | Forward | CAACGGCTCCGGCATGTGC |
| Reverse | CTCTTGCTCTGGGCCTCG | |
| CCND1 | Forward | GCACTTTTGGTCAGCTAGCT |
| Reverse | GACATGGCCCTAAACCTTCT | |
| Gli1 | Forward | ACTGGGGTGAGTTCCCTTCT |
| Reverse | AGGACTACCCAGCAAATCCT | |
| Mapt | Forward | AATGGAAGACCATGCTGGAG |
| Reverse | TCCCAATCTGAGTCCCAAAG | |
| Ret | Forward | TGGCACACCTCTGCTCTATG |
| Reverse | CTGTTCCCAGGAACTGTGGT | |
| Stmn3 | Forward | CCCGAACACCATCTACCAGT |
| Reverse | CTTCTGCAGCTCTTCCAAGG | |
| Ncan | Forward | GTGGCTGCTTCTCCTAGTGG |
| Reverse | AATGTCTCGCAGGGAGCTTA | |
| Ina | Forward | TTCGGGAATACCAGGACTTG |
| Reverse | GTGCTAAACCGCGTCTCTTC | |
| Islr2 | Forward | CTCTGCCTTTTCAAGGATGC |
| Reverse | CGCTGAGTTGAAAGGCCTAC | |
| Nell2 | Forward | CACAGTTGACCTTTCCTGCT |
| Reverse | CAGCACAAATGGCCATTCTT | |
| Stmn2 | Forward | GCAATGGCCTACAAGGAAAA |
| Reverse | GGTGGCTTCAAGATCAGCTC | |
| Gap43 | Forward | GTGCTGCTAAAGCTACCACT |
| Reverse | CTTCAGAGTGGAGCTGAGAA | |
| Nrn1 | Forward | CCAGGGGAATGACTTCAAGA |
| Reverse | TTTCGCTTTTCTGGAGGAGA | |
| Syt4 | Forward | TGTTGTAGGTGATGGTTTCA |
| Reverse | AGACCATGGTTCTTAGGTGA | |
| Dcx | Forward | ACAGATGTCAACCGGGAAAG |
| Reverse | TCGTTCGTCAAAATGTCCAA | |
| Pou4f1 | Forward | AGGCCTATTTGCCGTACAA |
| Reverse | CGTCTCACACCCTCCTCAGT | |
| Irx4 | Forward | GAGACCACCAGCACACTGAA |
| Reverse | AGGTGGAAACCTGTGTGAGG | |
| Cx3cr1 | Forward | AGCCCAGGGGAAGAAATAGA |
| Reverse | CTCTGTTGGCTCCAGTCTCC | |
| Capn3 | Forward | GCTTCTGGAGGAAGACGATG |
| Reverse | TTTGGGAACCTCGTAGATGG | |
| Igfbp7 | Forward | GGAAAATCTGGCCATTCAGA |
| Reverse | TGCGTGGCACTCATACTCTC | |
| Vit | Forward | GCGTCTACGCGTCTTACTCC |
| Reverse | CCCTTTTGGGGCTTACTTTC | |
| Nid1 | Forward | ACCATCACCTTCCAGGAGTG |
| Reverse | GCATAGCGCAAGATCCTCTC | |
| Stab1 | Forward | ACAAGATCTTCAGCCGCCTA |
| Reverse | AGTTTGTCACGGTGGTCCTC | |
| Spp1 | Forward | TGCACCCAGATCCTATAGCC |
| Reverse | CTCCATCGTCATCATCATCG | |
| Tgfbi | Forward | GGATGTCCTGAAGGGAGACA |
| Reverse | ATTGGTGGGAGCAAAAACAG | |
| Cox4i2 | Forward | AGCTGAGCCAAGCAGAGAAG |
| Reverse | GCCCATCACTGTCTTCCATT | |
| Idh1 | Forward | AGGTTCTGTGGTGGAGATGC |
| Reverse | GACGCCCACGTTGTATTTCT | |
| Dna2 | Forward | CGAAGTTCTGTGCATCCTGA |
| Reverse | TTCTCAGACACCGAATGCTG | |
| Gdap1 | Forward | CTGTGAGGCCACTCAGATCA |
| Reverse | TGAGCTCAGGATGCAAAATG | |
| Shmt2 | Forward | CTCTTTGCTTCGGACCACTC |
| Reverse | TTCTCCCTCTGCAGAAGCTC | |
| Cpt1a | Forward | CCAGGCTACAGTGGGACATT |
| Reverse | AAGGAATGCAGGTCCACATC | |
| Sardh | Forward | ACTCGGTTGTCTTCCCACAC |
| Reverse | CCTGTCGCTCTTGAAACACA | |
| Ucp2 | Forward | GCCACTTCACTTCTGCCTTC |
| Reverse | GAAGGCATGAACCCCTTGTA | |
| Pmaip1 | Forward | CCCAGATTGGGGACCTTAGT |
| Reverse | AGTTATGTCCGGTGCACTCC | |
| Rab32 | Forward | CTCTTCTCCCAGCACTACCG |
| Reverse | CAAATGCTCCAAGAGCTTCC | |
| Ldha | Forward | AGGCTCCCCAGAACAAGATT |
| Reverse | TCTCGCCCTTGAGTTTGTCT | |
| HK1 | Forward | GAAGCCAAATGGGACTGTGT |
| Reverse | CACGCACAGATTGGTTATGC | |
| Pfkp | Forward | GAAGCCAAATGGGACTGTGT |
| Reverse | CACGCACAGATTGGTTATGC | |
| Tpi1 | Forward | CCTGGCCTATGAACCTGTGT |
| Reverse | CAGGTTGCTCCAGTCACAGA | |
| Pgam2 | Forward | AGGAGCTGCCTACCTGTGAA |
| Reverse | GGGCTGCAATAAGCACTCTC | |
| Mfn1 | Forward | GCTGTCAGAGCCCATCTTTC |
| Reverse | CAGCCCACTGTTTTCCAAAT | |
| Mfn2 | Forward | GTCCTGGACGTCAAAGGGTA |
| Reverse | GCAGAACTTTGTCCCAGA | |
| Opa1 | Forward | GATGACACGCTCTCCAGTGA |
| Reverse | TCGGGGCTAACAGTACAACC |
Transmission electron microscopy
Eyeballs were fixed with 3% glutaraldehyde and 2% paraformaldehyde overnight at 4 °C. Retinas were washed and treated with 0.1% cacodylate-buffered tannic acid, post-fixed with 1% osmium tetroxide, stained en bloc with 1% uranyl acetate, and dehydrated with an ethanol gradient series. The samples were embedded in epon and sectioned with a JLB ultracut microtome (Leica, Wetzlar, Germany). Images were examined with a JEM 1010 REM (JEOL, Peabody, MA) and collected digitally. Fiji was used to analyze the size and circularity of inner segment (IS) and mitochondria [24].
Mitochondrial DNA quantitation
Quantification of the relative copy number of mitochondrial DNA present per nuclear genome was performed as previously described [25]. Mitochondrial DNA and genomic Pecam DNA were amplified and analyzed by quantitative PCR (ΔΔC(t) method). PCR primers used to amplify mitochondrial DNA were mtDNAf (5’-CCTATCACCCTTGCCATCAT-3′) and mtDNAr (5’-GAGGCTGTTGCTTGTGTGAC-3′). PCR primers to amplify nuclear DNA were Pecamf (5’-ATGGAAAGCCTGCCATCATG-3′) and Pecamr (5’-TCCTTGTTGTTCAGCATCAC-3′).
Electroretinography (ERG)
Mice were dark-adapted overnight and then anesthetized under infrared illumination by ketamine/xylazine/acepromazine through intraperitoneal injection (94/5/1 mg/kg), and pupils were dilated with 1% tropicamide and 2.5% phenylephrine topical eye drops (Bausch & Lomb, Tampa, FL). Body temperature was maintained at 35 °C to 37 °C by circulating 43.5 °C water through a plastic heating coil wrapped around the body. Stimulus-dependent transcorneal potential changes from both eyes were recorded simultaneously (UTAS BigShot system; LKC Technologies, Gaithersburg, MD) following the delivery of a white light flash with an intensity of 25 Candela sec m−2, as described previously [26]. The interstimulus interval was 120 s, and responses from 3 independent measurements were averaged and analyzed. Photopic ERG recordings ensued immediately after scotopic recordings by exposing the animals to a white background light of 30 Candela m−2 for 10 min. Transcorneal potential changes were then elicited by flashes of 25 Candela sec m−2 in intensity and presented at 1 Hz for 90 s, averaged, and then analyzed. A typical recording session lasted 1 h, and 5 μl of sterile-filtered PBS was applied every 20 min to ensure good electrical contact and delay the formation of corneal clouding and cataract.
Experimental design
Six E13.5 littermate embryos of each genotype (wildtype and Nrf1f/f;Six3-Cre) were used for counting BrdU+ and PH3+ cells’ RPCs. Four sections near the central retinas from each embryo were stained with BrdU or PH3. The most representative sections from each embryo were used for cell counting. Total RNAs were isolated and pooled from 9 pairs of E13.5 wildtype and Nrf1f/f;Six3-Cre retinas for RNA-seq. For RNA-seq data validation using qRT-PCR, 3 sets of 6 pairs of E13.5 wildtype and Nrf1f/f;Six3-Cre retinas were isolated and pooled, and 2 independent experiments were conducted. For each qRT-PCR experiment, total RNAs from 4 pairs of E13.5 wildtype and Nrf1f/f;Six3-Cre retinas were isolated and pooled, and 5 sets of independent experiments were conducted. The ratio of Nrf1f/f;Six3-Cre to wildtype expression was calculated for each experiment and averaged for further analysis. For counting photoreceptors, 20 littermates from each genotype (wildtype and Nrf1f/f;Rho-iCre) were used for the study. Five littermates per genotype were sacrificed, and 4 sections from the central area of each retina were stained. One representative section from each sample was used to count the number of rows of photoreceptors at each study time point (3, 6, 7, and 8 weeks). For electron microscopy analysis, 2 pairs of wildtype and Nrf1f/f;Rho-iCre littermates were used. Images were collected from both mouse retinas. Four images of the IS of each genotype were used to quantify the shape and size of the IS and number of mitochondria. For ERG, 3 pairs of wildtype and Nrf1f/f;Rho-iCre littermates were used.
Statistical analysis
All data are presented as mean ± standard deviation for each genotype. For all comparisons between genotypes, a two-tailed two-sample student’s t-test was used for all measurements. Results were considered significant when P < 0.05. Statistical tests were conducted using Excel (Microsoft, Redmond, WA).
Results
Nrf1 expression in the developing retina
To determine the expression and function of Nrf1 in the retina, we generated Nrf1LacZ and Nrf1flox targeted mouse lines (Fig. 1a, b). The Nrf1LacZ knock-in allele contains a LacZ cassette, which was used to trace the spatiotemporal expression of Nrf1. We first examined the expression of Nrf1 in developing and adult retinas. In E13.5 developing retinas, strong LacZ activity was detected near the apical and basal layers of the neural retina, suggesting Nrf1 is highly expressed in developing RGCs and photoreceptor precursor cells (Fig. 1c). Notably, weaker LacZ activity could also be detected in the neuroblast layer where proliferating RPCs and postmitotic precursor cells reside (Fig. 1c). Along the developmental progression, a similar pattern of LacZ expression was observed in E16.5 and P0 retinas (data not shown). In adult retinas, robust LacZ activity was observed in the ganglion cell layer and outer nuclear layer (ONL), where the metabolic activity is high (Fig. 1d) [27], while a moderate level of LacZ activity was observed in the inner nuclear layer (INL). The dynamic expression pattern of Nrf1 in both developing and mature retinas suggests that Nrf1 may play multiple roles in proliferating RPCs in the developing retina and in the differentiated retinal neurons.
To determine the functions of Nrf1 in RPCs and differentiated retinal neurons, we performed conditional knockout of Nrf1 by breeding the Nrf1flox allele with either Six3-Cre to delete Nrf1 in the proliferating RPCs or with Rho-iCre to delete Nrf1 in the rod PRs, respectively (Fig. 1e). During retinal development, the Six3-Cre transgenic line begins to activate Cre expression in the central retina at E11 [28], and the Rho-iCre transgenic line starts to activate Cre activity in differentiated rod PRs at P7 [26]. By immunostaining, Nrf1 protein was detected in developing RGCs and photoreceptor precursor cells, as well as in the neuroblast layer at E13.5 (Fig. 1f) and in cells in all nuclear layers in adult retinas (Fig. 1h). Consistent with the onset of Cre expression in both lines, the expression of Nrf1 protein was completely abolished in the central Nrf1f/f; Six3-Cre retina (compare Fig. 1f and g) and in the rod PRs of Nrf1f/f; Rho-iCre retinas (compare Fig. 1h and g) respectively, suggesting effective conditional deletion of Nrf1 in RPCs by Six3-Cre or in rod PRs by Rho-iCre.
Deleting Nrf1 in RPCs causes RGC loss and retinal degeneration
To determine whether deleting Nrf1 in embryonic retinas affects retinal development, we first examined the histology and morphology of Six3-Cre-mediated Nrf1 mutant retinas (Nrf1f/f;Six3-Cre) at different developmental stages. At E16.5, Nrf1f/f; Six3-Cre retinas were substantially smaller and thinner than those of the wildtype retinas (Fig. 2a, b), causing a large sub-retinal space between the retina and the pigmented epithelium. The central regions of Nrf1f/f;Six3-Cre retinas near the optic disc were completely disrupted and acellular (arrowheads in Fig. 2b). At P20, Nrf1f/f;Six3-Cre retinas were relatively thinner than those of wildtype retinas. Decreased cell numbers in all cellular layers were observed with near complete abolishment of RGCs. Although the stereotypic laminar structure was retained in Nrf1-mutant retinas, the cells in each laminar layer were not properly aligned as in control retinas (Fig. 2c, d). In 7-month-old Nrf1f/f;Six3-Cre retinas, the number of retinal cells was further reduced, and the laminar layers were completely disrupted (Fig. 2e, f). The surface of the whole retina from Nrf1f/f;Six3-Cre was much smaller, underlying only a limited area near the optic disc in the eyeball (Fig. 2g, h). There were no visible optic nerves or optic chiasms in Nrf1f/f;Six3-Cre mice (Fig. 2i, j). These data suggest that deleting Nrf1 in RPCs causes substantial RGC loss followed by the degeneration of the entire retina.
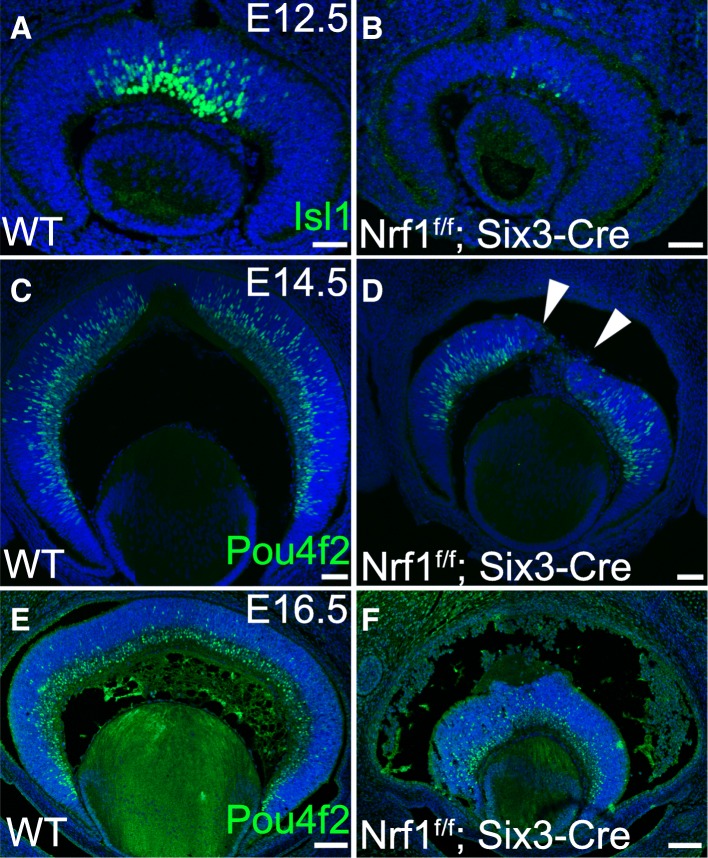
Fig. 2.
Loss of retinal ganglion cells and severe retinal degeneration in Nrf1-deficient RPCs. (a–f) Hematoxylin and eosin staining of retinal sections from wildtype (a, c and e) and Nrf1f/f;Six3-Cre (b, d, and f) at E16.5, P20, and 7 months old. (g, h) Eyeballs from wildtype (g) and Nrf1f/f;Six3-Cre (h) animals. The peripheral rim of underlying retinas is plotted with dotted lines. (i, j) Ventral view of the brains showing the optic nerve and optic chiasm in wildtype (i) and Nrf1f/f;Six3-Cre (j) animals. Scale bars: 50 μm. ONL: outer nuclear layer. INL: inner nuclear layer. GCL: ganglion cell layer. WT: wildtype. NBL: neural blast layer
Delayed RGC differentiation, defective RGC migration, and apoptotic RGCs in Nrf1f/f;Six3-Cre retinas
Because a significant loss of RGCs was seen in Nrf1f/f;Six3-Cre retinas, we examined whether and how the RGC differentiation program was affected. RGCs are the first retinal cell type to differentiate from Atoh7-expressing precursor cells during retinogenesis. RGC differentiation is marked by the onset of Pou4f2 and Isl1 expression in the central retina around E12 [29, 30]. To determine when RGCs began to differentiate in Nrf1f/f;Six3-Cre retina, we monitored Isl1 and Pou4f2 expression by immunostaining at different embryonic stages.
In E12.5 wildtype retinas, while the newly differentiated Isl1+ RGCs could be readily detected in the central retina (Fig. 3a), only a few Isl1+ cells were present in Nrf1 mutant retinas (Fig. 3b). At E14.5, while differentiating Pou4f2+ RGCs were widespread across the neuroblast and ganglion cell layers in wildtype retinas (Fig. 3c), fewer Pou4f2+ RGCs were detected in Nrf1 mutant retinas (Fig. 3d). No clear ganglion cell layer could be seen in Nrf1 mutant retinas. Furthermore, Pou4f2+ RGCs were distributed unevenly and formed patched clumps in the central region of the mutant retina (arrowheads in Fig. 3d). In E16.5 wildtype retinas, a distinct ganglion cell layer was formed, and newly differentiated Pou4f2+ RGCs were seen in the neuroblast layer (Fig. 3e). In contrast, a much thinner ganglion cell layer was observed in Nrf1f/f;Six3-Cre retinas, and Pou4f2+ RGCs were spread to the peripheral region (Fig. 3f). Together, these data suggest that RGC differentiation was delayed, and newly differentiated RGCs had defects in migrating toward the vitreous layer in the Nrf1f/f;Six3-Cre retinas.
Fig. 3.
Delayed onset of RGC differentiation in Nrf1f/f;Six3-Cre retina. (a–f) Immunostaining of wildtype (a, c, and e) and Nrf1f/f;Six3-Cre (b, d, and f) retinas. (a, b) E12.5 retinal sections labeled with anti-Isl1 antibody. (c, d) E14.5 and (e, f) E16.5 retinal sections labeled with anti-Pou4f2 antibody. Arrowheads indicate clumped Pou4f2+ cells in the central area of Nrf1f/f;Six3-Cre retina. Scale bars: 50 μm in a–d, 100 μm in e and f. WT: wildtype
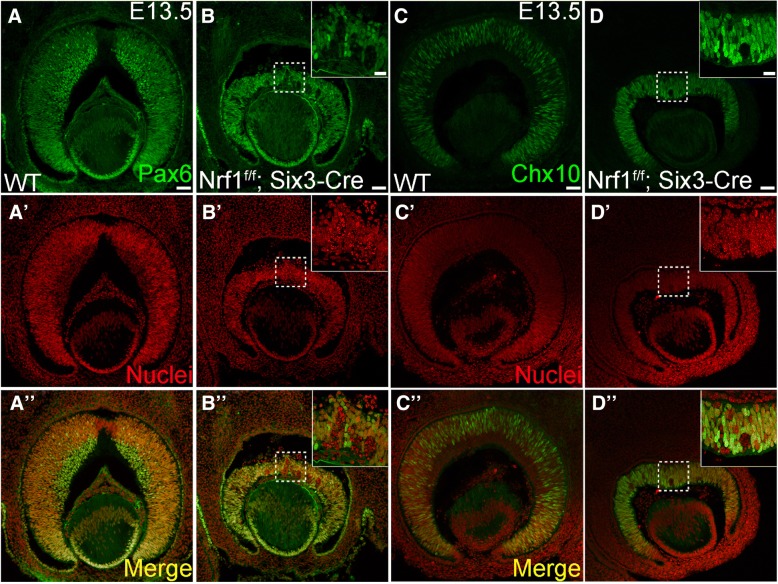
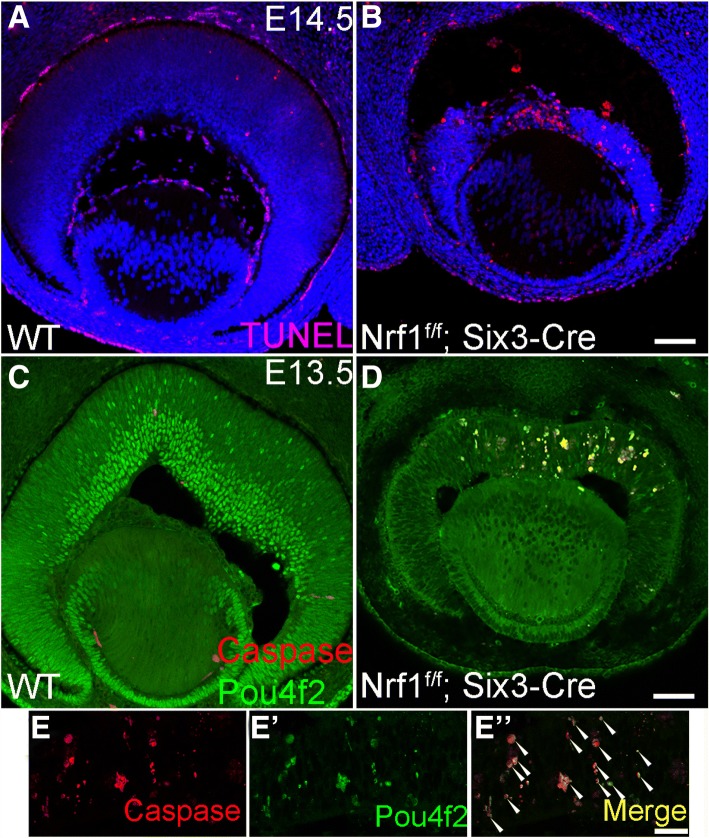
To detect RPCs, wildtype and Nrf1f/f;Six3-Cre retinal sections were immunolabeled with anti-Pax6 or Chx10 antibodies (Fig. 4a–d). Pax6 and Chx10 were expressed in both wildtype and Nrf1f/f;Six3-Cre retinas. However, Pax6+ or Chx10+ RPCs were unevenly distributed in the central region of Nrf1f/f retinas compared to the peripheral region (Fig. 4b, d). Interestingly, several clumps of RPCs lacking Pax6 expression were formed in Nrf1 mutant retinas, and the nuclei of these Pax6-negative cells appeared granulated, suggesting these were cells undergoing apoptosis (Fig. 4b, b’, b′′). Similarly, mis-patterned Chx10 expression and granular-shaped nuclei were observed in the central region of Nrf1f/f;Six3-Cre retinas (Fig. 4d, d’, d′′). Consistently, Nrf1f/f;Six3-Cre retinas contained significantly more apoptotic cells than wildtype retinas (Fig. 5a, b). The majority of apoptotic cells were found in the central region of Nrf1-mutant retinas. In addition, these apoptotic cells (marked by cleaved caspase 3 expression) were Pou4f2+ (Fig. 5c-e), indicating that ganglion cells had differentiated but could not migrate to the RGC layer and eventually died in situ.
Fig. 4.
Expression of progenitor cell markers in wildtype and Nrf1f/f;Six3-Cre retina. (a–d) Immunostaining of E13.5 wildtype (a, c) and Nrf1f/f;Six3-Cre (b, d) retinal sections using anti-Pax6 antibody (a, b) and anti-Chx10 antibody (c, d). (a′–d′) Nuclei staining. (a′′–d′′) Merged images. Insets show higher magnification images of indicated areas. Scale bars: 50 μm. WT: wildtype
Fig. 5.
Differentiated RGCs undergo apoptosis in Nrf1f/f;Six3-Cre retina. (a, b) TUNEL assay on E14.5 wildtype (a) and Nrf1f/f;Six3-Cre (b) retinal sections. (c-e) E13.5 wildtype (e) and Nrf1f/f;Six3-Cre (d, e) sections labeled with anti-Pou4f2 and anti-cleaved caspase 3 antibodies. (e, e′ and e′′) Higher magnification images of Nrf1f/f;Six3-Cre sections labeled with anti-caspase (e) and Pou4f2 (e′) antibodies. (e′′) Merged images. Arrowheads indicate cells that are double positive for caspase and Pou4f2. Scale bars: 50 μm in a and b, 100 μm in c and d. WT: wildtype
Severe reduction of proliferation in Nrf1f/f;Six3-Cre retina
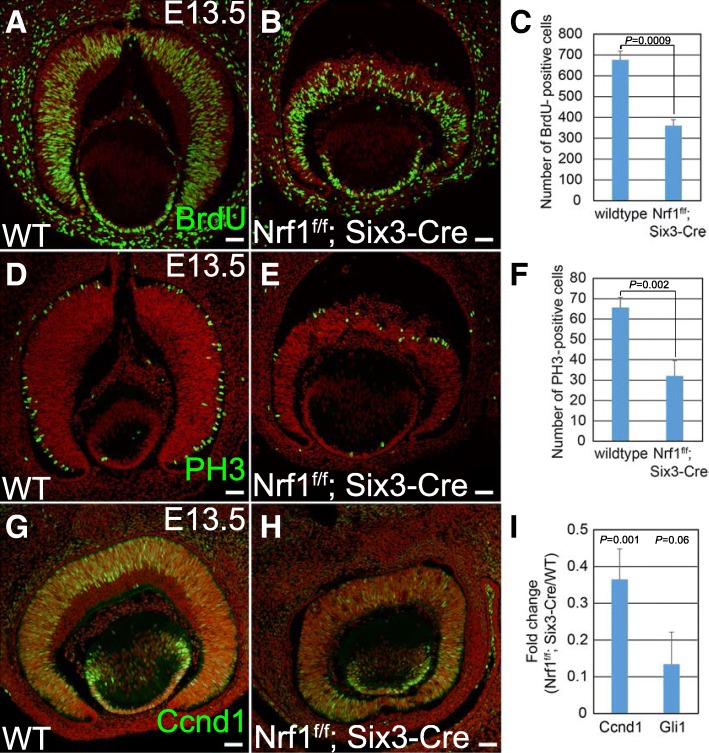
The Nrf1f/f;Six3-Cre retina was substantially smaller and thinner than the wildtype retina, suggesting that the proliferation of RPCs in the Nrf1-mutant retina was compromised. To examine this phenotype, wildtype and Nrf1f/f;Six3-Cre retinas were immuno-labeled with several cell cycle markers. To detect S-phase proliferating RPCs, we pulse-labeled E13.5 embryos with BrdU and then conducted immunostaining using anti-BrdU antibody on retinal sections. We counted the number of BrdU+ cells in sections and found that the number of BrdU+ S-phase RPCs were reduced to ~ 50% in Nrf1f/f;Six3-Cre retinas compared to wildtype retinas (Fig. 6a-c, P = 0.0009). We then conducted immunostaining using anti-PH3 and anti-cyclin D1 (Ccnd1) antibodies on retinal sections to detect RPCs in M-phase and G1-phase, respectively. The number of PH3+ M-phase RPCs per section from Nrf1f/f;Six3-Cre was also reduced to ~ 50% compared to wildtype retinas (Fig. 6d-f, P = 0.002), and Ccnd1+ RPCs were nearly absent in the central region of Nrf1-mutant retinas (Fig. 6g, h). The PH3+ RPCs in the Nrf1-mutants were not properly positioned at the apical side as in control retinas (Fig. 6d, e). In addition, using qRT-PCR, we found that the expression levels of Ccnd1 were reduced to ~ 10% in Nrf1-mutant retinas compared with wildtype retina, and Gli1, the key downstream effector of Shh pathway in RPCs [31, 32], was reduced to ~ 30% (Fig. 6i, Ccnd1: P = 0.001, Gli1: P = 0.006). Other Shh pathway genes, such as Shh and Ptch1, were also downregulated in Nrf1-mutant retinas (in GSE101550 dataset described in next section). Together, these data indicate that the RPC proliferation is reduced in Nrf1-mutant retinas.
Fig. 6.
Reduced cell proliferation in Nrf1f/f;Six3-Cre retina. (a, b) BrdU labeling of E13.5 wildtype (a) and Nrf1f/f;Six3-Cre (b) retinal sections. (c) The number of BrdU+ cells in wildtype and Nrf1f/f;Six3-Cre sections. (d, e) E13.5 wildtype and Nrf1f/f;Six3-Cre retinal sections labeled with anti-PH3 antibody. (f) The number of PH3+ cells in wildtype and Nrf1f/f;Six3-Cre retinal sections. (g, h) Immunostaining of E13.5 wildtype and Nrf1f/f;Six3-Cre retinal sections with anti-Ccnd1 antibody. (i) qRT-PCR analysis of Ccnd1 and Gli1 in E13.5 WT and Nrf1f/f;Six3-Cre retinas. Scale 50 μm. WT: wildtype
Identification of Nrf1-dependent retina-expressed genes at E13.5
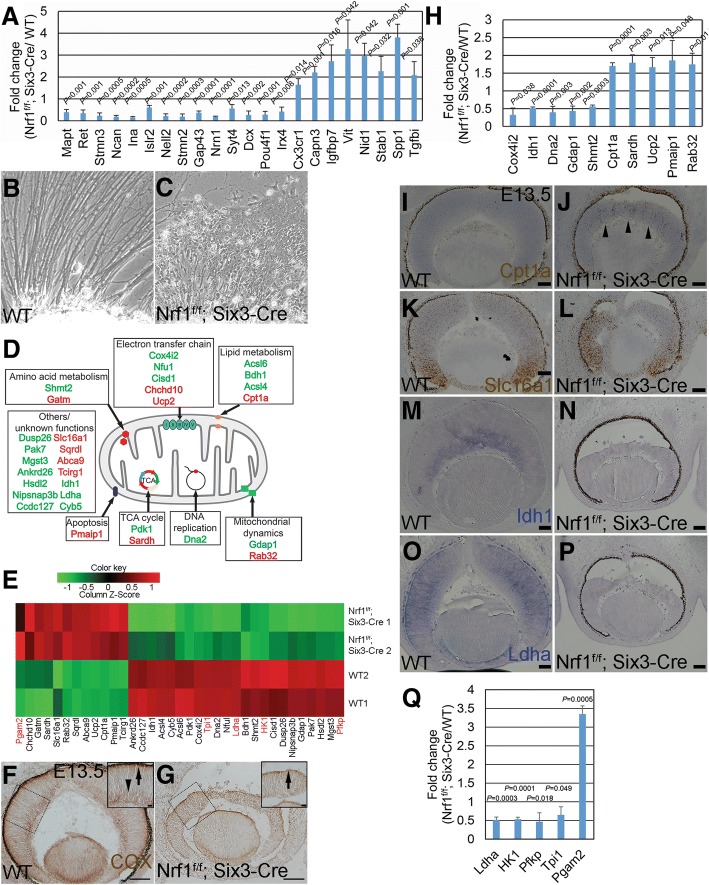
To further investigate how Nrf1 regulates retinal development, we performed RNA-seq analysis on E13.5 wildtype and Nrf1f/f;Six3-Cre retinas to identify genes whose expression was affected in the Nrf1f/f;Six3-Cre retinas. The data discussed here have been deposited in NCBI’s Gene Expression Omnibus [33] and are accessible through GEO Series accession number GSE101550 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE101550). The analysis revealed 488 downregulated and 595 upregulated genes in Nrf1-mutant retinas compared to control retinas. We first conducted qRT-PCR analysis on the 22 most affected genes (14 down- and 8 up-regulated) and found that their relative expression levels between the E13.5 control and Nrf1-mutant retinas are consistent with the RNA-seq output, demonstrating the reliability of the RNA-seq data (Fig. 7a). Using gene ontology for biological process analysis (GO-BP) of these gene lists, the top 5 categories of the downregulated genes are genes involved in nervous system development, neurogenesis, neuron differentiation, generation of neurons, and neuron projection development (Table 2), and the upregulated genes are involved in cell adhesion, biological adhesion, regulation of cell projection, angiogenesis, and positive regulation of developmental process (Table 3).
Fig. 7.
RNA-seq identifies genes involved in neurite outgrowth, mitochondrial functions, and energy production in Nrf1f/f;Six3-Cre retina. (a) qRT-PCR analysis of the 22 top affected genes identified in Nrf1f/f;Six3-Cre retinas, confirming changes in mRNA expression detected by RNA-seq. (b, c) Representative images of retinal explant cultures from E13.5 wildtype (b) and Nrf1f/f;Six3-Cre (c) embryos. (d) Schematic mapping of mitochondrial and functional annotation of upregulated (red) and downregulated (green) genes in Nrf1f/f;Six3-Cre retinas detected by RNA-seq. (e) Heatmap from Nrf1f/f; Six3-Cre RNA-seq showing 30 mitochondrial and 5 glycolytic genes whose expression changed. Mitochondrial genes are labeled in black, and glycolytic genes are labeled in red. (f, g) COX activity in E13.5 wildtype (f) and Nrf1f/f;Six3-Cre (g) retinas. Insets show higher magnification images of the indicated areas. Arrows indicate COX activities in the future photoreceptor layer; arrowheads indicate COX activities in ganglion cell layer. (h) qRT-PCR analysis of a subset of affected mitochondria genes between wildtype and Nrf1f/f;Six3-Cre retinas confirming changes of mRNA expression detected by RNA-seq. (i–p) In situ hybridization of mitochondrion-associated genes on E13.5 wildtype (i, k, m, and o) and Nrf1f/f;Six3-Cre (j, l, n, and p) retinal sections. Arrowheads indicate increased expression of Cpt1a in the central area of Nrf1f/f;Six3-Cre retina. (q) qRT-PCR analysis of glycolytic genes of wildtype and Nrf1f/f;Six3-Cre retinas. Scale bars: 100 μm in f and g, 20 μm in insets and 50 μm in i–j. WT: wildtype
Table 2.
Top 10 GO terms relevant to 488 downregulated genes in E13.5 Nrf1f/f; Six3-Cre retinas
| Rank | GO Category | GO ID | GO Term | Number of Focused Genes | P Value | FDR |
|---|---|---|---|---|---|---|
| 1 | GOTERM_BP_FAT | GO:0030182 | nervous system development | 156 | 1.70E-39 | 6.96E-36 |
| 2 | GOTERM_BP_FAT | GO:0048666 | neurogenesis | 126 | 1.80E-35 | 3.80E-32 |
| 3 | GOTERM_BP_FAT | GO:0031175 | neuron differentiation | 114 | 1.40E-34 | 2.00E-31 |
| 4 | GOTERM_BP_FAT | GO:0048667 | generation of neurons | 119 | 8.00E-34 | 8.40E-31 |
| 5 | GOTERM_BP_FAT | GO:0000904 | neuron projection development | 91 | 6.90E-33 | 5.80E-30 |
| 6 | GOTERM_BP_FAT | GO:0048812 | neuron development | 99 | 7.00E-33 | 4.90E-30 |
| 7 | GOTERM_BP_FAT | GO:0007409 | neuron projection morphogenesis | 68 | 5.00E-29 | 3.00E-26 |
| 8 | GOTERM_BP_FAT | GO:0030030 | cell morphogenesis involved in neuron differentiation | 62 | 5.50E-26 | 2.90E-23 |
| 9 | GOTERM_BP_FAT | GO:0019226 | axonogenesis | 55 | 2.70E-25 | 1.30E-22 |
| 10 | GOTERM_BP_FAT | GO:0048858 | cell projection organization | 99 | 1.40E-24 | 5.90E-22 |
| 1 | GOTERM_CC_FAT | GO:0030424 | axon | 54 | 4.00E-27 | 1.30E-24 |
| 2 | GOTERM_CC_FAT | GO:0045202 | synapse | 57 | 5.30E-23 | 8.90E-21 |
| 3 | GOTERM_CC_FAT | GO:0043025 | neuronal cell body | 58 | 1.40E-22 | 1.60E-20 |
| 4 | GOTERM_CC_FAT | GO:0043005 | neuron projection | 51 | 5.70E-22 | 4.80E-20 |
| 5 | GOTERM_CC_FAT | GO:0016020 | membrane | 243 | 9.40E-16 | 6.00E-14 |
| 6 | GOTERM_CC_FAT | GO:0030425 | dendrite | 46 | 1.90E-15 | 1.10E-13 |
| 7 | GOTERM_CC_FAT | GO:0010469 | postsynaptic density | 31 | 1.10E-14 | 1.70E-12 |
| 8 | GOTERM_CC_FAT | GO:0030054 | cell junction | 53 | 1.80E-13 | 7.50E-12 |
| 9 | GOTERM_CC_FAT | GO:0043195 | terminal bouton | 21 | 1.10E-12 | 4.10E-11 |
| 10 | GOTERM_CC_FAT | GO:0030426 | growth cone | 24 | 1.80E-12 | 6.10E-11 |
| 1 | GOTERM_MF_FAT | GO:0008092 | cytoskeletal protein binding | 52 | 2.40E-10 | 2.40E-07 |
| 2 | GOTERM_MF_FAT | GO:0022836 | gated channel activity | 26 | 6.10E-08 | 2.70E-05 |
| 3 | GOTERM_MF_FAT | GO:0005216 | ion channel activity | 29 | 2.60E-07 | 7.50E-05 |
| 4 | GOTERM_MF_FAT | GO:0022838 | substrate-specific channel activity | 29 | 4.90E-07 | 1.10E-04 |
| 5 | GOTERM_MF_FAT | GO:0015631 | tubulin binding | 23 | 7.60E-07 | 1.30E-04 |
| 6 | GOTERM_MF_FAT | GO:0005261 | cation channel activity | 23 | 1.20E-06 | 1.80E-04 |
| 7 | GOTERM_MF_FAT | GO:0022803 | passive transmembrane transporter activity | 29 | 2.00E-06 | 2.50E-04 |
| 8 | GOTERM_MF_FAT | GO:0015267 | channel activity | 29 | 2.00E-06 | 2.50E-04 |
| 9 | GOTERM_MF_FAT | GO:0019905 | syntaxin binding | 12 | 7.10E-06 | 7.80E-04 |
| 10 | GOTERM_MF_FAT | GO:0017075 | syntaxin-1 binding | 7 | 1.10E-05 | 1.00E-03 |
GO gene ontology, FDR false discovery rate
Table 3.
Top 10 GO terms relevant to 595 upregulated genes in E13.5 Nrf1f/f; Six3-Cre retinas
| Rank | GO Category | GO ID | GO Term | Number of Focused Genes | P Value | FDR |
|---|---|---|---|---|---|---|
| 1 | GOTERM_BP_FAT | GO:0007155 | cell adhesion | 70 | 1.40E-22 | 5.00E-33 |
| 2 | GOTERM_BP_FAT | GO:0022610 | biological adhesion | 70 | 1.60E-22 | 3.90E-33 |
| 3 | GOTERM_BP_FAT | GO:0042127 | regulation of cell projection | 57 | 4.80E-15 | 3.70E-31 |
| 4 | GOTERM_BP_FAT | GO:0001525 | angiogenesis | 23 | 2.40E-10 | 3.30E-31 |
| 5 | GOTERM_BP_FAT | GO:0051094 | positive regulation of developmental process | 29 | 9.60E-10 | 7.80E-31 |
| 6 | GOTERM_BP_FAT | GO:0007423 | sensory organ development | 31 | 1.00E-09 | 9.90E-31 |
| 7 | GOTERM_BP_FAT | GO:0048514 | blood vessel morphogenesis | 27 | 1.20E-09 | 1.10E-29 |
| 8 | GOTERM_BP_FAT | GO:0001568 | blood vessel development | 30 | 2.00E-09 | 1.10E-29 |
| 9 | GOTERM_BP_FAT | GO:0009611 | response to wounding | 36 | 2.10E-09 | 1.20E-28 |
| 10 | GOTERM_BP_FAT | GO:0001944 | vasculature development | 30 | 9.60E-09 | 1.20E-28 |
| 1 | GOTERM_CC_FAT | GO:0031012 | extracellular matrix | 77 | 7.30E-29 | 4.10E-26 |
| 2 | GOTERM_CC_FAT | GO:0005578 | proteinaceous extracellular matrix | 62 | 9.90E-27 | 2.80E-24 |
| 3 | GOTERM_CC_FAT | GO:0044421 | extracellular region part | 231 | 9.50E-24 | 1.80E-21 |
| 4 | GOTERM_CC_FAT | GO:0031982 | membrane-bounded vehicle | 206 | 5.60E-21 | 7.90E-19 |
| 5 | GOTERM_CC_FAT | GO:0005576 | extracellular region | 245 | 9.40E-21 | 1.10E-18 |
| 6 | GOTERM_CC_FAT | GO:0009986 | cell surface | 88 | 1.30E-20 | 1.20E-18 |
| 7 | GOTERM_CC_FAT | GO:0044420 | extracellular matrix component | 33 | 4.50E-19 | 3.70E-17 |
| 8 | GOTERM_CC_FAT | GO:1903561 | extracellular vesicle | 168 | 2.70E-18 | 1.90E-16 |
| 9 | GOTERM_CC_FAT | GO:0043230 | extracellular organelle | 168 | 3.50E-18 | 2.20E-16 |
| 10 | GOTERM_CC_FAT | GO:0070062 | extracellular exosome | 166 | 9.20E-18 | 5.20E-16 |
| 1 | GOTERM_MF_FAT | GO:0005212 | structural constituent of eye lens | 16 | 9.60E-18 | 1.10E-17 |
| 2 | GOTERM_MF_FAT | GO:0005515 | protein biding | 196 | 9.70E-14 | 1.60E-12 |
| 3 | GOTERM_MF_FAT | GO:0005178 | integrin binding | 22 | 1.20E-12 | 1.80E-11 |
| 4 | GOTERM_MF_FAT | GO:0005518 | collagen binding | 16 | 1.10E-10 | 4.50E-10 |
| 5 | GOTERM_MF_FAT | GO:0005509 | calcium ion binding | 52 | 2.60E-09 | 5.00E-09 |
| 6 | GOTERM_MF_FAT | GO:0005201 | extracellular matrix structural constituent | 12 | 1.90E-08 | 3.20E-08 |
| 7 | GOTERM_MF_FAT | GO:0008201 | heparin binding | 21 | 2.10E-08 | 6.10E-08 |
| 8 | GOTERM_MF_FAT | GO:0050840 | extracellular matrix binding | 10 | 6.80E-08 | 2.70E-07 |
| 9 | GOTERM_MF_FAT | GO:0004872 | receptor activity | 21 | 1.90E-07 | 4.00E-07 |
| 10 | GOTERM_MF_FAT | GO:0004714 | transmembrane receptor protein tyrosine kinase activity | 12 | 4.10E-07 | 6.10E-07 |
GO gene ontology, FDR false discovery rate
Since severe RGC loss was observed in Nrf1f/f;Six3-Cre retinas, we expected that RGC gene expression would be reduced in Nrf1-deficient retinas. Atoh7 is a key factor essential for RGC development. RNA-seq data revealed that Atoh7 expression was slightly reduced by ~ 19.5% in Nrf1-mutant retinas; however, Atoh7-expressing precursor cells can be readily detected in Nrf1-mutant retinas (data not shown), suggesting that the RGC loss phenotype is mainly due to a defective RGC differentiation process. Transcriptome analysis comparing Atoh7+ RPCs and Atoh7-negative cells in E13.5 has revealed 236 genes with altered expression levels [34]. We compared the 488 genes that are downregulated in Nrf1f/f;Six3-Cre retinas with the 236 genes enriched in Atoh7+ RPCs and found 121 common genes (Table 4). The majority of these genes were expressed in RGCs, including Pou4f1, Pou4f2, Isl1, and Myt1, which are known to be expressed in differentiating RGCs [35]. In addition, 41 genes involved in neuronal differentiation were found, such as neurofilament light chain (Nefl) and neurofilament middle chain (Nefm). We also compared the 488 downregulated genes in Nrf1f/f;Six3-Cre to the 49 significantly downregulated genes in the Pou4f2-/- retina [36] and found 18 common genes downregulated in both Nrf1f/f;Six3-Cre and Pou4f2-/- retinas (data not shown). Among them, 7 genes are enriched in Atoh7+ retinas. These results indicate Nrf1 depletion affects RGC gene expression.
Table 4.
Genes downregulated in Nrf1f/f; Six3-Cre that are enriched in Atoh7+ cells
| Gene name | FC | Spatial expression | |
|---|---|---|---|
| Transcription factor | Barhl2 | −1.63 | RGC |
| Ebf1 | −2.63 | RGC | |
| Ebf3 | −3.09 | RGC | |
| Irx2 | − 2.35 | RGC | |
| Irx3 | −1.97 | RGC | |
| Irx5 | −2.43 | RGC | |
| Irx6 | −2.18 | RGC | |
| Isl1 | −2.43 | RGC | |
| Myt1 | −2.14 | RGC | |
| Onecut3 | −2.15 | RGC | |
| Pou4f1 | −2.62 | RGC | |
| Pou4f2 | −1.89 | RGC | |
| Pou6f2 | −2.28 | RGC | |
| Ptf1a | −2.02 | retina | |
| Tub | −2.38 | RGC | |
| Neuron differentiation | Actl6b | −2.2 | RGC |
| Adcyap1 | −1.71 | RGC | |
| Bsn | −2.27 | RGC | |
| Celsr3 | −2.35 | RGC | |
| Cend1 | −1.89 | RGC | |
| Cntn2 | −3.09 | RGC | |
| Dcx | −2.87 | RGC | |
| Dner | −2.08 | RGC | |
| Dnm3 | −1.7 | unknown | |
| Dok5 | −1.52 | retina | |
| Dscam | −2.5 | retina | |
| Elavl3 | −1.52 | RGC | |
| Elavl4 | −2.67 | retina | |
| Gap43 | −3.02 | RGC | |
| Gprin1 | −2.51 | retina | |
| Ina | −3.56 | RGC | |
| Insc | −1.88 | unknown | |
| Islr2 | −3.49 | RGC | |
| Kif5a | −2.66 | RGC | |
| Klhl1 | −1.76 | RGC | |
| L1cam | −2.65 | RGC | |
| Mapt | −4.37 | RGC | |
| Mmp24 | −2.32 | RGC | |
| Myo16 | −1.61 | RGC | |
| Myt1l | −2.61 | RGC | |
| Nefl | −3.67 | RGC | |
| Nefm | −2.46 | RGC | |
| Nell2 | −3.49 | RGC | |
| Nptx1 | −1.63 | RGC | |
| Nrn1 | −2.91 | RGC | |
| Ret | −3.75 | RGC | |
| Scn3b | −2.77 | RGC | |
| Scrt1 | −2.11 | RGC | |
| Sez6l | −2.56 | RGC | |
| Slit1 | −1.81 | RGC | |
| Snap25 | −2.41 | RGC | |
| Stmn2 | −3.26 | RGC | |
| Stmn3 | −3.65 | RGC | |
| Th | −2.45 | retina | |
| Tnik | −1.63 | unknown | |
| Tubb3 | −2.38 | RGC | |
| Unc13a | −2.62 | RGC | |
| Others | 1810041L15Rik | −2.49 | RGC |
| A930011O12Rik | −2.08 | unknown | |
| Ajap1 | −1.66 | RGC | |
| Akap6 | −3.68 | RGC | |
| Apba2 | −1.93 | RGC | |
| Arg1 | −1.55 | RGC | |
| Atp1a3 | − 1.7 | retina | |
| Cacna1b | −2.28 | RGC | |
| Calb2 | −2.96 | RGC | |
| Ccnd1 | −1.83 | RPC | |
| Cda | −1.56 | RGC | |
| Celf3 | −2.19 | RGC | |
| Celf5 | −1.89 | retina | |
| Chga | −1.65 | RGC | |
| Chgb | −1.72 | RGC | |
| Chst8 | −1.95 | RGC | |
| Coro2a | −1.85 | RGC | |
| Crmp1 | −2.78 | RGC | |
| D930028M14Rik | −1.8 | RGC | |
| Disp2 | −2.82 | RGC | |
| Dnajc6 | −1.78 | RGC | |
| Dusp26 | −2.37 | RGC | |
| Eya2 | −1.59 | RGC | |
| Fam155a | −1.76 | RGC | |
| Fam78b | −1.73 | unknown | |
| Fgf15 | −1.61 | RPC | |
| Gabbr2 | −1.97 | unknown | |
| Gdap1l1 | −1.91 | RGC | |
| Grm2 | −2.67 | unknown | |
| Hecw1 | −2.5 | RGC | |
| Hspa12a | −2.21 | retina adult | |
| Igfbpl1 | −2.25 | RGC | |
| Iqsec3 | −1.53 | RGC | |
| Kcnq2 | −2.47 | unknown | |
| Mapk11 | −1.75 | RGC | |
| Mtus2 | −2.7 | RGC | |
| Nacad | −2.65 | RGC | |
| Nhlh2 | −2.68 | RGC | |
| Nmnat2 | −2.43 | RGC | |
| Nsg2 | −3.26 | RGC | |
| Pak7 | −2.04 | unknown | |
| Ppp2r2b | −1.8 | RGC | |
| Ppp2r2c | −1.88 | unknown | |
| Rab3c | −2.12 | RGC | |
| Rph3a | −2.58 | RGC | |
| Rtn1 | −2.59 | RGC | |
| Rundc3a | −2.27 | RGC | |
| Rusc1 | −1.92 | RGC | |
| Scg3 | −2.94 | RGC | |
| Scn3a | −2.4 | unknown | |
| Sez6l2 | −2.56 | RGC | |
| Slc17a6 | −2.88 | RGC | |
| Smpd3 | −1.97 | RGC | |
| Sncg | −4.74 | RGC | |
| Spire2 | −1.54 | RGC | |
| Srrm3 | −2 | unknown | |
| Sst | −2.14 | RGC | |
| Stk32a | −1.83 | RGC | |
| Svop | −2.17 | RGC | |
| Thsd7b | −2.45 | unknown | |
| Trim46 | −1.56 | unknown | |
| Trp53i11 | −1.79 | RGC | |
| Tubb2a | −2.35 | unknown | |
| Unc79 | −2.31 | unknown | |
| Vwa5b2 | −1.67 | RGC | |
| Xkr7 | −1.69 | unknown |
488 downregulated genes in Nrf1f/f; Six3-Cre retinas compared with 236 genes enriched in Atoh7+ retinal cells [34] identified 121 common genes. They are listed with official gene name, RNA-seq fold difference, and spatial expression pattern. Italicized gene names indicated downregulated genes in E14.5 Pou4f2−/− retinas [35]. FC fold change
Defective axon outgrowth in Nrf1f/f;Six3-Cre retinas
To determine whether retinal neurons were defective in neurite outgrowth, we cultured retinal explants from E13.5 wildtype and Nrf1f/f;Six3-Cre embryos to examine axonal outgrowth. Consistent with the RNA-seq analysis, we found that retinal explants from Nrf1f/f;Six3-Cre embryos failed to form and extend well-bundled axons as in wildtype explants (Fig. 7b, c), indicating an important function for Nrf1 in regulating genes involved in neurite outgrowth.
Altered expression of genes associated with mitochondrial function and energy production in Nrf1f/f;Six3-Cre retinas
Because Nrf1 is a key regulator of nuclear-encoded genes involved in mitochondrial functions, we then tested whether genes involved in mitochondrial functions were altered in Nrf1-deficient retinas. By comparing the gene list in MitoCarta 2.0 [37, 38], we revealed a subset of genes with altered expression levels in Nrf1-mutant retinas, and color-coded and mapped them to various functional subdomains in the mitochondria (Fig. 7d). In addition, 5 glycolysis genes with affected expression levels in Nrf1-deficient retinas were identified (Fig. 7e, q). For example, mRNA levels of cytochrome c oxidase subunit 4i2 (Cox4i2) in Nrf1-mutant retinas were reduced to ~ 44% of those in wildtype retinas. We tested mitochondrial respiratory activity in Nrf1f/f;Six3-Cre retinas by examining the histochemical activity of COX. Intense COX activity was detected in RGCs (arrowhead in Fig. 7f) and the outermost area of retina where photoreceptor precursors resided (arrow in Fig. 7f). In contrast, COX activity was diminished to background levels in the Nrf1f/f;Six3-Cre retina (Fig. 7g). We then performed qRT-PCR analysis on a small, selected set of these affected genes and found that the levels of expression of all of them were consistent with the RNA-seq data (Fig. 7h, Cox4i2: P = 0.038, Idh: P = 0.0001, Dna2: P = 0.003, Gdap1: P = 0.002, Shmt2: P = 0.003, Cpt1a: P = 0.0001, Sardh: P = 0.003, Ucp2: P = 0.013, Pmaip1: P = 0.048, Rab32: P = 0.014).
Furthermore, we performed in situ hybridization (ISH) for several genes whose expression was either upregulated (Cpt1a and Slc16a1) or downregulated (Idh1, Ldha) in Nrf1 mutants by RNA-seq analysis. Cpt1a, encoding carnitine palmitoyltransferase 1a, is involved in lipid transfer in mitochondria. In E13.5 wildtype retinas, Cpt1a was expressed at extremely low levels, barely detectable even by ultrasensitive RNAscope ISH (Fig. 7i). In Nrf1-mutant retinas, weak but detectable Cpta1 transcripts were visible in the central retina (arrowheads in Fig. 7j). Slc16a1, encoding solute carrier family 16 (monocarboxylic acid transporters) member 1, is involved in lactate/pyruvate transport in mitochondria. Slc16a1 was expressed in the peripheral retina in E13.5 wildtype retinas and upregulated in the central area of Nrf1f/f;Six3-Cre retinas (Fig. 7k, l). Idh1, encoding isocitrate dehydrogenase 1, was highly expressed in RGCs in wildtype retinas, whereas its expression was drastically reduced in Nrf1f/f;Six3-Cre retinas (Fig. 7m, n). Ldha, encoding lactate dehydrogenase A, which catalyzes the conversion of lactate to pyruvate in the glycolysis pathway, was highly expressed in RPCs in wildtype retinas but downregulated in Nrf1f/f;Six3-Cre retinas (Fig. 7o, p). To confirm the effect of Nrf1 deletion on the 5 genes involved in the glycolysis pathway, we performed qRT-PCR analysis for these 5 glycolysis-associated genes (Fig. 7q, Ldha: P = 0.0003, HK1: P = 0.0001, Pfkp: P = 0.018, Tpi1: P = 0.049, Pgam2: P = 0.0005) and confirmed that expression levels of these genes were indeed affected as revealed by RNA-seq analysis. These data indicate that Nrf1 is important in regulating various metabolic pathways, including lipid metabolism, glycolysis, and oxidative phosphorylation, during retinal development.
Deleting Nrf1 in rod photoreceptors caused complete rod degeneration
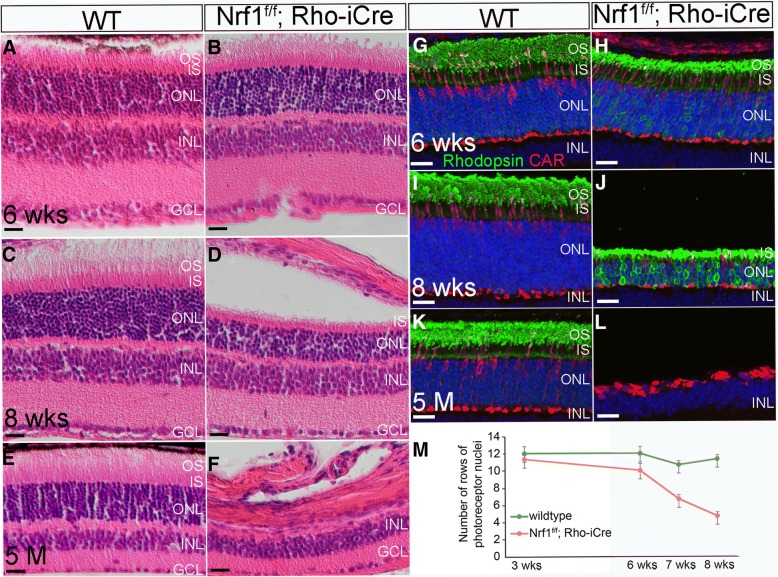
To investigate the in vivo function of Nrf1 in differentiated neurons, we choose to use rod PRs as a model system, because rod PRs are the major neuronal type in the retina, and a large number of genetic mutations causing PR degeneration have been identified [39]. We bred a Rho-iCre transgenic mouse line with mice harboring Nrf1flox allele to delete Nrf1 in the photoreceptor cells. Prior to 6 weeks of age, Nrf1f/f;Rho-iCre retinas did not show any sign of histological phenotype compared with wildtype retinas (Fig. 8a, b). Starting from 8 weeks, the thickness of the ONLs in the Nrf1f/f;Rho-iCre retinas was notably thinner than that in the control retinas (Fig. 8c, d), and the number of PRs decreased to 50% of that in wildtype retinas (Fig. 8m, 3 weeks: P = 0.373, 6 weeks: P = 0.070, 7 weeks: P = 0.001, 8 weeks: P = 0.0001). At 5 months, the ONLs had almost disappeared (Fig. 8e, f).
Fig. 8.
Nrf1 conditional knockout by Rho-iCre causes severe photoreceptor degeneration. (a–f) H&E staining of retinal sections from wildtype (a, c, and e) and Nrf1f/f;Rho-iCre (b, d, and f) at 6 weeks, 8 weeks, and 5 months old. (g–l) Immunostaining of wildtype (g, i, and k) and Nrf1f/f;Six3-Cre (h, j, and l) retinal sections with anti-rhodopsin and anti-cone arrestin antibodies at 6 weeks, 8 weeks, and 5 months old. (m) The number of rows of photoreceptor nuclei in wildtype and Nrf1f/f;Rho-iCre. Scale bars: 20 μm. OS: outer segment. IS: inner segment. ONL: outer nuclear layer. INL: inner nuclear layer. GCL: ganglion cell layer. WT: wildtype
To determine whether cone photoreceptors were also affected in rod-Nrf1-mutants, we immunolabeled wildtype and Nrf1f/f;Rho-iCre retinas with rod-specific rhodopsin and cone-specific arrestin (CAR). At 6 weeks of age, rhodopsin was enriched in the outer segments (OSs) of rod PRs. We observed slightly upregulated rhodopsin in Nrf1f/f;Rho-iCre retinas compared to wildtype retinas. There was no difference in numbers of CAR+ cells between control and mutant retinas. OSs and ISs were shorter in Nrf1f/f;Rho-iCre retinas than in wildtype retinas (Fig. 8g, h). In 8-week-old retinas, rhodopsin+ PRs in Nrf1f/f;Rho-iCre retinas were reduced to ~ 30% of wildtype retinas, while strong rhodopsin staining was detected in ONLs of Nrf1f/f;Rho-iCre retinas (Fig. 8i, j). At this stage, the number of cone photoreceptor cells was also reduced in Nrf1f/f;Rho-iCre retinas compared with wildtype retinas (Fig. 8i, j). In 5-month-old mutant retinas, no rhodopsin+ PRs were detected, while the few remaining cone photoreceptors formed a single column in the ONLs without the distinguishable normal cone-shaped morphology (Fig. 8k, l). Since rod PRs are required for cone survival [40, 41], the cone degeneration in Nrf1f/f;Rho-iCre retinas was likely secondary to rod PR degeneration. These results clearly indicate that Nrf1 is essential for the survival of photoreceptor cells.
Abnormal mitochondrial morphology and impaired mitochondrial functions in Nrf1f/f;Rho-iCre inner segments
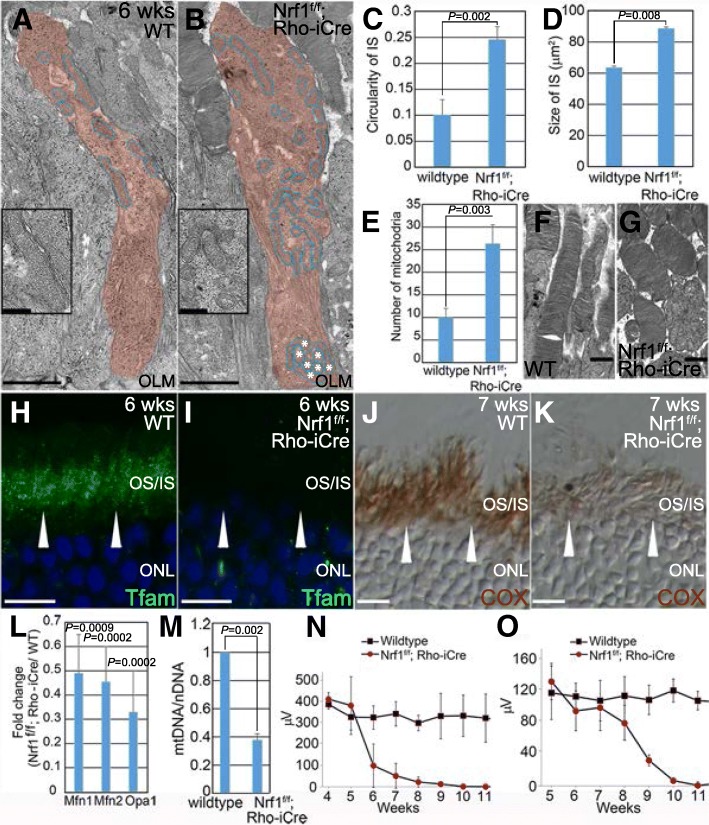
To examine how Nrf1 deletion affected mitochondria in rod PRs, we used transmission electron microscopy to inspect the morphology of mitochondria in 6-week-old Nrf1f/f;Rho-iCre retinas, when the ISs had not yet degenerated. We collected transmission electron microscopy images of ISs from wildtype Nrf1f/f;Rho-iCre photoreceptors and analyzed with Fiji for the circularity of IS, size of IS, and number of mitochondria. We found that the ISs in Nrf1f/f;Rho-iCre retinas were slightly wider than the wildtype ISs (Fig. 9a-c, P = 0.002), resulting in a ~ 40% increase in size compared to wildtype retinas (Fig. 9d, P = 0.008). The number of mitochondria in a Nrf1f/f;Rho-iCre IS section was 2.5 times than that of wildtype (Fig. 9e, P = 0.003). The mitochondria in the Nrf1f/f;Rho-iCre ISs were notably smaller and displayed a more rounded shape compared to mitochondria in the control retinas and were more widely distributed within the ISs (Fig. 9a, b). A cluster of mitochondria was observed near the outer limiting membranes while no mitochondria were present in the same area in the wildtype ISs (asterisks in Fig. 9b). We also noticed that the OSs in Nrf1f/f;Rho-iCre photoreceptors were shorter than that of the controls (Fig. 9f, g).
Fig. 9.
Defective mitochondria and ERG response in Nrf1f/f;Rho-iCre retina. (a, b) Transmission electron microscopy (TEM) images of the inner segments of wildtype and Nrf1f/f;Rho-iCre photoreceptors. Inner segment is color-labeled in red, and mitochondria are circled in blue. Asterisks indicate clustered mitochondria near the OLM in Nrf1f/f;Rho-iCre ISs. Insets show higher magnification images of indicated areas. (c–e) TEM images were analyzed with Fiji for the circularity of ISs (c), size of ISs (d) and the number of mitochondria (e). f, g TEM images of the outer segments of wildtype (f) and Nrf1f/f;Rho-iCre (g) photoreceptors. (h, i) Immunostaining of 6-week wildtype (h) and Nrf1f/f;Rho-iCre (i) retinal sections with anti-Tfam antibody. Arrowheads indicate Tfam staining in ISs. (j, k) COX activity of 7-week wildtype (j) and Nrf1f/f;Rho-iCre (k) retinal sections. Arrowheads indicate COX activities in ISs. (l) qRT-PCR analysis of genes involved in mitochondria fusion of wildtype and Nrf1f/f;Rho-iCre retinas. (m) Mitochondria copy number of 6-week-old wildtype and Nrf1f/f;Rho-iCre retinas. (n, o) ERGs of wildtype and Nrf1f/f;Rho-iCre littermates under dark-adapted (scotopic, n) and light-adapted (photopic, o) conditions. Scale bars: 1 μm in a, b, f and g, 10 μm in h–k. OLM: outer limiting membrane. OS: outer segment. IS: inner segment. ONL: outer nuclear layer. WT: wildtype
Nuclear-encoded mitochondrial transcription factor A (Tfam/mtTFA), a key regulator of mitochondrial transcription and mitochondrial genome replication, is a known downstream target of Nrf1 [42]. To examine whether Tfam was affected in Nrf1-deficient rod PRs, we inspected Tfam expression by immunostaining and found that Tfam expression was abolished in Nrf1f/f;Rho-iCre ISs whereas strong expression of Tfam was observed in the wildtype ISs (Fig. 9h, i). Abnormalities in the number, morphology, and distribution of mitochondria, and the downregulation of a key mitochondrial regulator Tfam in Nrf1f/f;Rho-iCre ISs prompted us to determine whether mitochondrial function was compromised. We performed a COX assay to examine the mitochondrial enzymatic activity. As expected, COX activity was weaker in Nrf1f/f;Rho-iCre ISs compared with wildtype ISs (Fig. 9j, k). Furthermore, we tested whether the expression levels of genes involved in mitochondrial fusion were affected in Nrf1f/f;Rho-iCre retinas. Mitofusion-1 (Mfn1), Mfn2, and Optic Atrophy 1 (Opa1) are key mitochondrial proteins mediating mitochondrial fusion [43–45]. Deletion of Mfn1 and Mfn2 in skeletal muscle results in reduction of mtDNA and respiratory deficiencies [25]. We performed qRT-PCR to compare mRNA expression levels of Mfn1, Mfn2, and Opa1 in 6-week-old wildtype and Nrf1f/f;Rho-iCre retinas. In Nrf1f/f;Rho-iCre retinas, Mfn1, Mfn2, and Opa1 levels decreased to ~ 50% of wildtype retinas (Fig. 9l, Mfn1: P = 0.0009, Mfn2: P = 0.0002, Opa1: P = 0.0002). In addition, the copy numbers of mtDNA in Nrf1f/f;Rho-iCre retinas was ~ 38% compared to that of wildtype retinas (Fig. 9m), consistent with Tfam’s role as a major regulator of mtDNA replication and mitochondrial transcription.
Because Nrf1f/f;Rho-iCre retinas displayed severe rod degeneration followed by cone degeneration, we set out to track outer retina function using electroretinography (ERG). Dark-adapted wildtype and Nrf1f/f;Rho-iCre mice were exposed to calibrated light flashes for ERG recording. The scotopic a-wave amplitudes of Nrf1f/f;Rho-iCre mice were similar to those of wildtype before 5 weeks of age, began to decline at 6 weeks, and had completely diminished by 3 months (Fig. 9n, 4 weeks: P = 0.3101, 5 weeks: P = 0.4548. Six weeks: P = 1.6988E-05, 7 weeks: P = 3.0756E-09, 8 weeks: P = 9.7899E-12, 9 weeks: P = 2.7743E-16, 10 weeks: P = 9.8167E-05, 11 weeks: P = 0.00497). Photopic ERG b-wave amplitudes from light-adapted wildtype and Nrf1f/f;Rho-iCre mice were similar before 7 weeks of age, started to decline noticeably at 8 weeks, and were undetectable beyond 10 weeks (Fig. 9o, 5 week: P = 0.7052, 6 weeks: P = 0.2420, 7 weeks: P = 0.4169, 8 weeks: P = 0.0522, 9 weeks: P = 8.0496E-05, 10 weeks: P = 0.0002, 11 weeks: P = 6.8768E-05). These data indicate that PR functional loss precedes morphological defects and further demonstrate that deleting Nrf1 in rod PRs causes abnormal mitochondria and impaired mitochondrial function, resulting in reduced outer retina activity and eventual complete photoreceptor loss.
Discussion
Functional mitochondrial biogenesis is essential for energy metabolism, calcium homeostasis, the biosynthesis of amino acids, cholesterol, and phospholipids, elimination of excessive reactive oxygen species, and apoptosis. Nrf1 was identified as a major transcriptional regulator that connects the regulation of nuclear-encoded genes and mitochondrial biogenesis and has been implicated in the pathology of several neurodegenerative diseases [16, 46]. However, little is known about its role in central nervous system development because of the lack of an appropriate animal model. To fill this gap of knowledge, we generated Nrf1 conditional knockout mouse models and used these mouse lines to conduct the first comprehensive in vivo study to delineate various roles of Nrf1 in proliferating neural progenitor cells, newly differentiated RGCs, and terminally differentiated rod PRs.
Previous studies have provided evidence for Nrf1’s role in cell growth and proliferation. For example, a genome-wide ChIP-chip study has revealed that Nrf1 binds and regulates a number of E2F-targeted genes involved in DNA replication and repair, mitosis and chromosome dynamics, and metabolism [47]. A ChIP-seq study using SK-N-SH human neuroblastoma cells has revealed that Nrf1 target genes contain genes associated with cell cycle regulation [16]. Cyclin D1-dependent kinase phosphorylates Nrf1 and inhibits its transcriptional activity [48]. Nrf1-deleted mouse embryos die during the peri-implantation stage between embryonic days 3.5 and 6.5 in part due to reduced cell proliferation [20]. In our study, we showed that deleting Nrf1 in the proliferating RPCs reduced cell proliferation indices in the developing retina. The few surviving RPCs that exited the cell cycle and differentiated into RGCs failed to migrate from the neuroblast layer to the ganglion cell layer. Using RNA-seq analysis, we discovered that genes involved in neurite outgrowth are significantly downregulated in Nrf1-deficient retinas. Consistent with the RNA-seq data, we demonstrated that neurite outgrowth activity was reduced in Nrf1-deleted retinal explants compared to control explants. Although we cannot exclude the possibility that the RGC migration and neurite outgrowth phenotypes seen in Nrf1-mutants are caused indirectly by defective mitochondria, a recent study on a RPC-specific knockout of Ronin, a key transcriptional regulator for mitochondrial gene expression and RPC proliferation, has shown that conditionally deleting Ronin in RPCs causes defective mitochondrial function and premature cell cycle exit in RPCs, leading to the generation of more RGCs [49]. Interestingly, these extra, newly differentiated RGCs survive and do not display any defects as observed in Nrf1-mutants, suggesting that Nrf1 directly regulates subsets of genes for RGC migration and neurite outgrowth during retinal development. Together this in vivo and ex vivo evidence supports the previous findings that Nrf1 is essential for cell growth, proliferation, and neurite outgrowth [50].
In the developing mouse retina, the proliferating RPCs and the terminally differentiated retinal neurons adopt different metabolic pathways for energy production. In RPCs, aerobic glycolysis is a predominant way to produce ATP, whereas oxidative phosphorylation is utilized in differentiated neurons [51]. Such a transition is observed in many developmental systems, suggesting that the reconfiguration of energy metabolic pathways is likely intricately mapped onto the regulatory networks controlling cell cycle progression and differentiation. In Nrf1-mutant retinas, Ldha, which encodes the enzyme that converts pyruvate to lactate and generates the nicotinamide adenine dinucleotide (NAD+) necessary for aerobic glycolysis [52], was significantly downregulated. Additionally, several glycolytic pathway genes were also downregulated in Nrf1-mutant RPCs, suggesting that Nrf1-mutant RPCs may shift to utilize oxidative phosphorylation to produce energy. Consistent with this, pyruvate dehydrogenase kinase isoenzyme 1 (Pdk1), a metabolic checkpoint enzyme that inactivates pyruvate dehydrogenase, was also downregulated in Nrf1 mutant RPCs. Hence the increased pyruvate dehydrogenase activity would enable pyruvate to enter the tricarboxylic acid cycle. Despite Nrf1’s known function as a transcriptional activator, a subset of genes carrying out various mitochondrial functions, and Pgam2, encoding phosphoglycerate mutase which is involved in glycolysis, are upregulated in Nrf1-mutant retinas. Among them we observed the upregulation of Cpt1a and Slc16a1 in mutant RPCs. It is currently unknown whether Nrf1 functions as a repressor that directly modulates the transcriptional levels of these genes or if deleting Nrf1 indirectly leads to reprogramming in their transcriptional regulatory regions in RPCs. Nevertheless, these data taken together implicate Nrf1 in a regulatory role to enable RPCs to alter their metabolic program and advance to a committed neuronal fate. Although the molecular mechanism regulating the metabolic transition is currently unclear, the potential roles of metabolites in epigenetic control at several levels, including DNA methylation/demethylation and histone modifications, could influence the cellular state and fate [53]. Interestingly, a recent study showed that in vivo Nrf1 binding to its target sites is inhibited by de novo DNA methylation, and active demethylation and obstruction of de novo methylation through the binding of methylation-insensitive transcription factors could de-methylate the nearby genome, thus restoring Nrf1 binding and transcriptional activity [54]. Future research is required to uncover and compare the in vivo occupancy of Nrf1 and the methylome in proliferating RPCs and differentiated rod photoreceptors to determine whether this novel mechanism is actively utilized by Nrf1 and co-regulators in regulating metabolic transition.
The discovery of nuclear-encoded mitochondrial transcription factor A (Tfam/mtTFA) as a target of Nrf1 established the regulatory link between nuclear and mitochondrial gene expression [42]. In wildtype retinal photoreceptors, Tfam was transported to and enriched in the ISs, but its expression was undetectable in 6-week-old Nrf1-mutant ISs, confirming that Tfam is a bona fide in vivo target of Nrf1. The small, rounded mitochondrial morphology and the increased number of mitochondria seen in the ISs in Nrf1-deficient rods suggest that the normal mitochondrial fusion/fission processes are defective in Nrf1-mutant rods. Continuous mitochondrial fusion and fission are essential for maintaining a functional mitochondrial network to ensure sufficient exchange of mitochondrial contents, which might be otherwise damaged under stressed environments [55, 56]. Several key molecular regulators for mitochondrial fusion, including Mfn1, Mfn2, and Opa1, were downregulated in 6-week-old Nrf1-deficient rods. Because loss of any of these genes causes defects in mitochondrial fusion, impairs mitochondrial oxidative phosphorylation, and eventually leads to apoptosis, it is likely that defective mitochondrial fusion in Nrf1-null rods is a major cause of rod degeneration. Consistent with this, ewg, the Drosophila homolog of Nrf1, has been shown to play a role in regulating mitochondrial fusion and expression of the Opa1-like gene during muscle growth in the fly [57]. It is noteworthy that mutations in human OPA1, a direct target of human NRF1, are the cause of autosomal dominant optic atrophy [58], which leads to retinal ganglion cell death. Thus, it would be interesting to test whether downregulation of Nrf1 contributes to RGC death in several glaucoma animal models.
For mammals with vascular retinas, mitochondria in the rod PRs migrate toward and localize in the outer part of the IS (the ellipsoid) for oxygen supplied from choriocapillaris [59, 60]. In Nrf1-null rods, however, mitochondria were often trapped near the base of the outer limiting membrane. Proper mitochondrial trafficking within a neuron is critical for clearing the older, damaged components and delivering the new materials encoded by nuclear genes [61]. It is therefore conceivable that mitochondrial trafficking defects in Nrf1-mutant retinas also contribute to the death of rod PRs.
Many mouse models of inherited retinal degenerative disease have been established to understand disease mechanisms and design treatment strategies for human diseases [62, 63]. In our study, we showed that Rho-iCre efficiently and specifically deleted Nrf1 in rod cells as early as P10; however, the Nrf1-deficient rods degenerated at a relatively slow pace. By 4 weeks of age, we did not find histological differences between the controls and mutants. The first sign of degeneration in rod-Nrf1 mutants was the slight thinning of the ONLs and OSs and the reduction of the scotopic a-wave amplitudes. It took approximately 3 months for the Nrf1-deficient rods to completely degenerate. The reason for such resiliency is currently unknown. It is possible that the glycolysis pathway partially supports the energy demand in Nrf1-deficient rods. Alternatively, other transcriptional factors and epigenetic memory may transiently compensate for the loss of Nrf1 to maintain the expression of Nrf1-regulated downstream genes. Nevertheless, the slow, progressive rod degeneration found in this new mouse model offers a unique opportunity to investigate how defective mitochondrial biogenesis affects different cellular processes whose defects frequently link to retinal degeneration. Furthermore, mitochondrial function declines with age and is associated with age-related disorders and cell death. It is of interest to test whether any of components in the Nrf1-regulated mitochondrial biogenesis pathway are associated with aging retinas and whether they can be used as therapeutic targets for ameliorating retinal degenerative diseases.
Conclusions
Our findings confirm some of the known functions of Nrf1 that were previously revealed mainly through in vitro studies. Additionally, we uncovered a novel role for Nrf1 in metabolic reprogramming, although the degree to which Nrf1 is involved in this process during neural development remains to be determined. Our data also shed new light on how dysfunctional mitochondrial biogenesis may be involved in various neurodegenerative diseases. For example, we have shown that RPCs and newly differentiated RGCs are very sensitive to Nrf1 deletion. In contrast, rod PRs, an energy demanding neuronal type, are much more tolerant of Nrf1 deletion. We also found that the terminally differentiated RGCs are less sensitive to Nrf1 deletion (data not shown). This difference may be in part due to the varying roles of Nrf1 in different cell types and developmental stages; however, it also suggests that different neuronal tissues and cell lineages may have diverse sensitivities to mitochondrial defects. Future experiments using tissue- and cell-specific Nrf1 deletions will be critical in directly addressing how dysfunctional mitochondrial biogenesis contributes to the pathology and disease progression in neurodegenerative diseases.
Acknowledgements
We are grateful to Dr. Yasuhide Furuta (RIKEN) for sharing the Six3-Cre mouse strain. We acknowledge the Genetically Engineered Mouse Facility, Kenneth Dunner, Jr. of the High Resolution Electron Microscope Facility, and the Sequencing and Microarray Facility at The University of Texas MD Anderson Cancer Center for their assistance, and Dr. Kimberly Mankiewicz at UTHealth for reading and editing the manuscript.
Funding
This work was supported by grants from the National Institutes of Health-National Eye Institute to C.-A.M. (EY024376), C.-K.C (EY013811, EY022228), and W.H.K. (EY011930) and from the National Institute of Allergy and Infectious Diseases to S.T. (AI057504). This work was also supported by National Eye Institute Vision Core Grant P30EY010608 (UTHealth).
Availability of data and materials
The RNA-seq data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE101550 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE101550).
Disclaimer
Part of this study was carried out by Dr. Shinako Takada in her previous capacity at the Department of Biochemistry and Molecular Biology, The University of Texas MD Anderson Cancer Center and her current personal capacity. The opinions expressed in this article are the authors’ own and do not reflect the views of the National Institutes of Health, the Department of Health and Human Services, or the United States government.
Abbreviations
- CAR
Cone-specific arrestin
- Ccnd1
Cyclin D1
- ChIP-seq
Chromatin immunoprecipitation sequencing
- COX
Cytochrome c oxidase
- ERG
Electroretinography
- ES
Embryonic stem
- ewg
Erect wing
- FRT
FLP recombinase target
- GCL
Ganglion cell layer
- INL
Inner nuclear layer
- IS
Inner segment
- ISH
In situ hybridization
- Mfn1
Mitofusion-1
- Mfn2
Mitofusion-2
- mtDNA
Mitochondrial DNA
- nefl
Neurofilament light chain
- nefm
Neurofilament middle chain
- Nrf1
Nuclear respiratory factor 1
- Nrf2/GABP
Nuclear respirator factor 2
- OCT
Optimal cutting temperature
- OLM
Outer limiting membrane
- ONL
Outer nuclear layer
- Opa1
Optic atrophy 1
- OS
Outer segments
- PARP-1
poly(ADP-ribose) polymerase 1
- PBS
Phosphate buffered saline
- PGC-1
Peroxisome proliferative activated receptor gamma coactivator 1
- PR
Photoreceptor
- qRT-PCR
Quantitative reverse transcriptase PCR
- RGC
Retinal ganglion cell
- RNA-seq
RNA sequencing
- RPC
Retinal progenitor cell
- TEM
Transmission electron microscopy
- Tfam
Mitochondrial transcription factor A
- TUNEL
Transferase dUTP nick end label
- WT
Wildtype
Authors’ contributions
TK., C-KC, SWW, ST, WHK and C-AM designed experiments. TK, C-KC, SWW, PP, ST and C-AM performed experiments. ZJ and JW conducted bioinformatics analysis. TK, C-KC, WHK and C-AM wrote the manuscript.
Ethics approval
All animal procedures followed the US Public Health Service Policy on Humane Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee at The University of Texas MD Anderson Cancer Center, Animal Welfare Committee at The University of Texas Health Science Center at Houston, and Animal Welfare Committee at Baylor College of Medicine.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Takae Kiyama, Email: Takae.Kiyama@uth.tmc.edu.
Ching-Kang Chen, Email: Ching-Kang.Chen@bcm.edu.
Steven W Wang, Email: SWang11@mdanderson.org.
Ping Pan, Email: Ping.Pan@uth.tmc.edu.
Zhenlin Ju, Email: zju@mdanderson.org.
Jing Wang, Email: jingwang@mdanderson.org.
Shinako Takada, Email: shinakotakada@gmail.com.
William H Klein, Email: whklein@mdanderson.org.
Chai-An Mao, Email: Chai-An.Mao@uth.tmc.edu, Email: chai-an.mao@uth.tmc.edu.
References
- 1.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 2.Virbasius CA, Virbasius JV, Scarpulla RC. NRF-1, an activator involved in nuclear-mitochondrial interactions, utilizes a new DNA-binding domain conserved in a family of developmental regulators. Genes Dev. 1993;7:2431–2445. doi: 10.1101/gad.7.12a.2431. [DOI] [PubMed] [Google Scholar]
- 3.Hock MB, Kralli A. Transcriptional control of mitochondrial biogenesis and function. Annu Rev Physiol. 2009;71:177–203. doi: 10.1146/annurev.physiol.010908.163119. [DOI] [PubMed] [Google Scholar]
- 4.Spiegelman BM. Transcriptional control of mitochondrial energy metabolism through the PGC1 coactivators. Novartis Found Symp. 2007;287:60–63. [PubMed] [Google Scholar]
- 5.Calzone FJ, Hoog C, Teplow DB, Cutting AE, Zeller RW, Britten RJ, Davidson EH. Gene regulatory factors of the sea urchin embryo. I. Purification by affinity chromatography and cloning of P3A2, a novel DNA-binding protein. Development. 1991;112:335–350. doi: 10.1242/dev.112.1.335. [DOI] [PubMed] [Google Scholar]
- 6.DeSimone SM, White K. The Drosophila erect wing gene, which is important for both neuronal and muscle development, encodes a protein which is similar to the sea urchin P3A2 DNA binding protein. Mol Cell Biol. 1993;13:3641–3649. doi: 10.1128/MCB.13.6.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becker TS, Burgess SM, Amsterdam AH, Allende ML. Hopkins N: not really finished is crucial for development of the zebrafish outer retina and encodes a transcription factor highly homologous to human nuclear respiratory factor-1 and avian initiation binding repressor. Development. 1998;125:4369–4378. doi: 10.1242/dev.125.22.4369. [DOI] [PubMed] [Google Scholar]
- 8.Schaefer L, Engman H, Miller JB. Coding sequence, chromosomal localization, and expression pattern of Nrf1: the mouse homolog of Drosophila erect wing. Mamm Genome. 2000;11:104–110. doi: 10.1007/s003350010021. [DOI] [PubMed] [Google Scholar]
- 9.Evans MJ, Scarpulla RC. NRF-1: a trans-activator of nuclear-encoded respiratory genes in animal cells. Genes Dev. 1990;4:1023–1034. doi: 10.1101/gad.4.6.1023. [DOI] [PubMed] [Google Scholar]
- 10.Scarpulla RC. Nuclear control of respiratory chain expression in mammalian cells. J Bioenerg Biomembr. 1997;29:109–119. doi: 10.1023/A:1022681828846. [DOI] [PubMed] [Google Scholar]
- 11.Gleyzer N, Vercauteren K, Scarpulla RC. Control of mitochondrial transcription specificity factors (TFB1M and TFB2M) by nuclear respiratory factors (NRF-1 and NRF-2) and PGC-1 family coactivators. Mol Cell Biol. 2005;25:1354–1366. doi: 10.1128/MCB.25.4.1354-1366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dhar SS, Ongwijitwat S, Wong-Riley MT. Nuclear respiratory factor 1 regulates all ten nuclear-encoded subunits of cytochrome c oxidase in neurons. J Biol Chem. 2008;283:3120–3129. doi: 10.1074/jbc.M707587200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dhar SS, Liang HL, Wong-Riley MT. Transcriptional coupling of synaptic transmission and energy metabolism: role of nuclear respiratory factor 1 in co-regulating neuronal nitric oxide synthase and cytochrome c oxidase genes in neurons. Biochim Biophys Acta. 2009;1793:1604–1613. doi: 10.1016/j.bbamcr.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dhar SS, Ongwijitwat S, Wong-Riley MT. Chromosome conformation capture of all 13 genomic loci in the transcriptional regulation of the multisubunit bigenomic cytochrome C oxidase in neurons. J Biol Chem. 2009;284:18644–18650. doi: 10.1074/jbc.M109.019976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scarpulla RC. Nucleus-encoded regulators of mitochondrial function: integration of respiratory chain expression, nutrient sensing and metabolic stress. Biochim Biophys Acta. 2012;1819:1088–1097. doi: 10.1016/j.bbagrm.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Satoh J, Kawana N, Yamamoto Y. Pathway analysis of ChIP-Seq-based NRF1 target genes suggests a logical hypothesis of their involvement in the pathogenesis of neurodegenerative diseases. Gene Regul Syst Bio. 2013;7:139–152. doi: 10.4137/GRSB.S13204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hossain MB, Ji P, Anish R, Jacobson RH, Takada S. Poly(ADP-ribose) polymerase 1 interacts with nuclear respiratory factor 1 (NRF-1) and plays a role in NRF-1 transcriptional regulation. J Biol Chem. 2009;284:8621–8632. doi: 10.1074/jbc.M807198200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herzig RP, Andersson U, Scarpulla RC. Dynein light chain interacts with NRF-1 and EWG, structurally and functionally related transcription factors from humans and drosophila. J Cell Sci. 2000;113(Pt 23):4263–4273. doi: 10.1242/jcs.113.23.4263. [DOI] [PubMed] [Google Scholar]
- 19.Hsiao HY, Jukam D, Johnston R, Desplan C. The neuronal transcription factor erect wing regulates specification and maintenance of Drosophila R8 photoreceptor subtypes. Dev Biol. 2013;381:482–490. doi: 10.1016/j.ydbio.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huo L, Scarpulla RC. Mitochondrial DNA instability and peri-implantation lethality associated with targeted disruption of nuclear respiratory factor 1 in mice. Mol Cell Biol. 2001;21:644–654. doi: 10.1128/MCB.21.2.644-654.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ross JM. Visualization of mitochondrial respiratory function using cytochrome c oxidase/succinate dehydrogenase (COX/SDH) double-labeling histochemistry. J Vis Exp. 2011:e3266. [DOI] [PMC free article] [PubMed]
- 22.Wang SW, Mu X, Bowers WJ, Klein WH. Retinal ganglion cell differentiation in cultured mouse retinal explants. Methods. 2002;28:448–456. doi: 10.1016/S1046-2023(02)00264-5. [DOI] [PubMed] [Google Scholar]
- 23.Mao CA, Kiyama T, Pan P, Furuta Y, Hadjantonakis AK, Klein WH. Eomesodermin, a target gene of Pou4f2, is required for retinal ganglion cell and optic nerve development in the mouse. Development. 2008;135:271–280. doi: 10.1242/dev.009688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen H, Vermulst M, Wang YE, Chomyn A, Prolla TA, McCaffery JM, Chan DC. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 2010;141:280–289. doi: 10.1016/j.cell.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li S, Chen D, Sauve Y, McCandless J, Chen YJ, Chen CK. Rhodopsin-iCre transgenic mouse line for Cre-mediated rod-specific gene targeting. Genesis. 2005;41:73–80. doi: 10.1002/gene.20097. [DOI] [PubMed] [Google Scholar]
- 27.Wong-Riley MT. Energy metabolism of the visual system. Eye Brain. 2010;2:99–116. doi: 10.2147/EB.S9078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Furuta Y, Lagutin O, Hogan BL, Oliver GC. Retina- and ventral forebrain-specific Cre recombinase activity in transgenic mice. Genesis. 2000;26:130–132. doi: 10.1002/(SICI)1526-968X(200002)26:2<130::AID-GENE9>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 29.Wu F, Kaczynski TJ, Sethuramanujam S, Li R, Jain V, Slaughter M, Mu X. Two transcription factors, Pou4f2 and Isl1, are sufficient to specify the retinal ganglion cell fate. Proc Natl Acad Sci U S A. 2015;112:E1559–E1568. doi: 10.1073/pnas.1421535112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan L, Deng M, Xie X, Gan L. ISL1 and BRN3B co-regulate the differentiation of murine retinal ganglion cells. Development. 2008;135:1981–1990. doi: 10.1242/dev.010751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang XM, Yang XJ. Regulation of retinal ganglion cell production by sonic hedgehog. Development. 2001;128:943–957. doi: 10.1242/dev.128.6.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dakubo GD, Wang YP, Mazerolle C, Campsall K, McMahon AP, Wallace VA. Retinal ganglion cell-derived sonic hedgehog signaling is required for optic disc and stalk neuroepithelial cell development. Development. 2003;130:2967–2980. doi: 10.1242/dev.00515. [DOI] [PubMed] [Google Scholar]
- 33.Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao Z, Mao CA, Pan P, Mu X, Klein WH. Transcriptome of Atoh7 retinal progenitor cells identifies new Atoh7-dependent regulatory genes for retinal ganglion cell formation. Dev Neurobiol. 2014;74:1123–1140. doi: 10.1002/dneu.22188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mu X, Fu X, Beremand PD, Thomas TL, Klein WH. Gene regulation logic in retinal ganglion cell development: Isl1 defines a critical branch distinct from but overlapping with Pou4f2. Proc Natl Acad Sci U S A. 2008;105:6942–6947. doi: 10.1073/pnas.0802627105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mu X, Beremand PD, Zhao S, Pershad R, Sun H, Scarpa A, Liang S, Thomas TL, Klein WH. Discrete gene sets depend on POU domain transcription factor Brn3b/Brn-3.2/POU4f2 for their expression in the mouse embryonic retina. Development. 2004;131:1197–1210. doi: 10.1242/dev.01010. [DOI] [PubMed] [Google Scholar]
- 37.Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Calvo SE, Clauser KR, Mootha VK. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2016;44:D1251–D1257. doi: 10.1093/nar/gkv1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wright AF, Chakarova CF, Abd El-Aziz MM, Bhattacharya SS. Photoreceptor degeneration: genetic and mechanistic dissection of a complex trait. Nat Rev Genet. 2010;11:273–284. doi: 10.1038/nrg2717. [DOI] [PubMed] [Google Scholar]
- 40.Ait-Ali N, Fridlich R, Millet-Puel G, Clerin E, Delalande F, Jaillard C, Blond F, Perrocheau L, Reichman S, Byrne LC, et al. Rod-derived cone viability factor promotes cone survival by stimulating aerobic glycolysis. Cell. 2015;161:817–832. doi: 10.1016/j.cell.2015.03.023. [DOI] [PubMed] [Google Scholar]
- 41.Cronin T, Raffelsberger W, Lee-Rivera I, Jaillard C, Niepon ML, Kinzel B, Clerin E, Petrosian A, Picaud S, Poch O, et al. The disruption of the rod-derived cone viability gene leads to photoreceptor dysfunction and susceptibility to oxidative stress. Cell Death Differ. 2010;17:1199–1210. doi: 10.1038/cdd.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Virbasius JV, Scarpulla RC. Activation of the human mitochondrial transcription factor a gene by nuclear respiratory factors: a potential regulatory link between nuclear and mitochondrial gene expression in organelle biogenesis. Proc Natl Acad Sci U S A. 1994;91:1309–1313. doi: 10.1073/pnas.91.4.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koshiba T, Detmer SA, Kaiser JT, Chen H, McCaffery JM, Chan DC. Structural basis of mitochondrial tethering by mitofusin complexes. Science. 2004;305:858–862. doi: 10.1126/science.1099793. [DOI] [PubMed] [Google Scholar]
- 44.Meeusen S, McCaffery JM, Nunnari J. Mitochondrial fusion intermediates revealed in vitro. Science. 2004;305:1747–1752. doi: 10.1126/science.1100612. [DOI] [PubMed] [Google Scholar]
- 45.Meeusen S, DeVay R, Block J, Cassidy-Stone A, Wayson S, McCaffery JM, Nunnari J. Mitochondrial inner-membrane fusion and crista maintenance requires the dynamin-related GTPase Mgm1. Cell. 2006;127:383–395. doi: 10.1016/j.cell.2006.09.021. [DOI] [PubMed] [Google Scholar]
- 46.Taherzadeh-Fard E, Saft C, Akkad DA, Wieczorek S, Haghikia A, Chan A, Epplen JT, Arning L. PGC-1alpha downstream transcription factors NRF-1 and TFAM are genetic modifiers of Huntington disease. Mol Neurodegener. 2011;6:32. doi: 10.1186/1750-1326-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cam H, Balciunaite E, Blais A, Spektor A, Scarpulla RC, Young R, Kluger Y, Dynlacht BD. A common set of gene regulatory networks links metabolism and growth inhibition. Mol Cell. 2004;16:399–411. doi: 10.1016/j.molcel.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 48.Wang C, Li Z, Lu Y, Du R, Katiyar S, Yang J, Fu M, Leader JE, Quong A, Novikoff PM, Pestell RG. Cyclin D1 repression of nuclear respiratory factor 1 integrates nuclear DNA synthesis and mitochondrial function. Proc Natl Acad Sci U S A. 2006;103:11567–11572. doi: 10.1073/pnas.0603363103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poche RA, Zhang M, Rueda EM, Tong X, McElwee ML, Wong L, Hsu CW, Dejosez M, Burns AR, Fox DA, et al. RONIN is an essential transcriptional regulator of genes required for mitochondrial function in the developing retina. Cell Rep. 2016;14:1684–1697. doi: 10.1016/j.celrep.2016.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang JL, Tong CW, Chang WT, Huang AM. Novel genes FAM134C, C3orf10 and ENOX1 are regulated by NRF-1 and differentially regulate neurite outgrowth in neuroblastoma cells and hippocampal neurons. Gene. 2013;529:7–15. doi: 10.1016/j.gene.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 51.Agathocleous M, Love NK, Randlett O, Harris JJ, Liu J, Murray AJ, Harris WA. Metabolic differentiation in the embryonic retina. Nat Cell Biol. 2012;14:859–864. doi: 10.1038/ncb2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 53.Mathieu J, Ruohola-Baker H. Metabolic remodeling during the loss and acquisition of pluripotency. Development. 2017;144:541–551. doi: 10.1242/dev.128389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Domcke S, Bardet AF, Adrian Ginno P, Hartl D, Burger L, Schubeler D. Competition between DNA methylation and transcription factors determines binding of NRF1. Nature. 2015;528:575–579. doi: 10.1038/nature16462. [DOI] [PubMed] [Google Scholar]
- 55.Westermann B. Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol. 2010;11:872–884. doi: 10.1038/nrm3013. [DOI] [PubMed] [Google Scholar]
- 56.Chen H, Chan DC. Emerging functions of mammalian mitochondrial fusion and fission. Hum Mol Genet. 2005;14 Spec No. 2:R283–R289. doi: 10.1093/hmg/ddi270. [DOI] [PubMed] [Google Scholar]
- 57.Rai M, Katti P, Nongthomba U. Drosophila Erect wing (Ewg) controls mitochondrial fusion during muscle growth and maintenance by regulation of the Opa1-like gene. J Cell Sci. 2014;127:191–203. doi: 10.1242/jcs.135525. [DOI] [PubMed] [Google Scholar]
- 58.Alexander C, Votruba M, Pesch UE, Thiselton DL, Mayer S, Moore A, Rodriguez M, Kellner U, Leo-Kottler B, Auburger G, et al. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat Genet. 2000;26:211–215. doi: 10.1038/79944. [DOI] [PubMed] [Google Scholar]
- 59.Stone J, van Driel D, Valter K, Rees S, Provis J. The locations of mitochondria in mammalian photoreceptors: relation to retinal vasculature. Brain Res. 2008;1189:58–69. doi: 10.1016/j.brainres.2007.10.083. [DOI] [PubMed] [Google Scholar]
- 60.Bentmann A, Schmidt M, Reuss S, Wolfrum U, Hankeln T, Burmester T. Divergent distribution in vascular and avascular mammalian retinae links neuroglobin to cellular respiration. J Biol Chem. 2005;280:20660–20665. doi: 10.1074/jbc.M501338200. [DOI] [PubMed] [Google Scholar]
- 61.Lovas JR, Wang X. The meaning of mitochondrial movement to a neuron's life. Biochim Biophys Acta. 2013;1833:184–194. doi: 10.1016/j.bbamcr.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chang B, Hawes NL, Hurd RE, Davisson MT, Nusinowitz S, Heckenlively JR. Retinal degeneration mutants in the mouse. Vis Res. 2002;42:517–525. doi: 10.1016/S0042-6989(01)00146-8. [DOI] [PubMed] [Google Scholar]
- 63.Veleri S, Lazar CH, Chang B, Sieving PA, Banin E, Swaroop A. Biology and therapy of inherited retinal degenerative disease: insights from mouse models. Dis Model Mech. 2015;8:109–129. doi: 10.1242/dmm.017913. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The RNA-seq data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE101550 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE101550).