Abstract
Background
COI (CORONATINE INSENSITIVE), an F-box component of the Skp1-Cullin-F-box protein (SCFCOI1) ubiquitin E3 ligase, plays important roles in the regulation of plant growth and development. Recent studies have shown that COIs are involved in pollen fertility. In this study, we identified and characterized COI genes in the wheat genome and analyzed expression patterns under abiotic stress.
Results
A total of 18 COI candidate sequences for 8 members of COI gene family were isolated in wheat (Triticum aestivum L.). Phylogenetic and structural analyses showed that these COI genes could be divided into seven distinct subfamilies. The COI genes showed high expression in stamens and glumes. The qRT-PCR results revealed that wheat COIs were involved in several abiotic stress responses and anther/glume dehiscence in the photoperiod-temperature sensitive genic male sterile (PTGMS) wheat line BS366.
Conclusions
The structural characteristics and expression patterns of the COI gene family in wheat as well as the stress-responsive and differential tissue-specific expression profiles of each TaCOI gene were examined in PTGMS wheat line BS366. In addition, we examined SA- and MeJA-induced gene expression in the wheat anther and glume to investigate the role of COI in the JA signaling pathway, involved in the regulation of abnormal anther dehiscence in the PTGMS wheat line. The results of this study contribute novel and detailed information about the TaCOI gene family in wheat and could be used as a benchmark for future studies of the molecular mechanisms of PTGMS in other crops.
Electronic supplementary material
The online version of this article (10.1186/s12864-018-5116-9) contains supplementary material, which is available to authorized users.
Keywords: Triticum aestivum L., COI, Gene family, JA signaling, Male sterile, Quantitative real-time PCR
Background
Jasmonates (JAs), including jasmonic acid (JA), methyl jasmonate (MeJA), and its derivatives, play important roles in the regulation of biotic and abiotic stresses as well as plant growth, development, and defense [1–4]. JA is also involved in the regulation of floral organ development, such as pollen maturation, anther dehiscence, and male fertility [5–8]. JA biosynthesis gene-deficient mutants (fad [9], opr3 [8, 10], dde1 [11], dad1 [12], and aos [13]) show abnormal anther growth and development in many plants. In rice, plants with mutant JA-amino acid synthetase osjar1–2 and osjar1–3 are insensitive to JA signaling, and the dehiscence of anthers filled with viable pollen is impaired, resulting in pollen sterility [14, 15]. In addition, JA plays a crucial role in sex determination during male flower development in maize [16].
JA signaling is mediated by CORONATINE INSENSITIVE 1 (COI1), which is an F-box component of the Skp1-Cullin-F-box protein (SCFCOI1) ubiquitin E3 ligase [17, 18]. The SCFCOI1 complex is predicted to target repressors of JA signaling to the 26S proteasome for degradation in Arabidopsis [19, 20]. In Arabidopsis, a single COI (At2939940) and 12 members of the JAZ family have been characterized. An Arabidopsis coi1 mutant exhibits a male sterile phenotype, including the inhibition of filament elongation and anther non-dehiscence [21]. COI is also involved in leaf senescence [22–25], apical dominance [26], inositol polyphosphates [27], and ethylene-induced root growth inhibition in the light in Arabidopsis thaliana [28]. In Arabidopsis, at least four of the 12 members of the AtJAZ family (JAZs1, 3, 9, and 10) interact with COI in a JA-Ile (jasmonoyl isoleucine)- or coronatine-dependent manner [29–33]. In Solanum nigrum, COI1 controls jasmonate metabolism and the production of a systemic signal against insect attacks [34]. The rice genome has three COI homologs, i.e., OsCOI1a (Os01g0853400; AK121543), OsCOI1b (Os05g0449500; AK101514), and OsCOI2 (Os03g0265500; AK100694). OsCOI1a and OsCOI1b share amino acid sequence identities of greater than 80%, and these loci share an identity of 63% with OsCOI2 [35]. OsCOI1a regulates OsbHLH148 expression in response to coronatine by forming an SCFCOI1 complex [36]. OsCOI1b is involved in leaf senescence [35]. Moreover, in addition to a MeJA-insensitive phenotype, OsCOI1 RNAi lines show an altered plant height, internode length, and grain length and increased susceptibility to chewing insects [37, 38].
Wheat (Triticum aestivum L.) is an important food crop worldwide. Plant fertility in photoperiod-temperature sensitive genic male sterile (PTGMS) lines is controlled by temperature and/or photoperiod; thus, the PTGMS line can be used as both a maintenance line and a male-sterile line for exploiting heterosis [39–42]. The discovery and application of PTGMS wheat series provided the basis for two-line hybrid wheat. The two-line hybrid system was quickly applied to wheat production owing to the potential for simple and low-cost seed production. Recently, it has been reported that male sterility of the female parent can be attributed to lack of anther development or dehiscence in the two line hybrid wheat system [43]. Our early research indicated that the anther of the PTGMS wheat line BS366 does not fully spill out, showing impaired anther dehiscence. The defective phenotype can be recovered by the application of MeJA in vitro [7]. Wheat genome sequencing is nearing completion (http://www.wheatgenome.org/), providing basic data for whole genome research and the identification of functional genes [44–47]. In wheat, the JA family is involved in anther dehiscence and male sterility [7, 48]. However, there are few studies of COI in the wheat jasmonic acid regulatory pathway. In this study, we first systematically identified and characterized the COI genes of wheat (TaCOI), and analyzed its architectural features, evolutionary history, and expression patterns in anther tissues in the PTGMS wheat line. In addition, the expression profiles of TaCOI genes in different tissues as well in response to several stresses were analyzed. Finally, the inducible expression of wheat COI genes was detected by qRT-PCR, and some COIs clearly responded to abiotic stress. The anther and glume dehiscence and the expression of TaCOI genes after treatment with MeJA and interactions between MeJA and SA were also analyzed to determine the role of COI in JA-mediated signaling pathways in anther dehiscence and male sterility.
Methods
Plant materials, growth conditions, and sample collection
The wheat (Triticum aestivum L.) PTGMS line BS366 (winter wheat) was used. The following treatments were evaluated. All plants were planted in experimental fields in Beijing (China, N 39°54′, E 116°18′) and managed conventionally.
Hormone cross-stimulation. After the four-leaf stage [42, 49], plants were transferred to an artificial climate incubator (CLC-BIV-M/CLC404-TV, MMM, Germany) at 20 °C (with 12-h day/12-h night) and a relative humidity of 60–80% for the entire reproductive period. After transferring samples to the artificial climate incubator, they will be incubated for about two weeks until head period [7, 48]. Then, the spikelets were sprayed with 0.5 mM, 2 mM, and 4 mM MeJA for each group at 20 °C (with 12-h day/12-h night) every day for 5 days. For the SA and MeJA interaction treatment, 0.5 mM, 2 mM, and 4 mM MeJA were applied every day for 5 days when spikelets were treated with 10 mM SA (20 °C, 12-h day/12-h night) in the artificial climate incubator. The glumes and anthers were collected for further analysis. Glume and anther dehiscence were detected using a dissecting microscope (Olympus SZX12; Tokyo, Japan).
Abiotic stresses. Two-week-old wheat seedlings (20 °C, 12-h day/12-h night cycle) were used for stress treatments. BS366 seedlings were sprayed with 2 mM SA, 100 mM MeJA, 100 mM GA3, 50 mM IAA, and 100 mM ABA. The control plants were treated with 0.1% (v/v) ethanol. For the high-salinity and drought treatments, the roots of wheat seedlings were soaked in 200 mM NaCl and PEG6000 (− 0.5 MPa). For low temperature (cold) stress, the seedlings were moved to an incubator at 10 °C (with a 12-h day/12-h night cycle) and a relative humidity of 60–80%. The leaf tissues from seedlings were collected at 0, 2, 4, 8, 12, and 24 h post-treatment.
Samples (root, stem, leaf, glume, stamen, and pistil) planted in beijing, were collected for tissue-specific expression analyses at the heading stage [7, 48].
Fertiliy analysis. The PTGMS wheat line BS366 shows thermo-photoperiod sensitivity in pollen fertiliy [49]. It shows anther indehiscent and pollen sterility (seed setting rate < 5%) in sterile condition (10 °C with 12-h day/12-h night for daily mean temperature during pollen development stages), but exhibits anther dehiscent and pollen fertility (40% < Seed setting rate < 55%) in fertile condition (20 °C with 12-h day/12-h night for daily mean temperature during pollen development stages) [42, 43, 48, 49]. In this part of the experiment, after the four-leaf stage [42, 49], plants were grouped two and transferred to artificial climate incubators at 20 °C and 10 °C (with 12-h day/12-h night), respectively, and a relative humidity of 60–80% for the entire reproductive period. Based on the leaf age index and anther length, spikelets for stage-specific expression analyses were collected from two fertility conditions for the PTGMS line BS366 at three different pollen developmental stages, which are stage 1: secondary sporogenous cells had formed; stage 2: all cell layers were present and mitosis had ceased, and stage 3: meiotic division stage [42, 49].
All samples were rapidly frozen in liquid nitrogen and stored in a − 80 °C freezer until RNA extraction.
Sources of sequence data
The COI protein sequences of Triticum aestivum L. (T. aestivum, Ta), Physcomitrella patens (Pp), Populus trichocarpa (Pt), Arabidopsis thaliana (At), Sorghum bicolor (Sb), Zea mays (Zm), Oryza sativa (Os), and Brachypodium distachyon (Bd) were obtained from the JGI database (http://genome.jgipsf.org/). Selaginella moellendorffii (Sm), Aegilops tauschii (Aet), Triticum urartu (Tu) and Hordeum vulgare L. (Hv) sequences were acquired from the Ensembl Plants database (http://plants.ensembl.org/index.html). All sequence information is provided in Additional file 2: Table S1.
Identification of COI gene family members in wheat
COI protein sequences were downloaded from the PLAZA database (http://hmmer.org/, ID: ORTHO001097) [50]. The hidden Markov model (HMM) profiles were constructed using the hmmbuild procedure (HMMER3.0) (http://hmmer.org/) for comparisons of the COI conserved protein structure domain in the wheat protein database (E-value ≤1.0E-10). Then, the COI gene family members were confirmed after removing non-COI conserved domains and repeats using the CDD (Conserved Domain Database) of NCBI and the SMART web server (http://smart.embl-heidelberg.de/). COI subgenomic copies were named A, B, and D according to their positions on chromosomes.
Analyses of gene characteristics
COI structures were analyzed using the Gene Structure Display Server (GSDS) program (http://gsds.cbi.pku.edu.cn/). Protein properties, including the relative molecular weight (MW) and isoelectric point (PI), were predicted using ExPASy (http://www.expasy.org/). WoLF PSORT (http://www.genscript.com/wolf-psort.html) was used for TaCOI gene family member subcellular localization predictions. Multiple Expectation Maximization for Motif Elicitation (MEME) was used to identify the motifs of COI proteins, with default settings. To analyze putative cis-acting elements in a promoter region, 2-kb promoter regions were selected and screened against the Plant CARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). The sequences of the COI promoters are listed in Additional file 2: Table S3.
Multiple sequence alignment and phylogenetic tree construction
To determine the roles of COIs in the JA signaling pathway, a multiple sequence alignment of the amino acid sequences of COI in selected plant genomes was generated using DNAMAN (ver. 6.0) with default settings. Subsequently, MEGA 6.0 was used to construct an unrooted phylogenetic tree based with the neighbor-joining method (with 1000 bootstrap replicates).
Chromosomal locations analysis
To map the corresponding COI loci on T. aestivum, T. urartu, Ae. Tauschii, and B. distachyon chromosomes, the genome annotation files for these plants were obtained from the JGI database (https://genome.jgi.doe.gov/portal/) and Ensembl Plants database (http://plants.ensembl.org/index.html). To detect synteny of the COI genes in T aestivum and selected plants, whole genome synteny block data were collected from the Plant Genome Duplication Database (http://chibba.agtec.uga.edu/duplication/) [51]. Chromosomal locations for the COI family genes were examined with default parameters to determine duplications and synteny relationships using Circos-0.69 (http://circos.ca/) [52].
Total RNA extraction, cDNA synthesis and qRT-PCR analysis
Total RNA was isolated for each sample using TRIzol Reagent (Invitrogen, Nottingham, UK) according to the manufacturer’s instructions. The purified RNA was stored at − 80 °C until subsequent analyses.
First-strand cDNA synthesis was performed using M-MLV Reverse Transcriptase according to the manufacturer’s instructions (Takara, Shiga, Japan). Quantitative real-time PCR (qRT-PCR) was performed using a SYBR Premix Ex Taq Kit (Takara) and a real-time PCR machine (CFX96; Bio-Rad, Hercules, CA, USA), following the manufacturer’s instructions. The procedure used for qRT-PCR was 10 min at 95 °C, followed by 38 cycles of 15 s at 95 °C and 60 s at 61–62 °C. β-Actin was used as a reference gene for the mRNA relative expression patterns analysis. The reactions were performed with three biological replicates and at least two technical replicates per sample. The data were analyzed using the 2-∆∆Ct method [53] and means ± standard errors (SE) of three biological replicates are presented. The primers for qRT-PCR are listed in Additional file 2: Table S2.
Results
Identification, phylogenetic analysis, and classification of wheat COI genes
After removing redundant reads, 18 COI candidate protein sequences were obtained by HMM searches against the T. aestivum genome sequence. These 18 COI protein sequences were clustered into eight groups based on high similarity (≥95%). These eight groups were referred to as TaCOI1–TaCOI8 according to naming conventions. In addition, These COI genes were distinguished by their positions in the wheat sub-genome (A, B, and D). Gene information is summarized in Table 1. To understand the role of gene functions, subcellular localization prediction for TaCOI family using WoLE PSORT (http://www.genscript.com/wolf-psort.html) was performed. It was showed that these TaCOI genes exhibited cytoplasmic, nuclear, and chloroplast localization. TaCOI-A, TaCOI2-A, TaCOI2-B, TaCOI3-B, TaCOI3-D, TaCOI4-D, TaCOI6-A, TaCOI6-B, TaCOI6-D, and TaCOI8-A were located in cytoplasmic fractions. TaCOI4-A and TaCOI4-B were located in the cytoplasm and nuclei. The remaining genes were located in the cytoplasm and chloroplasts. Moreover, the molecular weights (MW) and isoelectric points (pI) of TaCOI proteins were calculated using the Protparam tool in ExPASy. The MW of COIs varied from 20.39 to 66.36 kDa, and the pIs ranged from 5.41 to 8.79 (Table 1). Different copies of the same TaCOI showed different physicochemical properties, such as TaCOI3-B owing 3 times MW than that of TaCOI4-A.
Table 1.
Characteristics of TaCOI gene family members from wheat
| Gene name | Sequence ID | Location | Protein/AA | Molecular weight of decuced protein/KD | Isoelectric point | Subcellular Localization |
|---|---|---|---|---|---|---|
| TaCOI1-A | Traes_1AS_1E22B5174.1 | 1A:10791781:10793235 | 484 | 54.29 | 6.07 | Cytoplasm |
| TaCOI2-A | Traes_2AL_9EC359B65.2 | 2A:38684891:38689283 | 574 | 63.99 | 5.83 | Cytoplasm |
| TaCOI2-B | Traes_2BL_370AE211F.1 | 2B:111631663:111636013 | 574 | 64.21 | 6.25 | Cytoplasm |
| TaCOI3-B | TRAES3BF018800040CFD_t1 | 3B:443238454:443241608 | 594 | 66.36 | 6.58 | Cytoplasm |
| TaCOI3-D | Traes_3DL_845220DFC.1 | 3D:93268132:93270118 | 184 | 20.39 | 4.94 | Cytoplasm |
| TaCOI4-A | Traes_3AL_2240B0C5F.1 | 3A:163966748:163968217 | 191 | 20.75 | 6.91 | Cytoplasm Nuclear |
| TaCOI4-B | TRAES3BF021600080CFD_t1 | 3B:655287004:655289230 | 381 | 40.87 | 6.83 | Cytoplasm Nuclear |
| TaCOI4-D | Traes_3DL_3DB9F1EC5.1 | 3D:99011996:99014454 | 284 | 30.53 | 5.86 | Cytoplasm |
| TaCOI5-A | Traes_4AS_AD3F2B991.1 | 4A:48344426:48347184 | 575 | 64.29 | 5.96 | Cytoplasm Chloroplast |
| TaCOI5-B | Traes_4BL_499E7F095.1 | 4B:243711227:243714109 | 586 | 65.60 | 5.95 | Cytoplasm Chloroplast |
| TaCOI5-D | Traes_4DL_8C2C2ADD4.1 | 4D:19191547:19194445 | 586 | 65.57 | 6.01 | Cytoplasm Chloroplast |
| TaCOI6-A | Traes_5AL_27F194099.2 | 5A:77415870:77422532 | 569 | 62.09 | 8.79 | Cytoplasm |
| TaCOI6-B | Traes_5BL_7AE04C6B2.1 | 5B:189453532:189459589 | 572 | 62.14 | 8.57 | Cytoplasm |
| TaCOI6-D | Traes_5DL_4E8DD7C5F.3 | 5D:108986194:108992606 | 569 | 62.07 | 8.62 | Cytoplasm |
| TaCOI7-A | Traes_6AL_0F53490CA.1 | 6A:185429448:185432194 | 343 | 37.63 | 5.75 | Cytoplasm Chloroplast |
| TaCOI7-B | Traes_6BL_9E82CDBD1.1 | 6B:162540302:162543336 | 429 | 47.81 | 5.41 | Cytoplasm Chloroplast |
| TaCOI7-D | Traes_6DL_AD0DAD6D1.2 | 6D:140761611:140765528 | 343 | 37.54 | 5.60 | Cytoplasm Chloroplast |
| TaCOI8-A | Traes_7AS_78FEAE00C.1 | 7A:31411042:31413175 | 387 | 42.98 | 5.75 | Cytoplasm |
Phylogenetic analysis
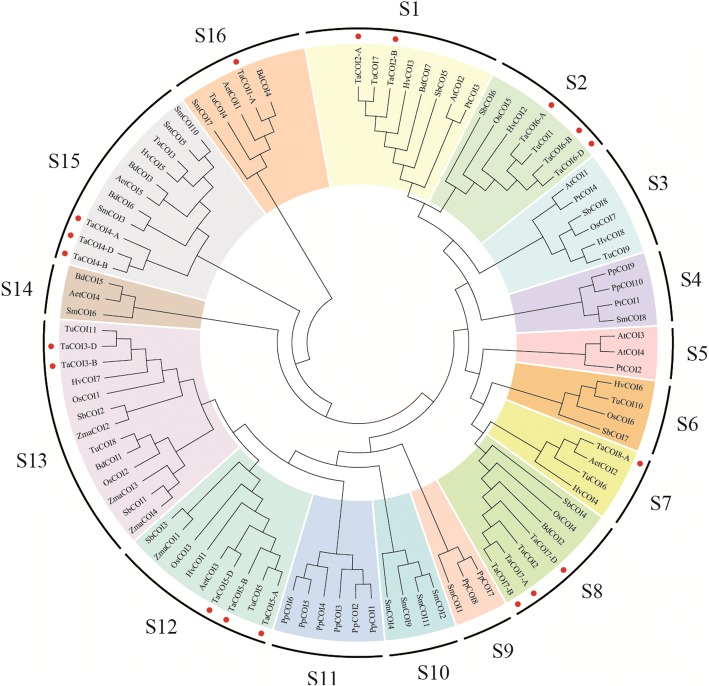
To study the phylogeny of COI proteins, a neighbor-joining phylogenetic tree was built using MEGA 6.0 with default parameters. As shown in Fig. 1, COI proteins were clustered into 16 sub-groups, S1–S16, and the 18 protein sequences from eight wheat TaCOI family members clustered into 8 sub-groups (Fig. 1). TaCOI1-A, TaCOI2 (TaCOI2-A and TaCOI2-B), TaCOI3 (TaCOI3-B and TaCOI3-D), TaCOI4 (TaCOI4-A, TaCOI4-B, and TaCOI4-D), TaCOI5 (TaCOI5-A, TaCOI5-B, and TaCOI5-D), TaCOI6 (TaCOI6-A and TaCOI6-B), TaCOI7 (TaCOI7-A, TaCOI7-B, and TaCOI7-D), and TaCOI8-A clustered into groups S16, S1, S12, S14, S11, S2, S8, and S7, respectively. The COI proteins clustered into the same clades with some OsCOI, SbCOI, BdCOI, PtCOI, AtCOI, and AetCOI proteins, indicating that the TaCOI proteins share high homology with the COI proteins of other plants (Fig. 1). Furthermore, we found that COI proteins from same species clustered into different clades. For example, PpCOI proteins were clustered into S4, S10, and S14, and SmCOI proteins were clustered into S4, S9, S13 S 14, S15, and S16, revealing that COI gene exhibited differences in evolution among species.
Fig. 1.
Bio Neighbor-Joining (BioNJ) phylogenetic tree (1000 bootstrap replicates), based on a protein alignment of COIs from T. aestivum (Ta), P. patens (Pp), P. trichocarpa (Pt), A. thaliana (At), S. bicolor (Sb), Z. mays (Zm), O. sativa (Os), B. distachyon (Bd), S. moellendorffii (Sm), Ae. tauschii (Aet), T. urartu (Tu), and H. vulgare (Hv). Each TaCOI protein is indicated by a red dot
Analysis of stress response-related cis-regulatory elements in the promoter regions of TaCOI genes
In plants, gene transcription is regulated by cis-acting regulatory elements that bind to target transcription factors [54]. Some cis-regulatory elements are involved in stress responses, such as dehydration and cold responses (DRE/CRT) [55], ABA responsive element (ABRE) [56], ARF binding sites (AuxRE) [57], and SA-responsive promoter elements (SARE) [58, 59]. To analyze how the expression levels of TaCOI genes responded to stress stimuli, 2.0-kb upstream promoter regions of TaCOI genes were scanned for stress-related cis-regulatory elements using the Plant CARE online service (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) [18]. As shown in Fig. 2 and Table 3, five hormone-responsive regulatory elements, ABRE, TGA-element, TATC-box, CGTCA/TGACG-motif, and TCA-element, associated with ABA, auxin (IAA), gibberellin (GA), methyl jasmonate (MeJA), and salicylic acid (SA) responses, were identified in the promotor region of TaCOIs. Additionally, two stress-responsive regulatory elements, TC-rich repeats and LTR, associated with defense/stress and low-temperature responses, respectively, were identified in the TaCOI promoter regions. Different types and numbers of regulatory elements were present in the distinct TaCOI promoters (Table 2), indicating that TaCOI genes might be involved in the response to various stress and hormone treatments via participating different regulatory mechanisms. The CGTCA/TGACG-motif, which was associated with a methyl jasmonate (MeJA) responsive element, was enriched in the promoters of TaCOI genes (Fig. 2, Additional file 1: Figure S1). Other than TaCOI1-A and TaCOI8-A, TaCOIs contained two or more CGTCA/TGACG-motifs; for example, TaCOI6-D contained three CGTCA/TGACG-motifs and TaCOI3-D contained four CGTCA/TGACG-motifs. It was also found that ABRE-motif was also enriched in the promoters of most of all TaCOI genes (Fig. 2 and Table 2). The numbers and types of stress-related cis-elements in the 2.0-kb upstream regions of TaCOI family genes are listed in Table 2 and Table 3.
Fig. 2.
Analysis of specific cis-elements in promoters. The 2 k-bp promoter sequences of corresponding COI genes were used to analyze five specific hormone-related cis-elements, including ABRE, TGA-element, TATC-box, CGTCA/TGACG-motif, TCA-element which respond to ABA, auxin (IAA), gibberellin (GA), methyl jasmonate (MeJA), salicylic acid (SA) and two stress-responsive regulatory elements, TC-rich repeats and LTR which respond to stress/defence and low temperature, which are color-coded
Table 3.
Functions of cis- acting regulatory elements in promoter regions of TaCOI genes
| Site Name | Organism | sequence | function |
|---|---|---|---|
| TC-rich repeats | Nicotiana tabacum | GTTTTCTTAC | cis-acting element involved in defense and stress responsiveness |
| TCA-element | Nicotiana tabacum | CCATCTTTTT | cis-acting element involved in salicylic acid responsiveness |
| CGTCA-motif | Hordeum vulgare | CGTCA | cis-acting regulatory element involved in the MeJA-responsiveness |
| TGACG-motif | Hordeum vulgare | TGACG | cis-acting regulatory element involved in the MeJA-responsiveness |
| LTR | Hordeum vulgare | CCGAAA | cis-acting element involved in low-temperature responsiveness |
| TATC-box | Oryza sativa | TATCCCA | cis-acting element involved in gibberellin-responsiveness |
| ABRE | Arabidopsis thaliana | TACGTG | cis-acting element involved in the abscisic acid responsiveness |
| TGA-element | Brassica oleracea | AACGAC | auxin-responsive element |
Table 2.
The number and composition of cis- acting regulatory elements of each TaCOI gene
| Gene | CGTCA-motif or TGACG-motif (MeJA) |
ABRE (ABA) |
TGA-element (IAA) |
TATC-box (GA) |
LTR (Cold) |
TC-rich repeats (Defense/stress) |
TCA-element (SA) |
|---|---|---|---|---|---|---|---|
| TaCOI1-A | 1 | 3 | 1 | 5 | |||
| TaCOI2-A | 4 | 5 | 2 | 1 | 2 | 1 | |
| TaCOI2-B | 3 | 3 | 1 | 1 | 2 | 1 | |
| TaCOI3-B | 3 | 8 | 1 | 1 | |||
| TaCOI3-D | 4 | 3 | 1 | 2 | 1 | 1 | |
| TaCOI4-A | 3 | 3 | 2 | 3 | 1 | 1 | |
| TaCOI4-B | 2 | 2 | 1 | 3 | 3 | ||
| TaCOI4-D | 3 | 5 | 1 | 4 | 3 | ||
| TaCOI5-A | 1 | 1 | 1 | ||||
| TaCOI5-B | 2 | 3 | 1 | ||||
| TaCOI5-D | 1 | 3 | 1 | 3 | 3 | 1 | |
| TaCOI6-A | 3 | 1 | 1 | 2 | |||
| TaCOI6-B | 4 | 2 | 5 | 1 | 1 | ||
| TaCOI6-D | 3 | 3 | 2 | 1 | 2 | 2 | |
| TaCOI7-A | 2 | 2 | 1 | 2 | 1 | ||
| TaCOI7-B | 3 | 1 | 2 | ||||
| TaCOI7-D | 2 | 1 | 1 | 1 | |||
| TaCOI8-A | 1 | 2 | 1 |
Structural analysis of wheat COI genes and proteins
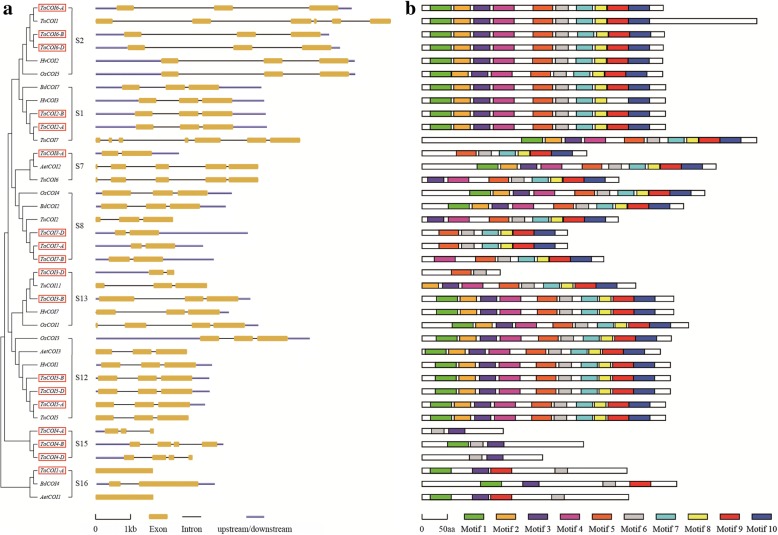
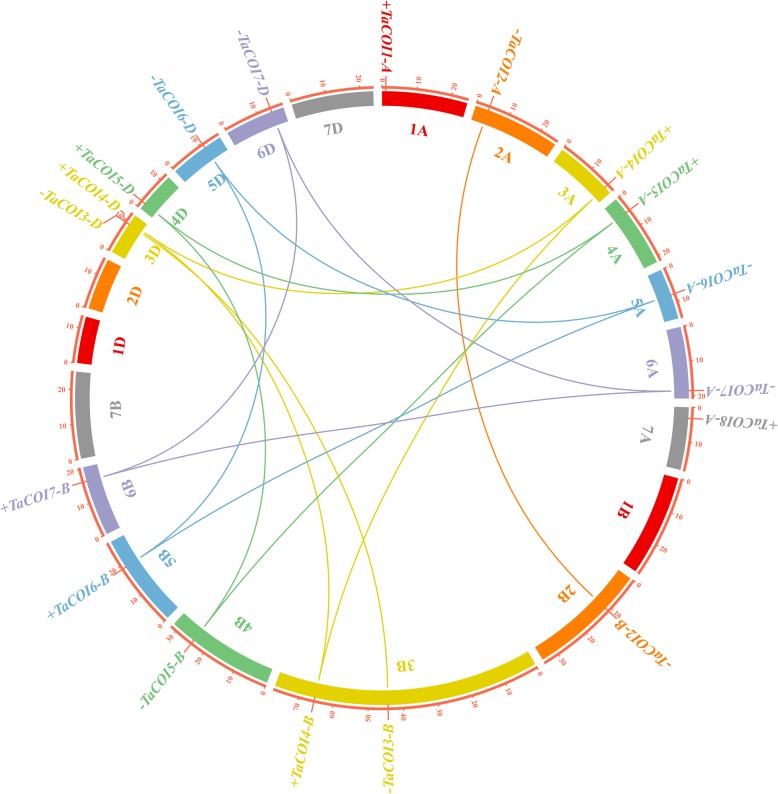
To further understand the function of TaCOI genes, the structural features of COIs were compared among families and species (O. sativa, Ae. tauschii, T. urartu, and H. vulgare) by aligning the predicted coding sequences (CDS) against the corresponding genomic sequences (Fig. 3a). The organization (number, length, and distribution) of exons and introns were variable (Fig. 3a). The sequence of TaCOI6 was longest among these TaCOIs, while TaCOI1-A and TaCOI4-A were the shortest. The majority of TaCOI genes contained two or three copies, and the number of exons ranged from one to four (Fig. 3a). The copies of TaCOI2, 5, 6, and 7 had the same gene structures, whereas TaCOI2 showed structural differences among copies. Furthermore, most COIs in the same sub-group shared the same structures; for example, BdCOI7 and HvCOI3 had the same structures as TaCOI2-A and B in S1, and AetCOI3, HvCOI1, and TuCOI5 had the same structures as TaCOI5-A, B, and D in S12 (Fig. 3a). However, some COIs showed divergence in gene structure within the same sub-group. TuCOI7 contained 7 exons, whereas other COIs in same sub-groups had three exons. To further investigate the function of TaCOIs, chromosomal locations and synteny analysis were performed. As shown in Fig. 4, 18 TaCOI members of 8 TaCOI families were located on 16 of the 21 chromosomes other than 1B, 1D, 2D, 7B, and 7D. The distribution of TaCOI family members in the wheat genome is depicted in Fig. 4. TaCOI3-B and TaCOI4-B as well as TaCOI3-D and TaCOI4-D were positioned on the same chromosomes, 3B and 3D, respectively. Each of the remaining chromosomes included a single TaCOI gene. The TaCOI genes showed a scattered chromosomal distribution. To analyze the relationships between the COI genes and gene duplications, COI gene synteny was analyzed (Fig. 4). The paralogous TaCOIs on different chromosomes were segmental duplication events and clustered together.
Fig. 3.
Exon-intron structures (a) of the COI genes in T. aestivum (Ta), O. sativa (Os), B. distachyon (Bd), T. urartu (Tu), and H. vulgare (Hv) and a motif distribution analysis (b). Exons are shown as yellow boxes, introns are shown as thin lines, and UTRs are shown as blue lines. The sizes of exons and introns can be estimated using the scale below. Ten motif types are shown as colored boxes
Fig. 4.
Chromosomal localizations and syntenic relationships among TaCOI genes in wheat. The positions of TaCOI genes are marked directly on chromosomes. “+” and “-” in front of gene names mean sense and anti-sense strand of genome
To characterize the architecture of COI proteins in wheat, motifs were analyzed by submitting the predicted amino acid sequences to the MEME website (Fig. 3b and Additional file 1: Figure S1). Most COIs in S1, S2, S12, and S13 possessed ten motifs, whereas COIs in S15 possessed motif3 and 6, and COIs in S16 possessed motif1, 3, 6, and 9. COIs in S7 and S8 possessed at least 6 motifs. Motif 1 was identified as an F-box domain, motif 2–6 were LRR domains (leucine-rich repeat), and motif 7–10 were also rich in leucine sequences. The motif logos and sequences are shown in Additional file 1: Figure S1.
Tissue/organ-specific and stage-specific expression profiles of TaCOIs
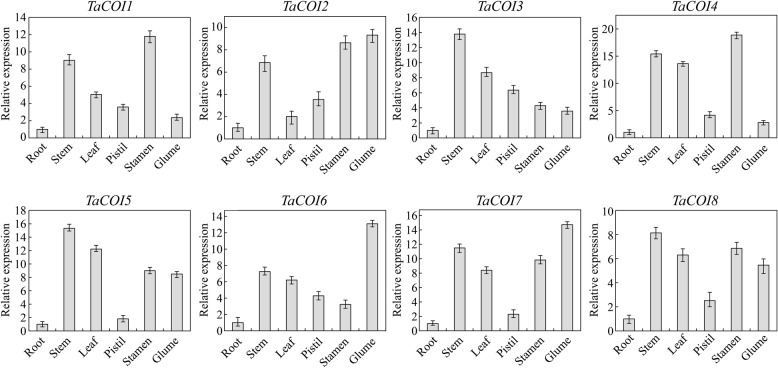
The COI gene family is primarily involved in plant growth [60], development [21, 35], and stress responses [61]. To study the biological functions of these genes in wheat, the expression profiles of the eight TaCOI genes were analyzed in six tissue types (i.e., root, stem, leaf, petal, pistil, stamen, and glume tissues) of PTGMS line BS366 at the heading stage by qRT-PCR. As shown in Fig. 5, eight TaCOI genes were expressed in different tissues, suggesting that the TaCOI genes were constitutively expressed in BS366. Compared with the expression of TaCOIs in root tissues, expression levels were higher in other aerial tissues (stem, leaf, petal, pistil, stamen, and glume). The expression levels of TaCOI2, TaCOI6, and TaCOI7 in glume tissues were markedly higher than those in other tissues. TaCOI1, TaCOI2, and TaCOI4 expression levels in the stamen were relatively highly than those in other tissues. Interestingly, expression levels were lower in important reproductive organs than in stem tissues.
Fig. 5.
Real-time PCR analysis of TaCOI genes in six wheat tissues (root, stem, leaf, pistil, stamen, and glume) in the heading stage. Actin was used as a reference control gene. The 2-∆∆Ct method was used to calculate the relative expression levels of the target genes. The error bars indicate the standard deviation obtained from three replicates
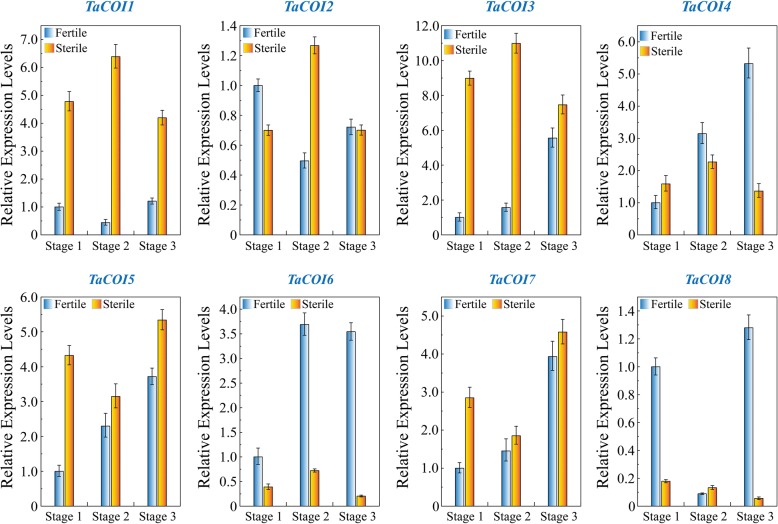
To further investigate the role of TaCOI genes in pollen fertility, the TaCOI expression profiles at three stages of stamen development in different fertility environments (fertile conditions and sterile condition) were analyzed. As shown in Fig. 6, TaCOI1, TaCOI3, TaCOI5, and TaCOI7 exhibited higher expression levels in all three stamen developmental stages in sterile condition than that in fertile condition, while the expression levels of TaCOI6 and TaCOI8 in the three stamen developmental stages were lower in sterile condition. TaCOI4 showed higher expression in early stamen developmental stages (stage1) in sterile condition, while TaCOI2 showed higher expression in later stamen developmental stages (stage2 and stage3) in sterile condition (stage1). These results suggested that these TaCOI genes may be involved in stamen development and participate in fertile transformation via some unknown pathway.
Fig. 6.
Expression profiling of TaCOI genes at three anther development stages in fertile (20 °C with 12-h day/12-h night for daily mean temperature during pollen development stages) and sterile conditions (10 °C with 12-h day/12-h night for daily mean temperature during pollen development stages). I, II and III are mean stage 1: secondary sporogenous cells had formed, stage 2: all cell layers were present and mitosis had ceased and stage 3: meiotic division stage, respectively. The error bars indicate the standard deviation of three replicates
Stress-induced expression profiles of TaCOIs
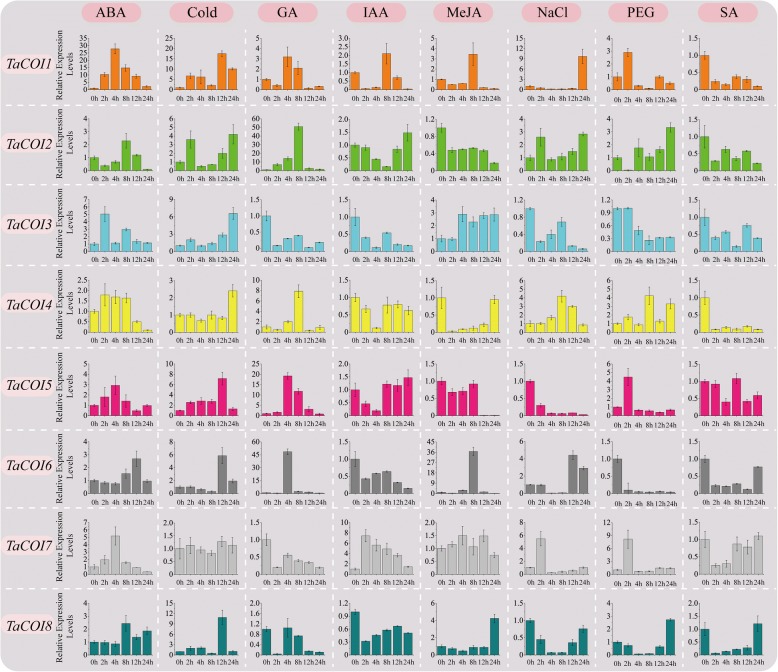
In plant, Auxin, ABA, SA, MeJA, and GA are major hormones involved in plant development and metabolism [8, 62, 63]. To determine the mechanisms involved in the responses of the wheat COI gene family to plant hormones, cold (10 °C), salt, and drought, qRT-PCR was performed to determine the relative expression pattern of each COI gene using 14-day-old BS366 seedlings (Fig. 7). The results showed that all TaCOI genes responded to plant hormones and stress (Fig. 7). Under ABA treatment, the transcript profiles of TaCOI genes showed the up-regulation of TaCOI1, 3, 4, 5, 7 and the down-regulation of TaCOI2, 6, 8 at early treatment time points (0-4 h post-treatment). TaCOI3 was induced 2 h post-treatment after ABA treatment and subsequently inhibited. In addition, we found that TaCOI1, 5, and 7 were peaked at 4 h post-treatment then inhibited (Fig. 7). Under GA treatment, TaCOI1 and 5 were up-regulated, while TaCOI7 and 8 were down-regulated during the treatment period; TaCOI2 and 4 were up-regulated at 2–8 h post-treatment, but were rapidly down-regulated thereafter (Fig. 7). Under auxin (IAA) treatment, only TaCOI7 was up-regulated during the treatment period. The remaining TaCOIs were down-regulated (Fig. 7). For MeJA treatment, the levels of TaCOI2, 4, 5, and 8 were down-regulated during the treatment period, while TaCOI 6 was up-regulated at 8 h post-treatment. TaCOI 3 was induced from 4 h post-treatment, following by a rapid up-regulation. Interestingly, all TaCOI genes were inhibited to different degrees under SA treatment. Notably, we found that the expressions of TaCOI 4, 6, 7 were decreased after treated by both MeJA and SA, while the expressions of TaCOI3 showed opposite pattern between MeJA and SA treaments, also between GA and ABA treaments, suggested that these hormones may play a antagonistic effect on expressions TaCOI3.
Fig. 7.
Expression profiling of TaCOI genes under five phytohormone (MeJA, ABA, GA, IAA, and SA), drought (PEG 6000), salt (NaCl), and cold (10 °C) treatments in BS366. Actin was used as a reference control gene. ABA (100 mM), Cold (10 °C), GA (100 mM), IAA (50 mM), MeJA (100 mM), NaCl (200 mM), PEG 6000 (− 0.5 MPa) and SA (2 mM), were preformed in this study. The 2-∆∆Ct method was used to calculate the relative expression levels of the target genes. There are three replicates
Under low temperature (cold) treatment, the expression levels of TaCOI1, 2, 3, and 5 were up-regulated during the whole treatment period (Fig. 7). For high salinity (NaCl) stress, the expression of TaCOI3, 5, and 8 showed down-regulation, whereas TaCOI4 showed up-regulation during the whole treatment period. The expression of TaCOI4 was increased at 4 h and peaked at 8 h. Under drought (PEG) stress, the expression levels of TaCOI1, 5, and 7 initially increased, and subsequently decreased. Under drought (PEG) stress, the expression levels of TaCOI3 and 6 were down-regulated at each time point after treatment. These results revealed that these TaCOI genes responded to various plant hormones and were involved in complex regulatory networks in the PTGMS wheat line BS366.
The regulation of COIs in wheat anther/glume dehiscence
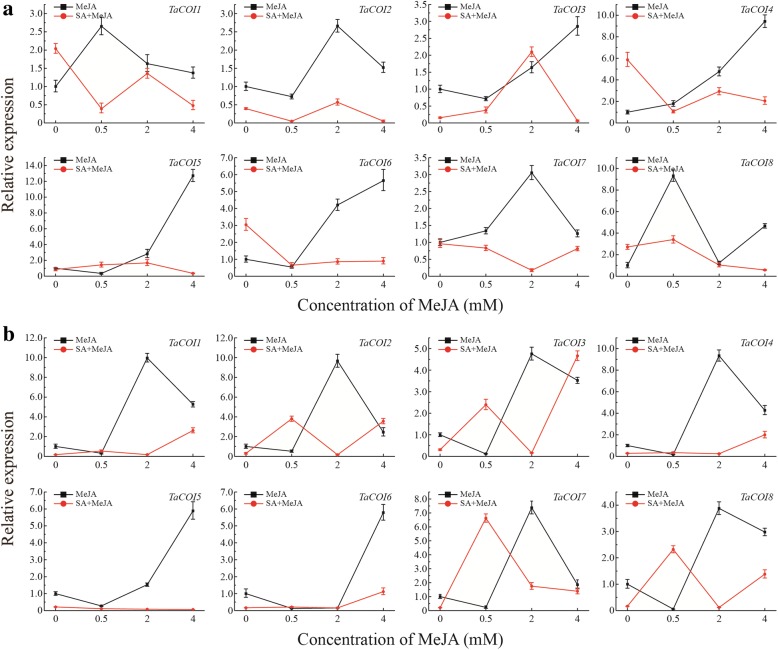
To investigated the regulation of COIs in the JA signaling pathway during anther dehiscence, wheat spikelets were treated with various concentrations of MeJA and SA during anther development. Glume dehiscence was induced by increasing concentrations of MeJA (Fig. 8a–d), and the angles of glume dehiscence for 0, 0.5, 2, and 4 mM MeJA were 10.4°, 11.1°, 13.1°, and 15.5°, respectively. After treatment of MeJA-treated plants with SA, glumes dehisced at a smaller angle compared with those of MeJA-treated plants. The angles were 7.7°, 8.4°, 10.0°, and 11.6°, respectively (Fig. 8e–h). Anther dehiscence was also induced by increasing concentrations of MeJA (Fig. 8i–l). The anthers exhibited little dehiscence before MeJA treatment, but dehisced slightly in 0.5 mM MeJA, and reached full dehiscence in 2 mM MeJA. The expression levels of COI genes in glumes and anthers under MeJA and SA treatments were also investigated (Fig. 8i–p). The expression levels of TaCOI3, 4, 5, and 6 were up-regulated with increasing MeJA concentrations in the glume (Fig. 9a). Additionally, TaCOI1 and 8 were induced and their expression levels peaked after treatment with 0.5 mM MeJA, while the expression levels of TaCOI2 and 7 peaked after treatment with 2 mM MeJA and subsequently decreased. After treatment of MeJA-treated plants with SA (10 mM), the expression of all TaCOI genes was repressed in glumes (Fig. 9a). In the anther, the expression levels of TaCOI5 and 6 were up-regulated as the MeJA concentration increased, and the levels of the remaining TaCOIs peaked after treatment with 2 mM MeJA and subsequently decreased (Fig. 9b). Furthermore, the expression levels of all TaCOIs in MeJA-treated plants were repressed by SA (10 mM), especially for plants treated with 2 mM MeJA (Fig. 9b).
Fig. 8.
The dehiscence of glumes (a–h) and anthers (i–p) treated with MeJA and SA. a–d and I–L represent the glumes and anthers treated with MeJA (0 mM, 0.5 mM, 2 mM, and 4 mM from left to right), respectively. e–h and m–p represent the glumes and anthers treated with MeJA (0 mM, 0.5 mM, 2 mM, and 4 mM from left to right) and SA (10 mM). MeJA: spikelets were treated with MeJA (0 mM, 0.5 mM, 2 mM, and 4 mM from left to right). MeJA+SA: SA (10 mM) and MeJA interaction treatment
Fig. 9.
Expression profiling of TaCOI genes in glumes (a) and anthers (b) treated with MeJA (0 mM 0.5 mM, 2 mM, and 4 mM) and SA (10 mM). Values on the x-axis indicate the MeJA concentration. MeJA: spikelets were treated with MeJA (0 mM, 0.5 mM, 2 mM, and 4 mM). MeJA + SA: SA (10 mM) and MeJA interaction treatment. The error bars indicate the standard deviation of three replicates
Discussion
JAs are oxylipin signaling molecules involved in the control of various aspects of plant developmental processes, such as plant fertility [5, 6, 8], anthocyanin accumulation [64], fruit ripening [65], root growth [66], and leaf senescence [24, 25]. In addition, they act as defense signals to mediate plant responses against abiotic and biotic stresses [18, 48, 60]. Upon exogenous or endogenous JA induction, the JA receptor CORONATINE INSENSITIVE 1 (COI1), which perceives bioactive molecules of JA [17, 21, 32, 67, 68], and triggers the degradation of the JASMONATE ZIM-domain (JAZ) proteins via the 26S-proteasome [18, 29]. The COI gene family has been characterized in many plants, such as A. thaliana [67], H. brasiliensis [69], G. hybridus [70], and O. sativa [35], including analyses of gene and protein characteristics and their functions. However, in wheat, little is known about COI genes. In the present study, 18 candidate COI copies clustered into seven sub-groups (Fig. 1) and eight members of COI gene family were newly isolated in wheat (Additional file 2: Table S4).
The analyses of TaCOI genes provided insight into the important roles underlying the roles of MeJA in pollen fertility in wheat. In plants, the expression levels of COI family genes are regulated by various hormones, especially MeJA [70, 71]. Phytohormone crosstalk is a complex regulatory network involved in plant growth and development. Recent studies have reported that the modulation of JA responses by JAZ proteins probably plays a pivotal role in phytohormone signaling pathways [48, 81]. For example, JA-GA crosstalk could be important for defense against necrotrophic pathogens [82] and acts synergistically during stamen development, as a deficiency in either hormone results in infertility [83]. To clarify the functions of TaCOI genes, cis-acting regulatory elements and expression profiles under various stresses were analyzed. As shown in Fig. 2, five hormone-responsive (ABA, IAA, GA, MeJA, and SA) and two stress-responsive (high salinity, drought, and cold) regulatory elements were found in the TaCOI promoter regions, suggesting that TaCOI genes have different regulatory mechanisms in response to various stress and hormone treatments. However, there were some differences between TaCOI genes with respect to the number and type of cis-acting regulatory elements. For example, most TaCOIs contained two or more CGTCA/TGACG-motifs, but not the promoters of TaCOI1-A and TaCOI8-A (Fig. 2, Table 3). These results indicated that different TaCOI members may respond to different stress and hormone stimulation. Therefore, we investigated the expression profiles of TaCOI genes in wheat seedlings under different stress conditions were evaluated. In plants, the expression levels of COI family genes were regulated by different hormones. In Arabidopsis, COI1 functions in the JA signaling pathway and is required for pollen development [9, 21] and defense against pests [80]. In this study, inducible expression analyses revealed that TaCOI expression could be induced by at least one hormone (Fig. 7). For example, both TaCOI2 and TaCOI6, with similar protein structures, were down-regulated by treatment with IAA, MeJA, SA and PEG (Fig. 7). TaCOI1 was up-regulated in response to GA and IAA. These TaCOI genes were also induced by some abiotic stresses (Fig. 7). TaCOI1 was up-regulated under salt and drought stresses, but TaCOI3, TaCOI6, and TaCOI8 were down-regulated. Based on the composition of cis-acting regulatory elements in promoter regions, TaCOI7-A, B, and D had two or more CGTCA/TGACG-motifs in their promoter regions (Fig. 2, Table 3), but the response to MeJA was not obvious for TaCOI7 (Fig. 7). Similar results were obtained for TaCOI 1 (Fig. 2 and Fig. 7). There was no obvious response to SA for TaCOI1, but there was a single TGA-element in its promoter region (Fig. 2 and Fig. 7). Therefore, these TaCOIs might be directly or indirectly involved in phytohormone crosstalk [48].
Recently, many studies have shown that the LRR domain (LXXLXXLXX LXLXXNXLVGXIP) may function in determining resistance specificity, suggesting that LRR domain-contained proteins are involved in stress resistance in plants [76, 77]. Function studies showed that LRR domain-containing proteins, such as CLAVATA1 and ERECTA, which are membrane receptor kinases, are involved in signal reception and transmission in plant development [78, 79]. FLOR1 is a flower-specific LRR protein involved in the development of floral organs in A. thaliana [6], suggesting that TaCOIs play a role in plant defense and floral organ development. In Arabidopsis, the sequence of the COIl protein contains a degenerate F-box motif [17, 21, 67, 72] and 16 imperfect leucine-rich repeats (LRRs) [73]. Some studies have shown that LRRs and F-box motifs are involved in protein–protein interactions [74, 75]. In our study, the structural features of TaCOI proteins in different families and species were also investigated. It was found that the most of COI proteins were enriched for the LRR domain and F-box domain, especially LRR domains (with 2–9 LRR domains per TaCOI) (Fig. 3 and Additional file 1: Figure S2). For example, TaCOI6 and TaCOI2 shared similar structures and contained the same ten motifs, and 9 out of 10 motifs were LRR domains or leucine-rich sequences, which was consistent with the conclusion obtained by the previous scholar [76, 77], suggesting that these TaCOI genes may play a role in plant defense.via these enriched domains.
The tissue expression profiles of TaCOI genes were also investigated to further examine TaCOI gene functions (Fig. 5). In this study, the expression levels of TaCOI2, TaCOI6, and TaCOI7 in the glume and TaCOI1 and TaCOI4 in the stamen were higher than those in other tissues (Fig. 5). Additionally, all TaCOI genes showed lower expression levels in root tissues than in other tissues (Fig. 5). These results suggested that TaCOI genes are involved in stamen and glume development. Plant hormones often play a key role in plant growth and development synergistic ally or antagonistically, through a series of complex networks [87]. Studies reported that GA and ABA showed antagonistic roles in plant growth and development [87]. Our results also showed that some TaCOI3 genes exhibited opposite expression pattern under GA and ABA treaments (Fig. 7). Recently, studies have indicated that JA and SA signaling act in a mutually antagonistic manner [84]. MeJA plays a positive regulatory role in glume opening, while SA inhibits glume opening in wheat [85]. It has been reported that JAs are involved in the regulation of anther dehiscence, filament elongation, and pollen fertility in plants [48, 83],. However, the mechanisms underlying JA–SA crosstalk and particularly the mechanisms by which JA signaling antagonizes SA signaling are still largely unknown. MeJA and SA may have antagonistic effect on expressions TaCOI genes in PTGMS wheat line (Fig. 7) Thus, the regulation of TaCOIs in JA–SA signaling crosstalk in anther dehiscence and glume opening was examined by evaluating the effects of various concentrations of MeJA and SA on wheat spikelets during anther development (Fig. 8). Glume dehiscence increased as the concentration of MeJA increased; the angles of glume dehiscence for 0, 0.5, 2, and 4 mM MeJA were 10.4°, 11.1°, 13.1°, and 15.5°, respectively (Fig. 8a–h). Changes were also detected in anthers (Fig. 8i-p). In addition, SA showed an inhibitory effect on anther and glume dehiscence. The glumes dehisced at a smaller angle compared with those of MeJA-treated plants (7.7°, 8.4°, 10.0°, and 11.6° respectively), consistent with the results of previous reports (Fig. 8a–h) [85, 86]. The expression of TaCOI genes in the glume and anther under various concentrations of MeJA and SA were also investigated (Fig. 9). COI is associated with jasmonate-regulated defense and fertility in Arabidopsis [67]. In the present study, some TaCOI genes were inhibited by SA and induced by MeJA (Fig. 9). For example, the expression levels of TaCOI 3, 4, 5, and 6 were up-regulated with increasing concentrations of MeJA in the glume and the expression levels of TaCOI 5 and 6 were up-regulated with increasing concentrations of MeJA in the anther (Fig. 9). In addition, all TaCOI genes were inhibited by SA in the glume and anther (Fig. 9). It was suggested that TaCOIs are involved in JA–SA signaling crosstalk, associating with anther and glume dehiscence. In addition, the expression results for TaCOI genes during anther development in different fertility environments showed that TaCOI1, TaCOI3, TaCOI5, and TaCOI7 exhibit higher expression, while the expression levels of TaCOI6 and TaCOI8 were lower at all three anther development stages in the sterile environment (Fig. 6), suggesting that TaCOI genes have different roles in fertility.
Taken together, these results demonstrated that TaCOIs may have multiple functions, in plants, including roles in abiotic stress resistance and pollen fertility. Structural analyses and the expression patterns of the TaCOI genes would provide a more comprehensive understanding of gene functions. Further studies are needed to confirm the conclusions presented in this study.
Conclusions
In this study, a comprehensive overview of the COI family in wheat, including gene structures, phylogenetic relationships, and expression profiles, was provided. The roles of TaCOI gene expression differences in anther development and in the responses to various abiotic stresses provide insight into the roles of MeJA signaling in the anther and glume dehiscence of wheat.
Additional files
Figure S1. Consensus sequence and logos of motifs from wheat COI proteins. (TIF 2339 kb)
Table S1. Summary of COI or COI-like genes in different plants. Table S2. Specific primers for TaCOI genes for qRT-PCR. Table S3. Promoter sequences of TaCOI genes. Table S4. Protein sequences, CDS sequences, and genomic sequences of TaCOI and COI or 12 COI-like loci in various plants. (DOCX 241 kb)
Acknowledgements
We are grateful to Beijing BioTrust Biotechnology Co. Ltd. (Beijing, China) for providing their support in bioinformatics analysis and language help. All authors also thank the reviewers for helpful suggestion.
Funding
In this work, qPCR-related kits and all consumable material equipments were supported by the National Key Research Project (Grant no. 2016YFD0101601), Natural Science Foundation of Beijing (Grant no. 6182014) and National Science and Technology Support Plan (2014BAD01B09); service charges for data analysis, travel expenses for sample collection and other fees were supported by National Natural Science Foundation of China (Grant no. 31661143018), National Natural Science Foundation of China (Grant No. 31872881) and International Cooperation Fund of Beijing Academy of Agricultural and Forestry Sciences (GJHZ2018–1).
Availability of data and materials
The data sets supporting the results of this article are included within the article and its additional files.
Abbreviations
- ABA
Abscisic acid
- COI
Coronatine insensitive
- GA
ibberellin
- IAA
Indole-3-acetic acid
- JA-Ile
Jasmonoyl isoleucine
- JAZ
Jasmonate-ZIM domain
- MeJA
Methyl jasmonate
- ORF
Open reading frame
- PTGMS
Photoperiod-Temperature sensitive Genic Male Sterile
- qRT-PCR
Quantitative real-time PCR
- SA
Salicylic acid
- SCFCOI1
Skp1-Cullin-F-box protein
Authors’ contributions
LZ and CZ designed the study. YW, PW and JB performed the experiments, analyzed the data, and drafted the manuscript. PW, WZ and WD assisted with bioinformatic analysis and aided in writing the manuscript. SY, JG, NW, MQ and FZ aided in performing the experiments. All authors carefully checked and approved this version of the manuscript.
Ethics approval and consent to participate
PTGMS wheat line BS366 seeds were provided by Beijing Engineering and Technique Research Center for Hybrid Wheat.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jian-fang Bai, Email: baijianfang1313@yahoo.com.
Yu-kun Wang, Email: 15652370148@163.com.
Peng Wang, Email: 931874319@qq.com.
Shao-hua Yuan, Email: keaidehuahua0830@126.com.
Jian-gang Gao, Email: jianganggao@qq.com.
Wen-jing Duan, Email: 1439158900@qq.com.
Na Wang, Email: wangna_00@sina.com.
Feng-ting Zhang, Email: lyezh@163.com.
Wen-jie Zhang, Email: lyezh@163.com.
Meng-ying Qin, Email: 18519194330@163.com.
Chang-ping Zhao, Email: cp_zhao@vip.sohu.com.
Li-ping Zhang, Email: lpzhang8@126.com.
References
- 1.Cheong JJ, Choi YD. Methyl jasmonate as a vital substance in plants. Trends Genet Tig. 2003;19(7):409. doi: 10.1016/S0168-9525(03)00138-0. [DOI] [PubMed] [Google Scholar]
- 2.Sasaki Y, Asamizu E, Shibata D, Nakamura Y, Kaneko T, Awai K, Amagai M, Kuwata C, Tsugane T, Masuda T. Monitoring of methyl jasmonate-responsive genes in Arabidopsis by cDNA macroarray: self-activation of jasmonic acid biosynthesis and crosstalk with other phytohormone signaling pathways. DNA Res. 2001;8(4):153. doi: 10.1093/dnares/8.4.153. [DOI] [PubMed] [Google Scholar]
- 3.Kim EH, Kim YS, Park SH, Koo YJ, Yang DC, Chung YY, Lee IJ, Kim JK. Methyl Jasmonate reduces grain yield by mediating stress signals to Alter spikelet development in Rice. Plant Physiol. 2009;149(4):1751–1760. doi: 10.1104/pp.108.134684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koo AJK, Howe GA, Hause B, Wasternack C, Strack D. The wound hormone jasmonate. Phytochemistry. 2009;70(13–14):1571. doi: 10.1016/j.phytochem.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turner JG, Ellis C, Devoto A. The jasmonate signal pathway. Plant Cell. 2002;14(Suppl 14):S153. doi: 10.1105/tpc.000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Acevedo FG, Gamboa A, Paezvalencia J, Jimenezgarcia LF, Izaguirresierra M, Alvarezbuylla ER. FLOR1, a putative interaction partner of the floral homeotic protein AGAMOUS, is a plant-specific intracellular LRR. Plant Sci. 2004;167(2):225–231. doi: 10.1016/j.plantsci.2004.03.009. [DOI] [Google Scholar]
- 7.Ma Hua, Zhang LP, Zhao CP, Xiao-Ping LU, Zhang FT, Yuan SH: Effects of Methyl-Jasmonic Acid on The Percentage of Anther Dehiscence and Seedset Rate of Photo-thermo-sensitive Male Sterile Wheat. Journal of Triticeae Crops 2011.
- 8.Wang Y, Yuan G, Yuan S, Duan W, Wang P, Bai J, Zhang F, Gao S, Zhang L, Zhao C. TaOPR2 encodes a 12-oxo-phytodienoic acid reductase involved in the biosynthesis of jasmonic acid in wheat (Triticum aestivum L.) Biochem Biophys ResCommun. 2016;470(1):233–238. doi: 10.1016/j.bbrc.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 9.Mcconn M, Browse J. The critical requirement for Linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell. 1996;8(3):403–416. doi: 10.1105/tpc.8.3.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stintzi A, Browse J. The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc Natl Acad Sci U S A. 2000;97(19):10625. doi: 10.1073/pnas.190264497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanders PM, Lee PY, Biesgen C, Boone JD, Beals TP, Weiler EW, Goldberg RB. The arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell. 2000;12(7):1041–1061. doi: 10.1105/tpc.12.7.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishiguro S, KawaiOda A, Ueda J, Nishida I, Okada K. The DEFECTIVE IN ANTHER DEHISCENCE1 gene encodes a novel phospholipase A1 catalyzing the initial step of Jasmonic acid biosynthesis, which synchronizes pollen maturation, anther DEHISCENCE, and flower opening in Arabidopsis. Plant Cell. 2001;13(10):2191–2209. doi: 10.1105/tpc.13.10.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park J, Halitschke R, Kim H, Baldwin I, Feldmann K, Feyereisen R. A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 2002;31(1):1–12. doi: 10.1046/j.1365-313X.2002.01328.x. [DOI] [PubMed] [Google Scholar]
- 14.Riemann M, Takano M. Rice JASMONATE RESISTANT 1 is involved in phytochrome and jasmonate signalling. Plant Cell Environ. 2008;31(6):783–792. doi: 10.1111/j.1365-3040.2008.01790.x. [DOI] [PubMed] [Google Scholar]
- 15.Xiao Y, Chen Y, Charnikhova T, Mulder PP, Heijmans J, Hoogenboom A, Agalou A, Michel C, Morel JB, Dreni L. OsJAR1 is required for JA-regulated floret opening and anther dehiscence in rice. Plant Mol Biol. 2014;86(1–2):19. doi: 10.1007/s11103-014-0212-y. [DOI] [PubMed] [Google Scholar]
- 16.Acosta IF, Laparra H, Romero SP, Schmelz E, Hamberg M, Mottinger JP, Moreno MA, Dellaporta SL. Tasselseed1 is a lipoxygenase affecting jasmonic acid signaling in sex determination of maize. Science. 2009;323(5911):262–265. doi: 10.1126/science.1164645. [DOI] [PubMed] [Google Scholar]
- 17.Yan J, Zhang C, Gu M, Bai Z, Zhang W, Qi T, Cheng Z, Peng W, Luo H, Nan F. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell. 2009;21(8):2220–2236. doi: 10.1105/tpc.109.065730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abuqamar S, Chai MF, Luo H, Song F, Mengiste T. Tomato protein kinase 1b mediates signaling of plant responses to necrotrophic fungi and insect herbivory. Plant Cell. 2008;20(7):1964. doi: 10.1105/tpc.108.059477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Devoto A, Nieto-Rostro M, Xie D, Ellis C, Harmston R, Patrick E, Davis J, Sherratt L, Coleman M, Turner JG. COI1 links jasmonate signaling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J. 2002;32(4):457–466. doi: 10.1046/j.1365-313X.2002.01432.x. [DOI] [PubMed] [Google Scholar]
- 20.Xu L, Liu F, Lechner E, Genschik P, Crosby W, Ma H, Peng W, Huang D, Xie D. The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell. 2002;14(8):1919. doi: 10.1105/tpc.003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feys B, Benedetti C, Penfold C, Turner J. Arabidopsis mutants selected for resistance to the Phytotoxin Coronatine are male sterile, insensitive to methyl Jasmonate, and Resistant to a bacterial pathogen. Plant Cell. 1994;6(5):751. doi: 10.1105/tpc.6.5.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castillo MC, León J. Expression of the β-oxidation gene 3-ketoacyl-CoA thiolase 2 (KAT2) is required for the timely onset of natural and dark-induced leaf senescence in Arabidopsis. J Exp Bot. 2008;59(8):2171–2179. doi: 10.1093/jxb/ern079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shan X, Wang J, Chua L, Jiang D, Peng W, Xie D. The role of Arabidopsis Rubisco Activase in Jasmonate-induced leaf senescence. Plant Physiol. 2011;155(2):751–764. doi: 10.1104/pp.110.166595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reinbothe C, Springer A, Samol I, Reinbothe S. Plant oxylipins: role of jasmonic acid during programmed cell death, defense and leaf senescence. FEBS J. 2009;276(17):4666. doi: 10.1111/j.1742-4658.2009.07193.x. [DOI] [PubMed] [Google Scholar]
- 25.Ueda J, Kato J. Isolation and identification of a senescence-promoting substance from wormwood (Artemisia absinthium L.) Plant Physiol. 1980;66(2):246–249. doi: 10.1104/pp.66.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim J, Dotson B, Rey C, Lindsey J, Bleecker AB, Binder BM, Patterson SE. New clothes for the Jasmonic acid receptor COI1: Delayed abscission, meristem arrest and apical dominance. PLoS One. 2013;8(4):e60505. doi: 10.1371/journal.pone.0060505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mosblech A, Thurow C, Gatz C, Feussner I, Heilmann I. Jasmonic acid perception by COI1 involves inositol polyphosphates in Arabidopsis thaliana. Plant J. 2011;65(6):949–957. doi: 10.1111/j.1365-313X.2011.04480.x. [DOI] [PubMed] [Google Scholar]
- 28.Adams E, Turner J. COI1, a jasmonate receptor, is involved in ethylene-induced inhibition of Arabidopsis root growth in the light. J Exp Bot. 2010;61(15):4373. doi: 10.1093/jxb/erq240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chini A, Fonseca S, Fernández G, Adie B, Chico J, Lorenzo O, GarcíaCasado G, LópezVidriero I, Lozano F, Ponce M. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448(7154):666. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- 30.Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature. 2007;448(7154):661–665. doi: 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- 31.Melotto M, Mecey C, Niu Y, Chung HS, Katsir L, Yao J, Zeng W, Thines B, Staswick P, Browse J. A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 2008;55(6):979–988. doi: 10.1111/j.1365-313X.2008.03566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fonseca S, Chini A, Hamberg M, Adie B, Porzel A, Kramell R, Miersch O, Wasternack C. Solano R: (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat Chem Biol. 2009;5(5):344. doi: 10.1038/nchembio.161. [DOI] [PubMed] [Google Scholar]
- 33.Chung HS, Howe GA. A critical role for the TIFY motif in repression of Jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell. 2009;21(1):131. doi: 10.1105/tpc.108.064097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vandoorn A, Bonaventure G, Schmidt DD, Baldwin IT. Regulation of jasmonate metabolism and activation of systemic signaling in Solanum nigrum: COI1 and JAR4 play overlapping yet distinct roles. New Phytol. 2011;190(3):640. doi: 10.1111/j.1469-8137.2010.03622.x. [DOI] [PubMed] [Google Scholar]
- 35.Sang-Hwa Y. Sakuraba, Taeyoung, Kyu-won, Gynheung, Yong, Nam-chon, Paek: mutation of Oryza sativa CORONATINE INSENSITIVE 1b (OsCOI1b) delays leaf senescence. J Integr Plant Biol. 2015;57(6):562. doi: 10.1111/jipb.12276. [DOI] [PubMed] [Google Scholar]
- 36.Seo JS, Joo J, Kim MJ, Kim YK, Nahm BH, Song SI, Cheong JJ, Lee JS, Kim JK, Choi YD. OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. 2011;65(6):907–921. doi: 10.1111/j.1365-313X.2010.04477.x. [DOI] [PubMed] [Google Scholar]
- 37.Ye M, Luo SM, Xie JF, Li YF, Xu T, Liu Y, Song YY, Zhu-Salzman K, Zeng RS. Silencing COI1 in rice increases susceptibility to chewing insects and impairs inducible defense. PLoS One. 2012;7(4):e36214. doi: 10.1371/journal.pone.0036214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang DL, He SY. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci U S A. 2012;109(19):E1192. doi: 10.1073/pnas.1201616109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jordaan JP. Hybrid wheat: advances and challenges. 1996. [Google Scholar]
- 40.Virmani SS, Ilyas-Ahmed M. Environment-sensitive genic male sterility (EGMS) in crops. Adv Agron. 2001;72(01):139–195. doi: 10.1016/S0065-2113(01)72013-5. [DOI] [Google Scholar]
- 41.Zhou H, Zhou M, Yang Y, Li J, Zhu L, Jiang D, Dong J, Liu Q, Gu L, Zhou L. RNase ZS1 processes UbL40 mRNAs and controls thermosensitive genic male sterility in rice. Nat Commun. 2014;5:4884. doi: 10.1038/ncomms5884. [DOI] [PubMed] [Google Scholar]
- 42.Tang Z, Zhang L, Xu C, Yuan S, Zhang F, Zheng Y, Zhao C. Uncovering small RNA-mediated responses to cold stress in a wheat Thermosensitive genie male-sterile line by deep sequencing. Plant Physiol. 2012;159(2):721–738. doi: 10.1104/pp.112.196048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li YF, Zhao CP, Zhang FT, Sun H, Sun DF. Fertility alteration in the photo-thermo-sensitive male sterile line BS20 of wheat ( Triticum aestivum L.) Euphytica. 2006;151(2):207–213. doi: 10.1007/s10681-006-9141-4. [DOI] [Google Scholar]
- 44.Mayer KFX, Rogers J, Doležel J, Pozniak C, Eversole K, Feuillet C, Gill B, Friebe B, Lukaszewski AJ, Sourdille P. A chromsome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science. 2014;345(6194):1251788. doi: 10.1126/science.1251788. [DOI] [PubMed] [Google Scholar]
- 45.Choulet F, Alberti A, Theil S, Glover N, Barbe V, Daron J, Pingault L, Sourdille P, Couloux A, Paux E. Structural and functional partitioning of bread wheat chromosome 3B. Science. 2014;345(6194):1249721. doi: 10.1126/science.1249721. [DOI] [PubMed] [Google Scholar]
- 46.Marcussen T, Sandve SR, Heier L, Spannagl M, Pfeifer M, Jakobsen KS, Wulff BBH, Steuernagel B, Mayer KFX, Olsen OA. Ancient hybridizations among the ancestral genomes of bread wheat. Science. 2014;345(6194):1250092. doi: 10.1126/science.1250092. [DOI] [PubMed] [Google Scholar]
- 47.Pfeifer M, Kugler KG, Sandve SR, Zhan B, Rudi H, Hvidsten TR, Mayer KF, Olsen OA. Genome interplay in the grain transcriptome of hexaploid bread wheat. Science. 2014;345(6194):1250091. doi: 10.1126/science.1250091. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y, Qiao L, Bai J, Peng W, Duan W, Yuan S, Yuan G, Zhang F, Zhang L, Zhao C. Genome-wide characterization of JASMONATE-ZIM DOMAIN transcription repressors in wheat ( Triticum aestivum L.) BMC Genomics. 2017;18(1):152. doi: 10.1186/s12864-017-3582-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bai JF, Wang YK, Wang P, Duan WJ, Yuan SH, Sun H, Yuan GL, Ma JX, Wang N, Zhang FT. Uncovering male fertility transition responsive miRNA in a wheat photo-Thermosensitive genic male sterile line by deep sequencing and Degradome analysis. Front Plant Sci. 2017;8:1370. doi: 10.3389/fpls.2017.01370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Proost S., Van Bel M., Sterck L., Billiau K., Van Parys T., Van de Peer Y., Vandepoele K. PLAZA: A Comparative Genomics Resource to Study Gene and Genome Evolution in Plants. The Plant Cell. 2009;21(12):3718–3731. doi: 10.1105/tpc.109.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee TH, Tang H, Wang X, Paterson AH. PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res. 2013;41(Database issue):D1152. doi: 10.1093/nar/gks1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krzywinski M, Schein J, Birol İ, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kenneth J, Livak TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 -ΔΔ C T method. Method. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 54.Butler JEF, Kadonaga JT. The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev. 2002;16(20):2583–2592. doi: 10.1101/gad.1026202. [DOI] [PubMed] [Google Scholar]
- 55.Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun. 2002;290(3):998–1009. doi: 10.1006/bbrc.2001.6299. [DOI] [PubMed] [Google Scholar]
- 56.Osakabe Y, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS. ABA control of plant macroelement membrane transport systems in response to water deficit and high salinity. New Phytol. 2014;202(1):35. doi: 10.1111/nph.12613. [DOI] [PubMed] [Google Scholar]
- 57.Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9(11):1963. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pieterse Corné MJ, Van Loon LC. NPR1: the spider in the web of induced resistance signaling pathways. Current Opinion in Plant Biology. 2004;7(4):456–464. doi: 10.1016/j.pbi.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 59.Liu K, Yuan C, Feng S, Zhong S, Li H, Zhong J, Shen C, Liu J. Genome-wide analysis and characterization of aux/IAA family genes related to fruit ripening in papaya (Carica papaya L.) BMC Genomics. 2017;18(1):351. doi: 10.1186/s12864-017-3722-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li L, Zhao Y, McCaig B, Wingerd B, Wang J, Whalon M, Pichersky E, Howe G. The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell. 2004;16(1):126. doi: 10.1105/tpc.017954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xiu Hong A, Hao YJ, Li EM, Xu K, Cheng CG. Functional identification of apple MdJAZ2 in Arabidopsis with reduced JA-sensitivity and increased stress tolerance. Plant Cell Rep. 2016:1–11. [DOI] [PubMed]
- 62.Li J, Tao X, Li L, Mao L, Luo Z, Khan ZU, Ying T. Comprehensive RNA-Seq analysis on the regulation of tomato ripening by exogenous Auxin. PLoS One. 2016;11(5):e0156453. doi: 10.1371/journal.pone.0156453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Iglesias-Fernández R, Matilla A. After-ripening alters the gene expression pattern of oxidases involved in the ethylene and gibberellin pathways during early imbibition of Sisymbrium officinale L. seeds. J Exp Bot. 2009;60(6):1645. doi: 10.1093/jxb/erp029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Y-m ZHANG, X-l NI, H-q MA. Qiu W: molecular mechanism for jasmonate-induction of anthocyanin accumulation in Arabidopsis. J Exp Bot. 2009;60(13):3849–3860. doi: 10.1093/jxb/erp223. [DOI] [PubMed] [Google Scholar]
- 65.Kondo S, Tomiyama A, Seto H. Changes of endogenous jasmonic acid and methyl jasmonate in apples and sweet cherries during fruit development. J Am Soc Hortic Sci. 2000;125(3):282–287. [Google Scholar]
- 66.Cheng Z, Li S, Qi T, Zhang B, Peng W, Liu Y, Xie D. The bHLH transcription factor MYC3 interacts with the Jasmonate ZIM-domain proteins to mediate Jasmonate response in Arabidopsis. Mol Plant. 2011;4(2):279. doi: 10.1093/mp/ssq073. [DOI] [PubMed] [Google Scholar]
- 67.Xie D, Feys B, James S, NietoRostro M, Turner J. COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science. 1998;280(5366):1091–1094. doi: 10.1126/science.280.5366.1091. [DOI] [PubMed] [Google Scholar]
- 68.Yan J, Li S, Gu M, Yao R, Li Y, Chen J, Yang M, Tong J, Xiao L, Nan F. Endogenous bioactive Jasmonate is composed of a set of (+)-7-iso-JA-amino acid conjugates. Plant Physiol. 2016;172(4):2154–2164. doi: 10.1104/pp.16.00906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peng SQ, Jing XU, Hui-Liang LI, Tian WM. Cloning and molecular characterization of HbCOI1 from Hevea brasiliensis. Biosci Biotechnol Biochem. 2009;73(3):665–670. doi: 10.1271/bbb.80721. [DOI] [PubMed] [Google Scholar]
- 70.Hybridus G, Insensitive C. Cloning and expression analysis of Coronatine insensitive gene encoding a Jasmonate receptor from Gladiolus hybridus. Acta Horticulturae Sinica. 2015.
- 71.Devoto A, Ellis C. Expression profiling reveals COI1 to be a key regulator of genes involved in wound- and methyl jasmonate-induced secondary metabolism, defence, and hormone interactions. Plant Mol Biol. 2005;58(4):497. doi: 10.1007/s11103-005-7306-5. [DOI] [PubMed] [Google Scholar]
- 72.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge J. S: SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86(2):263–274. doi: 10.1016/S0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 73.Buchanan S, Gay N. Structural and functional diversity in the leucine-rich repeat family of proteins. Prog Biophys Mol Biol. 1996;65(1–2):1. doi: 10.1016/S0079-6107(96)00003-X. [DOI] [PubMed] [Google Scholar]
- 74.Kobe B, Deisenhofer J. The leucine-rich repeat: a versatile binding motif. Trends Biochem Sci. 1994;19(10):415. doi: 10.1016/0968-0004(94)90090-6. [DOI] [PubMed] [Google Scholar]
- 75.Ceulemans H, De MM, Stalmans W, Bollen M. A capping domain for LRR protein interaction modules. FEBS Lett. 1999;456(3):349–351. doi: 10.1016/S0014-5793(99)00965-5. [DOI] [PubMed] [Google Scholar]
- 76.Parniske M, Hammondkosack KE, Golstein C, Thomas CM, Jones DA, Harrison K, Wulff BBH, Jones JDG. Novel disease resistance specificities result from sequence exchange between Tandemly repeated genes at the Cf-4/9Locus of tomato. Cell. 1997;91(6):821–832. doi: 10.1016/S0092-8674(00)80470-5. [DOI] [PubMed] [Google Scholar]
- 77.Ellis JG, Lawrence GJ, Luck JE, Dodds PN. Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell. 1999;11(3):495–506. doi: 10.1105/tpc.11.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Clark SE, Williams RW. Meyerowitz, M E: the CLAVATA1Gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell. 1997;89(4):575–585. doi: 10.1016/S0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- 79.Torii KU, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier RF, Komeda Y. The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular Leucine-rich repeats. Plant Cell. 1996;8(4):735. doi: 10.1105/tpc.8.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mcconn M, Creelman RA, Bell E, Mullet JE, Browse J. Jasmonate is essential for insect defense in Arabidopsis. Proc Natl Acad Sci U S A. 1997;94(10):5473–5477. doi: 10.1073/pnas.94.10.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kazan K, Manners JM. JAZ repressors and the orchestration of phytohormone crosstalk. Trends Plant Sci. 2012;17(1):22. doi: 10.1016/j.tplants.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 82.Kazan K, Manners JM. Linking development to defense: auxin in plant-pathogen interactions. Trends Plant Sci. 2009;14(7):373–382. doi: 10.1016/j.tplants.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 83.Cheng H, Song S, Xiao L, Soo HM, Cheng Z, Xie D, Peng J. Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet. 2009;5(3):e1000440. doi: 10.1371/journal.pgen.1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kazan K, Manners JM. Jasmonate signaling: toward an integrated view. Plant Physiol. 2008;146(4):1459–1468. doi: 10.1104/pp.107.115717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu S, Qin Z, Zhang F, Zhao C, Ma Q. Hormonal regulation of glume opening and closure of photo-thermo-sensitive male sterility wheat by methyl Jasmonate and salicylic acid. Journal of Triticeae Crops. 2007;27(4):593–596. [Google Scholar]
- 86.Song P, Xia K, Wu C, Bao D, Chen L, Zhou X, Cao X. Differential response of floret opening in male-sterile and male-fertile Rices to methyl Jasmonate. Acta Votanica Sinica. 2001;43(5):480–485. [Google Scholar]
- 87.Shu K, Zhou W, Yang W. APETALA 2-domain-containing transcription factors: focusing on abscisic acid and gibberellins antagonism. New Phytol. 2017;3:217. doi: 10.1111/nph.14880. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Consensus sequence and logos of motifs from wheat COI proteins. (TIF 2339 kb)
Table S1. Summary of COI or COI-like genes in different plants. Table S2. Specific primers for TaCOI genes for qRT-PCR. Table S3. Promoter sequences of TaCOI genes. Table S4. Protein sequences, CDS sequences, and genomic sequences of TaCOI and COI or 12 COI-like loci in various plants. (DOCX 241 kb)
Data Availability Statement
The data sets supporting the results of this article are included within the article and its additional files.