Abstract
Increase in global population and growing disease burden due to the emergence of infectious diseases (Zika virus), multidrug-resistant pathogens, drug-resistant cancers (cisplatin-resistant ovarian cancer) and chronic diseases (arterial hypertension) necessitate effective therapies to improve health outcomes. However, the rapid increase in drug development cost demands innovative and sustainable drug discovery approaches. Drug repositioning, the discovery of new or improved therapies by reevaluation of approved or investigational compounds, solves a significant gap in the public health setting and improves the productivity of drug development. As the number of drug repurposing investigations increases, a new opportunity has emerged to understand factors driving drug repositioning through systematic analyses of drugs, drug targets and associated disease indications. However, such analyses have so far been hampered by the lack of a centralized knowledgebase, benchmarking data sets and reporting standards. To address these knowledge and clinical needs, here, we present RepurposeDB, a collection of repurposed drugs, drug targets and diseases, which was assembled, indexed and annotated from public data. RepurposeDB combines information on 253 drugs [small molecules (74.30%) and protein drugs (25.29%)] and 1125 diseases. Using RepurposeDB data, we identified pharmacological (chemical descriptors, physicochemical features and absorption, distribution, metabolism, excretion and toxicity properties), biological (protein domains, functional process, molecular mechanisms and pathway cross talks) and epidemiological (shared genetic architectures, disease comorbidities and clinical phenotype similarities) factors mediating drug repositioning. Collectively, RepurposeDB is developed as the reference database for drug repositioning investigations. The pharmacological, biological and epidemiological principles of drug repositioning identified from the meta-analyses could augment therapeutic development.
Keywords: precision medicine, drug repositioning, translational bioinformatics, drug development, drug discovery, precision pharmacology, systems pharmacology
Introduction
Precision medicine, also known as stratified medicine, is a collective term that represents a new and evolving health care delivery model, which encompasses accurate diagnosis, personalized interventions and individualized recovery strategies for an individual patient [1–3]. The primary goal of precision medicine is precision therapeutics, which aims to provide optimized treatments with the highest efficiency and fewest side effects matching the unique disease signature of a patient. Drug repurposing (or drug repositioning), i.e. the development of new or improved therapies by reevaluation of approved or investigational compounds, is a promising strategy for precision pharmacology and, thus, may improve the productivity of drug development [4].
Recently, drug repositioning has emerged as a cost-effective and efficient approach to bring therapeutic discoveries from bench to bedside in a short span of time. Although traditional drug development relies on high-throughput screening of thousands of drug targets and millions of pharmaceutical compounds, drug repositioning focuses on the reuse of compounds with some degree of a priori knowledge. Although earlier examples of drug repurposing relied primarily on medicinal chemistry and clinical serendipity [5–7], more recent examples have successfully used diverse computational methods and open-access biomedical informatics resources [8–10]. The expanding catalog of drug, tissue, disease and gene expression signatures from cMAP [11] (https://www.broadinstitute.org/cmap/), LINCS (http://www.lincscloud.org/) and GEO (http://www.ncbi.nlm.nih.gov/geo/) is vital for implementing computational drug repurposing in the setting of precision medicine. One exemplary technique in computational repositioning is called connectivity mapping, where gene expression signatures of drugs and diseases are compared, positing that if a drug perturbs gene expression in opposition to disease perturbations, then that drug may be therapeutic for that disease. Combining genomic-based, transcriptomic-based and connectivity mapping-based approaches has also been used to recommend potential indications for different cancers, Zika virus, multidrug-resistant pathogens, cardiovascular diseases and psychiatric diseases [12–19].
Drug repositioning investigations are currently being used as a therapeutic development strategy for several common, chronic, rare and emerging diseases. As the number of drug repurposing investigations continues to increase, a new opportunity emerges from analyzing the universe of repositioned therapies to identify patterns that underlie successful drug repositioning. Several databases like PROMISCOUS and DMAP are also available (see Availability of related resources for drug repositioning in the Supplementary Materials) in the open access domain with drug repositioning and related content [20, 21]. However, such resources and previous analyses have so far been hampered by the lack of a centralized database as well as a lack of reporting standards for drug repositioning investigations. To address this gap, we developed RepurposeDB (http://repurposedb.dudleylab.org), a database of drug repositioning studies reported on public resources like PubMed and Food and Drug Administration (FDA) databases. The analyses of the repertoire of drugs, drug targets and associated disease indications from RepurposeDB reveal several factors associated with drug repurposing.
In this report, we discuss various features of the RepurposeDB (version 1) database and present collective insights obtained from the systematic analyses of the database content. For example, we generated a statistical summary of various physicochemical properties of repurposed compounds compared with various compound subsets from DrugBank. We also analyzed drug targets (proteins) of repurposed compounds, identifying over-represented patterns in the underlying biological activity (i.e. mechanisms of action of compounds, biological pathways of target genes and structural similarities of target proteins). Finally, we present a digital epidemiology analysis using electronic medical record (EMR) data, addressing the degree to which ‘repurposing disease pairs’ (i.e. disease pairs treated by the same drug) present as comorbidities. Together, findings from the systematic analyses of the data from RepurposeDB provide pharmacological, biological and epidemiological evidence to support data-driven drug repurposing strategies as an essential tool kit for drug discovery.
Methods
RepurposeDB (http://repurposedb.dudleylab.org) is a compendium of drugs (small molecules and biotech or protein drugs) and their associated primary and secondary diseases in which the compound was indicated as effective. Exploring these datasets using enrichment analysis helped us to understand key biological pathways, functional mechanisms, physicochemical features and side effects associated with successfully repositioned drugs, which can aid in designing better drug repositioning investigations in the future [5, 22]. Molecular function of proteins and biochemical pathways act in concert to perform a variety of functions in the illness and wellness states of human physiology [23]. Emerging evidence from pathway cross-talk studies indicates that the pathophysiology of mulitple diseases can be modulated by the same set of pathways [24, 25]. We have explored the proteins and gene sets from RepurposeDB using biological ontologies overlapped with a variety of gene set annotations to understand the functional and chemical promiscuity associated with repositioned compounds and their targets [26–28]. Findings from the meta-analyses of drugs, drug targets and disease phenotypes in RepurposeDB promote the inclusion of three additional data types and analytical strategies, namely pathway cross talks, shared genetic architectures (SGA) and prevalence of disease comorbidities into drug repurposing pipelines. Integrating these approaches to existing drug repurposing pipelines could help to identify new indications for existing compounds or alternate drugs for a disease with a known pathway or genetic associations.
RepurposeDB—data integration, design and database development
Data collection and curation
The catalog of repurposed drugs was compiled using a combination of text mining of the PubMed database [n = 23 million abstracts (www.ncbi.nlm.nih.gov/pubmed)] and manual curation of manuscripts that have reported drug repositioning. For the text mining, the initial searches in PubMed using combinations of ‘drug repositioning’ or ‘drug repurposing’ and semantic variations were implemented to generate a list of abstracts. Abstracts with more than one disease terms (e.g. ‘rheumatoid arthritis’ and ‘Crohn’s disease’) were filtered using NCBI E-utilities (http://www.ncbi.nlm.nih.gov/books/NBK25500/). Finally, for the manual biocuration, we examined curated research and review articles that report drug repositioning or repurposing investigations (Figure 1; n = 258; see Supplementary Data File: RepurposeDB_PubMed_Articles.xlsx).
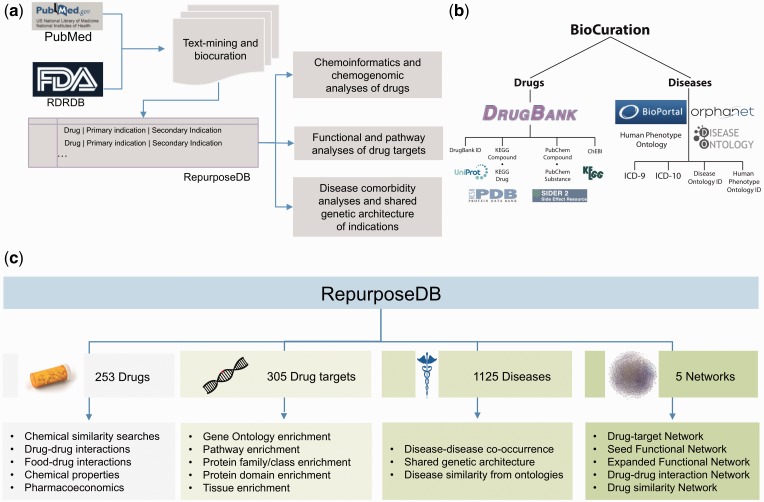
Figure 1.
Curation, mapping and analytics strategy of RepurposeDB. (A) Biocuration strategy leveraged to develop RepurposeDB. (B) Terminology mapping strategy used to compile disease dictionaries. (C) Analytics framework for analyzing medications (small molecules and biotech), drug targets, diseases and networks (drug–target, seed functional target network, expanded functional target network, drug–drug and drug similarity network).
Data processing and annotation
We collected data in the form of drugs, diseases and annotations from DrugBank [29], KEGG Drug and Compound databases (http://www.genome.jp/kegg/), PubChem (https://pubchem.ncbi.nlm.nih.gov/), Chemical Entities of Biological Interest (ChEBI; https://www.ebi.ac.uk/chebi/), SIDER (http://sideeffects.embl.de/) and US FDA Rare Disease Repurposing Database [30]. For disease and phenotype data, we manually curated terms and mapped them to three different disease ontologies, specifically International Classification of Diseases 9 (ICD-9) codes, Human Phenotype Ontology (HPO) and Disease Ontology (DO) using concept unique identifier (CUI) codes as intermediary identifiers. Finally, we integrated the phenotype data and drug data and generated an indexed resource using drugs and diseases. The final, nonredundant data set of drug repositioning investigations was compiled as triples in the format of ‘drug primary indication-secondary indications’. The entire database was finally mapped to the repertoire of biomedical ontologies (see Supplementary Data File: RepurposeDB_PanOntology_Mapping.xlsx). Primary indication refers to the original disease indication for which the drug is targeted, and secondary indication indicates any subsequent indications (see Limitations section).
Pan-ontology mapping of RepurposeDB knowledge corpus
We have mapped the entire RepurposeDB knowledge to all available biomedical ontologies from National Center for Biomedical Ontologies (NCBO) BioPortal using Annotator program (http://bioportal.bioontology.org/). Each term (drug, primary indication or secondary indication) in RepurposeDB was used as a query against BioPortal. We compiled the results in JSON formatted files using the REST interface of Annotator. We provide the statistics of term-level mapping in the Supplementary Material (see Pan-ontology mapping statistics and Supplementary Data File: RepurposeDB_PanOntology_Mapping.xlsx).
User interface design
The web interface of RepurposeDB, search utilities and Minimum Information About Drug Repositioning Investigations (MIADRI) standard was developed using HTML, CSS and JavaScript. We provide a technical summary of the database development methods, Web server architecture of various tools, database design and various features in the Supplementary Materials. We also provide a summary of the different user interfaces including ‘Drug’ page, ‘Disease’ page, browse utilities, search engines (keyword search, chemical similarity search and sequence similarity search), visual analytics tools and various files and data sets available for download in the Supplementary Materials.
Systematic analyses of drug repositioning investigations
Using the compendium of curated, nonredundant lists of drug repositioning examples, we performed extensive analyses to identify underlying properties that facilitate successful drug repositioning. Chemoinformatics features of the small molecules were computed using OpenBabel, Pybel [31], JOELib, JOELib2 [32–36] and Chemminer [35] services and custom Python and R scripts.
Pharmacological properties of small molecules in RepurposeDB
The small molecule analyses use the subset of small molecules in RepurposeDB (n = 188) after excluding protein drugs (n = 65). We observed that repurposed drugs from 19 different drug superclasses are represented in RepurposeDB (χ2, P < 0.001). Physicochemical features and chemical descriptors were computed using three different libraries and computed using SDF files. SDF files were also used for visual exploration of chemical structures in RepurposeDB. Precomputed chemical features using ChemAxon (https://www.chemaxon.com), absorption, distribution, metabolism, excretion and toxicity (ADMET) properties were aggregated from DrugBank.
Physicochemical features, chemical descriptors and ADMET values
We have compiled the physicochemical features computed using ALOGPS [37] and ChemAxon algorithms (https://www.chemaxon.com; see Supplementary Data: RepurposeDB_ChemicalProperties.xlsx). A total of 112 properties were computed using three different chemoinformatics libraries (Pybel, JOELib2 and Chemminer; see Supplementary Data: RepurposeDB_ChemicalProperties.xlsx).
ADMET data can help in filtering of individual small molecules as potential lead candidates for drug development [38]. We aggregated the predicted ADMET data from DrugBank and assessed whether repositioned drugs have any significant difference when compared with the approved drugs or compound repertoire in DrugBank.
Chemogenomic enrichment analysis of small molecules in RepurposeDB using Chemogenomic method
Chemogenomic enrichment analysis (CGEA) is a methodology (Manuscript in Preparation) that compares drug compounds with a variety of biological and chemical annotations similar to gene set enrichment analysis [39], metabolite set enrichment analysis [40] or compound set enrichment analysis [41]. We used a subset of 94 drugs from RepurposeDB to perform CGEA analysis (see Supplementary Data: RepurposeDB_CGEA.xlsx). Briefly, CGEA maps drug compound lists and genes to various annotation resources including gene sets, chemoinformatics annotations, drug targets, side effects and drug classes. It tests for over- and under-enrichment across various annotations and provides detailed enrichment results with ranked list of compounds, genes and annotation terms. CGEA facilitates interpretation of the ranked list of compounds that have been prioritized by similarity/dissimilarity between their transcriptional profile and a profile of interest.
Chemical ontology enrichment analysis of small molecules in RepurposeDB using BiNChE
We tested the compounds in RepurposeDB across both ‘structure’ and ‘role’ subsets of ChEBI ontology, which is a knowledge corpus of chemical compounds with biological roles [42]. A total of 145 compounds from RepurposeDB were mapped to ChEBI database. Lists of compounds mapped from RepurposeDB were tested against ChEBI ontology to understand biochemical properties of repositioned compounds using BiNChE [43] (see Supplementary Data: RepurposeDB_BinChe.xlsx).
Functional and pathway enrichment analysis
Gene ontology (GO) enrichment analyses were performed to identify significant categories of biological processes, molecular functions and cellular components associated with drug targets of repositioned compounds. Various protein-level enrichment analyses were performed using annotations from various protein-centric databases like Uniprot [44], Pfam [45], Structural Classification Of Proteins (SCOP) [46] and CATH [47]. Pathway enrichment analysis was performed using annotations from Reactome [48] and KEGG [49]. Biological functional enrichment and pathway enrichment analyses were performed using Enrichr [50] and DAVID [51]; both tools were used with the list of genes from the standard reference genome or the canonical list of proteins from human proteome as the back ground for enrichment tests. A Bonferroni threshold for multiple testing was defined to find statistically significant terms enriched among the target list.
Enrichment analysis using DAVID
We used the DAVID bioinformatics software package to test the functional association of drug targets in RepurposeDB with various annotation lists (listed in the Supplementary Materials; also, see Supplementary Data: RepurposeDB_DAVID.xlsx). We found statistically significant enrichments after multiple testing corrections for all except one-annotation resource (SCOP_CLASS).
Enrichment analysis using Enrichr
We used Enrichr (http://amp.pharm.mssm.edu/Enrichr/) to test for enrichment of targets in RepurposeDB using 56 gene lists. After multiple testing correction, 26 lists (listed in the Supplementary Materials; see Supplementary Data: RepurposeDB_Enrichr.xlsx) had significantly enriched annotations associated with list of targets in RepurposeDB.
Consensus pathway analysis using Consensus Pathway Annotations
Target proteins in RepurposeDB were tested for pathway-level enrichment using Consensus PathDB (CPDB) [52]. CPDB offers pathway enrichment over 4593 pathways integrated from 32 resources (see Supplementary Data: RepurposeDB_CPDB.xlsx). We defined pathway cross talk using pathway enrichment analyses results from CPDB. For example, the gene ADRA2A is part of the Reactome pathway ‘Adrenaline signalling through Alpha-2 adrenergic receptor’ and the drug target can bind and induce mechanistic action (inhibition or activation) via multiple drugs (apomorphine, aripiprazole, brimonidine, bromocriptine, guanfacine, phentolamine, pramipexole and ropinirole). Three targets (ADRA2A, ADRA2B and ADRA2C) from RepurposeDB are mapped to this pathway (P = 1.87E-05). Targets of aripiprazole, a drug that treats several psychiatric disorders, are enriched across 64 pathways. Targets of drugs like bromocriptine (menstrual problems, Parkinson’s disease and pituitary tumors), phentolamine (hypertension and impaired night vision), lapatinib (various cancers), bivalirudin (various cancers), arsenic (syphilis and leukemia), pemetrexed (lung cancer and mesothelioma), imatinib (chronic myeloid leukemia and gastrointestinal stromal tumor), sunitinib (various cancers), sorafenib (melanoma and various cancers), midazolam (seizure and epilepsy), nabumetone (rheumatoid arthritis and osteoarthritis), aminosalicylic acid (Crohn’s disease and ulcerative colitis), celecoxib (rheumatoid arthritis and various cancers), duloxetine (fibromyalgia and major depressive disorder), lenalidomide (various cancers), mazindol (obesity and Duchenne’s muscular dystrophy) and methylphenidate (eating disorder and attention-deficit hyperactivity syndrome) are all associated with >10 pathways (all observations P≤0.001).
Disease analysis of a compendium of 1125 diseases targeted by repositioned drugs
The relationship between diseases that are significantly comorbid is unexplored in the realm of drug repositioning. It remains unclear whether multiple indications of a drug are typically active because the diseases manifest as a comorbid condition in a population setting than random, such that a given heterogeneous patient population have higher prevalence a particular disease pair. Recent demonstrations of EMR-based phenomic analyses exemplify the secondary use of EMR data as a proxy for epidemiological observational studies to quantitatively estimate disease comorbidities using relative risk or standardized incident rates [53–55]. By consolidating data from RepurposeDB with publicly available genomic annotation databases and disease comorbidity data extracted from EHR, we tested whether shared genetic architecture or co-occurrence of comorbidity a pair of disease could assist in rational drug repositioning. We have also quantified the similarity between a pair of diseases using semantic similarity calculation using DO and HPO [56, 57]. To perform the disease analyses, individual disease terms from RepurposeDB were manually curated and mapped to the corresponding ICD-9 codes, HPO terms and DO terms. ICD-9 codes were used to aggregate EHR data and compute disease co-occurrences. Disease terms were used to compute shared genetic architecture (SGA), and both HPO and DO terms were used to compute semantic similarity of diseases.
Pair-wise disease comorbidity analyses using diagnosis data compiled from electronic health records
Manifestations of complex illnesses such as type 2 diabetes [58], peripheral arterial disease [59] or heart failure [60] often present with comorbid conditions in patient subpopulations. Estimating pair-wise disease comorbidity using EMR-wide disease prevalence data would help to understand whether drug repositioning is successful across two diseases if they are comorbid in patient population. To test this, we used data contained in the Mount Sinai Data Warehouse (MSDW) for disease co-occurrence analyses (https://msdw.mountsinai.org). MSDW hosts data from a large, tertiary care teaching hospital in the Greater New York City area. Health care and biomedical data from MSDW offer one of the most ethnically diverse, urban patient populations in the world (see: https://msdw.mountsinai.org/) because of the unique location of Mount Sinai Health System and affiliated hospitals. The MSDW, which houses all the clinical data, currently, has 2 125 468 unique patients (as of February 2015) with a minimum of one encounter, >16 million patient visits recorded, ∼1.7 billion patient encounters and >46 515 678 ICD-9-coded diagnoses documented. These >2 million patients were stratified by gender and self-reported ethnicity. For gender, the patient population consists of: 2 263 195 females (56.09%), 1 753 120 males (43.45%) and 18 609 other/unknown (0.47%). For self-reported ethnicity, the breakdown is as follows: 337 149 African American (8.36%), 92 447 Asian (2.29%), 943 742 Caucasian (23.38%), 363 447 Hispanic/Latino (9.01%), 6821 Native American (0.17%), 2103 (0.05%) Pacific Islander, 420 351 other (10.41%) and 1 868 864 unknown (46.32%). Within the MSDW, disease information is stored as ICD-9 codes. We used DO to map disease indications from RepurposeDB to ICD-9 codes, and we successfully mapped 887 unique disease terms to at least one ICD-9 code. Using these data, we performed comorbidity enrichment analysis between all unique combinations of primary and secondary indications per drug, resulting in 2970 total tests. To determine comorbidity enrichment, we performed a one-sided Fisher’s exact test comparing the number of instances of which a patient had both the primary and secondary disease with the number of instances of each disease separately to background of total patients in the EMR. To reduce the testing space, we restricted our disease pairing using a directional estimate of primary disease and the secondary and orphan disease pairs (see Supplementary Data: RepurposeDB_EHR_SGA.xlsx). It should be noted that some of the associations indicate inherent relationships across diseases observed in EMR. For example, both pediatric manifestation and adulthood form of the disease capture in EMR (e.g. juvenile growth hormone deficiency and adult growth hormone deficiency). Examples of disease recurrence as chronic presentation and acute disease or vice versa (e.g. acute intestinal amebiasis and chronic intestinal amebiasis) are also considered in our analyses without predicates (‘chronic’ or ‘recurrent’). Relative risk is computed using a number of patients diagnosed with both diseases and random expectation based on disease prevalence method explained in Hidalgo et al. [61, 62].
Genetic architectures shared between diseases treated by same drug
Emerging evidences indicate that disease and related clinical phenotypes could be driven by SGA [63]. For example, Li et al. [64] showed that a routinely measured laboratory test (mean corpuscular volume) was elevated in patients with acute lymphoblastic leukemia before the diagnosis; these two phenotypes shared a subset of genes and defined as the molecular basis of shared genetic architecture across clinical traits and diseases. Recently, we have discovered 19 novel disease relationships by leveraging disease comorbidities with genetic architectures [53, 64, 65]. We have compiled a list of disease–gene association data from various resources including Online Mendelian Inheritance in Man [66], GWAS-catalog [67], GWASdbv2 [68, 69] HuGENavigator [70] and a proprietary database built through text mining and manual curation (VarDi). We were able to map 755 diseases from RepurposeDB (67% of indications) to VarDi by mapping variants at the gene level using CUI codes as a bridge. We performed an identical Fisher’s exact analysis, as in the previous section, to test the significance of shared genes between diseases (see Supplementary Data: RepurposeDB_EHR_SGA.xlsx).
Phenomic similarities of primary and secondary indications in RepurposeDB
There has been a wide range of semantic measures developed for information extraction from diverse ontologies and structured data in the fields of bioinformatics, natural language processing, artificial intelligence and the Semantic Web [71, 72]. We first used an experimental approach to evaluate which of the measures are most robust for evaluating human phenotype ontologies (DO and HPO). Semantic measures of concept set similarity use combinations of multiple methods to characterize different ontology scales (i.e. from quantifying information content of a single node to summarizing the similarity between multiple pairs of nodes that were individually scored by a separate metric). We tested several distinct ontology evaluation methods including: (1) single node evaluation of intrinsic information content measures (three methods), (2) similarity of pairs of two nodes (three methods based on node set, three based on node information content and two based on edges), (3) group similarity of measures of pairs (five methods) and (4) group similarity of measures of many individual nodes (two methods based on connectivity and two methods based on information content). We generated 128 full combinatorial group similarity metrics by implementing each possible dependency combination. Semantic similarity of pair of diseases was computed using the Java-based Semantic Measures Library and Toolkit (SMLTK) [73]. Using SMLTK, 128 similarity scores of indications for each drug. We performed a transitive reduction and rerouting on both ontology hierarchies to maintain the network extensibility while eliminating potential biases in the depth of classification used for different phenotypes. We ranked each metric for robustness based on correlation between similarity scores in both phenotype ontologies for the 101 drugs with multiple indications. We evaluated the effect of the number of existing drug indications on similarity score by using an analysis of variance (ANOVA) test and found that different indirect methods are significantly biased in each direction. For specific applications within drug repositioning, these different indirect metrics provide different insights into the drug indication pleiotropy. For this initial study on semantic similarity to characterize drugs indication diversity and repurposing potential, we used an average of the most robust indirect metrics for each aspect of indication set similarity (the top measure of indication diversity, the top measure of indication clustering density and the top measure of balanced indication similarity; see Supplementary Data: RepurposeDB_Disease_Phenomic Similar ity.xlsx).
Reconstruction and analyses of repurposed drug–drug, drug–food and drug–target interaction network
Various factors influence repositioning strategies including side effects, network properties of the drug–targets, potential food–drug interactions [74, 75] and drug–drug interactions [76, 77]. We used the list of 298 proteins mapped to GeneMANIA [78, 79] as the query to understand putative interactions mediated by the target proteins of repositioned drugs and to generate two functional networks: the first using drug targets of repurposed compounds (Seed Functional Network) and the other network for finding targets that are functionally close to known repositioning targets from human protein interactome databases (Expanded Functional Network; see Supplementary Data files: RepurposeDB_Drug_Food_Target_Networks.xlsx and Repurpose DB_SFN_EFN_NA.xlsx). We provide details about the construction of the chemical similarity network, drug–target bipartite network, Seed Functional Network of targets, expanded functional network for novel drug target discovery and drug–drug interaction network of repositioned drugs in the Supplementary Materials. Drug–target networks were reconstructed using direct or inferred interactions derived from reference databases. We computed and visualized the chemical similarity network using chemical similarity distance computed using Tanimoto coefficients [80, 81].
Statistical analysis
Statistical analyses were performed using JMP 11 (SAS Institute Inc., Carey, NC) and R language (R Foundation for Statistical Computing, Vienna, Austria). Student’s t-test was used as appropriate to assess difference between two groups. Statistical significance was set at P < 0.05, using two-tailed distribution and two-sample equal/unequal variance. Three group comparisons were performed using two-way ANOVA. All enrichment P values were reported after multiple testing corrections using default setting of the respective analytical applications; corrected P-value threshold of P < 0.05 was used to define significance. No directionality is assumed during the enrichment analyses, network analytics or statistical testing. DrugBank is a compendium of all drugs ever marketed. However, some of these drugs could be retracted (because of safety or other post-market surveillance issues) or purged (e.g. because of patent issues). We used the entire DrugBank (designated as DrugBank-F) and a subset of DrugBank (designated as Drug Bank-A) with current approval status at the time of writing this manuscript in 2016 as our background set for various statistical comparisons. The rationale for this division is to test how the subset of repurposed drugs compare on not only the entire marketed but also the currently available drugs in the market. Our approach would also help to measure for bias, as the repurposed drugs are highly reused because they are also approved. By performing analyses in two levels, we will be able to control or adjust for such knowledge or market bias. For all annotation-based enrichment analyses, we have tested the enrichment across the human genome and human proteome to balance such biases.
Results
Building a reference data set of repositioned drugs, targets and diseases
Organization and content of the RepurposeDB database
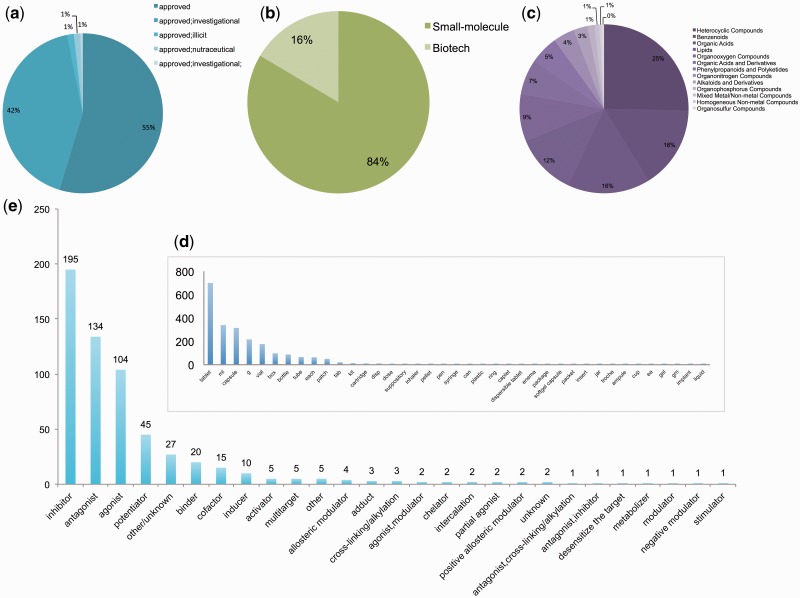
The current release of RepurposeDB (v1; as on 30 March 2016) contains 253 drugs, 1125 indications and 3660 data triples. The triples in RepurposeDB are annotated using 302 different biomedical and health care ontologies from NCBO-BioPortal (http://bioportal.bioontology.org/). We integrated 36 332 annotations using pan-ontology approaches, thus making RepurposeDB one of the most richly annotated biomedical reference databases currently available in the public domain. We organized and compiled RepurposeDB using ‘Drugs’ (n = 253) and ‘Disease’ (n = 1125) entry pages. Users can access individual pages by browsing or searching the database using the indexed keyword dictionary or search terms. We provide a detailed technological overview of the database development and various features (Figure 2) including tools for data visualizations and similarity searches (compound, drug target and protein–drug similarity) in the Supplementary Materials. RepurposeDB drugs with approval status from FDA consist of 84% small molecules and 16% biotech drugs (or protein drugs). Specific chemical classes have enrichment of repurposed compounds and depletion across others. For example, the drug superclasses like heterocyclic compounds, phenylpropanoids and organooxygen compounds have >10 drugs in RepurposeDB, representing around 39.5% of the compounds. The following superclasses had no representative drugs reported: lignans and norlignans, homogeneous metal compounds, organic halides, organometallic compounds and non-benzenoid aromatic compounds (tropones; see Figure 3 and Supplementary Data File for complete data: RepurposeDB_ChemicalProperties.xlsx).
Figure 2.
Database interface and features of RepurposeDB. (A) Web interface of RepurposeDB. (B) Plotting utility to compare and map various chemoinformatics features (n=112) and display on an interactive plot. (C) Web-based visualization to view drug–disease bipartite network. (D) Search service to compare a given small molecule in SMILE format across repositioned compounds in RepurposeDB using Tanimoto distance.
Figure 3.
Biochemical composition of medications in RepurposeDB a) Approval status b) Molecular types of medications in RepurposeDB c) Super-Classes of small molecules in RepurposeDB d) Distribution of units by which repositioned drugs are marketed e) Mode of drug-target interactions in RepurposeDB.
Minimum Information about Drug Repositioning Investigations
We propose MIADRI (see http://repurposedb.dudleylab.org/MIADRI) as a new standard for drug repurposing investigators to report their results to the community. We developed a dedicated interface to submit information about a new drug repositioning study not included in RepurposeDB. The absence of a common, community standard in reporting, aggregating and disseminating data hinders the impact of drug repositioning investigations and discovering new therapeutic indications for existing pharmaceutical agents. Submissions to RepurposeDB shall follow the MIADRI guidelines. We envisage that MIADRI will help in rapid aggregation and meta-analyses of drug repositioning investigations over the years. We further describe the various features and requirements of the new guidelines aiming to capture and reuse data from future drug repurposing investigations in the Supplementary Materials (also, see Supplementary sections under RepurposeDB—design and development, RepurposeDB—features, Submission of new drug repositioning investigations to RepurposeDB using MIADRI standard and Supplementary Table S1).
Pharmacological, biological and epidemiological factors of drug repurposing
Each entry (drug or disease) in RepurposeDB includes a drug (small molecule, bioactive, etc.), its primary and secondary disease indication and the PubMed identifier that reported the investigation. We aggregated various metadata to this list using the drug name as a query term input to other databases to retrieve mechanism of actions, biophysical and biochemical properties, side-effect profiles and target information. Drug compounds were assessed for statistical enrichment of various small molecule-related properties relative to all compounds (DrugBank-F; n = 7759) and approved subset (DrugBank-A; n = 1673) of compounds in DrugBank. We assessed the target proteins of repositioned compounds to find significantly enriched GO terms (biological processes, cellular compartment and molecular functions), gene sets and pathways. We have compiled data from three types of disease analyses: (1) disease comorbidities, (2) shared genetic architecture and (3) semantic similarity of diseases. Semantic similarity of diseases was computed using the two previously described disease ontologies.
Pharmacological profiling of small molecules in RepurposeDB
Integration of chemoinformatics and genomic (chemogenomic) approaches accelerates the drug target discovery cycle and the prioritization of new indications for existing or orphan compounds [82, 83]. Combining biological (genomic, proteomic and metabolomics) and chemical knowledge of the structure, activity and pharmacokinetic properties has been shown to provide better approaches for prediction and validation of new drug targets and aid in designing chemical entities against targets with functional roles [84, 85]. It is unclear, however, whether repositioned compounds share similar chemical features, descriptors or ADMET properties. To answer these questions, we evaluated various physicochemical characteristics (e.g. bond matrices of compounds, number of hydrogen donors and number of hydrogen acceptors) in RepurposeDB and compared them with the DrugBank-F and DrugBank-A. The subset of drug compounds (small molecule subset excluding protein or biotech drugs; n = 188) in RepurposeDB was analyzed to understand various physicochemical features, chemical descriptors and ADMET properties. We tested the RepurposeDB compounds for enrichment across various chemogenomic annotations using chemogenomic enrichment analyses (CGEA) and ontology-based enrichment analyses using ChEBI ontology.
Physicochemical features associated with repurposed compounds
We noted that mean values of various physicochemical features were different for repositioned compounds when compared with DrugBank-F and DrugBank-A. Mean values were lower than approved drugs for eight features (logP, logS, refractivity, polarizability, pKa [acidic], pKa [basic], physiological charge and the number of rings). Mean values were higher than approved drugs for hydrogen bond donor count suggesting a greater number of hydrogen bonds could contribute to the pluripotent drug–target binding mechanism across multiple disease phenotypes (Table 1; also, see Supplementary Data: RepurposeDB_ChemicalProperties.xlsx).
Table 1.
Chemical features of repositioned drugs
| Feature | DrugBank-F | DrugBank-A | RepurposeDB | P* |
|---|---|---|---|---|
| LogPa | 1.58 | 2.104 | 1.54 | <0.001 |
| Logsa | −3.137 | −3.492 | −3.119 | 0.003 |
| Molecular weight | 350.632 | 378.692 | 372.178 | 0.065 |
| Monoisotopic weight | 350.298 | 378.317 | 371.178 | 0.065 |
| PSA | 101.248 | 90.064 | 104.634 | 0.059 |
| Refractivity | 90.552 | 99.505 | 97.138 | 0.017 |
| Polarizability | 34.935 | 38.182 | 37.97 | 0.019 |
| Rotatable bond count | 5.65 | 5.66 | 5.12 | 0.43 |
| H-bond acceptor count | 5.197 | 4.931 | 5.734 | 0.224 |
| H-bond donor count | 2.75 | 2.246 | 2.713 | 0.018 |
| pKa (strongest acidic) | 8.08 | 9.501 | 9.453 | <0.001 |
| pKa (strongest basic) | 2.627 | 4.008 | 3.659 | <0.001 |
| Physiological charge | −0.195 | 0.209 | 0.144 | <0.001 |
| Number of rings | 2.442 | 2.814 | 2.663 | 0.003 |
Note. aFeature computed using ALOGPS, other features computed using ChemAxon (all values presented as mean). DrugBank-F=DrugBank Full; DrugBank-A=Approved subset of DrugBank.
Two-way ANOVA of feature across presence in RepurposeDB and approval status.
Chemical descriptors of repurposed compounds
We compiled a library of 110 chemical descriptors and identified a subset of 27 features significantly associated with drug repositioning. This library of descriptors includes different chemical classes including atomic, compositional and geometric descriptors (see Supplementary Data: Supplementary Table S2 and RepurposeDB_ChemicalProperties.xlsx).
ADMET properties
Nineteen different ADMET properties (listed in the Supplementary Materials) were extracted from DrugBank and compared against DrugBank and DrugBank-A (see Supplementary Data: RepurposeDB_ChemicalProperties.xlsx). Nine properties were significantly associated with RepurposeDB compounds (see Supplementary Data: Supplementary Table S3 and RepurposeDB_ChemicalProperties.xlsx).
Chemogenomic and side-effect enrichment analysis using CGEA
We used chemogenomic enrichment analysis (CGEA) package to analyze compounds in RepurposeDB to understand enrichment using a combined database of biochemical or genomic annotations (see Supplementary Data File: RepurposeDB_CGEA.xlsx). Two enzymes metabolize multiple repositioned drugs: CYP3A7 can metabolize 19 drugs, and CYP3A5 can metabolize 18 drugs, suggesting that enzymatic activity and metabolic modulation could be used as a possible feature of predicting drugs capable of drug repositioning. We observed that drug transporter genes including ABCC10, ABCB11 and ABCG2 were associated with the transport of multiple repositioned compounds, suggesting a vital role of drug transporters and their nonspecific binding affinity as a putative factor to assess the repurposability potential of a compound. Enrichment tests across different levels of Anatomical Therapeutic Chemical (ATC) Classification System (http://www.whocc.no/atc_ddd_index/) revealed that repositioned compounds are enriched for two features in ATC-3 levels (stomatological preparations and immunosuppressants) suggesting switching between formulation types (e.g. reformulation of a topical to systemic glucocorticoids) could represent new avenues for potential drug repositioning.
Using CGEA, we assessed the enrichment of side effects using the OFFSIDES database to understand the set of side effects associated drug repositioning (http://tatonettilab.org/resources/tatonetti-stm.html). Common side effects including pain, nausea and altered mental status could act as a proxy for multisystem interactions, and indicate repurposability. The most frequent molecular fragment in cMAP, c1ccccc1 (present in 354 of 1309 drugs), is under-represented in this subset suggesting compounds without the chemical moiety could be more likely to repositionable (Table 2; also, see Supplementary Data File RepurposeDB_CGEA.xlsx).
Table 2.
Top 20 side effects associated with repositioned drugs
| Side effects | Expected | Observed* | Pa | Adjusted Pb | FC |
|---|---|---|---|---|---|
| Pain | 13.86 | 51 | 5.09E-21 | 1.24E-17 | 3.68 |
| Nausea | 13.86 | 51 | 5.09E-21 | 1.24E-17 | 3.68 |
| Mental status changes | 11.27 | 44 | 9.98E-19 | 1.63E-15 | 3.90 |
| Nephrolithiasis | 7.90 | 37 | 1.92E-18 | 2.34E-15 | 4.68 |
| Abdominal discomfort | 13.28 | 47 | 3.16E-18 | 3.09E-15 | 3.54 |
| Aching joints | 12.35 | 45 | 7.12E-18 | 5.80E-15 | 3.64 |
| Dental abscess | 8.47 | 37 | 2.99E-17 | 2.09E-14 | 4.37 |
| Sinusitis | 11.71 | 43 | 4.18E-17 | 2.55E-14 | 3.67 |
| Periodontal disease | 5.96 | 31 | 8.28E-17 | 4.05E-14 | 5.20 |
| Diverticulum | 8.69 | 37 | 7.83E-17 | 4.05E-14 | 4.26 |
| Fatigue | 14.36 | 47 | 1.11E-16 | 4.95E-14 | 3.27 |
| Demyelination | 13.79 | 46 | 1.26E-16 | 5.15E-14 | 3.34 |
| Hypophagia | 8.33 | 36 | 1.41E-16 | 5.29E-14 | 4.32 |
| Loose tooth | 6.53 | 32 | 1.86E-16 | 6.48E-14 | 4.90 |
| Emesis | 14.58 | 47 | 2.17E-16 | 7.08E-14 | 3.22 |
| Anemia | 20.11 | 55 | 3.04E-16 | 9.28E-14 | 2.74 |
| Cellulitis | 13.50 | 45 | 3.52E-16 | 1.01E-13 | 3.33 |
| Abscess drainage | 6.25 | 31 | 4.09E-16 | 1.11E-13 | 4.96 |
| Atelectasis | 11.92 | 42 | 6.65E-16 | 1.62E-13 | 3.52 |
| Neuropathy peripheral | 12.50 | 43 | 6.50E-16 | 1.62E-13 | 3.44 |
Note. aFisher test. bBenjamini–Hochberg test FC=fold-change.
Tested using 94 compounds in RepurposeDB that mapped to Connectivity Map.
Chemical ontology enrichment analysis
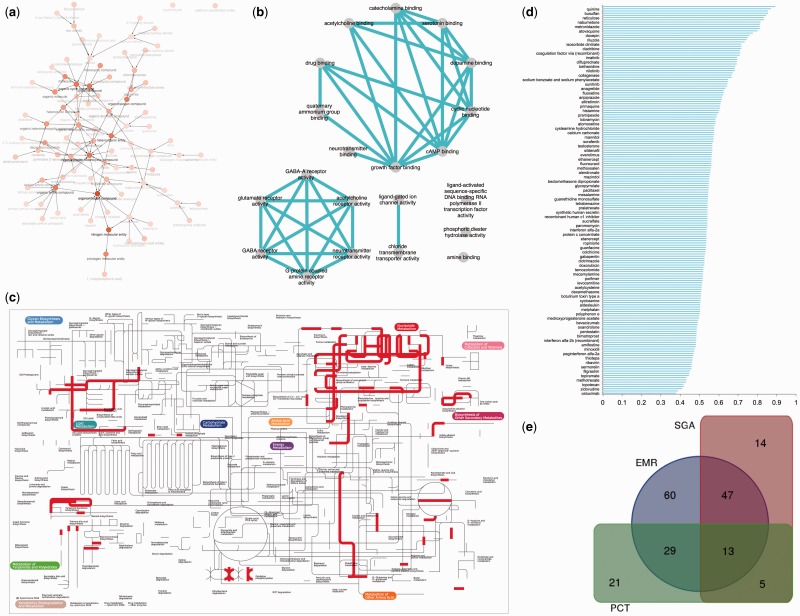
We have identified a set of 29 chemical ontology terms (Figure 4A and Table 3) associated with repositioned drugs (see Supplementary Data: RepurposeDB_BinChe.xlsx). Drugs annotated with pyrimidine 2′-deoxyribonucleoside, deoxyribonucleoside and nucleoside have ≥10-fold enrichment among repositioned drugs. Furthermore, repositioned drugs are enriched for hetero-organic chemical entities containing, at least, one carbon–nitrogen bond (organonitrogen compound, pnictogen molecular entity, nitrogen molecular entity and organonitrogen heterocyclic compound). Compounds were also enriched for terms indicating compounds with one carbon–halogen bond (heterocyclic compound, organic cyclic compound, cyclic compound, hetero-organic entity, organohalogen compound and organic amino compound).
Figure 4.
Chemical, biological, pathway-level and phenomic correlates of drug repositioning a) Chemical properties of repurposed drugs: Compounds in RepurposeDB mapped to ChEBI ontology (structure and role merged terminologies) b) Molecular function of repurposed drug targets: reduced representation of molecular function terms enriched among drug targets of repositioned drugs c) Targets of repurposed drugs mapped to KEGG metabolic pathways d) Distribution of semantic similarity of indications in RepurposeDB using Disease Ontology, Human Phenotype Ontology and combined scores. e) Overlap of posthoc validation of drug repositioning investigations in RepurposeDB using disease-comorbidity analyses (EHR), shared genetic architectures (SGA) and pathway cross-talks (PCT).
Table 3.
‘Structure’ and ‘Role’ terms from Chemical Entities of Biological Interest (ChEBI) ontology associated with repositioned drugs
| ChEBI_ID | ChEBI_Name | P a | Adjusted P b | FC |
|---|---|---|---|---|
| CHEBI:19255 | Pyrimidine 2'-deoxyribonucleoside | 9.22E-06 | 1.36E-04 | 77.40 |
| CHEBI:23636 | Deoxyribonucleoside | 4.35E-09 | 1.41E-07 | 47.85 |
| CHEBI:33838 | Nucleoside | 2.18E-07 | 5.05E-06 | 10.70 |
| CHEBI:35789 | Oxo steroid | 8.48E-06 | 1.31E-04 | 8.03 |
| CHEBI:21731 | N-glycosyl compound | 7.69E-07 | 1.46E-05 | 7.83 |
| CHEBI:50996 | Tertiary amino compound | 2.55E-07 | 5.70E-06 | 7.68 |
| CHEBI:23132 | Chlorobenzenes | 1.60E-06 | 2.87E-05 | 6.36 |
| CHEBI:26912 | Oxolanes | 2.11E-05 | 2.89E-04 | 6.06 |
| CHEBI:29347 | Monocarboxylic acid amide | 1.60E-05 | 2.29E-04 | 5.53 |
| CHEBI:36684 | Organohalogen compound | 9.89E-14 | 9.14E-12 | 5.19 |
| CHEBI:68452 | Azole | 2.14E-05 | 2.89E-04 | 4.81 |
| CHEBI:22712 | Benzenes | 6.12E-08 | 1.52E-06 | 4.22 |
| CHEBI:50047 | Organic amino compound | 1.21E-13 | 9.82E-12 | 3.85 |
| CHEBI:25693 | Organic heteromonocyclic compound | 2.66E-10 | 1.11E-08 | 3.45 |
| CHEBI:33661 | Monocyclic compound | 2.74E-10 | 1.11E-08 | 3.44 |
| CHEBI:38101 | Organonitrogen heterocyclic compound | 1.01E-14 | 1.63E-12 | 3.07 |
| CHEBI:33833 | Heteroarene | 4.58E-07 | 9.27E-06 | 3.00 |
| CHEBI:33597 | Homocyclic compound | 2.63E-06 | 4.37E-05 | 2.80 |
| CHEBI:35352 | Organonitrogen compound | 1.17E-19 | 7.59E-17 | 2.41 |
| CHEBI:51143 | Nitrogen molecular entity | 3.15E-17 | 1.02E-14 | 2.15 |
| CHEBI:33659 | Organic aromatic compound | 3.70E-08 | 9.96E-07 | 2.09 |
| CHEBI:24532 | Organic heterocyclic compound | 9.89E-10 | 3.55E-08 | 2.04 |
| CHEBI:5686 | Heterocyclic compound | 1.20E-09 | 4.07E-08 | 2.03 |
| CHEBI:33832 | Organic cyclic compound | 9.14E-15 | 1.63E-12 | 1.94 |
| CHEBI:33595 | Cyclic compound | 2.10E-14 | 2.72E-12 | 1.92 |
| CHEBI:33302 | Pnictogen molecular entity | 5.00E-12 | 3.59E-10 | 1.77 |
| CHEBI:72695 | Organic molecule | 3.81E-11 | 2.24E-09 | 1.56 |
| CHEBI:25367 | Molecule | 6.17E-11 | 3.07E-09 | 1.55 |
| CHEBI:33285 | Heteroorganic entity | 5.23E-14 | 5.64E-12 | 1.47 |
Note. aBinomial test. bBenjamini–Hochberg test FC=fold-change.
Tested using 145 compounds in RepurposeDB mapped to ChEBI database.
A new rule for drug repurposability
By compiling various physicochemical properties of small molecules, we compared the trends of compounds in RepurposeDB within the range of features used to defined ‘drug likeness’ or mass-logP space. A popular drug likeness estimation method, ‘Lipinski’s Rule of 5 (RO5)’, [86], can estimate whether a compound is suitable for drug development using a set of five chemical features. Lipinski’s rule of drug likeness is defined using the parameters and the range as follows: partition coefficient (computed; logP = −0.4 to + 5.6), molar refractivity (MR = 40–130), molecular weight (MW = 180–500), number of atoms (NA = 20–70) and polar surface area (PSA≤140 Å). Data from RepurposeDB suggest an equivalent molecular code could be used to predict drug repositioning potential. Computed physicochemical properties of compounds in RepurposeDB are in the following ranges: logP = 0.655–1.581, MR = 87.7–106.83, MW = 365–399, NA = 43–52 and PSA = 96.7–127. Leveraging this new rule of drug repurposability and assessing compounds in the physicochemical feature ranges represent the suboptimal space of compounds for repurposing for a different indication.
Combinations of features can also be used to prioritize of small molecules for drug development. For example, the correlation of determination of molecular mass (MW) and logP was defined as the ‘sweet spot’ [87] in compound space with an average molecular mass of 458.6 Da and average cLogP of 4.0. The results from chemoinformatics feature analytics suggest that compounds in RepurposeDB have higher correlation of MW-LogP (R2 = 0.569, n = 188) compared with the compounds from DrugBank-A (R2 = 0.328, n = 1673), DrugBank-F (R2 = 0.410, n = 7759) or compounds in Chemical Genomic Enrichment Analysis (CGEA) database (R2 = 0.527, n = 640; all observations P < 0.01; see Figure S). Multiple studies have reported various physicochemical properties of small molecule libraries and features including molecular mass, logP and number of atoms in a pharmacophore as key factors indicating the drug likeness of small molecules [88].
Biological function associations and pathway enrichment analysis of drug targets in RepurposeDB
Drugs in RepurposeDB were mapped to a target space of 305 targets genes/proteins using annotations from DrugBank. Enrichment analyses [89] using gene set and protein set databases revealed unique and shared functional mechanisms, molecular modules and pathways mediating drug repositioning.
Genomic features of drug repurposing
Targets in RepurposeDB were used to test for enrichment analysis of 56 different gene sets that span gene regulation, epigenetics, protein–protein interactions, GO terms, clinical or cellular phenotypes, functional annotations from gene set databases (GenSigdb, MSigdb, CCLE, etc.) protein expression and metabolomics databases. The enrichment associations show the relationship between repurposed drug targets with genomic elements (transcription factor binding sites and histone methylation patterns), protein annotations (signaling perturbations, protein complexes, protein–protein interaction networks), GO terms, phenotypes, pathways and tissues. After multiple testing correction, 26 reference gene sets had significantly enriched annotations associated with targets in RepurposeDB (see Supplementary Data File: RepurposeDB_Enrichr.xlsx).
Transcriptional regulation of drug repurposing
We identified several transcription factors (SUZ12, MTF2, EGR1, BACH1, SOX2, AR, JARID, RELA, HNF4A, TCF4, YAP1, LEF1, KLF11, KLF4, NFKB, CBEPA, MIB2, STAT3 and REST) as the common targets of repositioned drugs. Drugs (Supplementary Figure S2) that can regulate these transcription factors suggest the downstream gene expression changes could lead to a pluripotent effect (Figure 4B and Table 4; also, see Supplementary Data File: RepurposeDB_Enrichr.xlsx). We noted significant enrichment of various biological functions including regulatory, metabolic and transport processes among the targets of repurposed compounds. We observed significance for ligand binding, transmembrane receptors and signaling events associated with the drug targets. We also noted enrichment of cellular components including transmembrane regions, extracellular regions and different protein complexes (ion channel, chloride channel, sodium channel, acetylcholine-gated channel and N-methyl-D-aspartate selective glutamate receptor).
Table 4.
Gene ontology terms associated with targets of repositioned drugs
| Term | Overlap | P* |
|---|---|---|
| Biological processes | ||
| Synaptic transmission (GO:0007268) | 65/434 | <0.001 |
| Positive regulation of MAPK cascade (GO:0043410) | 51/395 | 1.16E-21 |
| Regulation of system process (GO:0044057) | 48/371 | 1.22E-20 |
| Behavior (GO:0007610) | 55/494 | 4.63E-21 |
| GPCR signaling pathway, coupled to cyclic nucleotide second messenger (GO:0007187) | 35/153 | 2.53E-21 |
| Single-organism behavior (GO:0044708) | 46/362 | 1.57E-19 |
| Response to drug (GO:0042493) | 44/354 | 1.94E-18 |
| Response to alkaloid (GO:0043279) | 30/111 | 4.73E-20 |
| Adenylate cyclase-modulating GPCR signaling pathway (GO:0007188) | 30/122 | 3.57E-19 |
| Regulation of amine transport (GO:0051952) | 24/60 | 3.57E-19 |
| Cellular components | ||
| Integral component of plasma membrane (GO:0005887) | 106/1066 | <0.001 |
| Postsynaptic membrane (GO:0045211) | 46/195 | 3.10E-29 |
| Synaptic membrane (GO:0097060) | 47/228 | 7.86E-28 |
| Transmembrane transporter complex (GO:1902495) | 49/286 | 5.50E-26 |
| Transporter complex (GO:1990351) | 49/291 | 7.36E-26 |
| Ion channel complex (GO:0034702) | 47/258 | 5.60E-26 |
| Synapse part (GO:0044456) | 53/395 | 6.44E-24 |
| Receptor complex (GO:0043235) | 41/272 | 6.22E-20 |
| Chloride channel complex (GO:0034707) | 19/50 | 3.47E-15 |
| Side of membrane (GO:0098552) | 28/235 | 3.30E-11 |
| Molecular functions | ||
| Extracellular ligand-gated ion channel activity (GO:0005230) | 39/74 | <0.001 |
| Ligand-gated channel activity (GO:0022834) | 45/145 | <0.001 |
| Ligand-gated ion channel activity (GO:0015276) | 45/145 | <0.001 |
| G-protein-coupled amine receptor activity (GO:0008227) | 27/41 | 2.28E-25 |
| Gated channel activity (GO:0022836) | 51/323 | 2.79E-24 |
| GABA-A receptor activity (GO:0004890) | 19/19 | 2.07E-20 |
| Ion channel activity (GO:0005216) | 53/396 | 2.15E-22 |
| Drug binding (GO:0008144) | 32/93 | 1.57E-23 |
| Substrate-specific channel activity (GO:0022838) | 53/406 | 5.40E-22 |
| GABA receptor activity (GO:0016917) | 19/22 | 1.25E-19 |
Note. *Adjusted P-values from Enrichr; only 10 terms per category are shown, full data are provided in the Supplementary File.
Epigenetic factors controlling repurposed drug targets
Epigenetic control of genomic region could help in developing novel therapies. However, direct evidence on how epigenetic factors may influence drug repositioning is unclear [90]. In total, 52 different data sets from ENCODE project (https://www.encodeproject.org/) have enrichment for the drug targets from RepurposeDB. For example, H3K27me3, a common methylation, had 69.23% of methylations in multiple tissues, suggesting that genes repressed by EZH2 may mediate an epigenomic target for drug repositioning. Among the remaining methylation indicators, H3K4me1 is associated with 19.23% of methylation in 10 cell lines or tissue types and H3K9me3 with four cell lines (CD14-positive monocyte, G1E-ER4, G1E and skeletal muscle myoblast), H3K4me3 with limb and H4K20me1 with H1-hESC (Supplementary Table S4; also, see Supplementary Data File: RepurposeDB_Enrichr.xlsx).
Repurposed drug targets are enriched across the circulatory system
Plasma, platelets, blood, placenta and liver have higher tissue-specific enrichment for the targets of repurposed drugs. Repurposed drugs may induce multiple effects. Drugs targeting the circulatory systems offer the convenience of perturbing other organ systems and, thus, may improve the polypharmacological impact of repurposed drugs [91].
Proteomic features of drug repositioning
We identified enrichment of protein annotation terms across 15 databases. Annotation enrichment using disease association data indicates that targets of repositioned compounds have associations with psychiatric and cardiovascular diseases. Repurposed drugs have enrichment for protein sequence features like G-protein coupled receptors (GPCRs), transmembrane regions, binding sites for adenosine triphosphate/carbohydrates, receptors, disulphide bonds, signal peptide, glycosylation sites, DNA-binding regions and zinc finger. Enrichment for hallmark molecular drug target classes including GPCRs, neurotransmitters, ion channels, kinases, acetylcholine receptors and nuclear hormone receptors was significant [88, 92–94]. Functional or chemical screenings of the proteins with repurposing-specific sequence and structural features are likely to yield compounds that could modulate various diseases (see Supplementary Data: RepurposeDB_DAVID.xlsx).
Conserved sequence domains encoded in repurposed drug targets
Conserved protein domains play a pivotal role in mediating function across various pathways and play a vital role in mediating functional and interaction promiscuity across protein families and aid in polypharmacology including drug repositioning [95]. We have also noted significant enrichment for protein sequence domains like ligand-binding domain of hormone receptors (HOLI domain), c4 zinc finger in nuclear hormone receptors (ZnF_C4 domain) and metal-dependent phosphohydrolases with conserved ‘HD’ motif (metal-dependent phosphohydrolases with conserved ‘HD’ motif; HDc domain) [96]. The human proteome contains 142 proteins with HOLI domains, 169 proteins with ZnF_C4 domains and 85 proteins with HDc domains (Supplementary Figure S3). HOLI and ZnF_C4 domains are hallmark features of a variety of receptors including members of steroid-thyroid hormone-retinoid receptor superfamily like glucocorticoid, retinoic acid, nuclear and androgen receptor molecules. HOLI domains, for example, are encoded in 20.21% of peroxisome proliferator-activated receptor (PPAR) signaling pathways [97]. PPAR pathways have several promiscuous drug targets (e.g. berberine) that treat diseases including hypolipidemia and diabetes [98]. Around 63% of proteins involved in purine metabolism encode HDc domains. Inhibition of purine metabolism is a primary pharmacological feature of azathioprine, a compound used for treating conditions such as transplant rejection and autoimmune disorders (e.g. rheumatoid arthritis and inflammatory bowel diseases) [99]. Exploring the remainder of poorly characterized proteins encoded in human proteome with ‘repositioned compounds associated protein domains’ could be potential targets for future repurposing opportunities. Based on the sequence-based evidence, these proteins could be preferentially prioritized targeted for developing compounds with multiple indications.
Structural domains of repurposed drug targets
Drug discovery relies on crystallography experiments to understand the structure, binding affinities and identifying pharmacophore moieties for precise ligand design. Nuclear receptor ligand-binding domain and Kringle modules are enriched across the repurposed drug targets. Both structural domains have mechanistic roles in mediating multiple functional pathways across human proteome and are targeted by ligands with varying degree of specificities [100, 101].
Pathway cross talks influence drug repositioning across multiple diseases
Pathway cross talk is a biological phenomenon where the components of a biological pathway (genes, proteins or small molecules) are shared across two or more pathways [24]. Pathway cross talks are essential components of functional promiscuity and, thus, may influence the success of drug repositioning [6, 20, 102]. Targets in RepurposeDB were enriched for 336 pathways across 10 different pathway databases (Figure 4C; also, see Supplementary Data: RepurposeDB_CPDB.xlsx; a subset of 30 highly enriched pathways listed in Table 5). After applying multiple testing threshold, pathways from 10 different pathway databases were significantly enriched across the drug repositioning target space: Small Molecule Pathway Database (SMPDB) [103], n = 97; Reactome[48], n = 82; KEGG [49], n = 50; WikiPathways [104], n = 37; BioCarta, n = 25; Pathway Interaction Database [105], n = 20; PharmGKB [106], n = 14; HumanCyc [107], n = 7; NetPath [107], n = 5; and Signalink [107], n = 1. We define a drug target as a mediator of a pathway cross talk when a drug target is involved in more than one pathway identified from pathway enrichment analyses. We define a recurrent target as a gene or a protein that participates in multiple pathways, providing evidence for pathway cross talk as a factor driving drug repositioning. Using comparative pathway analyses, we identified 64 recurrent targets that that may serve as drivers of pathway cross talks across 336 pathways. We noted that 37.5% of pathways (n = 126; ≥2 drug targets) participate in molecular cross talk events by sharing overlapping targets across different pathways.
Table 5.
Consensus pathways mediated by targets of repositioned drugs
| Pathway | q-value* | Source |
|---|---|---|
| Neuroactive ligand–receptor interaction—Homo sapiens (human) | 5.84E-65 | KEGG |
| Monoamine GPCRs | 6.72E-35 | Wikipathways |
| Amine ligand-binding receptors | 1.37E-33 | Reactome |
| Nicotine addiction—Homo sapiens (human) | 8.01E-30 | KEGG |
| Class A/1 (rhodopsin-like receptors) | 1.45E-22 | Reactome |
| GPCRs, Class A rhodopsin-like | 4.60E-22 | Wikipathways |
| Morphine addiction—Homo sapiens (human) | 2.69E-21 | KEGG |
| Defective ACTH causes Obesity and Pro-opiomelanocortinin deficiency | 1.59E-18 | Reactome |
| GPCR ligand binding | 1.59E-18 | Reactome |
| Neurotransmitter receptor binding and downstream transmission in the postsynaptic cell | 9.23E-18 | Reactome |
| Purine metabolism—Homo sapiens (human) | 1.54E-17 | KEGG |
| cAMP signaling pathway—Homo sapiens (human) | 4.78E-17 | KEGG |
| Metabolic disorders of biological oxidation enzymes | 9.02E-17 | Reactome |
| Transmission across chemical synapses | 1.14E-16 | Reactome |
| Integrated pancreatic cancer pathway | 3.47E-16 | Wikipathways |
| Pathway_PA165959425 | 1.79E-15 | PharmGKB |
| Ligand-gated ion channel transport | 8.73E-15 | Reactome |
| Calcium signaling pathway—Homo sapiens (human) | 4.03E-14 | KEGG |
| Neuronal system | 9.50E-14 | Reactome |
| GABA A receptor activation | 1.20E-13 | Reactome |
| Nalbuphine action pathway | 2.48E-13 | SMPDB |
| Signal transduction | 6.48E-13 | Reactome |
| Heroin action pathway | 7.38E-13 | SMPDB |
| Pathways in cancer—Homo sapiens (human) | 7.87E-13 | KEGG |
| Sorafenib pharmacodynamics | 1.47E-12 | PharmGKB |
| Highly calcium permeable postsynaptic nicotinic acetylcholine receptors | 1.47E-12 | Reactome |
| 3-Methylthiofentanyl action pathway | 1.47E-12 | SMPDB |
| Alfentanil action pathway | 1.47E-12 | SMPDB |
Note. *Adjusted q-values from ConsensusPathDB-Human; only 30 pathways are shown here, full data set is provided in the Supplementary File. Minimum overlap with input list was set to 2.
Altogether, our findings suggest additional evidence for using pathways cross talk as a useful metric to discover new indications for existing drugs or new indications for a disease. Our observation strengthens earlier findings that pluripotent modules of genes, pathways and molecular interactions that are active across multiple biological contexts may influence drug repositioning [6, 8, 108]. Such repurposing-associated modules may share regulatory roles, biological function, pathophysiological mechanisms and pathways.
Phenomics of disease pairs targeted by repositioned drugs
Understanding the relationship between two diseases at the gene, protein or pathway level and connecting with epidemiological evidence (e.g. SGA, GWAS [109, 110] or PheWAS-driven drug repositioning [111]) could improve drug repositioning capabilities of approved or investigational drugs. EMR-wide relative risk data were used to perform phenome-wide enrichment analyses (PheWAS) and to validate the off-label use of drugs for secondary indications. Emerging evidence from phenomics studies that leverage EMR data also suggest that phenotypic similarity between two conditions could aid in drug discovery and drug repurposing [57, 112] Disease comorbidity is correlated with age [113], but the impact of disease comorbidities or disease-pair prevalence for the success of drug repositioning is largely unknown [113–115]. To address this, we analyzed 1125 diseases in RepurposeDB using disease co-occurrence and SGA. Pair-wise comorbidity estimates and significant SGA associations for a subset of repurposed drugs (itraconazole, heparin, raloxifene, minoxidil and allopurinol) are provided in Table 6, and full data set is available Supplementary Data: RepurposeDB_EHR_SGA.xlsx; Figure 4.
Table 6.
Examples of pair-wise disease comorbidity estimates (itraconazole, heparin, raloxifene and allopurinol) and shared genetic architecture estimation (minoxidil and allopurinol)
| Pair-wise disease comorbidity estimation using an EMR-wide
prevalence estimation (n=21 25 468) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Primary indication | Secondary indication | PI∧SI (n) | PI (n) | SI (n) | OR | P | RR | |
| Rx00135 (itraconazole) | ||||||||
| Otomycosis | Cavitary pulmonary diseasea | 68 | 1402 | 12 121 | 8.93 | 2.67E-36 | 8.54 | |
| Otomycosis | Febrile neutropenia | 16 | 1402 | 5318 | 4.61 | 0.00262656 | 4.57 | |
| Fungal otitis externa | Cavitary pulmonary diseasea | 250 | 11 423 | 12 121 | 3.96 | 9.86E-66 | 3.89 | |
| Fungal otitis externa | Extrapulmonary aspergillosis | 15 | 11 423 | 448 | 6.41 | <0.001 | 6.4 | |
| Fungal otitis externa | Febrile neutropenia | 103 | 11 423 | 5318 | 3.67 | 7.03E-24 | 3.65 | |
| Fungal otitis externa | Immunodeficiency | 26 | 11 423 | 846 | 5.87 | 8.88E-09 | 5.86 | |
| Fungal otitis externa | Fungal infection | 1814 | 11 423 | 52 130 | 7.74 | <0.001 | 6.67 | |
| Fungal otitis externa | Pulmonary aspergillosis | 15 | 11 423 | 448 | 6.41 | 0.000106935 | 6.41 | |
| Rx00118 (heparin) | ||||||||
| Sickle cell disease | Thromboembolic disease | 12 | 1477 | 1383 | 12.67 | 1.47E-06 | 12.58 | |
| Sickle cell disease | Intravascular coagulationb | 28 | 1477 | 1307 | 32.06 | 1.64E-28 | 31.48 | |
| Sickle cell disease | Venous thrombosis | 61 | 1477 | 9840 | 9.31 | 1.92E-33 | 8.97 | |
| Sickle cell disease | Deep venous thrombosis | 33 | 1477 | 5588 | 8.71 | 1.55E-16 | 8.54 | |
| Sickle cell disease | Pulmonary embolism | 56 | 1477 | 6810 | 12.35 | 8.70E-37 | 11.92 | |
| Sickle cell disease | Consumptive coagulopathies | 76 | 1477 | 8875 | 13.03 | 5.78E-52 | 12.42 | |
| Cystic fibrosis | Consumptive coagulopathies | 11 | 314 | 8875 | 8.66 | 0.000383854 | 8.39 | |
| Rx00205 (raloxifene) | ||||||||
| Prostate cancer | Osteoporosis | 334 | 15 329 | 31 300 | 1.49 | 2.71E-08 | 1.48 | |
| Breast cancer | Osteoporosis | 2992 | 22 462 | 31 300 | 11.26 | <0.001 | 9.89 | |
| Rx00013 (allopurinol) | ||||||||
| Hyperuricemiab | Primary gout | 989 | 17 817 | 12 681 | 10.53 | <0.001 | 10.00 | |
| Hyperuricemiab | Secondary gout | 989 | 17 817 | 12 681 | 10.53 | <0.001 | 10.00 | |
| Hyperuricemiab | Leukemia | 94 | 17 817 | 709 | 18.17 | 1.18E-75 | 18.08 | |
| Hyperuricemia | Lymphoma | 270 | 17 817 | 4708 | 7.29 | 2.49E-127 | 7.19 | |
| Hyperuricemia | Primary gout | 989 | 17 817 | 12 681 | 10.53 | <0.001 | 10.00 | |
| Hyperuricemia | Secondary gout | 989 | 17 817 | 12 681 | 10.53 | <0.001 | 10.00 | |
| Hyperuricemia | Leukemia | 94 | 17 817 | 709 | 18.17 | 1.18E-75 | 18.08 | |
| Hyperuricemia | Lymphoma | 270 | 17 817 | 4708 | 7.293 | 2.49E-127 | 7.19 | |
| Renal calculid | Primary gout | 769 | 15 291 | 12 681 | 9.32 | <0.001 | 8.90 | |
| Renal calculi | Kidney transplantation | 364 | 15 291 | 13 091 | 4.01 | 1.29E-97 | 3.94 | |
| Renal calculi | Secondary gout | 769 | 15 291 | 12 681 | 9.32 | <0.001 | 8.90 | |
| Renal calculi | Leukemia | 22 | 15 291 | 709 | 4.42 | 6.21E-05 | 4.41 | |
| Renal calculi | Lymphoma | 121 | 15 291 | 4708 | 3.66 | 4.40E-28 | 3.64 | |
| Secondary gout | Kidney transplantation | 569 | 12 681 | 13 091 | 7.87 | 8.60E-285 | 7.57 | |
| Secondary gout | Leukemia | 31 | 12 681 | 709 | 7.63 | 9.34E-14 | 7.61 | |
| Secondary gout | Lymphoma | 177 | 12 681 | 4708 | 6.58 | 6.00E-77 | 6.50 | |
| Primary gout | Kidney transplantation | 569 | 12 681 | 13 091 | 7.87 | 8.60E-285 | 7.57 | |
| Primary gout | Leukemia | 31 | 12 681 | 709 | 7.63 | 9.34E-14 | 7.61 | |
| Primary gout | Lymphoma | 177 | 12 681 | 4708 | 6.58 | 6.00E-77 | 6.50 | |
|
| ||||||||
| Shared genetic architectures estimation using a reference database with 11 974 genes | ||||||||
|
| ||||||||
| Primary indication | Secondary indication | D1G∧D2G | D1G | D2G | OR | P | ||
|
| ||||||||
| Rx00165(minoxidil) | ||||||||
| Hypertension | Hair loss | 71 | 1777 | 137 | 3.49 | 3.46E-12 | ||
| Rx00013(allopurinol) | ||||||||
| Hyperuricemia | Visceral leishmaniasis | 6 | 75 | 29 | 33.03 | 5.19E-05 | ||
| Hyperuricemia | Cutaneous leishmaniasis | 9 | 75 | 38 | 37.81 | 1.32E-08 | ||
| Hyperuricemia | Leukemia | 31 | 75 | 1448 | 3.41 | 9.10E-05 | ||
| Hyperuricemia | Lymphoma | 34 | 75 | 1018 | 5.33 | 9.39E-10 | ||
Note. PI∧SI=number of patients with both primary indication and secondary indication; PI=number of patients with primary indications; SI=number of patients with secondary indications; P=Bonferroni correction applied; RR=relative risk for primary indication and secondary indication to present in the same patient estimated from same data set. Reference databases have predicates as follows:
aChronic; bdisseminated; cchemotherapy-induced; and drecurrent. D1G∧D2G=number of genes shared by primary indication and secondary indications of a compound; D1G=number of genes associated with primary indication; D2G=number of genes associated with secondary indication.
Improving drug repositioning efficiency using comorbidity risk estimation of disease pairs using EMR-wide analytics
Using EMR-wide comorbidity evaluation of pairs of primary and secondary conditions of a drug in RepurposeDB, we identified disease-pair comorbidities as a post hoc epidemiological evidence for repositioned drugs. For example, beclomethasone dipropionate (Rx00038) has therapeutic effects for multiple conditions like graft-versus-host disease (intestinal and gastrointestinal), Crohn’s disease, ulcerative colitis, rhinitis (perennial and allergic), nasal polyps and asthma. For this drug, we have tabulated all conditions (n = 9) and compiled all disease pairs (n = 20), i.e. we consider Crohn’s disease and perennial rhinitis as a disease pair and patients’ counts were derived after mapping disease name to best representative disease terminology. Next, we have computed the relative risk of these two conditions using the data compiled from EMR and found that the disease 10 of 20 pairs were significant. For example, Crohn’s disease and asthma have significant comorbidity with higher prevalence than expected when compared with the background population of (P = 7.81E-61, odds ratio = 1.81). Previous epidemiological surveys and genome-wide association studies also suggest that Crohn’s disease and asthma share etiological routes [116, 117].
Following the EMR-wide analyses of 2970 disease pairs, we identified 1548 significant disease pairs across 149 drugs after multiple testing corrections. We found EMR-wide disease comorbidity evidence for 58.9% of drugs in RepurposeDB, suggesting that systematic disease comorbidity and relative risk estimation analysis could help in developing rational drug repurposing methods and to prioritize compounds in the drug discovery pipeline. For example, a drug is more likely to repurpose across two diseases when they could share disease etiology and, thus, observable in EMR-based disease enrichment analyses (see Figure 5 for examples). Developing rational drug repositioning methods by considering prevalence rates of disease pairs in the target patient population may help to find develop precision repositioning therapies compared with traditional drug development approaches.
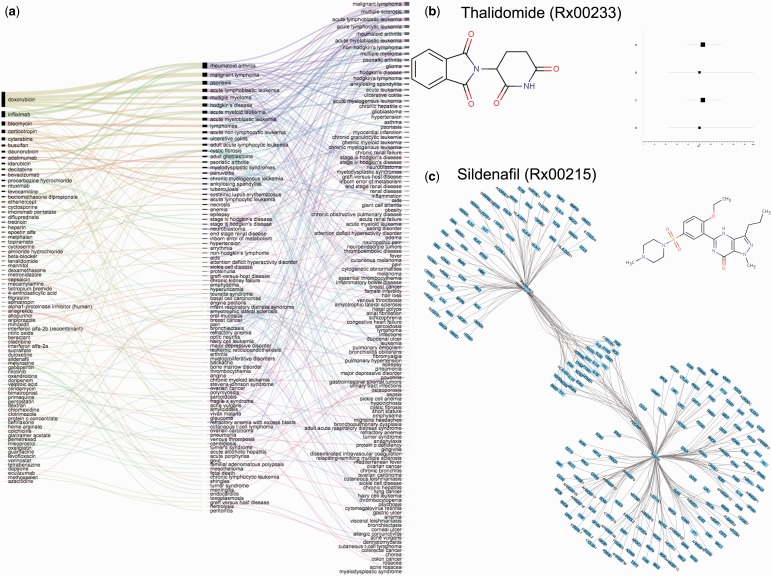
Figure 5.
Shared genetic architecture and pair-wise comorbidities of diseases targeted by repurposed drugs a) Shared genetic architecture of diseases targeted by same drug. Thickness of the lines between disease indicates number of shared genes across the diseases b) Distribution of semantic similarity of indications in RepurposeDB d) Overlap of validation of drug repositioning investigations in RepurposeDB using disease-comorbidity analyses, shared genetic architectures and pathway cross-talks b) Example of pair-wide disease comorbidity estimation: Thalidomide (Rx00233): 20 disease pairs were computed and the pairs significant after multiple testing correction are used to generate the plot. Disease pair #1=severe erythema nodosum leprosum and Crohn's disease; Disease pair #2=severe erythema nodosum leprosum and recurrent aphthous ulcers; Disease pair #3=moderate erythema nodosum leprosum and Crohn's disease and Disease pair #4=moderate erythema nodosum leprosum and recurrent aphthous ulcers c) Example of shared genetic architectures driving drug repurposing: Sildenafil (Rx00215): Three disease were associated with sildenafil (angina, erectile dysfunction and pulmonary hypertension). Reference database had 154 genomic associations for angina and 89 associations for pulmonary dysfunction; 26 genes were shared by both diseases suggesting the geneset as shared genetic architecture driving successful outcome of Sildenafil as a therapy for both diseases.
Role of shared genetic architecture in drug repositioning
We performed a systematic analysis to characterize the shared genetic architecture between the primary and secondary indications of all drugs in RepurposeDB. For example, the drug cyclosporine (Rx00075) is used for five indications with genomic associations (psoriasis, rheumatoid arthritis, amyotrophic lateral sclerosis, bronchiolitis obliterans and graft versus host disease). For this example, we have tabulated all diseases (n = 6) with their associated genes from an integrated disease–gene database [53]. After multiple testing corrections, five disease pairs had significant associations for shared genes. For example, rheumatoid arthritis and amyotrophic lateral sclerosis have 981 and 226 associated genes, respectively. We have computed the SGA of this disease pairs and identified 57 genes shared by two diseases (P = 2.35E-11). Shared genes across the two diseases (e.g. MMP12, IFNK, SERPINE1, MMP1, MMP3, MMP9, SH2B3, TGFB1, PPARG, TNF, HLA-B, F2, ATXN2 and VEGFA) suggest strong immune modulation of both diseases by a common subset of genes and, hence, suitable target for an immunosuppressant like cyclosporine (see Supplementary Data: RepurposeDB_EHR_SGA.xlsx). Similarly, if a drug can target the shared subset of genes associated with two diseases; they are more likely to be effective for both conditions. Using this approach, we computed SGA for 499 diseases pairs. A total of 235 disease and 79 (31.22%; Figure 5) drugs remain significant after multiple testing correction (see Supplementary Data: RepurposeDB_DiseaseSimilarity.xlsx).
Similarity of diseases target by repositioning drugs
Briefly, for each disease pair, we leveraged two phenotype ontologies (DO and HPO) to check how closely two diseases or their clinical phenotypes are related and assigned a phenomic similarity score based on the distance between the terms to each other using relationships derived from the ontologies. We have computed the phenomic similarity score for 176 drugs in RepurposeDB (0.572 ± 0.016). Drugs like cetuximab (score = 0.37; indications for multiple hematological cancers) and zidovudine (score = 0.409; indications for several cardio-metabolic diseases) have lower phenomic similarity scores compared with drugs like quinine (score = 0.869; malaria and leg cramps) and busulfan (score = 0.854; cancers of multiple organs). Our analyses provide a quantitative estimate of phenomic similarity (Figure 4D); Supplementary Figure S4) using clinical ontologies; such estimations could be a useful aid in developing future drug repositioning investigations.
Applications of RepurposeDB
Data compiled in RepurposeDB can be used to prioritize small molecules and drugs and targets for experimental or clinical evaluation. These data can further be extrapolated to identify new drug targets or new indications for existing compounds as well as to develop predictive models of repurposable drugs and targets. We have used RepurposeDB to assess post hoc validation of repurposability using three different data types (epidemiology, genetics and pathways), explore the pharmacoeconomics of repositioning space and develop networks to aid in the discovery of new targets for repurposing opportunities in the human proteome.
Validating drug repositioning investigations using disease comorbidities, genetic architectures and pathway cross talks
Pathway cross talk, SGA and epidemiological evidence provide evidence for 26.87, 31.22 and 58.89% of repositioned drugs, respectively. Furthermore, we found EMR and SGA evidence for 47 drugs, SGA and pathway cross talk evidence for 5 drugs and EMR and pathway cross talk evidence for 29 drugs. There were 13 drugs [interferon alfa-2b (recombinant), cladribine, bleomycin, anagrelide, dexamethasone, lenalidomide, aripiprazole, epoetin alfa, duloxetine, sucralfate, difluprednate, aminosalicylic acid and beclomethasone dipropionate) that have associations with all three approaches. Our analyses using three different data sets (comorbidity of disease pairs identified using EMR data, SGA captured using genetic modules shared by diseases and pathway cross talks by targets of repositioned compounds) provide validation for 69.16% of drugs in RepurposeDB (Figure 4E; see Supplementary Data: RepurposeDB_EvidenceTypes.xlsx).
Pharmacoeconomics of repurposed drugs
Drug development is an expensive process that requires significant capital investment to deliver a new drug from development to the market. Contradicting reports suggest that drug repositioning may improve revenue for pharmaceutical companies and help to use off-patent drugs for new indications. However, it is not clear about the costs of the repurposed compounds compared with the marketed drugs. Drug repositioning is often conflated with drug reformulations; analyses of pharmaceutical marketing data indicate that repositioned drugs are marketed as different drug reformulations (e.g. syrup reformulated as a capsule, capsule reformulated as injection) [118, 119].
We aggregated dispensing unit types and prices for all the drugs from DrugBank and compared the price of repositioned drugs in RepurposeDB with the rest of the drugs with the cost and unit formats (e.g. tablet, capsule and vial) of commercially marketed drugs. We extracted 11 813 drug description-cost-unit records, and 2273 annotations for 202 drugs (79.8% of drugs in RepurposeDB) sold as 39 different dispensing types from DrugBank. Average costs are higher for biotech drugs compared with small molecules, and the cost for various dispensing formats varies across the DrugBank. The average cost of drugs in DrugBank is also affected by the interaction of marketing type (vial, capsule, etc.) and value (10, 20 mg, etc.; all observations P < 0.001). Furthermore, we tested whether the cost was affected by the interaction between type (biotech drug or small molecule) and dispensing format (capsule, injection, syrup, etc.). The average cost of repositioned drugs is higher, but not significantly, compared with the rest of the drugs in DrugBank: the average cost of drugs differs between DrugBank ($339), RepurposeDB ($528) and the remainder of DrugBank excluding repurposed drugs ($297.7; P = 0.097; see Supplementary Data: RepurposeDB_Pharmacoeconomics.xlsx).
Network reconstruction and analyses
We constructed five networks (Supplementary Figure S4) using data from RepurposeDB, specifically: the networks including chemical similarity network of small molecules, drug–target network, a functional network using drug targets of repurposed compounds, expanded-target network of repurposed compounds, drug–drug interaction and drug–food interaction networks are compiled. The networks derived from RepurposeDB (Figure 6) are valuable tools for discovering chemical patterns and describing new targets to improve drug repositioning pipelines. We computed various network properties and prioritized hubs across the networks. These hubs can be further perturbed using functional experiments and high-throughput compound screening to find compounds that could be effective across multiple indications (see Methods and Supplementary Data files: RepurposeDB_SFN_EFN_NA.xlsx and RepurposeDB_Drug_Food_Target_Networks.xlsx).
Figure 6.
Chemical, biological and interaction networks compiled using data from RepurposeDB a) Chemical similarity network of small molecules in RepurposeDB: Histogram of Tanimoto similarity of small-molecules in RepurposeDB. Tanimoto similarity estimates the similarity of the two compounds based on the angle between the attribute vectors (fingerprint) of each compound. b) Tanimoto similarity network of small molecules in RepurposeDB was computed, and chemical similarity network was calculated and visualized using chemViz. Small molecule from RepurposeDB represents the nodes and edges are Tanimoto similarity (values between 0 to 1; threshold set at >=0.5 for visualization) and weighted by the Tanimoto similarity values. Inset highlights a section of the chemical similarity network of repositioned compounds and maximum common chemical substructures are indicated. c) Drug-target interaction network d) Drug-drug interaction network: Targets are colored according to the biochemical action (inhibitor, antagonist, agonist, potentiator and others) e) SFN: Seed Functional Network reconstructed using targets of repositioned drugs f) EFN: Expanded Functional Network reconstructed using targets of repositioned drugs as seed and adding 20% of genes shared by the nodes in SFN. Data to generate various networks and high-resolution versions of the network figures are provided in the Supplementary Data.
Chemical similarity of small molecules in RepurposeDB
We analyzed the chemical similarity network and identified H2N-CH3 as the maximum common substructure of the chemical repertoire, whereas the common substructure for DrugBank-A is H3C-CH2. The connectivity of the chemical similarity networks also indicates several closely connected modules with individual chemical signatures. Exploring these substructures and reformulating existing compounds with the repurposing-based substructure could enhance the success of future drug repurposing investigations.
Seed and expanded functional network of drug targets
The repurposed drug target network has a higher degree of connectivity compared with the human protein interactome, thus indicating close regulation of the repurposed target at the proteome level. For example, we tested the Seed Functional Network for enrichment of protein–protein interactions using a meta-database of protein–protein interactions (STRINGv10 [120]). When compared with the background of canonical human genome, the network was enriched for interactions (P≤0.001) with 2615 interactions using 299 proteins mapped to STRING database indicating that targeting this core network or its modulators will lead to effective molecules or molecules that can perturb multiple indications.
Drug–drug and drug–food interaction patterns of repurposed compounds
We compiled 305 experimentally validated drug targets for repurposed compounds from DrugBank. These drug–target interactions represent 27 different modes of actions (inhibitor, agonist, activator, etc.). The average number of drug targets per compound is higher for drugs in RepurposeDB (4.115 ± 0.621; t-test P≤0.001) compared with DrugBank-A (3.476 ± 0.237; t-test P≤0.001) and DrugBank-F (1.949 ± 0.092). The average number of reported drug–drug interactions per compound is higher for drugs in RepurposeDB (16.6 ± 3.542) compared with DrugBank-F (13.66 ± 1.91) and DrugBank-A (3.105 ± 0.291; P≤0.001).
We compiled 1186 food interactions for 649 drugs from DrugBank. There were 188 drugs in RepurposeDB that had, at least, one reported food interactions (74.3% of drugs in RepurposeDB). Food interactions vary by the type of medicine (biotech drug or a small molecule; P≤0.001), but food interactions and inclusion of a compound in RepurposeDB are independent. The average number of reported food–drug interaction per compound is higher but not significant for drugs in RepurposeDB (0.611 ± 0.16) compared with DrugBank-A (0.486 ± 0.054) DrugBank-F (0.11 ± 0.13; all observations P > 0.05).
Discussion
Systematic drug repurposing refers to the data-driven evaluation of the set of approved or investigational pharmaceutical compound databases as therapies for new indications. By leveraging systematic or targeted approaches to drug repurposing aims, we aim to expedite the recycling of existing drug compounds for new indications. We also hope to develop new leads for new combinations of drugs to increase treatment efficacy. Irrespective of the growing catalog of drug repositioning using approved, shelved or investigational compounds, there was previously no comprehensive collection or meta-analyses of drug repositioning examples from public biomedical and health care databases. From the first description of drug repurposing in the 1950s to the present day, >250 drug repurposing studies have been published [5, 121, 122]. The growing trend of repositioning investigations suggests that reuse of drugs are plausible and beneficial for patients. RepurposeDB fills a significant gap in the setting of drug repositioning by offering a unified database of drug repositioning database and collective insights. Our extensive meta-analyses of the data provide the first set of pharmacological, biological and disease-specific principles mediating drug repositioning. Drug repurposing studies and methodologies can use the RepurposeDB data set as a benchmarking or comparison resource. The new MIADRI standard will also help to streamline the process of reporting results from future drug repositioning studies.
Insights from RepurposeDB data set provide a collective set of principles of physicochemical features that contribute to drug repositioning, including chemical moieties, permeability properties and ADMET signatures. Analyses of small molecules in RepurposeDB reveal chemical compositions (9 experimentally characterized chemical features, 21 computationally estimated chemical properties and 9 ADMET properties compiled) and drug induced side effects associated with drug repositioning. Analyses of drug targets revealed transcription factors, epigenomic enrichments, functional roles and pathways associated with repurposed drugs and their targets. Using the RepurposeDB data set, we showed that pathways share targets of repositioned drugs (n = 68 compounds; 26.87% of RepurposeDB), and leveraging this knowledge could lead to better candidates for pathway-based drug repositioning. Genetic etiologies of various polygenic disorders have molecular level similarities. The expanding collection of genome-wide association studies and phenome-wide association studies further support these correlations and in the pleiotropic role of genetic variants in influencing multiple disease manifestations. Our analyses provide the first comorbidity-based evidence for drug repositioning investigations (n = 149; 58.89% of compounds in RepurposeDB) compounds and shared genomic evidence (n = 79; 31.22% of compounds in RepurposeDB). Together, these approaches have validated 74.7% of drugs in RepurposeDB. Small molecule filtering systems and high-throughput compound screening platforms and drug development pipelines can be redesigned to take the pharmacological, biological and epidemiological factors into account. Drug prioritization and drug likeness assessment algorithms can also use chemical features, molecular properties and target functions associated with drug repositioning to assess compounds for repurposing.
Conclusion
The burden of disease is increasing globally due to a variety of factors such as population growth, infectious disease outbreaks, and the emergence of antibiotic resistance. In combination with the ever-rising cost of drug-development, this demands innovative and robust approaches to drug discovery. Discovering the chemical, genetic and biological features associated with drug development is critical for rational drug and target discovery. Owing to the tremendous costs of successfully bringing new pharmaceutical compounds to market, repositioning of previously approved drugs is a popular method to increase the potential therapeutic space for human diseases. Drug repositioning can uniquely contribute to solving a significant gap in a public health setting by providing therapeutic options for complex, chronic or orphan diseases. The data compiled in RepurposeDB and the collective analytical insights presented in this article provide, for the first time, an understanding of the distinct physicochemical, biochemical and chemogenomic features of repurposed drugs. We are releasing RepurposeDB in the public domain under an open access license, with the hope that its high-quality content, user-friendly search engine and visual analytics tools will aid investigators hoping to conduct drug repositioning studies and discover repurposable compounds. Using analytical frameworks based on our findings for the assessment and prioritization of compounds, targets or pathways for drug repositioning experiments and clinical trials could help to develop drug repositioning pipelines. Availability of a reference database and chemical signatures and first set of principles of drug repositioning to evaluate new or existing compound could accelerate drug repositioning investigations. RepurposeDB aims to evolve through periodic updates, as an essential resource for the drug discovery community, we hope will contribute to unraveling novel indications for existing small molecule, protein or peptide drug collections. RepurposeDB could also be a useful resource for implementing personalized drug repositioning in the clinical setting to enable the delivery of precision medicine. For example, currently, no reference database of drug repositioning enables clinicians or researchers to submit evidence to a centralized resource readily available to other researchers and clinicians. Individual clinicians seeking treatments for their patients are perhaps the most common initiators of drug repurposing studies through the use of off-label prescriptions. It has been estimated that up to one-fifth of all available drugs are prescribed off-label; most of such off-label uses were initiated by clinicians and then shared after establishing efficacy. Observations of such off-label use, the first proof of drug repositioning, unless published as research papers or case reports are usually siloed in EHRs or sometimes not even publically shared, to the detriment of patient treatment. Furthermore, as the volume of published papers increases, it can be difficult for investigators to stay current with trends in drug repositioning that may apply to their field of research or clinical medicine. RepurposeDB provides an open, community-driven environment for drug repositioning evidence, and within few years, we hope will become the centralized resource for drug repositioning, by harnessing crowdsourcing methods. To conclude, the three most important developments of this article, namely (1) the presentation of RepurposeDB; (2) the statement of drug repositioning principles derived from our meta-analyses; and (3) the open access drug repositioning reporting standard (MIADRI) will be a valuable addition to drug development and repurposing investigations. Drug development companies and academic institutes can leverage these insights to develop new filtering methods to screen and prioritize compounds that may be suitable for new indications.
Limitations
RepurposeDB has various limitations and is not the only resource dedicated to drug repositioning. The limitations result from the methods of construction of the database, which may possibly introduce systematic biases into our results. We provide a detailed section on such limitations in the Supplementary Materials (see sections under: Availability of related resources for drug repositioning, Limitations of text mining and biocuration workflows and Limitations in biochemical inference based on enrichment analyses).
Availability
RepurposeDB database and all related data and source code are available in the public domain at the URL: http://repurposedb.dudleylab.org. The source code is available at the URL: https://bitbucket.org/dudleylab/repurposedb. Supplementary Data are available from the URL http://repurposedb.dudleylab.org/data and from the figshare URL: https://figshare.com/s/0364762ddd772076be31.
Key Points
Drug repositioning enables data-driven discovery of therapeutically actionable indications and offers a sustainable path to develop medicines for rare, common and chronic diseases in the setting of precision medicine.
Academic and industry drug discovery teams can leverage the knowledge from existing drug repurposing investigations to expand the knowledge-based drug repurposing efforts in the future.
We developed RepurposeDB: the first centralized open-access reference database of drug repositioning investigations and compiled data on drugs, disease indications, drug targets, disease comorbidities and various annotations.
Using systematic analyses of pharmacological properties of drugs, proteogenomic features of drug targets and epidemiological prevalence of disease indications from RepurposeDB, we have characterized the fundamental principles of drug repositioning.
RepurposeDB and its content could aid in rational drug repositioning and may accelerate the implementation of data-driven, precision therapeutics and help to implement systems pharmacology at the point of care in the near future.
Supplementary Data
Supplementary data are available online at http://bib.oxfordjournals.org/.
Supplementary Material
Acknowledgements
The authors would like to thank members of the Institute of Next Generation Healthcare, Icahn Institute for Genomics and Multiscale Biology (http://icahn.mssm.edu/departments-and-institutes/genomics), Department of Genetics and Genomic Sciences, Icahn School of Medicine at Mount Sinai, New York, NY and the Mount Sinai Data Ware House team of Mount Sinai Health System. The authors also acknowledge the Scientific Computing team, Icahn School of Medicine at the Mount Sinai (https://hpc.mssm.edu/).
Funding
This study was funded by the following grants from the National Institutes of Health: National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK, grant number R01-DK098242-03); National Cancer Institute (NCI, grant number U54-CA189201-02); Illuminating the Druggable Genome (IDG); Knowledge Management Center sponsored by NIH Common Fund; National Center for Advancing Translational Sciences (NCATS, grant number UL1TR000067); and Clinical and Translational Science Awards (CTSA) grant.
Khader Shameer is a senior biomedical and health care data scientist in the Dudley Laboratory and senior scientist at the Institute of Next Generation Healthcare, Mount Sinai Health System, New York, NY.
Benjamin S. Glicksberg is a PhD candidate in the Dudley and Chen Laboratories, Icahn School of Medicine at Mount Sinai, Mount Sinai Health System, New York, NY.
Rachel Hodos is a collaborative PhD candidate in the Dudley and Sontag Laboratories at the Icahn School of Medicine at Mount Sinai, Mount Sinai Health System, New York, NY and New York University, New York, NY.
Kipp W. Johnson is an MD-PhD student in the Dudley Laboratory at the Icahn School of Medicine at Mount Sinai, Mount Sinai Health System, New York, NY.
Marcus A. Badgeley is an MD-PhD student at the Icahn School of Medicine at Mount Sinai, Mount Sinai Health System, New York, NY.
Ben Readhead is a senior scientist in the Dudley Laboratory and Institute of Next Generation Healthcare, Mount Sinai Health System, New York, NY.
Max S. Tomlinson is a bioinformatician in the Dudley Laboratory and Institute of Next Generation Healthcare, Mount Sinai Health System, New York, NY.
Timothy O’Connor is a summer intern in the Dudley Laboratory and a student of Boston College, Boston, MA.
Riccardo Miotto is a data scientist in the Dudley Laboratory and Institute of Next Generation Healthcare, Mount Sinai Health System, New York, NY.
Brian A. Kidd is an assistant professor in the Dudley Laboratory and Institute of Next Generation Healthcare, Mount Sinai Health System, New York, NY.
Rong Chen is an assistant professor and the director of Clinical Genome Informatics at Icahn Institute of Genetics and Multiscale Biology, Mount Sinai Health System, New York, NY.
Avi Ma’ayan is a professor and the director of Mount Sinai Center for Bioinformatics, Mount Sinai Health System, New York, NY.
Joel T. Dudley is the executive director of Institute of Next Generation Healthcare, Associate Professor in the Department of Genetics and Genomic Sciences and Department of Population Health Science and Policy and Director of Biomedical Informatics, Icahn School of Medicine at Mount Sinai, Mount Sinai Health System, New York, NY.
References
- 1. Lyman GH, Moses HL.. Biomarker tests for molecularly targeted therapies–the key to unlocking precision medicine. N Engl J Med 2016;375:4–6. [DOI] [PubMed] [Google Scholar]
- 2. Collins FS, Varmus H.. A new initiative on precision medicine. N Engl J Med 2015;372:793–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mirnezami R, Nicholson J, Darzi A.. Preparing for precision medicine. N Engl J Med 2012;366:489–91. [DOI] [PubMed] [Google Scholar]
- 4. Xie L, Draizen EJ, Bourne PE.. Harnessing big data for systems pharmacology. Annu Rev Pharmacol Toxicol 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shameer K, Readhead B, Dudley JT.. Computational and experimental advances in drug repositioning for accelerated therapeutic stratification. Curr Top Med Chem 2015;15:5–20. [DOI] [PubMed] [Google Scholar]
- 6. Dudley JT, Deshpande T, Butte AJ.. Exploiting drug disease relationships for computational drug repositioning. Brief Bioinform 2011;12:303–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Readhead B, Dudley J.. Translational bioinformatics approaches to drug development. Adv Wound Care (New Rochelle) 2013;2:470–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Suthram S, Dudley JT, Chiang AP, et al. Network-based elucidation of human disease similarities reveals common functional modules enriched for pluripotent drug targets. PLoS Comput Biol 2010;6:e1000662.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sirota M, Dudley JT, Kim J, et al. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Sci Transl Med 2011;3:96ra77.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jahchan NS, Dudley JT, Mazur PK, et al. A drug repositioning approach identifies tricyclic antidepressants as inhibitors of small cell lung cancer and other neuroendocrine tumors, Cancer. Discov 2013;3:1364–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lamb J, Crawford ED, Peck D, et al. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science 2006;313:1929–35. [DOI] [PubMed] [Google Scholar]
- 12. Kidd BA, Readhead BP, Eden C, et al. Integrative network modeling approaches to personalized cancer medicine. Personalized Med 2015;12:245–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Blackwell AD. Measuring cognitive effects: cognition in drug development and repositioning. Drug Discov Today 2015;20:391–2. [DOI] [PubMed] [Google Scholar]
- 14. Power A, Berger AC, Ginsburg GS.. Genomics-enabled drug repositioning and repurposing: insights from an IOM Roundtable activity. JAMA 2014;311:2063–4. [DOI] [PubMed] [Google Scholar]
- 15. Wang W, Yang S, Zhang X, et al. Drug repositioning by integrating target information through a heterogeneous network model. Bioinformatics 2014;30:2923–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xu M, Lee EM, Wen Z, et al. Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat Med 2016;22:1101–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lau QY, Tan YY, Goh VC, et al. An FDA-Drug library screen for compounds with bioactivities against Meticillin-Resistant Staphylococcus aureus (MRSA). Antibiotics (Basel) 2015;4:424–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Irie H, Banno K, Yanokura M, et al. Metformin: a candidate for the treatment of gynecological tumors based on drug repositioning. Oncol Lett 2016;11:1287–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Amelio I, Gostev M, Knight RA, et al. DRUGSURV: a resource for repositioning of approved and experimental drugs in oncology based on patient survival information. Cell Death Dis 2014;5:e1051.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. von Eichborn J, Murgueitio MS, Dunkel M, et al. PROMISCUOUS: a database for network-based drug repositioning. Nucleic Acids Res 2011;39:D1060–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huang H, Nguyen T, Ibrahim S, et al. DMAP: a connectivity map database to enable identification of novel drug repositioning candidates. BMC Bioinformatics 2015;16 (Suppl 13):S4.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Keiser MJ, Irwin JJ, Shoichet BK.. The chemical basis of pharmacology. Biochemistry 2010;49:10267–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chan SY, Loscalzo J.. The emerging paradigm of network medicine in the study of human disease. Circ Res 2012;111:359–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li Y, Agarwal P, Rajagopalan D.. A global pathway crosstalk network. Bioinformatics 2008;24:1442–7. [DOI] [PubMed] [Google Scholar]
- 25. Natarajan M, Lin KM, Hsueh RC, et al. A global analysis of cross-talk in a mammalian cellular signalling network. Nat Cell Biol 2006;8:571–80. [DOI] [PubMed] [Google Scholar]
- 26. Hu Y, Bajorath J.. Monitoring drug promiscuity over time. F1000Res 2014;3:218.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tarcsay A, Keseru GM.. Contributions of molecular properties to drug promiscuity. J Med Chem 2013;56:1789–95. [DOI] [PubMed] [Google Scholar]
- 28. Basu MK, Carmel L, Rogozin IB, et al. Evolution of protein domain promiscuity in eukaryotes. Genome Res 2008;18:449–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Law V, Knox C, Djoumbou Y, et al. DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res 2014;42:D1091–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xu K, Cote TR.. Database identifies FDA-approved drugs with potential to be repurposed for treatment of orphan diseases. Brief Bioinform 2011;12:341–5. [DOI] [PubMed] [Google Scholar]
- 31. O'Boyle NM, Morley C, Hutchison GR.. Pybel: a Python wrapper for the OpenBabel cheminformatics toolkit. Chem Cent J 2008;2:5.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wegner J, JOELib, JOELib2: http://www-ra.informatik.uni-tuebingen.de/software/joelib/index.html 2005.
- 33. Steinbeck C, Han Y, Kuhn S, et al. The chemistry development kit (CDK): an open-source Java library for Chemo- and Bioinformatics. J Chem Inf Comput Sci 2003;43:493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Guha R, Howard MT, Hutchison GR, et al. The blue obelisk-interoperability in chemical informatics. J Chem Inf Model 2006;46:991–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cao Y, Charisi A, Cheng LC, et al. ChemmineR: a compound mining framework for R. Bioinformatics 2008;24:1733–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. O'Boyle NM, Banck M, James CA, et al. Open Babel: An open chemical toolbox. J Cheminform 2011; 3:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tetko IV, Gasteiger J, Todeschini R, et al. Virtual computational chemistry laboratory–design and description. J Comput Aided Mol Des 2005;19:453–63. [DOI] [PubMed] [Google Scholar]
- 38. van de Waterbeemd H, Gifford E.. ADMET in silico modelling: towards prediction paradise? Nat Rev Drug Discov 2003;2:192–204. [DOI] [PubMed] [Google Scholar]
- 39. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xia J, Wishart DS.. MSEA: a web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res 2010;38:W71–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Varin T, Gubler H, Parker CN, et al. Compound set enrichment: a novel approach to analysis of primary HTS data. J Chem Inf Model 2010;50:2067–78. [DOI] [PubMed] [Google Scholar]
- 42. Hastings J, de Matos P, Dekker A, et al. The ChEBI reference database and ontology for biologically relevant chemistry: enhancements for 2013. Nucleic Acids Res 2013;41:D456–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Moreno P, Beisken S, Harsha B, et al. BiNChE: a web tool and library for chemical enrichment analysis based on the ChEBI ontology. BMC Bioinformatics 2015;16:56.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. UniProt C. UniProt: a hub for protein information. Nucleic Acids Res 2015;43:D204–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Finn RD, Bateman A, Clements J, et al. Pfam: the protein families database. Nucleic Acids Res 2014;42:D222–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fox NK, Brenner SE, Chandonia JM.. SCOPe: structural classification of proteins–extended, integrating SCOP and ASTRAL data and classification of new structures. Nucleic Acids Res 2014;42:D304–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Das S, Lee D, Sillitoe I, et al. Functional classification of CATH superfamilies: a domain-based approach for protein function annotation. Bioinformatics 2016;32:2889.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Croft D, Mundo AF, Haw R, et al. The Reactome pathway knowledgebase. Nucleic Acids Res 2014;42:D472–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kanehisa M, Goto S.. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen EY, Tan CM, Kou Y, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 2013;14:128.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Huang da W, Sherman BT, Lempicki RA.. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- 52. Kamburov A, Stelzl U, Lehrach H, et al. The ConsensusPathDB interaction database: 2013 update. Nucleic Acids Res 2013;41:D793–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Glicksberg BS, Li L, Cheng WY, et al. An integrative pipeline for multi-modal discovery of disease relationships. Pac Symp Biocomput 2015;20:407–18. [PMC free article] [PubMed] [Google Scholar]
- 54. Glicksberg BS, Li L, Badgeley MA, et al. Comparative analyses of population-scale phenomic data in electronic medical records reveal race-specific disease networks. Bioinformatics 2016;32:i101–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chute CG. Invited commentary: Observational research in the age of the electronic health record. Am J Epidemiol 2014;179:759–61. [DOI] [PubMed] [Google Scholar]
- 56. Harispe S, Sanchez D, Ranwez S, et al. A framework for unifying ontology-based semantic similarity measures: a study in the biomedical domain. J Biomed Inform 2014;48:38–53. [DOI] [PubMed] [Google Scholar]
- 57. Menche J, Sharma A, Kitsak M, et al. Disease networks. Uncovering disease-disease relationships through the incomplete interactome. Science 2015;347:1257601.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Piette JD, Kerr EA.. The impact of comorbid chronic conditions on diabetes care. Diabetes Care 2006;29:725–31. [DOI] [PubMed] [Google Scholar]
- 59. Arain FA, Ye Z, Bailey KR, et al. Survival in patients with poorly compressible leg arteries. J Am Coll Cardiol 2012;59:400–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lang CC, Mancini DM.. Non-cardiac comorbidities in chronic heart failure. Heart 2007;93:665–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hidalgo CA, Blumm N, Barabasi AL, et al. A dynamic network approach for the study of human phenotypes. PLoS Comput Biol 2009;5:e1000353.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Robbins AS, Chao SY, Fonseca VP.. What's the relative risk? A method to directly estimate risk ratios in cohort studies of common outcomes. Ann Epidemiol 2002;12:452–4. [DOI] [PubMed] [Google Scholar]
- 63. Nelson MR, Tipney H, Painter JL, et al. The support of human genetic evidence for approved drug indications. Nat Genet 2015;47:856–60. [DOI] [PubMed] [Google Scholar]
- 64. Li L, Ruau DJ, Patel CJ, et al. Disease risk factors identified through shared genetic architecture and electronic medical records. Sci Transl Med 2014;6:234ra257.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Li L, Ruau D, Chen R, et al. Systematic identification of risk factors for Alzheimer's disease through shared genetic architecture and electronic medical records. Pac Symp Biocomput 2013;224–35. [PubMed] [Google Scholar]
- 66. Amberger JS, Bocchini CA, Schiettecatte F, et al. OMIM.org: Online Mendelian Inheritance in Man (OMIM(R)), an online catalog of human genes and genetic disorders. Nucleic Acids Res 2015;43:D789–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Welter D, MacArthur J, Morales J, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res 2014;42:D1001–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Li MJ, Liu Z, Wang P, et al. GWASdb v2: an update database for human genetic variants identified by genome-wide association studies. Nucleic Acids Res 2016;44:D869–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Li MJ, Wang P, Liu X, et al. GWASdb: a database for human genetic variants identified by genome-wide association studies. Nucleic Acids Res 2012;40:D1047–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yu W, Gwinn M, Clyne M, et al. A navigator for human genome epidemiology. Nat Genet 2008;40:124–5. [DOI] [PubMed] [Google Scholar]
- 71. Neves M, Leser U.. A survey on annotation tools for the biomedical literature. Brief Bioinform 2014;15:327–40. [DOI] [PubMed] [Google Scholar]
- 72. Hahn U, Cohen KB, Garten Y, et al. Mining the pharmacogenomics literature–a survey of the state of the art. Brief Bioinform 2012;13:460–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Harispe S, Ranwez S, Janaqi S, et al. The semantic measures library and toolkit: fast computation of semantic similarity and relatedness using biomedical ontologies. Bioinformatics 2014;30:740–2. [DOI] [PubMed] [Google Scholar]
- 74. Food-drug interactions could lower required dose of anticancer drug. Expert Rev Pharmacoecon Outcomes Res 2007;7:315–7. [DOI] [PubMed] [Google Scholar]
- 75. McCabe BJ. Prevention of food-drug interactions with special emphasis on older adults. Curr Opin Clin Nutr Metab Care 2004;7:21–6. [DOI] [PubMed] [Google Scholar]
- 76. Juurlink DN, Mamdani M, Kopp A, et al. Drug drug interactions among elderly patients hospitalized for drug toxicity. Jama 2003;289:1652–8. [DOI] [PubMed] [Google Scholar]
- 77. Vilar S, Uriarte E, Santana L, et al. Similarity-based modeling in large-scale prediction of drug drug interactions. Nat Protoc 2014;9:2147–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Warde-Farley D, Donaldson SL, Comes O, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 2010;38:W214–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Montojo J, Zuberi K, Rodriguez H, et al. GeneMANIA Cytoscape plugin: fast gene function predictions on the desktop. Bioinformatics 2010;26:2927–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Cao Y, Jiang T, Girke T.. A maximum common substructure-based algorithm for searching and predicting drug like compounds. Bioinformatics 2008;24:i366–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Chen X, Reynolds CH.. Performance of similarity measures in 2D fragment-based similarity searching: comparison of structural descriptors and similarity coefficients. J Chem Inf Comput Sci 2002;42:1407–14. [DOI] [PubMed] [Google Scholar]
- 82. Loging W, Harland L, Williams-Jones B.. High-throughput electronic biology: mining information for drug discovery. Nat Rev Drug Discov 2007;6:220–30. [DOI] [PubMed] [Google Scholar]
- 83. Bredel M, Jacoby E.. Chemogenomics: an emerging strategy for rapid target and drug discovery. Nat Rev Genet 2004;5:262–75. [DOI] [PubMed] [Google Scholar]
- 84. Keiser MJ, Roth BL, Armbruster BN, et al. Relating protein pharmacology by ligand chemistry. Nat Biotechnol 2007;25:197–206. [DOI] [PubMed] [Google Scholar]
- 85. Keiser MJ, Setola V, Irwin JJ, et al. Predicting new molecular targets for known drugs. Nature 2009;462:175–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Lipinski CA, Lombardo F, Dominy BW, et al. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 2001;46:3–26. [DOI] [PubMed] [Google Scholar]
- 87. Hann MM, Keseru GM.. Finding the sweet spot: the role of nature and nurture in medicinal chemistry. Nat Rev Drug Discov 2012;11:355–65. [DOI] [PubMed] [Google Scholar]
- 88. Waring MJ, Arrowsmith J, Leach AR, et al. An analysis of the attrition of drug candidates from four major pharmaceutical companies. Nat Rev Drug Discov 2015;14:475–86. [DOI] [PubMed] [Google Scholar]
- 89. Shameer K, Sowdhamini R.. Functional repertoire, molecular pathways and diseases associated with 3D domain swapping in the human proteome. J Clin Bioinforma 2012;2:8.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Mendez-Lucio O, Tran J, Medina-Franco JL, et al. Toward drug repurposing in epigenetics: olsalazine as a hypomethylating compound active in a cellular context. ChemMedChem 2014;9:560–5. [DOI] [PubMed] [Google Scholar]
- 91. Besnard J, Ruda GF, Setola V, et al. Automated design of ligands to polypharmacological profiles. Nature 2012;492:215–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Lappano R, Maggiolini M.. G protein-coupled receptors: novel targets for drug discovery in cancer. Nat Rev Drug Discov 2011;10:47–60. [DOI] [PubMed] [Google Scholar]
- 93. Moore JT, Collins JL, Pearce KH.. The nuclear receptor superfamily and drug discovery. ChemMedChem 2006;1:504–23. [DOI] [PubMed] [Google Scholar]
- 94. May AC, Johnson MS, Rufino SD, et al. The recognition of protein structure and function from sequence: adding value to genome data. Philos Trans R Soc Lond B Biol Sci 1994;344:373–81. [DOI] [PubMed] [Google Scholar]
- 95. Overington JP, Al-Lazikani B, Hopkins AL.. How many drug targets are there? Nat Rev Drug Discov 2006;5:993–6. [DOI] [PubMed] [Google Scholar]
- 96. Edwards DP. The role of coactivators and corepressors in the biology and mechanism of action of steroid hormone receptors. J Mammary Gland Biol Neoplasia 2000;5:307–24. [DOI] [PubMed] [Google Scholar]
- 97. Ahmadian M, Suh JM, Hah N, et al. PPARgamma signaling and metabolism: the good, the bad and the future. Nat Med 2013;19:557–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Kersten S, Desvergne B, Wahli W.. Roles of PPARs in health and disease. Nature 2000;405:421–4. [DOI] [PubMed] [Google Scholar]
- 99. Arnott ID, Watts D, Satsangi J.. Azathioprine and anti-TNF alpha therapies in Crohn's disease: a review of pharmacology, clinical efficacy and safety. Pharmacol Res 2003;47:1–10. [DOI] [PubMed] [Google Scholar]
- 100. Gronemeyer H, Gustafsson JA, Laudet V.. Principles for modulation of the nuclear receptor superfamily. Nat Rev Drug Discov 2004;3:950–64. [DOI] [PubMed] [Google Scholar]
- 101. Ji WR, Castellino FJ, Chang Y, et al. Characterization of kringle domains of angiostatin as antagonists of endothelial cell migration, an important process in angiogenesis. FASEB J 1998;12:1731–8. [DOI] [PubMed] [Google Scholar]
- 102. Pan Y, Cheng T, Wang Y, et al. Pathway analysis for drug repositioning based on public database mining. J Chem Inf Model 2014;54:407–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Jewison T, Su Y, Disfany FM, et al. SMPDB 2.0: big improvements to the small molecule pathway database. Nucleic Acids Res 2014;42:D478–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kelder T, van Iersel MP, Hanspers K, et al. WikiPathways: building research communities on biological pathways. Nucleic Acids Res 2012;40:D1301–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Schaefer CF, Anthony K, Krupa S, et al. PID: the pathway interaction database. Nucleic Acids Res 2009;37:D674–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Whirl-Carrillo M, McDonagh EM, Hebert JM, et al. Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther 2012;92:414–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Romero P, Wagg J, Green ML, et al. Computational prediction of human metabolic pathways from the complete human genome. Genome Biol 2005;6:R2.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Liu X, Zhu F, Ma XH, et al. Predicting targeted polypharmacology for drug repositioning and multi- target drug discovery. Curr Med Chem 2013;20:1646–61. [DOI] [PubMed] [Google Scholar]
- 109. Sanseau P, Agarwal P, Barnes MR, et al. Use of genome-wide association studies for drug repositioning. Nat Biotechnol 2012;30:317–20. [DOI] [PubMed] [Google Scholar]
- 110. Nelson MR, Johnson T, Warren L, et al. The genetics of drug efficacy: opportunities and challenges. Nat Rev Genet 2016;17:197–206. [DOI] [PubMed] [Google Scholar]
- 111. Rastegar-Mojarad M, Ye Z, Kolesar JM, et al. Opportunities for drug repositioning from phenome-wide association studies. Nat Biotechnol 2015;33:342–5. [DOI] [PubMed] [Google Scholar]
- 112. Vogt I, Prinz J, Campillos M.. Molecularly and clinically related drugs and diseases are enriched in phenotypically similar drug disease pairs. Genome Med 2014;6:52.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Piccirillo JF, Vlahiotis A, Barrett LB, et al. The changing prevalence of comorbidity across the age spectrum. Crit Rev Oncol Hematol 2008;67:124–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Starfield B, Lemke KW, Bernhardt T, et al. Comorbidity: implications for the importance of primary care in ′case′ management. Ann Fam Med 2003;1:8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. van Weel C, Schellevis FG.. Comorbidity and guidelines: conflicting interests. Lancet 2006;367:550–1. [DOI] [PubMed] [Google Scholar]
- 116. Hammer B, Ashurst P, Naish J.. Diseases associated with ulcerative colitis and Crohn's disease. Gut 1968;9:17–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Barrett JC, Hansoul S, Nicolae DL, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet 2008;40:955–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Kesselheim AS, Tan YT, Avorn J.. The roles of academia, rare diseases, and repurposing in the development of the most transformative drugs. Health Aff 2015;34:286–93. [DOI] [PubMed] [Google Scholar]
- 119. Murteira S, Ghezaiel Z, Karray S, Lamure M. . Drug reformulations and repositioning in pharmaceutical industry and its impact on market access: reassessment of nomenclature. J Mark Access Health Policy 2013;1:21131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Niedelman ML. Uses and abuses of drugs, new and old, in dermatology. Pa Med J 1954;57:333–8. [PubMed] [Google Scholar]
- 122. Hodos RA, Kidd BA, Shameer K, et al. In silico methods for drug repurposing and pharmacology. Wiley Interdiscip Rev Syst Biol Med 2016; [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.